Suppression of stem cell niche respecification in the shoot meristem periphery is mediated by a putative carboxypeptidase.
Abstract
Plants are able to reiteratively form new organs in an environmentally adaptive manner during postembryonic development. Organ formation in plants is dependent on stem cell niches (SCNs), which are located in the so-called meristems. Meristems show a functional zonation along the apical-basal axis and the radial axis. Shoot apical meristems of higher plants are dome-like structures, which contain a central SCN that consists of an apical stem cell pool and an underlying organizing center. Organ primordia are formed in the circular peripheral zone (PZ) from stem cell descendants in which differentiation programs are activated. One mechanism to keep this radial symmetry integrated is that the existing SCN actively suppresses stem cell identity in the PZ. However, how this lateral inhibition system works at the molecular level is far from understood. Here, we show that a defect in the putative carboxypeptidase ALTERED MERISTEM PROGRAM1 (AMP1) causes the formation of extra SCNs in the presence of an intact primary shoot apical meristem, which at least partially contributes to the enhanced shoot meristem size and leaf initiation rate found in the mutant. This defect appears to be neither a specific consequence of the altered cytokinin levels in amp1 nor directly mediated by the WUSCHEL/CLAVATA feedback loop. De novo formation of supernumerary stem cell pools was further enhanced in plants mutated in both AMP1 and its paralog LIKE AMP1, indicating that they exhibit partially overlapping roles to suppress SCN respecification in the PZ.
As sessile organisms, plants have evolved an indeterminate mode of development where organs are generated continuously and in an environmentally adaptive manner. The formation of aboveground organs is achieved by the activity of a stem cell harboring structure called the shoot apical meristem (SAM). In higher plants, the SAM has a dome-like structure and consists of different zones with distinct functions. The central zone (CZ) consists of an apical stem cell pool and an underlying organizing center (OC). Stem cell descendants, entering the peripheral zone (PZ), divide frequently and undergo transition to a determinate cell fate during the process of organ initiation (Barton, 2010). To ensure directional and coordinated growth of the shoot, it is essential that this structural organization is maintained. Intermeristematic gradients of molecular signals, which are readjusted in a continuous feedback, together with mechanical constraints, contribute to a self-organizing mode of SAM patterning (Aichinger et al., 2012; Murray et al., 2012).
One key player in the process of SAM formation and maintenance is the plant hormone cytokinin (CK). CK is sufficient to trigger de novo SAM formation (Skoog and Miller, 1957; Estruch et al., 1991). KNOTTED-like homeobox transcription factors, which are required to preserve the meristematic status in the SAM, mediate local synthesis of CK (Jasinski et al., 2005; Yanai et al., 2005). Epidermis-specific expression of CK-activating enzymes of the LONELY GUY family and presence of the CK receptor ARABIDOPSIS HISTIDINE KINASE4 (AHK4) in the central domain contribute to the apical-basal positioning of an area of high CK signaling activity in the SAM (Kurakawa et al., 2007; Gordon et al., 2009; Kuroha et al., 2009; Chickarmane et al., 2012). This CK response maximum reinforces expression of the homeodomain transcription factor WUSCHEL (WUS), which specifies OC identity (Gordon et al., 2009). By promoting WUS expression, CK also positively affects OC size and subsequently the dimension of the overlying stem cell pool. WUS maintains the identity of stem cells by entering them by intercellular movement, where it also directly induces transcription of CLAVATA3 (CLV3; Brand et al., 2000; Schoof et al., 2000; Yadav et al., 2011). The peptide CLV3, in turn, diffuses back to more basal regions of the meristem and limits lateral expansion of WUS expression (Müller et al., 2008). Thus, CK, WUS, and CLV3 interact to ensure the correct apical basal placement and size of the stem cell niche (SCN) in the SAM (Heidstra and Sabatini, 2014).
Less is known about the molecular factors, which maintain the radial integrity of the SCN by suppressing pluripotency in the PZ. Not surprisingly, chromatin assembly and remodeling play a key role in inducing and maintaining cell fate determination along this boundary. Mutants affected in nucleosome assembly, modification, or positioning have been shown to be associated with SAM disorganization and ectopic reappearance of stem cell/OC identity in the PZ (Kaya et al., 2001; Guyomarc’h et al., 2004; Suzuki et al., 2004; Takeda et al., 2004; Han et al., 2008; Graf et al., 2010). However, how changes in chromatin structure are imposed in the spatial context of the CZ/PZ boundary is not understood. It has been shown that microsurgical elimination of the CZ or only the OC results in de novo formation of a new SCN in the periphery of the meristem (Pilkington, 1929; Sussex, 1952; Loiseau, 1959; Reinhardt et al., 2003). Moreover, mutants affected in SCN maintenance also generate accessory SCNs in the primary SAM periphery usually after a certain lag phase (Laux et al., 1996; Würschum et al., 2006). These data indicate the existence of a lateral inhibition mechanism in which the primary SCN constantly communicates its presence to PZ cells and thereby actively suppresses reappearance of stem cell identity in the periphery. The molecular nature of the involved signals is unknown. It is also unclear to which extent the WUS/CLV3/CK regulatory module contributes to this mechanism (Reinhardt et al., 2003; Hohm et al., 2010).
Loss-of-function mutation of ALTERED MERISTEM PROGRAM1 (AMP1) in Arabidopsis (Arabidopsis thaliana) results in a pleiotropic developmental phenotype; most prominent is an exaggerated activity of the SAM during certain developmental stages. Mutant embryos form a larger SAM pole, which generates supernumerary cotyledons and true leaf primordia before germination (Conway and Poethig, 1997; Vidaurre et al., 2007). During the vegetative growth phase, the enlarged mutant SAM gives rise to a strongly enhanced leaf formation rate (Chaudhury et al., 1993; Nogué et al., 2000a). In contrast to other mutants with deregulated shoot meristem activity, amp1 SAM size normalizes after floral transition, and altered shoot meristem activity appears to be limited to decreased apical dominance (Vidaurre et al., 2007). Mutants of AMP1 orthologs in Zea mays, rice (Oryza sativa), and Lotus japonicus generate comparable SAM defects, indicating that the function of this factor in respect to shoot development is conserved among higher plants (Suzuki et al., 2008; Kawakatsu et al., 2009; Suzaki et al., 2013; Lv et al., 2014).
AMP1 encodes a putative Glu carboxypeptidase with unknown biochemical function (Helliwell et al., 2001). The best characterized structural homolog is human glutamate carboxypeptidase II (GCPII), which acts as an N-acetylated α-linked acidic dipeptidase important to modulate neurotransmitter concentrations in neuronal tissues (Robinson et al., 1987). The same enzyme also exerts folyl-poly-γ-Glu carboxypeptidase activity, which is thought to support cellular folate uptake (Pinto et al., 1996). Moreover, GCPII is used as a tumor marker because it is strongly up-regulated in most solid tumors, yet its role in cancer development is unclear (Hlouchová et al., 2012). To which extent these GCPII-specific catalytic activities are conserved in AMP1 has not been analyzed in detail; however, the limited level of structural conservation in the putative substrate-binding domains rather support different substrate specificities (Davis et al., 2005).
Originally, the altered SAM activity as well as additional phenotypes of amp1, including constitutive photomorphogenesis, enhanced shoot regeneration, and increased branching, were explained by the increased CK biosynthesis rate found in the mutant. (Chin-Atkins et al., 1996; Nogué et al., 2000b). The increased CK content has been suggested to also be responsible for the enhanced resistance against exogenous nitrite oxide treatment (Liu et al., 2013). However, other studies revealed that amp1 mutants show additional phenotypes, which are more difficult to explain by the elevated CK content. For example, amp1 alleles show alterations in seed dormancy and timing of phase transitions as well as a strongly enhanced leaf formation rate (Chaudhury et al., 1993; Conway and Poethig, 1997; Telfer et al., 1997; Griffiths et al., 2011). Other hormone pathways are also affected in amp1, including GA, ethylene, and abscisic acid, and AMP1 absence abrogates the requirement of the auxin-dependent transcription factor AUXIN RESPONSE FACTOR5 in the process of embryonic root formation (Saibo et al., 2007; Vidaurre et al., 2007; Shi et al., 2013a, 2013b; Yao et al., 2014). Recently, it has been shown that the translation rate of micro-RNA (miRNA)-controlled proteins in amp1 is specifically enhanced at endoplasmic reticulum (ER)-located polysomes by an unknown mechanism, suggesting that at least some of the amp1 phenotypes might be caused by deregulated activity of miRNA-controlled pathways (Li et al., 2013). Hence, AMP1 represents a key regulator of plant development and adaptive growth responses; however, how the diverse functions of this protein converge at the molecular level is not resolved.
How SAM architecture and activity is altered in detail in amp1 and to which extent these SAM-related defects can be directly attributed to the elevated CK biosynthesis rate found in the mutant are important unsolved questions to better position AMP1 function in the known regulatory network controlling SAM development. To this end, we dissected the causal relationship of amp1 phenotypes and altered CK accumulation and found that SAM size and SAM primordia formation changes are not directly dependent on CK levels. Moreover, we found that amp1 SAMs show ectopic SCN formation, which is also a largely CK-independent process. Finally, we also show that LIKE AMP1 (LAMP1) acts partially redundantly with AMP1 to suppress OC respecification in the PZ in the presence of an intact SCN.
RESULTS
Increased SAM Size in amp1 Coincides with the Formation of Ectopic Stem Cell Niches
Mutation of AMP1 causes a considerable expansion of SAM size during vegetative development (Nogué et al., 2000a; Saibo et al., 2007; Vidaurre et al., 2007). To specify which areas of the SAM contribute to this size gain, we analyzed longitudinal sections of 6-d-old amp1-1 seedlings (Fig. 1, A and B). At this stage, the amp1-1 shoot meristematic area was approximately three times bigger compared with the wild type (Fig. 1E), and it exhibited a broader and higher meristem dome (Fig. 1B). The L1 and L2 layers appeared to be normally arranged in amp1-1; however, the number of cell layers in the corpus was increased and the mutant SAM showed a clear lateral expansion by bending around a central area of differentiating pith tissue. To analyze whether increased meristem size correlated with altered OC organization, we performed WUS::GUS reporter analysis in amp1-1 median longitudinal SAM sections. WUS::GUS activity in the wild type is restricted to the CZ defining the OC (Fig. 1F). In amp1-1, the WUS::GUS signal appeared weaker and more diffuse (Fig. 1G) compared with the wild type.
Figure 1.
