Fruit monoterpene synthesis in kiwifruit is transcriptionally regulated by transcription factors that activate the terpene synthase promoter.
Abstract
Two kiwifruit (Actinidia) species with contrasting terpene profiles were compared to understand the regulation of fruit monoterpene production. High rates of terpinolene production in ripe Actinidia arguta fruit were correlated with increasing gene and protein expression of A. arguta terpene synthase1 (AaTPS1) and correlated with an increase in transcript levels of the 2-C-methyl-d-erythritol 4-phosphate pathway enzyme 1-deoxy-d-xylulose-5-phosphate synthase (DXS). Actinidia chinensis terpene synthase1 (AcTPS1) was identified as part of an array of eight tandemly duplicated genes, and AcTPS1 expression and terpene production were observed only at low levels in developing fruit. Transient overexpression of DXS in Nicotiana benthamiana leaves elevated monoterpene synthesis by AaTPS1 more than 100-fold, indicating that DXS is likely to be the key step in regulating 2-C-methyl-d-erythritol 4-phosphate substrate flux in kiwifruit. Comparative promoter analysis identified potential NAC (for no apical meristem [NAM], Arabidopsis transcription activation factor [ATAF], and cup-shaped cotyledon [CUC])-domain transcription factor) and ETHYLENE-INSENSITIVE3-like transcription factor (TF) binding sites in the AaTPS1 promoter, and cloned members of both TF classes were able to activate the AaTPS1 promoter in transient assays. Electrophoretic mobility shift assays showed that AaNAC2, AaNAC3, and AaNAC4 bind a 28-bp fragment of the proximal NAC binding site in the AaTPS1 promoter but not the A. chinensis AcTPS1 promoter, where the NAC binding site was mutated. Activation could be restored by reintroducing multiple repeats of the 12-bp NAC core-binding motif. The absence of NAC transcriptional activation in ripe A. chinensis fruit can account for the low accumulation of AcTPS1 transcript, protein, and monoterpene volatiles in this species. These results indicate the importance of NAC TFs in controlling monoterpene production and other traits in ripening fruits.
Fruit and flower volatiles are important signals to attract mutualists and repel antagonists involved in pollination, fruit predation, and seed dispersal. They have additional roles in protecting these tissues against pathogens and insects and may be involved in indirect trophic interactions such as attracting herbivore predators (Rodríguez et al., 2013). Flavor volatiles are also important targets in modern plant breeding programs in which specific combinations are selected to enhance the taste, aroma, and health properties of the fruit. Terpenoids form an important part of the aroma profile of many fruits. During fruit ripening, concentrations of many terpenoid compounds (including C10 monoterpene and C15 sesquiterpene volatiles) increase in conjunction with changes in texture, taste, and color. Fruit volatile terpenoids have been well characterized in Citrus spp. (Moufida and Marzouk, 2003), mango (Mangifera indica; MacLeod and Pieris, 1984), apple (Malus × domestica; Nieuwenhuizen et al., 2013), tomato (Solanum lycopersicum; Buttery et al., 1988), and grape (Vitis vinifera; Battilana et al., 2009; Duchêne et al., 2009). In kiwifruit (Actinidia spp.), volatile terpenoid concentrations also vary greatly, with some species such as Actinidia arguta showing very high concentrations of monoterpenes, whereas others, such as Actinidia chinensis and Actinidia deliciosa, show very low concentrations (Crowhurst et al., 2008).
In nature, the diverse array of terpene compounds essentially derives from the simple C5 prenyldiphosphate precursor isopentenyl diphosphate (IDP) and its allylic isomer dimethylallyl diphosphate (DMADP). In plants, IDP is synthesized via two compartmentally segregated substrate pathways: the mevalonate pathway located in the cytosol and the plastidial 2-C-methyl-d-erythritol 4-phosphate pathway (MEP; Rodríguez-Concepción and Boronat, 2002). In both compartments, IDP and DMADP are condensed by the action of short-chain prenyltransferases to generate the direct precursors for terpene synthase (TPS) enzymes, including the plastid-derived C10 mono-TPS substrate geranyl diphosphate (GDP) and the cytosol-derived C15 sesqui-TPS substrate farnesyl diphosphate. TPS enzymes are unique in their capacity to direct the formation of a myriad of products through subtle variations in their conserved catalytic fold (Yoshikuni et al., 2006; O’Maille et al., 2008; Chen et al., 2011) and in their propensity to generate multiple products from a single prenyldiphosphate substrate. Contrasting terpene profiles can also be caused by differences in the composition of TPS genes due to changes in copy number, variation of gene and protein expression, and variation in catalytic efficiency, as seen in Sitka spruce and the production of the monoterpene (+)-3-carene associated with white pine weevil (Pissodes strobi) resistance (Hall et al., 2011). In maize (Zea mays), the ability of roots or leaves to attract natural enemies upon herbivory was dependent on expression of an (E)-β-caryophyllene synthase allele that was lost in many maize lines during breeding (Köllner et al., 2008). For the scented species Clarkia breweri, floral linalool release likely evolved from its extant nonscented species Clarkia concinna through changes in expression of an S-linalool synthase (Dudareva et al., 1996).
The 1-deoxy-d-xylulose-5-phosphate synthase (DXS) gene has been shown to encode an important rate-limiting step in the biosynthesis of MEP-derived isoprenoids such as chlorophylls, tocopherols, carotenoids, abscisic acid, and GAs in Arabidopsis (Arabidopsis thaliana; Matthews and Wurtzel, 2000; Estévez et al., 2001) and other biological systems (Rodríguez-Concepción and Boronat, 2002; Cordoba et al., 2009). In tomato fruit, regulation of the MEP pathway by DXS has been extensively studied in relation to carotenoid biosynthesis (Lois et al., 2000; Enfissi et al., 2005). The DXS gene also appears to have an important role in regulating monoterpene biosynthesis in fruit, as quantitative trait loci for variation in concentrations of linalool, geraniol, nerol, citronellol, and α-terpineol colocate with candidate DXS genes in grape (Battilana et al., 2009; Duchêne et al., 2009). DXS is not the only gene regulating substrate flux through the MEP pathway, as later steps in the MEP pathway mediated by deoxyxylulose 5-phosphate reductoisomerase (DXR) and 4-hydroxy-3-methylbut-2-enyl diphosphate (HMBPP) reductase (HDR) have also been shown to be rate limiting in the leaves of Arabidopsis and peppermint (Mentha × piperita; Mahmoud and Croteau, 2001; Botella-Pavía et al., 2004). Overexpression of DXR increased biosynthesis of chlorophylls, carotenoids, and transgene-derived taxadiene in Arabidopsis as well as essential oil concentrations in peppermint (Mahmoud and Croteau, 2001). Overexpression of a tomato HDR gene in Arabidopsis gave similar results (Botella-Pavía et al., 2004). In Nicotiana tabacum, overexpression of a snapdragon (Antirrhinum majus) GDP synthase small subunit (ssGDPS) increased monoterpene emissions but at the expense of other isoprenoids such as carotenoids, chlorophyll, and GAs (Orlova et al., 2009).
In fruit crops such as tomato, apple, and banana (Musa acuminata), a rapid increase of ethylene production accompanied by a climacteric burst in respiration (Alexander and Grierson, 2002) initiates rapid fruit ripening. During climacteric fruit ripening, autocatalytic (type II) ethylene production initiates a transcriptional cascade that controls many ripening traits, including softening and flavor volatile production (Solano et al., 1998). The ETHYLENE-INSENSITIVE3-like (EIL) class of transcription factors (TFs) are the most downstream signaling components of the ethylene-signaling pathway and have been shown to modulate a multitude of downstream transcriptional cascades and hormone response pathways (Chang et al., 2013). Ethylene-induced stabilization of EIN3 protein is mediated by removal of EIN3-binding F-box proteins (EBFs) through the proteasome. In the absence of ethylene, EBFs are up-regulated and mediate EIN3 degradation (Solano et al., 1998). EIL TFs act on the promoters of early ethylene responsive genes such as ETHYLENE-RESPONSE-FACTOR1 (ERF1), which encodes a GCC-box-binding protein, by binding to the primary ethylene response element in the promoters of these genes (Yamasaki et al., 2005). EILs in kiwifruit were able to activate the 1-aminocyclopropane-1-carboxylic acid (ACC) OXIDASE1 (AdACO1) gene promoter, thus implicating them in a role during autocatalytic ethylene production (Yin et al., 2010). Antisense tomato ERF1 (LeERF1) tomatoes had longer fruit postharvest life (Li et al., 2007), and LeERF2 regulates ethylene production in tomato and tobacco by interaction with the promoters of LeACO3 and N. tabacum ACC SYNTHASE3 (Zhang et al., 2009). In banana, ERFs have also been shown to bind to the promoters of MaACO1 and ACC SYNTHASE1 (MaACS1) genes involved in ethylene biosynthesis (Xiao et al., 2013).
Transcriptional control of specific flavor and aroma genes is currently poorly understood with respect to development/senescence and ethylene production. In Petunia hybrida flowers, benzenoid scent production has been shown to be transcriptionally controlled through the ODORANT1 R2R3 MYB TF targeting the 5-enol-pyruvylshikimate-3-phosphate synthase promoter (Verdonk et al., 2005). Transgenic roses (Rosa hybrida) that overexpressed the PRODUCTION OF ANTHOCYANIN PIGMENT1 R2R3 MYB TF produced 6.5-fold higher concentrations of terpenoids than control flowers, which was partly explained by up-regulation of a GERMACRENE D SYNTHASE gene, although no direct evidence of promoter interaction was presented (Zvi et al., 2012). In cotton (Gossypium hirsutum), the action of a WRKY TF regulates the sesquiterpene synthase gene (+)-δ-CADINENE SYNTHASE-A involved in gossypol production (Xu et al., 2004), whereas in Arabidopsis inflorescences, the basic helix-loop-helix MYC2 directly binds and activates the promoter of the sesquiterpene synthase gene TPS21, inducing sesquiterpene release (Hong et al., 2012). In tomato, pleiotropic mutants such as ripening inhibitor (rin), nonripening (nor), and Colorless nonripening (Cnr) affect a wide range of ripening-related characters, including ethylene production and downstream volatile compound synthesis. The rin mutation has been shown to be encoded by a MADS-box TF (Vrebalov et al., 2002) and Cnr by a SQUAMOSA promoter-binding protein (Manning et al., 2006). An uncharacterized NAC (for no apical meristem [NAM], Arabidopsis transcription activation factor [ATAF], and cup-shaped cotyledon [CUC]-domain transcription factor)-domain TF mutation leads to the nor mutant phenotype (Martel et al., 2011), while SlNAC4 could interact with NOR and RIN proteins to affect fruit ripening and carotenoid accumulation (Zhu et al., 2014). Other tomato TFs shown to influence volatile production in fruit include the tomato homeobox protein (LeHB-1; Lin et al., 2008), which induces LeACO1 and ethylene production, and the MADS-box protein TOMATO AGAMOUS-LIKE1 (TAGL1; Itkin et al., 2009; Vrebalov et al., 2009) through direct activation of LeACS2. In Arabidopsis, the transcription factor gene Arabidopsis NAC domain protein (AtNAP) regulates silique senescence by affecting ethylene biosynthesis, perception, and signaling (Kou et al., 2012). Studies in banana have shown that expression levels of several NAC TFs are induced during fruit ripening, and that some of these can physically interact with EIL TFs (Shan et al., 2012). No information is available on the transcriptional regulation of monoterpenes in any plant species.
