Depleting galactose from the cell wall polysaccharide xyloglucan leads to a disruption of diverse cellular and physiological processes involved in normal plant growth.
Abstract
Xyloglucan is a polysaccharide that has important roles in the formation and function of the walls that surround growing land plant cells. Many of these plants synthesize xyloglucan that contains galactose in two different side chains (L and F), which exist in distinct molecular environments. However, little is known about the contribution of these side chains to xyloglucan function. Here, we show that Arabidopsis (Arabidopsis thaliana) mutants devoid of the F side chain galactosyltransferase MURUS3 (MUR3) form xyloglucan that lacks F side chains and contains much less galactosylated xylose than its wild-type counterpart. The galactose-depleted xyloglucan is dysfunctional, as it leads to mutants that are dwarfed with curled rosette leaves, short petioles, and short inflorescence stems. Moreover, cell wall matrix polysaccharides, including xyloglucan and pectin, are not properly secreted and instead accumulate within intracellular aggregates. Near-normal growth is restored by generating mur3 mutants that produce no detectable amounts of xyloglucan. Thus, cellular processes are affected more by the presence of the dysfunctional xyloglucan than by eliminating xyloglucan altogether. To identify structural features responsible for xyloglucan dysfunction, xyloglucan structure was modified in situ by generating mur3 mutants that lack specific xyloglucan xylosyltransferases (XXTs) or that overexpress the XYLOGLUCAN L-SIDE CHAIN GALACTOSYLTRANSFERASE2 (XLT2) gene. Normal growth was restored in the mur3-3 mutant overexpressing XLT2 and in mur3-3 xxt double mutants when the dysfunctional xyloglucan was modified by doubling the amounts of galactosylated side chains. Our study assigns a role for galactosylation in normal xyloglucan function and demonstrates that altering xyloglucan side chain structure disturbs diverse cellular and physiological processes.
There have been considerable advances in understanding how changes in specific amino acids or nucleotides affect the ability of a protein or an RNA to fold and to function (Wan et al., 2011; Sauer, 2013). By contrast, much less is known about how polysaccharide structure and function are affected by altering specific monosaccharides within its glycosyl sequence. Here, we report that xyloglucan, a plant cell wall polysaccharide, becomes dysfunctional rather than nonfunctional when its glycosyl sequence is altered. The term dysfunctional polysaccharide is used here to describe a structure that, in and of itself, has a deleterious effect on the growth and development of a plant.
XXXG-type xyloglucans (XyGs; Fig. 1A) are present in the cell walls of many flowering plants (Hoffman et al., 2005), gymnosperms (Hsieh and Harris, 2012), ferns and their allies, club and spike mosses, and the avascular hornworts (Peña et al., 2008). XyGs of this type are composed of subunits (Fig. 1, A and B) that have three consecutive β-d-glucopyranose (β-d-Glcp) backbone residues bearing a side chain followed by a single unbranched β-d-Glcp residue (Tuomivaara et al., 2015). Little is known about the contribution of these side chains to the role of XyG in wall structure and function, although the results of in vitro studies suggest that they modulate how an XyG interacts with cellulose (Whitney et al., 2006) and with itself (Shirakawa et al., 1998; de Freitas et al., 2011).
Figure 1.
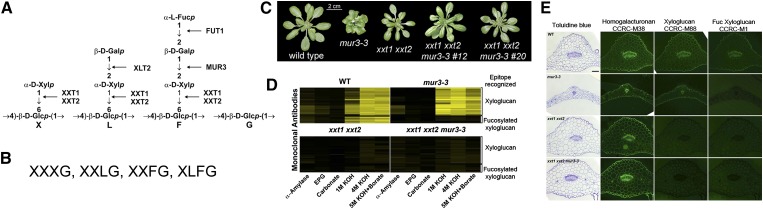
Eliminating xyloglucan suppresses the cabbage-like phenotype of mur3-3 plants. A, The major structural features of Arabidopsis XyG and the glycosyltransferases required for side chain formation. Side chains are represented by the letters X, L, and F. An unbranched Glc is designated by the letter G. B, The predominant subunits of Arabidopsis XyG. C, Eliminating XyG suppresses the dwarf phenotype of the mur3-3 mutant. mur3-3 was crossed with xxt1 xxt2, and two independent xxt1 xxt2 mur3-3 triple mutants (nos. 12 and 20) were identified by reverse transcription (RT)-PCR. Plants were grown with a 14-h-light (19°C) and 10-h-dark (15°C) cycle. D, The glycome profiles of the materials solubilized from the cell walls by enzymatic and KOH treatments. Each extract was probed with a series of monoclonal antibodies that recognize epitopes of XyG irrespective of the presence or absence of fucosylated side chains and monoclonal antibodies that bind only to XyG containing fucosylated side chains. Yellow, Strong binding; black, no binding. No XyG was detected in the xxt1 xxt2 double mutant or the xxt1 xxt2 mur3-3 triple mutants. E, Cross sections of wild-type and mutant leaves stained with toluidine blue (left column) or immunolabeled with monoclonal antibodies that recognize epitopes of deesterified homogalacturonan (CCRC-M38), XyG (CCRC-M88), or fucosylated XyG (CCRC-M1). WT, Wild type; EPG, endopolygalacturonase.
The Arabidopsis (Arabidopsis thaliana) genes that encode the glycosyltransferases required for the formation of the XyG side chains have been identified (see Fig. 1A). Five XYLOGLUCAN XYLOSYLTRANSFERASE (XXT1–XXT5) genes encode proteins that have been implicated in the formation of the α-d-xylopyranose-(1→6)-β-d-Glcp linkage (Cavalier et al., 2008; Zabotina et al., 2008; Vuttipongchaikij et al., 2012; Zabotina et al., 2012). Two side chain-specific galactosyltransferases have been characterized (Madson et al., 2003; Jensen et al., 2012). XYLOGLUCAN L-SIDE CHAIN GALACTOSYLTRANSFERASE2 (XLT2) catalyzes the addition of Gal to the middle Xyl (Jensen et al., 2012), whereas MURUS3 (MUR3) adds Gal to the Xyl adjacent to the unbranched Glc (Madson et al., 2003). The Gal added by MUR3 is itself the acceptor for FUCOSYLTRANSFERASE1 (FUT1), which adds a fucosyl residue to form the F side chain (Perrin et al., 1999). Up to 60% of the F side chains of Arabidopsis leaf XyG contain O-acetylated Gal (Perrin et al., 2003).
Arabidopsis fut1, xlt2, and xxt1 to xxt5 mutants that form structurally abnormal XyG (Vanzin et al., 2002; Madson et al., 2003; Perrin et al., 2003; Peña et al., 2004; Jensen et al., 2012; Schultink et al., 2013) or that have reduced (Zabotina et al., 2008; Vuttipongchaikij et al., 2012; Zabotina et al., 2012) or no discernible amounts of XyG (Cavalier et al., 2008) have no severe developmental or growth phenotypes. This has led plant scientists to question the biological role of XyG in wall assembly and architecture as well as its function in plant growth and development (Cavalier et al., 2008; Park and Cosgrove, 2012a, 2012b).
Arabidopsis mur3-1 and mur3-2 mutants, which carry different single-point mutations in MUR3, have been reported to form XyG that lacks F side chains (Madson et al., 2003; Peña et al., 2004), although they do produce discernible amounts of the MUR3 protein (Tamura et al., 2005). Both of these mutants are visibly similar to wild-type plants (Madson et al., 2003; Peña et al., 2004; Tamura et al., 2005). By contrast, mutant plants including katamari1 (kam1-1, kam1-2/mur3-7, and kam1-3/mur3-3; Tamura et al., 2005; Tedman-Jones et al., 2008), constitutive ICS1:luc expression1 (Tedman-Jones et al., 2008), and short root in salt medium (rsa3; Li et al., 2013), which are devoid of functional MUR3, have a dwarf cabbage-like growth phenotype with curled rosette leaves, short petioles, and endomembrane aggregates. Tamura et al. (2005) have proposed that the KAM1/MUR3 protein itself, rather than its galactosyltransferase activity, is required for cell elongation and normal endomembrane organization. This hypothesis is based in part on the assumption that the mur3-1, mur3-2, and mur3-3/kam1-3 mutants all lack MUR3 galactosyltransferase activity, but only the mur3-3/kam1-3 mutant lacks the MUR3 protein itself (Tamura et al., 2005).