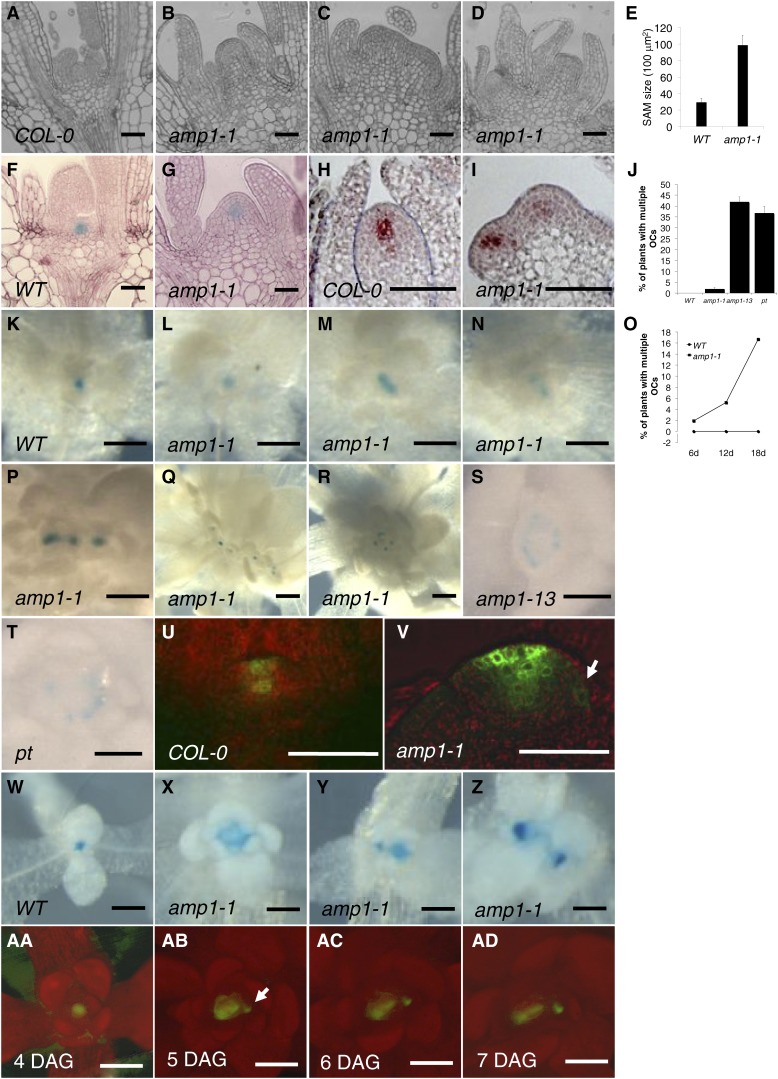
amp1 SAMs form ectopic SCNs. A to D, Median longitudinal SAM sections of seedlings at 6 DAG. A, The ecotype Columbia wild type (WT). B, amp1-1 single SAM with increased size. C, amp1-1 SAM forming an ectopic SCN. D, amp1-1 shoot apex with two independent SCNs. E, Quantification of SAM size from median longitudinal sections at 6 DAG (means ± se of the mean; n ≥ 3). F and G, WUS::GUS localization in median longitudinal SAM sections of the wild type (F) and amp1-1 (G) at 6 DAG. H and I, In situ hybridization showing WUS expression in median longitudinal SAM sections at 9 DAG. H, Wild-type SAM with the typical central WUS expression domain defining the OC. I, amp1-1 SAM showing two distinct WUS expression domains in close neighborhood. J, Percentage of plants with multiple OCs at 6 DAG in different amp1 alleles (means ± se of the mean from three experimental repeats; n ≥ 200 for each repeat). K to N and P to T, WUS::GUS localization in SAMs of different amp1 alleles at different developmental stages grown under short-day conditions to avoid floral transition. K, The wild type (6 DAG). L, amp1-1 (6 DAG) with a single diffuse OC. M, amp1-1 (6 DAG) showing band-like expansion of WUS domain. N, amp1-1 (6 DAG) with two distinct WUS foci. P, amp1-1 (12 DAG) showing three distinct OCs. Q and R, amp1-1 (18 DAG). S, amp1-13 (6 DAG). T, pt (6 DAG). O, Ontogenetic increase in OC numbers in amp1-1. Percentage of plants with multiple SAM OCs at the indicated developmental stage (n ≥ 200, per developmental stage). U and V, CLV3::GFP reporter analysis in median longitudinal cryo sections of 6-d-old SAMs. U, The wild type. V, amp1-1. The arrow indicates lateral expansion of CLV3::GFP signal in the L1 layer. W to Z, CLV3::GUS localization in SAMs of plants grown under short-day conditions to avoid floral transition at 12 DAG. W, The wild type. X, Overlapping stem cell pools in a radial constellation in amp1-1. Y, Formation of ectopic stem cell pool in close proximity of the primary SCN. Z, Twin SAM with similar-sized CLV3::GUS foci in amp1-1. AA to AD, CLV3::GFP time lapse analysis of a pt SAM between 4 and 7 DAG showing expansion of the original stem cell pool and de novo formation of ectopic stem cells (white arrow). Bars = 50 μm (A–D, F–I, U, and V) and 200 μm (K–N, P–T, and W–AD).
At this early growth stage, we observed that a significant number of amp1-1 seedlings developed adventitious SAM domes from the PZ of the primary SAM, which resulted in the presence of two independent functional meristems in one shoot apex (Fig. 1, C and D). This process was accompanied by the formation of two distinct WUS expression domains (Fig. 1, H and I). Importantly, we did not detect any signs of stem cell differentiation in the primary SCN, which has been described to cause de novo adventitious SAM formation. We quantified the frequency of this ectopic OC formation phenotype in amp1-1 plants by following WUS::GUS activity during vegetative development under short-day conditions to avoid floral transition. Six days after germination (DAG), 98% of amp1-1 seedlings showed a single WUS::GUS signal, which was broader but weaker compared with the wild type in the central area of the SAM (Fig. 1, K and L). The residual 2% showed two either overlapping or clearly separated WUS expression domains (Fig. 1, M–O). At day 12, the percentage of plants showing SAMs with multiple WUS::GUS expression domains raised to 5%, with the number of distinct foci per shoot apex ranging from two to five (Fig. 1, P and O). Finally, at day 18, 17% of amp1-1 plants showed more than one clearly distinguishable OCs, which were individually surrounded by leaf primordia and positioned either in a linear or circular manner (Fig. 1, Q, R, and O). Because amp1-1 has been described as a weak allele (Vidaurre et al., 2007), we analyzed WUS::GUS activity also in the potential null alleles amp1-13 and primordia timing (pt). Notably, for both strong mutant alleles, more than one-third of the plants exhibited ectopic OCs already at day 6, mainly in a circular constellation (Fig. 1, S, T, and J).
To test how this ectopic formation of OC-specified cell pools in amp1 affects stem cell behavior, we analyzed the expression domain of CLV3 transcripts by using GFP and GUS reporter lines. In amp1-1, the CLV3::GFP expression domain in the primary SCN was deeper and broader compared with the wild type, comprising around five cell layers in the basal direction (Fig. 1, U and V). Moreover, we also observed peripheral extension of CLV3::GFP expression along the L1 layer, most likely in areas where ectopic SAM formation started to occur (Fig. 1V, arrow). Whole-mount analysis of CLV3::GUS confirmed a substantial increase of the initial stem cell pool size in amp1-1, and we observed the formation of additional distinct CLV3::GUS positive domains with a similar frequency as it was found for WUS::GUS (7% at 12 DAG; Fig. 1, W–Z). However, in contrast to WUS::GUS, the ectopic CLV3::GUS-expressing foci had a stronger tendency to maintain areas of a partial overlap. To define the origin of additional stem cell pools in amp1, we made time lapse studies using the CLV3::GFP reporter in pt. Shoot apices were pictured every 24 h between 4 and 7 DAG (Fig. 1, AA–AD). In plants where SCN multiplication occurred, we observed de novo formation of CLV3::GFP-expressing foci as well as overlapping cloud-like expansion and subsequent division of the original stem cell pool. However, the later phenomenon could also be interpreted as de novo formation in close proximity. Taken together, amp1 primary SAMs show a strong tendency to generate fully active adventitious meristems, resulting in composite rosettes, which consist of multiple, hierarchically equivalent, vegetative shoot apices.
amp1 Shows Elevated IPT3 Expression Levels in Extrameristematic Tissues and Ectopic CK Response Maxima in the SAM
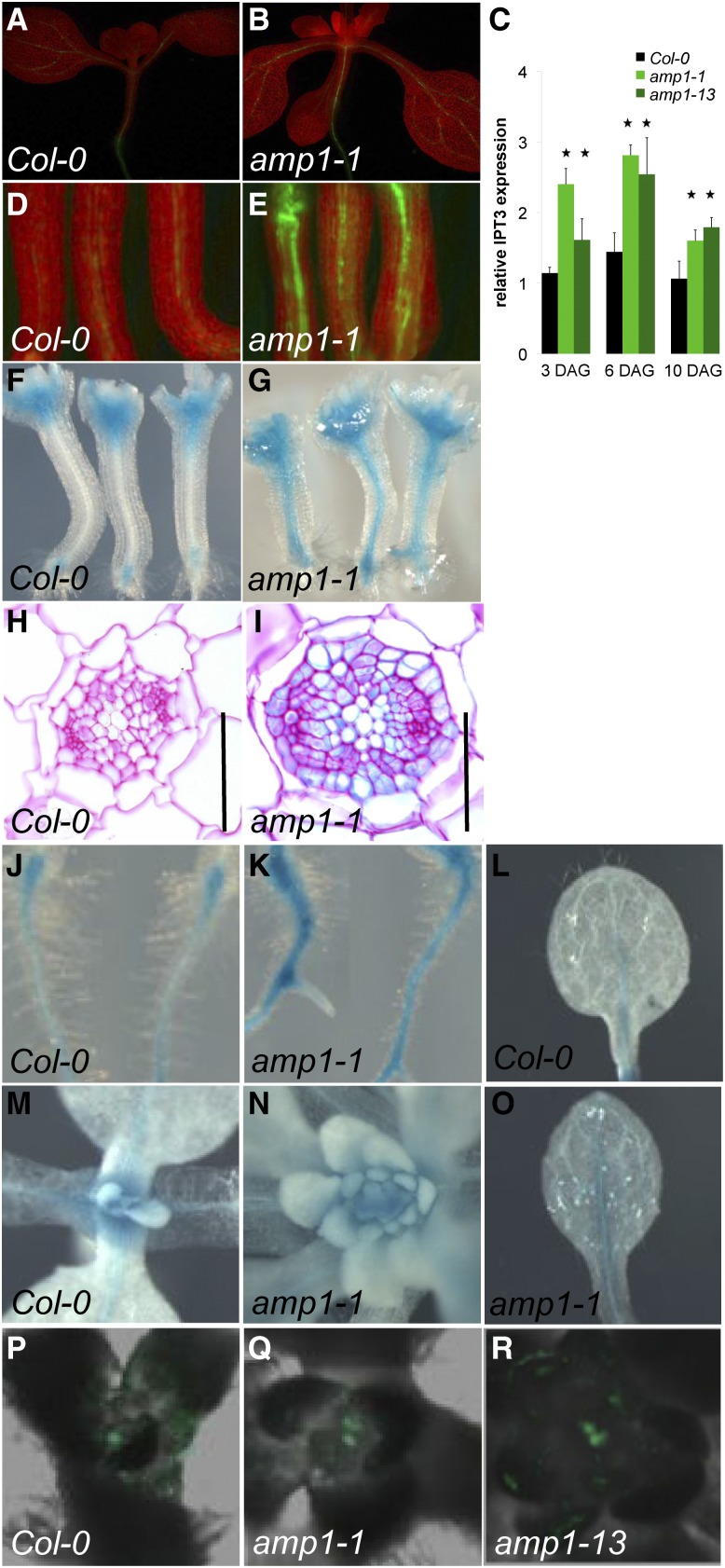
Total CK levels have been shown to be elevated in amp1 seedlings due to a higher CK biosynthesis rate; however, the molecular cause is not known (Nogué et al., 2000b). We compared the expression levels of the seven ISOPENTENYL TRANSFERASE (IPT) genes known to participate in CK biosynthesis by quantitative reverse transcription (qRT)-PCR and transcriptional reporter analysis and found that IPT3 was significantly up-regulated in amp1 seedlings (Fig. 2, A–E). Notably, AtIPT3::GFP levels in amp1 were increased but stayed restricted to extrameristematic tissues, mainly in vascular-associated cells throughout the plant (Fig. 2, B and E). Consistently, this increase of IPT3 expression spatially overlapped with elevated CK responses detected by ARABIDOPSIS RESPONSE REGULATOR5 (ARR5)::GUS analysis. This vascular bundle-specific enhancement of ARR5::GUS activity was visible in leaf and root tissues but was most striking in the hypocotyl stele (Fig. 2, F–O). The strong reporter activity in this tissue correlated with abnormal cell proliferation events, leading to an increase of procambial and pericycle cells surrounding the xylem and phloem strands (Fig. 2, H and I). ARR5::GUS was also expressed in the SAM, which is increased in amp1; however, there was no obvious difference in the GUS staining intensity when compared with the wild type. (Fig. 2, M and N). To evaluate CK responses in the SAM, we analyzed Two Component Signaling Sensor (TCS)::GFP reporter activity in amp1, which has been shown to visualize CK response maxima and colocalize with the OC in inflorescence meristems (Müller and Sheen, 2008; Gordon et al., 2009). Whereas, in the wild type, one central TCS::GFP positive domain was present, we detected the occurrence of multiple TCS::GFP foci in amp1-1 and amp1-13, with a frequency similar to that found for the WUS::GUS reporter (Fig. 2, P–R). Thus, amp1 SAMs show ectopic formation of distinct CK response maxima with matches with the supernumerary OC formation defect of the mutant.