In this study, we examine the terpenes and corresponding terpene synthases during fruit development and ripening in two kiwifruit species, A. chinensis and A. arguta, which vary greatly in their terpene volatile content. Our results reveal how natural variation in TF binding sites can influence a key ripening trait in fruit.
RESULTS
Volatile Terpenes in Ripe Actinidia Spp. Fruit
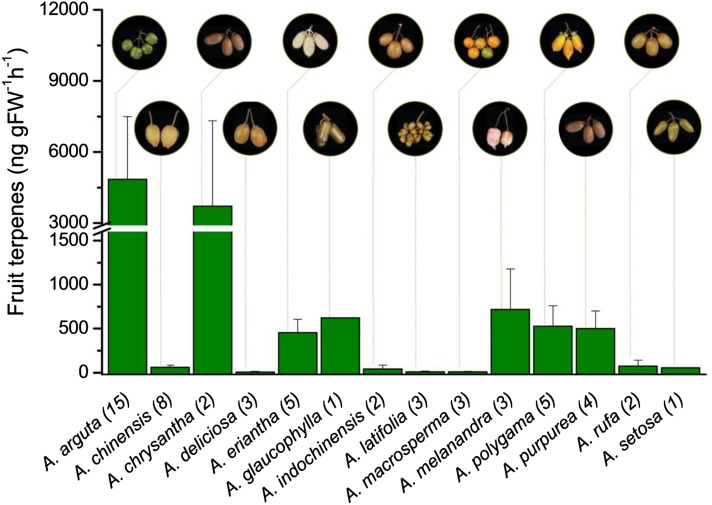
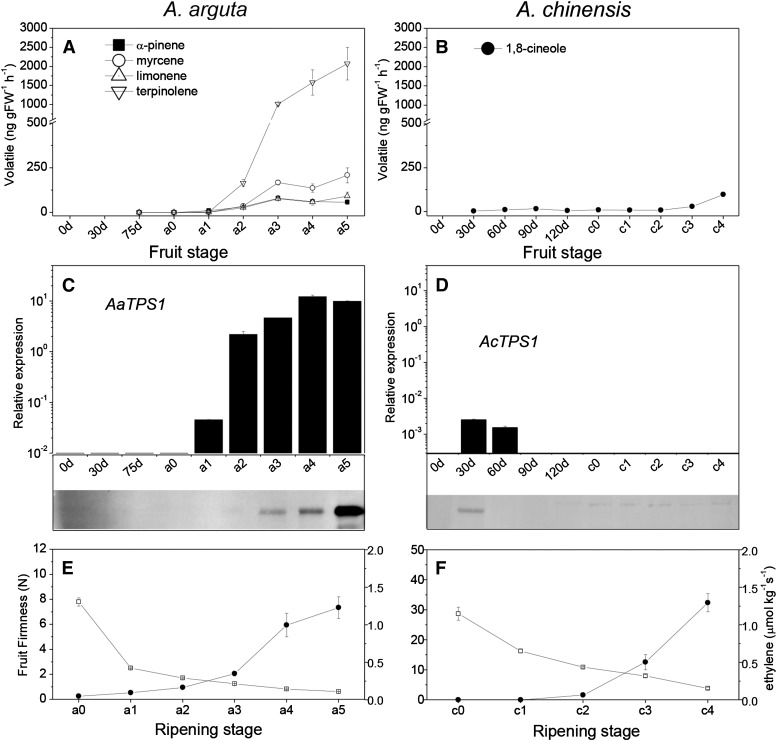
An extensive collection of wild kiwifruit (Actinidia spp.) germplasm was examined for variation in terpene production using a combination of volatile headspace sampling and gas chromatography-mass spectrometry (GC-MS) analysis. The concentrations of terpenes produced in ripe fruit varied greatly among the different kiwifruit species (Fig. 1). Fruit from some species, such as A. arguta and Actinidia chrysantha, produced very high concentrations of terpenes, whereas fruit from others, such as A. chinensis and A. deliciosa, produced much lower amounts. From this survey, we selected the small green-fleshed A. arguta ‘Hortgem Tahi’ and larger yellow-fleshed A. chinensis ‘Hort16A’ for detailed volatile analysis during fruit development and across different ripening stages (Fig. 2, A and B). A. arguta ‘Hortgem Tahi’ ripe fruit pulp released much higher concentrations of terpenes than A. chinensis ‘Hort16A’; 1,000 versus 7 ng g fresh weight−1 h−1) at the respective eating ripe stages (a3/c2), and in both species, the profile predominantly consisted of cyclic monoterpenes. In A. arguta, volatile terpene production increased rapidly during the fruit-ripening phase and correlated with large increases in ethylene release and fruit softening (Fig. 2E). The A. arguta headspace terpene profile was dominated by terpinolene (59% of terpene content at a3 eating ripe stage), myrcene (13%), α-pinene (11%), and limonene (10%). A. chinensis released relatively low concentrations of mainly 1,8-cineole (eucalyptol) at the overripe stage (Fig. 2, B and F).
Figure 1.
Total headspace terpene volatiles released from the ripe fruit of 14 Actinidia spp. obtained by purge and trap followed by thermal desorption and semiquantification by GC-MS. Average terpene concentrations are shown ±se. The number of accessions tested for each species is shown in brackets.
Figure 2.
Rates of volatile terpene release by A. arguta ‘Hortgem Tahi’ (A) and A. chinensis ‘Hort16A’ (B) during fruit development and ripening (stages a0–a5 for A. arguta ‘Hortgem Tahi’ and c0–c4 for A. chinensis ‘Hort16A’ fruit, ranging from harvest to overripe; see text). Measurements were made in triplicate ± se using a purge and trap method followed by thermal desorption and semiquantification by GC-MS against authentic standards. Only terpenes representing more than 1% of total terpene content are shown. D, Days after pollination. Real-time qPCR analysis of AaTPS1 (C, top) and AcTPS1 (D, top) gene expression levels was performed on pooled fruit samples using gene-specific primers given in Supplemental Table S2. Error bars are based on four technical replicates, and data were normalized against the housekeeping gene EF1α. Western analysis of AaTPS1 (C, bottom) and AcTPS1 (D, bottom) protein concentrations at equivalent time points using a polyclonal antibody raised against AcTPS1. Change in firmness (squares/left scale) and ethylene (circles/right scale) production in A. arguta ‘Hortgem Tahi’ (E) and A. chinensis ‘Hort16A’ (F) during fruit ripening.
Identification of AaTPS1 and AcTPS1 Terpene Synthase Genes
A single TPS candidate, AaTPS1, represented by multiple ESTs, was identified from an A. arguta ‘Hortgem Tahi’ ripe fruit EST library (n = 10,400 ESTs). No candidate TPS genes were identified in A. chinensis ‘Hort16A’ ripe fruit libraries (n = 7,900 ESTs). However, a highly homologous gene, AcTPS1, with 88.7% amino acid identity, was identified from an A. chinensis ‘Hort16A’ young fruit library (n = 10,200). In silico open reading frame prediction showed that the AaTPS1 and AcTPS1 genes encoded proteins of 603 and 604 amino acids, respectively, and that these proteins contained the expected divalent metal binding regions (i.e. DDXXD and NSE/DTE motifs) necessary for TPS activity (Cane and Kang, 2000; Rynkiewicz et al., 2001). AaTPS1 and AcTPS1 also possessed the RRX8W motif common to cyclic monoterpene-producing enzymes (Williams et al., 1998) and were both predicted to contain chloroplast-targeting peptides (Supplemental Fig. S1A).
AaTPS1 and AcTPS1 Likely Originate from a Common Ancestral Gene
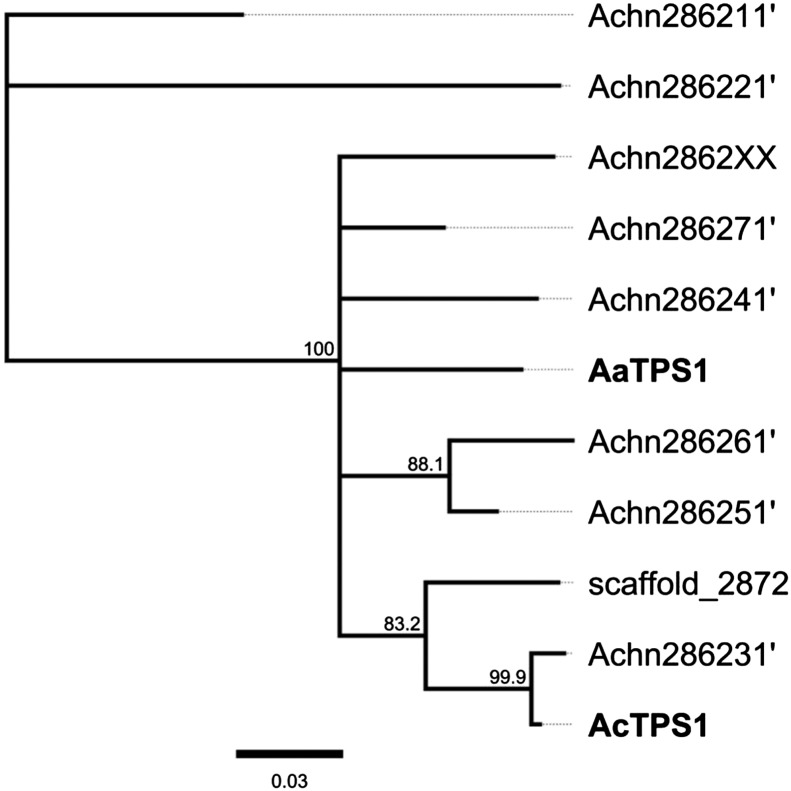
An analysis of the kiwifruit genome sequence of the A. chinensis ‘Hongyang’ (Huang et al., 2013) identified 35 terpene synthase gene models within the kiwifruit genome (Supplemental Fig. S1B). Phylogenetic clustering showed that AaTPS1 and AcTPS1 grouped with other mono-TPS genes in the Tps-b subfamily (Supplemental Fig. S1B; Bohlmann et al., 1998), and that the eight gene models most closely related to AaTPS1 and AcTPS1 were part of a single scaffold of tandemly repeated genes (Supplemental Fig. S2A) covering about 155 kb. The existing gene models were refined manually by identifying and extracting individual exons homologous to AcTPS1 exons 1 to 7. Phylogenetic analysis of the curated/translated proteins (Supplemental Fig. S2B) showed that AaTPS1 and AcTPS1 are likely to be orthologs originating from a common ancestral gene, and that A. chinensis paralogs arose because of multiple tandem duplication events (Fig. 3). These events were accompanied by a significant loss of exons and promoters, nonsense mutations, and insertions/deletions within the scaffold, resulting in only three gene models that could potentially encode functional TPS proteins (Achn286231, Achn286241, and Achn2862XX). Gene model Achn286231 represents the corresponding AcTPS1 sequence within the A. chinensis ‘Hongyang’ genome, sharing 99% amino acid identity in the coding region sequence. Other TPS scaffold members shared between 69% and 96% amino acid identity (Supplemental Fig. S2C).
Figure 3.
Phylogenetic analysis of the AaTPS1 and AcTPS1 homologs in A. chinensis ‘HongYang.’ Protein sequences extracted from the kiwifruit genome were aligned using ClustalW and manually curated (Supplemental Fig. S2, A and B).The evolutionary history was inferred using the neighbor-joining method with the Jukes-Cantor genetic distance model. The consensus tree is shown based on 80% support threshold. The percentage of trees in which the associated taxa clustered together is shown next to the branches based on 1,000 bootstrap replicates. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 11 amino acid sequences and no outgroups. Evolutionary analyses were conducted in Geneious 6.1.6.
