Viral replication complexes are found in phloem sieve elements and in xylem vessels.
Abstract
Plant viruses move systemically in plants through the phloem. They move as virions or as ribonucleic protein complexes, although it is not clear what these complexes are made of. The approximately 10-kb RNA genome of Turnip mosaic virus (TuMV) encodes a membrane protein, known as 6K2, that induces endomembrane rearrangements for the formation of viral replication factories. These factories take the form of vesicles that contain viral RNA (vRNA) and viral replication proteins. In this study, we report the presence of 6K2-tagged vesicles containing vRNA and the vRNA-dependent RNA polymerase in phloem sieve elements and in xylem vessels. Transmission electron microscopy observations showed the presence in the xylem vessels of vRNA-containing vesicles that were associated with viral particles. Stem-girdling experiments, which leave xylem vessels intact but destroy the surrounding tissues, confirmed that TuMV could establish a systemic infection of the plant by going through xylem vessels. Phloem sieve elements and xylem vessels from Potato virus X-infected plants also contained lipid-associated nonencapsidated vRNA, indicating that the presence of membrane-associated ribonucleic protein complexes in the phloem and xylem may not be limited to TuMV. Collectively, these studies indicate that viral replication factories could end up in the phloem and the xylem.
Plant viruses use the host preexisting transport routes to propagate infection to the whole plant. After replication in the initially infected cells, viruses move cell to cell through plasmodesmata (PD) and start a new round of replication in the newly infected cells. This cycle is repeated until viruses reach vascular tissues, where they enter into the conducting tubes for systemic movement. Several studies have indicated that plant viruses are passively transported along the source-to-sink flow of photoassimilates and thus are believed to move systemically through the phloem (for review, see Hipper et al., 2013).
The conducting tube of the phloem is the sieve element. The mature sieve element is enucleated and relies on the associated companion cells for the maintenance of its physiological function (Fisher et al., 1992). The specialized PD connecting one sieve element with one companion cell is called the pore plasmodesmal unit (PPU). Different from the other PDs, PPUs are always branched on the companion cell side but have only one channel on the sieve element side (Oparka and Turgeon, 1999). It is believed that the loading and uploading of viral material during phloem transport are through PPUs. Even though the size exclusion limit of PPUs (Kempers and Bel, 1997) is larger than that of the other PDs (Wolf et al., 1989; Derrick et al., 1990), PPUs should not allow, in their native state, virions or viral ribonucleoprotein (vRNP) complexes to pass through. It is thus believed that specific interactions between virus and host factors are required to allow the viral entity to go through. For instance, the movement protein of Cucumber mosaic virus (CMV) is targeted to PPUs (Blackman et al., 1998), suggesting that this viral protein modifies the size exclusion limit of PPUs and helps viral entry into sieve elements.
Most plant viruses are assumed to move systemically through the phloem as virions. This assumption is based on the observation that Coat Protein (CP) deletions debilitating virus assembly prevent systemic infection (Brault et al., 2003; Zhang et al., 2013; Hipper et al., 2014). Some investigations showed the actual presence of virions in sieve elements. This is the case for the icosahedral Tobacco ringspot virus (Halk and McGuire, 1973), Carrot red leaf virus (Murant and Roberts, 1979), Potato leaf roll virus (Shepardson et al., 1980), and Beet western yellows virus (Hoefert, 1984). In addition, virions also were observed in phloem sap, such as the icosahedral CMV (Requena et al., 2006) and the rigid rod-shaped Cucumber green mottle mosaic virus (Simón-Buela and García-Arenal, 1999). Some viruses also are believed to move as ribonucleic protein (RNP) complexes, since systemic movement was observed in CP mutants where virion assembly was hindered. For instance, Tobacco rattle virus, Potato mop-top virus, Brome mosaic virus, and Tomato bushy stunt virus can still move systemically when the CP gene has been deleted from the viral genome (Swanson et al., 2002; Savenkov et al., 2003; Gopinath and Kao, 2007; Manabayeva et al., 2013). For potyviruses, it is still not clear if long-distance transport involves exclusively viral particles or if vRNP complexes also are implicated (Dolja et al., 1994, 1995; Cronin et al., 1995; Schaad et al., 1997; Kasschau and Carrington, 2001; Rajamaki and Valkonen, 2002). But whether virions or vRNP complexes are involved in viral movement, the full nature of the viral entity being implicated has not been defined.
Xylem also is used for systemic infection of viruses, but its importance in viral transport generally has been overlooked. Vessel elements are the building blocks of xylem vessels, which constitute the major part of the water-upward-transporting system in a plant. The side walls of mature vessel elements contain pits, which are areas lacking a secondary cell wall; the end walls of the mature vessel elements are removed, and the openings are called perforation plates (Roberts and McCann, 2000). CP or virions of some plant viruses of all different shapes have been detected in the xylem vessels and/or guttation fluid, suggesting that these viruses may move systemically through xylem vessels. For example, the CP of the icosahedral Tomato bushy stunt virus (Manabayeva et al., 2013) and Rice yellow mottle virus (Opalka et al., 1998), the CP of the rigid rod-shaped Soilborne wheat mosaic virus (Verchot et al., 2001) and Cucumber green mottle mosaic virus (Moreno et al., 2004), and the flexuous rod-shaped Potato virus X (PVX; Betti et al., 2012) were detected in xylem vessels. Colocalization of anti-Rice yellow mottle virus antibodies and a cell wall marker for cellulosic β-(1-4)-d-glucans over vessel pit membranes suggests that the pit membranes might be a pathway for virus migration between vessels (Opalka et al., 1998). Moreover, flexuous rod-shaped virions of Zucchini yellow mosaic virus were found in both xylem vessels of root tissue and the guttation fluid (French and Elder, 1999). Finally, icosahedral Brome mosaic virus (Ding et al., 2001) and rigid rod-shaped Tomato mosaic virus and Pepper mild mottle virus (French et al., 1993) virions were found in guttation fluid. Guttation fluid originates from xylem exudate, indicating that these plant viruses can move through xylem within the infected plant. The above studies, however, mainly relied on electron microscopy and infection assays and may have missed the presence of other viral components that might be involved in transport.
Turnip mosaic virus (TuMV) is a positive-strand RNA virus belonging to the family Potyviridae, genus Potyvirus, which contains around 30% of the currently known plant viruses and causes serious diseases in numerous crops (Shukla et al., 1994). Potyviruses are nonenveloped, flexuous rod-shaped particles of 680 to 900 nm in length and 11 to 13 nm in diameter. The genomic approximately 10-kb RNA encodes a polyprotein, which is processed into at least 11 mature proteins. TuMV remodels cellular membranes into viral factories, which are intracellular compartments involved in viral replication and movement. These compartments take the form of vesicles of approximately 100 nm in diameter originating from the endoplasmic reticulum (Grangeon et al., 2012). These vesicles contain viral RNA (vRNA) and viral and host proteins involved in vRNA replication (Beauchemin et al., 2007; Beauchemin and Laliberté, 2007; Dufresne et al., 2008; Huang et al., 2010; Grangeon et al., 2012). The viral membrane 6K2 protein is involved in the membrane alterations and vesicle production (Beauchemin et al., 2007). The membrane-bound replication complexes can move intracellularly and cell to cell (Grangeon et al., 2013) at a rate of one cell being infected every 3 h (Agbeci et al., 2013). Intercellular trafficking of the replication complex is likely mediated by the PD-localized potyviral proteins Cytoplasmic Inclusion (CI) and P3N-PIPO (for N-terminal Half of P3 fused to the Pretty Interesting Potyviridae ORF; Carrington et al., 1998; Wei et al., 2010; Vijayapalani et al., 2012) as well as CP (Dolja et al., 1994, 1995), Viral Protein genome-linked (VPg; Nicolas et al., 1997; Rajamaki and Valkonen, 1999, 2002), and Helper Component-Proteinase (HC-Pro; Cronin et al., 1995; Kasschau et al., 1997; Rojas et al., 1997; Kasschau and Carrington, 2001), which are involved in both cell-to-cell and vascular movement.
It is expected that, ultimately, TuMV reaches the vascular tissues of the plant, but how and under what form it is released into the conducting tubes are not known. To further understand viral spread and systemic movement, we investigated the distribution of 6K2-tagged TuMV factories in all of the leaf and stem tissues other than the epidermal cells. We found TuMV factories in all tissues. Interestingly, we observed 6K2-tagged vesicles, containing vRNA and viral replication proteins, in both phloem sieve elements and xylem vessels. We confirmed that TuMV could move systemically through xylem by a so-called stem-girdling assay, which induces cell death of the phloem without affecting xylem integrity. Hence, our study indicates that membrane-associated TuMV replication complexes are involved in the systemic movement of the virus.
RESULTS
TuMV Factories Are Present in All Types of N. benthamiana Leaf and Stem Cells
TuMV factories in N. benthamiana epidermal cells are characterized by the presence of numerous 100-nm 6K2-tagged vesicles containing replication complexes, many of them aggregating to form a large irregularly shaped globe-like structure near the nucleus (Grangeon et al., 2012). Different types of cells have different structures and functions. Hence, it is of interest to investigate how TuMV factories appear in the other types of plant cells in addition to epidermal cells. To do this, we improved a cryohistological protocol (Knapp et al., 2012) to view TuMV factories in different types of cells. Instead of vacuum infiltration, the fixative was infiltrated in the leaf with a 3-mL syringe. Additionally, glutaraldehyde autofluorescence was quenched with Gly, allowing the glutaraldehyde concentration to be increased from 0.1% to 1%. This increase helped to maintain the integrity of the sections, especially for the fragile and large longitudinal stem sections.