Figure 2.
Tissue-specific expression analysis of IPT3 and CK response markers in amp1. A, B, D, and E, IPT3::GFP reporter analysis in 7-d-old wild-type (A and D) and amp1-1 (B and E) seedlings. D and E show hypocotyl region of three individual plants. C, Expression analysis of IPT3 in the wild type and amp1 by qRT-PCR. Relative transcript levels (mean ± sd) were calculated from triplicate qRT-PCR reactions of independent RNA samples prepared from three different sets of seedlings for each developmental stage. Relative expression is normalized to glyceraldehyde 3-phosphate dehydrogenase C2 (GAPC2). Asterisks indicate statistical significance (P < 0.05, n = 3). F to O, ARR5::GUS analysis in 7-d-old seedlings. Hypocotyls of the wild type (F) and amp1-1 (G). Transversal section of the vascular cylinder from the central hypocotyl of the wild type (H) and amp1-1 (I). J, Top root of the wild type. K, amp1-1. SAM area of the wild type (M) and amp1-1 (N). L, True leaf of the wild type. O, True leaf of amp1-1. P to R, TCS::GFP analysis in SAMs of 7-d-old seedlings. P, The wild type with one central TCS::GFP domain. amp1-1 (Q) and amp1-13 (R) show two distinct TCS::GFP foci in one SAM. Bars = 50 μm.
Exogenous CK Application or Endogenous Increase of CK Biosynthesis Do Not Phenocopy amp1 SAM Phenotypes
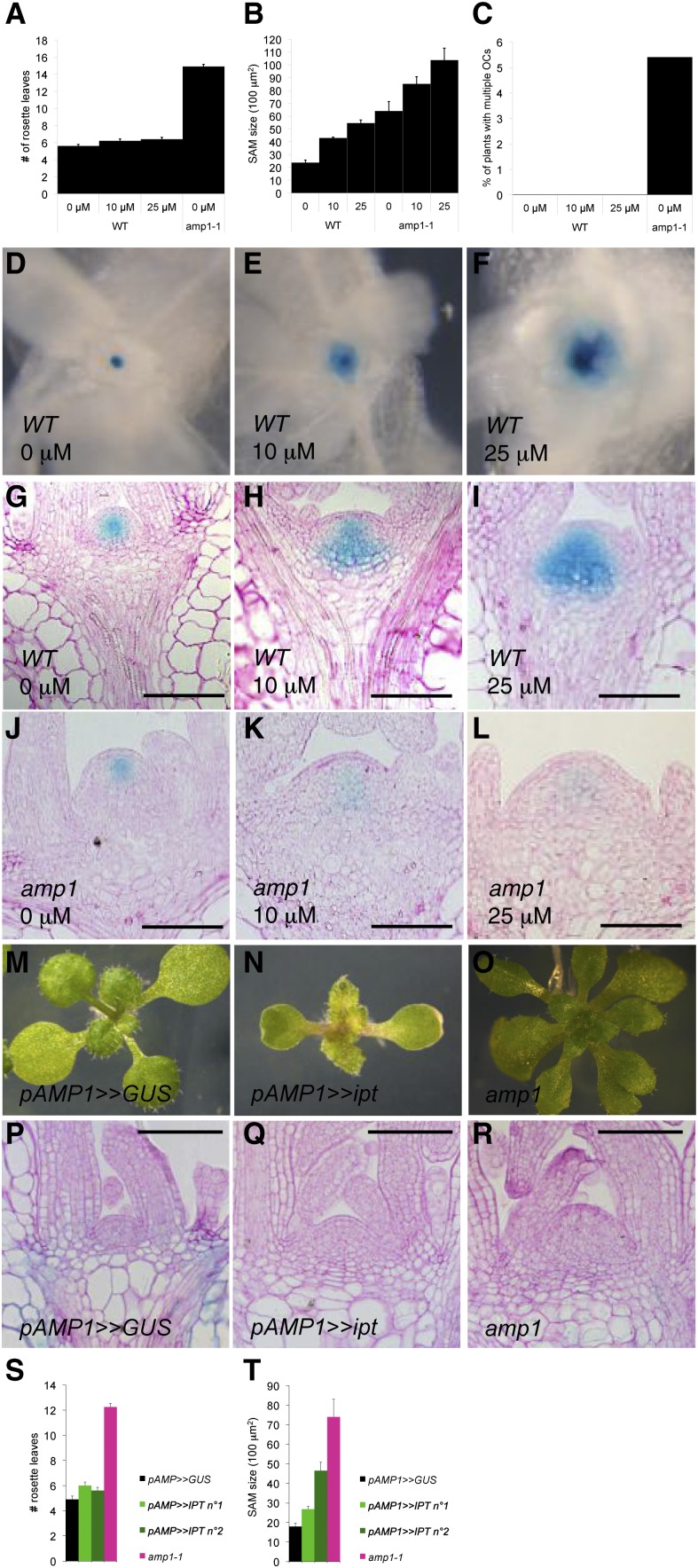
To further assess to which extent the elevated CK biosynthesis in amp1 is the cause for the increased SAM size, enhanced leaf formation rate, and ectopic SCN formation, we tested the effect of exogenous transzeatin on these processes. As reported before (Saibo et al., 2007), SAM size significantly increased upon CK treatment in a dose-dependent manner (Fig. 3, B and D–I) but not to the level found in untreated amp1-1. Interestingly, amp1-1 SAMs were able to further increase in size in response to exogenous CK to a similar extent as the wild type, suggesting that the SAM enlargement in amp1-1 might be a CK-independent effect (Fig. 3, B and J–L). Moreover, the CK-triggered increase in SAM size in the wild type did not coincide with an obvious change in the leaf formation rate under our growth conditions (Fig. 3A). To test the effect of exogenous CK on SCN integrity, we analyzed WUS::GUS activity. CK treatment in the wild type broadened and deepened the WUS expression domain in the vegetative SAM in a dose-dependent manner, similarly as already described for reproductive SAMs (Fig. 3, D–I; Gordon et al., 2009). However, this CK-driven expansion of the WUS expression domain in wild-type plants occurred in a strictly symmetric manner and never appeared to separate in distinct domains but stayed as one unit (Fig. 3C). Notably, CK treatment of amp1 provoked expansion of WUS expression in the initial OC but had, simultaneously, a strong negative effect on reporter activity, indicating that WUS expression is differently regulated in amp1 (Fig. 3, J–L).
Figure 3.
Exogenous CK application or endogenous increase of CK biosynthesis do not phenocopy amp1 SAM phenotypes A, Number of visible rosette leaves in wild-type (WT) plants treated with the indicated concentrations of transzeatin compared with untreated amp1-1 at 18 DAG under short-day conditions ( means ± se of the mean; n ≥ 10). B, Quantification of SAM size in the wild type and amp1-1 at 7 DAG after treatment with indicated concentrations of transzeatin (μm). Area of median longitudinal sections was measured (means ± se of the mean; n ≥ 5). C, Percentage of wild-type plants with multiple WUS::GUS foci after treatment with the indicated concentrations of transzeatin compared with untreated amp1-1 plants. Plants were grown under short-day conditions and were analyzed at 12 DAG (n ≥ 200). D to F, WUS::GUS expression domain in wild-type plants treated with 0 μm (D), 10 μm (E), or 25 μm (F) transzeatin at 7 DAG. G to L, WUS::GUS localization in median longitudinal SAM sections of the wild type (G–I) and amp1-1 (J–L) at 7 DAG. Plants were treated with 0 μm (G and J), 10 μm (H and K), or 25 μm (I and L) transzeatin. M to O, Shoot phenotypes of representative pAMP1>>GUS (M), pAMP1>>ipt (N), and amp1-1 (O) seedlings at 9 DAG. P to R, Median longitudinal SAM sections of pAMP1>>GUS (P), pAMP1>>ipt (Q), and amp1-1 (R) seedlings at 9 DAG. S, Quantification of rosette leaf number in pAMP1>>GUS (control), two independent pAMP1>>ipt lines, and amp1-1 at 9 DAG (means ± se of the mean; n ≥ 10). T, Quantification of SAM size in pAMP1>>GUS (control), two independent pAMP1>>ipt lines, and amp1-1 at 9 DAG (means ± se of the mean; n ≥ 3). Bars = 100 μm.
