In many cells, mRNAs containing inverted Alu repeats (IRAlus) in their 3′-untranslated regions (3′-UTRs) are inefficiently exported to the cytoplasm. Such nuclear retention correlates with paraspeckle-associated protein complexes containing p54nrb. Hu et al. show that disruption of arginine methyltransferase CARM1 enhances the nuclear retention of mRNAs containing IRAlus. CARM1 regulates this nuclear retention pathway at two levels: CARM1 methylates the coiled-coil domain of p54nrb, resulting in reduced binding of p54nrb to mRNAs containing IRAlus; CARM1 also acts as a transcription regulator to suppress NEAT1 transcription, leading to reduced paraspeckle formation.
Keywords: paraspeckles, p54nrb, nuclear retention, CARM1, NEAT1
Abstract
In many cells, mRNAs containing inverted repeated Alu elements (IRAlus) in their 3′ untranslated regions (UTRs) are inefficiently exported to the cytoplasm. Such nuclear retention correlates with paraspeckle-associated protein complexes containing p54nrb. However, nuclear retention of mRNAs containing IRAlus is variable, and how regulation of retention and export is achieved is poorly understood. Here we show one mechanism of such regulation via the arginine methyltransferase CARM1 (coactivator-associated arginine methyltransferase 1). We demonstrate that disruption of CARM1 enhances the nuclear retention of mRNAs containing IRAlus. CARM1 regulates this nuclear retention pathway at two levels: CARM1 methylates the coiled-coil domain of p54nrb, resulting in reduced binding of p54nrb to mRNAs containing IRAlus, and also acts as a transcription regulator to suppress NEAT1 transcription, leading to reduced paraspeckle formation. These actions of CARM1 work together synergistically to regulate the export of transcripts containing IRAlus from paraspeckles under certain cellular stresses, such as poly(I:C) treatment. This work demonstrates how a post-translational modification of an RNA-binding protein affects protein–RNA interaction and also uncovers a mechanism of transcriptional regulation of the long noncoding RNA NEAT1.
The mammalian nucleus is highly organized into chromosome territories and a number of distinct membrane-less nuclear bodies or subnuclear structures that can affect nuclear neighborhoods and gene regulation (Zhao et al. 2009). Distinct nuclear bodies contain specific protein and RNA components that define particular nuclear processes (Mao et al. 2011b).
Paraspeckles, first identified in 2002 (Fox et al. 2002), are composed of the long noncoding RNA (lncRNA) NEAT1, which confers structural integrity and multiple proteins for its potential functions (Prasanth et al. 2005; Chen et al. 2008; Chen and Carmichael 2009; Clemson et al. 2009; Sasaki et al. 2009; Sunwoo et al. 2009; Naganuma et al. 2012). There are two isoforms of NEAT1 lncRNAs, NEAT1_v1 and NEAT1_v2, produced by alternative 3′ end processing (Naganuma et al. 2012). A live-cell imaging system aiming to visualize the inducible transcription of NEAT1 lncRNAs and paraspeckle proteins revealed that both the active transcription of NEAT1 and NEAT1 lncRNAs regulate paraspeckle maintenance and dynamics (Mao et al. 2011a). In addition to several well-studied Drosophila behavior and human splicing (DBHS) family proteins (including PSP1α, p54nrb, and PSF, which were known to be localized to paraspeckles) (Fox et al. 2002, 2005; Bond and Fox 2009), new paraspeckle proteins (including many RNA-binding proteins) were recently identified (Naganuma et al. 2012; West et al. 2014).
Recent studies have revealed that both NEAT1 lncRNAs and paraspeckle proteins can mediate the function of paraspeckles in gene regulation, although the detailed mechanisms remain to be fully defined. For example, the abundant NEAT1 RNA was shown to regulate transcription by sequestrating the paraspeckle protein PSF (Hirose et al. 2014; Imamura et al. 2014). Such sequestration of the transcriptional repressor PSF resulted in the activation of IL8 expression upon immune stimuli (Imamura et al. 2014) and ADARB2 transcription (Hirose et al. 2014). Moreover, genome-wide analyses recently revealed that NEAT1 can associate with hundreds of active chromatin sites (West et al. 2014), consistent with the view that paraspeckles may be involved in transcription regulation.
In addition to NEAT1-mediated regulation, p54nrb is involved in the paraspeckle-mediated nuclear retention of mRNAs containing inverted repeats in their 3′ untranslated regions (UTRs) (Prasanth et al. 2005; Chen et al. 2008; Chen and Carmichael 2009; Mao et al. 2011a; Elbarbary et al. 2013). The mouse CTN-RNA, which contains a dsRNA structure resulting from inverted short interspersed nuclear elements (SINEs) in its 3′ UTR, is retained in the nucleus and at least partially localized to paraspeckles (Prasanth et al. 2005). In human cells, hundreds of genes contain inverted repeated SINEs (mainly Alu elements) in their 3′ UTRs. Alu elements are unique to primates and account for almost all of the human SINEs and >10% of the genome. Their abundance leads to the frequent occurrence of inverted repeat structures (inverted repeated Alu elements [IRAlus]) in gene regions (Chen and Carmichael 2008). We reported previously that mRNAs containing IRAlus in their 3′ UTRs are retained in the nucleus in paraspeckles and in association with p54nrb (Chen et al. 2008; Chen and Carmichael 2009). Such regulation at paraspeckles in human cells was further demonstrated by live-cell imaging (Mao et al. 2011a). Therefore, this nuclear retention pathway of IRAlus in 3′ UTRs of genes provides an additional layer of gene regulation by sequestering otherwise mature mRNAs within the nucleus.
Interestingly, we and others observed that the nuclear retention of transcripts containing IRAlus is variable, with some such mRNAs located in the nucleus, while others are in the cytoplasm (Chen and Carmichael 2008; Chen et al. 2008; Hundley et al. 2008; Elbarbary et al. 2013). How is such nuclear retention of mRNAs containing IRAlus at paraspeckles achieved and regulated? Earlier observations suggested that the mouse CTN-RNA is retained in the nucleus until cell stress occurs, resulting in the cleavage and removal of its 3′ UTR nuclear retention signal (inverted repeats of murine SINEs) by an unknown mechanism. The truncated message is then transported efficiently to the cytoplasm for translation (Prasanth et al. 2005). However, some mRNAs containing IRAlus were seen in the cytoplasm (Chen et al. 2008; Hundley et al. 2008; Elbarbary et al. 2013), implying that other mechanisms may be involved in the release of paraspeckle-mediated nuclear retention of mRNAs containing IRAlus. For instance, it had been shown recently that the dsRNA-binding protein Staufen 1 competed with p54nrb for the binding to 3′ UTR IRAlus, resulting in an enhanced nuclear export and translation of these RNAs (Elbarbary et al. 2013). Another hypothesis is that post-translational modifications on p54nrb could alter its binding activity to mRNAs containing IRAlus.
CARM1 (coactivator-associated arginine methyltransferase), also known as PRMT4, was the first identified nuclear receptor coactivator in a family of nine PRMT members (PRMT1–9) (Bedford and Clarke 2009). CARM1 methylates histone H3 at Arg17, generating a docking site for the recruitment of the methylarginine effector TDRD3 (Yang et al. 2010). CARM1 also methylates many nonhistone proteins that play important roles in a number of biological processes, including transcriptional regulation (Chen et al. 1999), mRNA splicing (Cheng et al. 2007), muscle differentiation (Chen et al. 2002), adipocyte differentiation (Yadav et al. 2008), and T-cell development (Li et al. 2013).
Here we describe a new mechanism of gene regulation by CARM1. Disruption of CARM1 significantly enhances the nuclear retention of mRNAs containing IRAlus. We demonstrate that CARM1 regulates the nuclear retention of mRNAs containing IRAlus in paraspeckles at two levels. On the one hand, CARM1 methylates p54nrb, resulting in the reduced binding capability to mRNAs containing IRAlus; on the other hand, CARM1 suppresses NEAT1 transcription and paraspeckle formation. Actions of CARM1 at these two levels synergistically work together to regulate the export of transcripts containing IRAlus from paraspeckles under certain cellular stresses, such as poly(I:C) treatment. This represents one of a few examples where post-translational modification of an RNA-binding protein affects protein–RNA interaction and gene regulation. In addition, it shows how transcriptional regulation of the lncRNA NEAT1 can occur.
Results
p54nrb is methylated by CARM1
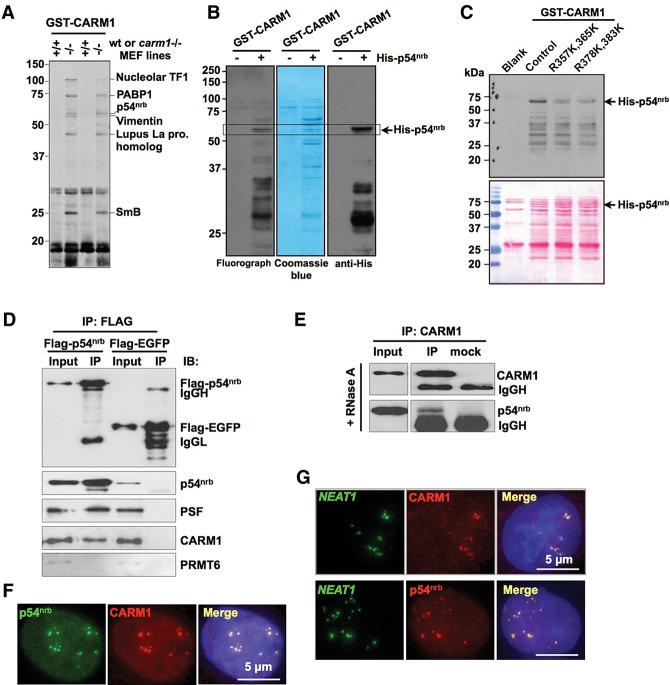
Very few substrates for CARM1 have been characterized, which limits the understanding of the roles of this enzyme. The identification of the repertoire of substrates for CARM1 is an essential step toward a complete understanding of its biological functions. CARM1 substrates are methylated in wild-type mouse embryonic fibroblast (MEF) cells but remain unmethylated in CARM1 knockout cells. Thus, the CARM1 knockout nuclear extracts are good substrates for in vitro methylation assays that would allow the identification of additional CARM1 substrates. We performed in vitro methylation assays using acid-extracted histones from the nuclei of wild-type and CARM1−/− MEFs as substrates and GST-CARM1 as an enzyme. The reaction mixtures were then run on an SDS-PAGE gel, and the indicated protein bands corresponding to potential CARM1 substrates were used for trypsin digestion and subsequent mass spectrometry (MS). Of the six identified substrates found in duplicated experiments, PABP1 (Lee and Bedford 2002) and SmB (Cheng et al. 2007) were previously described as CARM1 substrates. Interestingly, p54nrb, the well-known paraspeckle component, was also identified as a new CARM1 substrate and was chosen for further study (Fig. 1A).