RNA and Protein Expression Analysis
AaTPS1 expression increased sharply during fruit ripening, with the highest levels of expression being found in overripe fruit (stages a4/a5; Fig. 2C), whereas AcTPS1 expression was modest (less than 1/1,000 of AaTPS1) in developing fruit (30–60 d after pollination [DAP]) and not present in ripe fruit (Fig. 2D). Western analysis using a polyclonal antibody that detects both kiwifruit TPS enzymes (Fig. 2, C and D, bottom) showed that the AcTPS1 protein could be detected weakly in young fruit (30 DAP). In contrast, AaTPS1 protein was present at much higher concentrations, with accumulation first becoming apparent at the a2 stage, then increasing sharply and peaking in overripe fruit (a5 stage). The other two full-length TPS genes identified within the A. chinensis ‘Hongyang’ TPS scaffold (Achn286241 and Achn2862XX) were not expressed at harvest or in ripe A. chinensis ‘Hort16A’ fruit. TPS expression using primers universal for AaTPS1 and AcTPS1 was strongly ripening induced in three high-level terpene-producing species (A. arguta, Actinidia eriantha, and Actinidia macrosperma × melanandra) but undetectable in three low species (A. chinensis, A. deliciosa, and A. macrosperma; Supplemental Fig. S3A).
Functional Characterization of AaTPS1 and AcTPS1
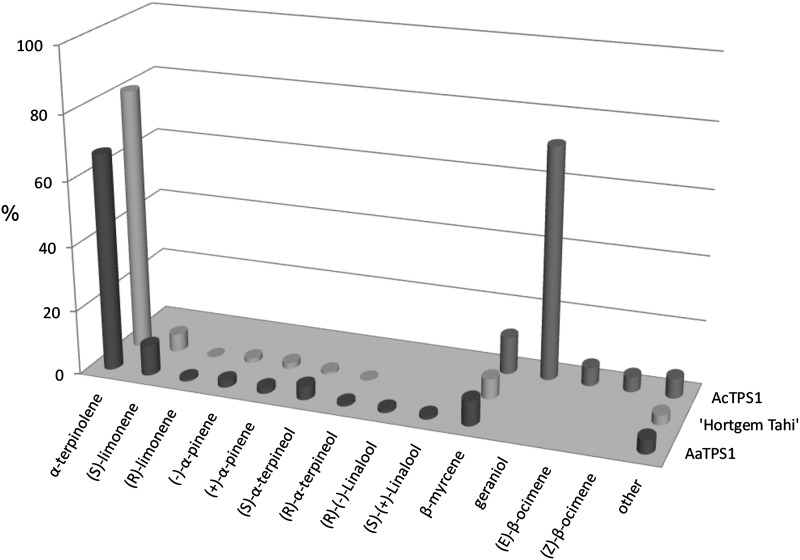
Recombinant proteins were expressed in Escherichia coli and purified using a combination of Ni2+ affinity and size exclusion chromatography. In vitro solvent extraction assays followed by enantioselective GC-MS showed that the recombinant AaTPS1 enzyme catalyzed the conversion of GDP to both cyclic and noncyclic monoterpene products. Specifically, α-terpinolene was the predominant terpene produced, accounting for approximately 67% of the total monoterpenes, followed by β-myrcene (10%), predominantly (S)-limonene (9%), and smaller amounts of α-pinene, linalool, and α-terpineol (Fig. 4). AcTPS1 produced the noncyclic monoterpenes geraniol and β-myrcene as the major products rather than 1,8-cineole, which is the predominant terpene associated with A. chinensis fruit. The enantiomeric composition of terpenes produced by AaTPS1 in vitro and those produced in ripe A. arguta ‘Hortgem Tahi’ fruit (stage a3/a4) were very similar (Fig. 4). To verify the likely in planta function of AaTPS1 and AcTPS1 further, both genes were transiently expressed in Nicotiana benthamiana leaves. Leaves transiently expressing AaTPS1 produced a monoterpene profile very similar to that seen in ripe A. arguta fruit and from the in vitro enzyme analysis (Supplemental Fig. S4A). Tobacco leaves expressing AcTPS1 produced β-myrcene in low amounts as the only gene-specific product (Supplemental Fig. S4B). Geraniol, which was observed in the AcTPS1 in vitro analysis, was not identified after transient expression in planta, likely due to glycosylation, which has been previously observed in N. tabacum with a transiently expressed geraniol synthase (Dong et al., 2013).
Figure 4.
Percentage of terpene volatiles produced by recombinant AaTPS1 and AcTPS1 enzymes in vitro and in ripe (stage a3/a4; see text) A. arguta ‘Hortgem Tahi’ fruit. The recombinant enzymes expressed in E. coli were purified by Ni2+ affinity and gel filtration chromatography and incubated with GDP as substrate. After solvent extraction, volatile terpenes were analyzed by enantioselective GC-MS. Extractions were performed in triplicate.
Kinetic characterization of the recombinant AaTPS1 and AcTPS1 enzymes confirmed their requirement for a divalent metal ion cofactor (Table I). AcTPS1 favored Mn2+ over Mg2+ (Km = approximately 20 μm and 1.5 mm, respectively) and showed a 2-fold increase in maximal velocity (Vmax) with Mn2+ compared with Mg2+. AaTPS1 binding constants for Mn2+ and Mg2+ were similar to AcTPS1, although a higher Vmax was observed with Mg2+. Despite an approximately 3-fold increase in AaTPS1 turnover compared with AcTPS1, catalytic efficiencies (kcat/Km) for both enzymes were similar. K+ had no effect on AaTPS1 and AcTPS1 activities, and neither enzyme used farnesyl diphosphate as a substrate.
Table I. Enzyme kinetic properties of recombinant AaTPS1 and AcTPS1.
Purified recombinant enzymes were obtained by Ni2+ affinity and gel filtration chromatography. Kinetic parameters were determined for GDP (1–50 µm) in the presence of 20 mm MgCl2 and for Mg2+ (0.2–25 mm) and Mn2+ (0–500 µm) in the presence of 25 µm GDP. All values represent the mean ± se, n = 3. kcat, Turnover; Vmax, maximum velocity.
| Enzyme | Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|
|
µm |
% |
s−1 |
s−1 mm−1 |
|
| AcTPS1 | ||||
| GDP (Mg2+) | 6.1 ± 0.2 | 0.0076 ± 0.0004 | 1.3 ± 0.08 | |
| Mg2+ (GDP) | 1478 ± 103 | 100 | ||
| Mn2+ (GDP) | 20.2 ± 2.0 | ∼200 | ||
| AaTPS1 | ||||
| GDP (Mg2+) | 13.1 ± 1.0 | 0.0195 ± 0.0020 | 1.5 ± 0.10 | |
| Mg2+ (GDP) | 1503 ± 208 | 100 | ||
| Mn2+ (GDP) | 28.6 ± 1.1 | ∼80 |
Substrate Flux of Terpene Precursors though the MEP Pathway
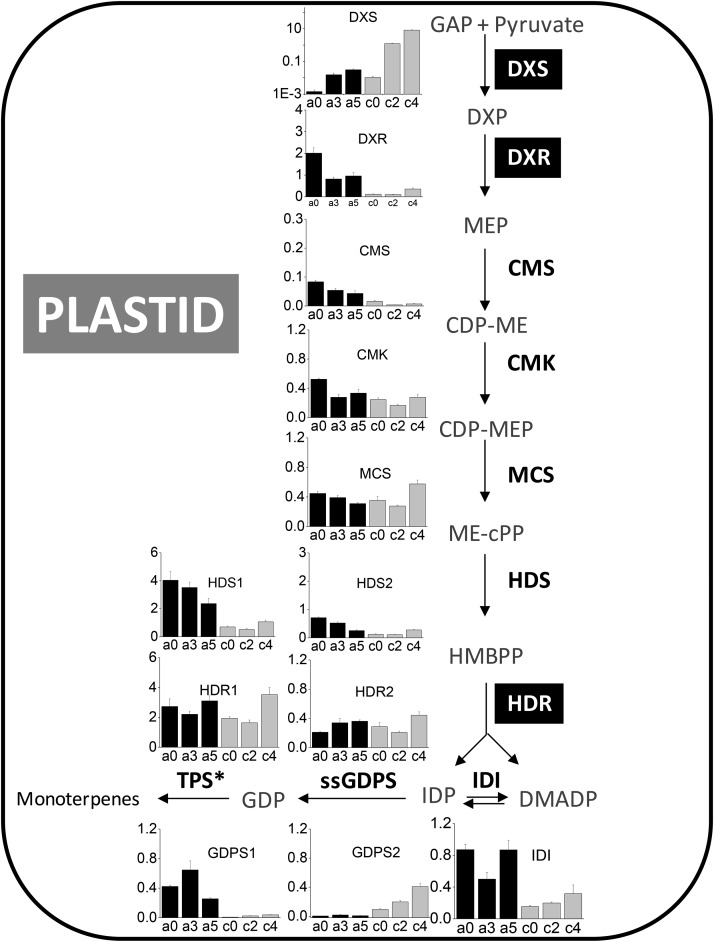
The principal route for monoterpene substrate synthesis is located in the plastids where the MEP pathway (see Fig. 5) produces IDP, which can be isomerized to DMADP through the action of IDP isomerase (IDI). To investigate the potential role of genes in the MEP pathway in controlling substrate flux in kiwifruit, gene candidates were identified in fruit EST collections for each of the MEP pathway enzymes. Patterns of gene expression were determined by quantitative PCR (qPCR) in both A. arguta and A. chinensis in unripe (stages a0/c0), eating ripe (a3/c2), and overripe (a5/c4) fruit (Fig. 5), and full-length candidate genes were cloned for in planta expression analysis. Results showed that DXS gene expression was strongly induced (greater than 20-fold) during fruit ripening in both kiwifruit species. Although the remaining MEP pathway genes showed small differences in gene expression, there was no discernible correlation with ripening and volatile release and little consistency in expression patterns across the two kiwifruit species.
Figure 5.
Expression of MEP pathway genes during fruit ripening in A. arguta (black bars, ripening stages a0, a3, and a5; see text) and A. chinensis (gray bars, ripening stages c0, c2, and c4; see text). Real-time qPCR analyses were performed on pooled fruit samples using the gene-specific primers given in Supplemental Table S2. Error bars are based on four technical replicates, and data were normalized against the housekeeping gene EF1α. Error bars represent ±se. MEP pathway intermediate abbreviations are: CDP-ME, 4-diphosphocytidyl-methylerythritol; CDP-MEP, 4-diphosphocytidyl-methylerythritol 2-phosphate; DXP, deoxyxylulose 5-phosphate; GAP, glyceraldehyde 3-phosphate; HBMPP, hydroxymethylbutenyl 4-diphosphate; and ME-cPP, methylerythritol 2,4-cyclodiphosphate. Enzyme abbreviations in bold are: CMK, CDP-ME kinase (EC 2.7.1.148); CMS, CDP-ME synthase (EC 2.7.7.60); DXR, DXP reductoisomerase (EC 1.1.1.267); DXS, DXP synthase (EC 2.2.1.7); HDR, HMBPP reductase (EC 1.17.1.2); HDS, HMBPP synthase (EC 1.17.4.3); IDI, IDP isomerase (EC 5.3.3.2); MCS, ME-cPP synthase (EC 4.6.1.12); and ssGDPS, geranyl diphosphate synthase small subunit (EC 2.5.1.1). Enzymes considered to be rate determining according to the literature are boxed. *, TPS data are shown in Fig. 2, C and D.