To test whether the amp1-specific SAM phenotypes are the result of elevated CK accumulation in a specific temporal and spatial manner, we overexpressed the bacterial ipt gene in the AMP1 expression domain using an established transactivation approach (Craft et al., 2005; Rutherford et al., 2005). Macroscopically, pAMP1>>ipt plants strongly resembled 35S-driven ipt overexpressors, showing typical CK overaccumulation phenotypes, including small serrated true leaves and increased anthocyanin accumulation (Fig. 3N; Craft et al., 2005). However, the leaf formation rate was not significantly affected in pAMP1>>ipt, indicating that localized enhancement of CK biosynthesis in the AMP1 expression domain might not be responsible for the shortened plastochron of amp1 (Fig. 3S). Like wild-type plants treated with exogenous CK, pAMP1>>ipt plants showed an obvious gain in SAM size, but not reaching the magnitude of untreated amp1 plants (Fig. 3, P–R and T). More importantly, they did not show any signs of ectopic SCN formation (for example, fused rosettes or twin shoots). Thus, based on these experiments, we conclude that the amp1-specific set of SAM defects is not primarily caused by the elevated CK biosynthesis found in the mutant.
Depletion of Endogenous CK Levels or CK Insensitivity Does Not Suppress amp1-Specific SAM Defects
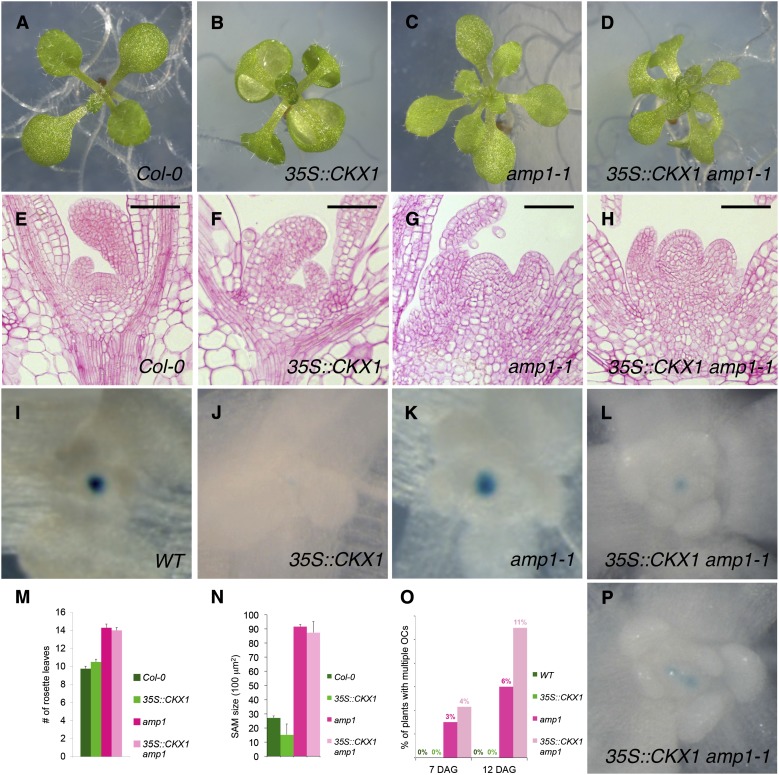
To further assess the functional relationship between increased CK levels and SAM defects in amp1, we analyzed the effect of endogenous CK depletion by overexpression of CYTOKININ OXIDASE1 (35::CKX1; Werner et al., 2003) on the mutant phenotypes. As previously described, 35S::CKX1 plants showed a reduced SAM size (Fig. 4, F and N); however, we did not observe any significant attenuation of leaf formation (Fig. 4, A, B, and M). Notably, in 35S::CKX1 amp1-1 plants, SAM size was only marginally reduced compared with amp1-1 plants, and also the leaf number was unchanged (Fig. 4, C, D, G, H, M, and N). The presence of the leaf epinastic phenotype indicated that 35S::CKX1 overexpression still takes place in these plants. We also monitored the effect of CK depletion on the development of ectopic WUS expression domains in amp1. In the wild type as well as in amp1-1, the presence of 35S::CKX1 led to reduced intensity of WUS::GUS activity, consistent with the described positive effect of CK on WUS expression (Fig. 4, I–L). However, 35S::CKX1 did not have any negative impact on the frequency of ectopic SCN formation events in amp1; it rather slightly increased the prevalence rate from 6% to 11% in 12-d-old seedlings (Fig. 4, O and P).
Figure 4.
Effect of reduced CK levels on amp1 shoot meristem phenotypes. A to D, Shoot phenotypes of representative Col-0 (A), 35S::CKX1 (B), amp1-1 (C), and 35S::CKX1 amp1-1 (D) seedlings at 9 DAG. E to H, Median longitudinal SAM sections of Col-0 (E), 35S::CKX1 (F), amp1-1 (G), and 35S::CKX1 amp1-1 (H) seedlings 9 DAG. I to L and P, WUS::GUS localization in SAMs of Col-0 (I), 35S::CKX1 (J), amp1-1 (K), and 35S::CKX1 amp1-1 (L and P) seedlings at 7 DAG. M, Quantification of rosette leaf number of indicated genotypes grown under short-day conditions at 12 DAG (means ± se of the mean; n ≥ 10). N, SAM size measurement of indicated genotypes at 9 DAG (means ± se of the mean; n ≥ 3). O, Quantification of ectopic OC formation in indicated genotypes at 7 and 12 DAG (n ≥ 200). WT, Wild type. Bars = 50 μm.
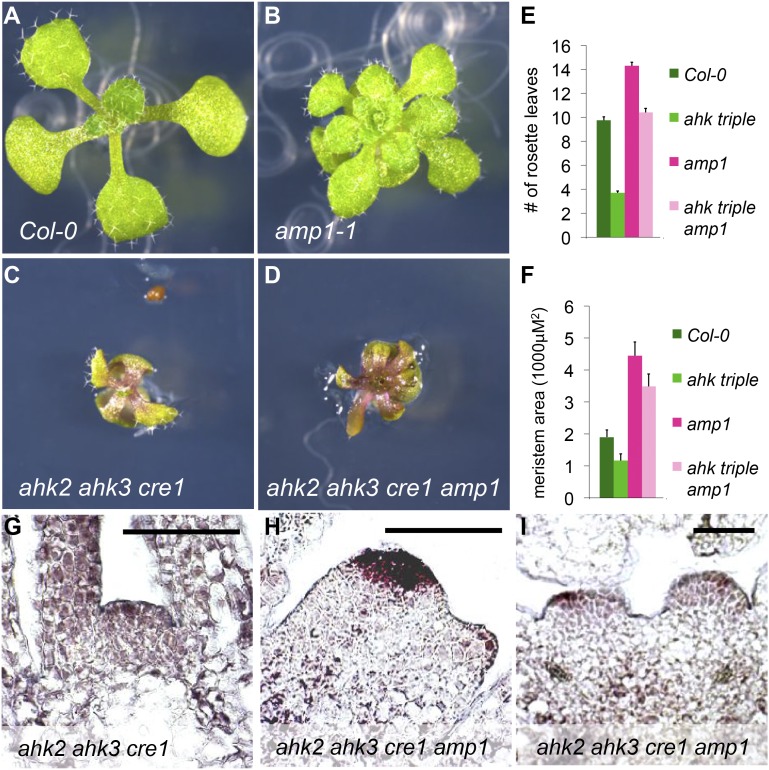
To further substantiate the apparent CK independence of amp1 SAM-related defects, we tested the phenotypic consequences of bringing amp1 into a CK-insensitive background by crossing with the CK receptor triple mutant ahk2 ahk3 cytokinin response1 (cre1). Similar to the receptor triple mutant, ahk2 ahk3 cre1 amp1 showed the described hallmarks of CK insensitivity in extrameristematic tissues, including strongly reduced organ size, epinasty, and drastic anthocyanin accumulation (Fig. 5, A–D). However, the amp1-dependent relative increase in leaf formation was even more pronounced in the CK-insensitive background (ahk2 ahk3 cre1 amp1/ahk2 ahk3 cre1: 2.7-fold increase) compared with the control (amp1/Columbia [Col-0]: 1.4-fold increase; Fig. 5, C–E). We observed a similar effect when we analyzed SAM size in amp1 plants with compromised CK perception, which still produced shoot meristems nearly 2 times bigger than wild-type plants (Fig. 5, F–H). The net reduction in SAM size triggered by CK sensitivity was very similar in the presence or absence of AMP1, indicating that shoot meristem hypertrophy in amp1 is mainly driven by a CK-independent process. Whereas, in the receptor triple mutant, CLV3 expression in the downsized SAM was hardly detectable, we observed a large and clearly defined CLV3 signal in ahk2 ahk3 cre1 amp1 meristems (Fig. 5, G and H). Moreover, we could still observe ectopic SCN formation events in the quadruple mutant (Fig. 5I). Hence, our genetic analyses strongly support the unexpected finding that increased leaf formation rate, SAM hypertrophy, and ectopic SCN formation in amp1 are essentially mediated by CK-unrelated processes.
Figure 5.
Effect of CK insensitivity on amp1 shoot meristem phenotypes. A to D, Shoot phenotypes of representative Col-0 (A), amp1-1 (B), ahk2-2 ahk3-3 cre1-12 (C), and ahk2-2 ahk3-3 cre1-12 amp1-1 (D) seedlings 9 DAG. E, Quantification of rosette leaf number of indicated genotypes grown under short day at 12 DAG (means ± se of the mean; n ≥ 10). F, SAM size measurement of indicated genotypes grown under short day at 11 DAG (means ± se of the mean; n ≥ 3). G to I, RNA in situ hybridization showing CLV3 expression in median longitudinal SAM sections (at 11 DAG) grown under short day. G, For ahk2-2 ahk3-3 cre1-12, CLV3 expression is barely detectable. H, ahk2-2 ahk3-3 cre1-12 amp1-1 SAM with a large central CLV3 expression domain and a peripheral CLV3 positive area. I, ahk2-2 ahk3-3 cre1-12 amp1-1 shoot apex with two CLV3-expressing SAM poles of similar size. Bars = 50 μm.