Figure 1.
p54nrb is methylated by CARM1. (A) p54nrb is a CARM1 substrate. Acid-extracted histones from CARM1 wild-type (wt; +/+) and knockout (−/−) MEFs were used as substrates, and GST-CARM1 was used as an enzyme to perform standard in vitro methylation assays. Reactions were done in duplicate, separated on SDS-PAGE, and transferred to PVDF membranes for fluorography and Ruby staining. The indicated methylated proteins were processed for protein identification using MS. (B) p54nrb is methylated in vitro by recombinant CARM1. In vitro methylation reactions were performed using recombinant His-tagged p54nrb with recombinant GST-CARM1 in the presence of [3H]AdoMet. Reactions were separated on SDS-PAGE and transferred to PVDF membranes for fluorography, Coomassie blue gel staining, and immunoblotting with anti-His antibody. (C) Lysine replacement of Arg357 and Arg365 (R357K, 365K) and of Arg378 and Arg383 (R378K, 383K) reduces p54nrb methylation. Mutants of p54nrb were made as indicated by MS results and expressed as His-tagged fusion proteins. Purified His-p54nrb mutants were incubated with GST-CARM1 in the presence of [3H]AdoMet. Methylated proteins were separated by SDS-PAGE and visualized by fluorography. The same membrane was subjected to Ponceau staining to verify equal loading. Note that R383 was not identified by MS but was included in the mutation assay. (D) p54nrb interacts with CARM1. HeLa cells expressing Flag-p54nrb or Flag-EGFP (control) were immunoprecipitated with anti-Flag antibody and then immunoblotted with anti-Flag, anti-p54nrb, anti-PSF, anti-CARM1, and anti-PRMT6. Note that p54nrb specifically interacted with CARM1 but not PRMT6 in vivo. (IgGH) IgG heavy chain; (IgGL) IgG light chain. (E) RNA-independent interaction of p54nrb and CARM1 in HeLa cells. Total extracts of HeLa cells treated with RNase A were immunoprecipitated with CARM1 antibody or mock antibody and then immunoblotted with anti-CARM1 and anti-p54nrb. (F) p54nrb and CARM1 colocalize in HeLa cells. HeLa cells were stained with anti-p54nrb and anti-CARM1 antibodies. DAPI was used to indicate DNA. (G) CARM1 is a new component of paraspeckles. RNA in situ hybridization (ISH) was performed with digoxigenin (Dig)-labeled antisense NEAT1 probe (green) in HeLa cells, and representative images are shown. CARM1 and p54nrb are in red. NEAT1 colocalizes with CARM1 (top panels) and p54nrb (bottom panels).
p54nrb as a substrate of CARM1 was further validated by CARM1 in vitro methylation of Escherichia coli-expressed histidine-tagged p54nrb (Fig. 1B). Furthermore, a pan-screening for the activity of arginine methylase family members on p54nrb revealed that PRMT1 and PRMT6 could also methylate p54nrb, but the reaction of CARM1 on p54nrb was the strongest (Supplemental Fig. S1A). Moreover, the in vitro methylation of p54nrb truncations revealed that multiple arginine methylation sites on p54nrb were enriched in the coiled-coil domain and the C-terminal region of the protein (Supplemental Fig. S1B).
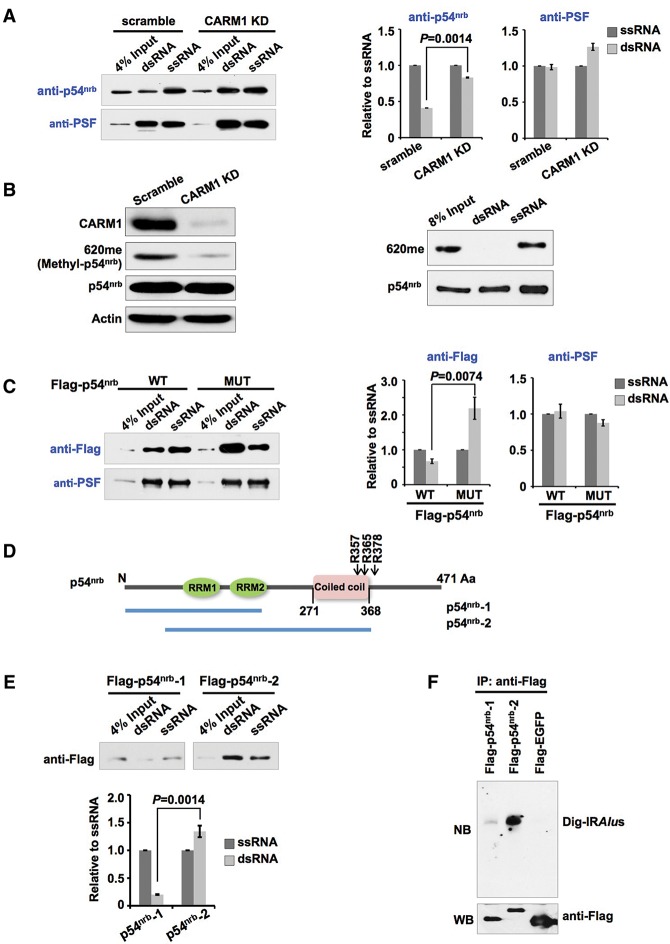
To identify the specific methylation sites catalyzed by CARM1, we developed CARM1 knockdown HeLa and HEK293 cell lines (Supplemental Fig. S2A). Flag-tagged p54nrb was expressed in scramble shRNA-treated (scramble) and CARM1 shRNA-treated (CARM1 knockdown) cell lines, and then immunoprecipitation by anti-Flag was performed (Supplemental Fig. S2B) followed by MS analysis. Multiple methylation sites in p54nrb were identified from MS (Supplemental Fig. S2C). Among these sites, R357, R365, and R378 were present in the anti-Flag precipitated complex in scramble cell lines but little in CARM1 knockdown cell lines (Supplemental Fig. S2D; data not shown), confirming that they are CARM1-methylated sites on p54nrb. In vitro methylation assays of p54nrb mutants in which these arginines were replaced with lysines further confirmed that R357, R365, and R378 were the major sites methylated by CARM1 (Fig. 1C).
The coimmunoprecipitation (co-IP) assays between p54nrb and CARM1 performed in HeLa (Fig. 1D,E) and HEK293 (data not shown) cells both revealed that they interacted with each other endogenously. CARM1 was detected in the Flag-p54nrb immunoprecipitation complexes and vice versa (Fig. 1D,E). Importantly, the co-IP was still detectable in the presence of RNase A (Fig. 1E). Colocalization of the endogenous p54nrb and CARM1 further revealed that they colocalized in the nucleus in HeLa cells (Fig. 1F) and largely within paraspeckles, as revealed by NEAT1 RNA in situ hybridization (ISH) and CARM1 immunostaining (Fig. 1G). Together, these results suggest that p54nrb is a new substrate for CARM1 and that p54nrb methylation by CARM1 may occur within paraspeckles.
CARM1 deficiency leads to enhanced nuclear retention of mRNAs containing IRAlus
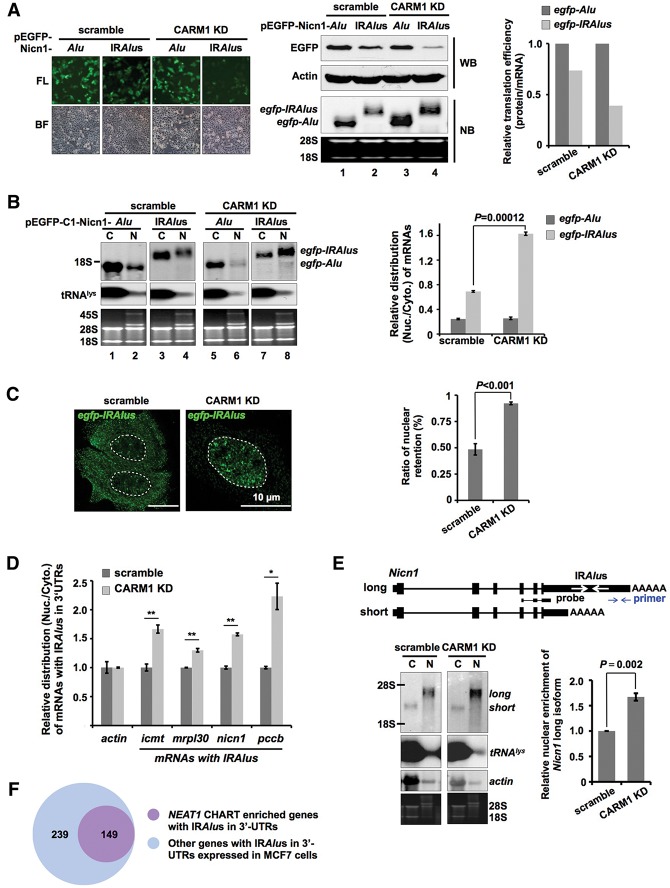
We next asked whether p54nrb methylation by CARM1 could alter paraspeckle-associated p54nrb function. Since p54nrb is involved in the nuclear retention of mRNAs containing IRAlus in their 3′ UTRs (mRNA-IRAlus) (Chen et al. 2008; Mao et al. 2011a), we first performed mRNA nuclear retention assays in the scramble and CARM1 knockdown cell lines with mRNA-IRAlus reporter constructs as previously described (Chen et al. 2008). Constructs with a single Alu element or IRAlus (originally from the 3′ UTR of the Nicn1 gene) in the 3′ UTR of egfp mRNA were individually transfected into the scramble and CARM1 knockdown HeLa cells. Consistent with previous reports (Chen et al. 2008; Mao et al. 2011a), the expression of EGFP protein was repressed by IRAlus, but not by a single Alu, in the 3′ UTR of egfp (Fig. 2A,B). This repressed EGFP expression was largely due to the nuclear retention of egfp-IRAlus (Fig. 2A,B).
Figure 2.