Expression of MEP Pathway Genes in a Transient System
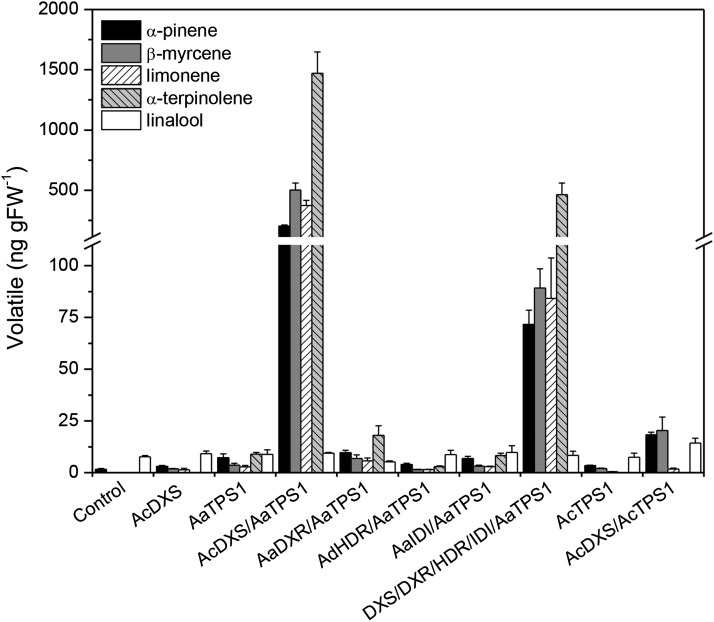
Transient overexpression of the DXS gene cloned from ripe A. chinensis fruit (AcDXS1) resulted in small but significant increases in endogenous terpene production from the infiltrated N. benthamiana leaves (Fig. 6). Coinfiltrating AcDXS1 with AaTPS1 resulted in a large increase (greater than 100-fold) in terpene production compared with infiltration of AaTPS1 alone. Combining transient expression of AaTPS1 and AcDXR1 resulted in a significant (α = 0.05) increase of about 2-fold in terpene production, whereas the other two genes tested (AcHDR1 and AcIDI1) had no stimulatory effect. Combining overexpression of all four MEP pathway genes with AaTPS1 did not enhance the production compared with transient expression of DXS alone (possibly because of a transgene dilution effect). These results demonstrate that the DXS gene is a rate-limiting step in the production of terpenes in planta by endogenous N. benthamiana terpene synthases, and support the hypothesis that DXS is the rate-limiting enzyme in terpene synthesis in ripe A. arguta fruit where DXS and TPS genes are both transcriptionally up-regulated.
Figure 6.
Monoterpene volatiles produced by transient overexpression of A. arguta (Aa), A. chinensis (Ac), and A. deliciosa (Ad) DXS, DXR, HDR, and IDI in combination with AaTPS1 and AcTPS1 in N. benthamiana leaves. Volatiles were sampled 7 d postinfiltration and analyzed by headspace sampling/GC-MS. Analyses were performed on six infiltrated leaves (approximately 3–4 g) in three biological replicates. Error bars represent ±se.
Upstream Regulatory Elements in the AaTPS1 and AcTPS1 Promoters
The promoter regions of AaTPS1 and AcTPS1 including 35 bp of the 5′-untranslated region (UTR; Supplemental Fig. S5A) were isolated to compare the upstream regulatory elements controlling transcription of both genes. A 911-bp AaTPS1 promoter fragment (AaTPS1pro-911) was obtained by inverse PCR, whereas a 3-kb promoter fragment of AcTPS1 (AcTPS1pro-3000) was obtained by PCR based on A. chinensis genome assemblies. The two promoters shared 68% identity (Supplemental Fig. S5B). Comparison of the AaTPS1 promoter region with the A. chinensis ‘Hongyang’ TPS scaffold members showed that only Achn286231 (the gene corresponding most closely to AcTPS1, 99% identity) and Achn286241 shared similar promoter identity (Supplemental Fig. S5B). Achn286241 contained a 1.2-kb insertion close to the predicted TATA box (Supplemental Fig. S5B), which would likely render the promoter inactive.
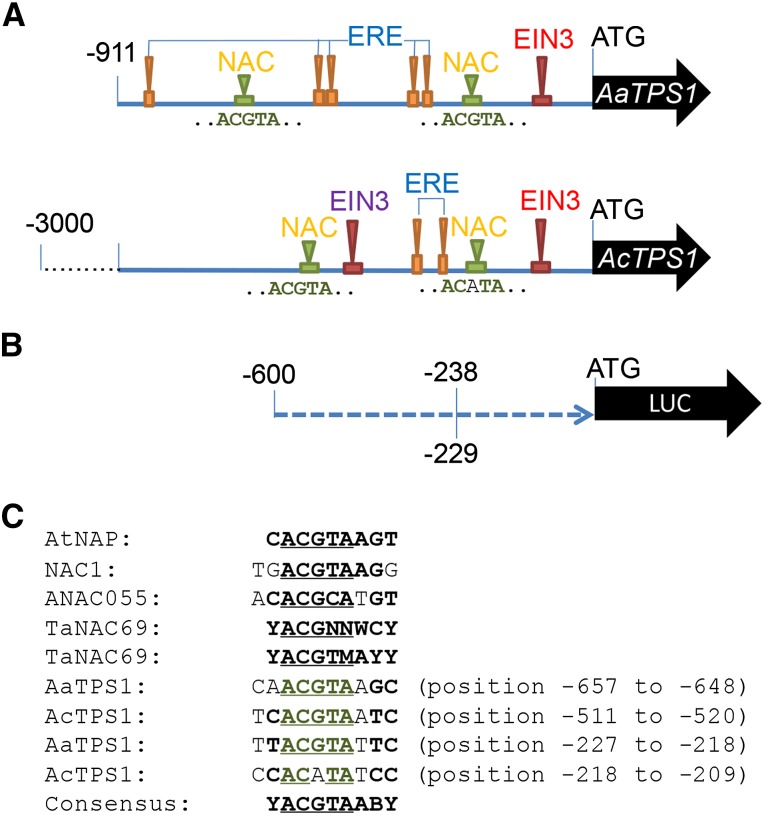
Potential NAC and EIL binding sites were identified manually within the first 911 bp of the AaTPS1 and AcTPS1 promoter fragments as well as multiple ethylene response elements (EREs) based on previously identified consensus binding sites (Fig. 7A; Supplemental Fig. S6). The AaTPS1pro-911 proximal region had five copies of the ERE, whereas the equivalent AcTPS1pro region only had two. Furthermore, AaTPS1pro-900 contained a potential proximal NAC binding site with a conserved ACGTA core motif, whereas this core was mutated in AcTPS1 (Supplemental Fig. S6).
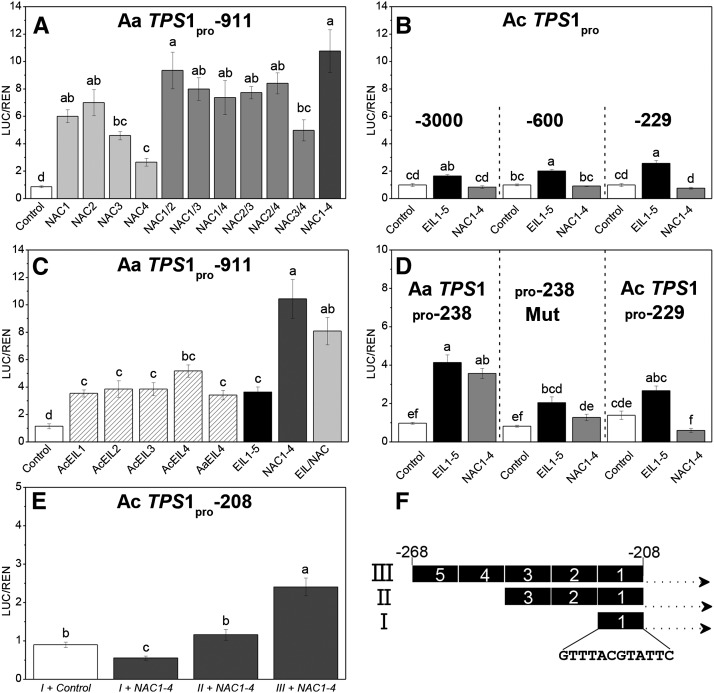
Figure 7.
A, Schematic showing the location of potential NAC and EIL TF binding sites and EREs in the AaTPS1pro-911 and AcTPS1pro-3000 promoter fragments. B, Schematic of the AcTPS1pro-600, AcTPS1pro-229, and AaTPS1pro-238 promoter truncation constructs cloned with the luciferase reporter gene of pGreenII 0800-LUC. C, Potential NAC TF binding sites in the AcTPS1 and AaTPS1 promoters aligned with NAC TF binding sites from Arabidopsis AtNAP (Zhang and Gan, 2012), Arabidopis NAC55 (ANAC055; Tran et al., 2004), NAC1 (Xie et al., 2000), and Triticum aestivum TaNAC69 (Xue, 2005). Numbers for AaTPS1 and AcTPS1 indicate base pairs upstream of the ATG.
Identification of NAC and EIL Transcription Factors in Ripe Kiwifruit
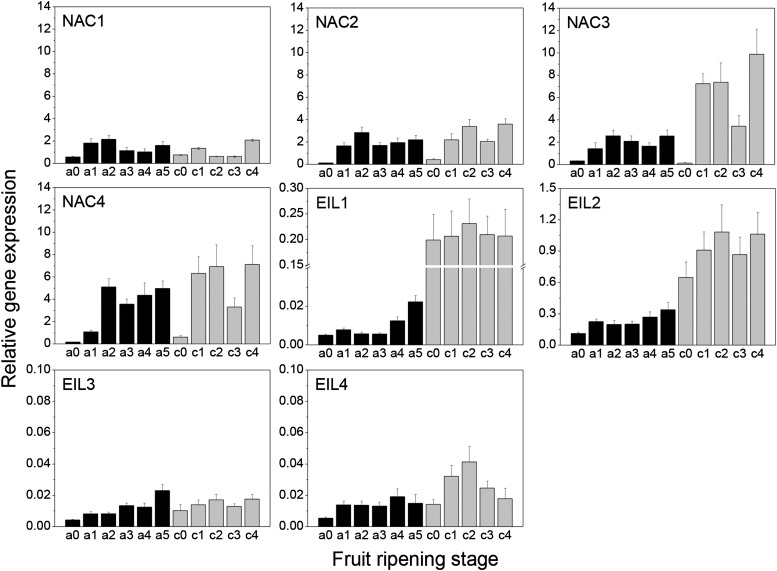
NAC and EIL TFs expressed in ripe A. chinensis and A. arguta fruit were identified by screening a wide range of ripe kiwifruit EST libraries (Crowhurst et al., 2008) supplemented with next-generation sequencing data. Five candidate EIL TFs (AcEIL1–AcEIL4/AaEIL4) and eight NAC TFs (AaNAC1–AaNAC4/AcNAC1–AcNAC4) were identified and cloned from ripe A. arguta and A. chinensis complementary DNA (cDNA). Within each class, there was high amino acid sequence identity (Supplemental Figs. S7 and S8). The four NAC TFs were all strongly induced during ripening (3- to 74-fold) and highly expressed relative to the reference gene ELONGATION FACTOR-1α (EF1α) in both A. chinensis and A. arguta (Fig. 8). Interestingly, there were also significant differences in the expression profiles between the two species. NAC1 showed the lowest induction in both species. NAC4 was the most up-regulated in A. arguta (32-fold), whereas NAC3 was most up-regulated in A. chinensis (74-fold). Overall, the five EIL TFs had lower expression levels than the NAC TF class, with EIL2 showing the highest level of expression, but all showed a degree of transcriptional induction (2- to 6-fold) over the course of ripening. To test whether the NAC1 to NAC4 transcriptional induction was a conserved phenomenon during kiwifruit fruit ripening, expression was tested on a taxonomically wider panel of Actinidia spp. In all cases, the NAC genes (1–4) were strongly induced in ripe fruit compared with fruit at harvest (Supplemental Fig. S3, B–E), strongly supporting the idea that NAC genes form an integral part of the fruit-ripening program within the Actinidia genus.