Genetic Interaction of amp1 and wus
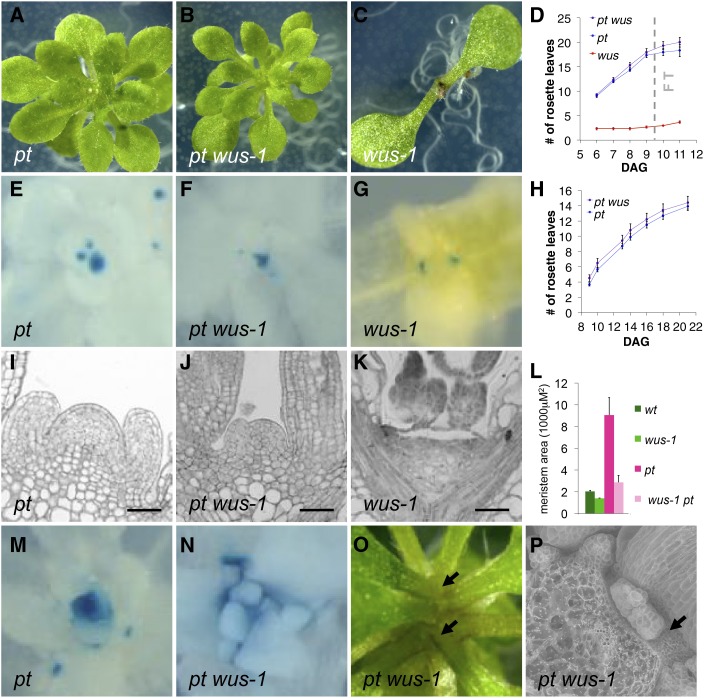
To test to what extend WUS is required for the formation of amp1-specific SAM phenotypes, we crossed the amp1 allele pt (ecotype Landsberg erecta [Ler]) with wus-1. In wus-1 seedlings, the primary SAM generates one or two true leaves before it differentiates and becomes inactive (Fig. 6C), until an accessory SAM forms from the periphery of the original SAM to generate again one or two leaves, resulting in a strongly decelerated on-off mode of organ formation (Laux et al., 1996). Notably, pt wus plants showed the same accelerated leaf formation rate as pt during the vegetative growth phase, indicating that amp1 stem cells can be maintained in a WUS-independent way (Fig. 6, A, B, D, and H). To understand this insensitivity of amp1 to the absence of WUS in respect to leaf formation, we followed the OC behavior by using WUS::GUS. In wus single mutants, the primary OC disappeared before day 7, and subsequent activation of WUS-expressing foci was evident close to the axils of the cotyledons (Fig. 6G). One of these secondary SCNs then produced one further leaf before terminating, and in the axil of this new primordium, again, a new SCN became activated. By contrast, the primary OC of pt wus appeared to sustain activity for a much longer period of time reflected by the presence of a typical primary SAM structure (Fig. 6, I–K) and continuous transcription from the WUS promoter (Fig. 6, E–G). However, compared with pt, the primary SAM of pt wus has a drastically reduced size (Fig. 6, I, J, and L), and the WUS::GUS activity appeared to be weaker and more diffuse (Fig. 6, E and F). Importantly, although WUS and CLV3 expression were still detected in the primary SAM, ectopic SCNs were formed in the areas between existing leaf primordia, which were able to form additional leaf anlagen in parallel with the primary SAM (Fig. 6, F, N, and O). At the onset of floral transition, pt wus plants abruptly stopped with organ formation and were unable to form a functional inflorescence meristem. At this stage, pt wus SAMs totally differentiated, resulting in a flat apex without any further primordia formation (Fig. 6, O and P). Taken together, the enhanced leaf formation rate typical for pt is not significantly affected by the absence of WUS. This appears to be caused by the ability of amp1 primary SAMs to maintain the stem cell pool of the primary SAM for an extended period of time during the vegetative growth phase. Moreover, whereas secondary SCNs became active in wus only after the primary SAM was fully differentiated, activity of the primary SAM and formation of adventitious SAMs overlapped temporarily in pt wus, resulting in no obvious decline in organ formation.
Figure 6.
Genetic interaction between AMP1 and WUS. A to C, Shoot phenotypes of pt (A), pt wus-1 (B), and wus-1 (C) seedlings at 10 DAG. D, Appearance of visible leaves in pt, pt wus-1, and wus-1 grown under long days (means ± se of the mean; n ≥ 10). Time point at which pt undergoes floral transition is indicated as dashed vertical line. E to G, WUS::GUS activity in 7-d-old shoots of pt (E), pt wus-1 (F), and wus-1 (G). H, Appearance of visible leaves in pt and pt wus-1 grown under short days (means ± se of the mean; n ≥ 10). I to K, Median longitudinal SAM sections of pt (I), pt wus-1 (J), and wus-1 (K) at 7 DAG. L, Quantification of SAM area in 7-d-old plants (means ± se of the mean; n ≥ 3). M and N, CLV3::GUS activity in 7-d-old shoots of pt (M) and pt wus-1 (N). O, Shoot apex of 15-d-old pt wus-1 grown under long days. An inflorescence meristem was not formed, and arrows point at leaves derived from two meristem poles. P, Scanning electron micrograph of shoot apex from 15-d-old pt wus-1 grown under long days. One leaf was removed for better visibility of differentiated areas between leaves. No obvious inflorescence meristem structures could be observed between the bases of the youngest leaves (arrow). Bars = 50 μm.
AMP1 and Its Homolog LAMP1 Act Together to Mediate SAM Integrity by Suppressing Ectopic SCN Formation
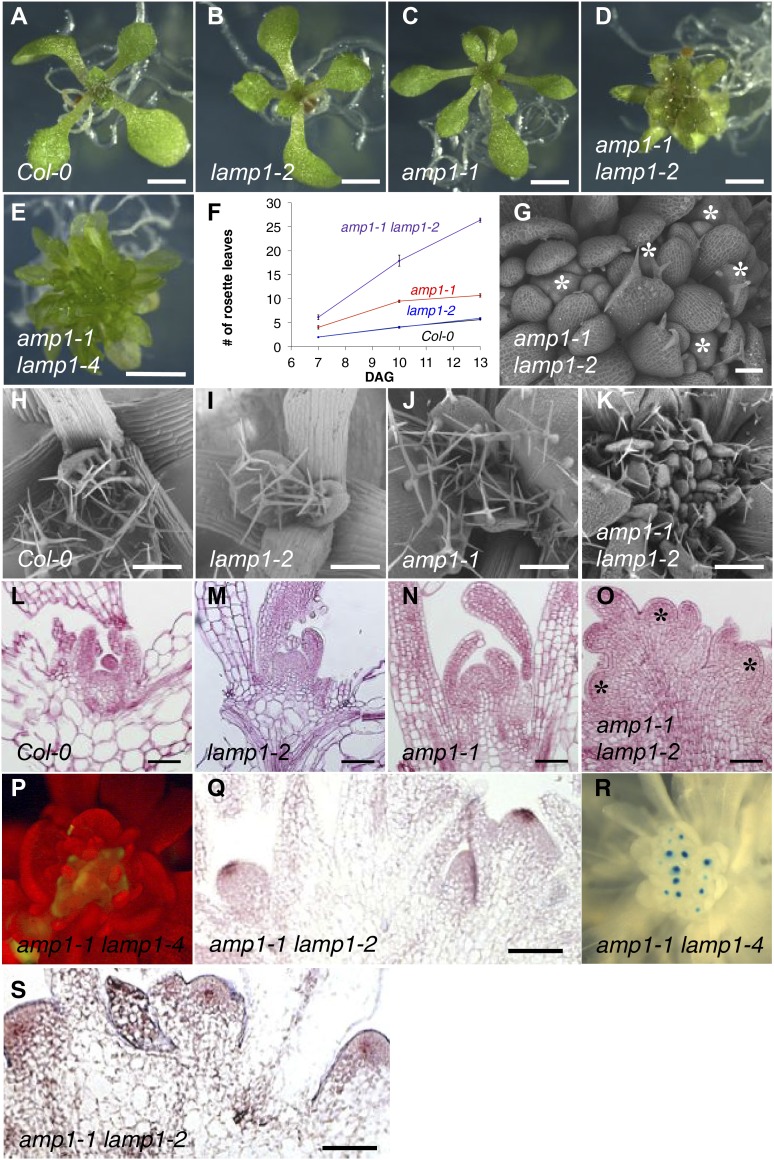
The Arabidopsis genome contains one homolog of AMP1, which was recently named LAMP1 (Li et al., 2013). LAMP1 (At5g19740) has a similar size to AMP1 and comprises the same set of protein motifs (overall predicted sequence identity of 44%). To learn more about the biological function of LAMP1, we characterized two independent transfer DNA insertion lines, which we called lamp1-2 and lamp1-4. Both transfer DNA insertions are located in the first exon of LAMP1 and result in the absence of a full-length LAMP1 transcript (Supplemental Fig. S1). Homozygous plants of both lamp1 alleles did not exhibit any obvious growth phenotypes in all developmental stages tested (Fig. 7, A and B; Supplemental Fig. S2). However, amp1 lamp1 double mutant seedlings showed a drastically expanded shoot apex area from which multiple leaf primordia were formed at the same time, resulting in a massively enhanced leaf formation rate compared with the wild type and amp1 (Fig. 7, A–F). The high numbers of leaf organs were formed from multiple independent meristem poles positioned in close proximity (Fig. 7, G–O). Tissue-specific expression analysis of CLV3 using CLV3::GFP and RNA in situ hybridization revealed the presence of multiple distinct stem cell pools in one shoot apex (Fig. 7, P and Q). Moreover, the multiple independent meristematic domes showed distinct WUS expression domains, indicating that these structures derived from massive ectopic SCN formation (Fig. 7, R and S). After floral transition, amp1 lamp1 plants stay dwarfed and produce numerous sterile inflorescences, which originate from the ectopic SAMs formed during the vegetative growth phase (Supplemental Fig. S2). Thus, amp1 lamp1 plants show a strong enhancement of the ectopic SCN formation phenotype present in amp1, indicating that both genes act together to ensure SAM integrity.
Figure 7.
Analysis of amp1 lamp1 phenotype. A to E, Shoot phenotypes of wild-type Col-0 (A), lamp1-2 (B), amp1-1 (C), amp1-1 lamp1-2 (D), and amp1-1 lamp1-4 (E) seedlings at 9 DAG. F, Number of visible rosette leaves quantified at days 7, 10, and 13 (means ± se of the mean; n ≥ 10). G to K, Scanning electron micrographs of SAM areas from 9-d-old wild-type Col-0 (H), lamp1-2 (I), amp1-1 (J), and amp1-1 lamp1-2 (K). G, Close-up of amp1-1 lamp1-2 shoot apex showing multiple independent functional SAM domes (asterisks). L to O, Median longitudinal SAM sections of seedlings at 7 DAG. Wild-type Col-0 (L), lamp1-2 (M), amp1-1 (N), and amp1-1 lamp1-2 (O) showing ectopic SCNs (asterisks). P, CLV3::GFP localization in amp1-1 lamp1-4 at 7 DAG. Q, In situ hybridization showing CLV3 expression in median longitudinal SAM sections of amp1-1 lamp1-2 at 9 DAG. R, WUS::GUS localization in 14-d-old amp1-1 lamp1-4 grown under short-day conditions. S, RNA in situ hybridization showing WUS expression in median longitudinal SAM sections of amp1-1 lamp1-2 at 9 DAG. Bars = 2,000 μm (A–G), 250 μm (H–K), and 50 μm (G, L–O, Q, and S).
