Epigenetic states defined by chromatin can be maintained through mitotic cell division. Alabert et al. tracked histone modifications and histone variants during DNA replication and across the cell cycle and found that post-translational modifications (PTMs) are transmitted with parental histones to newly replicated DNA. Di- and tri-methylation marks are diluted two-fold upon DNA replication, but within one cell cycle all PTMs are restored. H3K9me3 and H3K27me3 are propagated by continuous modification of parental and new histones, because the establishment of these marks extends over several cell generations.
Keywords: histone post-translational modifications, DNA replication, cell cycle, histone variants, epigenetics, histone recycling
Abstract
Epigenetic states defined by chromatin can be maintained through mitotic cell division. However, it remains unknown how histone-based information is transmitted. Here we combine nascent chromatin capture (NCC) and triple-SILAC (stable isotope labeling with amino acids in cell culture) labeling to track histone modifications and histone variants during DNA replication and across the cell cycle. We show that post-translational modifications (PTMs) are transmitted with parental histones to newly replicated DNA. Di- and trimethylation marks are diluted twofold upon DNA replication, as a consequence of new histone deposition. Importantly, within one cell cycle, all PTMs are restored. In general, new histones are modified to mirror the parental histones. However, H3K9 trimethylation (H3K9me3) and H3K27me3 are propagated by continuous modification of parental and new histones because the establishment of these marks extends over several cell generations. Together, our results reveal how histone marks propagate and demonstrate that chromatin states oscillate within the cell cycle.
Local chromatin environment is a major determinant of gene expression and underlies epigenetic phenomena such as X inactivation, gene silencing, and centromere function. Chromatin states can thus be maintained through cell division but, at the same time, must be sufficiently plastic to allow programmed changes in transcription patterns during development. Aberrant epigenetic regulation is prevalent in complex diseases, and cancer cells typically show widespread chromatin alterations. Thus, defining the mechanisms underpinning the propagation of chromatin states is important for understanding cellular memory and disease.
Histones are highly decorated with post-translational modifications (PTMs) that can directly affect chromatin accessibility or serve as a recognition site for specific proteins. Some histone marks are closely linked to gene activation, and others are required to maintain silencing (Kouzarides 2007; Pengelly et al. 2013). In addition, histone variants occupying distinct genomic regions also define chromatin structure (Skene and Henikoff 2013). For example, histones H3.3 and H2A.Z generally demarcate dynamic sites with high nucleosome turnover. During DNA replication, nucleosomes are disrupted ahead of the replication machinery and reassembled on the two newly synthesized DNA strands by the recycling of old histones and incorporation of new largely unmodified histones (Annunziato 2005; Probst et al. 2009; Margueron and Reinberg 2010; Alabert and Groth 2012). The recycling of modified parental histones offers an attractive epigenetic mechanism, as it ensures that histone marks are kept at their correct location on newly replicated DNA. Moreover, parental histones may also serve as a blueprint to modify neighboring new histones, given that modifications like H3K9 trimethylation (H3K9me3) and H3K27me3 can recruit their cognate enzyme and potentially be self-propagating (Aagaard et al. 1999; Hansen et al. 2008; Margueron et al. 2009). However, paradoxically, for several key modifications, mass spectrometry analysis showed that new histones had not acquired modifications to become identical to the old parental histones in the G1 phase of the next cell cycle (Scharf et al. 2009; Sweet et al. 2010; Xu et al. 2012). Besides, recent work in Drosophila embryos challenges the paradigm that histone marks are transmitted with the parental histones by suggesting that the marks are erased (Petruk et al. 2012, 2013). Thus, it remains unclear how histone PTM levels are restored after DNA replication to propagate epigenetic states through multiple rounds of cell divisions.
Results and Discussion
Histones are recycled with their PTMs at replication forks
We tracked newly synthesized and parental histones by quantitative mass spectrometry immediately after their deposition behind the replication fork using nascent chromatin capture (NCC). In NCC, biotin-dUTP labeling of newly replicated DNA and cross-linking of associated proteins allow the isolation of nascent chromatin (Supplemental Fig. S1A,B; Alabert et al. 2014). We combined NCC with pulsed SILAC (stable isotope labeling with amino acids in cell culture) to differentiate newly synthesized histones (heavy) and old recycled histones (light) (Fig. 1A; Scharf et al. 2009). The nascent chromatin purified by NCC contained approximately equal amounts of new and old histones (Fig. 1B). The slight overrepresentation of old histones likely reflects a contribution of recycled light amino acids during histone biosynthesis (Scharf et al. 2009; Xu et al. 2012; Zee et al. 2012). Thus, chromatin is reassembled by a combination of recycled parental histones and newly synthesized ones.
Figure 1.