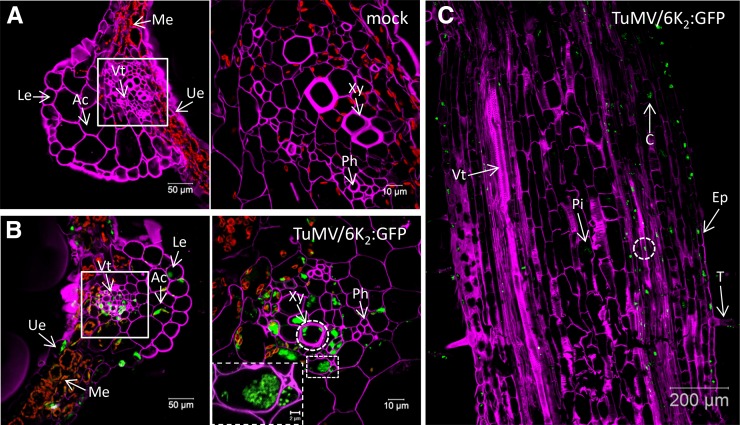
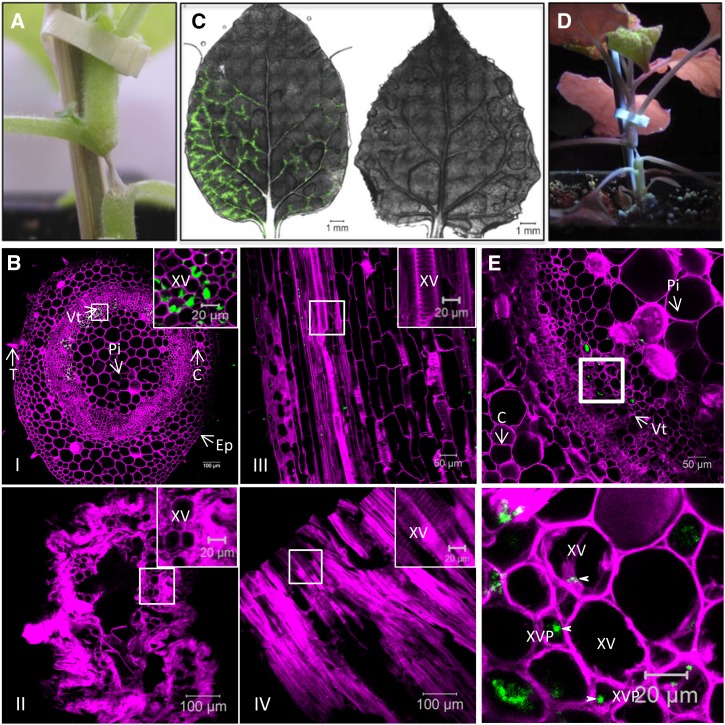
N. benthamiana plants were agroinfiltrated with the TuMV infectious clone pCambiaTuMV/6K2:GFP or with the mock empty vector pCambia 0390, and 6 d later the systemically infected and mock-infected leaf tissues were cryosectioned into 30-µm-thick cross-section samples and processed for confocal microscopy observation. To distinguish the different types of cells, the cell wall was labeled with Fluorescent Brightener 28 dye, which binds (1→4)-β-linked d-glucopyranosyl units (Wood, 1980). As shown in Figure 1A (left), the lower and upper epidermal cells, mesophyll cells, angular collenchyma cells, and vascular tissue were easily distinguishable. Notably, the chloroplasts (shown in red) are predominantly located in the mesophyll cells. At higher magnification (right), we could also distinguish the xylem and phloem easily. 6K2:GFP was present in all types of leaf cells of infected plants (Fig. 1B). We also detected 6K2:GFP-tagged signals in xylem vessels (Fig. 1B, right, dashed circle), suggesting that TuMV systemic movement involves these conducting tube elements. A closeup view of two vascular parenchyma cells (Fig. 1B, right, bottom dashed rectangle) showed that 6K2:GFP-tagged structures are approximately 0.5-µm vesicles. This is consistent with what is observed in live epidermal cells (Grangeon et al., 2012), which indicates that tissue fixation did not produce any apparent artifactual structures.
Figure 1.
Distribution of TuMV 6K2:GFP in Nicotiana benthamiana leaf and stem tissues. A and B, Cross sections of mock-infected (A) and TuMV/6K2:GFP systemically infected (B) N. benthamiana leaf midrib were observed by confocal microscopy. Samples were analyzed with a Zeiss LSM-780 confocal microscope using a 20× objective (left). White squares indicate the vascular tissue region observed with a 63× objective shown at right. The dashed rectangle at right in B shows a closeup view of two vascular parenchyma cells. C, Longitudinal section of a TuMV/6K2:GFP-infected N. benthamiana stem internode above the inoculated leaf imaged by confocal tile scanning. The tile scan was carried out by assembly two × four images with a 20× objective. The dashed circles indicate the presence of 6K2:GFP in xylem vessels. The Fluorescent Brightener 28-stained cell wall is shown in false-color magenta, 6K2:GFP is shown in green, and chloroplasts are shown in red. All of the images are single optical slices. Ac, Angular collenchyma cells; C, cortex; Ep, epidermal cells; Le, lower epidermis; Me, mesophyll cells; Ph, phloem; Pi, pith cells; T, trichome; Ue, upper epidermis; Vt, vascular tissue; Xy, xylem.
To further investigate if 6K2:GFP-tagged TuMV factories are found in vascular conducting tubes, we processed stem internodes just above the inoculated leaf at 6 d postinoculation to obtain 30-µm longitudinal section samples for confocal microscopy observation. In TuMV-infected stem, the 6K2:GFP-tagged TuMV factories were distributed in trichomes, epidermal cells, cortex, vascular tissue, and pith cells (Fig. 1C, arrows). As for leaf cross sections, we detected 6K2:GFP-tagged signals in xylem vessels (Fig. 1C, dashed circle; see below).
6K2-Associated Membrane Complexes Are Present in Sieve Elements
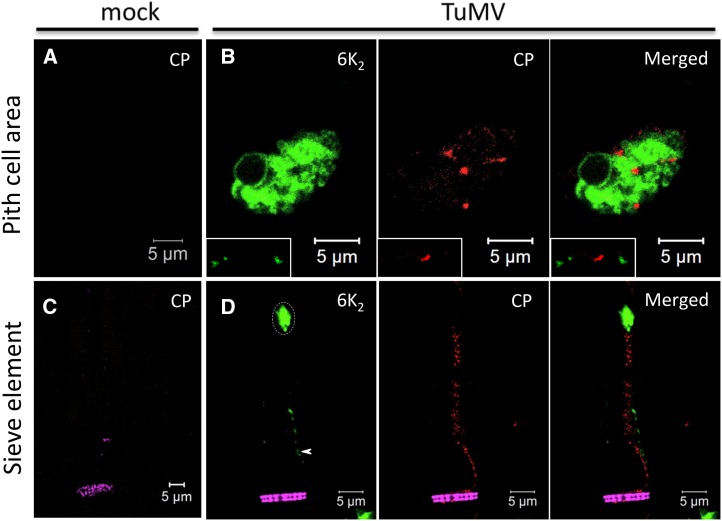
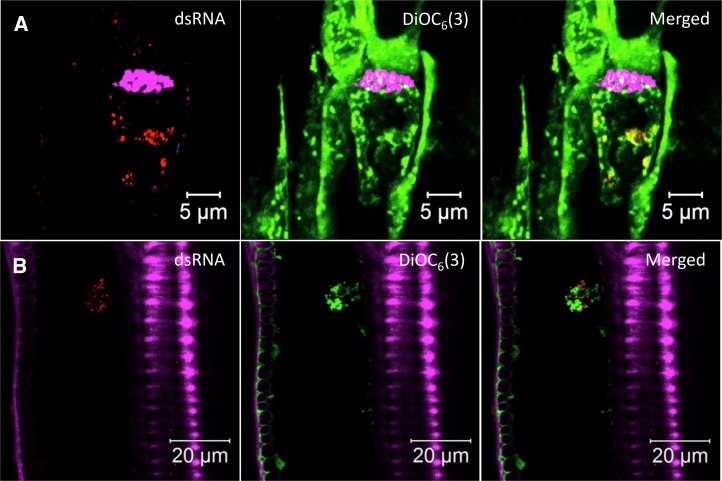
To examine more specifically the presence of 6K2:GFP in the phloem, sieve elements were highlighted by staining callose in the sieve plates with Aniline Blue (Evert, 1977; Koh et al., 2012). As shown in Figure 2A, the mock-infected stem longitudinal section was stained with Aniline Blue and the lipophilic dye 3,3′-dihexyloxacarbocyanine iodide [DiOC6(3)], which at high concentration stains all intracellular membranes in fixed cells (Terasaki and Reese, 1992). The Aniline Blue staining (shown in false-colored magenta) clearly highlighted the sieve plate (Fig. 2A, arrow) and labeled PPUs (Fig. 2A, dashed arrow) of one sieve element. DiOC6(3)-stained ring-like structures of about 1 µm in diameter were located at the parietal layer of sieve elements (Fig. 2A, arrowheads), which are sieve element plastids (Froelich et al., 2011). In TuMV-infected stems, 6K2:mCherry (shown in false-colored green) was observed as large aggregates (dashed ovals) throughout the sieve tube elements, and the aggregates contained lipids, as they were labeled with the lipophilic dye DiOC6(3) (Fig. 2B, shown in false-colored red). 6K2-labeled ring-like structures also were observed (Fig. 2B, arrowheads), and these may represent sieve element plastids whose membrane had incorporated the viral protein.
Figure 2.