Here, we provide genetic and chemical evidence that XyG deficient in extended side chains is dysfunctional and is responsible for the growth defects of mur3-3 and mur3-7 plants. The extent of XyG galactosylation is the major factor in determining these effects. The deleterious growth effects caused by this dysfunctional XyG are suppressed by introducing additional mutations that eliminate XyG formation altogether, by mutations that modify XyG structure, or by altering the plant's growth conditions. These observations provide new insight into XyG biosynthesis and the structural requirements that allow XyG to function normally in plant growth and development.
RESULTS AND DISCUSSION
Reducing XyG to below Detectable Levels Suppresses the Dwarf Phenotype of mur3-3 Plants
MUR3 encodes an XyG-specific galactosyltransferase responsible for most of the galactosylation of Arabidopsis XyG. To address the possibility that altered XyG structure leads to the phenotypes of Arabidopsis plants lacking functional MUR3, we took a genetic approach to reduce the amounts of XyG to undetectable levels in the mur3-3 mutant. Thus, we crossed mur3-3 with the xxt1 xxt2 double mutant, which has no detectable amounts of XyG in its cell wall (Cavalier et al., 2008; Zabotina et al., 2012). The xxt1 xxt2 mutant grows somewhat more slowly than wild-type plants but does not have a cabbage-like phenotype (Cavalier et al., 2008) or abnormal endomembrane aggregates (Uehara et al., 2014), making it visibly distinct from the mur3-3 mutant (Fig. 1C).
The xxt1 xxt2 mur3-3 triple mutant, which produces no discernible XXT1, XXT2, or MUR3 transcripts (Supplemental Fig. S1A), does not have a cabbage-like phenotype and is visibly indistinguishable from the xxt1 xxt2 mutant (Fig. 1C). Toluidine blue-stained cross sections of wild-type, xxt1 xxt2, and xxt1 xxt2 mur3-3 leaves are also visibly similar (Fig. 1E). Most of the mur3-3 cells are smaller than their wild-type counterparts, a result that is in agreement with the proposal that these cells have lost the ability to expand normally (Tamura et al., 2005). Glycome profiling (Fig. 1D) and immunocytochemistry (Fig. 1E) indicate that the XyG present in mur3-3 walls is not fucosylated, whereas wild-type walls contain fucosylated XyG. No discernible amounts of XyG were detected in xxt1 xxt2 mur3-3 leaf cell walls (Fig. 1, D and E), nor were xyloglucan oligosaccharides (XyGOs) detected after XyG-specific endoglucanase (XEG) treatment of the 1 and 4 n KOH-soluble materials from xxt1 xxt2 mur3-3 root, leaf, and stem cell walls. Thus, the presence of XyG is strongly correlated with the cabbage-like phenotype of the mur3-3 mutant. Our data suggest that the absence of the MUR3 galactosyltransferase leads to the formation of dysfunctional XyG, and that cellular processes are affected more by the presence of this dysfunctional xyloglucan than by eliminating XyG altogether.
The mur3-1, mur3-2, mur3-3, and mur3-7 Phenotypes Are Associated with Altered XyG Chemotypes
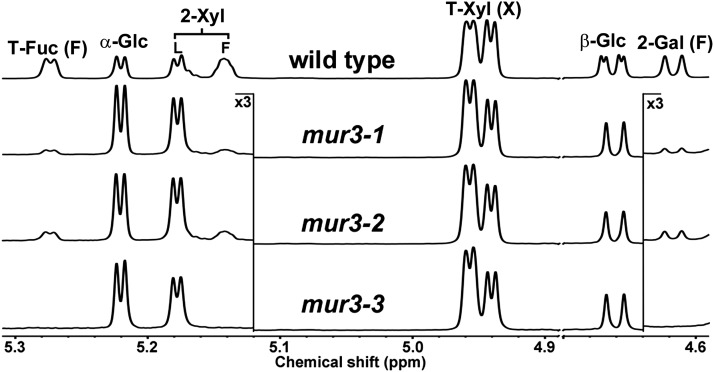
Since the presence of dysfunctional XyG is correlated with the cabbage-like phenotype of mur3-3 plants, we next determined the structural features of XyG that are associated with this phenotype. To this end, the XyGs from mur3-1, mur3-2, mur3-3, and mur3-7, each of which carries a different mutation in MUR3 (Supplemental Fig. S1B), were structurally characterized. The structures of these XyGs have not previously been compared, since only the distinguishing chemical structures of the mur3-1 XyG have been described (Madson et al., 2003; Peña et al., 2004). Here, the XyGOs generated by XEG fragmentation of leaf XyG obtained from each mutant grown at 19°C (Fig. 2) were characterized using 1H-NMR spectroscopy (Hoffman et al., 2005), matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) mass spectrometry (MS), and high-performance anion-exchange chromatography (HPAEC) with pulsed-amperometric detection (PAD; Tuomivaara et al., 2015).
Figure 2.

The phenotypes of Arabidopsis plants carrying mutations in one or more of the genes encoding the xyloglucan-specific galactosyltransferases MUR3 and XLT2. Plants were grown with a 14-h-light (19°C) and 10-h-dark (15°C) cycle or with a 14-h-light (28°C) and 10-h-dark (24°C) cycle. Images were taken when the plants were 4 weeks old.
Homozygous mur3-3 and mur3-7 plants, respectively, carry a transfer DNA insertion in the region encoding the exostosin-like domain and the protein’s N terminus (Tedman-Jones et al., 2008; Supplemental Fig. S1B). Both mutants produce no detectable MUR3 transcripts (Supplemental Fig. S1A) and have a cabbage-like phenotype when grown at 19°C in soil (Fig. 2). These mutants are visibly distinct from the mur3-1 and mur3-2 mutants (Fig. 2), both of which produce detectable amounts of the MUR3 protein (Tamura et al., 2005).
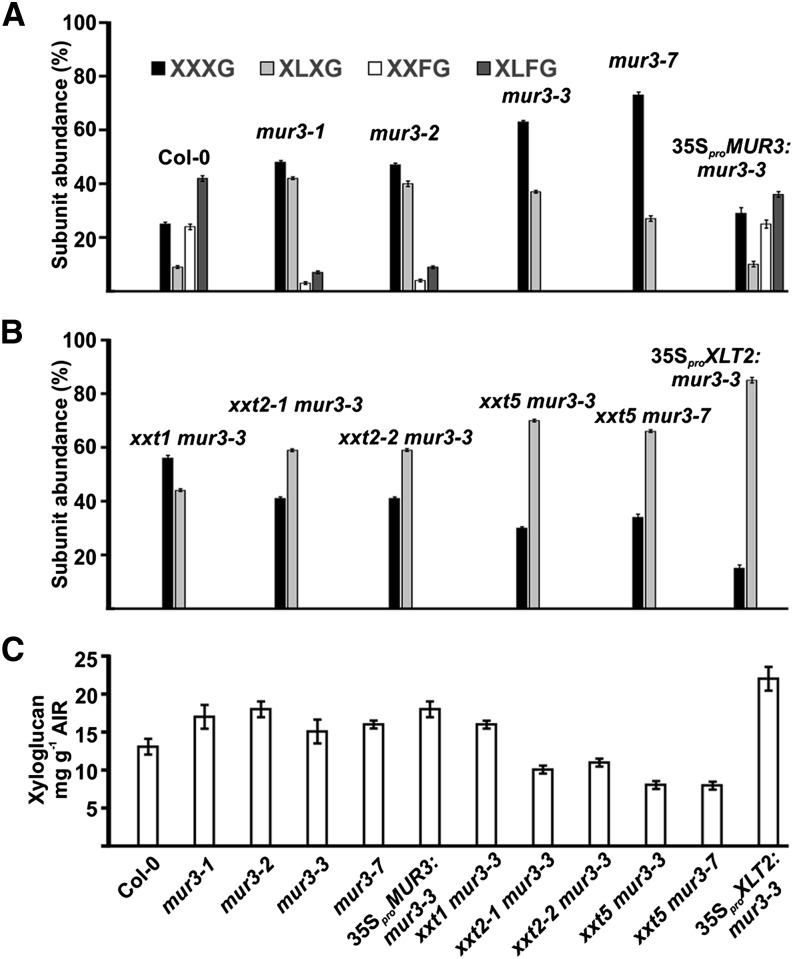
The 1H-NMR spectrum of the mur3-3 XyGOs contained no discernible signals corresponding to an F side chain at any position or to an L side chain adjacent to the unbranched Glc residue (Fig. 3; Supplemental Table S1). MALDI-TOF MS and HPAEC-PAD confirmed that the mur3-3 XyG is composed entirely of XXXG (63%) and XLXG (37%) subunits (Fig. 4A; Supplemental Fig. S2, A and B). The mur3-7 XyG is also composed of XXXG (73%) and XLXG (27%) subunits (Fig. 4A; Supplemental Fig. S3). Overexpressing MUR3 under the control of a 35S promoter in the mur3-3 mutant rescued the cabbage-like phenotype (Supplemental Fig. S4) and led to the synthesis of XyG containing near wild-type levels of XXFG and XLFG subunits (Fig. 4A; Supplemental Fig. S3). These analyses are in agreement with previous studies showing that MUR3 is a regiospecific galactosyltransferase that adds a Gal residue to the Xyl adjacent to the unbranched Glc in the XyG backbone (Madson et al., 2003; Peña et al., 2004).