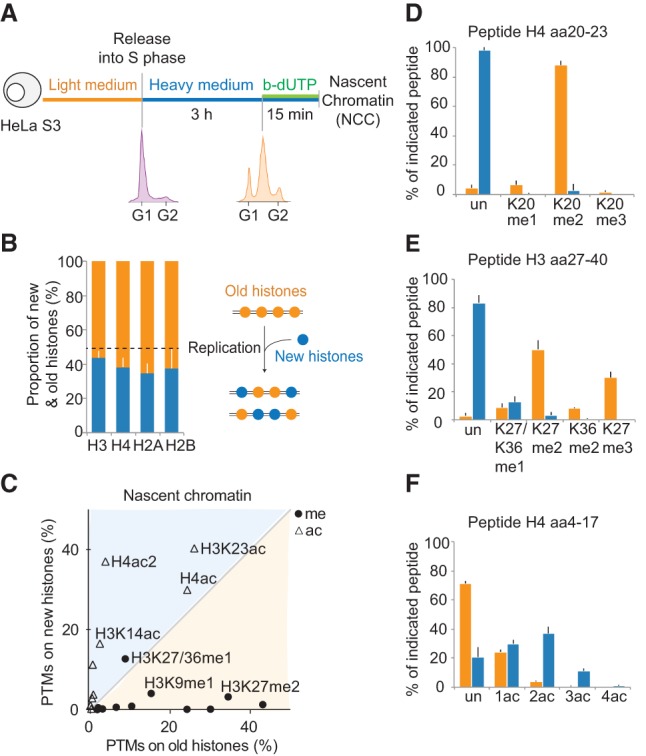
Histones are recycled together with their modifications on newly replicated DNA. (A) Experimental design. Cells were synchronized by thymidine and released in heavy SILAC medium. Newly replicated DNA was labeled with b-dUTP and isolated on streptavidin beads by NCC (Supplemental Fig. S1A). (B) Proportion of new and old histones on nascent chromatin. The dotted line indicates theoretical values. (C) Overview of how histone PTMs in nascent chromatin distribute on new and old histones. The mean of nine independent biological experiments is shown (n = 9). Old histones are mainly methylated (me), while new ones are acetylated (ac). Only shared marks are labeled. For values for all identified PTMs, see Supplemental Figure S1C. (D–F) Modifications on new (blue) and old (orange) histone H3 and H4 in nascent chromatin shown as a percentage of heavy or light peptides, respectively. The H4 amino acid 4–17 (aa4–17) peptide contains K5, K8, K12, and K16. (Un) Unmodified; (ac) acetylated; (ac2) diacetylated; (ac3) triacetylated; (ac4) quadriacetylated; (me1) monomethylated; (me2) dimethylated; (me3) trimethylated. K27/K36me1 is resolved in Supplemental Figure S1, E and F.
New and old histones remained markedly different within the first minutes following their assembly on newly replicated DNA (Fig. 1C; Supplemental Fig. S1C). Old recycled histones presented a large variety of PTMs, including mono-, di-, and trimethylation (Fig. 1D,E). Importantly, no marks were underrepresented on old histones in nascent chromatin compared with the whole genome (Supplemental Fig. S1D), arguing that histone marks are efficiently transmitted during DNA replication. Thus, in human somatic cells, there is no global replication-coupled erasure of parental histone PTMs, as proposed for Drosophila embryos (Petruk et al. 2012, 2013). New histone H4 showed high levels of diacetylation (Fig. 1F; Supplemental Fig. S1C), a hallmark of newly synthesized histone H4 (Sobel et al. 1995; Loyola et al. 2006; Jasencakova et al. 2010). New histone H3 carried H3K14ac, H3K18/K23ac, and H3K9me1 at levels closely mirroring the modification pattern of soluble histone H3 in predeposition complexes (Supplemental Fig. S1C; Loyola et al. 2006; Jasencakova et al. 2010), suggesting that these PTMs are maintained within the first minutes of incorporation. Notably, there is no enrichment of H3K56ac in the predeposition complex (Jasencakova et al. 2010) or in nascent chromatin (Supplemental Fig. S1C), arguing against a general function of this modification in chromatin assembly in human cells.