AMP1 and LAMP1 Both Reside in the ER and Show Distinct But Overlapping Tissue-Specific Expression Patterns
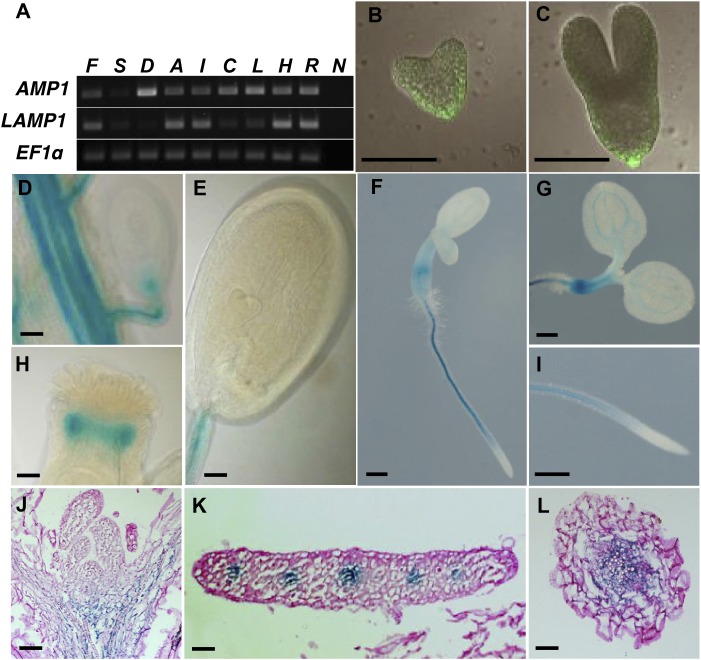
To better understand the synergistic genetic interaction of AMP1 and LAMP1, we compared their tissue-specific expression patterns during development. AMP1 transcripts are broadly expressed (Fig. 8A), and previous GFP reporter analysis revealed the presence of AMP1 in most seedling tissues, including apical meristems of shoot and root, vascular tissues, and the leaf mesophyll (Vidaurre et al., 2007). We analyzed AMP1p::AMP1:GFP distribution in the embryo and found that it is strongly expressed in the suspensor and the basal part of the embryo proper, where it appears to be enriched in the L1 layer (Fig. 8, B and C). We found earliest expression in the transition zone between the suspensor and the embryo, and from the late torpedo stage onwards, the GFP signal became weaker again. By contrast, we could not detect any LAMP1 expression in embryo or suspensor at different developmental stages when using the promoter-GUS reporter LAMP1::GUS (Fig. 8E). Reporter activity was only found after germination in vascular-associated tissues (Fig. 8, D–L). We could not detect any LAMP1 expression in the SAM or root apical meristem. In summary, AMP1 and LAMP1 expression overlap in the vasculature, whereas AMP1 is also present in embryonic and meristematic tissues. AMP1 has been shown to reside in the ER (Vidaurre et al., 2007; Li et al., 2013). To determine where LAMP1 is located in the cell, we generated C-terminal YELLOW FLUORESCENT PROTEIN (YFP) fusions of LAMP1 and AMP1 driven by the 35S promoter and transiently expressed them in epidermal cells of Nicotiana benthamiana. AMP1:YFP and LAMP1:YFP colocalized with RED FLUORESCENT PROTEIN (RFP) fused to the KDEL ER retention signal (RFPKDEL), indicating that the predominant fractions of both proteins reside in the ER (Supplemental Fig. S3, A–F).
Figure 8.
Tissue-specific expression patterns of AMP1 and LAMP1. A, Semiquantitative reverse transcription-PCR analysis comparing expression levels of AMP1 and LAMP1 in different tissues. ELONGATION FACTOR 1α (EF1α) was used as normalization control. B and C, AMP1::AMP1-GFP localization in heart stage embryo (B) and torpedo stage embryo (C). D to I, Whole-mount analysis of pLAMP1::GUS activity in the vascular tissue of the placenta and the funiculus and in the micropylar pole of the ovule (D), developing seed with heart stage embryo (E), 3-d-old seedling (F, G, and I), and style (H). J to L, pLAMP1::GUS localization in median longitudinal section of the shoot apex (J) and transversal sections of true leaf (K) and hypocotyl (L). F, Flowers; S, siliques; D, dry seeds; A, axillary buds; I, inflorescence stems; C, cauline leaves; L, rosette leaves; H, hypocotyl; R, roots; N, no cDNA added. Bars = 50 μm (B–E, H, and J–L) and 300 μm (F, G, and I).
DISCUSSION
amp1 SAMs Form Ectopic SCNs during Vegetative Development
Mutation of amp1 causes a very specific vegetative SAM phenotype combining increased SAM size with a strongly enhanced leaf formation rate, indicating that AMP1 plays a key role in limiting SAM activity (Chaudhury et al., 1993; Vidaurre et al., 2007). So far, however, it was unknown how SAM architecture is affected in detail in this mutant and which functional zones contribute to the elevated size and enhanced organ output. Here, we show that amp1 seedlings form distinct ectopic SCNs in the PZ of the initial SAM. Ectopic SCN formation in amp1 rather appears to be a stochastic process and occurs either very close to the initial OC, appearing like a division event, or clearly distant in a de novo manner. It results in the presence of multiple distinct stem cell pools in one shoot apex that generate organs in an abnormal pattern and speed. This phenomenon appears to further enhance the increased leaf formation rate already present in amp1 single shoot meristem poles. Our findings are consistent with earlier observations of double rosettes in amp1 seedlings and twin shoot formation in the rice plastochron3 mutant, indicating that the function of AMP1 in suppressing ectopic SCN formation is conserved between monocots and dicots (Vidaurre et al., 2007; Kawakatsu et al., 2009). Furthermore, we showed that AMP1 and LAMP1 act together in keeping the PZ in an OC-negative state.
It is rather unlikely that AMP1 directly acts in the WUS-CLV3 pathway controlling stem cell homeostasis. First, the production of supernumerary delimited stem cell pools in amp1 is different to the gradual lateral expansion of the initial stem cell pool in clv mutants (Brand et al., 2000; Schoof et al., 2000). Second, whereas SAM enlargement in clv mutants continues and gets more severe after floral transition, the SCN multiplication phenotype of amp1 is predominant during the vegetative growth phase. Third, it has been shown that pt clv3 mutants have an additive shoot meristem phenotype (Mordhorst et al., 1998). Analysis of the pt wus mutant revealed that the pt primary stem cell pool is maintained for an extended period of time in the absence of WUS in the vegetative growth phase. We think this might be caused by a bigger stem cell pool established during embryogenesis, which takes longer to be consumed, rather than by WUS-independent stem cell maintenance. This is supported by the observation that the primary SAM dome in pt wus during seedling development is clearly reduced in size compared with pt.
Our findings support a model in which ectopic SCN formation in amp1 is not a direct consequence of elevated CK levels found in the mutant. We could not phenocopy this defect in the wild type either by exogenous CK application at different concentrations or by elevated CK biosynthesis restricted to the AMP1 expression domain. This is in accordance with former studies where CK treatment of inflorescence meristems resulted in gradual expansion of the existing OC without any fragmentation events (Gordon et al., 2009; Buechel et al., 2010). Moreover, we could not suppress this phenomenon by reducing global CK levels in amp1. Finally, even in a nominal CK-insensitive background, this phenomenon could still be observed in amp1.
Classical microsurgery studies and modern laser ablation experiments revealed that the OC generates an unidentified short-range signal, which suppresses OC respecification in the PZ of the SAM (Pilkington, 1929; Sussex, 1952; Loiseau, 1959; Reinhardt et al., 2003). In amp1, this lateral inhibition of OC respecification (LIOR) mechanism appears to be absent because mutant SAMs generate peripheral accessory SCNs in the presence of a fully functional primary SCN. A role of AMP1 in this process is further supported by the relatively weak effect of the wus mutation on the organ formation rate of amp1. Because, in amp1, accessory SCNs are already formed when the primary SCN is still active, it does not lead to the typical stop-and-go growth mode of wus single mutants, where differentiation of the primary stem cell pool is a prerequisite of accessory SCN formation (Laux et al., 1996). Our study also revealed that the AMP1 homolog LAMP1 has a partially redundant role in mediating LIOR because amp1 lamp1 shows a strongly enhanced SCN proliferation phenotype, whereas lamp1 is aphenotypic. The different phenotypic strengths of single and double mutants might be explained by the partially overlapping expression patterns of the two genes, with AMP1 showing a much broader expression domain, including embryonic and meristematic tissues, whereas LAMP1 is coexpressed with AMP1 in vascular strands. Both proteins reside in the ER. Whether the biochemical function is fully conserved between both proteins will have to be determined. At the moment, we do not know how they influence LIOR at the molecular level. AMP1 seems to be broadly expressed in all SAM domains (Vidaurre et al., 2007), whereas LAMP1 is even absent from the SAM, indicating that their activity does not directly impose positional information in the communication between the CZ and PZ. In one scenario, AMP1/LAMP1 activities might be required to generate a mobile precursor for the LIOR signal emitted by the OC. Alternatively, they might provide a mobile cofactor essential for the formation/transport/signaling of the LIOR signal(s). Notably, amp1 appears to be also affected in the embryo-mediated lateral inhibition mechanism that prevents recovery of embryonic identity in suspensor cells. It has been shown that the mutant generates ectopic suspensor-derived embryos in the presence of an intact embryo proper, a process that only occurs in the wild type when the main embryo aborts (Vernon et al., 2001; Vidaurre et al., 2007). The communicative network involved is not characterized, but our findings indicate that components of it are probably shared with the LIOR pathway, including AMP1.
Functional Relationship between AMP1 and CK
As one of the first Arabidopsis mutants identified to have increased CK levels, long before the CK biosynthesis pathway in plants was characterized, amp1 was used in several studies as an exemplary CK overproduction line (Tantikanjana et al., 2001; Catterou et al., 2002). However, so far, the molecular cause for the increased CK biosynthesis rate in amp1 remained unexplored, as well as the extent to which it contributes to the elevated SAM size and leaf formation rate in the mutant. We found vascular tissue-specific up-regulation of IPT3 expression in amp1, which correlated with enhanced ARR5 reporter activity and overproliferation of procambial and pericycle cells, indicating that AMP1 limits CK biosynthesis by controlling IPT3 expression. This model has to be tested in future studies (for example, by analyzing CK contents in amp1 ipt3 mutants). Moreover, it will be important to dissect whether increased IPT3 expression is a direct cause or rather a consequence of the vascular overproliferation phenotype in amp1.
Importantly, we were not able to phenocopy any of the amp1-related SAM phenotypes in the wild type when we increased CK levels by different means, nor could we abrogate or clearly suppress them by introducing amp1 into CK-depleted or -insensitive genetic backgrounds. Thus, AMP1 does not only prevent ectopic SCN formation but also limits SAM size as well as the rate of leaf formation in a largely CK-independent manner. In this light, the increase in CK levels in amp1 appears to be a secondary effect and affects SAM phenotypes only slightly. This is in strong contrast to the traditional view that most of the SAM defects are CK related but consistent with later findings that amp1 also shows alterations in abscisic acid, GA, and ethylene responses (Saibo et al., 2007; Shi et al., 2013a, 2013b; Yao et al., 2014). However, based on the current knowledge, it is also difficult to coherently attribute the SAM syndrome of amp1 solely to altered activities in one of these hormonal pathways.