Phloem membrane aggregates contain viral replication complexes. Longitudinal sections of mock-infected (A, C, and E), TuMV/6K2:mCherry-infected (B), and TuMV/6K2:GFP-infected (D and F) N. benthamiana stem internodes above the inoculated leaf were observed with a Zeiss LSM-780 confocal microscope using a 63× objective. Aniline Blue-stained sieve plates are shown in false-color magenta, DiOC6(3)-stained lipids are shown in green in A and in red in the middle and right parts of B, and 6K2:mCherry has been false colored in green. Immunohistolocalization of dsRNA (C and D) and vRdRp (E and F) was performed on mock-infected (C and E) and TuMV/6K2:GFP-infected (D and F) longitudinal sections. 6K2:GFP is shown in green, and dsRNA and vRdRp are shown in red. Dashed ovals highlight the presence of 6K2 vesicle aggregates, and arrowheads point to the ring-like structures in sieve elements. Images are single optical slices. Sp, Sieve plate.
To test whether the 6K2 aggregates might be replication complexes, we performed immunohistolocalization with the double-stranded RNA (dsRNA)-specific monoclonal antibody J2, which specifically recognizes dsRNA of more than 40 bp in length in a sequence-nonspecific manner (Schönborn et al., 1991). No dsRNA signal was observed in noninfected samples (Fig. 2C). In infected samples, dsRNA was detected in the 6K2 aggregates (Fig. 2D, dashed oval) but not within the sieve element plastids (Fig. 2D, arrowheads). We also performed immunohistolocalization with a rabbit serum against the TuMV viral RNA-dependent RNA polymerase (vRdRp; Dufresne et al., 2008). No vRdRp signal was observed in mock samples (Fig. 2E). In the sieve elements of infected samples, vRdRp was associated with 6K2 aggregates (Fig. 2F, dashed ovals) but not with the 6K2-tagged sieve element plastids (Fig. 2F, arrowheads). Thus, 6K2-associated membrane complexes were present in phloem sieve elements and contained vRNA and vRdRp, suggesting that viral replication complexes were present in these conducting vessels.
Moreover, we performed immunohistolocalization with a rabbit serum against TuMV CP (Cotton et al., 2009). No CP signal was observed in noninfected samples (Fig. 3A). TuMV CP has been shown not to localize with dsRNA punctate structures in infected N. benthamiana protoplasts (Cotton et al., 2009). Likewise, CP was localized mainly at the periphery of 6K2 aggregates, as punctate and sometimes elongated structures, and was not associated with cortical 6K2 vesicles, as shown here for cells of the pith cell area (Fig. 3B, bottom left rectangle). These CP signals are likely TuMV virions, which have been observed adjacent to vesicle aggregates by transmission electron microscopy (TEM; J. Wan, unpublished data). We also detected CP in sieve elements (Fig. 3D), but it was not associated with 6K2 aggregates (dashed oval) or with sieve element plastids (arrowhead). Thus, the CP signal in sieve elements likely represents TuMV virions.
Figure 3.
CP distribution in TuMV-infected cells. Immunohistolocalization of CP was performed on mock-infected (A and C) and TuMV/6K2:GFP-infected (B and D) longitudinal sections of N. benthamiana stem internodes. Sections were observed with a Zeiss LSM-780 confocal microscope using a 63× objective. CP signal was detected in TuMV-infected pith cell area (B) as well as in sieve elements (D). The rectangles in B show 6K2 and CP at the cell periphery. Aniline Blue-stained sieve plates are shown in false-color magenta, 6K2:GFP is shown in green, and CP is shown in red. The images are single optical slices. The dashed oval highlights the presence of 6K2 vesicle aggregates, and the arrowhead points to the ring-like structures in sieve elements.
6K2-Associated Membrane Complexes Are Present in Xylem Vessels
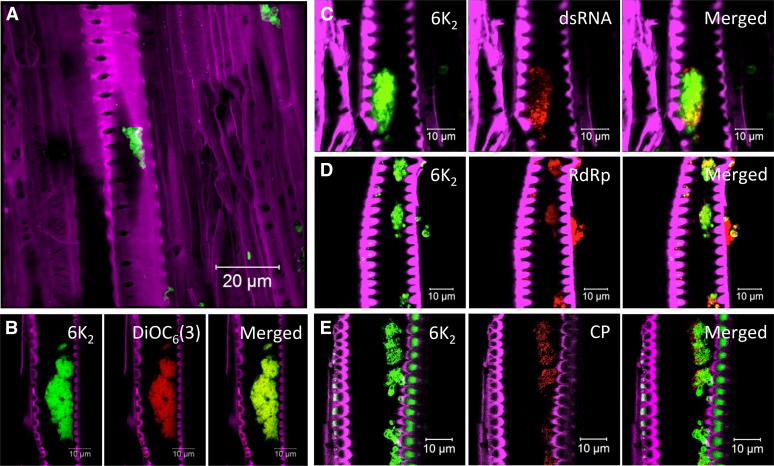
As described above, leaf cross sections pointed to the possible presence of 6K2:GFP in xylem vessels (Fig. 1B). Longitudinal sections of stem internodes just above the inoculated leaf confirmed the presence of 6K2:GFP in xylem vessels (Fig. 4A). Xylem vessels were easily discernible due to their characteristic perforated cell wall. The presence of 6K2:GFP within xylem vessels was not an experimental spillover contamination resulting from tissue damage during the sectioning process, as optical sectioning showed that 6K2:GFP-tagged aggregates were truly located inside the xylem vessels (see the three-dimensional reconstruction of an infected xylem vessel in Supplemental Movie S1). Moreover, 6K2:mCherry-tagged aggregates contained lipids, as they were labeled with the lipophilic dye DiOC6(3) (Fig. 4B).
Figure 4.
TuMV replication complexes in xylem vessel. Longitudinal sections of N. benthamiana stem internodes infected with TuMV expressing 6K2 fused to either GFP (A and C–E) or mCherry (B) were observed with a Zeiss LSM-780 confocal microscope with a 63× objective. Lipids were stained with the membrane dye DiOC6(3) (B). Immunohistolocalization of dsRNA (C), vRdRp (D), and CP (E) was performed as described in Figure 3. The Fluorescent Brightener 28-stained cell wall is shown in false-color magenta, 6K2:GFP is shown in green, 6K2:mCherry has been false colored in green, and DiOC6(3) has been false colored in red. dsRNA, vRdRp, and CP are shown in red. A is a three-dimensional image; the other confocal images are single optical slices.
Xylem vessels are dead cells normally used for water transportation. This raises the question of whether the 6K2:GFP aggregates are active for viral infection. To answer this question, we looked for the presence of dsRNA/vRNA, vRdRp, and CP via immunohistolocalization in the 6K2:GFP aggregates. dsRNA, vRdRp, and CP (Fig. 4, C–E) were detected in the 6K2:GFP aggregates. As for sieve elements (Figs. 2, C and E, and 3, A and C), no signal for the presence of dsRNA, vRdRp, and CP was observed in mock samples (data not shown). Taken together, these observations indicate that 6K2-associated membrane complexes in xylem vessels contain vRNA and viral replication and movement proteins (i.e. CP).
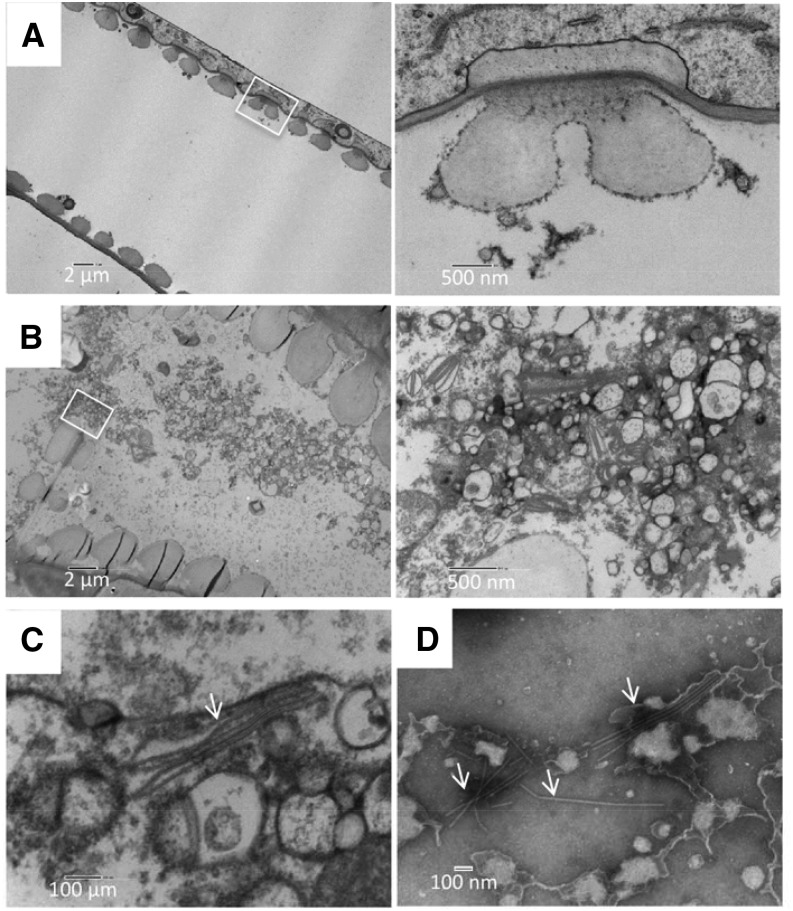
We then examined the ultrastructure of TuMV aggregates in xylem vessels by TEM. At 6 d postinoculation, TuMV-infected and mock-infected stem internodes just above the inoculated leaf were fixed, dehydrated, and embedded in Epon for structure observation or in LR White for immunogold labeling. In mock-infected stem, the xylem vessels contained only a few electron-opaque materials (Fig. 5A). In contrast, in TuMV-infected stem, the xylem vessels were full of vesicular structures, especially in the perforation plates between two xylem vessel elements (Fig. 5B), indicating that the vesicles might be moving through these pore structures. Interestingly, under higher magnification, we detected TuMV virion-like structures that were associated with the membrane vesicles (Fig. 5C, arrow). The presence of TuMV particles was confirmed by negative staining with 2% (w/v) phosphotungstate of the collected xylem sap (Fig. 5D, arrows).