New histone H3 also carries modifications that were not present in predeposition complexes: K27me1 (8%), K36me1 (4%), and K27me2 (3%) (Fig. 1E; Supplemental Fig. S1C). This suggests that these methylations are thus the first ones to be imposed on new histones after deposition onto newly replicated DNA. To confirm that H3K27me1 was imposed on new histones in nascent chromatin as also suggested previously (Benson et al. 2006), we blocked the production of new histones using cycloheximide treatment and measured H3K27me1 levels by NCC (Supplemental Fig. S2A). We found that H3K27me1 levels were reduced upon cycloheximide treatment, consistent with rapid acquisition of H3K27me1 on new histones (Supplemental Fig. S2B–E). We next performed similar experiments in early and late S phase in order to test whether our conclusions can be extended to all stages of S phase (Supplemental Fig. S3A). Indeed, since euchromatin is preferably replicated in early S phase and heterochromatin is preferably replicated in late S phase, it has been suggested that chromatin assembly may differ during S phase (Taddei et al. 1999; Zhang et al. 2002). We found that histone PTMs in nascent chromatin were largely similar in early and late S phase (Supplemental Fig. S3B–D). However, K27me1/36me1 was increased, and H4 diacetylation was reduced on new histones incorporated in late S phase (Supplemental Fig. S3B). These observations, together with previous work (Taddei et al. 1999; Zhang et al. 2002; Lande-Diner et al. 2009), suggest a faster onset of silencing in late S phase.
H2A.Z domains are challenged by the passage of replication forks
In parallel, we explored the maintenance of histone variants during DNA replication. We used unique peptides to differentiate the canonical histone H3.1 from the variant H3.3 and the canonical histone H2A from the two variants H2A.X and H2A.Z (Fig. 2A). We found that H2A.X and H3.3 were equally as abundant in newly replicated chromatin as over the whole genome (Fig. 2B,C), arguing that they are not lost during replication. Indeed, analysis of old histones revealed that H2A.X and H3.3 are efficiently recycled at replication forks (Supplemental Fig. S3E). However, newly replicated regions were depleted for the histone variant H2A.Z (Fig. 2D). The low abundance of H2A.Z on newly replicated chromatin was not due to the particular composition of the replicated region, since we observed the same trend in early, mid, and late S phase (Fig. 2D). H2A.Z domains are thus challenged by the passage of replication forks. In-depth analysis of old and new histones showed that H2A.Z loss is the result of both inefficient recycling and lack of new deposition (Supplemental Fig. S3E,F). Notably, H2A.Z levels increased within the next 10 h (Fig. 2E), consistent with replication-independent assembly (Luk et al. 2010; Nekrasov et al. 2012). Altogether, our data demonstrate that histone variants are not recycled with similar efficiency at replication forks and that H2A.Z marked sites may not be maintained after DNA replication.
Figure 2.
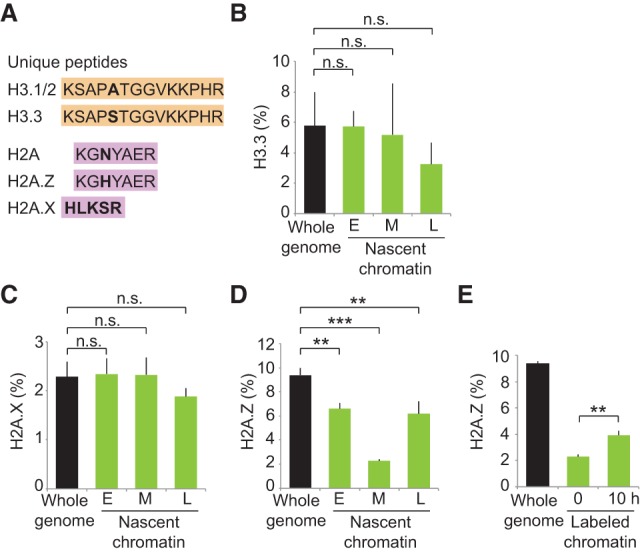
Recycling of histone variants at replication forks. Experimental design as in Figure 1A except that nascent chromatin was collected in early (E), mid- (M), and late (L) S phase (Supplemental Fig. S3A). (A) Unique peptides used to differentiate canonical histones H3.1 and H3.2 from the H3.3 variant as well as histone H2A from H2A.X and H2A.Z variants. (B) Enrichment of H3.3 relative to total H3 (H3.3+H3.1/2). (C,D) Enrichment of H2A.X and H2A.Z relative to total H2A (H2A+H2A.Z+H2AX). (E) Enrichment of H2A.Z in labeled chromatin at 0 h (nascent; mid-S phase) and 10 h later (see Supplemental Fig. S4A for experimental design). Error bars indicate SD. 3 < n < 9. Unpaired t-test: (***) P < 0.001; (**) P < 0.05; (n.s.) nonsignificant.