CARM1 deficiency enhances nuclear retention of mRNAs containing IRAlus. (A) CARM1 knockdown (KD) suppresses EGFP expression of egfp mRNA containing IRAlus in its 3′ UTR. (Left) IRAlus and Alu from the 3′ UTR of Nicn1 were inserted into the 3′ UTR of egfp mRNA (Chen et al. 2008). Stable HeLa cell lines with the scramble shRNA treatment and CARM1 knockdown were transfected with the indicated plasmids, and fluorescence was observed 24 h after transfection. (FL) Fluorescence; (BF) bright field. (Middle) The expression of EGFP and transcripts of egfp from the same batch of transfected HeLa cells as described in the left panels was investigated by Western blotting by probing with anti-EGFP antibody and by Northern blotting by probing with a Dig-labeled egfp fragment. Actin was used as loading control for Western blotting. Equivalent amounts of total RNAs were loaded for Northern blotting as indicated by 28S and 18S rRNAs. (Right) The relative translation efficiency (the relative intensity of each corresponding band from Western blotting and Northern blotting shown in the middle panels) of egfp-Alu mRNA and mRNA with egfp-IRAlus in scramble and CARM1 knockdown cells. (B) CARM1 deficiency significantly enhances the nuclear retention of egfp mRNA with IRAlus. (Left) Cytoplasmic and nuclear RNAs were isolated from the same batch of transfected HeLa cells used in A and then resolved on a denaturing agarose gel. Transcripts of egfp-tagged RNAs were probed with a Dig-labeled egfp fragment. tRNAlys and 45S rRNA were used as markers for cytoplasmic/nuclear RNA isolation. Equivalent amounts of RNAs from different subcellular compartments were loaded as indicated by 28S and 18S rRNAs. (Right) The relative subcellular distribution of RNAs with IRAlus or single Alu-containing RNAs was quantified from the left panels by ImageJ and normalized to the relative amount of tRNAlys. The ratio was obtained by comparing the value of each nuclear-retained fractionation with those of the cytoplasmic distributed RNAs. (C) Enhanced egfp mRNAs with IRAlus are retained in the nucleus upon CARM1 deficiency, as revealed by RNA ISH. (Left) The egfp mRNAs with IRAlus were probed with a Dig-labeled antisense egfp probe in HeLa cells transfected with the indicated plasmids, and representative images are shown. The white dotted line indicates the nucleus. (Right) Statistical analysis of the left panel. More than 300 transfected cells were analyzed after each transfection, and the P-value from a one-tailed t-test in the pairwise comparison is shown. (D) CARM1 knockdown enhances the nuclear retention of other endogenous mRNAs with IRAlus. RT-qPCR analyzed a number of other endogenous mRNAs with IRAlus using nuclear and cytoplasmic fractionated RNAs. Primers to detect RNAs with IRAlus were designed downstream from IRAlus in 3′ UTRs (Supplemental Fig. S5). The relative subcellular distribution of each RNA with IRAlus was normalized to the relative amount of actin mRNA. (E) CARM1 knockdown promotes the nuclear retention of endogenous nicn1 mRNA in HEK293 cells. (Top) A schematic drawing of transcripts of the Nicn1 gene containing two isoforms (Chen et al. 2008). (Bottom left) Note that the long isoform contains a pair of IRAlus in its 3′ UTR and is preferentially retained in the nucleus. Cytoplasmic and nuclear RNA fractionation of scramble and CARM1 knockdown cells followed by Northern blotting to detect nicn1 mRNAs. tRNAlys and actin revealed a successful cytoplasmic and nuclear fractionation. Equivalent amounts of RNA from different compartments were loaded as indicated by 28S and 18S rRNAs. (Bottom right) The relative subcellular distribution of the long isoform of nicn1 mRNA was quantified from the left panels by ImageJ and normalized to the relative amount of nuclear actin mRNA from the same stripped membrane. (F) Genes containing IRAlus in 3′ UTRs are enriched in the NEAT1-bound active chromatin sites. NEAT1 CHART-seq data in MCF7 cells were retrieved from West et al. (2014). Genes containing IRAlus in 3′ UTRs were identified from RefSeq by a home-brewed pipeline. In B, D, and E, error bars represent ±SD in triplicate experiments. (*) P < 0.05; (**) P < 0.01; n = 3.
Strikingly, the EGFP fluorescence in CARM1 knockdown cells transfected with the EGFP-IRAlus construct was much more significantly reduced than that in scramble cells (Fig. 2A). This enhanced repression effect was also confirmed by Western blotting with anti-GFP antibody (Fig. 2A, top right panels, lanes 2,4). Meanwhile, Northern blotting of transcripts of egfp from the same batch of transfected HeLa cells revealed that both transfections yielded comparable or higher levels of egfp mRNAs (Fig. 2A, bottom right panels, lanes 2,4), suggesting that the observed enhanced EGFP expression repression in CARM1 knockdown cells is post-transcriptional. We also observed similar phenomena using other constructs containing inserts of a single Alu element or IRAlus derived from the 3′ UTR of the Lin28 gene in the 3′ UTR of egfp mRNA (Supplemental Fig. S3A,B) and in other cell lines, such as in the scramble and CARM1 stable knockdown HEK293 cells (Supplemental Fig. S4A,B).
What mechanism accounts for reduced EGFP expression in CARM1 knockdown cells? Further analyses revealed that CARM1 knockdown strikingly enhanced the sequestration of mRNAs containing 3′ UTR IRAlus in the nucleus. First, nuclear/cytoplasmic (N/C) RNA fractionation analyses in scramble and CARM1 knockdown cells individually transfected with egfp-Alu or egfp-IRAlus constructs clearly showed that egfp-IRAlus mRNA was more efficiently retained in the nuclei of CARM1 knockdown cells than those in the nuclei of scramble cells (Fig. 2B, left panels [lanes 3,4,7,8] and right panel), while the N/C ratio of egfp-Alu mRNA was not altered (Fig. 2B, left panels [lanes 1,2,5,6] and right panel). Second, visualization of the subcellular distribution of egfp-IRAlus in scramble and CARM1 knockdown cells by RNA ISH at the single-cell level revealed that egfp mRNAs with IRAlus were more highly enriched in the nucleus in CARM1 knockdown cells than those in scramble cells (Fig. 2C). Third, if CARM1 were essential for the paraspeckle-associated mRNA-IRAlus nuclear retention, we would expect to observe that the endogenous mRNA-IRAlus would have a different fate in CARM1 depletion cells. There are hundreds of mRNAs containing IRAlus in their 3′ UTRs (Chen et al. 2008), and we chose a number of mRNAs that contain IRAlus in their 3′ UTRs and are expressed well in HeLa and HEK293 cells (nicn1, icmt, mrpl30, and pccb) (Supplemental Fig. S5). For each of these mRNAs, CARM1 knockdown resulted in more efficient retention in the nucleus in both cell lines (Fig. 2D,E; Supplemental Figs. S4D, S6), while the N/C distribution of actin mRNA remained unchanged. For experiments shown in these figures, we used PCR probes that specifically recognize mRNAs with the extended 3′ UTRs that contain IRAlus (Supplemental Fig. S5). Note that the longer isoform of each of the mRNAs containing IRAlus examined increased its N/C ratio after knockdown of CARM1 (Fig. 2D; Supplemental Figs. S4D, S6A). We further used Northern blotting to confirm the subcellular distribution of two such endogenous mRNAs: nicn1 (Fig. 2E) and icmt (Supplemental Fig. S6B). The long isoform of nicn1 contains one pair of IRAlus in its 3′ UTR, but the short isoform lacks IRAlus. Correspondingly, the nicn1 long isoform is preferentially localized to the nucleus, while the short one is almost exclusively cytoplasmic (Fig. 2E; Chen et al. 2008). Importantly, knockdown of CARM1 increased the amount of nicn1 long isoform retained in the nucleus (Fig. 2E), while the subcellular distribution of the nicn1 short isoform and actin mRNA remained unaltered (Fig. 2E). Such altered nuclear retention regulation by CARM1 was also seen in another examined endogenous mRNA, icmt (Supplemental Fig. S6B), by Northern blotting. Together, these results demonstrate that the absence of CARM1 leads to an enhanced nuclear retention of mRNAs containing 3′ UTR IRAlus.
Recently, NEAT1 was shown to bind to hundreds of active chromatin sites in MCF7 cells (West et al. 2014). We found that ∼40% of such genes contain at least one pair of IRAlus in their 3′ UTRs expressed in MCF7 cells (Fig. 2F), implying that these transcribed nascent RNAs and corresponding mRNAs with 3′ UTR IRAlus are located close to paraspeckles and thus are capable of association with paraspeckle protein p54nrb for their nuclear retention.
p54nrb methylation by CARM1 regulates nuclear retention of mRNAs containing IRAlus
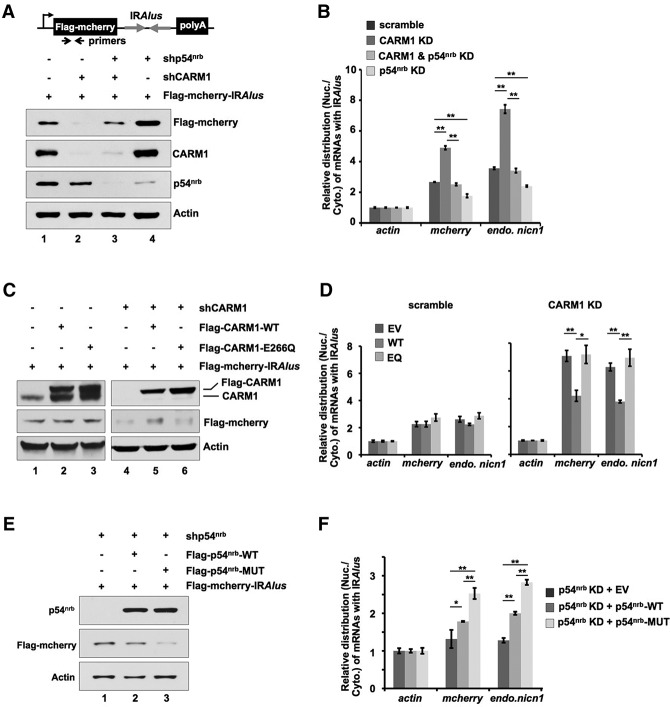
The next question is whether this CARM1-regulated nuclear retention is dependent on p54nrb. To answer this question, we knocked down CARM1 or p54nrb and then followed the expression and the subcellular distribution of the IRAlus-containing reporter plasmid Flag-mcherry-IRAlus. The pair of IRAlus used in this construct is from the 3′ UTR of the Nicn1 gene (Chen et al. 2008). While knockdown of CARM1 reduced Flag-mcherry expression (Fig. 3A, lane 2; Supplemental Fig. S7A) with an enhanced nuclear retention of Flag-mcherry-IRAlus mRNAs (Fig. 3B), as other assays revealed (Fig. 2; Supplemental Figs. S3, S4), knockdown of p54nrb enhanced Flag-mcherry expression (Fig. 3A, lane 4; Supplemental Fig. S7A) with a reduced nuclear retention of Flag-mcherry-IRAlus mRNAs (Fig. 3B). Double knockdown of CARM1 and p54nrb showed little change in both protein expression and mRNA nuclear retention (Fig. 3A [lane 3], B). These results confirm that the nuclear retention of mRNAs with 3′ UTR IRAlus is mediated by the paraspeckle-localized protein p54nrb and that CARM1 suppresses this nuclear retention pathway (Supplemental Fig. S7C).
Figure 3.