Figure 3.
Partial 600-MHz 1H-NMR spectra of the XyG oligosaccharides generated by XEG treatment of the 4 n KOH-soluble XyG from wild-type (Col-0), mur3-1, mur3-2, and mur3-3 leaf cell walls. Plants were grown with a 14-h-light (19°C) and 10-h-dark (15°C) cycle. The residues corresponding to each group of proton resonances (e.g. 2-Xyl) are indicated at the top of the wild-type spectrum. The location of these residues in a side chain (X, L, or F) is indicated.
Figure 4.
The amounts and subunit compositions of the xyloglucan present in the leaf cell walls of wild-type and selected mur3 mutant plants. A and B, The subunit compositions of XyG present in wild-type and mutant leaves. XyG oligosaccharides were generated by XEG treatment of the 4 n KOH-soluble materials from at least three individual plants and analyzed by MALDI-TOF MS. HPAEC-PAD (Supplemental Fig. S2) and 1H-NMR analyses (Supplemental Table S1) indicated that XXLG and XLLG account for between 6% and 9% of the subunits in Col-0 and 35SproMUR3:mur3-3 XyG. These subunits were not detected in any of the other plants. C, The amounts of Driselase-susceptible XyG in wild-type and mutant leaves determined from the amounts of isoprimeverose released by Driselase treatment of the cell walls.
In contrast to mur3-3, the 1H-NMR spectra of the XyGOs generated from mur3-1 and mur3-2 XyG contained low-intensity signals diagnostic of F side chains (Fig. 3; Supplemental Table S1). Subunits containing F side chains were also discernible in the MALDI-TOF MS and HPAEC-PAD profiles of the mur3-1 and mur3-2 XyGOs (Fig. 4A; Supplemental Figs. S2 and S4). Moreover, the MALDI-TOF MS of the XyGO subunits generated by XEG treatment of mur3-1 and mur 3-2 cell walls contained low-intensity ions corresponding to mono O-acetylated XXFG and XLFG subunits (Supplemental Fig. S5). No O-acetylated XyG subunits were generated by XEG treatment of mur3-3 cell walls (Supplemental Fig. S5). Such results are consistent with previous studies suggesting that the F side chain Gal accounts for the bulk of the O-acetylated glycoses in Arabidopsis leaf XyG (Pauly et al., 2001; Perrin et al., 2003). Our NMR data indicate that mur3-1 and mur3-2 XyGs contain approximately 7% and 14%, respectively, of the F side chain present in wild-type plants (Supplemental Table S1) and are consistent with the results of recent studies indicating that small amounts of the F side chain are present in the xyloglucan isolated from the mur3-1 mutant (Jensen et al., 2012; Schultink et al., 2013; Vinueza et al., 2013).
Our data show that the S470L amino acid substitution in mur3-1 has a more severe effect on xyloglucan galactosylation than the A290V substitution in mur3-2. Indeed, the mur3-1 and mur3-2 mutants are not identical, since the etiolated hypocotyls of mur3-1 are approximately 50% shorter than their mur3-2 counterparts (Supplemental Fig. S6). Li et al. (2013) have reported that the roots of the mur3-1 and mur3-2 plants are somewhat less sensitive to salt stress than their rsa3-1 and kam1-3 counterparts and concluded that RSA3/MUR3/KAM1 is important in the salt stress tolerance pathway. We extend this notion by proposing that the phenotype of each mur3 mutant is determined in large part by the extent of residual xyloglucan galactosylation at the MUR3-specific position.
Xyloglucan Side Chain Extension Is Increased and the Cabbage-Like Phenotype of mur3-3 and mur3-7 Plants Is Suppressed by Overexpression of XLT2
Our results indicated that reduced extension of XyG side chains is correlated with the cabbage-like phenotype of mur3-3 and mur3-7 plants. To determine if this is the case, we overexpressed XLT2 in the mur3-3 mutant (see Supplemental Fig. S7). The 35Spro:XLT2:mur3-3 plants (36 independent transgenic lines identified) are visibly indistinguishable from their wild-type counterparts (Fig. 5) and produce XyG containing less than 15% XXXG (Fig. 4B; Supplemental Fig. S4). Comparable results were obtained when XLT2 was overexpressed in mur3-7 plants. The ability of overexpressed XLT2 to significantly decrease the XXXG content of the XyG while suppressing the cabbage-like phenotype is again consistent with the notion that this phenotype is correlated with the presence of dysfunctional XyG. Indeed, the mild dwarf phenotype of the xlt2 mur3-1 mutant is also rescued by extending the XyG side chains with arabinofuranosyl residues (Schultink et al., 2013). Such data provide further evidence that XyG deficient in extended side chains is dysfunctional, and that the synthesis of dysfunctional XyG leads to abnormal growth.
Figure 5.
The phenotypes of Arabidopsis plants carrying mutations in one or more of the genes encoding the xyloglucan-specific galactosyltransferases MUR3 or the xylosyltransferases XXT1, XXT2, and XXT5. Plants were grown with a 14-h-light (19°C) and 10-h-dark (15°C) cycle. Images were taken when the plants were 4 weeks old.
The Phenotypes of mur3-3 and mur3-7 Plants Are Suppressed by Knocking out XXT2 or XXT5
Plants that had a wild-type phenotype, yet were genotyped as XXT1 xxt2 mur3-3, were obtained during the generation of the xxt1 xxt2 mur3-3 triple mutant, which led us to suspect that abolishing the expression of specific XXT genes suppressed the cabbage-like phenotype. To obtain experimental support for this hypothesis, we generated a series of double mutants by crossing mur3-3 and mur3-7 with the xxt1, xxt2, and xxt5 mutants (see Fig. 5), which themselves have wild-type phenotypes (Cavalier et al., 2008; Vuttipongchaikij et al., 2012; Zabotina et al., 2012).
The xxt2-1 mur3-3, xxt2-2 mur3-7, xxt5 mur3-3, and xxt5 mur3-7 mutants were phenotypically comparable with wild-type plants (Fig. 4). The walls of these four double mutants contain 35% to 50% less Driselase-susceptible XyG than the walls of the xxt1 mur3-3, mur3-3, and mur3-7 mutants (Fig. 4C; Supplemental Table S2). Notably, XLXG subunits accounted for at least 59% of the XyG in the normal-looking xxt2-1 mur3-3, xxt2-2 mur3-7, xxt5 mur3-3, and xxt5 mur3-7 mutant plants (Fig. 4B; Supplemental Fig. S4). By contrast, the XLXG subunit accounts for 44% of the XyG from the xxt1 mur3-3 double mutant (Fig. 4B; Supplemental Fig. S3), which retained the cabbage-like phenotype (Fig. 5). Such data are again consistent with the notion that this phenotype is a consequence of the synthesis of XyG deficient in extended side chains.