Histone PTM levels are restored by two distinct modes
Histone PTM levels must be maintained across the cell cycle (Probst et al. 2009; Margueron and Reinberg 2010; Alabert and Groth 2012; Zee et al. 2012). Previous analysis using SILAC labeling to differentiate new and old histones as well as methylation kinetics in total chromatin found that modification of new histones is a slow process extending into the next cell cycle (Pesavento et al. 2008; Scharf et al. 2009; Sweet et al. 2010; Xu et al. 2012; Zee et al. 2012). However, full restoration of the prereplication chromatin state could not be monitored, most likely due to limitations associated with analyzing total chromatin. Therefore, the mechanisms of restoration remain unclear. We thus investigated the kinetics of PTM establishment on new histones from the moment of their deposition and across several cell generations by combining NCC with double-pulsed (Supplemental Fig. S4A) and triple-pulsed (Supplemental Fig. S5A) SILAC. The triple-pulsed SILAC allowed us to follow one generation of histones through several cell cycles (Supplemental Fig. S5B,C). Using the PTM level of the parental histones as an indicator of the prereplication state, we found that most methylations marks were diluted by twofold immediately after replication (Fig. 3A, 0 h; Supplemental Fig. S5D). However, within one cell cycle, the level of all PTMs was fully restored (Fig. 3A, 24 h; Supplemental Fig. S5D). Thus, in this system, we can address how PTM levels are maintained across the cell cycle.
Figure 3.
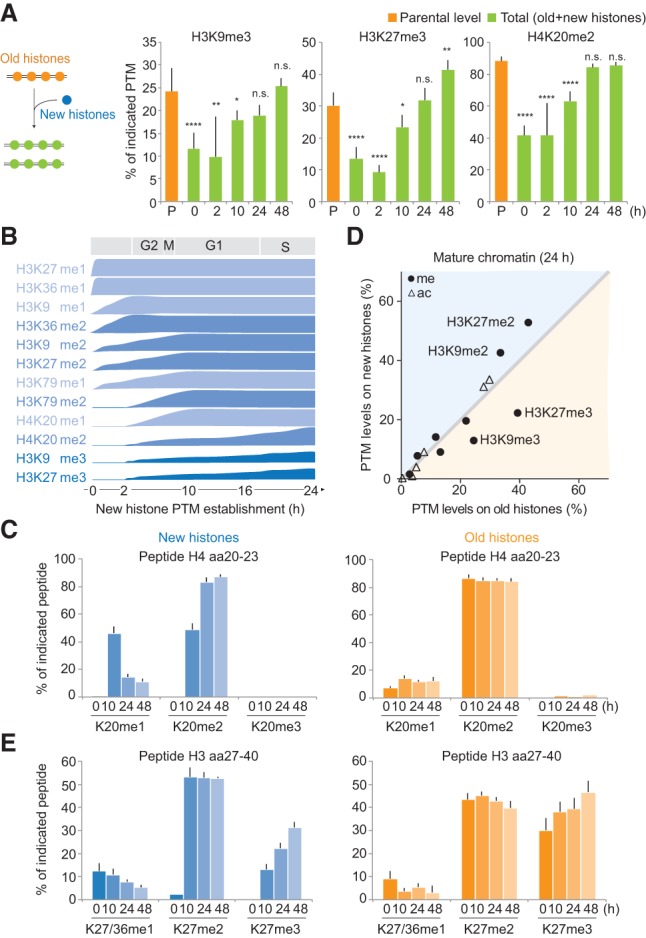
Propagation of histone PTMs across the cell cycle. Histone PTM propagation was determined by NCC coupled with double- and triple-SILAC labeling in a time-course analysis (Supplemental Figs. S4A, S5A). (A) Kinetics of PTM restoration. The total PTM level on replicated DNA (green) was compared with the PTM level of the old histones alone (P; orange), which, in nascent chromatin, represents the parental chromatin state that should to be reached by new and old histones together at the next round of replication, where they (new+old) constitute the parental histones of the daughter cells. After 24 h, the total PTM level of new+old histones (24 h; green) mirrors the parental histones of the previous round of replication (P; orange), showing that the chromatin state is propagated. Error bars indicate SD. n ≥ 3. Unpaired t-test identified differences from the parental state. A combination of double- and triple-SILAC time-course data is shown. (B) Kinetics of PTM establishment on new histones. (C) Dynamics of H4K20 methylation on new and old histones. Error bars indicate SD. n ≥ 3. (D) Overview of how histone PTMs in mature chromatin (24 h) distribute on new and old histones (n = 3). Old and new histones are identical except for the indicated marks. For values for all identified PTMs throughout the time course, C and E, and Supplemental Figure S6, A and B. (E) Dynamics of H3K27 methylation on new and old histones. Error bars indicate SD. n ≥ 3.