Figure 5.
Ultrastructure of TuMV-induced membrane alterations in xylem vessels. A and B, Longitudinal sections of mock-infected (A) and TuMV-infected (B) N. benthamiana stem internodes above the inoculated leaf were collected and observed by TEM. White squares indicate the areas that are shown at higher magnification at right. The higher magnification of the white square in B shows TuMV viral factories located in the perforation plate between two xylem vessel elements. C, TuMV virions (arrow) associated with vesicles. D, Xylem sap from TuMV-infected N. benthamiana was collected and observed by TEM following negative staining. TuMV virions are highlighted by arrows.
To further confirm the content of the membrane structures in xylem vessels, we performed immunogold labeling using the anti-dsRNA monoclonal antibody. Very few gold particles were found in xylem parenchyma cells (Fig. 6A) and xylem vessels (Fig. 6B) on a mock-infected section treated with primary antibody J2 and secondary antibodies. The detection of inclusion bodies and membrane vesicles in partially differentiated xylem vessels indicated that this cell was infected by TuMV (Fig. 6C). Under higher magnification, we detected gold particles (arrows) in the membrane vesicles in partially differentiated (Fig. 6C) and also in fully differentiated xylem vessels (Fig. 6D). Quantification of the gold particles per µm2 and relative labeling distribution were performed over mock-infected and TuMV-infected sections. The results revealed that, compared with TuMV-infected sections (0.42 ± 0.04 particles μm−2), the mock-infected sections did not show any significant labeling (0.07 ± 0.03 particles μm−2; Fig. 6E). In TuMV-infected sections, the gold particles were mainly found decorating TuMV-induced vesicles (76.25%). Only a few gold particles were found over the rest of the cytoplasm, corresponding to the nonspecific background signal from the antibody (Fig. 6F). Thus, TuMV virions in xylem vessels are associated with vesicular complexes that contain TuMV RNA.
Figure 6.
Immunoelectron microscopy localization of dsRNA in TuMV-infected xylem vessels. A to D, Labeling was performed by using the anti-dsRNA monoclonal antibody J2 on longitudinal sections of mock-infected (A and B) and TuMV-infected (C and D) N. benthamiana stem internodes. White squares indicate the areas that are shown at higher magnification at right. Arrows point to gold particles in vesicles. E and F, Number of gold particles per µm2 in mock-infected versus TuMV-infected cells (E) and relative labeling distribution in infected cells (F). Two different labeling experiments were considered, and 200 gold particles were counted per experiment. chl, Chloroplast; cw, cell wall; cyt, cytosol; ER, endoplasmic reticulum; In, inclusion bodies; mit, mitochondrion; N and nuc, nucleus; pm, plasma membrane; PW, primary cell wall; SW, secondary cell wall; vac, vacuole; Ve and ves, vesicles; XV, xylem vessel; XVP, xylem parenchymal cell.
Xylem Sap from TuMV-Infected Plants Contains Eukaryotic Initiation Factor (iso)4E and Is Infectious
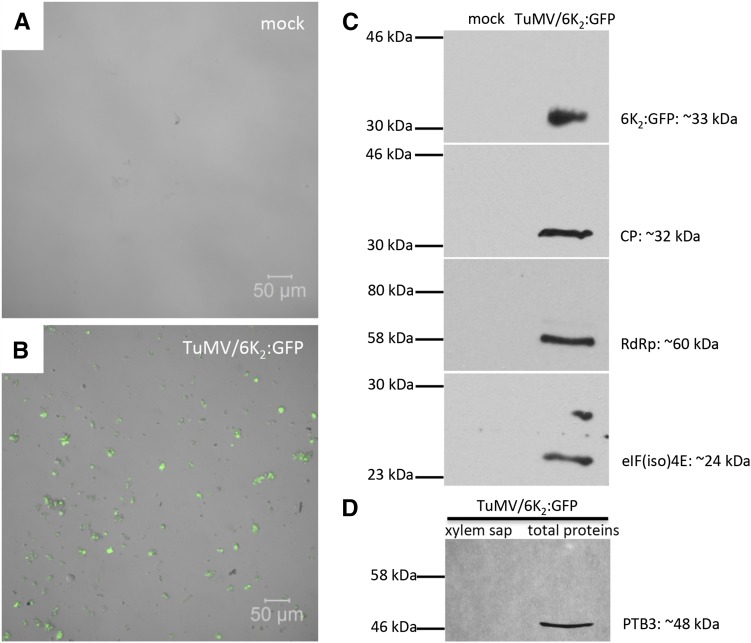
Analysis of xylem sap by confocal microscopy confirmed our previous finding in the fixed stem tissues. Using phase contrast, the xylem sap from mock-infected plants was essentially devoid of cellular material (Fig. 7A). In contrast, the xylem sap from TuMV-infected plants contained a large number of what appears to be vesicles, many of which were positively labeled with 6K2:GFP (Fig. 7B). We then looked for the presence of viral proteins and for the host translation eukaryotic initiation factor (iso)4E [eIF(iso)4E] in xylem sap by western-blot analysis. eIF(iso)4E was chosen because it has been shown to be part of the TuMV replication complex (Beauchemin et al., 2007). As expected, the viral proteins 6K2:GFP, CP, and vRdRp, as well as eIF(iso)4E, were detected in the xylem sap from TuMV-infected plants but not from mock-infected plants (Fig. 7C). Since eIF(iso)4E is an intracellular protein, its absence in mock-infected xylem sap indicates that the presence of the above proteins in infected xylem sap was not the consequence of contaminating cellular breakage during sap collection. As an indicator of phloem contamination in the collected xylem sap, we looked for the presence of Polypyrimidine Tract-Binding Protein3 (PTB3), a homolog to the pumpkin (Cucurbita maxima) phloem-mobile ribonucleoprotein complex RBP50 (Ham et al., 2009). This protein was detected in a total protein extract but not in an infected xylem sap from TuMV-infected plants (Fig. 7D), suggesting that there was no apparent contamination from companion cells and phloem exudates. The xylem sap from TuMV-infected plants also was tested for the infection of healthy N. benthamiana plants by mechanical inoculation. After 5 d, we detected TuMV systemic infection under UV light (data not shown), which indicated that the xylem sap was infectious. In conclusion, the xylem sap from TuMV-infected plants contains 6K2 vesicles and viral as well as host protein that are essential for TuMV replication and assembly.
Figure 7.
Western-blot protein analysis of xylem sap. A and B, Xylem sap was collected from mock-infected (A) and TuMV/6K2:GFP-infected (B) N. benthamiana plants and was observed by confocal microscopy. 6K2:GFP is shown in green. C, Western-blot analysis of xylem sap from mock- and TuMV-infected plants using rabbit sera against GFP, CP, vRdRp, or eIF(iso)4E. D, Western-blot analysis of xylem sap and total protein leaf extract from TuMV-infected plants using a rabbit serum against the phloem-specific PTB3 protein.
Stem Girdling Confirms That TuMV Can Spread Systemically through the Xylem
To further substantiate that xylem vessels are involved in TuMV systemic movement, we performed a stem-girdling experiment. N. benthamiana plants were agroinfiltrated with pCambiaTuMV/6K2:GFP. At 1 h following agroinfiltration with pCambiaTuMV/6K2:GFP, the stem internode just above the agroinfiltrated leaf was treated with a jet of steam for 15 s, until water soaking became apparent (Moreno et al., 2004). As a consequence, a segment of about 5 to 10 mm long was reduced to a very thin strand within 1 h and showed dry necrosis 1 d later (Fig. 8A). Since xylem vessels are dead tissues surrounded by a thick cell wall, their structural integrity should not be affected by the steam treatment. To confirm that this was the case, cross and longitudinal sections of girdled stems were examined by confocal microscopy, which showed that all cells had collapsed except for the xylem vessels (Fig. 8B). To further confirm that the phloem was totally destroyed after steam treatment, we used the phloem-loading dye 5-(and 6)-carboxyfluorescein diacetate (CFDA). It was reported previously that the phloem transport of CFDA was interrupted when phloem was destroyed following stem girdling (Grignon et al., 1989). Thus, CFDA was applied to an abraded source leaf below the girdled stem section, and its movement into upper sink leaves was assessed 6 and 24 h later by confocal microscopy. As expected for phloem movement, CFDA clearly marked the class I, II, and III vein network of a young sink leaf of nongirdled plants 6 h after CFDA application (Fig. 8C, left). On the other hand, no CFDA labeling was observed in young sink leaves of girdled plants (n = 10) 6 and 24 h after CFDA application (Fig. 8C, right). Normally, in TuMV-agroinfiltrated leaves, cell-to-cell movement starts at 3 d postinoculation and systemic infection is detected at 5 d postinoculation. Here, TuMV/6K2:GFP infection was observed above the steam-treated section by UV light between 7 and 10 d (Fig. 8D; Table I), suggesting that xylem vessels also are used by TuMV for its systemic infection, although at a slower rate. At the first indication of systemic infection taking place in girdled plants, we collected stem cross sections just above the girdled part for histological analysis. Figure 8E shows that 6K2:GFP was found only in vascular tissues at this early stage. Under higher magnification, we observed the 6K2:GFP-tagged vesicles in xylem vessels and xylem parenchymal cells (arrowhead) as well as other xylem-associated cells, indicating that once TuMV passed the girdled stem through xylem vessels, it can be unloaded in the xylem parenchymal cells just above the girdled stem.
Figure 8.
