Efficient light to heat dissipation in Chlamydomonas reinhardtii requires binding of a stress-related light-harvesting complex protein to photosystem II and associated light-harvesting complexes II.
Abstract
In Chlamydomonas reinhardtii, the LIGHT-HARVESTING COMPLEX STRESS-RELATED PROTEIN3 (LHCSR3) protein is crucial for efficient energy-dependent thermal dissipation of excess absorbed light energy and functionally associates with photosystem II-light-harvesting complex II (PSII-LHCII) supercomplexes. Currently, it is unknown how LHCSR3 binds to the PSII-LHCII supercomplex. In this study, we investigated the role of PHOTOSYSTEM II SUBUNIT R (PSBR) an intrinsic membrane-spanning PSII subunit, in the binding of LHCSR3 to PSII-LHCII supercomplexes. Down-regulation of PSBR expression diminished the efficiency of oxygen evolution and the extent of nonphotochemical quenching and had an impact on the stability of the oxygen-evolving complex as well as on PSII-LHCII-LHCSR3 supercomplex formation. Its down-regulation destabilized the PSII-LHCII supercomplex and strongly reduced the binding of LHCSR3 to PSII-LHCII supercomplexes, as revealed by quantitative proteomics. PHOTOSYSTEM II SUBUNIT P deletion, on the contrary, destabilized PHOTOSYSTEM II SUBUNIT Q binding but did not affect PSBR and LHCSR3 association with PSII-LHCII. In summary, these data provide clear evidence that PSBR is required for the stable binding of LHCSR3 to PSII-LHCII supercomplexes and is essential for efficient energy-dependent quenching and the integrity of the PSII-LHCII-LHCSR3 supercomplex under continuous high light.
Plant photosynthetic electron transfer is conducted by a series of reactions at the chloroplast thylakoid membrane, resulting in light-dependent water oxidation, NADP reduction, and ATP formation (Whatley et al., 1963). Two separate photosystems (PSI and PSII) and an ATP synthase catalyze these reactions. PSI and PSII are multiprotein complexes that are mainly embedded in unstacked and stacked regions of the thylakoid membrane, respectively. PSI consists of more than 10 subunits and a number of cofactors such as chlorophyll a, β-carotene, phylloquinone, and three iron-sulfur (4Fe-4S) clusters (Busch and Hippler, 2011). PSI catalyzes light-driven electron transfer from luminal plastocyanin to stromal ferredoxin. The latter reduces the ferredoxin-NADP reductase that, in turn, leads to the formation of NADPH. PSII catalyzes light-induced electron transfer from water to the plastoquinone pool by using chlorophyll a, carotenoids, as well as redox-active cofactors, causing the release of oxygen and protons (Pagliano et al., 2013). The core complex is organized as a dimer. Monomers are composed of the reaction center subunits PSBA (D1) and PSBD (D2), the inner antenna proteins PSBB (CP47) and PSBC (CP43), the α- and β-subunits (PSBE and PSBF) of cytochrome b559, as well as a number of intrinsic low-molecular-mass subunits. The core monomer is further associated with an inorganic Mn4O5Ca cluster and a few chloride ions (Rivalta et al., 2011; Umena et al., 2011) required for photosynthetic water oxidation. To optimize this process, the oxygen-evolving complex is formed at the luminal side by the extrinsic polypeptides PSBO, PSBP, PSBQ, and PSBR (for review, see Pagliano et al., 2013).
To enhance the light-harvesting capacity of PSII, various light-harvesting proteins bind to dimeric PSII core complexes (Dekker and Boekema, 2005). A common structure found for vascular plants and green algae is the C2S2 supercomplex, where two copies of monomeric Lhcb4 and Lhcb5 and two LHCII trimers (S-trimer; Boekema et al., 1995) bind to the dimeric PSII core. In vascular plants, larger but less stable PSII supercomplexes, known as C2S2M2, are composed of two extra copies of the monomeric Lhcb6 with two additional LHCII trimers (M-trimer) bound through Lhcb4 and Lhcb5 (Dekker and Boekema, 2005; Caffarri et al., 2009). Even larger complexes containing two additional LHCII trimers (L-trimer), bound via Lhcb6, are found and are known as C2S2M2L1–2 (Boekema et al., 1999). A recent study in Chlamydomonas reinhardtii identified PSII-LHCII supercomplexes with three LHCII trimers attached to each side of the core (C2S2M2L2; Tokutsu et al., 2012). Interestingly, such PSII-LHCII supercomplexes associate with LIGHT-HARVESTING COMPLEX STRESS-RELATED PROTEIN3 (LHCSR3; Tokutsu and Minagawa, 2013), an ancient light-harvesting protein required for efficient energy-dependent (qE) quenching in the alga (Peers et al., 2009). The qE component of nonphotochemical quenching (NPQ) is an energy-dependent constituent of NPQ and regulates the thermal dissipation of excess absorbed light energy (Li et al., 2000; Peers et al., 2009). The qE capacity in C. reinhardtii increases proportionally to the light-dependent accumulation of the LHCSR3 protein (Peers et al., 2009). In contrast, in vascular plants, qE is constitutively active and dependent on PSBS, a PSII polypeptide (Li et al., 2000). Mass spectrometric analyses of isolated C2S2M2 PSII supercomplexes revealed the presence of extrinsic subunits PSBP, PSBQ, and PSBR, while PSBS was not identified, suggesting that PSBS does not influence the association of the PSII core with the outer light-harvesting complex system (Pagliano et al., 2014). In line with the proteomic findings, recent data suggest that subunits PSBP, PSBQ, and PSBR contribute to the stability of PSII-LHCII supercomplexes in vascular plants (Caffarri et al., 2009; Ifuku et al., 2011; Allahverdiyeva et al., 2013). A recent quantitative proteomic study performed with C. reinhardtii identified PSBR as the only PSII subunit to be induced upon the shift from photoheterotrophic to photoautotrophic growth conditions similar to LHCSR3 (Höhner et al., 2013).
In vascular plants and green algae, PSBR is nucleus encoded and has a mass of about 10 kD. The mature protein has a predicted 70-amino acid luminal N-terminal part and a C-terminal transmembrane span (Ljungberg et al., 1986; Lautner et al., 1988; Webber et al., 1989). An association of PSBR with the oxygen-evolving complex has been suggested, as its presence is required for the stable assembly of PSBP with the PSII core and its absence also impacts the binding of PSBQ to the core (Suorsa et al., 2006; Liu et al., 2009). For stable association with the PSII core complex, PSBR needs the presence of PSBJ (Suorsa et al., 2006). Functionally, the depletion of PSBR protein expression decreased rates of oxygen evolution (Allahverdiyeva et al., 2007, 2013) and quinone reoxidation (Allahverdiyeva et al., 2007). PSBR phosphorylation is known for Arabidopsis (Arabidopsis thaliana; Reiland et al., 2009, 2011; Nakagami et al., 2010) and in the green alga C. reinhardtii (Turkina et al., 2006), although phosphorylation sites are not conserved between the alga and the vascular plant.
In this work, we addressed the question of whether down-regulation of PSBR expression would affect LHCSR3 binding to the PSII-LHCII supercomplex in C. reinhardtii. To this end, we took advantage of artificial microRNA (amiRNA) technology to down-regulate PSBR expression and investigated the impact of PSBR down-regulation on photosynthetic performance as well as on PSII-LHCII-LHCSR3 supercomplex formation. Our data provide evidence that PSBR is required for the stable binding of LHCSR3 to PSII-LHCII supercomplexes.
RESULTS
PSBR Is a PSII Subunit in C. reinhardtii
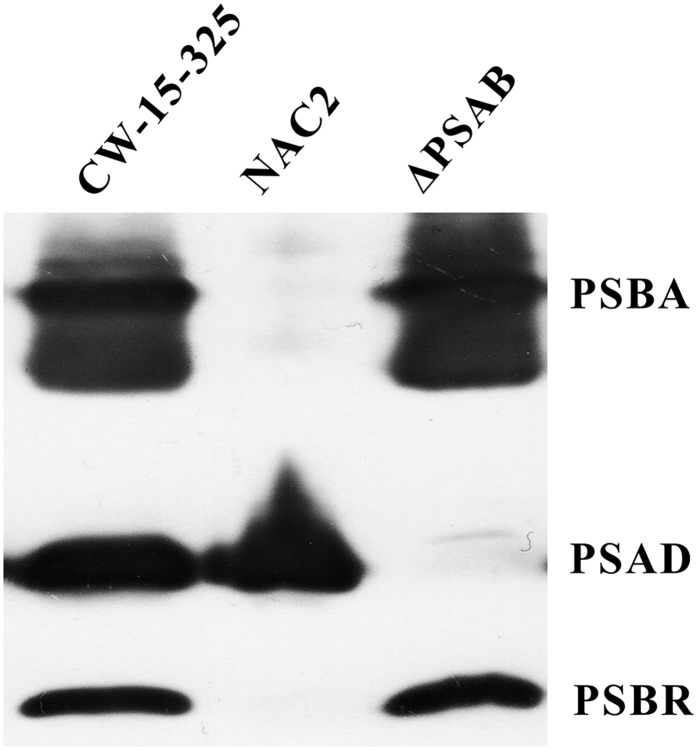
To address the question of whether PSBR is associated with PSII, we analyzed the presence of the protein in wild-type, PSII-deficient (nac2; Kuchka et al., 1989), and PSI-deficient (ΔpsaB; Redding et al., 1998) cells using immunoblotting after SDS-PAGE fractionation (Fig. 1). Immunoblot analyses revealed that the PSBR signal is absent in the PSII-deficient mutant (nac2). The absence of PSII is proven by the lack of a PSBA immunoblot signal. In contrast, PSBR is present in the absence of PSI. The lack of PSI is confirmed by the absence of a PSAD signal in ΔpsaB cells. In conclusion, the appearance of PSBR is linked to the presence of PSII; thus, PSBR is likely to be a genuine PSII subunit as described for vascular plants (see above).
Figure 1.
Immunoblot analysis of the wild type (cw15-325), NAC2, and ΔPSAB. The wild type and NAC2 were grown in Tris-acetate-phosphate (TAP) medium in low light (LL; approximately 30 μE m−2 s−1). ΔPSAB was grown in TAP in very LL (approximately 6 μE m−2 s−1) conditions. Two micrograms of chlorophyll of the whole-cell extracts was loaded on a 13% SDS-PAGE gel and analyzed by immunoblotting using anti-PSBA, anti-PSAD, and anti-PSBR antibodies.