Figure 8.
Expression of NAC and EIL TFs during fruit ripening in A. arguta (black bars, ripening stages a0–a5; see text) and A. chinensis (gray bars, ripening stages c0–c4; see text). Real-time qPCR analyses were performed on pooled fruit samples using gene-specific primers given in Supplemental Table S2. Error bars are based on four technical replicates, and data were normalized against the housekeeping gene EF1α. Error bars represent ±se.
Functional Testing of Kiwifruit NAC and EIL Transcription Factors
Promoter activation was assessed by comparison of firefly (Photinus pyralis):renilla (Renilla reniformis) luciferase luminescence ratios determined 3 to 4 d after Agrobacterium tumefaciens infiltration into N. benthamiana leaves. AaTPS1pro-911 was activated 3- to 6-fold by all individual AaNAC and AcEIL/AaEIL TFs tested (Fig. 9, A and C). Infiltration with an equimolar mixture of all four AaNAC TFs activated the AaTPS1pro-911 more strongly than any of the individual AaNAC TFs, but the difference was only significantly higher (α = 0.01) than with AaNAC3 and AaNAC4. Infiltration of AaNAC TFs in paired combinations typically (five of the six combinations tested) led to higher and more consistent promoter activation than activation by individual TFs, suggesting that heterodimers might activate more favorably (Fig. 9A). In contrast, AcTPS1pro-3000 and two promoter deletion constructs (AcTPS1pro-600 and AcTPS1pro-229) were all weakly activated (less than 2-fold) by the equimolar mixtures of AcEIL/AaEIL TFs, and no significant activation was observed for the mixture of NAC TFs (Fig. 9B).
Figure 9.
A, Promoter activation assay of AaTPS1pro-911 by A. arguta fruit NAC TF combinations. NAC1 to NAC4, Single TF; NAC1/2, NAC1/3, etc., mix of two NAC TFs; NAC1-4, mix of four TFs. A. tumefaciens harboring 35S-NAC1 to NAC4 cloned from ripe A. arguta fruit cDNA were coinfiltrated into N. benthamiana leaves with AaTPS1pro-911 fused to the firefly luciferase gene. Activation is given by the ratio of LUC/REN. The same vector containing a CaMV 35S promoter-driven GUS reporter gene was used as a baseline activation control. B, Promoter activation of AcTPS1pro-3000, AcTPS1pro-600, and AcTPS1pro-229 by a mix of EIL (n = 5) or a mix of NAC (n = 4) fruit TFs. C, Promoter activation assay of AaTPS1pro-911 by A. arguta and A. chinensis fruit EIL and NAC combinations. EIL1 to EIL4, Single TF; EIL1-5, mix of five EILs; NAC1-4, mix of four NACs; EIL/NAC, mix of five EIL and four NAC TFs. D, Promoter activation of a minimal AaTPS1pro-238, pro-238Mut, and AcTPS1pro-229 promoter with mixtures of EIL and NAC TFs. Nucleotide substitutions in AaTPS1pro-238Mut (underlined) and the proximal NAC core binding motif (boxed) are shown in Supplemental Figure S9. E, Promoter activation assay by NAC TFs of AcTPS1pro-208 (Supplemental Fig. S9) containing one (I), three (II), and five (III) tandem copies of the AaTPS1 NAC binding region (…GTTTACGTATTC…) as shown in F. Experiments are based on eight biological replicates, except C and D (n = 4). All data were log10 transformed prior to statistical analysis to stabilize the residual variation. Pairwise comparisons were performed by Tukey's honestly significant difference test at α = 0.01. Means with the same letter are not significantly different at the 0.01 level.
For the EIL TFs, there was little difference in activation of AaTPS1pro-911 between individual TFs versus a mix of all five EIL TFs (Fig. 9C). Combining all EIL and NAC TFs did not further activate AaTPS1pro-911 compared with NAC TFs alone, suggesting that there are no interactive or additive activation effects between these two classes of TFs.
A 238-bp minimal AaTPS1 promoter clone (AaTPS1pro-238) was generated to compare activation directly with that of the equivalent region in AcTPS1 (AcTPS1pro-229), which contains a substitution in the conserved NAC core-binding motif (ACGTA→ACATA). An AaTPS1pro-238 mutant (pro-238Mut) was also generated that contained five nucleotide substitutions around the NAC core (Supplemental Fig. S9) compared with AaTPS1pro-238. Neither the pro-238Mut nor the AcTPS1pro-229 promoter was activated by AaNAC TFs, whereas the pro-238Mut still showed activation by AcEIL/AaEIL TFs (albeit slightly less than AaTPS1pro-238). These data strongly suggest that a proximal NAC binding region is present in the AaTPS1 promoter and is important for NAC activation, which has been lost in the AcTPS1 promoter.
To test the importance of the NAC binding region further, 12 bp (spanning the conserved core-binding motif) of the NAC binding site in the AaTPS1 promoter was reinserted into the AcTPS1 promoter at position −208 to generate a minimal promoter that contained intact NAC binding sites (Supplemental Fig. S9). Inserting this repeat rescued the activation of AcTPS1pro-208 by AaNAC TFs. Five tandem copies of the repeat restored NAC activation of the minimal 208-bp promoter, whereas one and three copies were insufficient (Fig. 9E). Enhanced autoactivation of the apple MYB10 promoter containing multiple repeats of a MYB binding site has been previously observed (Espley et al., 2009).
In Vitro Binding of AaNACs to the Proximal AaTPS1 NAC Binding Site
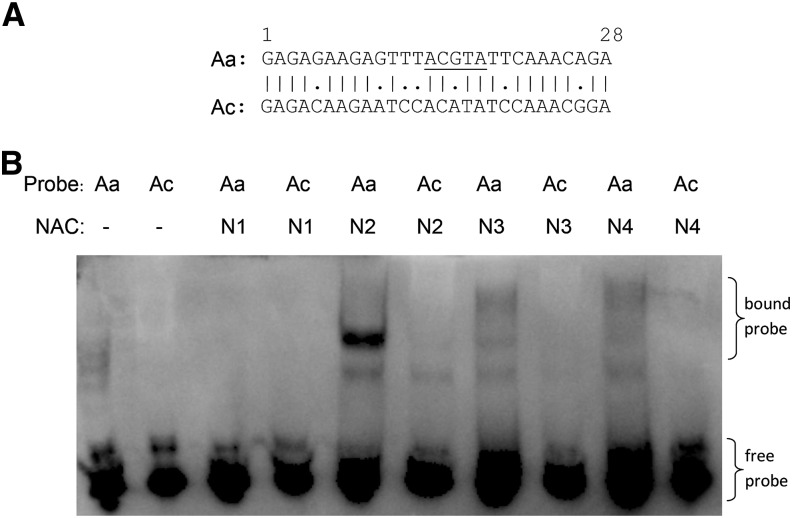
Electrophoretic mobility shift assays (EMSAs) were undertaken to directly test for binding of the four AaNAC TFs to the proximal NAC binding site in the AaTPS1 promoter. The predicted NAC DNA binding domains of all four AaNACs were overexpressed in E. coli and purified by Ni2+ affinity and size exclusion chromatography to more than 95% purity. Based on size exclusion chromatography, all four proteins ran as dimers. The EMSAs demonstrated strong binding between a biotin-labeled 28-bp double-stranded DNA fragment of the proximal AaTPS1 NAC binding site (spanning the conserved core-binding motif) and AaNAC2 and weaker binding to AaNAC3 and AaNAC4 (Fig. 10). No binding was observed using the binding domain of AaNAC1. The equivalent 28-bp fragment from the AcTPS1 promoter with the mutated NAC core-binding motif showed essentially no capacity to bind to the NAC TFs. Together with the transient expression analyses, these results demonstrate the requirement for the proximal NAC binding site and NAC TF binding for controlling monoterpene production in kiwifruit.
Figure 10.
EMSAs show that AaNAC2 to AaNAC4 (N2–N4) bind to a 28-bp double-stranded DNA fragment of the AaTPS1 promoter (Aa) but not to the AcTPS1 promoter (Ac). A, Sequence alignment of the DNA probes used for the EMSA with the NAC core underlined. B, EMSAs of 3′-biotin-labeled double-stranded DNA probes with the AaNAC1-4 DNA binding domain proteins. Lane 1, Aa DNA probe only, no protein; lane 2, Ac DNA probe only, no protein; lanes 3 to 10, Aa or Ac DNA probe incubated with AaNAC1-4 protein combinations.
DISCUSSION
Kiwifruit have only very recently been domesticated, and different accessions and cultivars show wide natural variation in the types and quantity of terpenes they produce during ripening (Crowhurst et al., 2008; Fig. 1; Supplemental Table S3). This variation provides an excellent resource for studying the mechanisms that control terpene production in fruit. Here, we report on the mechanisms that lead to differential monoterpene production in the fruit of two species of kiwifruit: A. arguta ‘Hortgem Tahi,’ which produces high concentrations of terpinolene during the final climacteric ripening stages (Fig. 2), and A. chinensis ‘Hort16A,’ which produces low concentrations of 1,8-cineole during the equivalent stages. In A. arguta ‘Hortgem Tahi,’ the AaTPS1 gene can account for the qualitative variation in ripe fruit monoterpene production (Fig. 4), whereas in A. chinensis ‘Hort16A’ fruit, the AcTPS1 gene is expressed at low levels and makes β-myrcene and geraniol, which are found in negligible amounts during fruit development and ripening. The production of 1,8-cineole in A. chinensis ‘Hort16A’ fruit indicates that a second TPS is expressed during ripening in this cultivar, but the corresponding gene was not identified in ripe fruit EST libraries or genome sequences. Considering the inherent functional plasticity within TPS enzymes (Yoshikuni et al., 2006; Keeling et al., 2008; O’Maille et al., 2008), it is not surprising that AcTPS1 and AaTPS1, despite sharing approximately 90% amino acid homology (excluding chloroplast transit peptide), have very different product specificities.
MEP Pathway Analysis
Quantitative variation in monoterpene production can be controlled by substrate flux through the MEP pathway (Muñoz-Bertomeu et al., 2006). The MEP pathway is subject to multiple levels of regulation, including transcriptional, posttranscriptional, product feedback inhibition, and retrograde chloroplast to nuclear signaling (Cordoba et al., 2009; Banerjee et al., 2013). In A. arguta, expression of DXS increased dramatically during fruit ripening, whereas the transcript levels of other MEP pathway genes were induced to a much lesser extent (Fig. 5). Overexpression of the AcDXS gene in N. benthamiana leaves resulted in significantly increased concentrations of AaTPS1-derived monoterpenes (more than 100-fold), whereas transient overexpression of DXR only resulted in a 2-fold increase in monoterpene production. These results suggest that transcriptional control of the DXS gene is likely to be a key step in controlling terpene substrate flux in A. arguta fruit (Fig. 6). These data contrast with results from Arabidopsis, where overexpression of DXS, DXR, and HDR all led to only moderate (3- to 13-fold) increases in transgenic taxadiene production (Botella-Pavía et al., 2004; Carretero-Paulet et al., 2006), probably because modifying the MEP flux leads to deleterious phytohormone concentrations (abscisic acid/GAs) and alters amounts of chlorophyll and carotenoids in stably transformed transgenic plants.