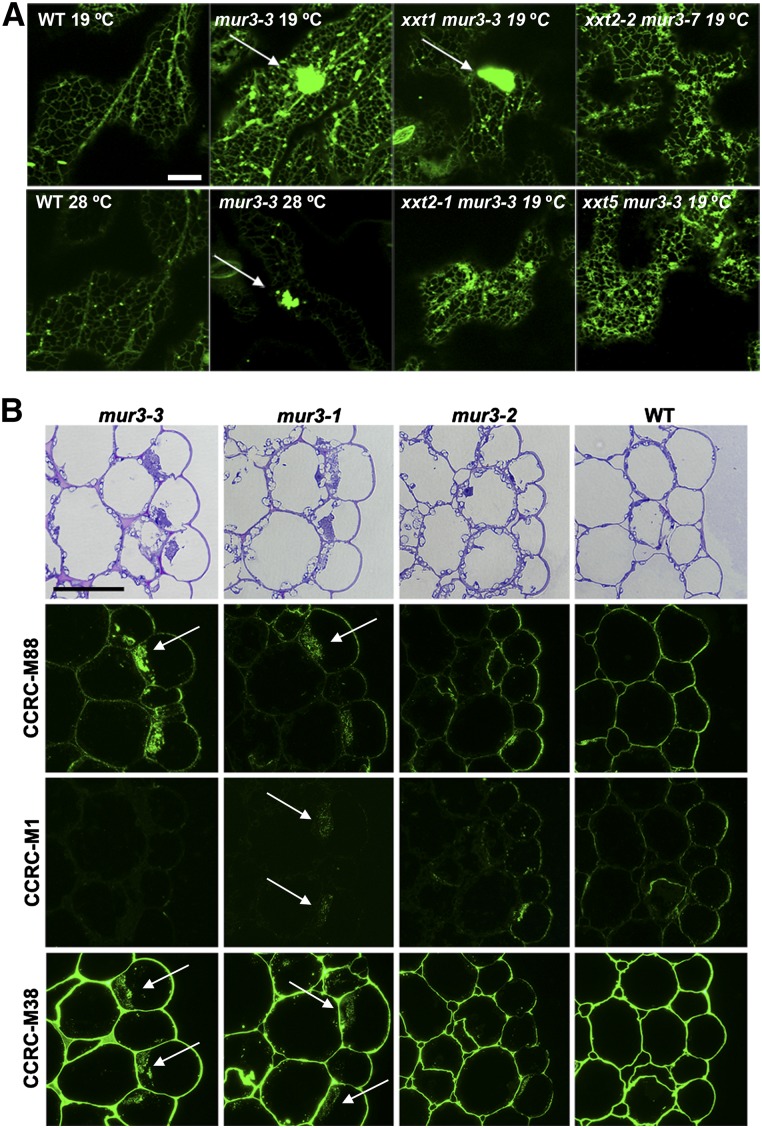
Tamura et al. (2005) have proposed that the cabbage-like phenotype of kam1/mur3 mutant plants is a consequence of abnormal endomembrane organization resulting from the absence of the MUR3 protein. However, the endomembrane aggregation phenotype observed in mur3-3 plants was largely suppressed in the xxt2-1 mur3-3 and xxt5 mur3-3 mutant plants (Fig. 6A) but not in the xxt1 mur3-3 mutant. Our data are thus consistent with the notion that XyG with a high proportion of unsubstituted XXXG subunits is dysfunctional, and that its presence, rather than the absence of a functional MUR3 protein, is strongly correlated with the presence of endomembrane aggregates. Swollen epidermal cells with endomembrane aggregates have been observed in the roots of plants lacking an isoform of UDP-Glc 4-epimerase (uge4; Uehara et al., 2014). The uge4 root XyG contains approximately 50% less Gal than wild-type XyG (Nguema-Ona et al., 2006; Rösti et al., 2007). Membrane aggregation and cell bulging were suppressed by growing uge4 in the presence of Gal, which led to the suggestion that reduced galactosylation of XyG or arabinogalactan protein is responsible for endomembrane disorganization (Uehara et al., 2014). Nevertheless, a direct causal relationship between endomembrane aggregation and plant growth is unlikely, as Arabidopsis plants carrying a mutation in the coat promoter complex II coat protein SECRETORY 24A have endomembrane aggregates comparable to mur3-3 but are not dwarfed (Faso et al., 2009).
Figure 6.
Endomembrane aggregation and intracellular accumulation of cell wall polysaccharides in mur3-3 plants. A, Endomembrane aggregation in mur3-3 plants is suppressed by knocking out XXT2 or XXT5. Col-0 (wild-type [WT]) and mutant plants were individually transformed with the SP-2SC-GFP (for signal peptide of pumpkin 2S albumin followed by a linker of GGG and the C-terminal 18-amino acid sequence of pumpkin 2S albumin) construct and grown with a 14-h-light (19°C) and 10-h-dark (15°C) cycle or a 14-h-light (28°C) and 10-h-dark (24°C) cycle. Images of epidermal cells of 14-d-old cotyledons expressing the endomembrane-targeted SP-2SC-GFP were obtained using an inverted laser scanning confocal microscope. Bar = 10 µm. B, Immunocytochemistry reveals intracellular aggregates (see arrows) containing XyG and pectin in mur3 hypocotyls. Top, Toluidine blue-stained cross sections. CCRC-M88 recognizes XyG, CCRC-M1 recognizes fucosylated XyG, and CCRC-M38 recognizes deesterified homogalacturonan. Bar = 50 µm.
Intracellular Aggregates Containing Xyloglucan and Pectin Accumulate in the Cells of mur3-3 Hypocotyls
Pectic and hemicellulosic polysaccharides are believed to be synthesized in the Golgi apparatus and are then transferred from the trans-Golgi network in vesicles that move to and fuse with the plasma membrane (Bashline et al., 2014). The polysaccharides are then released into the apoplast and incorporated into the existing cell wall. Defects in trans-Golgi network-dependent secretion of polysaccharides in the echidna mutant have been shown to result in the accumulation of intracellular aggregates containing pectin and xyloglucan (Gendre et al., 2013). Thus, we examined polysaccharide secretion in mur3-3, as its cells contain abnormal endomembrane aggregates (Fig. 6A). Intracellular aggregates were clearly visible in the epidermal cells of a toluidine blue-stained cross section of the mur3-3 hypocotyl (Fig. 6B; Supplemental Fig. S8A). Smaller amounts of aggregates were also visible in mur3-1 hypocotyls (Fig. 6B). These aggregates were labeled with monoclonal antibodies that recognize xyloglucan and deesterified homogalacturonan (Fig. 6B). Few, if any, aggregates were discernible in mur3-2, wild-type (Fig. 6B), or 35SproMUR3:mur3-3 hypocotyls (Supplemental Fig. S8B). Taken together, these data provide evidence that, in mur3-3 hypocotyls and to a lesser extent in mur3-1 hypocotyls, cell wall matrix polysaccharides, including xyloglucan and pectins, are not properly secreted and instead accumulate within intracellular aggregates. These data again support the notion that the phenotype of each mur3 mutant is determined in large part by the amount of catalytically functional MUR3 protein it produces.
Temperature Affects the Growth of mur3 Plants and the Catalytic Activity of the Mutated MUR3 Protein
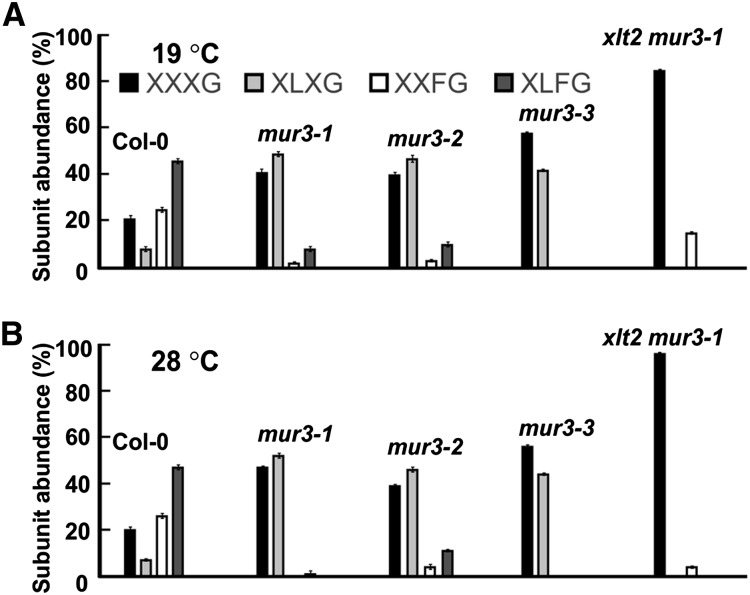
Early studies reported that mur3-1 XyG lacks F side chains (Madson et al., 2003; Peña et al., 2004), although subsequent analyses indicated that fucosylated subunits account for up to 5% of the mur3-1 and xlt2 mur3-1 XyGs (Jensen et al., 2012; Schultink et al., 2013; Vinueza et al., 2013). Nevertheless, those values are lower than the values we obtained (Fig. 4A). As our plants were grown at 19°C rather than at 22°C or 26°C as in the earlier studies, we determined whether temperature affects the extent of XyG fucosylation or plant growth.
