Nitric oxide positively regulates a key Arabidopsis peroxidase through S-nitrosylation and enhances resistance to oxidative stresses.
Abstract
Nitric oxide (NO) and reactive oxygen species (ROS) are two classes of key signaling molecules involved in various developmental processes and stress responses in plants. The burst of NO and ROS triggered by various stimuli activates downstream signaling pathways to cope with abiotic and biotic stresses. Emerging evidence suggests that the interplay of NO and ROS plays a critical role in regulating stress responses. However, the underpinning molecular mechanism remains poorly understood. Here, we show that NO positively regulates the activity of the Arabidopsis (Arabidopsis thaliana) cytosolic ascorbate peroxidase1 (APX1). We found that S-nitrosylation of APX1 at cysteine (Cys)-32 enhances its enzymatic activity of scavenging hydrogen peroxide, leading to the increased resistance to oxidative stress, whereas a substitution mutation at Cys-32 causes the reduction of ascorbate peroxidase activity and abolishes its responsiveness to the NO-enhanced enzymatic activity. Moreover, S-nitrosylation of APX1 at Cys-32 also plays an important role in regulating immune responses. These findings illustrate a unique mechanism by which NO regulates hydrogen peroxide homeostasis in plants, thereby establishing a molecular link between NO and ROS signaling pathways.
When challenged by biotic and abiotic stresses, plants respond by rapid activation of specific signaling pathways to cope with these unfavorable conditions. During the onset of these responses, a common feature is to induce the burst of several signaling molecules, the most important of which are reactive oxygen species (ROS) and nitric oxide (NO). NO is an important cellular signal molecule and plays an important role in many physiological and pathological processes in animals and plants (Besson-Bard et al., 2008; Leitner et al., 2009; Groß et al., 2013; Wendehenne et al., 2014; Yu et al., 2014). In plants, NO is involved in the regulation of various processes, including seed germination, root development, stomatal closure, flowering, hormone signaling, stress responses, and cell death (Besson-Bard et al., 2008; Leitner et al., 2009; Yu et al., 2014). In particular, NO plays a central role in redox signaling and is involved in responses to both biotic and abiotic stresses. In all living organisms, S-nitrosoglutathione (GSNO) is a major bioactive NO species. The homeostasis of GSNO is largely regulated by GSNO reductase (GSNOR) that catalyzes irreversibly degrading GSNO (Liu et al., 2001). In Arabidopsis (Arabidopsis thaliana), allelic mutations in GSNOR1/HOT TEMPERATURES5 (HOT5)/PARAQUAT RESISTANT2 (PAR2) cause a dramatic increase in intracellular GSNO and S-nitrosothiols (SNOs), altered stress responses, and various developmental defects (Feechan et al., 2005; Lee et al., 2008; Chen et al., 2009; Kwon et al., 2012).
A major bioactivity of NO is executed by regulating the activity of proteins through S-nitrosylation, a redox-based posttranslational modification mechanism that covalently links an NO group to the reactive Cys thiol of a protein to form an SNO (Hess et al., 2005; Wang et al., 2006; Astier et al., 2011; Gupta, 2011; Hess and Stamler, 2012). In plants, several proteins have been characterized as being modified by S-nitrosylation, which directly regulates the activity of the target proteins by various mechanisms (Astier et al., 2011; Gupta, 2011; Yu et al., 2014). In plant immune responses, S-nitrosylation has been found as an important mechanism controlling the activity of several key regulators. S-Nitrosylation of NONEXPRESSOR OF PATHOGEN-RELATED1 (NPR1), a central regulator of salicylic acid (SA)-mediated defense signaling, enhances the formation of NPR1 oligomers that are sequestered in the cytoplasm as an inactive form, whereas denitrosylation positively regulates the SA-induced conversion of NPR1 oligomers into the active monomers (Tada et al., 2008). S-Nitrosylation of TGACG motif-binding factor (TGA1), a transcription factor that directly controls the expression of downstream defense genes, facilitates the NPR1-TGA interaction in the nucleus and enhances DNA binding activity of TGA1 in the presence of NPR1 (Lindermayr et al., 2010). Interestingly, the intracellular level of SA is also modulated through S-nitrosylation of Salicylic Acid Binding Protein3 (SABP3), a key enzyme in SA metabolism (Wang et al., 2009), suggesting that the SA pathway is extensively regulated by the cellular redox status. In addition, NO has been shown to regulate the homeostasis of ROS through S-nitrosylation. S-Nitrosylation of peroxiredoxin II E inhibits its peroxynitrite reductase activity that catalyzes the formation of peroxynitrite (ONOO−) from NO and O2−, resulting in the increased level of toxic O2− and consequently inducing cell death (Romero-Puertas et al., 2007). On the other hand, S-nitrosylation of NADPH oxidase impairs its enzymatic activity, resulting in the reduction in ROS biosynthesis during immune responses (Yun et al., 2011).
In response to various stresses, whereas the burst of ROS activates downstream signaling pathways, the rapidly increased intracellular level of ROS is also toxic to plants (Mittler, 2002; Mittler et al., 2004, 2011). To minimize the toxic effects of ROS, plants are equipped with efficient scavenging mechanisms to remove the excess ROS. The best known ROS-scavenging systems include ascorbate peroxidase (APX), superoxide dismutase, glutathione peroxidase, peroxidase, and catalase (Mittler, 2002; Mittler et al., 2011). Of these enzymes, APX catalyzes the reduction of toxic hydrogen peroxide (H2O2) into water using ascorbate as a substrate, and has been extensively studied in Arabidopsis and other plant species. APXs are present in most, if not all, subcellular compartments, including the chloroplast, cytosol, mitochondria, and peroxisomes (Mittler et al., 2004). The Arabidopsis genome contains nine genes encoding APX, of which APX1, APX2, and APX6 are cytosolic enzymes (APX6 is also localized in the chloroplast and mitochondria), and the other six enzymes are found in various subcellular compartments (Mittler et al., 2004). The Arabidopsis APX1 gene is one of the best studied members of this small gene family. Knockout mutations in APX1 cause defective growth and development, impaired stomatal responses, and increased sensitivity to heat shock during light stress (Pnueli et al., 2003). In addition, apx1 mutations also cause severe deficiency of the entire chloroplastic H2O2-scavenging system, resulting in an increased H2O2 level and protein oxidation, illustrating a critical role of APX1 in the intracellular ROS homeostasis (Davletova et al., 2005).
The signaling pathways mediated by ROS and NO interact actively, and these interactions play critical roles in regulating various physiological and pathological processes (Beligni and Lamattina, 1999a, 1999b; Beligni et al., 2002; Delledonne, 2005; Guo and Crawford, 2005; Zaninotto et al., 2006; Chen et al., 2009; Yun et al., 2011; Groß et al., 2013). One of the mechanisms modulating the interaction is attributed to the reciprocal regulation of intracellular ROS and NO levels (Zaninotto et al., 2006; Yun et al., 2011; Groß et al., 2013). As mentioned earlier, NO inhibits the NADPH oxidase activity through S-nitrosylation, resulting in a reduction in ROS biosynthesis during hypersensitive response (Yun et al., 2011). In response to abiotic stresses, mutations in the Arabidopsis GSNOR1/HOT5/PAR2 gene lead to resistance to the oxidative damage induced by the herbicide paraquat, accompanied by dramatically increased intracellular NO (Chen et al., 2009). Similarly, NO donors effectively protect potato (Solanum tuberosum) leaves from oxidative stresses and cell death induced by diquat and paraquat, suggesting that NO acts as an antioxidant during the stress response (Beligni and Lamattina, 1999a, 1999b). The NO donor sodium nitroprusside (SNP) also protects cucumber (Cucumis sativus) seedlings from salt-induced oxidative damage, possibly through the enhanced ROS-scavenging enzyme activities (Shi et al., 2007). Notably, this NO-mediated antioxidation mechanism appears to be conserved, as SNP effectively protects Cyanobacterium Spirulina platensis from the UVB-induced oxidative damage associated with the increased ROS-scavenging enzyme activities (Xue et al., 2007).
Consistent with these observations, NO was proposed to reduce the ROS level by activating or enhancing the ROS scavenging enzymes, such as APX, catalase, and superoxide dismutase, during stress responses (Beligni et al., 2002; Xue et al., 2007; Keyster et al., 2011; Begara-Morales et al., 2014). In particular, treatments of pea (Pisum sativum) APX recombinant protein with peroxynitrite and GSNO reduce and increase enzymatic activities, respectively. Moreover, pea APX is characterized as being nitrated at two Tyr residues and S-nitrosylated at Cys-32 (Begara-Morales et al., 2014). However, it remains unknown whether nitration and S-nitrosylation play a role in directly regulating the activity of pea APX. In contrast with these results, it has been reported that NO inhibits the activity of cytosolic APX in tobacco (Nicotiana tabacum) Bright Yellow-2 suspension cells through S-nitrosylation (de Pinto et al., 2013). Here, we show that S-nitrosylation of Arabidopsis APX1 at Cys-32 positively and directly regulates its activity during stress responses, thereby enhancing the tolerance of plants to oxidative damages.