In A. chinensis, expression of DXS also increased strongly during fruit ripening, and transient overexpression of DXS with AcTPS1 resulted in a 10-fold increase in production of myrcene. This increase was not as large as that observed with AaTPS1, suggesting that structural differences in the two enzymes may influence their respective capacities to use the increased amounts of substrate present. This may also reflect the 3-times reduced turnover rate of AcTPS1 compared with AaTPS1. These results suggested that lack of MEP substrate flux was not a major contributor to the low terpene production observed in ripe A. chinensis ‘Hort16A’ fruit, but rather reduced terpene synthase activity.
Terpene Synthase Transcriptional Regulation
Ethylene biosynthesis is tightly controlled during fruit development, and two modes of regulation have been proposed (Barry et al., 2000). During vegetative growth and in immature fruit, system I ethylene is autoinhibitory, whereas during climacteric ripening and senescence, system II ethylene is autocatalytic and can be induced by ethylene application. In kiwifruit, studies on ethylene biosynthesis (ACO) knockdown A. chinensis ‘Hort16A’ lines indicated that volatile production was tightly associated with system II/climacteric ethylene production (Atkinson et al., 2011). Here, we show that several NAC and EIL TFs are induced during climacteric ripening and expressed at high levels in ripe A. arguta and A. chinensis fruit (Fig. 7). EIL TFs have been shown to be regulated posttranslationally because of stabilization of the protein in the presence of ethylene (Guo and Ecker, 2003; Potuschak et al., 2003; Yanagisawa et al., 2003). The AaTPS1 promoter, which contains several predicted NAC and EIN3 binding sites (Figs. 7 and 9), was activated by all the kiwifruit NAC and EIL TFs tested. EMSA showed that a 28 bp fragment of the AaTPS1 promoter (spanning the conserved core-binding motif), but not the AcTPS1 fragment (containing a mutated form of the core-binding motif), physically interacts with the AaNAC2-4 DNA binding domain. AaNAC1 does not bind to the fragment, so it likely binds to a different site in the AaTPS1 promoter. The EIL activation was similar for all of the class members. Notably, for the NAC class, our data suggest that heterodimers might activate the AaTPS1 promoter more favorably than homodimers, and more strongly than EIL activation. Plant NAC TFs have previously been shown to act as homo- and heterodimers (Hegedus et al., 2003; Jeong et al., 2009). This allows for increased combinatorial potential, as NACs are one of the largest families of plant-specific TFs, with an estimated more than 100 members in various plant species (Ooka et al., 2003; Rushton et al., 2008). Recent work suggests that some NAC members may also physically interact with EIL TFs (Shan et al., 2012), although the biological relevance of this is still unclear.
For most NACs, the biochemical and functional specificity is associated with both the DNA binding domain and the transcriptional regulation domain (Jensen et al., 2010). Furthermore, NAC TFs act not only as activators, since they carry the NAC repression domain (Hao et al., 2010). It is conceivable that, prior to fruit ripening, other flavor-related gene promoters and promoters involved in ethylene biosynthesis and softening are actively repressed. Such complexity has been observed in tomato, where the action of the APETALA2 (AP2) class TF SlAP2a/AP2a (Chung et al., 2010; Karlova et al., 2011) negatively regulates fruit ripening, whereas the action of positive signals such as LeMADS-RIN (Vrebalov et al., 2002), CLEAR NON-RIPENING/Cnr (Manning et al., 2006), TAGL1 (Vrebalov et al., 2009), LeHB-1 (Lin et al., 2008), and the NAC TFs SlNAC4 and NOR (Martel et al., 2011; Zhu et al., 2014) promotes ripening.
By exploring natural variation in TPS1 gene promoters, we show that high terpene production in A. arguta ripe fruit is mediated through NAC activation of the AaTPS1 promoter, whereas low terpene production in A. chinensis ‘Hort16A’ ripe fruit is due to lack of AcTPS1 expression/NAC activation. NAC TF levels are highly up-regulated in ripening in all kiwifruit species tested, whereas TPS up-regulation is restricted to species producing high levels of terpenes (Supplemental Fig. S3). These results clearly identify that NAC TFs are important in controlling monoterpene production in kiwifruit and are likely to activate or repress ripening traits in many fruits.
MATERIALS AND METHODS
Plant Material and Headspace Volatile Analysis
Fruit were ripened at room temperature without exogenous ethylene application before headspace sampling, with the exception of Actinidia chinensis ‘Hort16A.’ Headspace volatiles from ripe fruit were obtained from a 1- to 1.5-g sample of pulped tissue from five to 10 whole fruit (including skin) and collected for 20 min by a purge and trap method onto Chromosorb105 tubes (Matich et al., 2003). Thermal desorption and GC-MS peak identification were performed as described in Wang et al. (2011). Volatiles were semiquantitatively analyzed by comparison with the average response factor of a standard containing methyl butanoate, ethyl butanoate, hexanol, and methyl benzoate using flame ionization detection (Wang et al., 2011).
Actinidia arguta ‘Hortgem Tahi’ is derived directly from a cross between two wild (noncultivated) parents. Fruit volatiles were trapped for 3 h using sliced fruit at 75 DAP and for 15 min using pulped fruit at 120 DAP (harvest, stage a0) and during ripening (stages a1–a5). Soluble solids content (Supplemental Table S1) and ethylene production were monitored using standard protocols (Atkinson et al., 2011). Fruit firmness was monitored nondestructively as described in Wang et al. (2011). Fruit were distributed into ripening stages based on firmness: a0, harvest (firmness, 8 Newtons [N]); a1, unripe (3.0–2.0 N); a2, near ripe (1.95–1.50 N); a3, eating ripe (1.45–1.00 N); a4, near overripe (0.95–0.70 N); and a5, overripe (less than 0.5 N).
A. chinensis ‘Hort16A’ is derived from a cross between a wild (noncultivated) male parent and a female parent that is one cross from wild material. Fruit volatiles were trapped monthly during development from 30 to 120 DAP for 3 h using sliced fruit and during ripening (stages based on days postethylene treatment: 0 d [c0], 1 d [c1], 2 d [c2], 4 d [c3], and 6 d [c4]) for 15 min using pulped fruit. After harvest (175 DAP), fruit were held at room temperature, then coordinately ripened with ethylene (100 µL L−1 for 20 h). Soluble solids (Supplemental Table S1) content and ethylene production were monitored as described previously. Fruit firmness was measured using a 7.9-mm-diameter probe attached to a GUSS Fruit Texture Analyzer (GUSS Manufacturing Ltd.) with probe speed, trigger force, and measurement distance set to 10 mm s−1, 50 g, and 8 mm, respectively. Fruit were sampled at five ripening stages, c0 to c4, as described earlier. In parallel, material from both cultivars was stored at −80°C for RNA and protein isolation.
Gene Identification and Sequence Alignments
AaTPS1 was identified in an A. arguta ‘Hortgem Tahi’ ripe fruit EST library and AcTPS1 in an A. chinensis ‘Hort16A’ young fruit (14–30 DAP) EST library (Crowhurst et al., 2008) using known TPS sequences in GenBank as query sequences and cloned using the primers and vectors described in Supplemental Table S2A. Kiwifruit MEP pathway genes were identified in available EST assemblies (Crowhurst et al., 2008) using Arabidopsis (Arabidopsis thaliana) MEP pathway genes (Phillips et al., 2008), AT5G16440 (IDI), and AF182827 (geranyl diphosphate synthase small subunit) as query sequences. Full-length AcDXS, AaDXR, and AaIDI genes were cloned from ripe A. arguta and A. chinensis fruit cDNA (stages a3 and c2, respectively) as described in Supplemental Table S2A. AdHDR was obtained directly from an Actinidia deliciosa EST library. Putative NAC-like TFs were identified from predicted gene models based on an in-house, low-coverage, A. chinensis genome assembly and from the kiwifruit genome assembly for A. chinensis ‘Hongyang’ (Huang et al., 2013). Deep-sequencing data of an A. arguta fruit library (stage a3) were then read mapped to identify TFs highly expressed in ripe fruit and to map the likely transcriptional start site. AaNAC1 to AaNAC4/AcNAC1 to AcNAC4 were cloned from ripe cDNA (a3/c2 stage) using the primers and vectors described in Supplemental Table S2B. AaEIL1 to AaEIL4/AcEIL1 to AcEIL4 were also cloned from ripe fruit cDNA (a3/c2 stage) using the primers and vectors described in Supplemental Table S2B, based on available EIL sequences from A. deliciosa (Yin et al., 2010). Chloroplast signal peptide predictions were carried out using ChloroP (Emanuelsson et al., 1999), and sequences were aligned using Geneious (www.geneious.com).
Gene Expression Analysis
Total RNA was extracted according to Nieuwenhuizen et al. (2007) from a mixture of three or more representative fruit and treated with 10 units of DNaseI (Roche Applied Science) before cDNA synthesis. First strand cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit according to the manufacturer’s instructions and diluted 50-fold before use. Relative quantification real-time gene expression analyses were performed on a LightCycler 480 platform using the LightCycler 480 SYBR Green master mix in quadruplicate technical replicates, and results were analyzed using the LightCycler 480 software (Roche) with PCR efficiency corrections for all primer pairs (program: 5 min at 95°C; 40 cycles of 10 s at 95°C, 10 s at 60°C, and 20 s at 72°C; followed by melting curve analysis: 5 s at 95°C, 60 s at 65°C, then ramping at 0.18°C s−1 to 95°C). Gene-specific primers used in PCR are given in Supplemental Table S2C. The data were analyzed using the target-reference ratio calculated with the LightCycler 480 software 1.5 as described in Montefiori et al. (2011), enabling a comparison of the level of expression of two different genes to a stably expressed reference gene (EF1α).
Expression of Putative Terpene Synthases in Vitro and Western Analysis
For functional studies, N-terminal truncated AcTPS1 and AaTPS1 open reading frames were amplified to remove the chloroplast-targeting peptide while retaining the RRX8W motif crucial for cyclic mono-TPS activity and TOPO (Invitrogen) cloned into a bacterial expression vector. Primers and vectors used are given in Supplemental Table S2D. Recombinant N-terminal His-tagged proteins were expressed by autoinduction in Escherichia coli BL21 Star (DE3; Invitrogen) and purified by Ni2+ affinity and size exclusion chromatography as described previously (Nieuwenhuizen et al., 2013). AcTPS1 purified recombinant protein (500 µg) was used to raise a polyclonal antibody in rabbit (AgResearch).
For western analysis, kiwifruit tissue (100 mg) was ground to a fine powder in liquid nitrogen and boiled with 0.4 mL of 4× SDS loading buffer (Schägger and von Jagow, 1987) for 5 min. Protein extracts (10 µL) were separated by reducing SDS-PAGE and transferred to polyvinylidene difluoride membrane (Bio-Rad Laboratories) as previously described (Nieuwenhuizen et al., 2007). Blots were incubated with 1:250 diluted rabbit anti-AcTPS1 primary polyclonal antibody and 1:1,000 diluted secondary Qdot 655 goat anti-rabbit IgG conjugate (Invitrogen) according to Mansfield (1995), and visualized on a Typhoon 9400 scanner (GE Healthcare).