TuMV infection following the stem-girdling experiment. A, Closeup view of an N. benthamiana stem internode that was treated with steam for 15 s the day before. The internode is above the leaf that was infiltrated with A. tumefaciens containing pCambiaTuMV/6K2:GFP 1 h before the steam treatment. B, Cross sections and longitudinal sections of TuMV-infected stem internode above the inoculated leaf were collected by cryosectioning and observed by confocal microscopy. Cross-sections of TuMV-infected stem internode are shown before (I) and after (II) steam treatment. Longitudinal sections of TuMV-infected stem internode are shown before (III) and after (IV) steam treatment. Top right squares show at a high magnification the xylem vessels and surrounding cells before steam treatment (I and III) and intact xylem vessels and dead surrounding cells after steam treatment (II and IV). C, Upper young leaves of stem nongirdled (left) and girdled (right) plants in which the source leaves were treated with CFDA. D, The steam-treated plant from A shows TuMV systemic infection under UV light at 10 d after agroinoculation. E, A stem cross-section just above the girdled stem section at a very early stage of systemic infection. The white square indicates the area of higher magnification shown at bottom. C, Cortex; Ep, epidermal cell; Pi, pith cells; T, trichome; Vt, vascular tissue; XV, xylem vessel; XVP, xylem parenchymal cell.
Table I. Analysis of TuMV infection in steam-treated N. benthamiana plants.
N, Number of inoculated plants; p.i., postinoculation; TuMV steamed 1/2/3/4, four separate experiments.
Membrane-Associated vRNA of PVX Is Present in Both Phloem and Xylem
The observations that TuMV factories are present in both phloem and xylem led us to ask whether this is a unique feature of potyviruses. PVX virions can enter, move through, and exit the phloem, as demonstrated by grafting experiments (Cruz et al., 1998). In addition, the CP of PVX was detected in xylem vessels (Betti et al., 2012). We thus analyzed the vascular tissues of PVX-infected plants. N. benthamiana plants were agroinfiltrated with the PVX infectious clone PVX-GFP (Peart et al., 2002), and 8 d later the stem internodes just above the inoculated leaf were processed to collect 30-µm longitudinal section samples for confocal microscopy observation. Sieve elements were stained with Aniline Blue and xylem vessels were stained with Fluorescent Brightener 28, and both are shown in magenta. We detected dsRNA in the sieve elements via immunohistolocalization, which was associated with lipids (Fig. 9A), suggesting that those structures could be PVX replication complexes. A similar pattern was observed in the xylem vessels of PVX-infected plants (Fig. 9B). Thus, PVX RNA was present in both phloem and xylem and was associated with lipids.
Figure 9.
Membrane-associated vRNA in PVX-infected sieve elements and xylem vessels. Longitudinal sections of PVX-infected N. benthamiana stem internode above the inoculated leaf were collected by cryosectioning and observed by confocal microscopy. Immunohistolocalization of dsRNA and lipid staining with DiOC6(3) in one sieve element (A) and one xylem vessel (B) of a PVX-infected plant are shown. Aniline Blue-stained callose and Fluorescent Brightener 28-stained cell wall are shown in false-color magenta, DiOC6(3) is shown in green, and dsRNA is shown in red. All images are single optical slices.
DISCUSSION
Viruses use the vascular tissues, in particular the phloem, for the systemic infection of plants. Viral particles are apparently the transmission agent responsible for transmitting the disease in the whole plant, but vRNP complexes also have been implicated. The nature of this trafficking vRNA complex, however, has not been defined.
In this study, we showed the presence of membrane-associated complexes centered on the 6K2 protein of TuMV, which also contained vRNA and vRdRp, in sieve elements and xylem vessels. Since 6K2 is detected as a 6K2-VPg-Proteinase (Pro) fusion in infected cells (Léonard et al., 2004), it is likely that VPg and Pro also are present. The vRNA associated with the 6K2:GFP aggregates is probably the replicative form of the vRNA, since the dsRNA punctate structures observed in xylem vessels are identical to those observed in the cytoplasm of infected epidermal cells. Moreover, the dsRNA recognized by the monoclonal antibody is not likely to be encapsidated vRNA, since the immunohistolocalization protocol does not involve protease treatment, and gold particles are associated with vesicles and not viral particles in immunogold electron microscopy images following incubation with the dsRNA monoclonal antibody. The vesicles in sieve elements and xylem vessels are reminiscent to those found in the cytoplasm of TuMV (Grangeon et al., 2012) and are very similar to other plant and mammalian virus-infected cells (for review, see Miller and Krijnse-Locker, 2008; Laliberté and Sanfaçon, 2010). These intracellular vesicles are the building blocks of viral factories, which contain viral replication complexes. In the case of TuMV, these same vesicles also are the vehicles for the intracellular and intercellular movement of vRNA (Grangeon et al., 2012, 2013). It thus appears that the TuMV RNP complexes found in vascular conducting tubes are, in fact, membrane-associated viral replication complexes. The observation that the host translation initiation factor eIF(iso)4E, which is part of the potyvirus replication complex (Beauchemin et al., 2007), was found in infected xylem sap supports the idea that viral factories end up in vascular conducting tubes. It will be interesting to see if CI and P3N-PIPO are involved in phloem loading from the companion cells and if they are associated with the 6K2 structures found in sieve elements and xylem vessels. Consequently, the long-distance transducing agent for TuMV is likely to contain membrane-bound replication complexes closely associated with particles. The presence of viral factories in the phloem and xylem may not be limited to TuMV, since membrane-associated, nonencapsidated vRNA also was observed in PVX-infected plants.
To our knowledge, the possibility that viral replication complexes could end up in, and could travel through, the phloem and the xylem has never been considered. Limited evidence for their presence, on the other hand, can be found in some previous investigations. For instance, the membrane association of Turnip yellow mosaic virus and CMV particles in xylem has been reported (Hitchborn and Hills, 1965). Likewise, Potato virus Y particles along with cytoplasmic inclusions as well as vesicles were documented in xylem vessels (Otulak and Garbaczewska, 2009, 2012). Finally, large aggregates of Triple Gene Block Protein3 (TGBp3) of CMV were found in sieve elements and were associated with CMV particles (Blackman et al., 1998). Recently, TGBp2 and TGBp3 of PVX were shown to induce the formation of endoplasmic reticulum-derived vesicles at PD orifices. These vesicles contained the vRdRp and nonencapsidated vRNA (Tilsner et al., 2013). So, it may well be that the TGBp3 aggregates are, in fact, CMV replication complexes associated with viral particles.
Traditionally, the phloem has been considered as a canal for distributing water, nutrients, and hormones essential for the growth and development of various organs of the plant. Recent studies have shown, however, that the phloem has more elaborate functions (for review, see Lucas et al., 2013). For instance, it has been demonstrated that PDs and the phloem sieve tube system function cooperatively for the long-distance delivery of proteins (Xoconostle-Cázares et al., 2000; Aoki et al., 2002; Gómez et al., 2005), mRNA (Sasaki et al., 1998; Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999; Haywood et al., 2005; Roney et al., 2007), and small RNA (Yoo et al., 2004; Buhtz et al., 2008; Lin et al., 2008; Pant et al., 2008). Furthermore, the identification of ribosomal and associated components involved in mRNA translation in the sieve tube system of the cucurbits (Lin et al., 2009; Ma et al., 2010) is challenging the general belief that mature sieve elements lack the capacity for protein synthesis (Ham and Lucas, 2014). Sieve elements thus have the capacity of enclosing intricate structures such as membrane-bound vRNP complexes that might be active in RNA synthesis, as evidenced by the presence of dsRNA, a marker for vRNA replication, in the 6K2 aggregates. This hypothesis could be tested by collecting phloem exudate and subjecting it to in vitro metabolic labeling to see if vRNA synthesis and translation are indeed taking place.
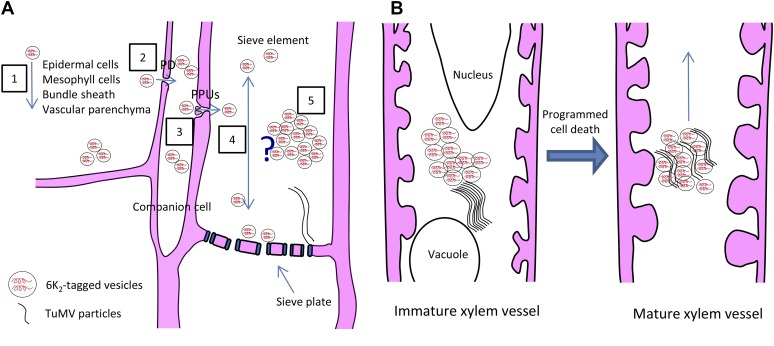
Based on our observation, we suggest a schematic model for the movement of TuMV into small vein sieve elements (Fig. 10A). In this scheme, 6K2-tagged vesicles, which contain vRNA and vRdRp, move from epidermal cells to mesophyll cells, bundle sheath, vascular parenchyma, and companion cells through PDs. Once in companion cells, the 6K2-tagged vesicles are loaded into sieve elements through PPUs. Either individual 6K2 vesicles move through sieve elements or they amalgamate to form a membrane support for virion assembly, which would be released for phloem transport, as proposed by Blackman et al. (1998) for CMV.
Figure 10.
Model for TuMV long-distance movement. A, Schematic model for TuMV moving into small vein sieve elements. First, 6K2-tagged vesicles, which contain vRNA and vRdRp, move from epidermal cells to mesophyll cells, bundle sheath, vascular parenchyma (1), and companion cells through PDs (2). Then, the 6K2-tagged vesicles are loaded into sieve elements through PPUs (3). Once in the sieve element, there are two possibilities. One possibility is that individual 6K2 vesicles move through sieve elements (4). Another possibility is that the 6K2 vesicles amalgamate together to form membrane aggregates, which stay in sieve elements for virion assembly, and the assembled virions are then released for phloem transport (5). B, Schematic model for TuMV movement into xylem vessels. 6K2-tagged vesicles enter into the immature xylem elements through pit membranes and replicate in the cytoplasm before programmed cell death occurs and then move upward in the plant after xylem becomes a hollow vessel.