IRAlus mRNA nuclear retention is achieved through p54nrb and requires the catalytic activity of CARM1. (A) IRAlus mRNAs nuclear retention is achieved through p54nrb. (Top) A schematic drawing of Flag-mcherry-containing IRAlus in its 3′ UTR (Flag-mcherry-IRAlus). (Bottom) Knockdown of CARM1 (lane 2) or p54nrb (lane 4) or double knockdown of CARM1 and p54nrb (lane 3) in HeLa cells expressing Flag-mcherry-IRAlus, followed by Western blotting. Note that knockdown of CARM1 suppresses Flag-mcherry expression, while knockdown of p54nrb increases Flag-mcherry expression. (B) The altered Flag-mcherry protein expression in A corresponds to altered nuclear retention of transcripts of Flag-mcherry-IRAlus. The relative subcellular distribution of transcripts of Flag-mcherry-IRAlus from the same batch of transfected cells as described in A was investigated by RT-qPCR by probing mcherry. The level of nuclear retention was presented as the ratio of nuclear-retained transcripts of Flag-mcherry-IRAlus to those in the cytoplasm after normalization to the relative amount of actin mRNA in each fraction. The endogenous mRNA with 3′ UTR IRAlus was also assayed and normalized in the same way. (C) IRAlus mRNA nuclear retention is dependent on the catalytic activity of CARM1. Reintroduction of wild-type (WT) CARM1 or the catalytic inert E266Q-CARM1 into scramble (lanes 2,3) or CARM1 knockdown (KD) (lanes 5,6) HeLa cells expressing Flag-mcherry-IRAlus, followed by Western blotting. Note that expression of wild-type CARM1 but not the E266Q-CARM1 in CARM1 knockdown cells could rescue the suppressed expression of Flag-mcherry-IRAlus. (D) Altered Flag-mcherry protein expression in C corresponds to the altered nuclear retention of transcripts of Flag-mcherry-IRAlus. The relative subcellular distribution of transcripts of Flag-mcherry-IRAlus and the endogenous mRNA with 3′ UTR IRAlus from the same batch of transfected cells as described in C were investigated by RT-qPCR. See B for details. (E) Mutation of CARM1 methylation sites on p54nrb enhances the p54nrb -mediated mRNA nuclear retention. Reintroduction of the lysine replacement of Arg357, Arg365, and Arg378 (R357K, R365K, and R378K) of p54nrb (p54nrb-MUT; lane 3) into p54nrb knockdown cells resulted in an enhanced mCherry suppression compared with that of wild-type p54nrb (lane 2), as revealed by Western blotting. (F) Altered Flag-mcherry expression in E corresponds to the altered nuclear retention of transcripts of Flag-mcherry-IRAlus. The relative subcellular distribution of transcripts of Flag-mcherry-IRAlus and the endogenous mRNA with 3′ UTR IRAlus from the same batch of transfected cells as described in E were investigated by RT-qPCR. See for details. In B, D, and F, error bars represent ±SD in triplicate experiments. (*) P < 0.05; (**) P < 0.01; n ≥ 3.
We next asked whether the catalytic activity of CARM1 is required for the observed nuclear retention. We expressed the wild-type or the catalytically inactive E266Q mutant of CARM1 (Lee et al. 2002) in scramble or CARM1 stable knockdown cell lines followed by the reporter assays described above (Fig. 3A,B). We found that reintroduction of the wild-type CARM1, but not the E266Q-CARM1, into CARM1 knockdown cells in which p54nrb is largely unmethylated due to CARM1 knockdown could largely restore the reduced Flag-mcherry expression (Fig. 3C, lanes 4–6; Supplemental Fig. S7B), the enhanced nuclear retention of Flag-mcherry-IRAlus mRNAs, and the endogenous mRNA with 3′ UTR IRAlus (Fig. 3D). However, reintroduction of either wild-type or E266Q-CARM1 into scramble treated cells in which CARM1 exists and p54nrb is methylated had little effect on the reporter assay or the endogenous mRNA with 3′ UTR IRAlus (Fig. 3C [lanes 1–3], D).
Moreover, we set up to examine whether the p54nrb variant carrying the lysine replacement of Arg357, Arg365, and Arg378 (R357K, 365K, and R378K) that showed a reduced p54nrb methylation by CARM1 (Fig. 1C) could enhance the p54nrb-mediated mRNA nuclear retention. Reintroduction of the p54nrb variant that carries these three point mutants (R357K, 365K, and R378K) into p54nrb knockdown cells not only was able to restore the p54nrb function but also exhibited a stronger effect on protein expression suppression and the corresponding mRNA nuclear retention in the reporter assay (Fig. 3E,F). Meanwhile, a similar observation was also seen in the examined endogenous mRNA with 3′ UTR IRAlus (Fig. 3F). Together, these results strongly suggest that CARM1 methylation on p54nrb attenuates the p54nrb-mediated mRNA nuclear retention.
The absence of CARM1 enhances the association of p54nrb and mRNAs with IRAlus
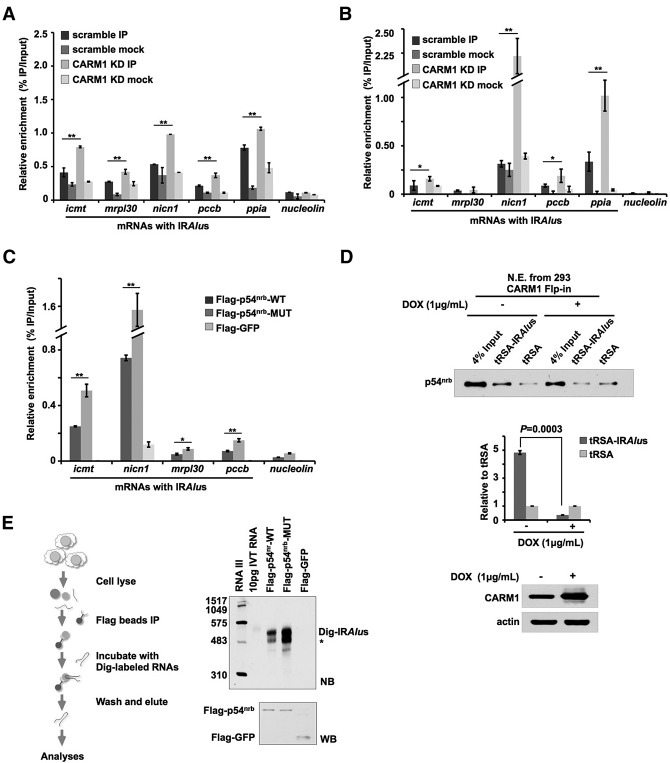
How does p54nrb methylation by CARM1 alter mRNA nuclear retention? Since p54nrb can bind to IRAlus mRNAs and subsequently retain them in the nucleus (Prasanth et al. 2005; Chen et al. 2008; Mao et al. 2011a), we performed p54nrb RNA immunoprecipitation (RIP) assays to examine whether the methylation of p54nrb could alter its binding activity with the IRAlus mRNAs. Both formaldehyde cross-linking RIP and UV cross-linking RIP with the anti-p54nrb antibody in scramble and CARM1 knockdown cell lines revealed that the absence of CARM1 augmented the association of p54nrb and mRNAs with IRAlus (Fig. 4A,B). Importantly, the lysine replacement of Arg357, Arg365, and Arg378 (R357K, 365K, and R378K) of Flag-p54nrb that showed a reduced p54nrb methylation by CARM1 (Fig. 1C) also increased its ability to bind to mRNAs with IRAlus compared with the wild-type Flag-p54nrb (Fig. 4C), with an anti-Flag antibody UV cross-linking RIP in HeLa cells.
Figure 4.
p54nrb methylation reduces its ability to bind to IRAlus mRNAs. (A) CARM1 deficiency increases the association of p54nrb and the endogenous IRAlus mRNAs. The association between p54nrb and mRNAs with IRAlus was assayed by formaldehyde cross-linking RIP from scramble or CARM1 knockdown HeLa cells using anti-p54nrb and anti-IgG followed by RT-qPCR. Bar plots represent the fold enrichments of RNAs immunoprecipitated by anti-p54nrb or anti-IgG over the same amount of input across different samples, and error bars represent SD in triplicate experiments. Nucleolin mRNA was a control that does not bind to p54nrb. (B) CARM1 deficiency increases the association of p54nrb and endogenous IRAlus mRNAs, as assayed by UV cross-linking RIP from scramble or CARM1 knockdown HeLa cells using anti-p54nrb and anti-IgG followed by RT-qPCR. See A for details. (C) Lysine replacement of Arg357, Arg365, and Arg378 (R357K, R365K, and R378K) of p54nrb (p54nrb-MUT) increases the association of p54nrb and endogenous IRAlus mRNAs. The interaction of p54nrb and endogenous IRAlus mRNAs was assayed by RIP in HeLa cells expressing Flag-p54nrb, Flag-p54nrb-MUT, or Flag-EGFP followed by RT-qPCR. See A for details. (D) CARM1 overexpression attenuates the interaction between p54nrb and IRAlus RNA. (Top) tRSA-RNA pull-downs (Iioka et al. 2011) using in vitro transcribed tRSA-tagged IRAlus RNAs were performed in nuclear extracts of 293 CARM1 Flp-in cells with or without 1 μg/mL doxycycline (DOX) induction. (Middle) The quantitative analyses of the top panel from three independent experiments. (Bottom) CARM1 expression was induced upon the DOX treatment, as revealed by Western blotting. (E) Partially purified triple mutations of R357K, R365K, and R378K of p54nrb (p54nrb-MUT) exhibit increased ability to bind to IRAlus RNAs. (Left) A schematic drawing of the experimental flow. (Right) Flag-p54nrb-wt, but not Flag-EGFP, exhibited an ability to bind to Dig-labeled IRAlus RNA; Flag-p54nrb-MUT showed an enhanced ability to bind to IRAlus RNA.
To further confirm that p54nrb unmethylated in its coiled-coil domain has an increased ability to bind to mRNA-IRAlus, we then performed a reciprocal pull-down assay by tagging an aptamer tRSA (Iioka et al. 2011) to IRAlus RNA (tRSA-IRAlus) (Fig. 4D). Following tRSA immunoprecipitation with nuclear extracts, we found that tRSA-IRAlus, but not the tRSA alone, was preferentially associated with p54nrb (Fig. 4D). Overexpression of CARM1 resulted in reduced binding of tRSA-IRAlus with p54nrb (Fig. 4D), confirming that methylation by CARM1 on p54nrb reduced its ability to bind to RNA-IRAlus. Moreover, the partially purified Flag-p54nrb-MUT carrying the lysine replacements of R357K, 365K, and R378K significantly increased its ability to bind to the in vitro transcribed IRAlus compared with the wild-type protein (Fig. 4E). Since R357 and R365 are located at the coiled-coil domain and R378 is located very close to this domain at the C terminus of p54nrb (Supplemental Fig. S2D), these results strongly suggest that methylation on p54nrb at or near the coiled-coil domain by CARM1 reduces its capability to associate with RNAs containing IRAlus.
p54nrb methylation at or near the coiled-coil domain by CARM1 alters its binding to dsRNAs
How does unmethylated p54nrb bind to mRNAs with IRAlus? Since p54nrb is known to bind to inosine-containing RNAs (Zhang and Carmichael 2001), one possibility is that p54nrb binds to edited mRNA-IRAlus (Prasanth et al. 2005; Chen et al. 2008). However, some extensively adenosine (A)-to-inosine (I) edited mRNA-IRAlus were also exported to the cytoplasm (Prasanth et al. 2005; Chen et al. 2008; Hundley et al. 2008; Elbarbary et al. 2013), and knockdown of ADAR1 did not alter the nuclear retention of these RNAs (Elbarbary et al. 2013), indicating that p54nrb can bind to RNAs by recognizing other RNA structures, such as dsRNAs formed by the IRAlus at the 3′ UTR of mRNAs. We therefore examined whether p54nrb could bind to dsRNAs and whether p54nrb methylation by CARM1 could selectively alter its binding to these molecules.