What other regulatory pathway(s) might come into consideration? In a recent study, AMP1 has been described to be required together with RNA-DEPENDENT RNA POLYMERASE6 and LAMP1 for miRNA-dependent translation control specifically at the ER by an unknown molecular mechanism (Li et al., 2013). Moreover, the authors claimed that some of the amp1 phenotypes are similar to plants with a general block in miRNA processing, such as defects in leaf polarity, vascular differentiation, and suspensor fate control, and they postulated the interesting theory that amp1-related phenotypes at least partially derive from a general overaccumulation of miRNA-regulated factors. Some of the SAM-specific processes altered in amp1 are also controlled by specific miRNAs, including leaf initiation rate and shoot meristem activity. However, in this respect, amp1 is more reminiscent of plant lines with enhanced activity of individual miRNAs than those with a general loss of miRNA function. For instance, it has been shown that overexpression of miRNA156 results in a strongly shortened plastochron, and mutants with a general defect in miRNA activity such as argonaute1, serrate, and hasty have significantly extended plastochron lengths (Clarke et al., 1999; Bollman et al., 2003; Wang et al., 2008). Furthermore, vegetative SAM splitting has been described in the miRNA166g-overexpressing line jabba-D, which causes reduced expression of the class III HOMEODOMAIN LEUCINE ZIPPER transcription factors PHABULOSA, PHAVOLUTA, and ARABIDOPSIS THALIANA HOMEOBOX GENE15/CORONA/INCURVATA (Williams et al., 2005). Thus, it appears that there might be no simple relationship between the observed SAM defects and the described general overaccumulation of miRNA-targeted factors in amp1. Next to identifying the biochemical function of AMP1 in miRNA-dependent translation control, further studies will be required to analyze to which extent the amp1-related SAM phenotypes can be attributed to specific misregulation of a subset of miRNA targets, which will help to clarify the potential existence of miRNA-independent activities of AMP1.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Plants were grown under long days (16-h photoperiod) or short days (8-h photoperiod) with cool-white fluorescent light at a photon flux of 120 μmol m–2 s–1 and temperatures of 18°C to 22°C on Arabidopsis (Arabidopsis thaliana) salts medium containing 0.8% (w/v) agar and 1% (w/v) Suc (Lincoln et al., 1990) or in soil. All transgenic lines and mutants are in Col-0 background unless stated otherwise. Arabidopsis lines used in this research are amp1-1 (Chaudhury et al., 1993), amp1-13 (Vidaurre et al., 2007), pt (Ler; Mordhorst et al., 1998), lamp1-2 (SM_3.22750), lamp1-4 (SALK_117012), wus-1 (Ler; Laux et al., 1996), ahk2-2 ahk3-3 cre1-12 (Higuchi et al., 2004), WUS::GUS (Ler) and CLV3::GUS (Ler; Gross-Hardt et al., 2002), pCLV3:GFP-ER (Ler; Lenhard and Laux, 2003), AtIPT3::GFP (Takei et al., 2004), ARR5::GUS (D’Agostino et al., 2000), TCS::GFP (Müller and Sheen, 2008), pOp-GUS and pV-ipt (Craft et al., 2005), 35S::CKX1 (Werner et al., 2003), AMP1::AMP1-GFP (Vidaurre et al., 2007), and 35S::RFPKDEL (Thomas et al., 2008). Combinations of mutants and reporter lines were obtained by crossing individual lines, and genotypes were verified phenotypically and by PCR genotyping (primer listed in Supplemental Table S1). Homozygosity of reporters was determined by unanimous GUS and GFP signal presence (n ≥ 30).
Gene Constructs and Transformation
PCR was performed with proofreading Taq polymerase, and all clones were confirmed by sequencing. A 1,855-bp genomic sequence upstream of the LAMP1 open reading frame (ORF) was amplified with primers AMP1-RELATED1 (AMR1):GUSF/AMR1::GUSR and subcloned into pGEM-T Easy (Promega). The fragment was excised via SalI and ligated into the SalI site of pPZP-GUS-1 (Diener et al., 2000), resulting in pLAMP1::GUS. To create 35S::AMP1:YFP and 35S::LAMP1:YFP, the ORFs of AMP1 and LAMP1 were amplified by PCR using AMP1cDNAF(EcoRV)/AMP1cDNAR-ST(EcoRV) and AMR1cDNAF(EcoRV)/AMR1cDNAR-ST(EcoRV), respectively. The fragments were subcloned into pGEM-T Easy. Subsequently, each ORF was transferred via EcoRV into pGWR8-YFP (Rozhon et al., 2010). To generate pAMP1::LHG4, a 1,722-bp genomic sequence upstream of the AMP1 ORF was amplified with primers AMP1pF(BamHI)/AMP1pR(KpnI/XhoI) and subcloned into pGEM-T Easy. The fragment was excised and ligated into pBIN+LhG4 via BamHI/KpnI (Rutherford et al., 2005).
Flowering Arabidopsis plants were transformed with Agrobacterium tumefaciens by the floral dip method (Clough and Bent, 1998). Resulting T2 lines were confirmed for single-transgene insertion sites and propagated for further analysis, and at least 10 independent transformants were characterized for each line. Infiltration assays on Nicotiana benthamiana leaves were performed with A. tumefaciens strain GV3101 harboring the appropriate binary vectors as described (Winter et al., 2007).
Growth Assays
The sequential appearance of leaves was recorded by visual observation of the shoot apex area under the stereomicroscope (3× magnification). In each experiment, the mean number of appeared leaves from at least 10 plants was calculated at each observation date.
GUS Reporter Analysis
For detection of GUS activity, plant material was first fixed in 90% (v/v) acetone on ice for 30 min and washed with GUS buffer {100 mm sodium phosphate, pH 7.0, 0.5 mm K3[Fe(CN)6], 0.5 mm K4[Fe(CN)6], 10 mm EDTA, and 0.1% [v/v] Triton X-100}. Seedlings were incubated at 37°C in GUS reaction buffer (GUS buffer with 1 mm 5-bromo-4-chloro-3-indoyl-β-d-glucuronid [cyclohexylammonium salt]) until the required blue staining was reached (Jefferson et al., 1987). Samples were dehydrated in ethanol, mounted in chloral hydrate:glycerol:water (8:3:1, w/v/v), and photographed with a MZ16FA binocular equipped with a DFC300FX camera (Leica).
Tissue Sectioning
For plastic sections, fresh or GUS-stained samples were fixed in an ethanol-formaldehyde-acetic acid solution overnight and embedded in Technovit 7100 (Heraeus) as described (Leroux et al., 2007). Sections of 7 µm were cut with a Microtome (Leica RM 2155), stained with 0.1% (w/v) ruthenium red, and mounted with Roti-Histokitt (Roth). Sections were photographed using an Axio Imager MI microscope fitted with a MRC5 camera (Zeiss). SAM size and WUS::GUS expression domain area were quantified on median longitudinal sections using the ImageJ software (Schneider et al., 2012).
Tissue sectioning for GFP analysis was performed as described (Goldshmidt et al., 2008). After fixation, shoot apices of 7-d-old seedlings were embedded in low-melting point agarose 7% (w/v) and mounted on a prechilled microtome block with Tissue-Tek (Sakura Finetek). Shoot apices were cut longitudinally to 20- to 35-μm thickness with a Leica 2000 freezing microtome.
Fluorescence Microscopy
CLV3::GFP and IPT3::GFP whole-mount samples were analyzed with a MZ16FA equipped with a DFC300FX camera (Leica). CLV3::GFP, TCS::GFP, and AMP1::AMP1-GFP analysis in Arabidopsis and 35S::AMP1:YFP, 35S::LAMP1:YFP, and 35S::RFPKDEL analysis in N. benthamiana were done with a Zeiss LSM510 confocal laser-scanning microscope. For imaging, we used the following excitation conditions: 488 nm (GFP), 514 nm (YFP), and 543 nm (RFP). Confocal laser-scanning microscope settings were kept constant in individual sets of experiments to allow for comparison of reporter proteins. The images were assembled using the Zeiss LSM image browser software.
Scanning Electron Microscopy
Plant material was fixed on a metal support rack with Tissue-Tek (Sakura Finetek) and shock frozen in liquid nitrogen, and tissue surfaces were subsequently visualized with a T-1000 scanning electron microscope (Hitachi).
RNA Expression Analysis
Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen) and treated with ribonuclease-free DNase I. Complementary DNA (cDNA) was synthesized using RevertAid H Minus First-Strand cDNA Synthesis Kit (Fermentas) with oligo(dT)18 primers. Semiquantitative reverse transcription-PCR was done as described using EF1a transcript levels as standard (Bennett et al., 2006). qRT-PCR was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems) using GoTaq qPCR Master Mix Kit (Promega). Cycling and quantification were performed as recommended by the manufacturer (initial denaturation, 94°C for 10 min; 40 cycles at 94°C for 15 s and 60°C for 1 min); finally, a melting curve was recorded. A dilution series of cloned cDNA was run under the same conditions, and the results were used to plot a calibration curve, which served to calculate the relative transcript abundance in the samples. The relative expression levels were calculated from four replicates and by normalization to GAPC2. Shown sds were deduced from three biological repeats. Student’s t test was used for comparison of means (significantly different at P < 0.05). The primers used are listed in Supplemental Table S1.
RNA in Situ Hybridization
Sample preparations and in situ hybridizations of 7-µm sections were done as described (Greb et al., 2003). To generate WUS and CLV3 antisense and sense probes, the respective ORFs were amplified from cDNA using the oligos WUSisF/WUSisR and CLV3isF/CLV3isR, subcloned into pGEM-T Easy, and used as a template for transcription from the T7 or SP6 promoters.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AMP1 (AT3G54720) and LAMP1 (AT5G19740).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genotypic characterization of lamp1 and amp1 lamp1 alleles.
Supplemental Figure S2. Adult phenotypes of amp1 lamp1.
Supplemental Figure S3. Subcellular localization of 35S::AMP1:YFP and 35S::LAMP1:YFP in N. benthamiana leaf epidermal cells.
Supplemental Table S1. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Thomas Berleth, Tatsuo Kakimoto, Fritz Kragler, Thomas Laux, Ian Moore, Hitoshi Sakakibara, and Thomas Werner for providing published material.
Glossary
- SAM
shoot apical meristem
- SCN
stem cell niche
- CZ
central zone
- OC
organizing center
- PZ
peripheral zone
- CK
cytokinin
- ER
endoplasmic reticulum
- DAG
days after germination
- qRT
quantitative reverse transcription
- Ler
ecotype Landsberg erecta
- LIOR
lateral inhibition of organizing center respecification
- ORF
open reading frame
- cDNA
complementary DNA
Footnotes
This work was supported by the Austrian Science Fund (FWF; grant no. P19935 to T.S.), an APART fellowship from the Austrian Academy of Sciences (ÖAW; grant no. 11300 to T.S.), and the Vienna Science and Technology Fund (WWTF; grant no. LS2009–055 to T.S.).
Articles can be viewed without a subscription.