RESULTS
APX1 Is a Positive Regulator of NO-Modulated Resistance to Oxidative Stress
In a previous study, we found that the Arabidopsis par2-1 mutant was resistant to paraquat, an herbicide inducing the generation of ROS, and the treatment of wild-type plants with GSNO and the NO donor SNP enhanced the paraquat resistance (Chen et al., 2009; Supplemental Fig. S1), suggesting that NO acts as an antioxidant to negatively regulate oxidative stress. NO has been proposed to reduce the H2O2 level by activating the ROS-scavenging enzyme APX (Keyster et al., 2011; Lin et al., 2011; Begara-Morales et al., 2014). We reasoned that the tolerance of the par2-1 mutant to oxidative stress might be partly attributed to the NO-induced APX activity. To test this possibility, we first identified and analyzed an apx1 allelic mutant, because the cytosol-localized APX1 was shown to substantially impair the stress response and cause defective growth and development (Pnueli et al., 2003; Davletova et al., 2005).
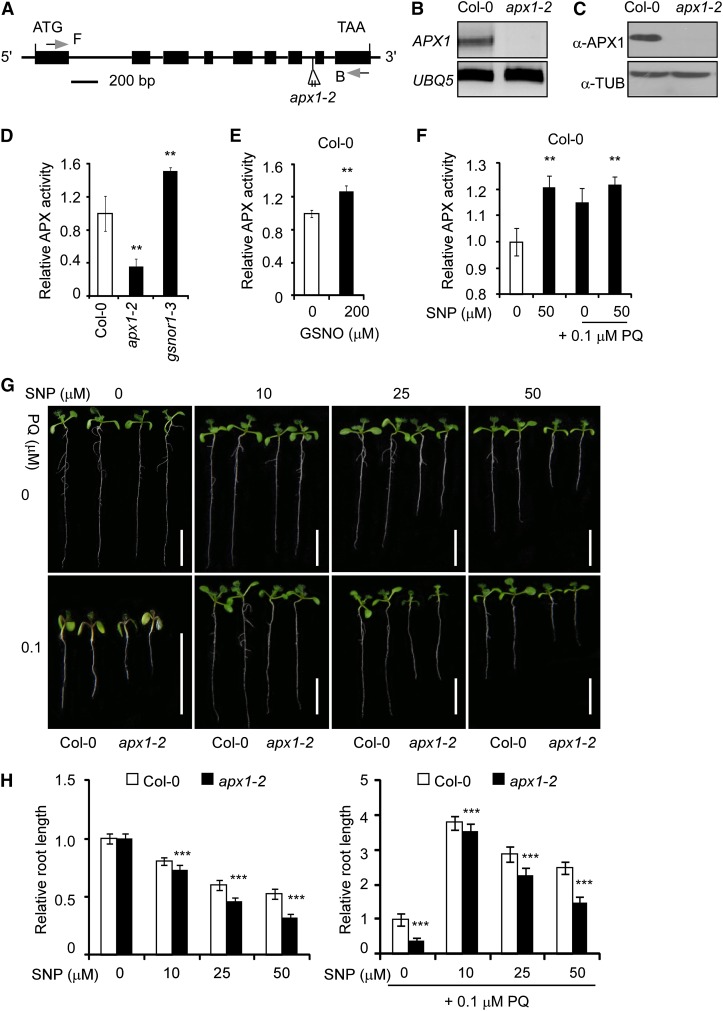
We identified the mutant SALK_000249, which contained a transfer DNA insertion in intron 7 of APX1 (At1g07890; Fig. 1A). The transfer DNA insertion caused undetectable APX1 messenger RNA and APX1 protein (Fig. 1, B and C), indicating that it is a null mutation. We designated this mutant as apx1-2. Similar to the apx1-1 mutant (Pnueli et al., 2003; Davletova et al., 2005), the apx1-2 mutant showed various developmental defects, including reduced seedling size, plant height, and abnormal siliques (Supplemental Fig. S2, A–C). These and other defects were fully rescued by an APX1 transgene (see below). The apx1-2 mutant showed more than 70% decrease in total APX activity compared with wild-type plants (Fig. 1D), indicating that APX1 represents the major APX activity in Arabidopsis. Correlated to its resistance to paraquat and the accumulation of the excessive amount of NO (Feechan et al., 2005; Lee et al., 2008; Chen et al., 2009), the gsnor1-3 mutant showed a remarkable increase in total APX activity (Fig. 1D). Moreover, treatment with GSNO or SNP enhanced the APX enzymatic activity in wild-type seedlings (Fig. 1, E and F). Paraquat marginally increased the APX activity (Fig. 1F). Under the assay conditions, NO had no detectable effect on the accumulation of APX1 protein (Supplemental Fig. S3). These results suggest that NO positively regulates APX1 enzymatic activity, which is correlated to the paraquat resistance of gsnor1-3/par2. Consistently, the apx1-2 mutant was hypersensitive to paraquat and displayed a compromised response to the SNP-enhanced paraquat resistance (Fig. 1, G and H; Supplemental Fig. S4). Taken together, these results suggest that APX1 plays a critical role in the NO-modulated resistance against oxidative stresses.
Figure 1.
NO positively regulates APX activity and resistance to oxidative stress. A, A schematic diagram of the apx1-2 mutant genome. Exons and introns are indicated by black boxes and lines, respectively. The positions and orientation of the PCR primers used in B are shown (F and B). B, Analysis of APX1 expression in the wild-type (Col-0) and apx1-2 mutant seedlings by RT-PCR. C, The accumulation of APX1 protein in the wild-type (Col-0) and apx1-2 mutant plants analyzed by immunoblotting. α-TUB, α-TUBULIN. D to F, Analysis of the APX activity in 10-d-old seedlings of wild-type (Col-0), apx1-2, and gsnor1-3 (D), Col-0 seedlings treated with 200 μm GSNO for 6 h (E), and Col-0 seedlings germinated and grown on one-half-strength Murashige and Skoog agar plates supplemented with various combinations of SNP and paraquat (PQ) as indicated (F). The relative APX enzymatic activity of Col-0 seedlings grown under the normal growth condition was set as 1.0. G, Ten-day-old wild-type (Col-0) and apx1-2 seedlings germinated and grown on one-half-strength Murashige and Skoog agar plates with various combinations of PQ (0.1 μm) and SNP. Scale bars = 1 cm. H, A quantitative analysis of the relative length of the primary roots shown in G. The relative root length of Col-0 seedlings grown under normal growth condition is set at 1.0. At least 30 seedlings were analyzed for each sample. Error bars in D to F and H indicate sd from three independent experiments (biological replicates). Two-tailed Student’s t test; **, P < 0.01; ***, P < 0.001.
The cellular redox status, marked by the intracellular glutathione pool, plays an important role in regulating the oxidative stress response (Dubreuil-Maurizi et al., 2011; Aquilano et al., 2014). We found that treatment with SNP had no apparent effect on the glutathione pool (reduced glutathione [GSH] + oxidized glutathione [GSSG]) and the GSH/GSSG ratio (Supplemental Fig. S5, A and B). However, treatment with paraquat substantially increased the cellular glutathione pool (GSH+GSSG) and the GSH/GSSG ratio in an APX1-independent manner (Supplemental Fig. S5, A and B). These results suggest that the increased sensitivity of apx1-2 to paraquat is not directly related to the cellular redox homeostasis.
APX1 Is S-Nitrosylated at Cys-32 and Cys-49
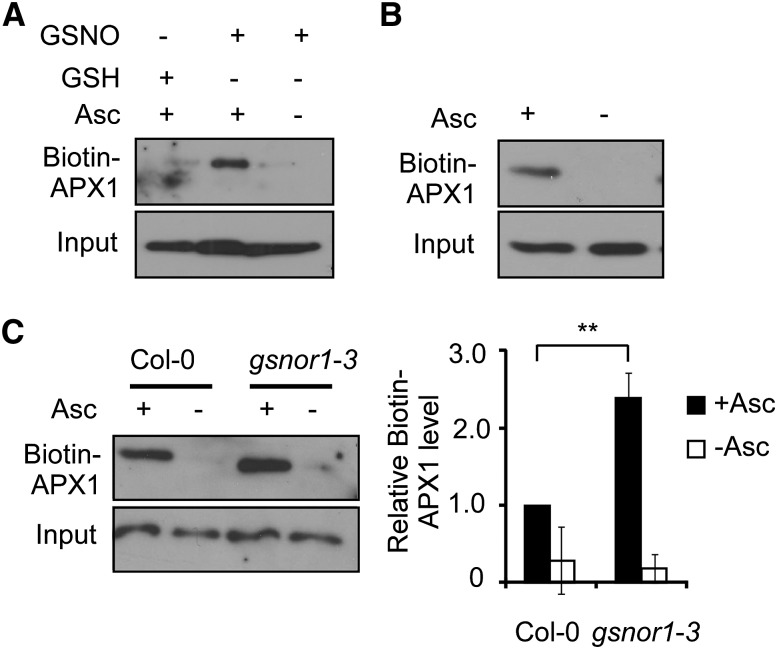
One of the major physiological roles of NO is executed through S-nitrosylation. We reasoned that the NO-enhanced APX1 enzymatic activity might be partly attributed to S-nitrosylation. Consistent with this notion, APX1 was identified as a putative S-nitrosylated protein by nitrosoproteomic studies (Fares et al., 2011; Hu et al., 2015). By using the biotin-switch method (Jaffrey and Snyder, 2001), we found that APX1 recombinant protein was efficiently S-nitrosylated in vitro (Fig. 2A). Moreover, APX1 was specifically S-nitrosylated in planta under normal growth conditions (Fig. 2B). Notably, a higher level of S-nitrosylated APX1 protein was found in gsnor1-3 compared with wild-type plants (Fig. 2C), which correlated to the observation that the gsnor1-3 mutant has a higher APX enzymatic activity and is resistant to paraquat (see Fig. 1).