The in vitro binding assays revealed that full-length of p54nrb bound to both dsRNAs and ssRNAs (Fig. 5A); importantly, knockdown of CARM1 altered the binding ability of p54nrb to only dsRNAs but not to ssRNAs (Fig. 5A). With a newly developed antibody (620me) that specifically recognizes the methylated R357 and R365 on a p54nrb peptide (Fig. 5B, left panels), we found that the methylated p54nrb (620me) was enriched in ssRNA but was barely detected in dsRNA pull-downs (Fig. 5B, right panels). This observation further suggests that CARM1 methylation on p54nrb reduces its binding capacity to dsRNAs. Moreover, the partially purified Flag-p54nrb-MUT carrying the lysine replacements of R357K, 365K, and R378K promoted its binding capability to the in vitro transcribed dsRNAs but showed little binding alteration to ssRNAs (Fig. 5C). These results thus support the notion that p54nrb methylation by CARM1 alters its binding to dsRNAs and that the unmethylated p54nrb binds more strongly to dsRNAs.
Figure 5.
p54nrb methylation by CARM1 alters its binding to dsRNAs. (A) CARM1 knockdown affects p54nrb binding to dsRNAs but has little effect on ssRNAs. Biotin-labeled dsRNAs (double-stranded-egfp 76–495) or ssRNAs (single-stranded-egfp 1–798) were made from in vitro transcription (IVT). Nuclear extracts of scramble or CARM1 knockdown (KD) HeLa cells were incubated with biotin-dsRNA or biotin-ssRNA followed by anti-biotin pull-down and Western blotting with antibodies for anti-p54nrb and anti-PSF. (Right) The quantitative analyses of the left panels from three independent experiments. (B) Methylated p54nrb binds to few dsRNAs. (Left) The 620me antibody was developed to specifically recognize the methylated p54nrb. Western blotting analyzed the specificity of 620me antibody purified from rabbit serum immunized with methylated peptides of R357 and R365. p54nrb and Actin antibodies were used as controls. (Right) The methylated p54nrb (620me) was enriched by biotin-ssRNA but not by biotin-dsRNA pull-downs from wild-type HeLa cell nuclear extracts. See A for details. (C) Lysine replacement of Arg357, Arg365, and Arg378 (R357K, 365K, and R378K) of p54nrb (p54nrb-MUT) increases the association of p54nrb and dsRNAs. (Right) The quantitative analyses of the left panels from three independent experiments. See A for details. (D) A schematic drawing of different truncations of p54nrb used in E and F. (E) A different domain of p54nrb selectively binds to ssRNAs or dsRNAs. (Top) Nuclear extracts of HeLa cells transfected with different p54nrb truncations were incubated with biotin-dsRNAs or biotin-ssRNAs followed by anti-biotin pull-down and Western blotting with anti-Flag. (Bottom) The quantitative analyses of the top panels from three independent experiments. (F) Partially purified truncation of p54nrb selectively binds to IRAlus RNAs. Different p54nrb truncations shown in D were purified and incubated with Dig-labeled IRAlus RNAs. See Figure 4E for details. In A, C, and E, error bars represent ±SD in triplicate experiments. P-values from one-tailed t-test in the pairwise comparison are shown.
As it has been reported that p54nrb bound to ssDNAs (and RNAs) through its N terminus and to DNA through its C terminus (Yang et al. 1993), we speculated that the coiled-coil domain at the C terminus of p54nrb might mediate its binding to dsRNAs, such as mRNAs containing IRAlus. This is indeed the case. Incubation of ssRNAs or dsRNAs with different Flag-tagged p54nrb truncations individually expressed in HeLa cells revealed that the truncation of p54nrb containing two RRM domains preferred to bind to ssRNAs, and the truncation of p54nrb with the addition of the coiled-coil domain exhibited a much higher capacity to bind to dsRNAs (Fig. 5D,E). Importantly, while the partially purified Flag-tagged p54nrb carrying only RRM domains showed little association to the in vitro transcribed IRAlus, the inclusion of the coiled-coiled domain to this truncation significantly increased its ability to bind to IRAlus (Fig. 5F), confirming that the coiled-coil domain of p54nrb mediates its binding to dsRNAs, including mRNA-IRAlus. Together, these results strongly suggest that p54nrb methylation by CARM1 at or near the coiled-coil domain alters the interaction between dsRNAs and p54nrb via the coiled-coil domain.
CARM1 inhibits NEAT1 transcription and reduces paraspeckle formation
We showed that CARM1 methylated the coiled-coil domain of p54nrb, resulting in the reduced binding of p54nrb to mRNAs containing IRAlus. CARM1 is also known for its activity in transcription regulation (Chen et al. 1999). CARM1 largely functions as a transcriptional coactivator (Chen et al. 1999) but also acts as a transcriptional cosuppressor (Xu et al. 2001). We then asked whether the paraspeckle-localized CARM1 could also affect NEAT1 transcription.
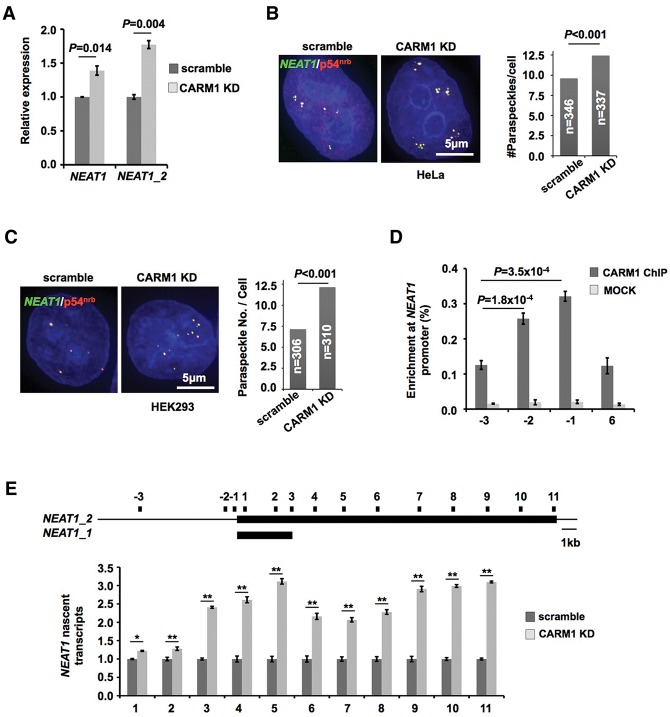
Surprisingly, we found that in CARM1 knockdown stable cell lines, the expression of both isoforms of NEAT1 was increased (Fig. 6A). Correspondingly, the number of paraspeckles was also modestly increased in both CARM1 stable knockdown HeLa cells (Fig. 6B) and HEK293 cells (Fig. 6C). CARM1 chromatin immunoprecipitation (ChIP) revealed that CARM1 was enriched at the promoter of the NEAT1 gene (Fig. 6D). Further studies by nuclear run-on (NRO) assays showed that the transcription of nascent transcripts of NEAT1 was increased upon CARM1 knockdown, confirming the notion that CARM1 could regulate NEAT1 transcription as a transcriptional cosuppressor.
Figure 6.
CARM1 is enriched at the NEAT1 promoter and inhibits NEAT1 transcription and paraspeckle formation. (A) CARM1 deficiency promotes the expression of both isoforms of NEAT1. (B) CARM1 knockdown increases paraspeckle formation. (Left) Colocalization of NEAT1 RNA (green) and p54nrb protein (red) revealed paraspeckles in scramble and CARM1 knockdown (KD) HeLa cells. (Right) Statistical analysis of numbers of paraspeckles from >300 cells in scramble and CARM1 knockdown HeLa cells. The P-value from a one-tailed t-test in the pairwise comparison is shown. (C) CARM1 knockdown increases paraspeckle formation in HEK293 cells. (D) CARM1 is enriched on the promoter region of NEAT1. CARM1 ChIP followed by qPCR analyzed the occupation of CARM1 on the NEAT1 gene. The positions of examined primers are shown in E. (E) CARM1 deficiency promotes the transcription of nascent NEAT1. (Top) A schematic drawing indicates the primer sets used. (Bottom) A crude preparation of nuclei was subjected to NRO assays under the indicated conditions in HeLa cells. Nascent transcription of NEAT1 detected from scramble nuclei was defined as one. In A, D, and E, error bars represent ±SD in triplicate experiments. (*) P < 0.05; (**) P < 0.01; n ≥ 3.
Collectively, we propose a model for CARM1-regulated nuclear retention of mRNAs containing IRAlus at paraspeckles. p54nrb methylation by CARM1 at the coiled-coil domain reduces its ability to bind to mRNAs containing IRAlus (Figs. 2–5). This is in coordination with the transcriptional suppression of CARM1 at the NEAT1 promoter (Fig. 6). Thus, CARM1 can synergistically act at two levels within paraspeckles to regulate its function for mRNA nuclear retention (Fig. 7I).
Figure 7.