Diminishment of PSBR Expression Decreases the Magnitude of Light-Induced NPQ But Does Not Impact the Extent of LHCSR3 Expression
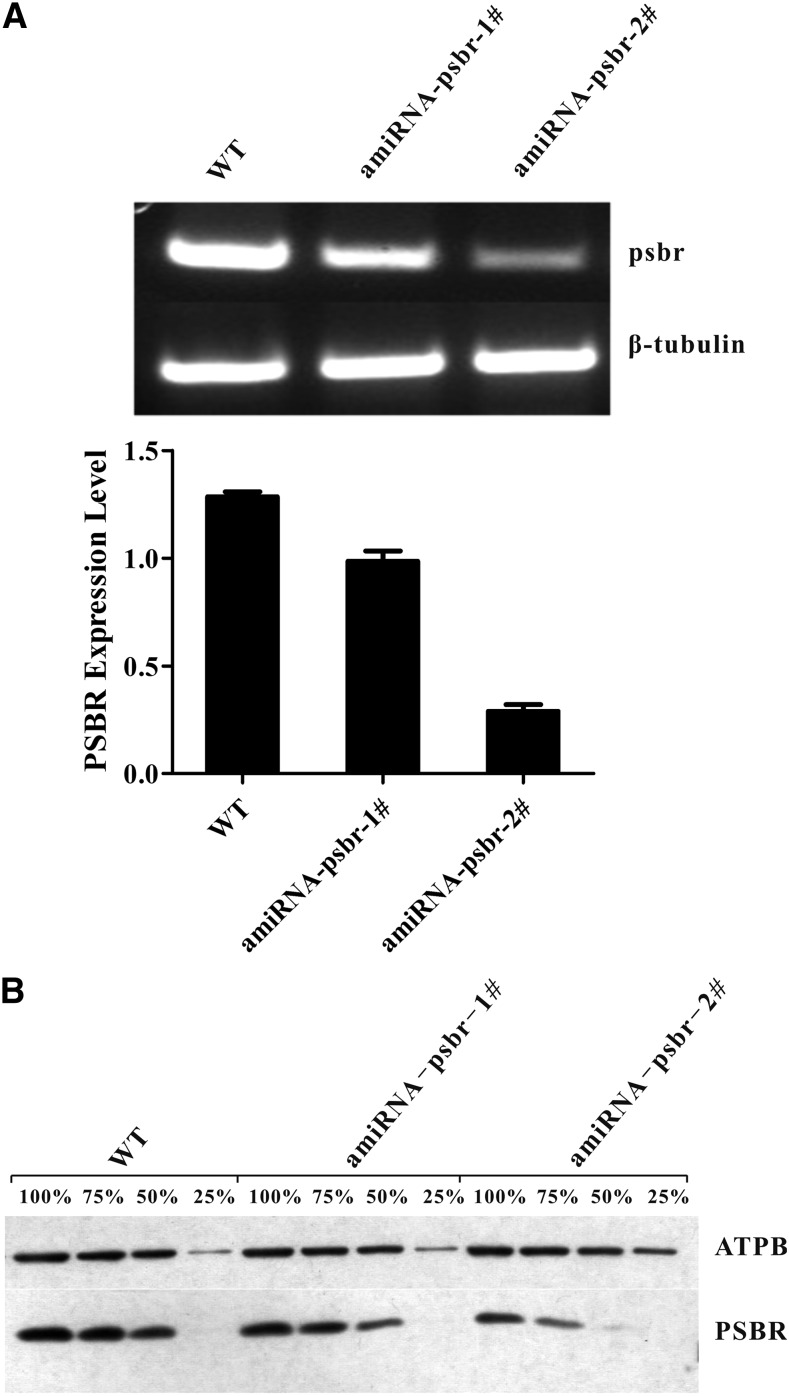
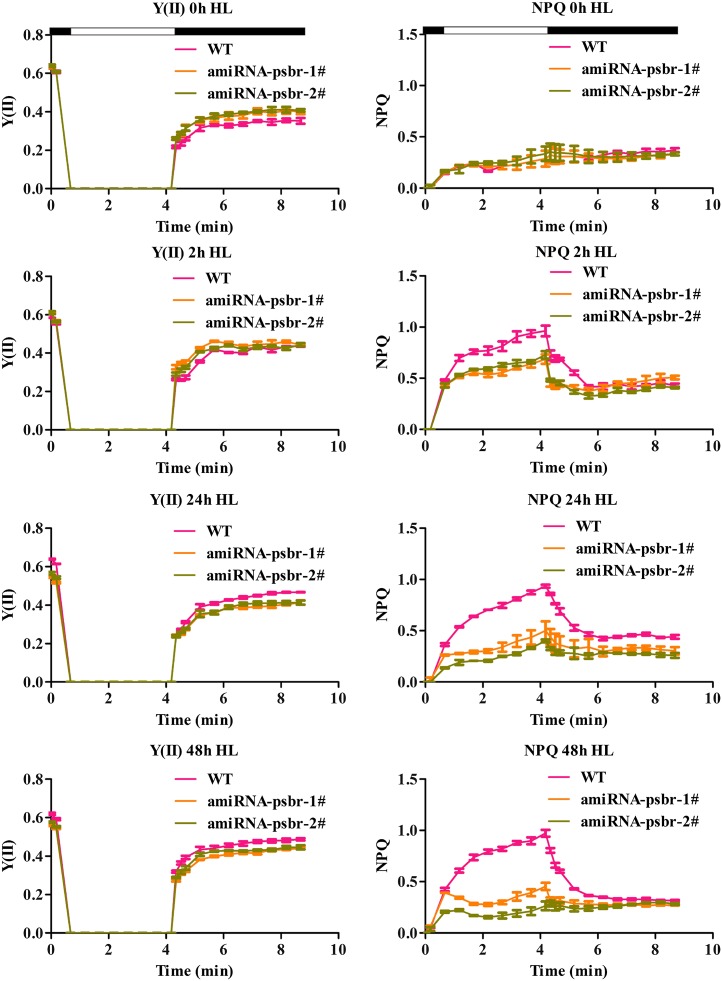
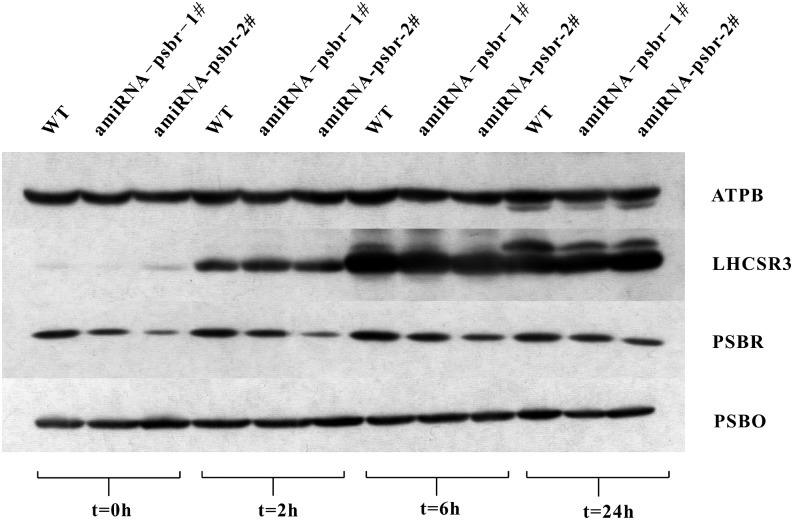
To determine the function of the PSBR protein in C. reinhardtii, an amiRNA approach was employed. We isolated several amiRNA strains in which the accumulation of the psbr RNA (Fig. 2A) and protein (Fig. 2B), in comparison with the wild type, was reduced to different degrees. The amounts of PSBR were tested in thylakoids that were isolated from cells shifted from TAP, LL to high-salt medium (HSM), HL (200 μE m−2 s−1) for 24 h to ensure the development of NPQ in the green alga. The reduction in PSBR protein amounts ranged from 1.25-fold (ami-RNA-psbr-1) and 1.5-fold (ami-RNA-psbr-44; Supplemental Fig. S1A) to 2-fold (ami-RNA-psbr-2; Fig. 2B). To assess the photosynthetic performance of the ami-RNA-psbr strains in regard to the wild type, cells were shifted from TAP LL to HSM HL for 48 h, and the PSII-dependent quantum efficiency and NPQ were measured after 0, 2, 24, and 48 h (Figure 3). These data showed that the quantum efficiency remained similar over 48 h. The NPQ increased in the wild type, ami-RNA-psbr-1, and ami-RNA-psbr-2 after 2 h but then increased further for the wild type but decreased in the ami-RNA-psbr strains. Here, the decrease was more enhanced in ami-RNA-psbr-2 as compared with ami-RNA-psbr-1, correlating with the respective degree of PSBR diminishment in the two knockdown strains. A clear reduction of NPQ in regard to the wild type is also seen in ami-RNA-psbr-44 after 24 h of HL in HSM (Supplemental Fig. S1B). In the next step, we monitored the extent of LHCSR3 expression after a shift from TAP LL to HSM HL after 0, 2, 6, and 24 h in the wild-type and ami-RNA-psbr-1 as well as ami-RNA-psbr-2 strains (Fig. 4). Cells were harvested at the respective time points and fractionated by SDS-PAGE, immunoblotted, and proteins of interest were assayed by immunodetection. As expected, the immunoblot data revealed an induction of LHCSR3 expression in the wild type (Peers et al., 2009; Petroutsos et al., 2011). Interestingly, the extent of LHCSR3 induction was similar in the two ami-RNA-psbr strains. Notably, the expression of PSBR also was induced slightly by HL and HSM growth conditions. Neither PSBO, another luminal PSII subunit, nor ATP synthase subunit β (ATPB) displayed this trend, but they revealed equal loading of the examined protein samples.
Figure 2.