CP or particles for some viruses have been found in the xylem (French et al., 1993; Andrianifahanana et al., 1997; Opalka et al., 1998; French and Elder, 1999; Ding et al., 2001; Verchot et al., 2001; Moreno et al., 2004; Betti et al., 2012; Manabayeva et al., 2013). This conducting vessel would not be a dead end for infection, since the stem-girdling experiment (Fig. 8) demonstrated that TuMV spreading through xylem vessels caused systemic infection. This investigation has shown that, in addition to viral particles, membrane-bound replication complexes also end up in xylem vessels. TuMV vesicles containing dsRNA were found in immature xylem vessels. This suggests that TuMV replication vesicles may enter into immature xylem vessels and replicate in the cytoplasm before programmed cell death occurs and then move upward in the plant after the xylem becomes a hollow vessel (Fig. 10B), as proposed by Opalka et al. (1998) for Rice yellow mosaic virus. The question that follows is how the replication complex is unloaded from xylem vessels. The fluid and content that are exchanged between xylem vessels and xylem parenchymal cells move through pit membranes. Small and large solutes, such as the membrane-impermeant fluorophores CFDA and Texas Red-labeled dextran (10 kD), are unloaded from xylem via the xylem vessel-xylem parenchyma pit membranes, likely by endocytosis. They are then symplasmically transported through PDs to other parenchyma elements and ultimately to sieve elements (Botha et al., 2008). This may also be the case for TuMV, since 6K2 is observed in xylem parenchymal cells just above the girdled stem section in which only xylem vessels are intact. This has been suggested for Soilborne wheat mosaic virus, which enters and moves systemically through xylem vessels and also laterally between adjacent xylem vessels through pit membranes (Verchot et al., 2001). Displacement of the Ca2+ from pit membranes by some viral factors may contribute to the disruption of the pit membranes and may facilitate systemic virus transport (Opalka et al., 1998).
MATERIALS AND METHODS
Plasmid DNA and Plant Inoculation
Turnip mosaic virus infectious clone was engineered to coproduce 6K2 as a fluorescence protein fusion at its C-terminal end to form the plasmids pCambiaTuMV/6K2:GFP and pCambiaTuMV/6K2:mCherry (Cotton et al., 2009). The 6K2:GFP or 6K2:mCherry coding region was inserted between P1 and HC-Pro cistrons of the TuMV genome. The junctions flanking P1 and HC-Pro contained residues recognized by the P1 and Pro proteinases, respectively. 6K2:GFP or 6K2:mCherry is thus released from the polyprotein when the latter is processed during infection. TuMV infectious clones and the mock empty vector pCambia 0390 were electroporated into Agrobacterium tumefaciens strain AGL1 and selected on Luria-Bertani ampicillin-kanamycin plates. The pellet of an overnight culture was resuspended in water supplemented with 10 mm MgCl2 and 150 μm acetosyringone and kept at room temperature for 2 to 4 h. The solution was then diluted to an optical density at 600 nm of 0.2. Agroinfiltration was performed in 4-week-old Nicotiana benthamiana plants, which were grown under a 16-h-light/8-h-dark photoperiod with 22°C day/20°C night temperatures.
Antibodies
The mouse monoclonal anti-dsRNA J2 antibody (stock solution is 1 mg mL−1; English and Scientific Consulting) was used at the following dilutions: for immunohistolocalization, 1:500; and for immunogold labeling, 1:40. The rabbit polyclonal antibodies were used at the following dilutions for immunolocalization: for anti-vRdRp (Dufresne et al., 2008), 1:50; and for anti-CP (Cotton et al., 2009), 1:50. The dilutions for western-blot analysis were as follows: anti-GFP, 1:20,000; anti-vRdRp, 1:6,000; anti-eIF(iso)4E (Léonard et al., 2004), 1:1,000; anti-CP, 1:2,500; and anti-PTB3 (Rühl et al., 2012), 1:5,000.
For immunohistolocalization, goat anti-mouse and goat anti-rabbit conjugated to Alexa Fluor 568 secondary antibodies (Molecular Probes) were used at a dilution of 1:500. For immunogold labeling, secondary goat anti-mouse antibodies conjugated to 10-nm gold particles (Sigma-Aldrich) were used at a dilution of 1:20. For western-blot analysis, the secondary goat anti-rabbit antibodies conjugated to horseradish peroxidase (Kirkegaard & Perry Laboratories) were used at a dilution of 1:20,000.
Cryohistological Preparation and Immunohistolocalization
Histological preparation and immunohistolocalization were done as described previously (Grangeon et al., 2013). Briefly, one N. benthamiana leaf was agroinfiltrated with mock empty vector or TuMV infectious clones or PVX-GFP (Peart et al., 2002). Systemically infected leaves and stems were fixed at 6 d postinoculation followed by a Suc gradient and cryosectioning as described (Knapp et al., 2012). In addition, before Suc gradient treatment, the samples were treated with 100 mm Gly in phosphate-buffered saline (PBS) for 1 h to reduce the background fluorescence. The sections were collected on Superfrost Plus Microscope Slides (Fisher Scientific) pretreated with 0.01% (w/v) poly-l-Lys (Sigma-Aldrich). After drying for 2 h, the sections were incubated in PBS for 20 min. They were then incubated for 1 h in a blocking solution of 5% (w/v) bovine serum albumin (BSA) and 0.3% (v/v) Triton X-100 in PBS. Thereafter, the sections were incubated for 1 h with the primary antibody in an incubation solution containing 5% (w/v) BSA and 0.05% (v/v) Tween 20 in PBS, washed three times with a washing solution of 0.05% (v/v) Tween 20 in PBS for 10 min, incubated for 1 h with secondary antibody, followed by washing three times with washing solution for 10 min. Both the primary and secondary antibodies were pretreated with mock sections for 1 h to reduce the nonspecific binding. SlowFade Gold (Molecular Probes) was mounted on the sections after they dried, and cover glasses (VWR) were sealed with nail polish.
Fluorescent Brightener 28, Aniline Blue, and DiOC6(3) Staining
Fluorescent Brightener 28 (Calcofluor White M2R; Sigma-Aldrich) staining was done as described previously (Knapp et al., 2012). For staining sieve plate callose with Aniline Blue (Methyl Blue; Sigma-Aldrich), sections were incubated for 15 min in 0.05% (w/v) Aniline Blue in 50 mm sodium phosphate, pH 7.2, and then washed three times with PBS for 10 min. The stock solution and the buffer were mixed just before use. For DiOC6(3) staining (Molecular Probes), DiOC6(3) was used at a final concentration of 1 µm in PBS. Sections were incubated for 30 min in DiOC6(3) solution and washed three times with PBS for 10 min. When Aniline Blue and DiOC6(3) staining were coupled with immunohistolocalization, DiOC6(3) staining was processed after immunohistolocalization, and Aniline Blue staining was the last step. All of the staining steps were performed in the dark.
Confocal Microscopy
The sections were observed using a 20× (numerical aperture = 0.8) objective and a 63× (numerical aperture = 1.4) oil-immersion objective on an LSM-780 confocal microscope (Zeiss). Zen 2011 (Zeiss) was used for image acquisition. Solid-state and argon ion lasers were used for LSM-780 microscope experiments. Excitation/emission wavelengths were 405/410 to 440 nm for Fluorescent Brightener 28 and Aniline Blue, 488/490 to 560 nm for GFP and CFDA, 561/600 to 630 nm for mCherry, 561/570 to 640 nm for Alexa Fluor 568 goat anti-mouse and anti-rabbit IgG, and 561/650 to 700 nm for chloroplasts. Data from blue, green, and red channels were collected separately.
TEM
For TEM, the stems at 6 d postinoculation were fixed in 2.5% (w/v) glutaraldehyde in 0.1 m sodium cacodylate buffer, pH 7.4, postfixed in 1% (w/v) osmium tetroxide with 1.5% (w/v) potassium ferrocyanide in sodium cacodylate buffer, stained with 2% (w/v) tannic acid, dehydrated in a graded series of acetone, and embedded in Epon resin. After polymerization, 90- to 100-nm ultrathin sections were obtained and stained with 4% (w/v) uranyl acetate for 8 min and Reynolds lead citrate for 5 min. Then, the sections were examined in a Tecnai T12 transmission electron microscope (FEI) operating at 120 kV. Images were recorded using an AMT XR80C CCD camera system (FEI).
Immunogold Labeling
The stems obtained at 6 d postinoculation were fixed in 4% (w/v) formaldehyde and 0.25% (w/v) glutaraldehyde in 0.1 m Sorensen’s phosphate buffer, pH 7.4, dehydrated in a graded series of alcohol, and embedded in LR White resin. Ninety- to 100-nm sections were treated with 0.02 m Gly in Dulbecco’s phosphate-buffered saline (DPBS; 137 mm NaCl, 2.7 mm KCl, 1.5 mm KH2PO4, 6.5 mm Na2HPO4, 1 mm CaCl2, and 0.5 mm MgCl2, pH 7.4) to inactivate residual aldehyde groups for 10 min, blocked with DPBS plus 2% (w/v) BSA, 2% (w/v) casein, and 0.5% (w/v) ovalbumin (DPBS-BCO) for 5 min, and then incubated for 1 h with the primary antibody that was diluted in DPBS-BCO. Grids were washed six times for 5 min in DPBS, incubated for 1 h with the secondary antibody that was diluted in DPBS-BCO, and washed six times for 5 min in DPBS and then six times for 5 min in water. The sections were stained with 4% (w/v) uranyl acetate for 5 min and Reynolds lead citrate for 3 min. Background labeling was determined using mock-infected stem sections. Quantification of the distribution of gold particles per µm2 and relative labeling distribution were performed over mock-infected and TuMV-infected sections according to Lucocq et al. (2004).