We next examined the PTM levels on new and old histones during the maturation process. Since high histone turnover at gene bodies and regulatory elements was suggested to impact histone PTM maintenance (Deal et al. 2010), we first estimated the histone turnover in our system. We detected a turnover rate of ∼0.4% histone H4 per hour (Supplemental Fig. S5C), likely having a limited role in histone PTM maintenance for the replicated regions examined in our study. We found that for the majority of the histone PTMs, the prereplication level was restored by gradual modification of new histones until they became identical to the old histones (Fig. 3B; Supplemental Fig. S6A). Depending on the modification, the kinetics of PTM establishment varied from 2 to 24 h (Fig. 3B,C; Supplemental Figs. S4B–E, S6A), arguing against a general rapid replication-coupled restoration of histone PTMs. Notably, H3K9me3 and H3K27me3 remained underrepresented on new histones after 24 h (Fig. 3D), supporting previous findings (Xu et al. 2012) and indicating that restoration of these marks must occur by a different mechanism. Consistent with this, we found that both new and old histones acquired H3K27me3 and H3K9me3 (Fig. 3E; Supplemental Fig. S6B), together restoring the prereplication PTM level (Fig. 3A). This provides direct evidence for the model proposed by Zhu and colleagues (Xu et al. 2012; Huang et al. 2013) and argues against a simple copy–paste mechanism for propagation of histone PTMs. Strikingly, the addition of these trimethylation marks continues for up to three cell generations after histone incorporation (Fig. 3E; Supplemental Fig. S6B), and, consequently, H3K9me3 and H3K27me3 accumulate with histone age. Altogether, our findings define two distinct modes of PTMs maintenance: (1) For most PTMs (Fig. 3B), levels are maintained according to the simple paradigm that new histones acquire modifications to become identical to the old ones. (2) H3K9me3 and H3K27me3, marks central for cellular memory (Kouzarides 2007; Probst et al. 2009; Margueron and Reinberg 2010; Apostolou and Hochedlinger 2013), propagate by continuous modification of both new and old histones.
Cell cycle withdrawal impacts histone PTM levels
Collectively, our data argue that due to replication-coupled dilution of histone PTMs, chromatin states oscillate within the cell cycle. We thus investigated how active cycling impacts histone PTM levels. First, we blocked cells by nocodazole (Supplemental Fig. S7A) to determine whether PTM establishment on new histones requires passage through mitosis, as shown for incorporation of the histone variant CENP-A (Silva et al. 2012). We found that H3K9me3 and H3K27me3 acquisition was impeded in nocodazole (Fig. 4A; Supplemental Fig. S7B), suggesting that cell cycle regulation partly accounts for the slow establishment of these marks. Next, we tested whether exit from the cell cycle impacts histone methylation levels, as predicted from (1) the continuous PTM dilution in proliferating cells (Fig. 3A) and (2) the fact that older histones in general show a higher methylation state than new ones (Fig. 3E; Supplemental Fig. S6B). We used primary human TIG-3 fibroblasts to compare histone methylation levels in proliferating and G0-arrested cells (Fig. 4B; Supplemental Fig. S7C). H3K9 methylation did not accumulate further in G0-arrested cells, suggesting that accumulation of this modification requires an S-phase arrest as previously reported (Di Micco et al. 2011; Xu et al. 2012). However, for several other methylation marks, including H3K27me2/3, H3K79me1/me2, and H4K20me2/me3, withdrawal from the cell cycle was indeed accompanied by a significant increase (Fig. 4B; Supplemental Fig. S7D). This suggests that histone PTM levels and possibly the stability of chromatin states are linked to cell cycle progression.
Figure 4.
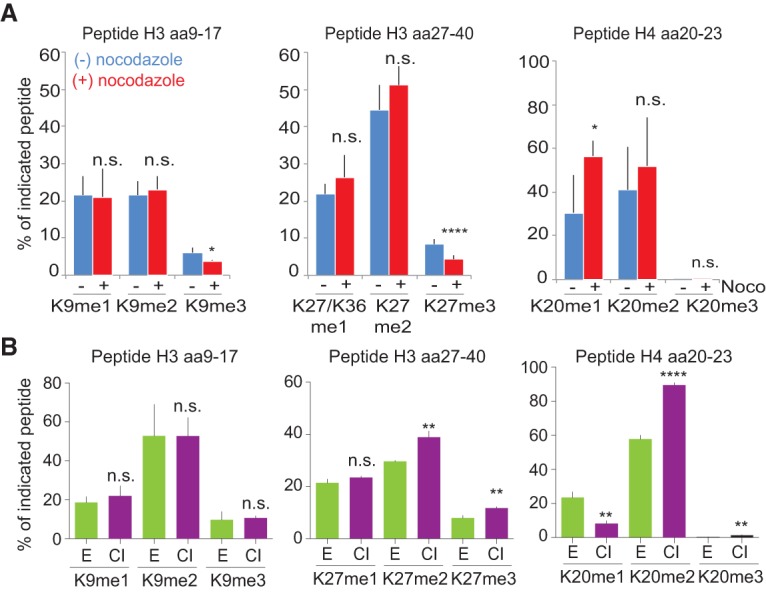
Cell cycle control of histone PTM levels. (A) After b-dUTP labeling, cells were treated with (+) or without (−) nocodazole for an additional 10, 16, or 18.5 h (Supplemental Fig. S7A). The mean of three time points is shown, with error bars indicating SD. n ≥ 3. Unpaired t-test: (****) P < 0.0001; (*) P < 0.05. (B) Analysis of histone PTM levels in exponentially growing (green) and contact-inhibited (purple) primary TIG-3 fibroblasts. Mean is shown, with error bars indicating SD. n = 3. Unpaired t-test: (****) P < 0.0001; (**) P < 0.01. For an outline of the experimental design, see Supplemental Figure S7C.