Enantioselective GC-MS and Kinetic Analysis
Purified recombinant AaTPS1 and AcTPS1 were incubated with 100 µm GDP (Keller and Thompson, 1993) at room temperature for 1 h. Reactions were overlaid with a 1:1 mixture of pentane:ether to extract volatile terpenes, according to Green et al. (2012b). Volatile terpenes were solvent extracted from eating ripe A. arguta ‘Hortgem Tahi’ fruit (a3/a4) according to Matich et al. (2003). Enantioselective analysis of terpene products was performed as described in Nieuwenhuizen et al. (2013).
All kinetic analyses were performed in quadruplicate as described in Green et al. (2012b) in buffer containing 50 mm Bis-Tris propane (pH 7.5). For GDP kinetics, 20 mm MgCl2 was included. Cofactor determinations for Mg2+ and Mn2+ were performed in the presence of 50 µM 3H1-GDP. Kinetic constants were calculated from Becquerel data by nonlinear regression of the Michaelis-Menten equation using the Origin 7.5 (Microcal Software Inc.) graphics package.
Transient Expression of TPS, NAC, EIL, and MEP Pathway Genes in Nicotiana benthamiana
Kiwifruit TPS genes and AdHDR were cloned directionally from the original EST plasmids (Crowhurst et al., 2008) using gene-specific primers into pENTR/D-TOPO (Invitrogen). The additional MEP pathway genes (AaDXR, AcDXS, and AaIDI) and NAC and EIL genes were cloned from ripe fruit cDNA by Gateway BP cloning into pDONR221 (Invitrogen) using the primers described in Supplemental Table S2, A and B, and then transferred by Gateway LR reactions as recommended by the manufacturer (Invitrogen) into the binary destination vector pHEX2. pHEX2 contains the Cauliflower mosaic virus (CaMV) 35S promoter and octopine synthase terminator (Hellens et al., 2005). Agrobacterium tumefaciens strain GV3101 infiltration of the previous constructs, including a pHEX2-GUS control and subsequent headspace volatile analysis of N. benthamiana, was carried out as described previously (Green et al., 2012a).
Cloning of AaTPS1 and AcTPS1 Promoters
The AaTPS1 promoter was cloned by inverse PCR (Triglia, 2000). A. arguta genomic DNA was isolated using a DNeasy Plant Mini Kit (Qiagen) and digested with HindIII for 5 h. The reaction was column purified and the DNA ligated overnight at 4°C in 200 µL of reaction buffer using 6 units of T4 DNA ligase (New England Biolabs) according to the manufacturer’s instructions. Promoter fragments were PCR amplified using iProof polymerase (Bio-Rad Laboratories) in 50-µL reactions using 5 µL of the ligation as the template, 98°C denaturation, 65°C annealing, and 1-min extension at 72°C for 35 cycles of amplification using primers F1_717/R1_131 (Supplemental Table S2E). A second, nested round of PCR was performed using 1 µL of the first PCR reaction as the template with primers F2_757/ R2_101 for 20 cycles. Primers AaProF/R containing BamHI/NcoI sites were then used to amplify AaTPS1pro-911. AcTPS1pro-3000 was identified by a BLASTn search of an in-house A. chinensis genomic sequence database. Primers AcProF/R (also containing BamHI/NcoI sites; Supplemental Table S2E) were used to amplify the promoter from A. chinensis ‘Hort16A’ genomic DNA and confirmed by sequencing.
Promoter deletion constructs and mutants were generated by PCR using the primers in Supplemental Table S2E. The 238-bp AaTPS1pro-238 promoter and mutant were generated by PCR using primers F-238 and F-238mut, respectively, in combination with AaProR. The 600-bp AcTPS1pro-600 and 229-bp AcTPS1pro-229 were generated by PCR using primers F-600 and F-229, respectively, in combination with AcProR. The AcTPS1pro-208 constructs (I–III) were generated by PCR using forward primers 208 (I)F, (II)F, and (III)F in combination with AcProR. All fragments (including 5′-UTR) were cloned by BamHI/NcoI digestion into pGreenII 0800-LUC upstream of the LUC reporter gene NcoI site/ATG start, then transferred into A. tumefaciens strain GV3101 harboring pSOUP (Hellens et al., 2005).
Promoter Activation Assays
A. tumefaciens infiltrations of 3- to 4-week-old N. benthamiana leaves were carried out as described previously (Green et al., 2012a) with single promoter:TF combinations in a 1:4 ratio, with the starting concentration for each component having an absorbance at 600 nm of 0.5. A. tumefaciens infiltrations with two TFs were carried out in a 1:2:2 (promoter:TF1:TF2) ratio. Infiltrations with more than two TFs (equimolar mix) were carried out at a 1:4 (promoter:TF mix) ratio. Promoter activation was assessed by comparison of firefly:renilla luciferase luminescence ratios (LUC/REN) determined 3 to 4 d after infiltration according to Hellens et al. (2005).
EMSA
The DNA binding domains of AaNAC1 to AaNAC4 were amplified by PCR (Supplemental Table S2F) and Gateway cloned into the E. coli expression vector pET300 according to the manufacturer’s instructions (Invitrogen), then expressed and purified as described for the terpene synthase enzymes. Complementary 28-bp oligonucleotides for EMSA were 3′-end biotin labeled (Macrogen) and slowly annealed at room temperature after heat denaturation. For the binding assay, 1.5 µg of recombinant NAC protein was mixed with 0.3 pmol of double-stranded DNA probe in binding buffer [0.2 mm dithiothreitol, 0.02 mm EDTA, 5 mm HEPES-KOH, pH 7.6, 30 mm sodium chloride, 0.8 μg of salmon sperm DNA (sheared), 0.2 μg of poly(dI-dC)] in a 20-µL reaction at room temperature for 20 min. The bound complexes were resolved by electrophoresis on native 4% (w/v) polyacrylamide gels in 0.5% (w/v) Tris-borate EDTA buffer containing 5% (v/v) glycerol, pH 8.3, at 200 V for 25 min at 4°C. The gels were electroblotted onto positively charged Hybond N+ membrane (GE Healthcare; 25 V/15 min) and cross-linked using a UVC500 (Hoefer) at 120 mJ cm−2. Blots were blocked in Tris-buffered saline (TBS) containing 3% (w/v) bovine serum albumin for 1 h and incubated with 1:1,000 Streptavidin-Peroxidase Polymer (Sigma) for 1 h in TBS with 0.05% (v/v) Tween 20. All washes (six × 5 min) were in TBS with 0.05% (v/v) Tween 20. Imaging was conducted with ECL Select substrate (GE Healthcare) using a ChemiDoc MP imager (Bio-Rad).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KF319035 (AcTPS1), KF319036 (AaTPS1), KF319037 (AcDXS1), KF319038 (AaDXR1), KF319039 (AdHDR1), KF319040 (AaIDI1), KF319041 (AcEIL1), KF319042 (AcEIL2), KF319043 (AcEIL3), KF319044 (AcEIL4), KF319045 (AaEIL4), KF319046 (AaNAC1), KF319047 (AaNAC2), KF319048 (AaNAC3), KF319049 (AaNAC4), KF319050 (AcNAC1), KF319051 (AcNAC2), KF319052 (AcNAC3), KF319053 (AcNAC4), KF319054 (AcTPS1 promoter), and KF319055 (AaTPS1 promoter).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of the deduced amino acid sequences of AaTPS1 and AcTPS1 and other Tps-b subfamily terpene synthases and an unrooted maximum likelihood tree of AcTPS1 and AaTPS1 with other terpene synthases from kiwifruit, grape (Vitis vinifera), apple (Malus × domestica), tomato (Solanum lycopersicum), and poplar (Populus spp.).
Supplemental Figure S2. Mapping of AcTPS1 exons 1 to 7 onto scaffolds in the kiwifruit genome: amino acid alignment of manually curated kiwifruit TPS1-like gene models and a sequence identity table of aligned kiwifruit TPS1-like amino acid sequences.
Supplemental Figure S3. Real-time qPCR analysis of TPS1 and NAC1-4 gene expression levels in harvest and ripe fruit for six Actinidia spp.
Supplemental Figure S4. GC-MS analysis of the monoterpenes produced by transient expression of AaTPS1 and AcTPS1 in planta.
Supplemental Figure S5. Identifying the transcriptional start site in the 5′-UTR of AaTPS1 and a sequence alignment of AaTPS1, AcTPS1, and Achn286241 promoter regions.
Supplemental Figure S6. Binding sites in the AaTPS1 and AcTPS1 promoter regions.
Supplemental Figure S7. ClustalW alignment of the A. chinensis and A. arguta NAC1 to NAC4 proteins and an unrooted maximum likelihood tree of selected Arabidopsis, A. arguta, and A. chinensis NACs.
Supplemental Figure S8. ClustalW alignment of the A. deliciosa, A. chinensis, and A. arguta EIL1 to EIL4 proteins and an unrooted maximum likelihood tree of A. deliciosa, A. arguta, and A. chinensis EIN3-like TFs compared to Arabidopsis.
Supplemental Figure S9. Sequence alignment of minimal AaTPS1pro and AcTPS1pro promoter fragments.
Supplemental Table S1. Characteristics of A. arguta ‘Hortgem Tahi’ and A. chinensis ‘Hort16A’ ripening fruit.
Supplemental Table S2. Cloning and PCR primers.
Supplemental Table S3. Ripe fruit headspace terpene compounds in 14 kiwifruit species.
Supplementary Material
Acknowledgments
We thank Monica Dragulescu and Wade Wadasinghe for maintaining the plants; Ric Broadhurst (AgResearch) for antibody production; Mark Wohlers for help with statistics; Tim Holmes and Darren Snaith for help with imaging; Robert Schaffer, Kevin Davies, and William Laing for critically reviewing the manuscript; Andrew Gleave and the cloning team for help with vector construction; and Lesley Beuning for preliminary gene mining.
Glossary
- IDP
isopentenyl diphosphate
- DMADP
dimethylallyl diphosphate
- MEP
2-C-methyl-d-erythritol 4-phosphate
- GDP
geranyl diphosphate
- GDPS
geranyl diphosphate synthase
- TF
transcription factor
- GC-MS
gas chromatography-mass spectrometry
- DAP
days after pollination
- qPCR
quantitative PCR
- UTR
untranslated region
- ERE
ethylene response element
- cDNA
complementary DNA
- EMSA
electrophoretic mobility shift assay
- TBS
Tris-buffered saline
Footnotes
This work was supported by the Foundation for Research, Science and Technology of New Zealand (grant nos. CO6X0403 and C11X1007) and the Ministry of Science and Innovation.