Xylem Sap Collection and Western-Blot Analysis
The collection of xylem sap was based on a protocol that was described previously (Metzner et al., 2010). Briefly, stems from N. benthamiana were harvested 7 d postinoculation by agroinfiltration with pCambiaTuMV/6K2:GFP. The stems were then cut, washed under water to avoid sampling contamination from injured cells, and dried on tissue papers. They were next placed in different 3-mL syringes that were connected with centrifugation tubes following by centrifugation at 3,000 rpm for 10 min at 4°C. Western-blot analysis was done as described previously (Cotton et al., 2009).
Surface Application of CFDA
The dye CFDA (Sigma-Aldrich) was made up as a 6 mg mL−1 stock in acetone and diluted with water to 60 μg mL−1 for use. Approximately 1 cm2 of adaxial mature source leaf surfaces was gently abraded with fine sandpaper, and 20 μL of CFDA was applied to both steam-treated and control non-steam-treated N. benthamiana leaf surfaces. The treated leaves were then covered with a thin polyethylene film to ensure an even coverage of the dye across the leaf surface and to prevent evaporation. After 6 and 24 h, young sink leaves were observed with a confocal microscope.
Supplemental Data
The following supplemental materials are available.
Supplemental Movie S1. Three-dimensional rotation of an intact xylem vessel that contains 6K2:GFP aggregates.
Supplementary Material
Acknowledgments
We thank Dr. Andreas Wachter (University of Tübingen) for the serum against PTB3, Dr. Peter Moffett (Université de Sherbrooke) for the plasmid PVX-GFP and critical reading of the article, Dr. Hojatollah Vali and Jeannie Mui (Facility of Electron Microscopy Research, McGill University) for TEM technical support, and Jessy Tremblay (Institut National de la Recherche Scientifique-Institut Armand-Frappier) for support with the Zeiss LSM-780 confocal microscope.
Glossary
- PD
plasmodesmata
- PPU
pore plasmodesmal unit
- vRNP
viral ribonucleoprotein
- CMV
Cucumber mosaic virus
- RNP
ribonucleic protein
- PVX
Potato virus X
- TuMV
Turnip mosaic virus
- vRNA
viral RNA
- dsRNA
double-stranded RNA
- vRdRp
viral RNA-dependent RNA polymerase
- TEM
transmission electron microscopy
- CFDA
5-(and 6)-carboxyfluorescein diacetate
- PBS
phosphate-buffered saline
- BSA
bovine serum albumin
- DPBS
Dulbecco’s phosphate-buffered saline
- DPBS-BCO
DPBS plus 2% (w/v) BSA, 2% (w/v) casein, and 0.5% (w/v) ovalbumin
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada and Le Fonds Québécois de Recherche sur la Nature et les Technologies (to H.Z. and J.-F.L.).
References
- Agbeci M, Grangeon R, Nelson RS, Zheng H, Laliberté JF (2013) Contribution of host intracellular transport machineries to intercellular movement of turnip mosaic virus. PLoS Pathog 9: e1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianifahanana M, Lovins K, Dute R, Sikora E, Murphy JF (1997) Pathway for phloem-dependent movement of pepper mottle potyvirus in the stem of Capsicum annuum. Phytopathology 87: 892–898 [DOI] [PubMed] [Google Scholar]
- Aoki K, Kragler F, Xoconostle-Cazares B, Lucas WJ (2002) A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proc Natl Acad Sci USA 99: 16342–16347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin C, Boutet N, Laliberté JF (2007) Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J Virol 81: 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin C, Laliberté JF (2007) The poly(A) binding protein is internalized in virus-induced vesicles or redistributed to the nucleolus during turnip mosaic virus infection. J Virol 81: 10905–10913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betti C, Lico C, Maffi D, D’Angeli S, Altamura MM, Benvenuto E, Faoro F, Baschieri S (2012) Potato virus X movement in Nicotiana benthamiana: new details revealed by chimeric coat protein variants. Mol Plant Pathol 13: 198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman LM, Boevink P, Cruz SS, Palukaitis P, Oparka KJ (1998) The movement protein of Cucumber mosaic virus traffics into sieve elements in minor veins of Nicotiana clevelandii. Plant Cell 10: 525–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botha CE, Aoki N, Scofield GN, Liu L, Furbank RT, White RG (2008) A xylem sap retrieval pathway in rice leaf blades: evidence of a role for endocytosis? J Exp Bot 59: 2945–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V, Bergdoll M, Mutterer J, Prasad V, Pfeffer S, Erdinger M, Richards KE, Ziegler-Graff V (2003) Effects of point mutations in the major capsid protein of beet western yellows virus on capsid formation, virus accumulation, and aphid transmission. J Virol 77: 3247–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J (2008) Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J 53: 739–749 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Jensen PE, Schaad MC (1998) Genetic evidence for an essential role for potyvirus CI protein in cell-to-cell movement. Plant J 14: 393–400 [DOI] [PubMed] [Google Scholar]
- Cotton S, Grangeon R, Thivierge K, Mathieu I, Ide C, Wei T, Wang A, Laliberté JF (2009) Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J Virol 83: 10460–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin S, Verchot J, Haldeman-Cahill R, Schaad MC, Carrington JC (1995) Long-distance movement factor: a transport function of the potyvirus helper component proteinase. Plant Cell 7: 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz SS, Roberts AG, Prior DAM, Chapman S, Oparka KJ (1998) Cell-to-cell and phloem-mediated transport of potato virus X: the role of virions. Plant Cell 10: 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrick PM, Barker H, Oparka KJ (1990) Effect of virus infection on symplastic transport of fluorescent tracers in Nicotiana clevelandii leaf epidermis. Planta 181: 555–559 [DOI] [PubMed] [Google Scholar]
- Ding XS, Boydston CM, Nelson RS (2001) Presence of brome mosaic virus in barley guttation fluid and its association with localized cell death response. Phytopathology 91: 440–448 [DOI] [PubMed] [Google Scholar]
- Dolja VV, Haldeman R, Robertson NL, Dougherty WG, Carrington JC (1994) Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. EMBO J 13: 1482–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja VV, Haldeman-Cahill R, Montgomery AE, Vandenbosch KA, Carrington JC (1995) Capsid protein determinants involved in cell-to-cell and long distance movement of tobacco etch potyvirus. Virology 206: 1007–1016 [DOI] [PubMed] [Google Scholar]
- Dufresne PJ, Thivierge K, Cotton S, Beauchemin C, Ide C, Ubalijoro E, Laliberté JF, Fortin MG (2008) Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 374: 217–227 [DOI] [PubMed] [Google Scholar]
- Evert RF. (1977) Phloem structure and histochemistry. Annu Rev Plant Physiol 28: 199–222 [Google Scholar]
- Fisher DB, Wu Y, Ku MSB (1992) Turnover of soluble proteins in the wheat sieve tube. Plant Physiol 100: 1433–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CJ, Elder M (1999) Virus particles in guttate and xylem of infected cucumber (Cucumis sativus L.). Ann Appl Biol 134: 81–87 [Google Scholar]
- French CJ, Elder M, Skelton F (1993) Recovering and identifying infectious plant viruses in guttation fluid. HortScience 28: 746–747 [Google Scholar]
- Froelich DR, Mullendore DL, Jensen KH, Ross-Elliott TJ, Anstead JA, Thompson GA, Pélissier HC, Knoblauch M (2011) Phloem ultrastructure and pressure flow: sieve-element-occlusion-related agglomerations do not affect translocation. Plant Cell 23: 4428–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez G, Torres H, Pallás V (2005) Identification of translocatable RNA-binding phloem proteins from melon, potential components of the long-distance RNA transport system. Plant J 41: 107–116 [DOI] [PubMed] [Google Scholar]
- Gopinath K, Kao CC (2007) Replication-independent long-distance trafficking by viral RNAs in Nicotiana benthamiana. Plant Cell 19: 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeon R, Agbeci M, Chen J, Grondin G, Zheng H, Laliberté JF (2012) Impact on the endoplasmic reticulum and Golgi apparatus of turnip mosaic virus infection. J Virol 86: 9255–9265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeon R, Jiang J, Wan J, Agbeci M, Zheng H, Laliberté JF (2013) 6K2-induced vesicles can move cell to cell during turnip mosaic virus infection. Front Microbiol 4: 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignon N, Touraine B, Durand M (1989) 6(5) Carboxyfluorescein as a tracer of phloem sap translocation. Am J Bot 76: 871–877 [Google Scholar]
- Halk EL, McGuire JM (1973) Translocation of tobacco ringspot virus in soybean. Phytopathology 63: 1291–1300 [Google Scholar]
- Ham BK, Brandom JL, Xoconostle-Cázares B, Ringgold V, Lough TJ, Lucas WJ (2009) A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21: 197–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham BK, Lucas WJ (2014) The angiosperm phloem sieve tube system: a role in mediating traits important to modern agriculture. J Exp Bot 65: 1799–1816 [DOI] [PubMed] [Google Scholar]
- Haywood V, Yu TS, Huang NC, Lucas WJ (2005) Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J 42: 49–68 [DOI] [PubMed] [Google Scholar]
- Hipper C, Brault V, Ziegler-Graff V, Revers F (2013) Viral and cellular factors involved in phloem transport of plant viruses. Front Plant Sci 4: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipper C, Monsion B, Bortolamiol-Bécet D, Ziegler-Graff V, Brault V (2014) Formation of virions is strictly required for turnip yellows virus long-distance movement in plants. J Gen Virol 95: 496–505 [DOI] [PubMed] [Google Scholar]
- Hitchborn JH, Hills GJ (1965) The use of negative staining in the electron microscopic examination of plant viruses in crude extracts. Virology 27: 528–540 [DOI] [PubMed] [Google Scholar]
- Hoefert LL. (1984) Beet western yellows virus in phloem of pennycress. J Ultrastruct Res 88: 44–54 [Google Scholar]