Figure 2.
APX1 protein is S-nitrosylated. A, Detection of the S-nitrosylated His-APX1 recombinant protein (biotin-APX1; biotinylated proteins; for technical details, see “Materials and Methods”) in vitro. Treatments with GSH in the presence or absence of ascorbate sodium (Asc) are served as negative controls. B, Analysis of S-nitrosylation of APX1 in vivo. C, Accumulation of the S-nitrosylated APX1 in Col-0 and gsnor1-3 seedlings. A quantitative analysis of the data is shown on the right. Error bars = sd from three independent experiments (biological replicates). Two-tailed Student’s t test; **, P < 0.01.
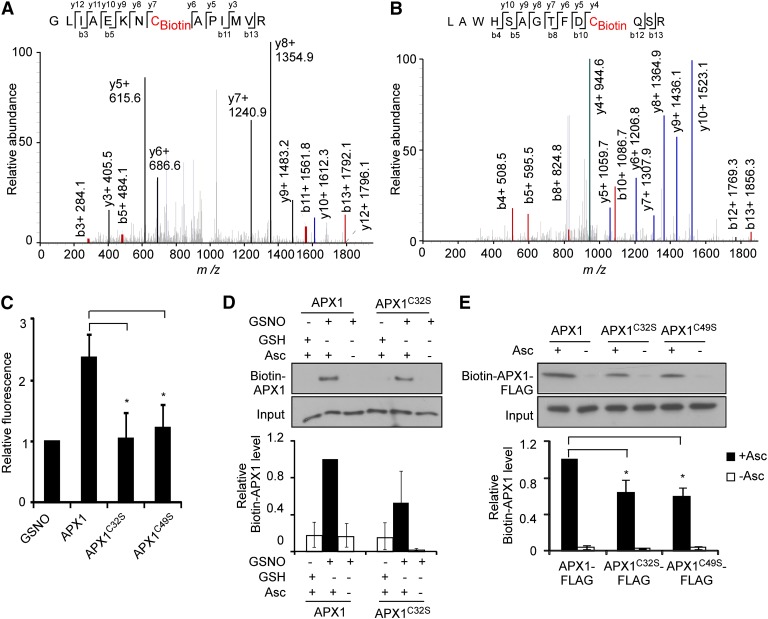
APX1 contains five Cys residues, all in sequence context that perfectly or partially matched the S-nitrosylation consensus characterized as an acid-base/hydrophobic motif (Hess et al., 2005; Xue et al., 2010; Supplemental Figure S6). By using liquid chromatography (LC)-tandem mass spectrometry (MS/MS), we identified Cys-32 and Cys-49 as S-nitrosylated residues from the tryptic fragments derived from GSNO-treated APX1 recombinant protein, but not from the nontreated sample (Fig. 3, A and B; Supplemental Fig. S7). Consistently, in an assay for the conversion ratio of 2,3-diaminonaphthalene (DAN) into fluorescent 2,3-naphthyltriazole (NAT) catalyzed by NO released from the thiol group (Cho et al., 2009), APX1 recombinant protein was found to contain two S-nitrosylated Cys residues, whereas both APX1C32S and APX1C49S mutant proteins had only one S-nitrosylated Cys residue (Fig. 3C). Similarly, the substitution of Cys-32 with Ser residue (APX1C32S) caused nearly 50% reduction of S-nitrosylation of APX1 recombinant proteins (Fig. 3D). Attempts for the expression of APX1C32S APX1C49S double mutant protein were unsuccessful, possibly owing to the instability or toxic effects of the recombinant protein. Nevertheless, the level of the S-nitrosylated APX1C32S-FLAG and APX1C49S-FLAG proteins was also reduced compared with that of the wild-type APX1-FLAG protein in planta (Fig. 3E). Finally, a structure-modeling analysis showed that Cys-32 was located in an α-helix of APX1 and surrounded by Lys-30, Phe-34, and Leu-81, whereas Cys-49 was located in a flexible loop region surrounded by Pro-47, Asp-48, and Arg-52 (Supplemental Fig. S8, A and B). These structural features largely matched the S-nitrosylation consensus motif (Hess et al., 2005). Taken together, these results demonstrate that Cys-32 and Cys-49 are S-nitrosylated in planta.
Figure 3.
APX1 is S-nitrosylated at Cys-32 and Cys-49. A and B, Mass spectrometric analysis of the tryptic fragments of GSNO-treated His-APX1 recombinant protein. Cys-32 (A) and Cys-49 (B) charged with biotin were identified as S-nitrosylated residues. C, DAN analysis of S-nitrosylated residues in GSNO tripeptide, His-APX1, His-APX1C32S, and His-APX1C49S recombinant proteins. D, S-Nitrosylation of His-APX1 and His-APX1C32S recombinant proteins in vitro. A quantitative analysis of the data is shown below the blot. E, S-Nitrosylation of APX1-FLAG, APX1C32S-FLAG, and APX1C49S-FLAG proteins in vivo. A quantitative analysis of the data is shown below the blot. Error bars in C to E = sd from three independent experiments (biological replicates). Two-tailed Student’s t test; **, P < 0.01; Asc, ascorbate sodium.
S-Nitrosylation of Cys-32 Positively Regulates APX1 Enzymatic Activity
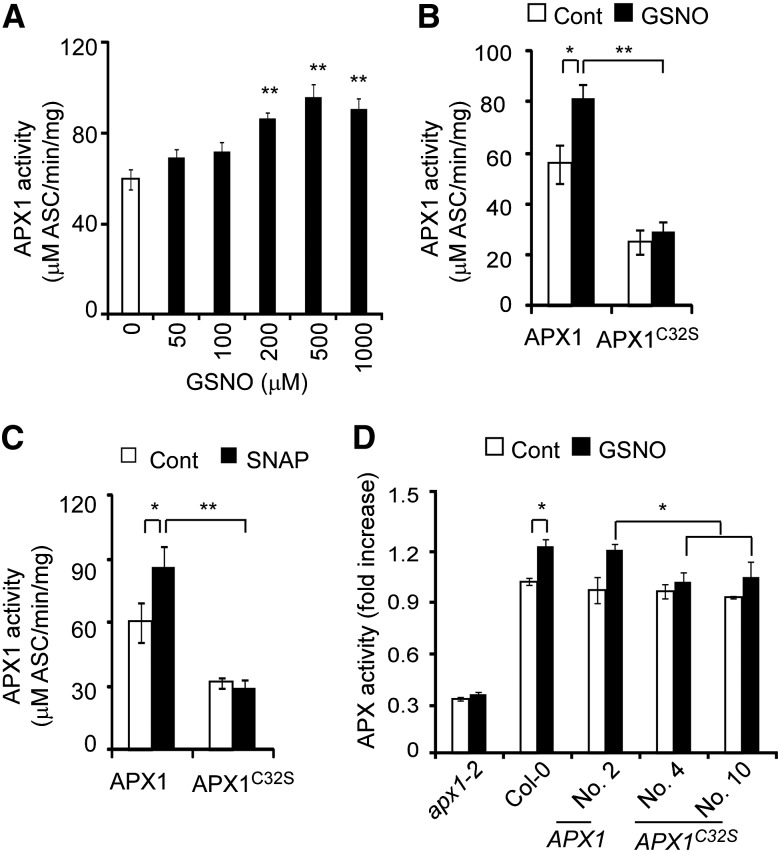
S-nitrosylation has been shown to regulate various aspects of a modified protein, including the conformational changes, stability, subcellular localization, and the biochemical activity of target proteins (Hess et al., 2005; Astier et al., 2011; Gupta, 2011). We found that the stability of the APX1 protein was not affected by NO donors or mutations that altered the intracellular NO level (Supplemental Fig. S3). Instead, APX1 activity was enhanced by the gsnor1-3 mutation and GSNO (see Fig. 1, D and E). We then investigated the potential regulatory role of the S-nitrosylated Cys-32 and Cys-49 on the APX1 enzymatic activity. GSNO enhanced the enzymatic activity of APX1 recombinant protein in a dose-dependent manner (Fig. 4A). The enzymatic activity of APX1C49S recombinant protein was similar to that of wild-type APX1 recombinant protein in the absence or presence of GSNO (Supplemental Fig. S9A), indicating that the S-nitrosylation of Cys-49 does not play a major role in regulating APX1 activity. However, whereas the enzymatic activity of APX1C32S protein was reduced more than 50% compared to that of APX1 recombinant protein, the GSNO-enhanced enzymatic activity was nearly abolished by the APX1C32S mutation (Fig. 4B). Similarly, treatment with S-nitroso-N-acetylpenicillamine also enhanced the activity of APX1 recombinant protein, but not that of APX1C32S protein (Fig. 4C).