Comparative quantitative reverse transcription (RT)-PCR and immunoblot analysis of psbr RNA and protein complement in the wild type (WT) and PSBR knockdown mutants. A, PSBR expression is significantly down-regulated in PSBR knockdown mutants as revealed by RT-PCR. β-Tubulin was used as a loading control. PSBR expression level is expressed as mean PSBR/β-tubulin ± sd; n = 3. B, Thylakoids isolated from high light (HL)-treated (24 h) cells were separated on a 13% (w/v) SDS-PAGE gel and analyzed by immunoblotting using anti-PSBR antibodies. Six micrograms of chlorophyll equals 100%, and ATPB served as a loading control.
Figure 3.
PSII quantum yield [Y(II)] and NPQ induction of the wild-type (WT) and ami-RNA-psbr strains. Knockdown of PSBR leads to a decrease in NPQ induction. Cells were grown in TAP medium under LL (20 μE m−2 s−1) and then shifted to HL (200 μE m−2 s−1) and HSM for 2, 24, and 48 h.
Figure 4.
Immunoblot analysis of PSBR and LHCSR3 protein abundance of whole-cell extracts from HL-treated cultures. Cell cultures (the wild type [WT], ami-RNA-psbr-1, and ami-RNA-psbr-2) were grown photoheterotrophically in LL and then shifted to HL and photoautotrophic growth for 2, 6, and 24 h. Six micrograms of chlorophyll of the whole-cell extracts was loaded on a 13% (w/v) SDS-PAGE gel and analyzed by immunoblotting using anti-LHCSR3 and anti-PSBR antibodies. ATPB and PSBO were used as loading controls.
Diminishment of PSBR Expression Destabilizes the Binding of LHCSR3 to a PSII-LHCII Supercomplex
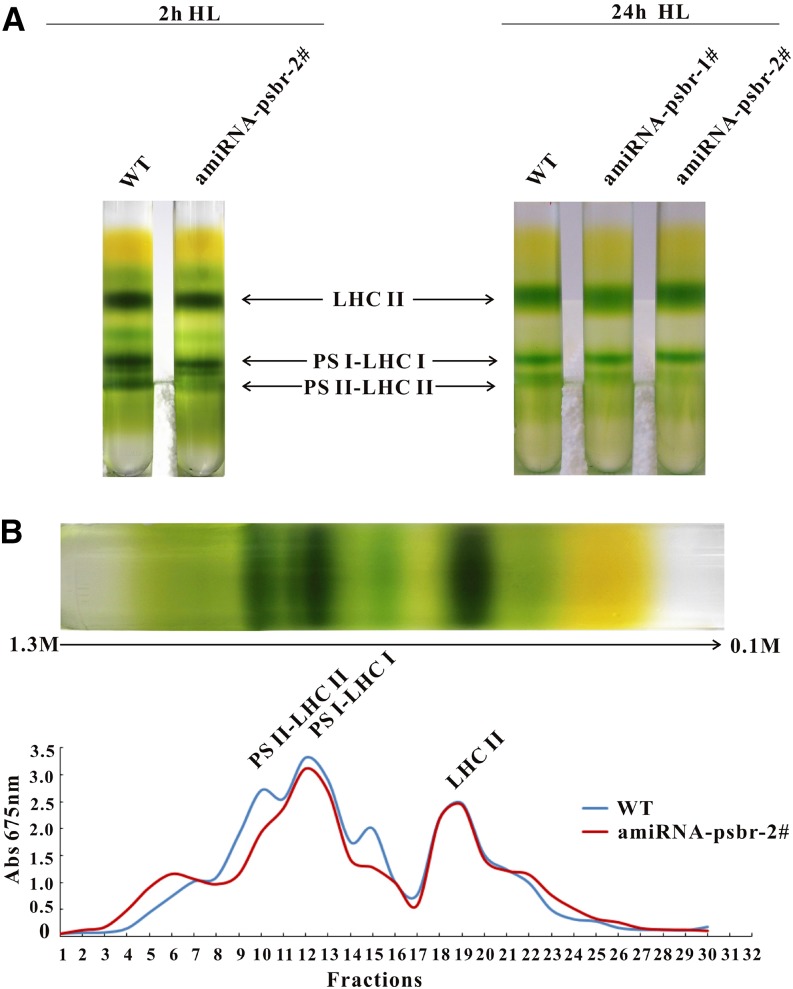
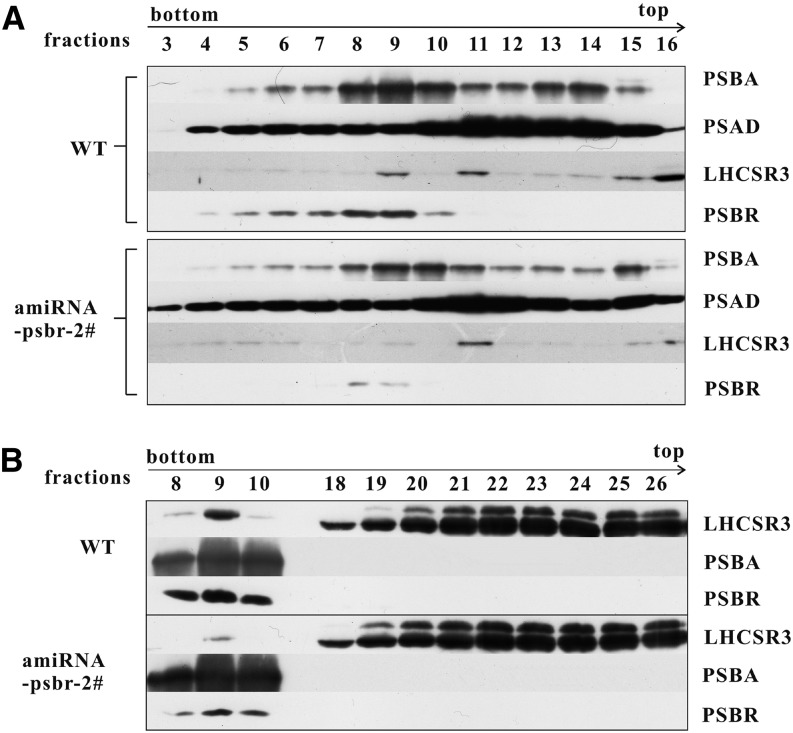
As the diminishment of PSBR expression impacts the magnitude of NPQ but not the extent of LHCSR3 induction, we rationalized that PSBR reduction might affect the formation of an LHCSR3-PSII supercomplex that was described previously (Tokutsu and Minagawa, 2013). To this end, C. reinhardtii cells (the wild type and ami-RNA-psbr-2) were grown in TAP LL and shifted to HL in HSM for 2 and 24 h. C. reinhardtii ami-RNA-psbr-1 cells were additionally analyzed after 24 h of HL. Thylakoids purified from these cells were solubilized with 1% (w/v) n-dodecyl α-d-maltoside (α-DM) and separated by sucrose density gradient (SDG) centrifugation (Fig. 5; Supplemental Fig. S2). The SDG separation displayed three green bands for the wild type, designated as LHCII, PSI-LHCI, and PSII-LHCII according to preceding work (Tokutsu and Minagawa, 2013). For ami-RNA-psbr-1 and ami-RNA-psbr-2 strains, visual inspection of the Suc gradients indicated that the PSII-LHCII bands were less pronounced in comparison with the wild-type SDG separation. For ami-RNA-psbr-2, this trend also can be observed after 2 h of HL (Fig. 5). Measuring the absorbance of collected Suc fractions at 675 nm confirmed this observation and showed that the PSII-LHCII complex is already reduced after 2 h of HL in ami-RNA-psbr-2 followed by ami-RNA-psbr-1 after 24 h of HL, while the other bands are comparable in absorbance to the wild type (Fig. 5B; Supplemental Fig. S2A). Suc fractions were collected, and fractions 3 to 16 were separated by SDS-PAGE and subjected to immunoblotting (Fig. 6A; Supplemental Fig. S2B). These data revealed that, in the wild type, PSBA, as a marker for PSII, peaked in fractions 9 and 10 after 2 h of HL, while PSAD, as a marker for PSI, peaked in fractions 11 to 13, in agreement with the designation shown in Figure 5 (Fig. 6A). Moreover, LHCSR3 comigrated with both PSII and PSI, in agreement with a recent report (Allorent et al., 2013), while PSBR peaked with PSBA, in accordance with being a PSII complex subunit.
Figure 5.
SDGs of thylakoid membranes from the wild-type and PSBR knockdown strains. A, Cultures (the wild type [WT], ami-RNA-psbr-1, and ami-RNA-psbr-2) were initially grown in TAP medium under LL (20 μE m−2 s−1) and then treated with HL (200 μE m−2 s−1) in HSM for 2 and 24 h. Isolated thylakoid membranes (200 μg of chlorophyll) were solubilized with 1% (w/v) α-DM and separated by SDG ultracentrifugation. B, SDGs of 2 h of HL were fractionated from the bottom to the top (fractions 1–30), and absorption at 675 nm was measured. SDG fractions are labeled according to their protein complex composition. The green SDG fraction obtained after 2 h of HL between LHCII and PSI-LHCI is probably composed of PSII core particles.
Figure 6.
Down-regulation of PSBR affects PSII-LHCII-LHCSR3 supercomplex formation. A, Polypeptides from the 2-h HL SDG fractions 3 to 16 of the wild type (WT) and ami-RNA-psbr-2 were analyzed by immunoblotting with antibodies against LHCSR3, PSAD, PSBA, and PSBR. Sixty microliters of each fraction was loaded per lane. B, Two-hour fractions (8–10 and 18–26) of the wild type and ami-RNA-psbr-2 were subjected to immunoblotting with an anti-LHCSR3, anti-PSBA, or anti-PSBR antibody. Sample preparation was performed as in A.