There was an approximately 85% decrease in the relative abundance of the fucosylated subunits in the XyG from mur3-1 and xlt2 mur3-1 plants grown at 28°C (Fig. 7, A and B; Supplemental Fig. S9). xlt2 mur3-1 mutants grown at 19°C or 28°C do not have a severe cabbage-like phenotype but do have short petioles and slightly curved leaves, making them visibly distinct from wild-type plants (Fig. 2). The relative abundance of the fucosylated subunits in mur3-2 XyG was comparable at both growth temperatures (Fig. 7; Supplemental Fig. S9). These data confirm that mur3-1 and mur3-2 plants retain a residual amount of MUR3 activity and show that, in plants with the allelic (S470L) MUR3-1 protein, XyG galactosylation at the MUR3 position is diminished by increasing growth temperature.
Figure 7.
The effect of growth temperature on the xyloglucan subunit compositions of selected mur3 and wild-type plants. A, XyG subunit compositions of plants grown with a 14-h-light (19°C) and 10-h-dark (15°C) cycle. B, XyG subunit compositions of plants grown with a 14-h-light (28°C) and 10-h-dark (24°C) cycle. XyG oligosaccharides were generated by XEG treatment of the 4 n KOH-soluble materials from at least three individual plants and analyzed by MALDI-TOF MS. HPAEC-PAD analyses indicated that XXLG and XLLG accounted for approximately 6% of the subunits in Col-0 XyG (see Supplemental Fig. S9).
Somewhat surprisingly, the cabbage-like phenotype is suppressed when mur3-3 and mur3-7 plants are grown at 28°C (Fig. 2; Supplemental Fig. S10), even though the cell walls of the mutants still contain XyG, in which the XXXG subunit predominates (Fig. 7B; Supplemental Fig. S9), and the endomembrane aggregates still exist (Fig. 6A). That mur3-3 and mur3-7 exhibit near-normal growth at 28°C is consistent with the temperature-dependent changes in the rheological properties of Gal-depleted XyG (Shirakawa et al., 1998). Alternatively, the protective mechanisms activated in plants in response to heat stress (Bita and Gerats, 2013) may suppress the effects of the dysfunctional mur3-3 XyG. Nevertheless, the mechanisms by which elevated temperature suppresses the cabbage-like phenotype remain to be determined. It may well be the case that this phenotype results from an accumulation of temperature- and XyG structure-dependent modifications that affect both cell wall structure and remodeling and endomembrane organization.
Dysfunctional Xyloglucan and Plant Growth
Our data show that the extent of side chain galactosylation in mur3-3 and mur3-7 XyG is reduced by approximately 70% compared with the wild type. A probability analysis (see Supplemental Protocol S1) using the XXXG subunit abundance in the XyG from mur3-3 (63%), mur3-7 (73%), the wild type (25%), mur3-1 (48%), mur3-2 (47%), and 35SproMUR3:mur3-3 (29%) plants predicts that domains consisting of five or more tandemly repeated XXXG sequences comprise a much higher proportion of the XyG in mur3-3 and mur3-7 plants than in XyG from the other plants (Supplemental Fig. S11). The existence of large tandemly repeated XXXG domains may lead to the aggregation or otherwise aberrant interactions of XyG with itself and/or with other wall polymers (Shirakawa et al., 1998; Whitney et al., 2006; de Freitas et al., 2011). Such interactions together with the inability to extend the side chain adjacent to the unbranched Glc appear to be major factors in the dysfunctional properties of the mur3-3 and mur3-7 XyG and the cabbage-like phenotype of these plants. The aggregation of the nascent mur3-3 and mur3-7 XyGs may prevent the normal movement of XyG through the Golgi and the packaging of XyG in vesicles for delivery to the apoplast, and may also account for the abnormal membrane organization and the accumulation of intracellular aggregates containing wall polysaccharides in mur3-3 plants.
The differences in subunit composition of the mur3-3 and xlt2 mur3-1 XyGs (Fig. 7) indicate that failure to extend the side chains next to the unbranched Glc is especially detrimental to XyG function and to plant growth. Most, if not all, of the galactosyl residues adjacent to the unbranched Glc are further extended with Fuc in the XyG present in the wall of xlt2 mur3-1 plants. Although XyG fucosylation is not in itself required for the normal growth of laboratory-grown Arabidopsis plants (Vanzin et al., 2002), there is evidence that fucosylation does have a role, albeit less than galactosylation, in the formation of XyG-cellulose composites (Whitney et al., 2006).
Dysfunctional xyloglucan may disrupt wall remodeling that is required for plant cells to expand and grow (Peña et al., 2004; Park and Cosgrove, 2012a, 2012b). For example, the results of in vitro studies suggest that the decreased solubility of Gal-depleted XyG reduces its accessibility to the action of XyG endotransglucosylases/hydrolases (XTHs) that are believed to be involved in wall remodeling during cell growth (Peña et al., 2004). Li et al. (2013) have reported that the expression of XTH18 is increased 2-fold in rsa3-1. Thus, the cabbage-like phenotype may result at least in part from the reduced susceptibilities of the XyG to XTH. These authors also reported a 12-fold increase in the expression of the gene encoding the wall-associated kinase WAK1 (Li et al., 2013). Since WAKs are likely involved in wall sensing, cell expansion, and responses to pathogens and environmental signals (Kohorn and Kohorn, 2012), they may have a role in communicating changes in wall properties to the cells of the mur3 mutants.
Our data favor a model that invokes a dysfunctional xyloglucan structure as the most direct cause of the growth defects of plants carrying loss-of-function mutations in MUR3. Nevertheless, the possibility cannot be discounted that other cellular mechanisms are responsible. For example, the phenotypes observed in plants lacking the MUR3 protein may be a consequence of an XyG biosynthesis complex (Chou et al., 2015; Lund et al., 2015) that becomes dysfunctional when no MUR3 protein is present. In this scenario, the mur3-1 and mur3-2 mutants are not cabbage like because the residual MUR3 may still be capable of forming a nondysfunctional XyG biosynthetic complex. However, to be consistent with our data, XLT2, which has 42% amino acid sequence identity with MUR3, must be able to substitute for MUR3 to form a nondysfunctional complex when XLT2 is expressed at sufficiently high levels. XLT2 does not replace MUR3 when expressed at normal levels. This model also requires that the direct effects of the putative dysfunctional biosynthetic complex are suppressed by eliminating the XXT2 or the XXT5 protein in mur3-3 or mur3-7 plants. The xxt2 mur3-3 and xxt5 mur3-3 double mutants have near wild-type morphology and produce xyloglucan with less than 50% unsubstituted XXXG. Models that invoke a dysfunctional biosynthetic complex as the direct cause of the mur3-3 phenotypes are thus complicated by several difficult-to-reconcile requirements.