Figure 4.
S-Nitrosylation at Cys-32 enhances APX1 enzyme activity. A, GSNO enhances the enzymatic activity of His-APX1 recombinant protein. Purified His-APX1 recombinant protein was treated with various concentrations of GSNO for 1 h and then assayed for the enzymatic activities. B and C, Analysis of the enzymatic activity of His-APX1 and His-APX1C32S recombinant proteins. The purified recombinant proteins treated with 200 μm GSNO (B) or 100 μm S-nitroso-N-acetylpenicillamine (C) for 1 h were used for the enzymatic analysis. Cont, Control (not treated). D, Analysis of the APX activity of 7-d-old seedlings of the indicated genotypes. The seedlings were treated with 200 μm GSNO for 6 h or not treated (Cont). Transgenic plants were generated in the apx1-2 mutant background (numbers refer to the transgenic lines). Error bars = sd from three independent experiments (biological replicates). Two-tailed Student’s t test: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
To further analyze the regulatory role of Cys-32 on APX1 activity, we generated transgenic plants in the apx1-2 background, which carried wild-type APX1 or APX1C32S genomic DNA fragments fused to a FLAG tag under the control of the native APX1 promoter. Both APX1:APX1-FLAG and APX1:APX1C32S-FLAG transgenes fully rescued the developmental defects of apx1-2 (Supplemental Fig. S10), indicating that they are physiologically functional under the normal growth condition. Under the normal growth condition, the APX1:APX1-FLAG and APX1:APX1C32S-FLAG transgenic plants showed APX activity similar to that of wild-type plants. However, the GSNO-enhanced APX activity was abolished by the APX1C32S mutation in planta (Fig. 4D). We noticed that the APX1C32S recombinant protein showed a considerably lower activity compared with the wild-type APX1 and APX1C32S-FLAG proteins in the absence of GSNO, which might be caused by structural alterations of APX1C32S recombinant protein or other unknown reasons. We also noticed that the APX activity was not altered by GSNO in the apx1-2 mutant seedlings (Fig. 4D), suggesting that, unlike APX1, other APX proteins may not be regulated by NO. Alternatively, the enzymatic activity of some of APX proteins may be regulated by NO to a lesser degree, and the altered activity is below the detection limit under our assay condition, considering the fact that APX1 accounts for the majority of APX activity (approximately 70%; see Fig. 1D). Currently, we are unable to distinguish these possibilities. Nevertheless, these results suggest that S-nitrosylation of Cys-32 plays a critical role in positively regulating APX1 activity, consistent with the observation that the highly conserved Cys-32 is located adjacent to the enzymatic active center of soybean (Glycine max) cytosolic APX (Sharp et al., 2003; Supplemental Fig. S8C).
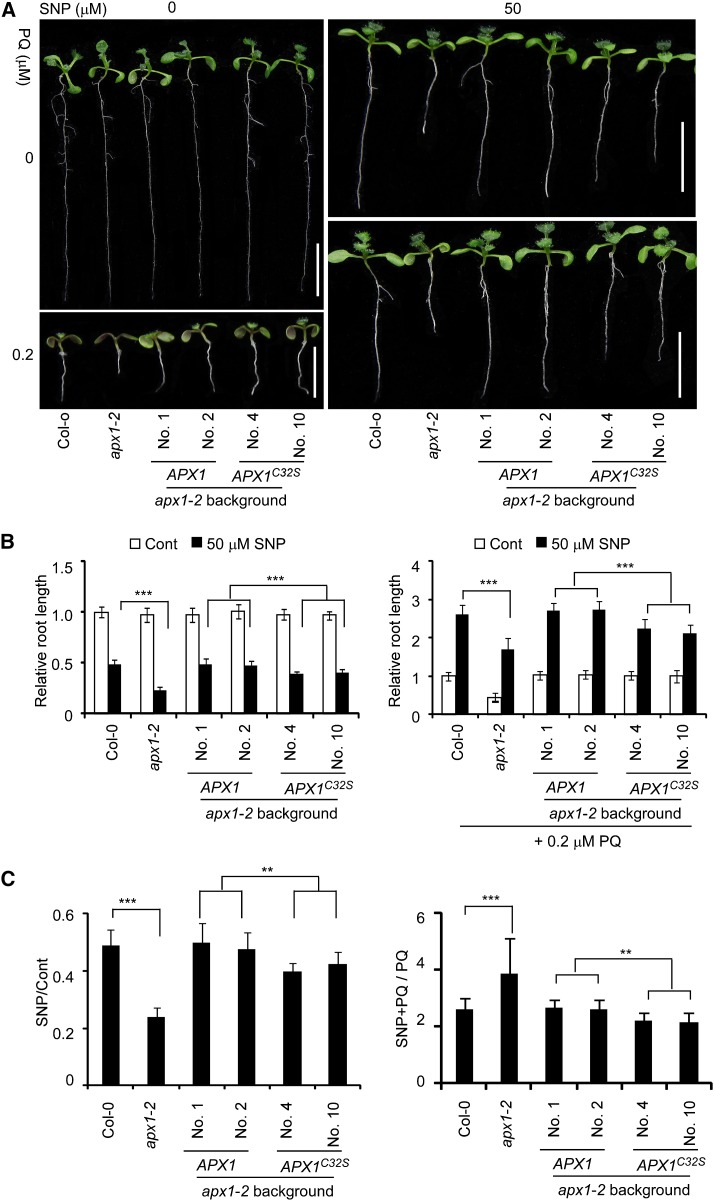
S-Nitrosylation of APX1 at Cys-32 Enhances the Resistance to Oxidative Stress
The previous data indicate that S-nitrosylation at Cys-32, but not Cys-49, positively regulates the APX1 activity. To assess the physiological relevance of S-nitrosylation of APX1, we analyzed the phenotype of the APX1:APX1-FLAG, APX1:APX1C32S-FLAG, and APX1:APX1C49S-FLAG transgenic plants in response to oxidative stress. Under normal growth conditions, both the APX1:APX1-FLAG and APX1:APX1C32S-FLAG transgenes rescued the developmental defects and paraquat sensitivity of the apx1-2 mutant (Fig. 5; Supplemental Fig. S10). SNP substantially enhanced the paraquat resistance in the wild-type and APX1:APX1-FLAG transgenic plants. However, this SNP-enhanced resistance was reduced in the APX1:APX1C32S-FLAG transgenic plants (Fig. 5), which correlated to the reduction of S-nitrosylation and the enzymatic activity of APX1C32S. Contrary to the APX1:APX1C32S-FLAG transgenic plants, the APX1:APX1C49S-FLAG transgenic plants showed a phenotype similar to that of the wild type in response to the paraquat and SNP (Supplemental Fig. S9B). These results indicate that S-nitrosylation of Cys-32 positively regulates the H2O2-scavenging activity of APX1, thereby protecting plants from oxidative stress.
Figure 5.
S-Nitrosylation at Cys-32 enhances plants resistance to oxidative stress. A, Ten-day-old seedlings germinated and grown on one-half-strength Murashige and Skoog agar plates with various combinations of 50 μm SNP and 0.2 μm paraquat (PQ). Scale bars = 1 cM. B, A quantitative analysis of the relative length of the primary roots (n = 30 seedlings for each genotype). The relative root length of Col-0 seedlings grown under normal growth condition is set at 1.0. C, The ratio of the primary root length shown in B. Error bars in C and D = sd from three independent experiments (biological replicates). Two-tailed Student’s t test; **, P < 0.01; and ***, P < 0.001.