After 24 h of HL, the PSII peak fraction numbers changed in the wild type, ami-RNA-psbr-1, and ami-RNA-psbr-2 to 9 to 11, 10 and 11, and 10 to 12, respectively (Supplemental Fig. S2B). In ami-RNA-psbr-1 in comparison with the wild type (Supplemental Fig. S2B), the amounts of PSBR as well as LHCSR3 in the PSII peak fraction 10 are diminished. Notably, the amount of LHCSR3 with PSI is increased in regard to the wild type. In ami-RNA-psbr-2, the quantity of PSBR signal in the PSII peak fraction was even more diminished than for ami-RNA-psbr-1, as revealed by the immunoblot analyses (Supplemental Fig. S2B). Already after 2 h of HL as well as under 24 h of HL, only traces of LHCSR3 were found associated with PSII, while it still comigrated, although to a lesser extent, with PSI (Fig. 6A; Supplemental Fig. S2B). These data were confirmed by an analysis of thylakoid membranes from ami-RNA-psbr-44 after 24 h of HL and showed that, after detergent solubilization, Suc density centrifugation, and SDS-PAGE separation and immunoblotting of collected Suc fractions, LHCSR3 was strongly depleted from PSII-LHCII supercomplexes (Supplemental Fig. S1, C and D). Separation of the LHCII band by SDS-PAGE and subsequent immunoblotting of Suc fractions 18 to 26 (fractions 20–28 after 24 h of HL), here shown in comparison with PSII-LHCII supercomplex peak fractions, revealed that the amounts and distribution of LHCSR3 are similar between the wild type and the PSBR knockdown strain ami-RNA-psbr-2 (Fig. 6B; Supplemental Fig. S2C). Moreover, the data confirmed the diminishment of LHCSR3 association with PSII-LHCII supercomplexes when PSBR amounts are reduced (ami-RNA-psbr-2 strain, 2 and 24 h of HL). Importantly, the fractionation results observed for ami-RNA-psbr-2 in regard to PSBA, PSBR, and LHCSR3 could be confirmed independently with strain ami-RNA-psbr-44 (Supplemental Fig. S1D), where PSBR amounts also were more decreased, as in ami-RNA-psbr-1.
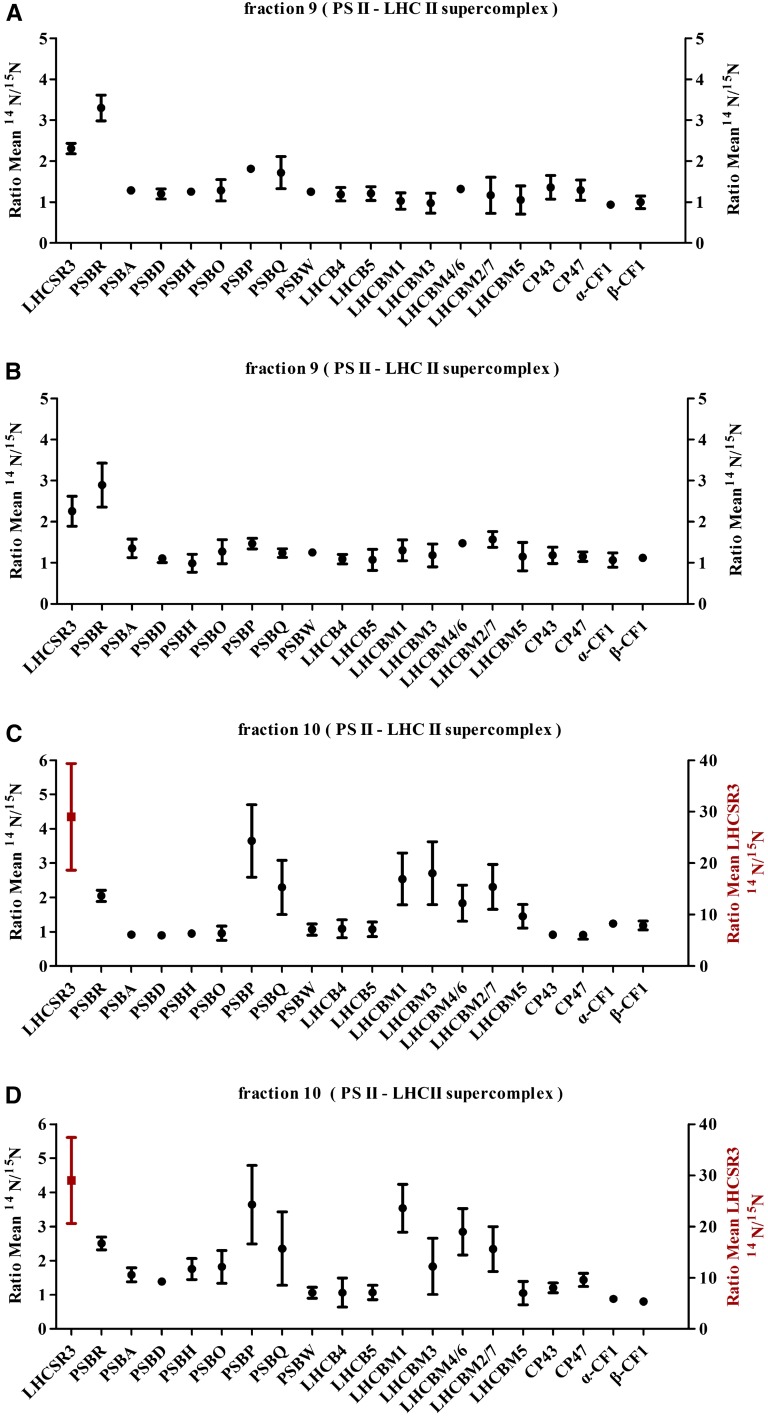
To extend the analyses of the collected Suc fractions to other subunits of PSII, we took advantage of isotopic labeling using differentially labeled 14N and 15N C. reinhardtii cell cultures. Comparative quantitative proteomics data were collected from 14N-labeled wild-type and 15N-labeled ami-RNA-psbr-2 strains after 2 and 3 h of HL and from 14N-labeled wild-type and 15N-labeled ami-RNA-psbr-1 and ami-RNA-psbr-2 strains after 24 h of HL (Fig. 7, A–D; Supplemental Tables S1–S4). Samples for mass spectrometric analyses were prepared by combining equal volumes of the PSII-LHCII supercomplex peak fractions from the wild type and one mutant followed by tryptic digestion according to the filter-aided sample preparation protocol (Wiśniewski et al., 2009). The mass spectrometric data were gathered from SDS-PAGE-fractionated, 14N/15N-isolated PSII-LHCII supercomplexes (Fig. 7). Quantitative data (Fig. 7) are presented as ratios of 14N to 15N (mean values and their respective sd), thereby showing protein amounts of wild-type versus ami-RNA-psbr supercomplex subunits.
Figure 7.
Comparative quantitative proteomic analysis of polypeptides of PSII supercomplex and ATPase from the wild-type and PSBR knockdown strains. The same volume (100 μL) of PSII supercomplex fractions from 2-h (A), 3-h (B), and 24-h (D) HL SDGs of the wild type and ami-RNA-psbr-2, respectively, and 24-h (C) HL SDGs of the wild type and ami-RNA-psbr-1 were analyzed by mass spectrometry. The ratios of 14N-labeled (cw15-325) and 15N-labeled (PSBR knockdown strains) polypeptides are presented (mean values and their respective sd).
The comparative proteomics data confirmed the down-regulation of PSBR expression in ami-RNA-psbr-1 (2-fold, 24 h of HL; Fig. 7B) and ami-RNA-psbr-2 (3.3-, 2.9-, and 2.5-fold after 2, 3, and 24 h of HL, respectively; Fig. 7, A, B, and D) strains and the depletion of LHCSR3 in the isolated PSII supercomplexes from the PSBR knockdown strains. In particular, after 2 and 3 h of HL, LHCSR3 was 2.3-fold diminished in ami-RNA-psbr-2 versus the wild type, independently confirming the immunoblot data obtained for Suc density fractions (Fig. 6). Moreover, PSII subunits PSBP and PSBQ appear to be slightly reduced in comparison with the PSII core subunits PSBA, PSBD, CP43, and CP47 in ami-RNA-psbr-2. On the other hand, subunits of the minor and major PSII light-harvesting complexes are not diminished with regard to PSII core subunits.
After 24 h of HL, LHCSR3 was about 25-fold depleted from PSII-LHCII supercomplexes in ami-RNA-psbr-1 and ami-RNA-psbr-2. Likewise, the data revealed that subunits PSBP and PSBQ were both diminished in amounts in conjunction with PSBR, in agreement with data obtained for Arabidopsis PSBR-deficient plants (Suorsa et al., 2006; Liu et al., 2009; Allahverdiyeva et al., 2013). On the other hand, other PSII core subunits or the minor core PSII light-harvesting proteins, LHCB4 and LHCB5, were not significantly altered in their protein ratios in both ami-RNA-psbr strains. Notably, LHCBM1, LHCBM3, LHCBM4/6, and LHCBM2/7, which are part of trimeric LHCII complexes (Stauber et al., 2003), were found to be decreased between 3.5- and 2-fold, where LHCBM1 was most affected, with 3.5-fold reduction in the ami-RNA-psbr-2 strain. In contrast, LHCBM5, involved in state transitions in C. reinhardtii (Takahashi et al., 2006), appeared to be unchanged.