Our data demonstrate that histones PTMs are transmitted with high efficiency at replication forks. This explains the observation that H3K27me3 can be transmitted through several cell generations in the absence of the enzyme (Gaydos et al. 2014). We show that chromatin states oscillate within the cell cycle due to replication-coupled dilution of histone PTMs. Whether the drop in histone PTM levels after DNA replication affects gene expression and could provide an opportunity to modify the transcription program remains an open question (Steffen and Ringrose 2014). We envision that it could provide a window of opportunity to rewire cell fate (Fisher and Mechali 2003; Tsubouchi et al. 2013). By following histones from the moment of their incorporation, we found that most PTMs are not imposed on new histones in a rapid replication-coupled manner. Instead, we identified two modes for PTM transmission across the cell cycle (Fig. 5). For H3K9me3 and H3K27me3, our data question the simple self-propagation mechanism in which PTMs on old histones are duplicated to neighboring new histones (Probst et al. 2009; Margueron and Reinberg 2010; Alabert and Groth 2012). The establishment of these trimethylation marks is slow and continues beyond several cell generations after histone deposition. However this does not lead to dilution or loss of H3K27me3 and H3K9me3, as the total levels are maintained across cell generations by the combined methylation of both new and old histones (Fig. 3A). The slow establishment of H3K9me3 and H3K27me3 on new histones could reflect that dimethylation-to-trimethylation conversion is not processive and may require recruitment of enzyme cofactors (Alabert et al. 2014; Kalb et al. 2014). Such a tight regulation of PTM establishment could limit unwarranted silencing, which is jeopardized in cancers carrying EZH2-activating mutations (McCabe et al. 2012). Evidently, it will be important to consider the kinetics of PTM restoration in current models of epigenetic inheritance (Alabert and Groth 2012; Huang et al. 2013; Campos et al. 2014). We further envision that this mode of PTM maintenance could explain how cell cycle speed and withdrawal impact on cellular plasticity. DNA replication could be instrumental to reset epigenetic states (Tsubouchi et al. 2013), and, by revealing how histone PTMs are maintained across the cell cycle, our findings pave the way to understanding how cellular identity and reprogramming potential are connected with cell proliferation (Guo et al. 2014).
Figure 5.
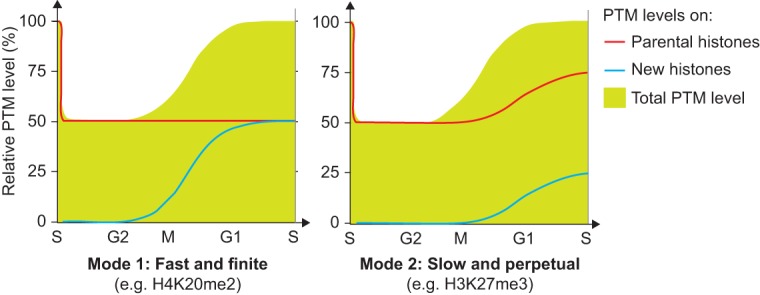
Illustration of the two basic principles for histone PTM propagation. The contribution of new and parental histones (lines) to the total PTM level (green) is shown. (Mode 1) New histones acquire PTMs to become identical to the parental histones within one cell cycle (H3K9me1, H3K27/36me1, H3K36me2, H3K9me2, H3K27me2, H3K79me1, H3K79me2, H4K20me1, and H4K20me2); (Mode 2) propagation relies on progressive modification of both new and parental histones (H3K9me3 and H3K27me3).
Materials and methods
NCC
Cells were synchronized by thymidine in light medium and released into S phase in heavy medium for 3 h. Newly synthesized DNA was pulse-labeled for 15 min with b-dUTP, and NCC was performed as described (Alabert et al. 2014). In brief, cells were labeled with b-dUTP for 5 min in hypotonic buffer (50 mM KCl, 10 mM HEPES) and fixed 15 min later in 2% formaldehyde. Nuclei were isolated in sucrose buffer (0.3 M sucrose, 10 mM HEPES-NaOH at pH 7.9, 1% Triton X-100, 2 mM MgOAc), and chromatin was solubilized by sonication using a Bioruptor. b-dUTP–labeled chromatin was purified on streptavidin-coated magnetic beads. Total chromatin (input) and isolated nascent chromatin was decross-linked by boiling for 40 min in LSB (50 mM Tris-HCl at pH 6.8, 100 mM DTT, 2% SDS, 8% glycerol, Bromphenol blue).