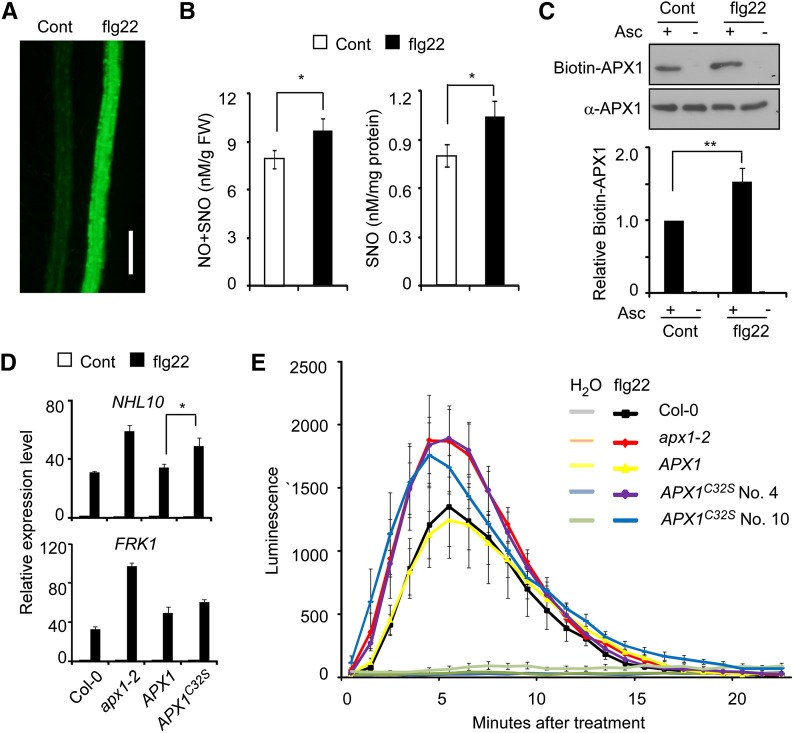
S-Nitrosylation of APX1 at Cys-32 Negatively Regulates the Immune Response
We next analyzed the potential role of S-nitrosylation of APX1 in immune responses. We found that treatment with flagellin elicitor peptide (flg22), a pathogen-associated molecular pattern derived from bacterial flagellin, caused an NO burst in wild-type plants (Fig. 6, A and B). Consistent with this result, a higher level of the S-nitrosylated APX1 protein was detected when treated with flg22 (Fig. 6C). Compared to wild-type plants, the flg22-induced expression of NONRACE-SPECIFIC DISEASE RESISTANCE1/HIN1-LIKE10 (NHL10) and FLAGELLIN-RESPONSIVE RECEPTOR-LIKE KINASE1 (FRK1), two marker genes in the immune signaling pathway, was substantially enhanced in apx1-2, and this phenotype was rescued by an APX1:APX1-FLAG transgene, but not by an APX1:APX1C32S-FLAG transgene (Fig. 6D). Moreover, the flg22-induced H2O2 burst was enhanced in apx1-2, and the increased H2O2 level was restored to the wild-type level by an APX1:APX1-FLAG transgene (Fig. 6E). However, the APX1:APX1C32S-FLAG transgene only partially rescued the apx1-2 phenotype in response to flg22 (Fig. 6E). These results suggest that S-nitrosylation of APX1 at Cys-32 is functionally important for H2O2 homeostasis during immune responses. When inoculated with various bacteria pathogens, apx1-2 and related transgenic plants showed a phenotype similar to that of wild-type plants (Supplemental Fig. S11).
Figure 6.
S-Nitrosylation of APX1 negatively regulates immune signaling. A, Flg22 induces NO burst. Five-day-old seedlings treated with or without (Cont) 10 μm flg22 for 10 min were stained with 4,5-diaminofluorescein diacetate, and the accumulation of NO in the root was visualized under a fluorescent microscope. Scale bar = 200 μm. B, The accumulation of NO and SNO in 4-week-old Col-0 seedlings treated with or without (Cont) 10 μm flg22. C, Accumulation of the S-nitrosylated APX1 protein in Col-0 seedlings treated with or without (Cont) 10 μm flg22. A quantitative analysis of the data is shown below the blot. D, Quantitative RT-PCR analysis of the NHL10 and FRK1 expression levels in 10-d-old seedlings treated with 10 μm flg22. The relative expression level of each gene in untreated Col-0 seedlings is set as 1.0. E, Analysis of flg22-induced H2O2 production in 4-week-old seedlings with the indicated genotypes. Error bars in B to D = sd from three independent experiments (biological replicates). Error bars in E = sd from four technical replicates. Two-tailed Student’s t test; *, P < 0.05; and **, P < 0.01.
DISCUSSION
As a common strategy to cope with stresses and environmental changes, plants usually respond by modulating the homeostatic redox balance, which is executed in part by regulating intracellular ROS and reactive nitrogen species, especially H2O2 and NO. The interaction between ROS- and NO-mediated signaling pathways has long been appreciated as a critical mechanism regulating both biotic and abiotic stress responses. However, the underpinning molecular mechanism remains poorly understood. In this study, we present multiple lines of evidence showing that NO positively regulates the enzymatic activity of APX1 by S-nitrosylation, which consequently controls the intracellular ROS homeostasis during stress responses. Consistent with our observation, the APX activity of pea is correlated with the increased level of S-nitrosylated APX protein when under the saline stress growth condition (Begara-Morales et al., 2014). Moreover, the S-nitrosylation-regulated APX activity also appears to play an important role in various developmental processes. When treated with the NO donor 2,2'-(hydroxynitrosohydrazono)bis-ethanimine, the soybean root nodules showed enhanced activity of three APX isoforms (Keyster et al., 2011). In Arabidopsis, auxin regulates root development partly by inducing APX1 denitrosylation that causes the partial inhibition of APX1 activity (Correa-Aragunde et al., 2013). Similarly, NO enhances the desiccation tolerance of recalcitrant Anticaria toxicaria seeds by reducing the H2O2 level through enhancing the activities of several antioxidant enzymes, including APX, presumably via protein S-nitrosylation and carbonylation (Bai et al., 2011). Therefore, the S-nitrosylation-regulated APX activity is likely a conserved mechanism in plants. In contrast with the results highlighted earlier (see below for additional examples), a recent study found that S-nitrosylation of cytosolic APX in tobacco Bright Yellow-2 suspension cells induces its ubiquitination and subsequent proteasomal degradation, resulting in a reduction of the tobacco APX activity (de Pinto et al., 2013). Under our assay condition, the stability of Arabidopsis APX1 protein remains largely unaltered by NO donors or in mutants that have an elevated intracellular NO level (see Supplemental Fig. S3), suggesting that NO does not play a major role in regulating the turnover of APX1 in Arabidopsis. It may be that tobacco uses a unique mechanism different from Arabidopsis and other plant species to regulate the NO-modulated APX activity.
Arabidopsis APX1 contains five Cys residues, two of which (Cys-32 and Cys-49) were identified as S-nitrosylated by our LC-MS/MS and mutagenesis studies. Homology modeling predicts that Cys-32 and Cys-49 are both exposed to the surface of APX1, thereby increasing their accessibility to bioactive NO and thus facilitating S-nitrosylation (Supplemental Fig. S8C). Cys-32 is highly conserved in APX1-like proteins across the plant kingdom, ranging from the unicellular red alga Galdieria partita to higher plants (see Supplemental Fig. S6). Moreover, this residue has also been identified to be S-nitrosylated in Arabidopsis by proteomics studies and in pea by a mass spectrometric study, respectively (Fares et al., 2011; Begara-Morales et al., 2014; Hu et al., 2015). The importance of Cys-32 is evident by the observation that soybean APX1 contains a single Cys residue (accession no. NP_001237785.1; see also Sharp et al., 2003), suggesting that Cys-32 may be functionally indispensable. In agreement with this notion, we found that S-nitrosylation of Arabidopsis APX1 at Cys-32 is not only important for its activity under normal growth conditions, but also plays a key role in response to the increased intracellular level of NO. Interestingly, the unicellular red alga G. partita APX-B recombinant protein has been found to be oxidized and glutathionylated, and the oxidation of APX-B at Cys-25, a residue equivalent to Cys-32 of the Arabidopsis APX1 and pea cytosolic APX, causes rapid inactivation of the enzymatic activity (Kitajima et al., 2008). These observations indicate that Cys-32 is a target for various forms of posttranslational modifications, through which the activity of APX1 is regulated in response to distinctive signals. In this regard, we propose that Cys-32 of APX1 may act to sense and monitor the intracellular redox changes.
Although it remains unclear how the activity of APX1 is regulated by S-nitrosylation and other modifications at Cys-32, the analysis of the three-dimensional structure of soybean APX1 protein has provided valuable clues for understanding the regulatory mechanism (Sharp et al., 2003). Cys-32 is near the propionate side chain of the haem group, and the distance between Cys-32 and the APX1 substrate ascorbate is approximately 3.8 Å (Sharp et al., 2003; see also Supplemental Fig. S8C). Presumably, specific modifications at Cys-32, such as S-nitrosylation and oxidation, may cause local conformational changes around the haem group, which may directly regulate the binding affinity of APX1 with ascorbate. On the other hand, Cys-49 was also identified as an S-nitrosylated residue in this study. However, the APX1C49S mutation does not appear to have an apparent effect on the APX1 activity under our assay condition. Currently, we cannot exclude the possibility that the S-nitrosylation of Cys-49 may play a regulatory role in the APX1 activity under other conditions.