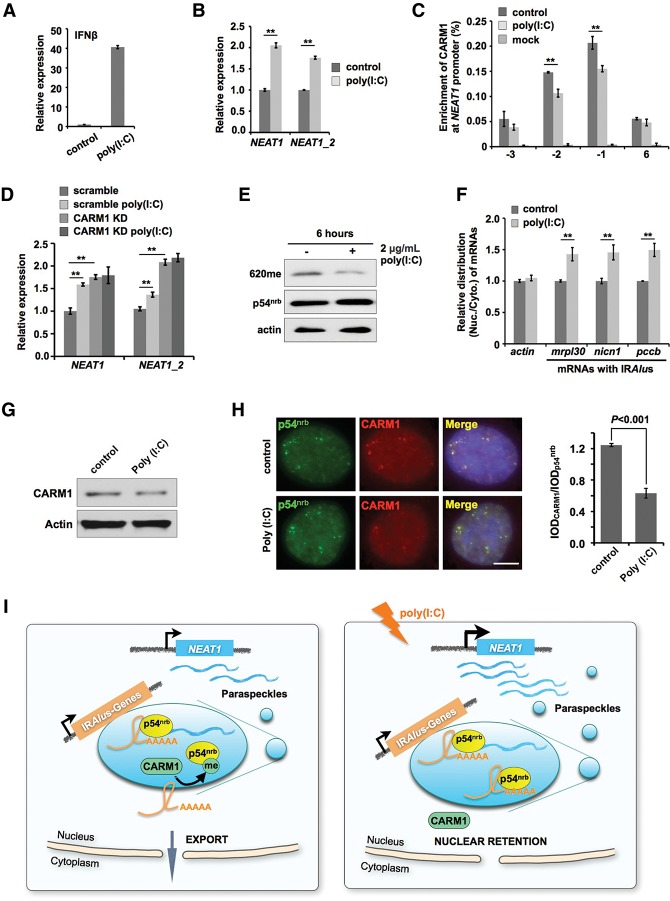
CARM1 is involved in the poly(I:C)-stimulated enhancement of nuclear retention. (A) Poly(I:C) treatment induces the expression of IFNβ. HeLa cells were transfected with 2 μg/mL poly(I:C) for 6 h and then harvested for analysis. (B) Poly(I:C) treatment induces NEAT1 expression. (C) Poly(I:C) treatment reduces the occupancy of CARM1 at the NEAT1 promoter. HeLa cells were transfected with poly(I:C) for 6 h and then harvested for CARM1 ChIP followed by qPCR. Data are presented as the percentage of CARM1 coprecipitating DNAs along the NEAT1 gene versus input under each indicated condition. (D) Poly(I:C) treatment induces NEAT1 expression in scramble-treated HeLa cells but not in CARM1 knockdown (KD) HeLa cells. (E) Poly(I:C) treatment reduces p54nrb methylation. HeLa cells were transfected with 2 μg/mL poly(I:C) for 6 h and then harvested for Western blotting analyses. (F) Poly(I:C) treatment enhances the nuclear retention of mRNAs containing IRAlus. HeLa cells were transfected with poly(I:C) for 6 h and then harvested for the nuclear and cytoplasmic fractionations. The relative subcellular distribution of transcripts of endogenous transcripts with IRAlus was investigated by RT-qPCR, normalized to the relative amount of actin mRNA in each fractionation, and compared with control HeLa cells. (G) The expression of CARM1 remains unchanged upon poly(I:C) treatment, as revealed by Western blotting. (H) The poly(I:C) treatment attenuates CARM1 localization to paraspeckles. (Left) Control and poly(I:C) transfected HeLa cells were stained with anti-p54nrb and anti-CARM1 antibodies, and representative images are shown. (Right) The statistics of p54nrb and CARM1 colocalization. The IODs (integrated optical densities) of punctuated anti-CARM1 and anti-p54nrb signals were measured by Image-Pro Plus from images taken with the same parameters (n > 100 double-positive staining cells). The ratio of IODCARM1 to IODp54 nrb was used to evaluate the extent of colocalization. The P-value from a one-tailed t-test in the pairwise comparison is shown. (I) A model of how the nuclear retention of IRAlus mRNAs at paraspeckles is regulated. (Left) Under normal conditions, CARM1 suppresses NEAT1 transcription and paraspeckle formation and also methylates p54nrb, resulting in the reduced ability to bind to mRNAs containing IRAlus. (Right) Upon appropriate stimulation, such as upon poly(I:C) treatment, actions of CARM1 are attenuated, resulting in an increased expression of NEAT1 RNA, unmethylated p54nrb, and enhanced nuclear retention of IRAlus mRNAs at paraspeckles. See the text for details. In B–D and F, error bars represent ±SD in triplicate experiments. (**) P < 0.01; n = 3.
CARM1 is involved in the poly(I:C)-stimulated enhancement of nuclear retention regulation
Finally, it will be of interest to identify conditions that affect CARM1-regulated mRNA nuclear retention within paraspeckles. It is known that the transcription of NEAT1 RNAs and the formation of paraspekles are induced upon virus infection or poly(I:C) treatment (Saha et al. 2006; Zhang et al. 2013a; Imamura et al. 2014). We therefore investigated whether CARM1-regulated nuclear retention is affected by poly(I:C) treatment. In our experiments, transfection of poly(I:C) led to increased expression of interferon β and NEAT1 RNAs (Fig. 7A,B). Importantly, we found that the enrichment of CARM1 at the NEAT1 promoter was reduced upon poly(I:C) treatment (Fig. 7C), corresponding to the observed up-regulation of NEAT1 RNAs (Fig. 7B). Furthermore, the expression of NEAT1 RNAs remained at high levels in CARM1 knockdown cells, and NEAT1 expression could not be further induced upon poly(I:C) treatment (Fig. 7D). In addition, we found that the treatment of cells with poly(I:C) led to a strong reduction of p54nrb methylation on these arginine residues (Fig. 7E), while the expression of p54nrb remained unchanged. Finally, poly(I:C) treatment also resulted in an enhanced nuclear retention of a number of examined mRNAs containing IRAlus (Fig. 7F). Interestingly, we found that upon poly(I:C) treatment, the expression of CARM1 protein remained largely unchanged (Fig. 7G), while the paraspeckle-localized CARM1 was significantly reduced (Fig. 7H), indicating that CARM1 may rapidly change its subcellular localization with an unknown mechanism upon the stress treatment. Taken together, these results suggest that actions of CARM1 at p54nrb methylation and NEAT1 transcription synergistically work together to regulate the export of transcripts containing IRAlus from paraspeckles under certain cellular stresses, such as the poly(I:C) treatment (Fig. 7I).
Discussion
Paraspeckles play a role in gene regulation through nuclear retention mediated by the association of their key protein component, p54nrb, with mRNAs containing inverted repeats (Alu repeats in humans) in their 3′ UTRs (Prasanth et al. 2005; Chen et al. 2008; Chen and Carmichael 2009; Mao et al. 2011a). Such nuclear-retained mRNAs are inefficiently exported to the cytoplasm, resulting in silencing of gene expression. However, the level of nuclear retention of transcripts containing IRAlus is variable, with some such mRNAs located in the nucleus, while others are in the cytoplasm (Prasanth et al. 2005; Chen et al. 2008; Hundley et al. 2008; Chen and Carmichael 2009; Elbarbary et al. 2013). How the nuclear retention of mRNAs containing IRAlus at paraspeckles is regulated has remained elusive. Here we demonstrate that CARM1 is a novel component of paraspeckles (Fig. 1). Disruption of CARM1 significantly enhances the nuclear retention of mRNAs containing 3′ UTR IRAlus and represses gene expression (Fig. 2; Supplemental Figs. S3, S4, S6). To achieve this regulation, CARM1 methylates p54nrb (Fig. 1; Supplemental Figs. S1, S2) and reduces its ability to associate with dsRNAs, such as mRNAs with IRAlus (Figs. 4, 5). On the other hand, CARM1 suppresses NEAT1 transcription and inhibits paraspeckle formation (Fig. 6).
While p54nrb is required for nuclear retention of mRNAs with 3′ UTR IRAlus, the catalytic activity of CARM1 is also required for this effect (Fig. 3). How is binding of p54nrb to mRNA-IRAlus achieved? Since it is known that p54nrb binds to inosine-containing RNAs (Zhang and Carmichael 2001) and that a strong correlation between A-to-I RNA editing and retention was seen in mRNAs containing inverted repeats (Prasanth et al. 2005; Chen and Carmichael 2008; Chen et al. 2008), it has been thought that one of the consequences of Alu RNA editing is to retain edited mRNAs within nuclear paraspeckles. However, some mRNAs with edited IRAlus in their 3′ UTRs were also observed in the cytoplasm (Chen et al. 2008; Hundley et al. 2008). Knockdown of ADAR1, which is responsible for A-to-I RNA editing, has little effect on the export of nuclear-retained IRAlus mRNAs (Elbarbary et al. 2013). These results thus suggested that another nuclear retention signal is required for p54nrb-mediated mRNA nuclear retention in addition to RNA editing. Another formal possibility is that long imperfect duplexes formed by IRAlus in the 3′ UTRs of genes might influence gene regulation even in the absence of editing. We showed that such unique hairpin structures can directly bind to unmethylated p54nrb (Figs. 4, 5), which in turn could lead to the nuclear retention of mRNAs containing IRAlus within paraspeckles. Moreover, it has been reported that p54nrb can bind DNA through its C terminus (Yang et al. 1993). Consistent with this view, we demonstrated that the selective binding of dsRNAs to p54nrb also requires its coiled-coil domain (Fig. 5).
Post-translational modifications of proteins play key roles in the regulation of many cellular processes by altering their associated effectors, including both proteins and a few reported RNAs. For instance, methylation/demethylation of Polycomb 2 protein could modulate its interaction with different lncRNAs (either TUG1 or MALAT1) with an unknown mechanism, resulting in the coordinated gene expression program in distinct subnuclear architectural compartments in response to growth signals (Yang et al. 2011). We observed that CARM1 methylation on p54nrb occurs at or near the dsRNA-binding coiled-coil domain and that knockdown of CARM1 significantly alters p54nrb-binding activity to dsRNAs but not ssRNAs (Fig. 5), resulting in enhanced nucleocytoplasmic export of mRNAs containing inverted Alu repeats. Thus, this regulation pathway represents one of a few examples where post-translational modification of an RNA-binding protein affects protein–RNA interaction and gene expression. Furthermore, although our data support the view that the reduced binding capability of methylated p54nrb to dsRNAs could result from a direct conformational change of p54nrb methylation at the coiled-coil domain, we cannot exclude the possibility that these methyl sites may recruit other effector proteins to facilitate the release of mRNA-IRAlus from methylated p54nrb (Yang et al. 2014). Finally, the dsRNA-binding protein STAU1 (Wickham et al. 1999) was recently shown to compete with p54nrb for the binding of 3′ UTR IRAlus, independent of editing (Elbarbary et al. 2013). It will be of interest to examine whether the binding of 3′ UTR IRAlus with STAU1 occurs after the release of mRNAs containing IRAlus from methylated p54nrb.
Although many lncRNAs have been implicated in gene regulation and mammalian development (Ulitsky and Bartel 2013), how the expression of lncRNAs is regulated has remained poorly understood. Interestingly, we found that paraspeckle-localized CARM1 also plays a role in the transcription regulation of NEAT1 RNAs and affects paraspeckle formation (Fig. 6). CARM1 is recognized as a transcriptional coactivator (Chen et al. 1999; Bedford and Clarke 2009) but also acts as a transcriptional cosuppressor, such as in the cyclic adenosine monophosphate signaling pathway (Xu et al. 2001). In the case of NEAT1 regulation, we found that CARM1 is enriched at the NEAT1 promoter and acts as a transcriptional repressor (Fig. 6). Conspicuously, this “negative” regulation of NEAT1 transcription and paraspeckle formation by CARM1 could in fact lead to a “positive” gene expression output by allowing the export of more mRNAs with IRAlus to the cytoplasm for protein translation. This final output in gene expression is therefore consistent with a general role of CARM1 in promoting gene transcription (Chen et al. 1999; Bedford and Clarke 2009). However, we do not yet know what directs CARM1 to the NEAT1 promoter and what other factors are involved in this NEAT1 transcription regulation by CARM1. It had been reported recently that NEAT1 RNAs can sequester the transcriptional regulator PSF to regulate gene expression (Hirose et al. 2014; Imamura et al. 2014). Since PSF is largely localized to paraspeckles, it will be of interest to examine whether PSF can autoregulate NEAT1 transcription in coordination with CARM1.
The identification of CARM1 functioning at two levels within paraspeckles is particularly interesting. On the one hand, CARM1 methylates p54nrb, resulting in the reduced ability to binding to mRNAs containing IRAlus (Figs. 3–5). On the other hand, CARM1 suppresses NEAT1 transcription and paraspeckle formation (Figs. 6, 7I). Actions of CARM1 at these two levels synergistically work together to regulate the export of transcripts containing IRAlus from paraspeckles. In response to certain cellular stressors, such as poly(I:C) treatment, we observed the reduced p54nrb methylation (enhanced mRNA nuclear retention) and the decreased binding of CARM1 to the NEAT1 promoter (enhanced transcription of NEAT1 and paraspeckle formation) (Fig. 7A–F). Thus, this paraspeckle-mediated nuclear retention was enhanced upon poly(I:C) stimulation, leading to less translation of mRNAs containing IRAlus (Fig. 7I).