CONCLUSION
We have shown that Arabidopsis mutants devoid of the F side chain galactosyltransferase MURUS3 form xyloglucan that lacks F side chains and contains much less galactosylated Xyl than wild-type xyloglucan. This xyloglucan structure is dysfunctional, as it leads to mutants that exhibit numerous growth defects. Near-normal growth is restored by eliminating the xyloglucan altogether or by increasing the extent of its galactosylation. The altered growth of the mur3 family of mutants and their responses to temperature, pathogens (Tedman-Jones et al., 2008), and salt stress (Li et al., 2013) reveal how modifying one element of a biochemical network, in this case, xyloglucan biosynthesis, may disturb diverse cellular and physiological processes. Such data provide additional evidence in support of the notion that altering wall structure affects numerous aspects of plant life (Seifert and Blaukopf, 2010). Thus, the mur3 mutants provide a unique opportunity to increase our understanding of how changes in cell wall biosynthesis and structure affect plant growth and development as well as disease resistance and responses to environmental stimuli.
MATERIALS AND METHODS
All primers used in this study are listed in Supplemental Table S3.
Plant Lines
Arabidopsis (Arabidopsis thaliana) plants were in the Columbia (Col-0) background. Col-0, mur3-3 (At2g20370; Salk_141953), mur3-7 (SALK_127057), xxt1 (At3g62720; SAIL_785_E02), xxt2-1 (At4g02500; SALK_101308), and xxt2-2 (SALK_150920) were obtained from the Arabidopsis Biological Resource Center (www.arabidopsis.org). Homozygous plants were identified by PCR. mur3-1 and mur3-2 were from a mutant collection described previously (Reiter et al., 1997). xxt5 (Zabotina et al., 2008) and xxt1 xxt2 (Cavalier et al., 2008) mutant seeds were obtained from Olga Zabotina (Iowa State University, Ames) and Kenneth Keegstra (Michigan State University, East Lansing), respectively. xlt2 mur3-1 mutant (Jensen et al., 2012) seeds were obtained from Marcus Pauly (University of California, Berkeley).
Plant Growth
Plants were grown in controlled-environmental growth chambers (Adaptis 2000, Conviron). Seeds were germinated and grown on Fafard 3B potting soil at 19°C and 15°C with a 14-h-light and 10-h-dark cycle, respectively, with a light intensity of 120 µmol quanta m−2 s−1 and 70% relative humidity. In a second series of experiments, wild-type and mutant seeds were germinated and grown for 10 d on Fafard 3B potting soil at 19°C and 15°C with a 14-h-light and 10-h-dark cycle, respectively. One-half of the plants was then transferred to a second controlled environmental growth cabinet operated at 28°C and 24°C with a 14-h-light and 10-h-dark cycle, respectively. Images of plants were obtained using a Panasonic Lumix DMC-FZ28 digital camera. All image manipulations were performed using Adobe Photoshop CS4 software.
Double and Triple Mutant Generation
The xxt1 xxt2-1 double mutant was crossed with the mur3-3 mutant. Segregating progenies homozygous for xxt1 mur3-3, xxt2-1 mur3-3, and xxt1 xxt2 mur3-3 were identified in the subsequent F2 and F3 generations. Double mutants were generated by crossing mur3-3 or mur3-7 with xxt1, xxt2-1, xxt2-2, or xxt5. Allele-specific primers for each line were used to identify homozygous mutant alleles by PCR.
RT-PCR
Total RNA was isolated from the leaves of wild-type and mutant plants using an RNeasy plant mini kit with an on-column DNase I treatment to remove contaminating genomic DNA, according to the manufacturer's instructions (Qiagen). One microgram of total RNA was reverse transcribed using Superscript III Reverse Transcriptase (Invitrogen), and one-twentieth of the first-strand complementary DNA was used for PCR amplification with gene-specific primers. Actin-2 was used as an internal control with primers to amplify a complementary DNA fragment.
Plasmid Construction and Plant Transformation
The full-length XLT2 open reading frame was PCR amplified with primers XLT2-OE F and XLT2-OE R and then inserted downstream of the cauliflower mosaic virus 35S promoter in the pCam35tl:egfps2#4 vector (Pattathil et al., 2005). The construct was transformed into Agrobacterium tumefaciens strain GV3101, which was then used to transform the mur3-3 mutant using a floral dip procedure (Clough and Bent, 1998). Transgenic plants were selected by growth on one-half-strength Murashige and Skoog medium containing hygromycin (15 mg L−1). The presence of the transgene was confirmed by PCR using the gene-specific primer XLT2 F paired with vector-specific primer P1300 R.
Preparation of the Alcohol-Insoluble Residues and Solubilization of Xyloglucan
Cell walls from the aerial portions of wild-type and mutant plants were prepared as their alcohol-insoluble residues (AIRs). The tissues were homogenized in aqueous 80% (v/v) ethanol using a Polytron blender (Kinematica). The suspension was centrifuged and the insoluble residue washed with absolute ethanol and acetone and then vacuum dried at room temperature. The AIR (0.5–1.0 g) was then treated sequentially for 16 h at room temperature with 50 mm ammonium oxalate (50 mL), 1 n KOH containing 0.5% (w/v) sodium borohydride, and then with 4 n KOH containing 0.5% (w/v) sodium borohydride. The 4 n KOH extract, which contains most of the xyloglucan, was adjusted to pH 5 with glacial acetic acid, dialyzed against several changes of deionized water, and freeze dried.
Structural Characterization of Xyloglucan Oligosaccharides
Xyloglucan oligosaccharides were generated by treating suspensions of the AIR and solutions of the 4 n KOH-soluble materials in 50 mm ammonium formate, pH 5, with an XEG as described (Pauly et al., 1999). Ethanol was added to 70% (v/v) and the mixture kept for 24 h at 4°C. The precipitate that formed was removed by centrifugation and the soluble fraction concentrated to dryness. The residue was dissolved in 500 μL of water and then freeze dried to remove residual ammonium formate. Positive-ion MALDI-TOF mass spectra were recorded using a Bruker Microflex LT mass spectrometer and workstation. Solutions (5 μL) of the oligosaccharides (approximately 1 mg mL−1 in water) were mixed with an equal volume of 10 mm NaCl. A portion of this mixture (1 μL) was then added to a matrix solution (0.1 m 2,5-dihydroxybenzoic acid in aqueous 50% [v/v] acetonitrile, 1 μL) on the stainless steel MALDI target plate and concentrated to dryness using a flow of warm air from a hair dryer. Spectra from at least 200 laser shots were summed up to generate each mass spectrum.
1H-NMR spectra were recorded with a Varian Inova NMR spectrometer (Agilent Technologies) operating at 600 MHz using a 3-mm cold probe and a sample temperature of 25°C. XyGOs (0.5–1 mg) were dissolved in D2O (0.7 mL, 99.9% [v/v]; Cambridge Isotope Laboratories). All two-dimensional spectra were recorded using standard Varian pulse programs. Chemical shifts were measured relative to internal acetone (δ 2.225). Data were processed using MestReNova software (Universidad de Santiago de Compostela).
Xyloglucan Quantification
The amounts of xyloglucan present in the AIR were determined by measuring the amounts of isoprimeverose released by Driselase treatment of the AIR (Fry, 1982). Suspensions of the AIR (approximately 1 g) from mutant and wild-type plants in aqueous 80% (v/v) ethanol (100 mL) were ball milled for 16 h at 4°C and 90 rpm in ceramic milling jars (0.3 L) containing a mixture of one-fourth- and one-half-inch zirconia milling beads (U.S. Stoneware). The milled AIR was collected by centrifugation, washed with absolute ethanol and then with acetone, and then vacuum dried at room temperature. Suspensions of the milled AIR (250–500 mg) in 50 mm sodium acetate, pH 5.2 (30 mL), were then treated for 24 h at 45°C with Spirizyme (30 μL, Novozymes A/S) and Liquizyme (150 μL, Novozymes A/S) to remove starch. The destarched AIR was collected by centrifugation, washed thoroughly with deionized water, and freeze dried.