The gsnor1-3/hot5/par2 mutations substantially increase the intracellular level of NO (Feechan et al., 2005; Lee et al., 2008; Chen et al., 2009). Consistently, we found that a higher level of S-nitrosylated APX1 protein in the gsnor1-3 mutant correlated to the increased APX enzymatic activity in gsnor1-3. Moreover, the APX1C32S mutation abolishes the responsiveness to NO-induced enhancement of APX1 activity. Together, these observations suggest that the S-nitrosylation-mediated APX1 regulation plays a critical role in protecting against the oxidative stress, which partly explains the paraquat-resistant phenotype of the par2 mutant allele (Chen et al., 2009). Intriguingly, S-nitrosylation has been shown to inhibit the H2O2-scavenging activity of peroxiredoxins in mammals (Fang et al., 2007; Engelman et al., 2013), further supporting the notion that the redox-based S-nitrosylation modification may play an important role in the regulation of the H2O2-scavenging enzymatic activity to cope with various stresses in both animals and plants. The important role of S-nitrosylation in the regulation of the intracellular ROS homeostasis has been reinforced by the finding that S-nitrosylation of NADPH oxidase causes the decreased enzymatic activity, resulting in the reduction of superoxide during immune responses in Arabidopsis (Yun et al., 2011). Therefore, NO coordinates the intracellular level of ROS during stress responses through S-nitrosylation-mediated boosting of the H2O2-scavenging activity of APX1 and attenuation of the ROS synthesis activity of NADPH oxidase.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) wild-type accession Columbia-0 (Col-0) and related mutants were used in this study. The gsnor1-3 (Feechan et al., 2005) and apx1-2 (SALK_000249) mutant seeds were obtained from Dr. Gary Loake (University of Edinburgh) and the Arabidopsis Biological Resource Center, respectively.
Seeds were germinated on one-half-strength Murashige and Skoog medium agar plates containing 1% (w/v) Suc. The plate was imbibed at 4°C for 2 d and then cultured at 22°C under continuous white light. For flg22-induced assays, plants were grown under a cycle of 8 h of light and 16 h of dark at 22°C.
The generation of transgenic Arabidopsis plants was performed by Agrobacterium tumefaciens-mediated transformation (Bechtold and Pelletier, 1998). For all transgenic studies, two to four independent transgenic lines were analyzed, and similar results were obtained. Representative results were presented. The GV3101 strain of A. tumefaciens was used in all genetic transformation experiments.
For analysis of the response to SNP and paraquat, seedlings were grown vertically on agar plates with or without various concentrations of SNP and/or paraquat, and the root length was measured 10 d postgermination.
Plasmid Construction
The coding sequences of APX1 were PCR amplified from Arabidopsis complementary DNA (the stop codon was eliminated during PCR), and the resulting complementary DNA fragment was cloned into the BamHI and SalI sites of pET28a (Novagen) to generate the pET28a-APX1 expression vector.
To make APX1:APX1-FLAG, a genomic DNA fragment of APX1 was amplified by PCR (primer pairs APX1F2 and APX1B2), which included a 1,260-bp promoter and a 1,428-bp coding region. The fragment was ligated to pBlueScript SK(−) (Stratagene), fused to a single copy of FLAG tag at the C termini. The APX1:APX1-FLAG fusion gene was cloned into the XhoI/SpeI sites of the binary vector pER8 (Zuo et al., 2000). Site-directed mutagenesis was performed using the Easy Mutagenesis System (TransGen Biotech) according to the manufacturer’s instructions.
All PCR-amplified fragments in the constructs described were confirmed by DNA sequencing analysis, and all constructs were verified by extensive restriction digests. All PCR primers used in this study are listed in Supplemental Table S1.
Quantitative Reverse Transcription-PCR
Total RNA was prepared by the RNAprep Pure Plant RNA Purification Kit (Tiangen Biotech) according to the manufacturer’s instructions. Reverse transcription (RT)-PCR analyses were carried out as described previously, and Ubiquitin5 (UBQ5; At3g62250) was used as an internal control (Mu et al., 2008). Real-time PCR was performed using the UltraSYBR Mixture (CWBIO) according to the manufacturer’s instructions using the CFX96 Real-Time PCR Detection System (Bio-Rad). The relative expression level of the target genes was analyzed with the delta-delta cycle threshold method and normalized to the expression level of ACTIN7 (At5g09810). All of the experiments were repeated for at least three times with two to three technical repeats for each experiment. All primers used in the RT-PCR and quantitative RT-PCR analyses are listed in Supplemental Table S1.
Expression and Purification of Recombinant Proteins
The pET28a-(6XHis tag)-based expression vectors were transformed into Escherichia coli BL21. The bacterial cultures were grown in liquid Luria-Bertani medium at 37°C for about 3 h. When optical density at 600 nm reached around 0.9 to 1.0, isopropyl-β-d-thiogalactoside was added to a final concentration of 0.2 mm. The cultures were incubated for an additional 6 h at 37°C. Recombinant proteins were purified using 3S-NTA resin (Biocolor Bioscience and Technology Company) following the manufacturer’s instructions. Purified proteins were exchanged for HEN buffer (250 mm HEPES, pH 7.7, 1 mm EDTA) containing 10% (v/v) glycerol or potassium-phosphate buffer (50 mm, pH 7.0) with Pierce Desalting Cartridges (Pierce Biotechnology) following the manufacturer’s instructions.
Generation of Antibodies and Immunoblotting
Purified His-APX1 recombinant protein (approximately 1 mg) was used to immunize rabbits, and the resulting antiserum was used directly for immunoblotting (dilution 1:5,000). Cytosolic TUBULIN was served as a loading control using anti-TUBULIN antibody (Sigma-Aldrich, catalog no. T5168, dilution 1:5,000). Immunoblotting was carried out as previously described (Chen et al., 2009).
S-Nitrosylation Analysis
S-Nitrosylated proteins were analyzed by the biotin-switch assay as described previously (Feng et al., 2013). In brief, approximately 100 to 200 μL of the protein extracts in HEN (250 mm HEPES, pH 7.7, 1 mm EDTA) buffer was blocked using blocking buffer 1 (250 mm HEPES, pH 7.7, 1 mm EDTA, 0.1 mm neocuproine, 2.5% [w/v] SDS, and 25 mm S-methylmethane thiosulfonate) at 50°C for 1 h. The sample was precipitated with cold acetone, washed three times with 70% (v/v) acetone, and dissolved in 170 μL of HEN buffer supplemented with 1% (w/v) SDS. After adding 10 μL of 1 m sodium ascorbate and 20 μL of 4 mm biotin-HPDP, the sample was labeled for 1 h at room temperature and then precipitated with acetone. The pellet was dissolved in 400 μL of 250 mm HEPES, pH 7.7, 1 mm EDTA, and 1% (w/v) SDS and neutralized with 800 μL of neutralization buffer (25 mm HEPES, pH 7.7, 100 mm NaCl, 1 mm EDTA, and 0.5% [v/v] Triton X-100). The sample was then mixed with 30 μL of streptavidin beads (Thermo Scientific, catalog no. 29202) and incubated at 4°C overnight. All of the previous manipulations were performed in a dark room with occasional dim light. The beads were washed four times with washing buffer (25 mm HEPES, pH 7.7, 600 mm NaCl, 1 mm EDTA, and 0.5% [v/v] Triton X-100). The proteins were then eluted and analyzed by immunoblotting with the appropriate antibodies. The in vitro S-nitrosylation assay was performed as described previously (Feng et al., 2013).
Fluorometric Measurement of S-Nitrosylated Proteins by the DAN Assay
Measurement of the S-nitrosylated Cys residue numbers was performed by the DAN assay as described (Romero-Puertas et al., 2007; Cho et al., 2009), with modifications. In this assay, the released NO from the thiol group is detected by the conversion of DAN into fluorescent NAT. The released NO moiety per protein molecule, which represents the number of the S-nitrosylated residues, is calculated by the DAN-NAT conversion ratio (Cho et al., 2009).