Mass spectrometry analysis of solubilized chromatin
Histone bands were separated by SDS-PAGE, stained with Coomassie (Brilliant blue G-250), and excised at appropriate heights. Bands were destained in 50% acetonitrile/50% 100 mM ammonium bicarbonate. Histones were chemically modified by propionylation (30 min at room temperature, 2.5% propionic anhydride [Sigma] in ammonium bicarbonate at pH 7.5) to prevent tryptic cleavage. Histone proteins were then digested with 200 ng of trypsin (Promega) in 50 mM ammonium bicarbonate overnight, and the supernatant was desalted by carbon TopTips (Glygen) according to the manufacturer's instructions. The peptides were injected into an Ultimate 3000 high-performance liquid chromatography (HPLC) system (LC Packings Dionex) and separated with a gradient from 5% to 60% acetonitrile in 0.1% formic acid over 40 min at 300 nL/min on a 75-µm ID × 10-cm ReproSil-Pur C1-AQ analytical column (2.4-µm) (Dr. Maisch GmbH). The effluent from the HPLC was directly electrosprayed into the LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). The mass spectrometry instrument was operated in the data-dependent mode to automatically switch between full-scan mass spectrometry and tandem mass spectrometry acquisition. Survey full-scan spectra (m/z 250–2000) were acquired in the Orbitrap with resolution R = 60,000 at m/z 400. For all measurements with the Orbitrap detector, three lock mass ions from ambient air (m/z = 371.10123, 445.12002, and 519.13882) were used for internal calibration as described (Olsen et al. 2005). The six most intense peptide ions with charge states between 2 and 5 were sequentially isolated (window = 2.0 m/z) to a target value of 10,000 and fragmented in the linear ion trap by collision-induced dissociation (CID). Fragment ion spectra were recorded in the linear trap of the instrument. A dynamic exclusion time of 180 sec was applied. Typical mass spectrometric conditions were spray voltage 1.4 kV, no sheath and auxiliary gas flow, heated capillary temperature 200°C, and normalized collision energy 35% for CID in the linear ion trap. An activation Q = 0.25 and activation time of 30 msec were used. Data analysis was performed with XCalibur Qual browser software (Thermo Fisher Scientific) by using doubly and triply charged peptide masses for extracted ion chromatograms (XICs). XICs were checked manually, and values were exported to Excel for further calculations. The mass spectrometry raw data were deposited to the ProteomeXchange Consortium with the data set identifier px-submission number 37929.
Histone variants
Unique heavy and light peptides were used to identify histone variants. H3.1 and H3.3 differ in amino acids 31, 87, 89, and 90. The tryptic peptide containing amino acids 87, 89, and 90 (amino acids 84–116) is difficult to detect; therefore, the unmodified peptide comprising amino acids 27–40 was taken for quantitation of the H3.3 variant. H2A was identified by the peptide KGNYAER (amino acids 38–44), H2A.X was identified by the peptide KGHYAER (38–44), and H2A.Z was identified by the peptide HLKSR (37–41). The proportion of each canonical and variant histone is expressed as a percentage of all detected variants (H2A: H2A+H2A.X+H2A.Z; and H3: H3.1+H3.3 or the common peptide H3 41–49).
Statistical analysis
All diagrams show the mean of biological independent experiments, with error bars indicating standard deviation. For statistical analysis, unpaired t-test was performed using Prism.6. P-values are indicated by asterisks (P < 0.0001 [****], P < 0.001 [***], P < 0.01 [**], and P < 0.05 [*]), and n.s. indicates nonsignificant.