References
- Aichinger E, Kornet N, Friedrich T, Laux T (2012) Plant stem cell niches. Annu Rev Plant Biol 63: 615–636 [DOI] [PubMed] [Google Scholar]
- Barton MK. (2010) Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev Biol 341: 95–113 [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16: 553–563 [DOI] [PubMed] [Google Scholar]
- Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS (2003) HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493–1504 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Buechel S, Leibfried A, To JP, Zhao Z, Andersen SU, Kieber JJ, Lohmann JU (2010) Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur J Cell Biol 89: 279–284 [DOI] [PubMed] [Google Scholar]
- Catterou M, Dubois F, Smets R, Vaniet S, Kichey T, Van Onckelen H, Sangwan-Norreel BS, Sangwan RS (2002) hoc: An Arabidopsis mutant overproducing cytokinins and expressing high in vitro organogenic capacity. Plant J 30: 273–287 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Letham S, Craig S, Dennis ES (1993) amp1: a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J 4: 907–916 [Google Scholar]
- Chickarmane VS, Gordon SP, Tarr PT, Heisler MG, Meyerowitz EM (2012) Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc Natl Acad Sci USA 109: 4002–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-Atkins A, Craig S, Hocart C, Dennis E, Chaudhury A (1996) Increased endogenous cytokinin in the Arabidopsis amp1 mutant corresponds with de-etiolation responses. Planta 198: 549–556 [DOI] [PubMed] [Google Scholar]
- Clarke JH, Tack D, Findlay K, Van Montagu M, Van Lijsebettens M (1999) The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J 20: 493–501 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conway LJ, Poethig RS (1997) Mutations of Arabidopsis thaliana that transform leaves into cotyledons. Proc Natl Acad Sci USA 94: 10209–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft J, Samalova M, Baroux C, Townley H, Martinez A, Jepson I, Tsiantis M, Moore I (2005) New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J 41: 899–918 [DOI] [PubMed] [Google Scholar]
- D’Agostino IB, Deruère J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MI, Bennett MJ, Thomas LM, Bjorkman PJ (2005) Crystal structure of prostate-specific membrane antigen, a tumor marker and peptidase. Proc Natl Acad Sci USA 102: 5981–5986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR (2000) STEROL METHYLTRANSFERASE 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch JJ, Prinsen E, VAN Onckelen H, Schell J, Spena A (1991) Viviparous leaves produced by somatic activation of an inactive cytokinin-synthesizing gene. Science 254: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y (2008) Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell 20: 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf P, Dolzblasz A, Würschum T, Lenhard M, Pfreundt U, Laux T (2010) MGOUN1 encodes an Arabidopsis type IB DNA topoisomerase required in stem cell regulation and to maintain developmentally regulated gene silencing. Plant Cell 22: 716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Clarenz O, Schafer E, Muller D, Herrero R, Schmitz G, Theres K (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Barrero JM, Taylor J, Helliwell CA, Gubler F (2011) ALTERED MERISTEM PROGRAM 1 is involved in development of seed dormancy in Arabidopsis. PLoS ONE 6: e20408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt R, Lenhard M, Laux T (2002) WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev 16: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyomarc’h S, Vernoux T, Traas J, Zhou DX, Delarue M (2004) MGOUN3, an Arabidopsis gene with TetratricoPeptide-Repeat-related motifs, regulates meristem cellular organization. J Exp Bot 55: 673–684 [DOI] [PubMed] [Google Scholar]
- Han P, Li Q, Zhu YX (2008) Mutation of Arabidopsis BARD1 causes meristem defects by failing to confine WUSCHEL expression to the organizing center. Plant Cell 20: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Sabatini S (2014) Plant and animal stem cells: similar yet different. Nat Rev Mol Cell Biol 15: 301–312 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Chin-Atkins AN, Wilson IW, Chapple R, Dennis ES, Chaudhury A (2001) The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13: 2115–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al. (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlouchová K, Navrátil V, Tykvart J, Sácha P, Konvalinka J (2012) GCPII variants, paralogs and orthologs. Curr Med Chem 19: 1316–1322 [DOI] [PubMed] [Google Scholar]
- Hohm T, Zitzler E, Simon R (2010) A dynamic model for stem cell homeostasis and patterning in Arabidopsis meristems. PLoS ONE 5: e9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Taramino G, Itoh J, Allen J, Sato Y, Hong SK, Yule R, Nagasawa N, Kojima M, Kusaba M, et al. (2009) PLASTOCHRON3/GOLIATH encodes a glutamate carboxypeptidase required for proper development in rice. Plant J 58: 1028–1040 [DOI] [PubMed] [Google Scholar]
- Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T (2001) FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131–142 [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655 [DOI] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H (2009) Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21: 3152–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Laux T (2003) Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130: 3163–3173 [DOI] [PubMed] [Google Scholar]
- Leroux O, Van der Kinderen G, Viane RL (2007) A sandwich-embedding method for oriented sectioning. J Microsc 227: 79–82 [DOI] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, et al. (2013) MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WZ, Kong DD, Gu XX, Gao HB, Wang JZ, Xia M, Gao Q, Tian LL, Xu ZH, Bao F, et al. (2013) Cytokinins can act as suppressors of nitric oxide in Arabidopsis. Proc Natl Acad Sci USA 110: 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau JE. (1959) Observations et experimentation sur la phyllotaxie et le fonctionnement du sommet vegetatif chez quelques Balsaminacees. Ann. Sci. Nat. Bot. Ser. 11: 201–214 [Google Scholar]
- Lv H, Zheng J, Wang T, Fu J, Huai J, Min H, Zhang X, Tian B, Shi Y, Wang G (2014) The maize d2003, a novel allele of VP8, is required for maize internode elongation. Plant Mol Biol 84: 243–257 [DOI] [PubMed] [Google Scholar]
- Mordhorst AP, Voerman KJ, Hartog MV, Meijer EA, van Went J, Koornneef M, de Vries SC (1998) Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Sheen J (2008) Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Bleckmann A, Simon R (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JA, Jones A, Godin C, Traas J (2012) Systems analysis of shoot apical meristem growth and development: integrating hormonal and mechanical signaling. Plant Cell 24: 3907–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogué F, Grandjean O, Craig S, Dennis S, Chaudhury M (2000a) Higher levels of cell proliferation rate and cyclin CycD3 expression in the Arabidopsis amp1 mutant. Plant Growth Regul 32: 275–283 [Google Scholar]
- Nogué N, Hocart H, Letham DS, Dennis ES, Chaudhury AM (2000b) Cytokinin synthesis is higher in the Arabidopsis amp1 mutant. Plant Growth Regul 32: 267–273 [Google Scholar]
- Pilkington M. (1929) The regeneration of the stem apex. New Phytol 28: 37–53 [Google Scholar]
- Pinto JT, Suffoletto BP, Berzin TM, Qiao CH, Lin S, Tong WP, May F, Mukherjee B, Heston WD (1996) Prostate-specific membrane antigen: a novel folate hydrolase in human prostatic carcinoma cells. Clin Cancer Res 2: 1445–1451 [PubMed] [Google Scholar]
- Reinhardt D, Frenz M, Mandel T, Kuhlemeier C (2003) Microsurgical and laser ablation analysis of interactions between the zones and layers of the tomato shoot apical meristem. Development 130: 4073–4083 [DOI] [PubMed] [Google Scholar]
- Robinson MB, Blakely RD, Couto R, Coyle JT (1987) Hydrolysis of the brain dipeptide N-acetyl-L-aspartyl-L-glutamate. Identification and characterization of a novel N-acetylated α-linked acidic dipeptidase activity from rat brain. J Biol Chem 262: 14498–14506 [PubMed] [Google Scholar]
- Rozhon W, Mayerhofer J, Petutschnig E, Fujioka S, Jonak C (2010) ASKtheta, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J 62: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S, Brandizzi F, Townley H, Craft J, Wang Y, Jepson I, Martinez A, Moore I (2005) Improved transcriptional activators and their use in mis-expression traps in Arabidopsis. Plant J 43: 769–788 [DOI] [PubMed] [Google Scholar]
- Saibo NJ, Vriezen WH, De Grauwe L, Azmi A, Prinsen E, Van der Straeten D (2007) A comparative analysis of the Arabidopsis mutant amp1-1 and a novel weak amp1 allele reveals new functions of the AMP1 protein. Planta 225: 831–842 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Shi H, Ye T, Wang Y, Chan Z (2013a) Arabidopsis ALTERED MERISTEM PROGRAM 1 negatively modulates plant responses to abscisic acid and dehydration stress. Plant Physiol Biochem 67: 209–216 [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang Z, Meng P, Tian S, Zhang X, Yang S (2013b) The glutamate carboxypeptidase AMP1 mediates abscisic acid and abiotic stress responses in Arabidopsis. New Phytol 199: 135–150 [DOI] [PubMed] [Google Scholar]
- Skoog F, Miller CO (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol 11: 118–130 [PubMed] [Google Scholar]
- Sussex IM. (1952) Regeneration of the potato shoot apex. Nature 170: 755–757 [PubMed] [Google Scholar]
- Suzaki T, Kim CS, Takeda N, Szczyglowski K, Kawaguchi M (2013) TRICOT encodes an AMP1-related carboxypeptidase that regulates root nodule development and shoot apical meristem maintenance in Lotus japonicus. Development 140: 353–361 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Latshaw S, Sato Y, Settles AM, Koch KE, Hannah LC, Kojima M, Sakakibara H, McCarty DR (2008) The Maize Viviparous8 locus, encoding a putative ALTERED MERISTEM PROGRAM1-like peptidase, regulates abscisic acid accumulation and coordinates embryo and endosperm development. Plant Physiol 146: 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Inagaki S, Nakajima S, Akashi T, Ohto MA, Kobayashi M, Seki M, Shinozaki K, Kato T, Tabata S, et al. (2004) A novel Arabidopsis gene TONSOKU is required for proper cell arrangement in root and shoot apical meristems. Plant J 38: 673–684 [DOI] [PubMed] [Google Scholar]
- Takeda S, Tadele Z, Hofmann I, Probst AV, Angelis KJ, Kaya H, Araki T, Mengiste T, Mittelsten Scheid O, et al. (2004) BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev 18: 782–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H (2004) AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol 45: 1053–1062 [DOI] [PubMed] [Google Scholar]
- Tantikanjana T, Yong JW, Letham DS, Griffith M, Hussain M, Ljung K, Sandberg G, Sundaresan V (2001) Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev 15: 1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ (2008) Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon DM, Hannon MJ, Le M, Forsthoefel NR (2001) An expanded role for the TWN1 gene in embryogenesis: defects in cotyledon pattern and morphology in the twn1 mutant of Arabidopsis (Brassicaceae). Am J Bot 88: 570–582 [PubMed] [Google Scholar]
- Vidaurre DP, Ploense S, Krogan NT, Berleth T (2007) AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development 134: 2561–2567 [DOI] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC (2005) Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132: 3657–3668 [DOI] [PubMed] [Google Scholar]
- Winter N, Kollwig G, Zhang S, Kragler F (2007) MPB2C, a microtubule-associated protein, regulates non-cell-autonomy of the homeodomain protein KNOTTED1. Plant Cell 19: 3001–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T, Gross-Hardt R, Laux T (2006) APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N (2005) Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol 15: 1566–1571 [DOI] [PubMed] [Google Scholar]
- Yao Y, Dong CH, Yi Y, Li X, Zhang X, Liu J (2014) Regulatory function of AMP1 in ABA biosynthesis and drought resistance in Arabidopsis. J Plant Biol 57: 117–126 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.