Triplicate suspensions of the AIR (6.25 mg) in 50 mm ammonium formate, pH 4.7 (500 μL), containing 0.5% (w/v) chlorobutanol were treated for 24 h at 22°C with 0.25% (w/v) Driselase (100 μL, Sigma-Aldrich) prepared as described (Fry, 1982). A second portion of Driselase (50 μL) was added and the suspension kept for a further 24 h at 22°C. Ethanol (1.5 mL) was added and the mixture kept at 4°C overnight. The suspension was centrifuged and the soluble fraction concentrated to dryness. The residue was dissolved in water (1 mL) and freeze dried to remove residual ammonium formate. The residue was dissolved in water (1 mL), and a portion (250 μL) was filtered using a 0.45-μm centrifugal microfilter. Portions (10 μL) of the soluble material were then analyzed using a Dionex ICS-3000 HPAEC system with pulsed amperometric detection and a CarboPac PA1 column (250 × 4.6 mm, Thermo Fisher). The column was eluted at 1.0 mL min−1 for 5 min with 0.1 n NaOH, then with a gradient of sodium acetate (0–100 mm) in 0.1 n NaOH for 30 min. The column was washed for 15 min with 1 m sodium acetate in 0.1 n NaOH and then reequilibrated for 15 min in 0.1 n NaOH. The chromatography system was calibrated using known amounts of isoprimeverose (Megazyme).
Xyloglucan Subunit Composition Analysis Using HPAEC with Pulsed Amperometric Detection
Solutions (10 μL) of XyGOs in water (approximately 200 μg mL−1) released by XEG treatment of AIR or the 4 n KOH-soluble materials were analyzed using a Dionex ICS-3000 system and CarboPac PA1 column (250 × 4.6 mm; Thermo Fisher). The column was eluted at 1.0 mL min−1 for 3 min with 0.1 n NaOH, from 3 to 5 min with a gradient of sodium acetate (0–40 mm) in 0.1 n NaOH and from 5 to 38 min with a gradient of sodium acetate (40–100 mm) in 0.1 n NaOH. The column was washed with 1 m sodium acetate in 0.1 n NaOH for 7 min and reequilibrated for 15 min in 0.1 n NaOH. The column was calibrated using known amounts (0.5–2 μg injected) of homogeneous XXXG, XXLG, XLXG, XXFG, XLLG, and XLFG purified from XEG digests of tamarind and sycamore cell wall xyloglucan (Tuomivaara et al., 2015).
Glycome Profiling of Plant Cell Wall Extracts
AIR generated from wild-type and mutant plants (approximately 500 mg) was treated sequentially with α-amylase, endopolygalacturonase, 50 mm sodium carbonate, 1 n KOH, 4 n KOH, and 5 n KOH containing 3% (w/v) boric acid to generate fractions enriched in pectic and hemicellulosic polysaccharides. The soluble extracts were adjusted to pH 5 if required, dialyzed (3,500 Mr cutoff) against deionized water, and freeze dried. Solutions (50 µL) of each wall extract (60 µg total sugar mL−1 water) were then used for enzyme-linked immunosorbent assays using monoclonal antibodies that recognize xyloglucan epitopes (see Supplemental Table S4) as described (Pattathil et al., 2010).
Confocal Imaging of Endomembrane Organization
Col-0, mur3-3, xxt1 mur3-3, xxt2-1 mur3-3, and xxt5 mur3-3 plants were individually transformed with the SP-2SC-GFP construct (Mitsuhashi et al., 2000; Tamura et al., 2005). Transgenic plants were obtained by screening the seeds on kanamycin plates. Epidermal cells of 14-d-old cotyledons were analyzed using an inverted laser scanning confocal microscope (LSM510 META, Zeiss) as described (Faso et al., 2009).
Light Microscopy
Leaves from 4-week-old soil-grown Col-0, mur3-3, xxt1 xxt2, and xxt1 xxt2 mur3-3 plants and hypocotyls from Col-0, mur3-1, mur3-2, mur3-3, and mur3-3:35SproMUR3 seedlings grown for 4 d under constant light at 22°C were fixed for 3 h in 25 mm sodium phosphate, pH 7.1, containing 2.5% (v/v) glutaraldehyde. The fixed leaves and hypocotyls were processed for light microscopy as described previously (Avci et al., 2012).
Immunocytochemistry
Hypocotyl cross sections approximately 3 to 4 mm away from the shoot meristem and leaf cross sections (250 nm thick) approximately 3 to 4 mm away from the leaf tip were mounted on ColorFrost Plus glass slides (Thermo Fisher Scientific) and blocked for 30 min with 10 mm potassium phosphate, pH 7.1, containing 0.5 m NaCl and 3% (w/v) nonfat dry milk. Hypocotyl and leaf cross sections were processed for immunolabeling with monoclonal antibodies CCRC-M1, CCRC-M38, or CCRC-M88 as described (Avci et al., 2012).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RT-PCR analysis of gene transcripts in the mutant lines.
Supplemental Figure S2. The MALDI-TOF mass spectra and HPAEC-PAD profiles of the XyGOs generated by XEG treatment of the 4 n KOH-soluble materials from Col-0, mur3-1, mur3-2, and mur3-3 mutant plants.
Supplemental Figure S3. The MALDI-TOF mass spectra and HPAEC-PAD profiles of the XyGOs generated by XEG treatment of the 4 n KOH-soluble materials from Col-0, mur3, xxt, and mur3 xxt mutant plants.
Supplemental Figure S4. The phenotypes and MALDI-TOF mass spectra of the XyGOs generated by XEG treatment of the 4 n KOH-soluble materials from individual 35SproMUR3:mur3, mur3-1, and mur3-2 plants.
Supplemental Figure S5. The MALDI-TOF mass spectra of the XyGOs released by XEG treatment of Col-0, mur3-1, mur3-2, and mur3-3 AIR.
Supplemental Figure S6. The effect of the absence of light on the hypocotyl length of seedlings carrying single or combined mutations in genes encoding xylosyltransferases XXT1, XXT2, and XXT5 and galactosyltransferases MUR3 and XLT2.
Supplemental Figure S7. RT-PCR analyses of XLT2 transcripts in the rosette leaves of mur3-3:35Spro:XLT2 plants.
Supplemental Figure S8. Intracellular accumulation of cell wall polysaccharides in mur3 hypocotyls.
Supplemental Figure S9. The MALDI-TOF mass spectra and HPAEC-PAD profiles of the XyGOs generated by XEG treatment of the 4 n KOH-soluble materials from Col-0 and selected mutant plants grown at 19°C and 28°C.
Supplemental Figure S10. The effects of temperature on the growth and development of mur3-3 plants.
Supplemental Figure S11. Abundance of tandemly repeated XXXG domains in the xyloglucan from transgenic plants.
Supplemental Table S1. Side chain compositions of the xyloglucans isolated from the cell walls of Arabidopsis wild-type, mur3-1, mur3-2, and mur3-3 plants.
Supplemental Table S2. The amounts and subunit compositions of the xyloglucan present in the leaf cell walls of wild-type and selected xxt Arabidopsis mutants.
Supplemental Table S3. Primers used in this study.
Supplemental Table S4. Monoclonal antibodies used for glycome profiling.
Supplemental Protocol S1. Calculating the fraction of a polysaccharide that is composed of domains containing n or more tandemly repeated copies of a sequence.
Supplementary Material
Acknowledgments
We thank Ikuko Hara-Nishimura (Kyoto University) for providing the 2sc-GFP construct and Maor Bar-Peled (Complex Carbohydrate Research Center) for comments on the manuscript.
Glossary
- XyG
XXXG-type xyloglucan
- XyGO
xyloglucan oligosaccharide
- XEG
XXXG-type xyloglucan-specific endoglucanase
- MALDI-TOF
matrix-assisted laser-desorption ionization time of flight
- MS
mass spectrometry
- HPAEC
high-performance anion-exchange chromatography
- PAD
pulsed-amperometric detection
- Col-0
Columbia
- AIR
alcohol-insoluble residue
- RT
reverse transcription
Footnotes
This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, U.S. Department of Energy (grant nos. DE–FG02–96ER20220 to A.G.D., M.J.P., M.G.H., M.A.O., and W.S.Y.; DE–FG02–93ER20097 to A.G.D.; DE–FG02–12ER16324 to W.S.Y., M.A.O., and M.J.P.; and DE–FG02–91ER20021 to F.B.); the U.S. Department of Energy Great Lakes Bioenergy Research Center (grant nos. BER DE–FC02–07ER64494 to F.B. and DE–FG02–08ER20203 to W.-D.R.); and the National Science Foundation (Plant Genome grant no. ISO–0923992 to M.G.H.).