Supplementary Material
Acknowledgments
We thank Carl Wu, Konrad Hochedlinger, Kristian Helin, and Peter Becker for critical reading of this manuscript, and Zuzana Jasencakova and Cristina Gonzalez Aguilera for technical support. This work was supported by a European Research Council Starting Grant (ERC2011StG, no. 281,765 to A.G.), the Danish National Research Foundation to the Center for Epigenetics (DNRF82 to S.S., O.N.J., and A.G.), the Danish Cancer Society (A.G.), and the Lundbeck Foundation (A.G.). A.G. is an EMBO Young Investigator. The work in A.I.’s laboratory is supported by grants from the European Union (EpiGeneSys 2570829) and the German Research Council (DFG SFB 1064-Z3). C.A. was supported by fellowships from Human Frontier Science Program and the Danish Medical Research Council.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.256354.114.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J 18: 1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C, Groth A. 2012. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 13: 153–167. [DOI] [PubMed] [Google Scholar]
- Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. 2014. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 16: 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato AT. 2005. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem 280: 12065–12068. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. 2013. Chromatin dynamics during cellular reprogramming. Nature 502: 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, Cook RG, Mizzen CA, Annunziato AT. 2006. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J Biol Chem 281: 9287–9296. [DOI] [PubMed] [Google Scholar]
- Campos EI, Stafford JM, Reinberg D. 2014. Epigenetic inheritance: histone bookmarks across generations. Trends Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. 2010. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328: 1161–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Sulli G, Dobreva M, Liontos M, Botrugno OA, Gargiulo G, dal Zuffo R, Matti V, d'Ario G, Montani E, et al. 2011. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol 13: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D, Mechali M. 2003. Vertebrate HoxB gene expression requires DNA replication. EMBO J 22: 3737–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos LJ, Wang W, Strome S. 2014. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science 345: 1515–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Zi X, Schulz VP, Cheng J, Zhong M, Koochaki SH, Megyola CM, Pan X, Heydari K, Weissman SM, et al. 2014. Nonstochastic reprogramming from a privileged somatic cell state. Cell 156: 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. 2008. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300. [DOI] [PubMed] [Google Scholar]
- Huang C, Xu M, Zhu B. 2013. Epigenetic inheritance mediated by histone lysine methylation: maintaining transcriptional states without the precise restoration of marks? Philos Trans R Soc Lond B Biol Sci 368: 20110332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z, Scharf AN, Ask K, Corpet A, Imhof A, Almouzni G, Groth A. 2010. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell 37: 736–743. [DOI] [PubMed] [Google Scholar]
- Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Muller J. 2014. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 21: 569–571. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Lande-Diner L, Zhang J, Cedar H. 2009. Shifts in replication timing actively affect histone acetylation during nucleosome reassembly. Mol Cell 34: 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. 2006. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell 24: 309–316. [DOI] [PubMed] [Google Scholar]
- Luk E, Ranjan A, Fitzgerald PC, Mizuguchi G, Huang Y, Wei D, Wu C. 2010. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell 143: 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2010. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet 11: 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ III, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, et al. 2012. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci 109: 2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Amrichova J, Parker BJ, Soboleva TA, Jack C, Williams R, Huttley GA, Tremethick DJ. 2012. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat Struct Mol Biol 19: 1076–1083. [DOI] [PubMed] [Google Scholar]
- Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. 2005. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 4: 2010–2021. [DOI] [PubMed] [Google Scholar]
- Pengelly AR, Copur O, Jackle H, Herzig A, Muller J. 2013. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339: 698–699. [DOI] [PubMed] [Google Scholar]
- Pesavento JJ, Yang H, Kelleher NL, Mizzen CA. 2008. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. Mol Cell Biol 28: 468–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, Mazo A. 2012. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell 150: 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Black KL, Kovermann SK, Brock HW, Mazo A. 2013. Stepwise histone modifications are mediated by multiple enzymes that rapidly associate with nascent DNA during replication. Nat Commun 4: 2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G. 2009. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10: 192–206. [DOI] [PubMed] [Google Scholar]
- Scharf AN, Barth TK, Imhof A. 2009. Establishment of histone modifications after chromatin assembly. Nucleic Acids Res 37: 5032–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. 2012. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell 22: 52–63. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2013. Histone variants in pluripotency and disease. Development 140: 2513–2524. [DOI] [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. 1995. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci 92: 1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen PA, Ringrose L. 2014. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol 15: 340–356. [DOI] [PubMed] [Google Scholar]
- Sweet SM, Li M, Thomas PM, Durbin KR, Kelleher NL. 2010. Kinetics of re-establishing H3K79 methylation marks in global human chromatin. J Biol Chem 285: 32778–32786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Roche D, Sibarita JB, Turner BM, Almouzni G. 1999. Duplication and maintenance of heterochromatin domains. J Cell Biol 147: 1153–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Soza-Ried J, Brown K, Piccolo FM, Cantone I, Landeira D, Bagci H, Hochegger H, Merkenschlager M, Fisher AG. 2013. DNA synthesis is required for reprogramming mediated by stem cell fusion. Cell 152: 873–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Wang W, Chen S, Zhu B. 2012. A model for mitotic inheritance of histone lysine methylation. EMBO Rep 13: 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee BM, Britton LM, Wolle D, Haberman DM, Garcia BA. 2012. Origins and formation of histone methylation across the human cell cycle. Mol Cell Biol 32: 2503–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H. 2002. Establishment of transcriptional competence in early and late S phase. Nature 420: 198–202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