- Huang TS, Wei T, Laliberté JF, Wang A (2010) A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol 152: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Carrington JC (2001) Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285: 71–81 [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Cronin S, Carrington JC (1997) Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology 228: 251–262 [DOI] [PubMed] [Google Scholar]
- Kempers R, Bel AE (1997) Symplasmic connections between sieve element and companion cell in the stem phloem of Vicia faba L. have a molecular exclusion limit of at least 10 kDa. Planta 201: 195–201 [Google Scholar]
- Knapp E, Flores R, Scheiblin D, Modla S, Czymmek K, Yusibov V (2012) A cryohistological protocol for preparation of large plant tissue sections for screening intracellular fluorescent protein expression. Biotechniques 52: 31–37 [DOI] [PubMed] [Google Scholar]
- Koh EJ, Zhou L, Williams DS, Park J, Ding N, Duan YP, Kang BH (2012) Callose deposition in the phloem plasmodesmata and inhibition of phloem transport in citrus leaves infected with “Candidatus Liberibacter asiaticus”. Protoplasma 249: 687–697 [DOI] [PubMed] [Google Scholar]
- Laliberté JF, Sanfaçon H (2010) Cellular remodeling during plant virus infection. Annu Rev Phytopathol 48: 69–91 [DOI] [PubMed] [Google Scholar]
- Léonard S, Viel C, Beauchemin C, Daigneault N, Fortin MG, Laliberté JF (2004) Interaction of VPg-Pro of turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J Gen Virol 85: 1055–1063 [DOI] [PubMed] [Google Scholar]
- Lin MK, Lee YJ, Lough TJ, Phinney BS, Lucas WJ (2009) Analysis of the pumpkin phloem proteome provides insights into angiosperm sieve tube function. Mol Cell Proteomics 8: 343–356 [DOI] [PubMed] [Google Scholar]
- Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ (2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ, Groover A, Lichtenberger R, Furuta K, Yadav SR, Helariutta Y, He XQ, Fukuda H, Kang J, Brady SM, et al. (2013) The plant vascular system: evolution, development and functions. J Integr Plant Biol 55: 294–388 [DOI] [PubMed] [Google Scholar]
- Lucocq JM, Habermann A, Watt S, Backer JM, Mayhew TM, Griffiths G (2004) A rapid method for assessing the distribution of gold labeling on thin sections. J Histochem Cytochem 52: 991–1000 [DOI] [PubMed] [Google Scholar]
- Ma Y, Miura E, Ham BK, Cheng HW, Lee YJ, Lucas WJ (2010) Pumpkin eIF5A isoforms interact with components of the translational machinery in the cucurbit sieve tube system. Plant J 64: 536–550 [DOI] [PubMed] [Google Scholar]
- Manabayeva SA, Shamekova M, Park JW, Ding XS, Nelson RS, Hsieh YC, Omarov RT, Scholthof HB (2013) Differential requirements for Tombusvirus coat protein and P19 in plants following leaf versus root inoculation. Virology 439: 89–96 [DOI] [PubMed] [Google Scholar]
- Metzner R, Schneider HU, Breuer U, Thorpe MR, Schurr U, Schroeder WH (2010) Tracing cationic nutrients from xylem into stem tissue of French bean by stable isotope tracers and cryo-secondary ion mass spectrometry. Plant Physiol 152: 1030–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J (2008) Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6: 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno IM, Thompson JR, García-Arenal F (2004) Analysis of the systemic colonization of cucumber plants by Cucumber green mottle mosaic virus. J Gen Virol 85: 749–759 [DOI] [PubMed] [Google Scholar]
- Murant AF, Roberts IM (1979) Virus-like particles in phloem tissue of chervil (Anthriscus cerefolium) infected with carrot red leaf virus. Ann Appl Biol 92: 343–346 [Google Scholar]
- Nicolas O, Dunnington SW, Gotow LF, Pirone TP, Hellmann GM (1997) Variations in the VPg protein allow a potyvirus to overcome va gene resistance in tobacco. Virology 237: 452–459 [DOI] [PubMed] [Google Scholar]
- Opalka N, Brugidou C, Bonneau C, Nicole M, Beachy RN, Yeager M, Fauquet C (1998) Movement of rice yellow mottle virus between xylem cells through pit membranes. Proc Natl Acad Sci USA 95: 3323–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka KJ, Turgeon R (1999) Sieve elements and companion cells: traffic control centers of the phloem. Plant Cell 11: 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otulak K, Garbaczewska G (2009) Ultrastructural events during hypersensitive response of potato cv. Rywal infected with necrotic strains of potato virus Y. Acta Physiol Plant 32: 635–644 [Google Scholar]
- Otulak K, Garbaczewska G (2012) Cytopathological potato virus Y structures during solanaceous plants infection. Micron 43: 839–850 [DOI] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Cook G, Feys BJ, Parker JE, Baulcombe DC (2002) An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J 29: 569–579 [DOI] [PubMed] [Google Scholar]
- Rajamäki ML, Valkonen JP (1999) The 6K2 protein and the VPg of potato virus A are determinants of systemic infection in Nicandra physaloides. Mol Plant Microbe Interact 12: 1074–1081 [DOI] [PubMed] [Google Scholar]
- Rajamäki ML, Valkonen JP (2002) Viral genome-linked protein (VPg) controls accumulation and phloem-loading of a potyvirus in inoculated potato leaves. Mol Plant Microbe Interact 15: 138–149 [DOI] [PubMed] [Google Scholar]
- Requena A, Simón-Buela L, Salcedo G, García-Arenal F (2006) Potential involvement of a cucumber homolog of phloem protein 1 in the long-distance movement of Cucumber mosaic virus particles. Mol Plant Microbe Interact 19: 734–746 [DOI] [PubMed] [Google Scholar]
- Roberts K, McCann MC (2000) Xylogenesis: the birth of a corpse. Curr Opin Plant Biol 3: 517–522 [DOI] [PubMed] [Google Scholar]
- Rojas MR, Zerbini FM, Allison RF, Gilbertson RL, Lucas WJ (1997) Capsid protein and helper component-proteinase function as potyvirus cell-to-cell movement proteins. Virology 237: 283–295 [DOI] [PubMed] [Google Scholar]
- Roney JK, Khatibi PA, Westwood JH (2007) Cross-species translocation of mRNA from host plants into the parasitic plant dodder. Plant Physiol 143: 1037–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühl C, Stauffer E, Kahles A, Wagner G, Drechsel G, Rätsch G, Wachter A (2012) Polypyrimidine tract binding protein homologs from Arabidopsis are key regulators of alternative splicing with implications in fundamental developmental processes. Plant Cell 24: 4360–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Xoconostle-Cázares B, Lucas WJ (1999) Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development 126: 4405–4419 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Chino M, Hayashi H, Fujiwara T (1998) Detection of several mRNA species in rice phloem sap. Plant Cell Physiol 39: 895–897 [DOI] [PubMed] [Google Scholar]
- Savenkov EI, Germundsson A, Zamyatnin AA Jr, Sandgren M, Valkonen JPT (2003) Potato mop-top virus: the coat protein-encoding RNA and the gene for cysteine-rich protein are dispensable for systemic virus movement in Nicotiana benthamiana. J Gen Virol 84: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Schaad MC, Lellis AD, Carrington JC (1997) VPg of tobacco etch potyvirus is a host genotype-specific determinant for long-distance movement. J Virol 71: 8624–8631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönborn J, Oberstrass J, Breyel E, Tittgen J, Schumacher J, Lukacs N (1991) Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res 19: 2993–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepardson S, Esau K, McCrum R (1980) Ultrastructure of potato leaf phloem infected with potato leafroll virus. Virology 105: 379–392 [DOI] [PubMed] [Google Scholar]
- Shukla DD, Ward CW, Brunt AA (1994) The Potyviridae. CAB International, Wallingford, UK [Google Scholar]
- Simón-Buela L, García-Arenal F (1999) Virus particles of cucumber green mottle mosaic tobamovirus move systemically in the phloem of infected cucumber plants. Mol Plant Microbe Interact 12: 112–118 [DOI] [PubMed] [Google Scholar]
- Swanson M, Barker H, Macfarlane SA (2002) Rapid vascular movement of tobraviruses does not require coat protein: evidence from mutated and wild-type viruses. Ann Appl Biol 141: 259–266 [Google Scholar]
- Terasaki M, Reese TS (1992) Characterization of endoplasmic reticulum by co-localization of BiP and dicarbocyanine dyes. J Cell Sci 101: 315–322 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Linnik O, Louveaux M, Roberts IM, Chapman SN, Oparka KJ (2013) Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J Cell Biol 201: 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot J, Driskel BA, Zhu Y, Hunger RM, Littlefield LJ (2001) Evidence that soilborne wheat mosaic virus moves long distance through the xylem in wheat. Protoplasma 218: 57–66 [DOI] [PubMed] [Google Scholar]
- Vijayapalani P, Maeshima M, Nagasaki-Takekuchi N, Miller WA (2012) Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog 8: e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang A (2010) Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog 6: e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Deom CM, Beachy RN, Lucas WJ (1989) Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246: 377–379 [DOI] [PubMed] [Google Scholar]
- Wood PJ. (1980) Specificity in the interaction of direct dyes with polysaccharides. Carbohydr Res 85: 271–287 [Google Scholar]
- Xoconostle-Cázares B, Ruiz-Medrano R, Lucas WJ (2000) Proteolytic processing of CmPP36, a protein from the cytochrome b(5) reductase family, is required for entry into the phloem translocation pathway. Plant J 24: 735–747 [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cázares B, Xiang Y, Ruiz-Medrano R, Wang HL, Monzer J, Yoo BC, McFarland KC, Franceschi VR, Lucas WJ (1999) Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283: 94–98 [DOI] [PubMed] [Google Scholar]
- Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ (2004) A systemic small RNA signaling system in plants. Plant Cell 16: 1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao X, Zhang Y, Niu S, Qu F, Zhang Y, Han C, Yu J, Li D (2013) N-terminal basic amino acid residues of beet black scorch virus capsid protein play a critical role in virion assembly and systemic movement. Virol J 10: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.