Recombinant His-APX1 protein was purified using the 3S-NTA-resin as described earlier. One hundred microliters of purified 3S-NTA-resin-His-APX1 was diluted with 1.1 mL of PBS buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 2 mm KH2PO4), and aliquots of the sample (200, 400, and 600 μL, respectively) were washed twice with phosphate-buffered saline (PBS) buffer. The pellets were resuspended in 1 mL of PBS, followed by adding GSNO at a final concentration of 100 μm. The reaction was carried out at room temperature in the dark for 1 h, and the resin was washed three times with PBS buffer to remove residual GSNO. The sample was mixed with 300 μL of 200 μm DAN and 200 μm HgCl2 and incubated in the dark for 30 min with continuous vortex. The reaction was stopped by adding 15 μL of 2.8 m NaOH. The supernatant and the pellet separated by centrifugation were used for the analysis of the fluorescent signal and the protein concentration, respectively. The fluorescent signal emitted from NAT was measured at an excitation spectrum of UV (365 ± 10 nm) and an emission wavelength of 450 nm using a NanoDrop 3300 fluorospectrometer (Thermo Scientific). The resin was mixed with Coomassie Brilliant Blue G250 to determine the protein concentration by the Bradford method. Serial GSNO dilutions were treated with DAN as described earlier to construct a standard curve. The relative DAN-NAT conversion rate was calculated as the fluorescent intensity of NAT per micromolar protein or peptide.
Mass Spectrometric Analysis of S-Nitrosylated Proteins
Mass spectrometry was performed as described with minor modifications (Huang and Chen, 2010; Feng et al., 2013). In brief, purified His-APX1 was first labeled with biotin-maleimide (Sigma-Aldrich, catalog no. B1267) instead of biotin-HPDP. The labeled sample was digested in gel using trypsin (Promega, catalog no. V5280) and then analyzed by LC-MS/MS using a Thermo Fisher Finnigan linear ion trap quadrupole mass spectrometer in line with a Thermo Fisher Finnigan Surveyor MS Pump Plus HPLC system. The digested sample was loaded onto a trap column (300SB-C18, 5 × 0.3 mm, 5-μm particle; Agilent Technologies) that was connected through a zero dead volume union to the self-packed analytical column (C18, 100-μm i.d. × 100 mm, 3-μm particle; SunChrom). The peptides were then eluted over a gradient (0%–45% B in 55 min, 45%–100% B in 10 min; where B = 80% [v/v] acetonitrile, 0.1% [v/v] formic acid) at a flow rate of 500 nL min−1 and introduced online into the linear ion trap mass spectrometer (Thermo Fisher) using nanoelectrospray ionization. Data-dependent scanning was incorporated to select the five most abundant ions (one microscan per spectra; precursor isolation width 1.0 mass-to-charge ratio, 35% collision energy, 30-ms ion activation; exclusion duration, 90s; repeat count, 1) from a full-scan mass spectrum for fragmentation by collision-induced dissociation.
Mass spectrometry data were analyzed using SEQUEST Bioworks version 3.2 (Thermo Scientific; http://fields.scripps.edu/sequest/) against the National Center for Biotechnology Information Arabidopsis protein database, and the results were filtered, sorted, and displayed using Bioworks version 3.2. Peptides with +1, +2, or +3 charge states were accepted if they were fully enzymatic and had a cross correlation of >1.90, >2.5, and >3.0, respectively. The following residue modifications were allowed in the search: carboxyamidomethylation, propionamidation, and biotinylation on Cys and oxidation on Met. SEQUEST was searched with a peptide tolerance of 3 atomic mass units and a fragment ion tolerance of 1.0 atomic mass units.
Analysis of the APX Enzymatic Activity
Analysis of the enzymatic activity of APX1 recombinant protein was performed as described (Mittler and Zilinskas, 1991; Dalton et al., 1996) with minor modifications. Approximately 100 μg of purified His-APX1, His-APX1C32S, and His-APX1C49S recombinant proteins was treated with various concentrations of GSNO for 1 h in the dark. Excessive GSNO was removed using Zeba Spin Desalting Columns (Thermo Scientific, catalog no. 89883) equilibrated with potassium-phosphate buffer (50 mm, pH 7.0). Ten percent excess over the stoichiometric amount of bovine hemin (Sigma-Aldrich, catalog no. 51280) was slowly added to the APX1 protein solution with gentle stirring in the dark, and the sample was incubated at 4°C for 15 min with gentle shaking. The protein concentration of the reconstitution products was adjusted to 0.1 mg mL−1 with 50 mm sodium phosphate (pH 7.0). Typically, 500 μL of the reconstitution products was used for the assay by adding sodium ascorbate and hydrogen peroxide to final concentrations of 0.4 and 1 mm, respectively. After the initiation of the reaction by adding hydrogen peroxide, the sample was immediately analyzed with SpectraMax Paradigm (Molecular Devices) by measuring the absorbance at A290 every 15 s for a 2-min recording time.
Analysis of total APX activity was performed with the inhibition of the ascorbate-dependent reduction of nitroblue tetrazolium (NBT) as described previously (Mittler and Zilinskas, 1993). To prepare total proteins, plant materials were ground in liquid nitrogen and extracted in grinding buffer (100 mm potassium-phosphate buffer, pH 7.0, 5 mm ascorbate, and 1 mm EDTA) supplied with the protease inhibitor cocktail (Sigma-Aldrich, catalog no. P9599). Approximately 30 μg of proteins was incubated with 0.8 mm ascorbate and 0.4 mm H2O2 in 1-mL reaction volume for 20 min at 25°C with gentle shaking. Afterward, 0.5 mL of 28 mm N,N,N′,N′-tetramethylethylenediamine and 2.45 mm NBT was added, and the reaction was incubated for 10 min. All of the previous manipulations were performed in a dark room with occasional dim light. The degree of NBT reduction was then analyzed at the isobestic point of formazan formation (585 nm) using SpectraMax Paradigm (Molecular Devices).
Analysis of NO and Oxidative Burst
Confocal imaging of NO in Arabidopsis roots was performed by 4,5-diaminofluorescein diacetate staining as previously described (Chen et al., 2009). The SNO level in the total cellular lysate was measured by photolysis chemiluminescence as described previously (Liu et al., 2001). Analysis of oxidative burst was performed as previously described (Li et al., 2014), and the luminescence was recorded by a GLOMAX96 Luminometer (Promega). Bacterial growth assay was performed as previously described (Li et al., 2014).
Analysis of GSH and GSSG Level
Total glutathione (GSH+GSSG) was measured by the Glutathione Assay Kit (Sigma-Aldrich, catalog no. CS0260) according to the manufacturer’s instructions. To analyze the GSSG level, the sample was treated with 2-vinylpyridine for 1 h and centrifuged at 13,000 rpm for 10 min. The supernatant was used for glutathione determination by the Glutathione Assay Kit (Sigma-Aldrich, catalog no. CS0260) according to the manufacturer’s instructions.
Computational Modeling
The prediction of putative S-nitrosylated Cys residues was performed by a manual search combined with the use of GPS-SNO software (Xue et al., 2010) with the threshold set to high. Modeling of the three-dimensional structure of APX1 was performed online at the SwissModel server (http://swissmodel.expasy.org) using the structure of soybean (Glycine max) cytosolic APX (PDB:1OAF; Sharp et al., 2003) as a template. Molecular graphics were prepared using PyMOL software (http://www.pymol.org/).
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the accession numbers At1g07890 (APX1), At5g09810 (ACTIN7), At2g19190 (FRK1), At5g43940 (GSNOR1), At2g35980 (NHL10), At5g33320 (NITRIC OXIDE OVERPRODUCER1), At1g20010 (TUBULIN5), and At3g62250 (UBQ5).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. NO enhances the resistance to oxidative stress.
Supplemental Figure S2. The apx1-2 mutant phenotype.
Supplemental Figure S3. Analysis of the accumulation of APX1 protein.
Supplemental Figure S4. SNP enhances the resistance to oxidative stress.
Supplemental Figure S5. Analysis of GSH and GSSG level.
Supplemental Figure S6. Sequence alignment of APX proteins from various species.
Supplemental Figure S7. Mass spectra of APX1 peptides.
Supplemental Figure S8. Structural modeling of Arabidopsis APX1.
Supplemental Figure S9. Functional characterization of Cys-49 of APX1.
Supplemental Figure S10. Characterization of APX1 transgenic plants.
Supplemental Figure S11. Analysis of bacterial growth in apx1-2 and transgenic plants.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center and Dr. Gary Loake for mutant seeds.
Glossary
- ROS
reactive oxygen species
- NO
nitric oxide
- GSNO
S-nitrosoglutathione
- SNO
S-nitrosothiols
- SA
salicylic acid
- SNP
sodium nitroprusside
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- MS/MS
tandem mass spectrometry
- DAN
2,3-diaminonaphthalene
- NAT
2,3-naphthyltriazole
- Col-0
Columbia-0
- RT
reverse transcription
- PBS
phosphate-buffered saline
- MS
mass spectrometry
- NBT
nitroblue tetrazolium
- H2O2
hydrogen peroxide
- LC
liquid chromatography
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31130014 and 91217302) and State Key Laboratory of Plant Genomics (grant no. SKLPG2011A0210).