Materials and methods
Cell culture, plasmids, transfection, and knockdown with shRNAs
HeLa, HEK293, and MEF cell lines were cultured using standard protocols. HEK293 CARM1 Flp-in stable cell line, wide-type, and CARM1−/− MEFs have been described (Cheng et al. 2007). Transfection was carried out with either X-tremeGENE 9 (Roche) or Lipofectamine 2000 (Invitrogen) according to the manufacturers’ protocols. To generate Flag-p54nrb-wt (MUT) or Flag-EGFP stable cell lines, pcDNA3.1 (+)-Flag-p54nrb-wt (MUT) or pcDNA3.1 (+)-Flag-EGFP was transfected into HEK293 CARM1 Flp-in cells followed by G418 selection. The plasmids pEGFP-SC-Nicn1-Alu (IRAlus) and pEGFP-SC-Lin28-Alu (IRAlus) have been described (Chen et al. 2008). CARM1 knockdown was carried out as described (Ou et al. 2011), and stable HeLa and HEK293 lines were generated. To knock down p54nrb, the sequence “GCAGGCGAAGTCTTCATTCAT” was inserted into pLVTHM vector with Mlu1 and Cla1, and the construct was packaged into lentivirus to infect HeLa cells. All plasmids used are listed in the Supplemental Material.
In vitro methylation assay
In vitro methylation assay was carried out as described (Cheng et al. 2007). GST-CARM1 and His-p54nrb were overexpressed and purified from E. coli. In vitro methylation reactions were performed in a final volume of 30 µL of PBS (pH 7.4). The reaction contained 0.5–1.0 µg of substrates and 0.2–0.4 µg of recombinant enzymes. All methylation reactions were carried out in the presence of 0.5 μCi S-adenosyl-L-[methyl-3H] methionine (85 Ci/mmol from a 0.5 mCi/mL stock solution; Perkin-Elmer). The reaction was incubated for 1 h at 30°C and then subjected to fluorography by separation on SDS-PAGE (12% gel), transferred to a PVDF membrane, treated with Enhance (Perkin-Elmer), and exposed to film overnight. After in vitro methylation followed by fluorography, the same membrane was subjected to Coomassie blue staining, immunoblotting with anti-His tag, or Ponceau staining. We overlaid the fluorograph with the stained membrane and signals from immunoblots and verified that the stained protein bands matched with the methylated bands.
Immunoprecipitation
HeLa cells (107) were harvested and suspended in immunoprecipitation buffer (50 mM HEPES at pH 7.6, 250 mM NaCl, 5 mM EDTA at pH 8.0, 0.1% NP-40, 1 mM PMSF, protease inhibitor cocktail) followed by sonication. After centrifuging at 13,000 rpm for 15 min at 4°C, the supernatant was transferred into a new tube and precleared with 10 μL of Dynabeads G. Next, the precleared supernatant was incubated with 20 μL of Dynabeads G with antibodies for p54nrb (BD) or IgG (Sigma) for 4 h at 4°C followed by washing with immunoprecipitation buffer. To harvest the protein complex, 50 μL of 1× SDS loading buffer (62.4 mM Tris at pH 6.8, 10% glycerol, 2% SDS, 0.0012% bromophenol blue) was added, incubated for 10 min at 95°C, and analyzed by Western blotting.
MS for methylation
pcDNA3.1 (+)-Flag- p54nrb was transfected into scramble and CARM1 knockdown stable HeLa cells for 24 h, and cells transfected with Flag-EGFP were used as control. Cells (107) were harvested as described above, and 20 μL of anti-Flag M2 beads (Sigma) was used in each reaction. One out of 10 immunoprecipitated beads was saved for Western blotting and silver staining, and the others were used for MS. Beads were washed with PBS and then aliquoted into three parts (one each for Glu-C, substilisin, and trypsin digestion) followed by incubation with DTT and urea for 30 min at 60°C. Next, 15 mM iodoacetamide was added into the beads and incubated for 20 min at room temperature. Digestion was performed using sequencing grade Glu-C, substilisin, and trypsin for 4 h at 37°C. Liquid chromatography-tandem MS (LC-MS/MS) was carried out on the Thermo Q Exactive Hybrid quadrupole orbitrap mass spectrometer.
Nuclear and cytoplasmic RNA fractionation, RNA isolation, qRT–PCR, and Northern blotting
Nuclear and cytoplasmic fractionation was carried out as described (Chen et al. 2008) with slight modifications. Cell pellets were suspended by gentle pipetting in 200 μL of lysis buffer (10 mM Tris at pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% Igepal, 2 mM vanadyl ribonucleoside complex [VRC]) and incubated for 5 min on ice. One-fifth of the lysate was saved as total RNA. The rest of the lysate was centrifuged at 1000g for 3 min at 4°C to pellet the nuclei, and the supernatant was the cytoplasmic fraction. To obtain pure cytoplasmic RNA, the supernatant fraction was further centrifuged at 13,000 rpm for 10 min at 4°C and then collected carefully to a new tube, and RNA was extracted with Trizol. To obtain pure nuclear RNA, the nuclear pellets were subjected to two additional washes with 160 µL of lysis buffer and one additional wash by adding 0.5% deoxycholic acid into the lysis buffer. Finally, the purified nuclei were resuspended in 100 µL of lysis buffer followed by extraction with Trizol. The RNA was extracted per standard protocols. For Northern blotting, equal amounts of RNAs from different fractionations were loaded. Northern blotting was performed using the DIG Northern starter kit (Roche). The probes used were produced by digoxigenin (Dig)-labeled in vitro transcription (IVT) of specific PCR products. Primers used for amplifying templates for IVT are listed in the Supplemental Material. For qRT–PCR, after treatment with DNase I (Ambion), equal amounts of RNAs from different fractionations were reverse-transcribed into cDNAs with SuperScript II (Invitrogen). β-Actin was used as an endogenous control.
RNA ISH and immunofluorescence microscopy
Simultaneous RNA ISH and immunofluorescence were performed as described (Yin et al. 2012) with slight modifications. Hybridization was performed with in vitro transcribed Dig-labeled probes. For colocalization studies, cells were costained with mouse anti-p54nrb (BD) and/or rabbit anti-CARM1 (Bethyl Laboratories). The nuclei were counterstained with DAPI. Images were taken with a Zeiss LSM 510 microscope or an Olympus IX70 DeltaVision RT deconvolution system microscope. For statistical analysis, >300 cells from each group were observed and calculated. Image analyses of signal intensity were carried out by Image-Pro Plus according to standard protocols and were described previously (Yin et al. 2015).
Formaldehyde cross-linking RIP
HeLa cells (107) were washed twice with 5 mL of PBS and cross-linked with 1% formaldehyde for 10 min at room temperature. Cross-linking was stopped by the addition of glycine to a final concentration of 0.25 M followed by incubation for 5 min at room temperature. After washing twice with 5 mL of cold PBS, cells were collected and suspended in 1 mL of RIP buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 1 mM PMSF, 2 mM VRC, protease inhibitor cocktail). The cells were homogenized by sonication and then centrifuged at 13,000 rpm for 10 min at 4°C to remove the insoluble material. Fifty microliters of supernatant was saved as input. The rest of the supernatant was precleared by applying 10 μL of Dynabeads G (Invitrogen) with 20 μg/mL yeast tRNA for 1 h at 4°C. Next, the precleared lysate was incubated with Dynabeads G that were precoated with 2 μg of antibodies for p54nrb (BD) or IgG (Sigma) for 4 h at 4°C. The beads were washed three times for 5 min with washing buffer I (50 mM Tris-HCl at pH 7.5, 1 M NaCl; 1% NP40, 1% sodium deoxycholate, 2 mM VRC) and three times for 5 min with washing buffer II (50 mM Tris-HCl at pH 7.5, 1 M NaCl, 1% NP40, 1% sodium deoxycholate, 2 mM VRC, 1 M urea). The immunoprecipitated complex was eluted from Dynabeads G by adding 100 μL of elution buffer (100 mM Tris HCl at pH 8.0, 10 mM EDTA, 1% SDS). Proteinase K (0.2 μg/μL) and 200 mM NaCl were added into the RNA sample and incubated for 1 h at 42°C followed by 1 h at 65°C. RNA was then extracted, digested with DNase I (Ambion), and used to synthesize cDNA using random hexamers (SuperScript III, Invitrogen) followed by qPCR analysis (AceQ, Vazyme).
UV cross-linking RIP
UV cross-linking RIP was carried out as described (Zhang et al. 2013b). HeLa cells (107) were washed twice with 5 mL of cold PBS and irradiated at 150 mJ/cm2 at 254 nm in a Stratalinker. Cells were collected and resuspended in 1 mL of RIP buffer. The cells were homogenized by sonication and then centrifuged at 13,000 rpm for 10 min at 4°C to remove the insoluble material. Fifty microliters of supernatant was saved as input. The rest of the supernatant was precleared by applying 10 μL of Dynabeads G (Invitrogen) with 20 μg/mL yeast tRNA for 1 h at 4°C. Next, the precleared lysate was incubated with Dynabeads G that were precoated with 2 μg of antibodies for p54nrb (BD) or IgG (Sigma) for 4 h at 4°C. The beads were washed three times for 5 min with washing buffer I and three times for 5 min with washing buffer II. The immunoprecipitated complex was eluted from Dynabeads G by adding 100 μL of elution buffer (100 mM Tris-HCl at pH7.0, 5 mM EDTA, 10 mM DTT, 1% SDS). Five microliters of 10 mg/mL proteinase K was added into the RNA sample and incubated for 30 min at 55°C. RNA was then extracted, digested with DNase I (Ambion), and used to synthesize cDNA using random hexamers (SuperScript III, Invitrogen) followed by qPCR analysis.
ChIP
ChIP was carried out as described (Zhang et al. 2013b). HeLa cells (107) were cross-linked in 1% formaldehyde for 5 min at room temperature, quenched by adding 0.25 M glycine, and then collected by cell scraper. After being suspended in 1 mL of ChIP lysis buffer (1% Triton X-100, 0.1% sodium deoxycholate, 50 mM Tris at pH 8.0, 150 mM NaCl, 5 mM EDTA), cells were sonicated until the majority of DNA fragments was 300–500 base pairs (bp). Supernatants were collected and subjected to preclearing with Dynabeads G (Invitrogen) with a supplement of 100 μg of ssDNA. Next, the precleared lysates were used for ChIP with 2 μg of CARM1 antibody (Bethyl Laboratories). ChIP was carried out overnight at 4°C. The beads were washed with 600 μL of ChIP lysis buffer, 600 μL of high-salt wash buffer (1% Triton X-100, 0.1% deoxycholate, 50 mM Tris at pH 8.0, 500 mM NaCl, 5 mM EDTA), and 600 μL of LiCl immune complex wash buffer (0.25 M LiCl, 0.5% Igepal, 0.5% deoxycholate, 10 mM Tris at pH 8.0, 1 mM EDTA) sequentially followed by two washes with 600 μL of 1× TE buffer (10 mM Tris at pH 8.0, 1 mM EDTA) at 4°C. The immunoprecipitated complex was eluted from Dynabeads G by adding 200 μL of fresh-prepared elution buffer (1% SDS, 0.1 M NaHCO3) with rotation for 15 min at room temperature. Next, the reverse cross-linking was carried out by adding 8 μL of 5 M NaCl and incubation for 4 h at 65°C followed by the addition of 4 μL of 0.5 M EDTA and 10 μL of 10 mg/mL proteinase K for 2 h at 55°C. DNA was then extracted and analyzed by qPCR.