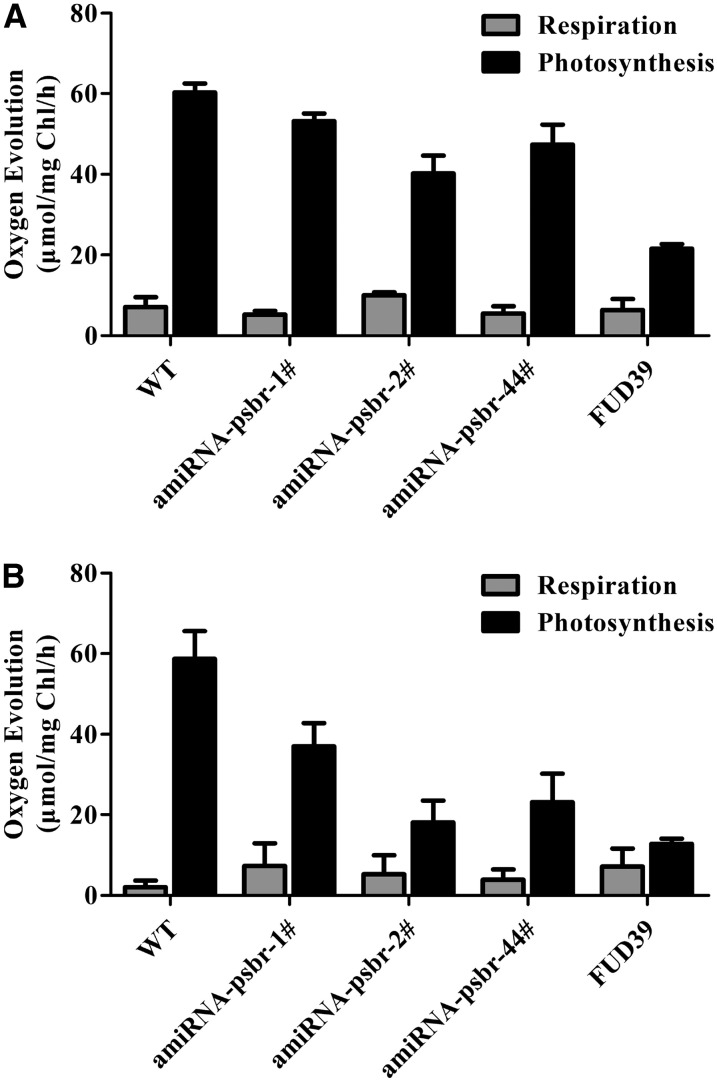
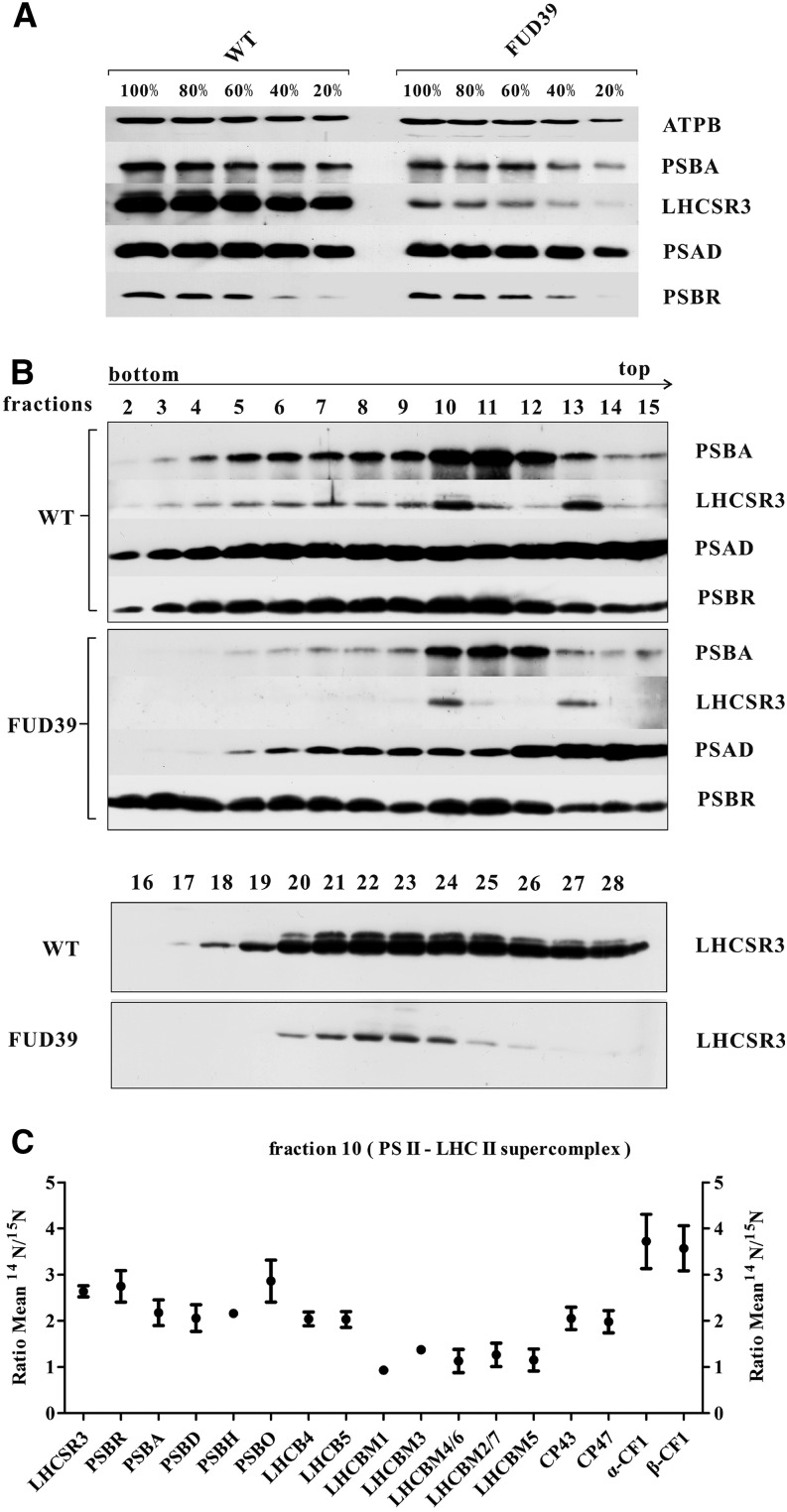
As oxygen-evolving complex subunits PSBP and PSBQ were decreased in amounts in ami-RNA-psbr-1 and ami-RNA-psbr-2 strains, we decided to measure in vivo oxygen evolution and uptake using a Clarke-type electrode. For comparison, we also included the FUD39 mutant that lacks PSBP (de Vitry et al., 1989) and has been shown to possess a low oxygen-evolving activity (de Vitry et al., 1989; Rova et al., 1994). Before measurements, cell cultures were incubated in LL TAP or for 24 h under HL in HSM. FUD39 was incubated for 24 h under HL in TAP. Oxygen evolution of cells was determined in the presence of 2-phenyl-1,4-benzoquinone (Fig. 8). In LL TAP, oxygen evolution of FUD39 was 3-fold diminished, while among the ami-RNA-psbr strains, ami-RNA-psbr2 was affected most strongly and had a 1.5-fold reduction in oxygen evolution. In HL HSM, the impact on oxygen evolution was more pronounced for the ami-RNA-psbr strains. Oxygen evolution in ami-RNA-psbr-1, ami-RNA-psbr-2, and ami-RNA-psbr-44 cells were 1.6-, 3.2-, and 2.5-fold lower compared with the wild type. In FUD39, oxygen evolution dropped 5.4-fold in HL TAP with regard to the wild type. Thus, as expected, the decrease in oxygen evolution correlated with the extent of PSBR down-regulation but was less pronounced as compared with the PSBP deletion strain.
Figure 8.
Comparison of oxygen evolution rates of the wild type (WT), PSBR knockdown strains, and FUD39. A, Cells were grown with LL in TAP medium. Oxygen evolution measurements were carried out with cells incubated in TAP medium containing 0.15 μm 2-phenyl-1,4-benzoquinone. Values are means ± sd of three replicates. B, Cell culture of the wild-type and ami-RNA-psbr lines were harvested in HSM at HL (200 μE m−2 s−1) for 24 h. FUD39 was first treated with HL in TAP medium for 24 h. Oxygen evolution measurements were carried out with cells incubated in HSM and TAP medium containing 0.12 μm 2-phenyl-1,4-benzoquinone. Values are means ± sd of three replicates. Chl, Chlorophyll.
Deletion of PSBP Does Not Impact the Binding of PSBR or LHCSR3 to PSII-LHCII Supercomplexes
Deletion of PSBP (FUD39) diminished the accumulation of PSBA and PSBR in whole-cell extracts about 1.5-fold as assessed by immunoblotting (Fig. 9A), while the accumulation of ATPB and PSAD was also diminished slightly. However, the induction of LHCSR3 was reduced drastically in FUD39. This could be explained by a lower electron transfer capacity due to the strong impact in oxygen evolution, known to be required for the induction of LHCSR3 protein expression in response to HL (Petroutsos et al., 2011). To assess the capacity for LHCSR binding to PSII-LHCII supercomplexes in FUD39, wild-type and FUD39 cells were grown for 24 h in TAP and HL. Isolated thylakoids were then solubilized with α-DM, as described above, and separated by Suc density ultracentrifugation. The SDGs were fractionated, and Suc fractions were separated by SDS-PAGE followed by immunoblotting (Fig. 9B). As demonstrated above (Fig. 6A), in the wild type, LHCSR3 comigrated with PSBA and PSAD in PSII-LHCII and PSI-LHCI supercomplexes, respectively. For FUD39, a similar result was obtained, showing comigration of LHCSR3 with PSII-LHCII and PSI-LHCI supercomplexes. In comparison with the wild type, PSBA, PSBR, and LHCSR3 amounts were diminished in FUD39, suggesting that the overall amount of PSII-LHCII supercomplexes is reduced. The comparison of LHCSR3 amounts in fractions 16 to 28 revealed lower amounts of LHCSR3 in FUD39 with regard to the wild type. To confirm the absence of PSBP from FUD39, PSII-LHCII supercomplexes from the wild type and FUD39 were subjected to trypsin digestion and analyzed by mass spectrometry. These tandem mass spectrometry data demonstrated that not only PSBP but also PSBQ was absent in the PSII-LHCII supercomplexes from FUD39 (Supplemental Table S5), in accordance with a previous characterization (de Vitry et al., 1989). Moreover, to comparatively quantify PSII from the wild type and FUD39, we took advantage of isotopic labeling using differentially labeled 14N wild-type and 15N FUD39 cell cultures. The comparative quantitative proteomics data, collected from isolated PSII-LHCII supercomplexes, revealed that PSII core subunits such as PSBA-D and PSBH as well as the minor light-harvesting complex subunits LHCB4 and LHCB4 were about 2-fold diminished in FUD39 in comparison with the wild type (Fig. 9C; Supplemental Table S6). This is in accordance with the immunoblot data (Fig. 9B) and confirms that PSII-LHCII supercomplexes were reduced in FUD39. PSBO, PSBR, and LHCSR3 were slightly less abundant and about 2.5-fold diminished in FUD39 with regard to the wild type. No quantitative data could be gathered for PSBP and PSBQ, supporting the notion that these subunits are absent in the PSII-LHCII supercomplexes from FUD39. Interestingly, the amounts of LHCBM subunits were basically the same in the wild type and FUD39. These data clearly demonstrate that PSBR and LHCSR3 comigrate with PSII core subunits and that LHCSR3 is linked to the PSII core complex rather than to the mobile LHCBM fraction.
Figure 9.