References
- Aquilano K, Baldelli S, Ciriolo MR. (2014) Glutathione: new roles in redox signalling for an old antioxidant. Front Pharmacol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astier J, Rasul S, Koen E, Manzoor H, Besson-Bard A, Lamotte O, Jeandroz S, Durner J, Lindermayr C, Wendehenne D (2011) S-nitrosylation: an emerging post-translational protein modification in plants. Plant Sci 181: 527–533 [DOI] [PubMed] [Google Scholar]
- Bai X, Yang L, Tian M, Chen J, Shi J, Yang Y, Hu X (2011) Nitric oxide enhances desiccation tolerance of recalcitrant Antiaris toxicaria seeds via protein S-nitrosylation and carbonylation. PLoS ONE 6: e20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Begara-Morales JC, Sánchez-Calvo B, Chaki M, Valderrama R, Mata-Pérez C, López-Jaramillo J, Padilla MN, Carreras A, Corpas FJ, Barroso JB (2014) Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J Exp Bot 65: 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Fath A, Bethke PC, Lamattina L, Jones RL (2002) Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol 129: 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Lamattina L (1999a) Nitric oxide protects against cellular damage produced by methylviologen herbicides in potato plants. Nitric Oxide 3: 199–208 [DOI] [PubMed] [Google Scholar]
- Beligni MV, Lamattina L (1999b) Nitric oxide counteracts cytotoxic processes mediated by reactive oxygen species in plant tissues. Planta 208: 337–344 [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D (2008) New insights into nitric oxide signaling in plants. Annu Rev Plant Biol 59: 21–39 [DOI] [PubMed] [Google Scholar]
- Chen R, Sun S, Wang C, Li Y, Liang Y, An F, Li C, Dong H, Yang X, Zhang J. , et al (2009) The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res 19: 1377–1387 [DOI] [PubMed] [Google Scholar]
- Cho D-H, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA (2009) S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science 324: 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Aragunde N, Foresi N, Delledonne M, Lamattina L (2013) Auxin induces redox regulation of ascorbate peroxidase 1 activity by S-nitrosylation/denitrosylation balance resulting in changes of root growth pattern in Arabidopsis. J Exp Bot 64: 3339–3349 [DOI] [PubMed] [Google Scholar]
- Dalton DA, Diaz del Castillo L, Kahn ML, Joyner SL, Chatfield JM (1996) Heterologous expression and characterization of soybean cytosolic ascorbate peroxidase. Arch Biochem Biophys 328: 1–8 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M. (2005) NO news is good news for plants. Curr Opin Plant Biol 8: 390–396 [DOI] [PubMed] [Google Scholar]
- de Pinto MC, Locato V, Sgobba A, Romero-Puertas MdelC, Gadaleta C, Delledonne M, De Gara L (2013) S-nitrosylation of ascorbate peroxidase is part of programmed cell death signaling in tobacco Bright Yellow-2 cells. Plant Physiol 163: 1766–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil-Maurizi C, Vitecek J, Marty L, Branciard L, Frettinger P, Wendehenne D, Meyer AJ, Mauch F, Poinssot B (2011) Glutathione deficiency of the Arabidopsis mutant pad2-1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol 157: 2000–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman R, Weisman-Shomer P, Ziv T, Xu J, Arnér ESJ, Benhar M (2013) Multilevel regulation of 2-Cys peroxiredoxin reaction cycle by S-nitrosylation. J Biol Chem 288: 11312–11324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA (2007) S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci USA 104: 18742–18747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares A, Rossignol M, Peltier JB (2011) Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem Biophys Res Commun 416: 331–336 [DOI] [PubMed] [Google Scholar]
- Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ (2005) A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA 102: 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Wang C, Chen Q, Chen H, Ren B, Li X, Zuo J (2013) S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat Commun 4: 1529. [DOI] [PubMed] [Google Scholar]
- Groß F, Durner J, Gaupels F (2013) Nitric oxide, antioxidants and prooxidants in plant defence responses. Front Plant Sci 4: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Crawford NM (2005) Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell 17: 3436–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KJ. (2011) Protein S-nitrosylation in plants: photorespiratory metabolism and NO signaling. Sci Signal 4: jc1. [DOI] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS (2005) Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6: 150–166 [DOI] [PubMed] [Google Scholar]
- Hess DT, Stamler JS (2012) Regulation by S-nitrosylation of protein post-translational modification. J Biol Chem 287: 4411–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Huang X, Chen L, Sun X, Lu C, Zhang L, Wang Y, Zuo J (2015) Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol 167: 1731–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Chen C (2010) Detection of protein S-nitrosation using irreversible biotinylation procedures (IBP). Free Radic Biol Med 49: 447–456 [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001: pl1. [DOI] [PubMed] [Google Scholar]
- Keyster M, Klein A, Egbichi I, Jacobs A, Ludidi N (2011) Nitric oxide increases the enzymatic activity of three ascorbate peroxidase isoforms in soybean root nodules. Plant Signal Behav 6: 956–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S, Kurioka M, Yoshimoto T, Shindo M, Kanaori K, Tajima K, Oda K (2008) A cysteine residue near the propionate side chain of heme is the radical site in ascorbate peroxidase. FEBS J 275: 470–480 [DOI] [PubMed] [Google Scholar]
- Kwon E, Feechan A, Yun BW, Hwang BH, Pallas JA, Kang JG, Loake GJ (2012) AtGSNOR1 function is required for multiple developmental programs in Arabidopsis. Planta 236: 887–900 [DOI] [PubMed] [Google Scholar]
- Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E (2008) Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20: 786–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner M, Vandelle E, Gaupels F, Bellin D, Delledonne M (2009) NO signals in the haze: nitric oxide signalling in plant defence. Curr Opin Plant Biol 12: 451–458 [DOI] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y. , et al (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338 [DOI] [PubMed] [Google Scholar]
- Lin CC, Jih PJ, Lin HH, Lin JS, Chang LL, Shen YH, Jeng ST (2011) Nitric oxide activates superoxide dismutase and ascorbate peroxidase to repress the cell death induced by wounding. Plant Mol Biol 77: 235–249 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Sell S, Müller B, Leister D, Durner J (2010) Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22: 2894–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS (2001) A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410: 490–494 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Mittler R, Zilinskas BA (1991) Purification and characterization of pea cytosolic ascorbate peroxidase. Plant Physiol 97: 962–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Zilinskas BA (1993) Detection of ascorbate peroxidase activity in native gels by inhibition of the ascorbate-dependent reduction of nitroblue tetrazolium. Anal Biochem 212: 540–546 [DOI] [PubMed] [Google Scholar]
- Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ. , et al (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Liang H, Rozenberg M, Mittler R (2003) Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 34: 187–203 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Laxa M, Mattè A, Zaninotto F, Finkemeier I, Jones AME, Perazzolli M, Vandelle E, Dietz KJ, Delledonne M (2007) S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell 19: 4120–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp KH, Mewies M, Moody PCE, Raven EL (2003) Crystal structure of the ascorbate peroxidase-ascorbate complex. Nat Struct Biol 10: 303–307 [DOI] [PubMed] [Google Scholar]
- Shi Q, Ding F, Wang X, Wei M (2007) Exogenous nitric oxide protect cucumber roots against oxidative stress induced by salt stress. Plant Physiol Biochem 45: 542–550 [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yun BW, Kwon E, Hong JK, Yoon J, Loake GJ (2006) S-nitrosylation: an emerging redox-based post-translational modification in plants. J Exp Bot 57: 1777–1784 [DOI] [PubMed] [Google Scholar]
- Wang YQ, Feechan A, Yun BW, Shafiei R, Hofmann A, Taylor P, Xue P, Yang FQ, Xie ZS, Pallas JA. , et al (2009) S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J Biol Chem 284: 2131–2137 [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Gao QM, Kachroo A, Kachroo P (2014) Free radical-mediated systemic immunity in plants. Curr Opin Plant Biol 20: 127–134 [DOI] [PubMed] [Google Scholar]
- Xue L, Li S, Sheng H, Feng H, Xu S, An L (2007) Nitric oxide alleviates oxidative damage induced by enhanced ultraviolet-B radiation in cyanobacterium. Curr Microbiol 55: 294–301 [DOI] [PubMed] [Google Scholar]
- Xue Y, Liu Z, Gao X, Jin C, Wen L, Yao X, Ren J (2010) GPS-SNO: computational prediction of protein S-nitrosylation sites with a modified GPS algorithm. PLoS ONE 5: e11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Lamattina L, Spoel SH, Loake GJ (2014) Nitric oxide function in plant biology: a redox cue in deconvolution. New Phytol 202: 1142–1156 [DOI] [PubMed] [Google Scholar]
- Yun BW, Feechan A, Yin M, Saidi NB, Le Bihan T, Yu M, Moore JW, Kang JG, Kwon E, Spoel SH. , et al (2011) S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478: 264–268 [DOI] [PubMed] [Google Scholar]
- Zaninotto F, La Camera S, Polverari A, Delledonne M (2006) Cross talk between reactive nitrogen and oxygen species during the hypersensitive disease resistance response. Plant Physiol 141: 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.