References
- Avci U, Pattathil S, Hahn MG (2012) Immunological approaches to plant cell wall and biomass characterization: immunolocalization of glycan epitopes. InHimmell ME, ed, Biomass Conversion: Methods and Protocols, Vol 908 Humana Press, New York, pp 73-82 [DOI] [PubMed] [Google Scholar]
- Bashline L, Lei L, Li S, Gu Y (2014) Cell wall, cytoskeleton, and cell expansion in higher plants. Mol Plant 7: 586–600 [DOI] [PubMed] [Google Scholar]
- Bita CE, Gerats T (2013) Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci 4: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M. , et al (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YH, Pogorelko G, Young ZT, Zabotina OA (2015) Protein–protein interactions among xyloglucan-synthesizing enzymes and formation of Golgi-localized multiprotein complexes. Plant Cell Physiol 56: 255–267 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Freitas RA, Busato AP, Mitchell DA, Silveira JL (2011) Degalactosylation of xyloglucan: effect on aggregation and conformation, as determined by time dependent static light scattering, HPSEC–MALLS and viscosimetry. Carbohydr Polym 83: 1636–1642 [Google Scholar]
- Faso C, Chen YN, Tamura K, Held M, Zemelis S, Marti L, Saravanan R, Hummel E, Kung L, Miller E. , et al (2009) A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. Plant Cell 21: 3655–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. (1982) Phenolic components of the primary cell wall: feruloylated disaccharides of D-galactose and L-arabinose from spinach polysaccharide. Biochem J 203: 493–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre D, McFarlane HE, Johnson E, Mouille G, Sjödin A, Oh J, Levesque-Tremblay G, Watanabe Y, Samuels L, Bhalerao RP (2013) Trans-Golgi network localized ECHIDNA/Ypt interacting protein complex is required for the secretion of cell wall polysaccharides in Arabidopsis. Plant Cell 25: 2633–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M, Jia Z, Peña MJ, Cash M, Harper A, Blackburn AR II, Darvill A, York WS (2005) Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res 340: 1826–1840 [DOI] [PubMed] [Google Scholar]
- Hsieh YSY, Harris PJ (2012) Structures of xyloglucans in primary cell walls of gymnosperms, monilophytes (ferns sensu lato) and lycophytes. Phytochemistry 79: 87–101 [DOI] [PubMed] [Google Scholar]
- Jensen JK, Schultink A, Keegstra K, Wilkerson CG, Pauly M (2012) RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol Plant 5: 984–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Kohorn SL (2012) The cell wall-associated kinases, WAKs, as pectin receptors. Front Plant Sci 3: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Guan Q, Wang ZY, Wang Y, Zhu J (2013) A bi-functional xyloglucan galactosyltransferase is an indispensable salt stress tolerance determinant in Arabidopsis. Mol Plant 6: 1344–1354 [DOI] [PubMed] [Google Scholar]
- Lund CH, Bromley JR, Stenbæk A, Rasmussen RE, Scheller HV, Sakuragi Y (2015) A reversible Renilla luciferase protein complementation assay for rapid identification of protein-protein interactions reveals the existence of an interaction network involved in xyloglucan biosynthesis in the plant Golgi apparatus. J Exp Bot 66: 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter WD (2003) The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15: 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi N, Shimada T, Mano S, Nishimura M, Hara-Nishimura I (2000) Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol 41: 993–1001 [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E, Andème-Onzighi C, Aboughe-Angone S, Bardor M, Ishii T, Lerouge P, Driouich A (2006) The reb1-1 mutation of Arabidopsis: effect on the structure and localization of galactose-containing cell wall polysaccharides. Plant Physiol 140: 1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2012a) Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol 158: 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ (2012b) A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol 158: 1933–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. (2010) A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Harper AD, Bar-Peled M (2005) Biosynthesis of UDP-xylose: characterization of membrane-bound AtUxs2. Planta 221: 538–548 [DOI] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kauppinen S, Kofod LV, York WS, Albersheim P, Darvill A (1999) A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Pauly M, Eberhard S, Albersheim P, Darvill A, York WS (2001) Effects of the mur1 mutation on xyloglucans produced by suspension-cultured Arabidopsis thaliana cells. Planta 214: 67–74 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Darvill AG, Eberhard S, York WS, O’Neill MA (2008) Moss and liverwort xyloglucans contain galacturonic acid and are structurally distinct from the xyloglucans synthesized by hornworts and vascular plants. Glycobiology 18: 891–904 [DOI] [PubMed] [Google Scholar]
- Peña MJ, Ryden P, Madson M, Smith AC, Carpita NC (2004) The galactose residues of xyloglucan are essential to maintain mechanical strength of the primary cell walls in Arabidopsis during growth. Plant Physiol 134: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979 [DOI] [PubMed] [Google Scholar]
- Perrin RM, Jia Z, Wagner TA, O’Neill MA, Sarria R, York WS, Raikhel NV, Keegstra K (2003) Analysis of xyloglucan fucosylation in Arabidopsis. Plant Physiol 132: 768–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter WD, Chapple C, Somerville CR (1997) Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J 12: 335–345 [DOI] [PubMed] [Google Scholar]
- Rösti J, Barton CJ, Albrecht S, Dupree P, Pauly M, Findlay K, Roberts K, Seifert GJ (2007) UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana. Plant Cell 19: 1565–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT. (2013) Mutagenic dissection of the sequence determinants of protein folding, recognition, and machine function. Protein Sci 22: 1675–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultink A, Cheng K, Park YB, Cosgrove DJ, Pauly M (2013) The identification of two arabinosyltransferases from tomato reveals functional equivalency of xyloglucan side chain substituents. Plant Physiol 163: 86–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Blaukopf C (2010) Irritable walls: the plant extracellular matrix and signaling. Plant Physiol 153: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa M, Yamatoya K, Nishinari K (1998) Tailoring of xyloglucan properties using an enzyme. Food Hydrocoll 12: 25–28 [Google Scholar]
- Tamura K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I (2005) KATAMARI1/MURUS3 Is a novel golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedman-Jones JD, Lei R, Jay F, Fabro G, Li X, Reiter WD, Brearley C, Jones JDG (2008) Characterization of Arabidopsis mur3 mutations that result in constitutive activation of defence in petioles, but not leaves. Plant J 56: 691–703 [DOI] [PubMed] [Google Scholar]
- Tuomivaara ST, Yaoi K, O’Neill MA, York WS (2015) Generation and structural validation of a library of diverse xyloglucan-derived oligosaccharides, including an update on xyloglucan nomenclature. Carbohydr Res 402: 56–66 [DOI] [PubMed] [Google Scholar]
- Uehara M, Wang S, Kamiya T, Shigenobu S, Yamaguchi K, Fujiwara T, Naito S, Takano J (2014) Identification and characterization of an Arabidopsis mutant with altered localization of NIP5;1, a plasma membrane boric acid channel, reveals the requirement for D-galactose in endomembrane organization. Plant Cell Physiol 55: 704–714 [DOI] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99: 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinueza NR, Gallardo VA, Klimek JF, Carpita NC, Kenttämaa HI (2013) Analysis of xyloglucans by ambient chloride attachment ionization tandem mass spectrometry. Carbohydr Polym 98: 1203–1213 [DOI] [PubMed] [Google Scholar]
- Vuttipongchaikij S, Brocklehurst D, Steele-King C, Ashford DA, Gomez LD, McQueen-Mason SJ (2012) Arabidopsis GT34 family contains five xyloglucan α-1,6-xylosyltransferases. New Phytol 195: 585–595 [DOI] [PubMed] [Google Scholar]
- Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY (2011) Understanding the transcriptome through RNA structure. Nat Rev Genet 12: 641–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SEC, Wilson E, Webster J, Bacic A, Reid JSG, Gidley MJ (2006) Effects of structural variation in xyloglucan polymers on interactions with bacterial cellulose. Am J Bot 93: 1402–1414 [DOI] [PubMed] [Google Scholar]
- Zabotina OA, Avci U, Cavalier D, Pattathil S, Chou YH, Eberhard S, Danhof L, Keegstra K, Hahn MG (2012) Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol 159: 1367–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, van de Ven WTG, Freshour G, Drakakaki G, Cavalier D, Mouille G, Hahn MG, Keegstra K, Raikhel NV (2008) Arabidopsis XXT5 gene encodes a putative α-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J 56: 101–115 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.