Immunoblot analysis of whole cells and SDG fractions 2 to 28 from 24 h of HL and comparative quantitative proteomic analysis of polypeptides of the PSII supercomplex and ATPase from the wild type (WT) and FUD39. A, Cell cultures of the wild type and FUD39 were grown in TAP in HL for 24 h. Whole-cell extracts were loaded on a 13% (w/v) SDS-PAGE gel and analyzed by immunoblotting using anti-PSBA, anti-LHCSR3, anti-PSAD, and anti-PSBR antibodies. Three micrograms of chlorophyll equals 100%, and ATPB served as a loading control. B, SDG fractions 2 to 28 from 24 h of HL (the wild type and FUD39) were subjected to immunoblotting with anti-PSBA, anti-LHCSR3, anti-PSAD, and anti-PSBR antibodies. Sixty microliters of each fraction was loaded per lane. C, The same volume (100 μL) of PSII supercomplex fractions from 24 h of HL SDGs (the wild type and FUD39) was analyzed by mass spectrometry. The ratios of 14N-labeled wild-type and 15N-labeled FUD39 polypeptides are presented (mean values ± sd).
Phosphorylation of PSBR
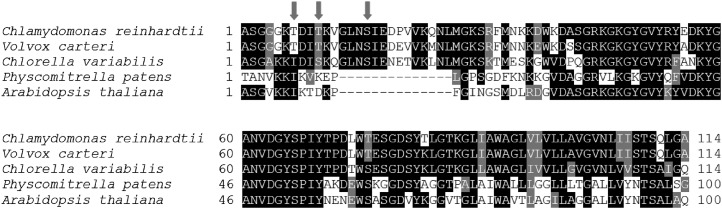
PSBR phosphorylation is known for Arabidopsis (Reiland et al., 2009, 2011; Nakagami et al., 2010) and C. reinhardtii (Turkina et al., 2006). To validate PSBR phosphorylation in C. reinhardtii, we performed mass spectrometric analyses of thylakoid membranes using sequential elution from immobilized metal ion affinity chromatography (SIMAC) for phosphopeptide enrichment (Thingholm et al., 2008). Using this approach, three distinct phosphorylation sites could be detected on PSBR (Table I): two T phosphorylation sites were unknown, while the phosphorylation of S40 has already been published (Turkina et al., 2006), referred to as Ser-43. All three phosphorylation sites differ from those known for Arabidopsis as visualized in a ClustalW (Larkin et al., 2007) multiple sequence alignment (Fig. 10).
Table I. Analysis of PSBR phosphorylation revealed three distinct phosphorylation sites.
To investigate PSBR phosphorylation in C. reinhardtii, we performed mass spectrometric analyses of thylakoid membranes using SIMAC for phosphopeptide enrichment (Thingholm et al., 2008). Subscript numbers indicate positions of phosphorylation sites within the unprocessed polypeptide. Phosphorylation site validation was done with phosphoRS (Taus et al., 2011).
| Peptide | No. of Phosphorylations | Observed Mass | Charge | Theoretical Mass | Posterior Error Probability | phosphoRS |
||
|---|---|---|---|---|---|---|---|---|
| Sequence Probability | Peptide Score | Site Probabilities | ||||||
| m | m | |||||||
| TDITKVGLNs40IEDPVVK | 1 | 1,906.969 | 2 | 1,906.965 | 3.99E-05 | 1 | 183.87 | T(1), 0.0; T(4), 0.0; S(10), 100.0 |
| t31DITKVGLNSIEDPVVK | 1 | 1,906.973 | 2 | 1,906.965 | 1.18E-09 | 0.95 | 192.88 | T(1), 94.5; T(4), 5.5; S(10), 0.0 |
| t31DITKVGLNs40IEDPVVK | 2 | 1,986.935 | 2 | 1,986.931 | 5.42E-06 | 0.94 | 181.60 | T(1), 93.9; T(4), 6.1; S(10), 100.0 |
| TDIt34KVGLNSIEDPVVK | 1 | 1,906.965 | 2 | 1,906.965 | 8.62E-16 | 1 | 202.86 | T(1), 0.4; T(4), 99.6; S(10), 0.0 |
| TDIt34KVGLNs40IEDPVVK | 2 | 1,986.932 | 2 | 1,986.931 | 8.62E-16 | 1 | 136.01 | T(1), 0; T(4), 100.0; S(10), 100.0 |
| VGLNs40IEDPVVK | 1 | 1,348.665 | 2 | 1,348.663 | 8.84E-16 | 1 | 149.95 | S(5), 100.0 |
Figure 10.
PSBR sequence alignment. Mature PSBR amino acid sequence alignment of PSBR in C. reinhardtii (Merchant et al., 2007), Volvox carteri (Prochnik et al., 2010), Chlorella variabilis (Blanc et al., 2010), Physcomitrella patens (Rensing et al., 2008), and Arabidopsis (Gil-Gómez et al., 1991) was performed using ClustalW (Larkin et al., 2007) and shaded with version 3.21 of BOXSHADE. Protein sequences were taken from the National Center for Biotechnology Information (reference sequences XP_001696588.1, XP_002945815.1, XP_005847921.1, XP_001760191.1, and NP_178025.1). Transit peptide cleavage sites were deduced by Emanuelsson et al. (2000) and PredAlgo (Tardif et al., 2012). Arrows indicate identified phosphorylation sites.
DISCUSSION
In C. reinhardtii, the LHCSR3 protein is crucial for efficient qE (Peers et al., 2009) and functionally associates with PSII-LHCII supercomplexes (Tokutsu and Minagawa, 2013). Currently, it is unknown how LHCSR3 binds to the PSII-LHCII supercomplex. In this study, we investigated the impact of PSBR down-regulation on photosynthetic performance through measurements of PSII quantum efficiency, oxygen evolution, and NPQ as well as on PSII-LHCII-LHCSR3 supercomplex formation. We provide evidence that PSBR is required for the stable binding of LHCSR3 to PSII-LHCII supercomplexes.
PSBR Is Involved in Forming a Recognition Site for the Binding of LHCSR3 to PSII-LHCII Supercomplexes
Down-regulation of PSBR expression decreased the extent of NPQ and affected the binding of LHCSR3 to PSII-LHCII supercomplexes. On the other hand, the reduction of PSBR expression had no impact on the extent of LHCSR3 expression (Figs. 4 and 6B; Supplemental Fig. S2C), thus indicating that differences in the amounts of LHCSR3 cannot explain the decrease in NPQ. As already described for a PSBR-deficient mutant in Arabidopsis (Allahverdiyeva et al., 2007, 2013), the depletion of PSBR diminished PSII quantum efficiency and decreased rates of oxygen evolution after 24 h of HL (Figs. 3 and 8). Consequently, less efficient electron transfer from PSII into the plastoquinone pool could explain the impact on NPQ observed in the C. reinhardtii PSBR knockdown strains after 24 h of HL. Additionally, detachment of LHCSR3 from the PSII-LHCII supercomplex could contribute to the lower NPQ in PSBR knockdown mutants, as the PSII-LHCII-LHCSR3 supercomplex formed in the wild type (Fig. 5) is competent in energy dissipation (Tokutsu and Minagawa, 2013). The diminishment of PSBR expression after 24 h of HL greatly destabilized the binding of LHCSR3 to the PSII-LHCII supercomplex, while binding of LHCBM5 and of the minor core PSII light-harvesting proteins, LHCB4 and LHCB5, was not affected significantly (Fig. 7, C and D). On the other hand, the major light-harvesting proteins LHCBM1 and LHCBM3 were slightly diminished in the PSBR knockdown strains, suggesting that PSII-LHCII complex formation is impaired by PSBR depletion. In line with this, it was reported that in PSBR-deficient Arabidopsis plants, the formation of PSII-LHCII supercomplexes was marginally affected (Allahverdiyeva et al., 2013). Interestingly, deletion of LHCBM1 resulted in a significant decrease of NPQ (Elrad et al., 2002) in C. reinhardtii. The fact that, besides LHCSR3, LHCBM1 was most affected by PSBR knockdown in strain ami-RNA-psbr-2 might suggest that this protein is directly or indirectly linked to LHCSR3 and PSBR, which might explain the NPQ phenotype in the LHCBM1-deficient mutant. Strikingly, the 2.5-fold diminishment of PSBR expression destabilized the binding of LHCSR3 to the PSII-LHCII supercomplex almost completely after 24 h of HL (ami-RNA-psbr-2; Figs. 6A and 7D). It is important to note that, already after 2 and 3 h of HL, LHCSR3 is, besides PSBR, the only other PSII-LHCII supercomplex-associated protein, which is more than 2-fold depleted from the complex in ami-RNA-psbr-2 (Figs. 6A and 7, A and B). In contrast, the major PSII light-harvesting proteins did not change with regard to PSII core subunits. Moreover, diminishment in PSBP and PSBQ as observed after 24 h of HL was not as pronounced after 2 and 3 h of HL in ami-RNA-psbr-2. After 2 and 3 h of HL, the reduction in NPQ was not accompanied by lower PSII quantum efficiency, suggesting that the impact seen in NPQ is due to less stable binding of LHCSR3 to PSII (Fig. 3; Supplemental Fig. S3). The data suggest that (1) stable LHCSR3 binding to the PSII-LHCII supercomplex is dependent on PSBR enabling efficient qE quenching and (2) the absence of PSBR destabilizes the PSII-LHCII supercomplex in continued HL. The data obtained for FUD39 (Fig. 9, B and C) strongly indicate that PSBP and PSBQ do not affect LHCSR3 binding to PSII-LHCII supercomplexes, as LHCSR3 association with these supercomplexes is clearly recognized, although the particles are lacking PSBP and PSBQ polypeptides.
Intriguingly, Ido et al. (2014) identified two cross-linked sites between PSBP and PSBR in PSII-enriched membranes from spinach (Spinacia oleracea) using chemical cross-linking and mass spectrometry. Moreover, those authors identified cross-links between PSBP and CP26. The tight interaction between PSBP and PSBR, as suggested earlier (Suorsa et al., 2006; Liu et al., 2009), is also confirmed in our study, as depletion of PSBR impacts the association of PSBP with the PSII-LHCII supercomplex (Fig. 7). On the other hand, deletion of PSBP did not strongly impact the binding of PSBR to PSII, suggesting that the PSBP presence is not critical for PSBR but the absence of PSBR destabilizes the association of PSBP and PSBQ with PSII. The cross-linking between PSBP and CP26 demonstrates the close interaction of PSBR/PSBP with the light-harvesting protein network in the PSII-LHCII supercomplex and suggests that binding of LHCSR3 to PSBR enables energetic coupling between LHCSR3, the core of the PSII complex, and associated LHCII for efficient light-to-heat dissipation. The fact that the absence of PSBP in FUD39 clearly diminished PSBR and LHCSR3 as well as PSII core subunits but not the LHCBM proteins (Fig. 9C) strongly indicates that LHCSR3 is indeed coupled via PSBR and the PSII core complex and not via the LHCBM interaction network. Thus, the presence of PSBR is required for the binding of LHCSR3 to the PSII-LHCII supercomplex and for efficient qE quenching.