References
- Alexander L, Grierson D (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53: 2039–2055 [DOI] [PubMed] [Google Scholar]
- Atkinson RG, Gunaseelan K, Wang MY, Luo L, Wang T, Norling CL, Johnston SL, Maddumage R, Schröder R, Schaffer RJ (2011) Dissecting the role of climacteric ethylene in kiwifruit (Actinidia chinensis) ripening using a 1-aminocyclopropane-1-carboxylic acid oxidase knockdown line. J Exp Bot 62: 3821–3835 [DOI] [PubMed] [Google Scholar]
- Banerjee A, Wu Y, Banerjee R, Li Y, Yan H, Sharkey TD (2013) Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway. J Biol Chem 288: 16926–16936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, Llop-Tous MI, Grierson D (2000) The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol 123: 979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battilana J, Costantini L, Emanuelli F, Sevini F, Segala C, Moser S, Velasco R, Versini G, Stella Grando M (2009) The 1-deoxy-D: -xylulose 5-phosphate synthase gene co-localizes with a major QTL affecting monoterpene content in grapevine. Theor Appl Genet 118: 653–669 [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella-Pavía P, Besumbes O, Phillips MA, Carretero-Paulet L, Boronat A, Rodríguez-Concepción M (2004) Regulation of carotenoid biosynthesis in plants: evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J 40: 188–199 [DOI] [PubMed] [Google Scholar]
- Buttery RG, Teranishi R, Ling LC, Flath RA, Stern DJ (1988) Quantitative studies on origins of fresh tomato aroma volatiles. J Agric Food Chem 36: 1247–1250 [Google Scholar]
- Cane DE, Kang I (2000) Aristolochene synthase: purification, molecular cloning, high-level expression in Escherichia coli, and characterization of the Aspergillus terreus cyclase. Arch Biochem Biophys 376: 354–364 [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Cairó A, Botella-Pavía P, Besumbes O, Campos N, Boronat A, Rodríguez-Concepción M (2006) Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Mol Biol 62: 683–695 [DOI] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SS, Schmitz RJ, Urich MA, Kuo D, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66: 212–229 [DOI] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Cordoba E, Salmi M, León P (2009) Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J Exp Bot 60: 2933–2943 [DOI] [PubMed] [Google Scholar]
- Crowhurst RN, Gleave AP, MacRae EA, Ampomah-Dwamena C, Atkinson RG, Beuning LL, Bulley SM, Chagne D, Marsh KB, Matich AJ, et al. (2008) Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics 9: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Miettinen K, Goedbloed M, Verstappen FW, Voster A, Jongsma MA, Memelink J, van der Krol S, Bouwmeester HJ (2013) Characterization of two geraniol synthases from Valeriana officinalis and Lippia dulcis: similar activity but difference in subcellular localization. Metab Eng 20: 198–211 [DOI] [PubMed] [Google Scholar]
- Duchêne E, Butterlin G, Claudel P, Dumas V, Jaegli N, Merdinoglu D (2009) A grapevine (Vitis vinifera L.) deoxy-D: -xylulose synthase gene colocates with a major quantitative trait loci for terpenol content. Theor Appl Genet 118: 541–552 [DOI] [PubMed] [Google Scholar]
- Dudareva N, Cseke L, Blanc VM, Pichersky E (1996) Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enfissi EM, Fraser PD, Lois LM, Boronat A, Schuch W, Bramley PM (2005) Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotechnol J 3: 17–27 [DOI] [PubMed] [Google Scholar]
- Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP. , et al (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21: 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez JM, Cantero A, Reindl A, Reichler S, León P (2001) 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem 276: 22901–22909 [DOI] [PubMed] [Google Scholar]
- Green SA, Chen X, Matich AJ (2012a) In planta transient expression analysis of monoterpene synthases. Methods Enzymol 515: 43–61 [DOI] [PubMed] [Google Scholar]
- Green SA, Chen X, Nieuwenhuizen NJ, Matich AJ, Wang MY, Bunn BJ, Yauk YK, Atkinson RG (2012b) Identification, functional characterization, and regulation of the enzyme responsible for floral (E)-nerolidol biosynthesis in kiwifruit (Actinidia chinensis). J Exp Bot 63: 1951–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Hall DE, Robert JA, Keeling CI, Domanski D, Quesada AL, Jancsik S, Kuzyk MA, Hamberger B, Borchers CH, Bohlmann J (2011) An integrated genomic, proteomic and biochemical analysis of (+)-3-carene biosynthesis in Sitka spruce (Picea sitchensis) genotypes that are resistant or susceptible to white pine weevil. Plant J 65: 936–948 [DOI] [PubMed] [Google Scholar]
- Hao YJ, Song QX, Chen HW, Zou HF, Wei W, Kang XS, Ma B, Zhang WK, Zhang JS, Chen SY (2010) Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232: 1033–1043 [DOI] [PubMed] [Google Scholar]
- Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53: 383–397 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Ding J, Deng D, Tang W, Sun H, Liu D, Zhang L, Niu X, Zhang X, Meng M, et al. (2013) Draft genome of the kiwifruit Actinidia chinensis. Nat Commun 4: 2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A (2009) TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K (2010) The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J 426: 183–196 [DOI] [PubMed] [Google Scholar]
- Jeong J, Park Y, Jung H, Park SH, Kim JK (2009) Rice NAC proteins act as homodimers and heterodimers. Plant Biotechnol Rep 3: 127–134 [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling CI, Weisshaar S, Lin RPC, Bohlmann J (2008) Functional plasticity of paralogous diterpene synthases involved in conifer defense. Proc Natl Acad Sci USA 105: 1085–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller RK, Thompson R (1993) Rapid synthesis of isoprenoid diphosphates and their isolation in one step using either thin layer or flash chromatography. J Chromatogr A 645: 161–167 [DOI] [PubMed] [Google Scholar]
- Köllner TG, Held M, Lenk C, Hiltpold I, Turlings TC, Gershenzon J, Degenhardt J (2008) A maize (E)-beta-caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most American maize varieties. Plant Cell 20: 482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou X, Watkins CB, Gan SS (2012) Arabidopsis AtNAP regulates fruit senescence. J Exp Bot 63: 6139–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhu B, Xu W, Zhu H, Chen A, Xie Y, Shao Y, Luo Y (2007) LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep 26: 1999–2008 [DOI] [PubMed] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D (2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois LM, Rodríguez-Concepción M, Gallego F, Campos N, Boronat A (2000) Carotenoid biosynthesis during tomato fruit development: regulatory role of 1-deoxy-D-xylulose 5-phosphate synthase. Plant J 22: 503–513 [DOI] [PubMed] [Google Scholar]
- MacLeod AJ, Pieris NM (1984) Comparison of the volatile components of some mango cultivars. Phytochemistry 23: 361–366 [Google Scholar]
- Mahmoud SS, Croteau RB (2001) Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc Natl Acad Sci USA 98: 8915–8920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Mansfield MA. (1995) Rapid immunodetection on polyvinylidene fluoride membrane blots without blocking. Anal Biochem 229: 140–143 [DOI] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ (2011) The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol 157: 1568–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matich AJ, Young H, Allen JM, Wang MY, Fielder S, McNeilage MA, MacRae EA (2003) Actinidia arguta: volatile compounds in fruit and flowers. Phytochemistry 63: 285–301 [DOI] [PubMed] [Google Scholar]
- Matthews PD, Wurtzel ET (2000) Metabolic engineering of carotenoid accumulation in Escherichia coli by modulation of the isoprenoid precursor pool with expression of deoxyxylulose phosphate synthase. Appl Microbiol Biotechnol 53: 396–400 [DOI] [PubMed] [Google Scholar]
- Montefiori M, Espley RV, Stevenson D, Cooney J, Datson PM, Saiz A, Atkinson RG, Hellens RP, Allan AC (2011) Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J 65: 106–118 [DOI] [PubMed] [Google Scholar]
- Moufida S, Marzouk B (2003) Biochemical characterization of blood orange, sweet orange, lemon, bergamot and bitter orange. Phytochemistry 62: 1283–1289 [DOI] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Arrillaga I, Ros R, Segura J (2006) Up-regulation of 1-deoxy-d-xylulose-5-phosphate synthase enhances production of essential oils in transgenic spike lavender. Plant Physiol 142: 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuizen NJ, Beuning LL, Sutherland PW, Sharma NN, Cooney JM, Bieleski LRF, Schröder R, MacRae EA, Atkinson RG (2007) Identification and characterisation of acidic and novel basic forms of actinidin, the highly abundant cysteine protease from kiwifruit. Funct Plant Biol 34: 946–961 [DOI] [PubMed] [Google Scholar]
- Nieuwenhuizen NJ, Green SA, Chen X, Bailleul EJ, Matich AJ, Wang MY, Atkinson RG (2013) Functional genomics reveals that a compact terpene synthase gene family can account for terpene volatile production in apple. Plant Physiol 161: 787–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Maille PE, Malone A, Dellas N, Andes Hess B Jr, Smentek L, Sheehan I, Greenhagen BT, Chappell J, Manning G, Noel JP (2008) Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat Chem Biol 4: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Orlova I, Nagegowda DA, Kish CM, Gutensohn M, Maeda H, Varbanova M, Fridman E, Yamaguchi S, Hanada A, Kamiya Y, et al. (2009) The small subunit of snapdragon geranyl diphosphate synthase modifies the chain length specificity of tobacco geranylgeranyl diphosphate synthase in planta. Plant Cell 21: 4002–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MA, León P, Boronat A, Rodríguez-Concepción M (2008) The plastidial MEP pathway: unified nomenclature and resources. Trends Plant Sci 13: 619–623 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Rodríguez A, Alquézar B, Peña L (2013) Fruit aromas in mature fleshy fruits as signals of readiness for predation and seed dispersal. New Phytol 197: 36–48 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Boronat A (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol 130: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Bokowiec MT, Han S, Zhang H, Brannock JF, Chen X, Laudeman TW, Timko MP (2008) Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol 147: 280–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynkiewicz MJ, Cane DE, Christianson DW (2001) Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proc Natl Acad Sci USA 98: 13543–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Shan W, Kuang JF, Chen L, Xie H, Peng HH, Xiao YY, Li XP, Chen WX, He QG, Chen JY. , et al (2012) Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J Exp Bot 63: 5171–5187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T. (2000) Inverse PCR (IPCR) for obtaining promoter sequence. Methods Mol Biol 130: 79–83 [DOI] [PubMed] [Google Scholar]
- Verdonk JC, Haring MA, van Tunen AJ, Schuurink RC (2005) ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17: 1612–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJ, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J. , et al (2009) Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang MY, Wohlers M, Chen X, Marsh KB (2011) Changes in volatile production and sensory quality of Actinidia arguta fruit during fruit maturation. In G Costa, AR Ferguson, eds, Proceedings of the VIIth International Symposium on Kiwifruit, Faenza, Italy. Acta Hortic 913: 677–684 [Google Scholar]
- Williams DC, McGarvey DJ, Katahira EJ, Croteau R (1998) Truncation of limonene synthase preprotein provides a fully active ‘pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry 37: 12213–12220 [DOI] [PubMed] [Google Scholar]
- Xiao YY, Chen JY, Kuang JF, Shan W, Xie H, Jiang YM, Lu WJ (2013) Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. J Exp Bot 64: 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14: 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YH, Wang JW, Wang S, Wang JY, Chen XY (2004) Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol 135: 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP. (2005) A CELD-fusion method for rapid determination of the DNA-binding sequence specificity of novel plant DNA-binding proteins. Plant J 41: 638–649 [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, Terada T, et al. (2005) Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3. J Mol Biol 348: 253–264 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Yin XR, Allan AC, Chen KS, Ferguson IB (2010) Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol 153: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikuni Y, Ferrin TE, Keasling JD (2006) Designed divergent evolution of enzyme function. Nature 440: 1078–1082 [DOI] [PubMed] [Google Scholar]
- Zhang K, Gan SS (2012) An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol 158: 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang H, Quan R, Wang XC, Huang R (2009) Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol 150: 365–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Chen G, Zhou S, Tu Y, Wang Y, Dong T, Hu Z (2014) A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol 55: 119–135 [DOI] [PubMed] [Google Scholar]
- Zvi MM, Shklarman E, Masci T, Kalev H, Debener T, Shafir S, Ovadis M, Vainstein A (2012) PAP1 transcription factor enhances production of phenylpropanoid and terpenoid scent compounds in rose flowers. New Phytol 195: 335–345 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.