Phosphorylation of PSBR
The ClustalW multiple sequence alignment of PSBR revealed that the HL/state 2-induced (Turkina et al., 2006) phosphorylation site Ser-40 of C. reinhardtii PSBR is located in a protein segment that appears to be absent in land plants (as seen for P. patens and Arabidopsis PSBR), while C-terminal sequences are more conserved (Fig. 10). On the other hand, the phosphorylation site of Arabidopsis PSBR (Ser-58) is neither conserved in green algae nor in the moss P. patens. The second N-terminal phosphorylation site appears to be conserved in algae (T in C. reinhardtii and V. carteri, S in C. variabilis). It is tempting to speculate that the protein sequence harboring the Ser-40 phosphorylation site in conjunction with the N-terminal Thr phosphorylation sites in C. reinhardtii might be involved in protein-protein interaction with LHCSR3. In P. patens, both PSBS and LHCSR proteins are utilized to manage qE (Alboresi et al., 2010). As the PSII subunit PSBS is operating in qE in the moss in contrast to green algae, this might have altered the binding of LHCSR3 to the PSII-LHCII supercomplex and additionally adapted the functional requirement of PSBR, thereby promoting the evolution of a land plant PSBR amino acid sequence type.
MATERIALS AND METHODS
Strains and Growth Conditions
Wild-type Chlamydomonas reinhardtii Arg auxotrophic strain (cw15-325) and ΔpsaB were gifts from Michael Schroda (Max Planck Institute of Molecular Plant Physiology). nac2 was kindly provided by Joerg Nickelsen (Ludwig Maximilian University, Munich). FUD39 was obtained from the Institut de Biologie Physico-Chimique, CNRS (Paris). For the photoheterotrophic phase, the wild type, FUD39, nac2, and ami-RNA-psbr knockdown cells were cultivated in TAP medium (Harris, 1989) at 25°C with 120 rpm shaking and continuous low light (20–40 μE m−2 s−1). ΔpsaB was grown in TAP at very low light (approximately 6 μE m−2 s−1). For the growth of cw15-325, the medium was supplemented with l-Arg (100 μg mL−1). For HL experiments, cells were grown in photoheterotrophic (in TAP) conditions until the cell density reached approximately 4 × 106 cells mL−1. Wild-type and ami-RNA-psbr knockdown cells were diluted to 1 × 106 cells mL−1 in HSM (Harris, 1989). For the FUD39 experiment, wild-type and FUD39 cells were diluted to 1 × 106 cells mL−1 in TAP medium. Afterward, cells were transferred to HL (approximately 185 μE m−2 s−1) conditions and harvested at the designated time.
Generation of amiRNA Mutants
Plasmid generation of ami-RNA-psbr lines was performed as described in the literature (Molnar et al., 2009; Petroutsos et al., 2011). The oligonucleotides used for ami-RNA-psbr lines were amiFor_ psbr (5′-ctagtCCGTCGTTAAGCAGAGTCTAAtctcgctgatcggcaccatgggggtggtggtgatcagcgctaTTAGGTTCTGCTTAACGACGGg-3′) and amiRev_psbr (5′-ctagcCCGTCGTTAAGCAGAACCTAAtagcgctgatcaccaccacccccatggtgccgatcagcgagaTTAGACTCTGCTTAACGACGGa-3′). The generated plasmid ami-RNA-psbr was transformed into cw15-325 strain using glass beads according to established protocols (Kindle et al., 1989). The vector pMS539 (Schmollinger et al., 2010) was used in this study to express amiRNA constructs targeting psbr.
RNA Analysis
Total RNA was extracted using TRIZOL reagent and treated with DNase from Invitrogen. Complementary DNA synthesis was carried out with the iScript cDNA Synthesis Kit (Bio-Rad). For each sample, 800 μg of purified RNA was used for a 20-μL reaction according to the manufacturer’s instructions. RT-PCR was performed using MangoTaq Polymerase (Bio-21078-82-83). The psbr product was amplified with primers psbr _for (5′-CCCCTCTGCGCCCCCGCGTGT-3′) and psbr _rev (5′-ACCAGGCCAGCCCAGGCGATCAGG-3′). Amplification of a β-tubulin product was used as an endogenous control using the following set of oligonucleotides: β-tubulin _for (5′-TGCCTGCAGGGCTTCCAGG-3′) and β-tubulin _rev (5′-GGGATCCACTCGACGAAGTA-3′).
Isolation of Thylakoid Membranes
Thylakoid membranes were isolated as described previously (Harris, 1989).
SDG Ultracentrifugation
Discontinuous SDGs were prepared by sequential layering of 1.3, 1, 0.7, 0.4, and 0.1 m Suc (from bottom to top). Suc gradient centrifugation of isolated thylakoid membranes was performed as described in the literature (Tokutsu et al., 2012) but with some modifications. An amount of thylakoid membranes corresponding to 200 µg of chlorophyll was resuspended in 5 mm HEPES and 10 mm EDTA (pH 7.5) buffer and solubilized in 1% (w/v) α-DM on ice in the dark for 5 min. Solubilized thylakoid membranes were loaded on top of prepared SDGs and centrifuged at 29,000 rpm for 22 h using Rotor SW41Ti (Beckman) at 4°C.
SDS-PAGE and Immunoblotting
Whole cells were centrifuged at 5,000 rpm for 5 min at 4°C and resuspended in 5 mm HEPES (pH 7.5) buffer. The entire gradient was fractionated from bottom to top (approximately 300 µL per fraction). The same volume of each fraction and the same amount of chlorophyll of whole cells and thylakoid membranes in the wild type and knockdown mutants were separated by 13% (w/v) SDS-PAGE (Laemmli, 1970). Proteins were blotted on nitrocellulose membranes (Amersham) and analyzed by specific antibodies. Antipeptide antibody against PSBR (ASGGGKTDITKVGLN) was purchased from Eurogentec and used in a 1:2,000 dilution. Antibody against PSBA and ATPB were obtained from Agrisera and used as 1:10,000 and 1:6,000 dilutions, respectively. Antibody against PSBO (1:1,000) was a gift from F. Koenig (University of Bremen). Antibodies against PSAD (Naumann et al., 2005) and LHCSR3 (Naumann et al., 2007) were used at 1:2,000 dilutions.
Mass Spectrometry
Isotopic 15N and 14N labeling was performed for ami-RNA-psbr mutant/FUD39 and wild-type cells, respectively. Isolated thylakoids from 14N-labeled wild type and 15N-labeled psbr knockdown strains and FUD39 cells were solubilized and separated by SDG centrifugation as described (Tokutsu et al., 2012). Separated LHCII, PSII, and PSI fractions from 14N-labeled wild-type and 15N-labeled psbr knockdown strains and FUD39 cells were mixed at equal volume and digested with trypsin according to the filter-aided sample preparation protocol (Wiśniewski et al., 2009). Peptide identification, determination of false discovery rates, and protein quantification were performed as described (Höhner et al., 2013), with the only exception being that carbamidomethylation of Cys was set as a fixed modification during the database search.
PSBR phosphopeptides were identified by shaving thylakoid membranes, which were isolated in the presence of 10 mm sodium fluoride, with trypsin as described (Turkina et al., 2006). Phosphopeptides were enriched using the SIMAC protocol (Thingholm et al., 2008). Liquid chromatography-mass spectrometry analysis of phosphopeptides was performed as described (Pan et al., 2011). Peptide identification and determination of false discovery rates were carried out as described (Höhner et al., 2013) with the following modifications: oxidation of Met and phosphorylation of Ser, Thr, and Tyr were allowed as variable modifications during the database search. Phosphorylation site assignments were validated by phosphoRS (Taus et al., 2011) with fragment ion mass accuracy set to 0.5 D.
Chlorophyll Fluorescence
Chlorophyll fluorescence measurements of cells were performed using a pulse amplitude-modulated chlorophyII fluorometer (Heinz Walz). Cultures were prepared as described in the literature (Petroutsos et al., 2011). Effective PSII quantum yield and NPQ were calculated as (Fm′ − F)/Fm′ and (Fm − Fm′)/Fm′, respectively, where Fm′ is the maximum fluorescence yield in the light-acclimated state, F is the fluorescence yield, and Fm is the maximum fluorescence yield measured during a brief, saturating flash of light.
Oxygen Evolution Measurements
Samples were prepared and measured using a Clark-type oxygen electrode as described in the literature (Naumann et al., 2007) with the following modifications. C. reinhardtii cells were grown under LL and HL for 24 h. Whole cells were centrifuged at 5,000 rpm for 5 min at 4°C and resuspended in TAP and HSM. Cells were adapted in the dark for 20 min before oxygen evolution measurement. Measurement of oxygen evolution was performed at 25°C and 1,000 μE m−2 s−1 in the presence of 0.15 μm (in TAP) and 0.12 μm (in HSM) 2-phenyl-1,4-benzoquinone.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Immunoblot analysis of thylakoid membrane and Suc density gradients and PSII quantum yield and NPQ induction of the wild type and amiRNA-psbr-44.
Supplemental Figure S2. Absorbance measurement and immunoblot analysis of SDG fractions from 24-h HL wild-type and PSBR knockdown strains.
Supplemental Figure S3. PSII quantum yield and NPQ induction of wild-type and ami-RNA-psbr-2 from 3-h HL cells.
Supplemental Table S1. Quantitative proteomics data of Figure 7A.
Supplemental Table S2. Quantitative proteomics data of Figure 7B.
Supplemental Table S3. Quantitative proteomics data of Figure 7C.
Supplemental Table S4. Quantitative proteomics data of Figure 7D.
Supplemental Table S5. Proteomics analysis of polypeptides of PSII-LHCII supercomplex from wild-type and FUD39.
Supplemental Table S6. Quantitative proteomics data of Figure 9A.
Supplementary Material
Acknowledgments
We thank Sandrine Bujaldon (Institut de Biologie Physico-Chimique) for providing the FUD39 C. reinhardtii strain and Susan Hawat for excellent technical assistance.
Glossary
- NPQ
nonphotochemical quenching
- amiRNA
artificial microRNA
- TAP
Tris-acetate-phosphate
- HSM
high-salt medium
- LL
low light
- HL
high light
- α-DM
n-dodecyl α-d-maltoside
- SIMAC
sequential elution from immobilized metal ion affinity chromatography
- RT
reverse transcription
- SDG
sucrose density gradient
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. HI 739/8.1 to M.H.), the Chinese Scholarship Council (to H.X.), and the Japan Society for the Promotion of Science (grant no. 26251033 to J.M.).
References
- Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T (2010) Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc Natl Acad Sci USA 107: 11128–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdiyeva Y, Mamedov F, Suorsa M, Styring S, Vass I, Aro EM (2007) Insights into the function of PsbR protein in Arabidopsis thaliana. Biochim Biophys Acta 1767: 677–685 [DOI] [PubMed] [Google Scholar]
- Allahverdiyeva Y, Suorsa M, Rossi F, Pavesi A, Kater MM, Antonacci A, Tadini L, Pribil M, Schneider A, Wanner G, et al. (2013) Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J 75: 671–684 [DOI] [PubMed] [Google Scholar]
- Allorent G, Tokutsu R, Roach T, Peers G, Cardol P, Girard-Bascou J, Seigneurin-Berny D, Petroutsos D, Kuntz M, Breyton C, et al. (2013) A dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell 25: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, et al. (2010) The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22: 2943–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema EJ, Hankamer B, Bald D, Kruip J, Nield J, Boonstra AF, Barber J, Rögner M (1995) Supramolecular structure of the photosystem II complex from green plants and cyanobacteria. Proc Natl Acad Sci USA 92: 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema EJ, van Roon H, Calkoen F, Bassi R, Dekker JP (1999) Multiple types of association of photosystem II and its light-harvesting antenna in partially solubilized photosystem II membranes. Biochemistry 38: 2233–2239 [DOI] [PubMed] [Google Scholar]
- Busch A, Hippler M (2011) The structure and function of eukaryotic photosystem I. Biochim Biophys Acta 1807: 864–877 [DOI] [PubMed] [Google Scholar]
- Caffarri S, Kouril R, Kereïche S, Boekema EJ, Croce R (2009) Functional architecture of higher plant photosystem II supercomplexes. EMBO J 28: 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker JP, Boekema EJ (2005) Supramolecular organization of thylakoid membrane proteins in green plants. Biochim Biophys Acta 1706: 12–39 [DOI] [PubMed] [Google Scholar]
- de Vitry C, Olive J, Drapier D, Recouvreur M, Wollman FA (1989) Posttranslational events leading to the assembly of photosystem II protein complex: a study using photosynthesis mutants from Chlamydomonas reinhardtii. J Cell Biol 109: 991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrad D, Niyogi KK, Grossman AR (2002) A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14: 1801–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Gil-Gómez G, Marrero PF, Haro D, Ayté J, Hegardt FG (1991) Characterization of the gene encoding the 10 kDa polypeptide of photosystem II from Arabidopsis thaliana. Plant Mol Biol 17: 517–522 [DOI] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- Höhner R, Barth J, Magneschi L, Jaeger D, Niehues A, Bald T, Grossman A, Fufezan C, Hippler M (2013) The metabolic status drives acclimation of iron deficiency responses in Chlamydomonas reinhardtii as revealed by proteomics based hierarchical clustering and reverse genetics. Mol Cell Proteomics 12: 2774–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ido K, Nield J, Fukao Y, Nishimura T, Sato F, Ifuku K (2014) Cross-linking evidence for multiple interactions of the PsbP and PsbQ proteins in a higher plant photosystem II supercomplex. J Biol Chem 289: 20150–20157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifuku K, Ido K, Sato F (2011) Molecular functions of PsbP and PsbQ proteins in the photosystem II supercomplex. J Photochem Photobiol B 104: 158–164 [DOI] [PubMed] [Google Scholar]
- Kindle KL, Schnell RA, Fernández E, Lefebvre PA (1989) Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol 109: 2589–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchka MR, Goldschmidt-Clermont M, van Dillewijn J, Rochaix JD (1989) Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell 58: 869–876 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lautner A, Klein R, Ljungberg U, Reiländer H, Bartling D, Andersson B, Reinke H, Beyreuther K, Herrmann RG (1988) Nucleotide sequence of cDNA clones encoding the complete precursor for the “10-kDa” polypeptide of photosystem II from spinach. J Biol Chem 263: 10077–10081 [PubMed] [Google Scholar]
- Li XP, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Liu H, Frankel LK, Bricker TM (2009) Characterization and complementation of a psbR mutant in Arabidopsis thaliana. Arch Biochem Biophys 489: 34–40 [DOI] [PubMed] [Google Scholar]
- Ljungberg U, Akerlund HE, Andersson B (1986) Isolation and characterization of the 10-kDa and 22-kDa polypeptides of higher plant photosystem 2. Eur J Biochem 158: 477–482 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D (2009) Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J 58: 165–174 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Sugiyama N, Mochida K, Daudi A, Yoshida Y, Toyoda T, Tomita M, Ishihama Y, Shirasu K (2010) Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol 153: 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann B, Busch A, Allmer J, Ostendorf E, Zeller M, Kirchhoff H, Hippler M (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7: 3964–3979 [DOI] [PubMed] [Google Scholar]
- Naumann B, Stauber EJ, Busch A, Sommer F, Hippler M (2005) N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J Biol Chem 280: 20431–20441 [DOI] [PubMed] [Google Scholar]
- Pagliano C, Nield J, Marsano F, Pape T, Barera S, Saracco G, Barber J (2014) Proteomic characterization and three-dimensional electron microscopy study of PSII-LHCII supercomplexes from higher plants. Biochim Biophys Acta 1837: 1454–1462 [DOI] [PubMed] [Google Scholar]
- Pagliano C, Saracco G, Barber J (2013) Structural, functional and auxiliary proteins of photosystem II. Photosynth Res 116: 167–188 [DOI] [PubMed] [Google Scholar]
- Pan J, Naumann-Busch B, Wang L, Specht M, Scholz M, Trompelt K, Hippler M (2011) Protein phosphorylation is a key event of flagellar disassembly revealed by analysis of flagellar phosphoproteins during flagellar shortening in Chlamydomonas. J Proteome Res 10: 3830–3839 [DOI] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Petroutsos D, Busch A, Janssen I, Trompelt K, Bergner SV, Weinl S, Holtkamp M, Karst U, Kudla J, Hippler M (2011) The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell 23: 2950–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, et al. (2010) Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329: 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, MacMillan F, Leibl W, Brettel K, Hanley J, Rutherford AW, Breton J, Rochaix JD (1998) A systematic survey of conserved histidines in the core subunits of photosystem I by site-directed mutagenesis reveals the likely axial ligands of P700. EMBO J 17: 50–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Finazzi G, Endler A, Willig A, Baerenfaller K, Grossmann J, Gerrits B, Rutishauser D, Gruissem W, Rochaix JD, et al. (2011) Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc Natl Acad Sci USA 108: 12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S (2009) Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Rivalta I, Amin M, Luber S, Vassiliev S, Pokhrel R, Umena Y, Kawakami K, Shen JR, Kamiya N, Bruce D, et al. (2011) Structural-functional role of chloride in photosystem II. Biochemistry 50: 6312–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rova M, Franzén LG, Fredriksson PO, Styring S (1994) Photosystem II in a mutant of Chlamydomonas reinhardtii lacking the 23 kDa psbP protein shows increased sensitivity to photoinhibition in the absence of chloride. Photosynth Res 39: 75–83 [DOI] [PubMed] [Google Scholar]
- Schmollinger S, Strenkert D, Schroda M (2010) An inducible artificial microRNA system for Chlamydomonas reinhardtii confirms a key role for heat shock factor 1 in regulating thermotolerance. Curr Genet 56: 383–389 [DOI] [PubMed] [Google Scholar]
- Stauber EJ, Fink A, Markert C, Kruse O, Johanningmeier U, Hippler M (2003) Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot Cell 2: 978–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suorsa M, Sirpiö S, Allahverdiyeva Y, Paakkarinen V, Mamedov F, Styring S, Aro EM (2006) PsbR, a missing link in the assembly of the oxygen-evolving complex of plant photosystem II. J Biol Chem 281: 145–150 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Iwai M, Takahashi Y, Minagawa J (2006) Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 103: 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugière S, Hippler M, Ferro M, Bruley C, Peltier G, et al. (2012) PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol 29: 3625–3639 [DOI] [PubMed] [Google Scholar]
- Taus T, Köcher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K (2011) Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res 10: 5354–5362 [DOI] [PubMed] [Google Scholar]
- Thingholm TE, Jensen ON, Robinson PJ, Larsen MR (2008) SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol Cell Proteomics 7: 661–671 [DOI] [PubMed] [Google Scholar]
- Tokutsu R, Kato N, Bui KH, Ishikawa T, Minagawa J (2012) Revisiting the supramolecular organization of photosystem II in Chlamydomonas reinhardtii. J Biol Chem 287: 31574–31581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutsu R, Minagawa J (2013) Energy-dissipative supercomplex of photosystem II associated with LHCSR3 in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 110: 10016–10021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkina MV, Kargul J, Blanco-Rivero A, Villarejo A, Barber J, Vener AV (2006) Environmentally modulated phosphoproteome of photosynthetic membranes in the green alga Chlamydomonas reinhardtii. Mol Cell Proteomics 5: 1412–1425 [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Webber AN, Packman LC, Gray JC (1989) A 10 kDa polypeptide associated with the oxygen-evolving complex of photosystem II has a putative C-terminal non-cleavable thylakoid transfer domain. FEBS Lett 242: 435–438 [DOI] [PubMed] [Google Scholar]
- Whatley FR, Tagawa K, Arnon DI (1963) Separation of the light and dark reactions in electron transfer during photosynthesis. Proc Natl Acad Sci USA 49: 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.