NRO assay
The NRO in HeLa cells was performed as described (Zhang et al. 2013b). HeLa cells (107) were washed with cold PBS three times and incubated in swelling buffer (10 mM Tris at pH 7.5, 2 mM MgCl2, 3 mM CaCl2) for 5 min on ice. Cells were collected, suspended in 1.5 mL of lysis buffer (10 mM Tris at pH 7.5, 2 mM MgCl2, 3 mM CaCl2, 0.5% Igepal, 10% glycerol, 2 U/mL RNasin ribonuclease inhibitor [Promega]), gently pipetted, and then centrifuged at 1500g for 10 min at 4°C. The pellets were collected and subjected to another lysis to obtain purer nuclei. The resulting nuclear pellets were resuspended in 100 μL of NRO buffer (50 mM Tris at pH 7.5, 5 mM MgCl2, 150 mM KCl, 0.1% sarkosyl, 2 U/mL RNase inhibitor, 10 mM DTT) containing 0.1 mM ATP, GTP, CTP, and BrUTP (Sigma). Transcription was performed for 3 min on ice and then 5 min at room temperature. The reaction was stopped by addition of 600 μL of Trizol reagent, and RNA was extracted followed by the DNase I (Ambion) treatment to remove genomic DNA. The purified RNAs were incubated with 2 μg of anti-BrdU antibody (Sigma) or an equal amount of IgG antibody (Sigma) for 2 h at 4°C and then immunoprecipitated for 1 h with Dynabeads Protein G (Invitrogen) precoated with yeast tRNA (Sigma). Precipitated RNAs were extracted by Trizol reagent and used for cDNA synthesis and qPCR analysis. All measured samples were normalized to β-actin transcription.
Biotin-labeled RNA pull-down
Biotinylated RNA pull-down assays were performed as described (Zhang et al. 2013b). DNA fragments of full-length EGFP with T7 promoter on its 5′ end and fragment EGFP (76–495) with T7 promoters on both ends were in vitro transcribed with the biotin RNA-labeling mix (Roche) and T7 transcription kit (Promega). HeLa cells (107) were collected and suspended in 1 mL of RIP buffer (25 mM Tris at pH 7.5, 150 mM KCl, 0.5 mM DTT, 0.5% NP40, 1 mM PMSF, 2 mM VRC, protease inhibitor cocktail) followed by sonication. After being centrifuged at 13,000 rpm for 15 min at 4°C, the supernatant was transferred into a new tube and precleared with 40 μL of streptavidin Dynabeads for 20 min at 4°C. Next, 20 μg/mL yeast tRNA was added to block unspecific binding and incubated for 20 min at 4°C. The precleared lysate was divided into two parts, and each was supplemented with 2 μg of biotin-labeled ss_egfp (full length of egfp) or ds_egfp (an inverted repeated fragment of egfp) and incubated for 1.5 h followed by addition of 40 μL of streptavidin Dynabeads and incubation for another 1.5 h at room temperature. Beads were washed four times for 5 min with RIP buffer containing 0.5% sodium deoxycholate and boiled in 1× SDS loading buffer for 10 min at 100°C. The retrieved proteins were analyzed by Western blotting with anti-p54nrb (BD), anti-methyl-p54nrb (620me), and anti-PSF (Sigma).
tRSA RNA pull-down
tRSA RNA pull-down assays were carried out as described (Iioka et al. 2011) with modifications. IRAlus from the 3′ UTR of Nicn1 were cloned into pcDNA3 with the tRSA tag at its 5′end. tRSA-IRAlus or tRSA was in vitro transcribed using the T7 transcription kit (Promega). Ten micrograms per reaction of synthetic RNAs was denatured for 5 min at 65°C and cooled to room temperature in the presence of PA buffer (10 mM Tris at pH 7.4, 10 mM MgCl2, 100 mM NH4Cl). Next, the RNAs were incubated with 40 μL of streptavidin Dynabeads for 20 min at 4°C in the presence of 2 U/mL RNasin (Promega). HeLa cells (107) were collected and resuspended in 1 mL of lysis buffer (25 mM Tris at pH 7.4, 150 mM KCl, 0.5 mM DTT, 0.5% NP40, 1 mM PMSF, 2 mM VRC, protease inhibitor cocktail) followed by sonication. After centrifuging at 13,000 rpm for 15 min 4°C, the supernatant was transferred into a new tube and precleared with 40 μL of streptavidin Dynabeads for 20 min at 4°C followed by the addition of 20 μg/mL yeast tRNA for 20 min at 4°C. Beads prebound with tRSA-IRAlus RNA or tRSA RNA were incubated with precleared lysates for 4 h at 4°C followed by washing with lysis buffer containing 0.5% sodium deoxycholate. To harvest the protein complex, 50 μL of 1× SDS loading buffer was added and incubated for 10 min at 95°C and then analyzed by Western blotting.
Dig-labeled RNA pull-down
Dig-RNA pull-down assays were carried out as described (Arab et al. 2014) with modifications. Two 10-cm dishes of cells expressing Flag-p54nrb-wt (MUT) or Flag-EGFP were used to immunoprecipitate with anti-Flag M2 (20 μL for each reaction) (Sigma). After immunoprecipitation and washing, one out of five beads was saved for Western blot. The rest were equilibrated in binding buffer (50 mM Tris at pH 8.0, 10% glycerol, 100 mM KCl, 5 mM MgCl2, 10 mM β-mercaptoethanol, 0.1% NP-40) and incubated with 200 ng of Dig-labeled RNA in binding buffer for 1.5 h at room temperature. The Dig-labeled RNA was produced by Dig-labeled IVT of an IRAlus DNA fragment, which was amplified from the 3′ UTR of Nicn1. After washing with immunoprecipitation buffer containing 500 mM NaCl, the bound RNA was extracted and analyzed by Northern blotting.
Computational pipeline for the analysis of genes containing 3′ UTR IRAlus enriched in the NEAT1 DNA CHART
A stringent pipeline was developed to identify 3′ UTR IRAlus genes enriched by NEAT1 DNA CHART. To identify 3′ UTR IRAlus genes, IRAlus were identified by RepeatMasker (Tarailo-Graovac and Chen 2009). The 3′ UTRs were defined in a RefSeq gene file. Next, the overlap region between IRAlus and the 3′ UTR was calculated to ensure that IRAlus were located in the 3′ UTRs. In total, 545 genes containing 3′ UTR IRAlus were obtained. In MCF7 cells, 388 genes containing 3′ UTR IRAlus were expressed (reads per kilobase per million mapped reads [RPKM] ≥ 2) by analyzing the available RNA sequencing data sets (MCF7 RNA-seq; Gene Expression Omnibus [GEO]: GSE47042) (Janky et al. 2014). To analyze the NEAT1 DNA CHART-enriched IRAlus-containing genes, the NEAT1 DNA CHART-seq data sets carried out in MCF7 cells (GEO: GSE47042) (West et al. 2014) were used to call peaks by MACS (version 1.4.2, 20120305) (Feng et al. 2012). One-hundred-forty-nine MCF cells expressed genes that contain 3′ UTR IRAlus enriched by the NEAT1 DNA CHART.
Supplementary Material
Acknowledgments
We are grateful to G. Carmichael for critical reading of the manuscript, M.R. Stallcup for CARM1 shRNA, and M. Person for MS analysis at the Protein and Metabolite Analysis Facility (University of Texas, Austin) supported by RP110782 (Cancer Prevention Research Institute of Texas). This work is supported by grants 91440202 and 31322018 from the Natural Science Foundation of China (NSFC), and XDA01010206 from Chinese Academy of Sciences (CAS) to L.-L.C.; 31271390 from NSFC to L.Y; and DK062248 from National Institutes of Health to M.T.B. M.T.B. is a cofounder of EpiCypher.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.257048.114.
References
- Arab K, Park YJ, Lindroth AM, Schafer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et al. 2014. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell 55: 604–614. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. 2009. Protein arginine methylation in mammals: who, what, and why. Mol Cell 33: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CS, Fox AH. 2009. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol 186: 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. 2008. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle 7: 3294–3301. [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. 1999. Regulation of transcription by a protein methyltransferase. Science 284: 2174–2177. [DOI] [PubMed] [Google Scholar]
- Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE. 2002. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J Biol Chem 277: 4324–4333. [DOI] [PubMed] [Google Scholar]
- Chen LL, DeCerbo JN, Carmichael GG. 2008. Alu element-mediated gene silencing. EMBO J 27: 1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Cote J, Shaaban S, Bedford MT. 2007. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell 25: 71–83. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, Li W, Tian B, Maquat LE. 2013. STAU1 binding 3′ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev 27: 1495–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. 2002. Paraspeckles: a novel nuclear domain. Curr Biol 12: 13–25. [DOI] [PubMed] [Google Scholar]
- Fox AH, Bond CS, Lamond AI. 2005. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell 16: 5304–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, et al. 2014. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 25: 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Krauchuk AA, Bass BL. 2008. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. RNA 14: 2050–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iioka H, Loiselle D, Haystead TA, Macara IG. 2011. Efficient detection of RNA–protein interactions using tethered RNAs. Nucleic Acids Res 39: e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, et al. 2014. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell 53: 393–406. [DOI] [PubMed] [Google Scholar]
- Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D, et al. 2014. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 10: e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Bedford MT. 2002. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep 3: 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Koh SS, Zhang X, Chen X, Stallcup MR. 2002. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol 22: 3621–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhao Z, Carter C, Ehrlich LI, Bedford MT, Richie ER. 2013. Coactivator-associated arginine methyltransferase 1 regulates fetal hematopoiesis and thymocyte development. J Immunol 190: 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL. 2011a. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13: 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. 2011b. Biogenesis and function of nuclear bodies. Trends Genet 27: 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. 2012. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J 31: 4020–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou CY, LaBonte MJ, Manegold PC, So AY, Ianculescu I, Gerke DS, Yamamoto KR, Ladner RD, Kahn M, Kim JH, et al. 2011. A coactivator role of CARM1 in the dysregulation of β-catenin activity in colorectal cancer cell growth and gene expression. Mol Cancer Res 9: 660–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. 2005. Regulating gene expression through RNA nuclear retention. Cell 123: 249–263. [DOI] [PubMed] [Google Scholar]
- Saha S, Murthy S, Rangarajan PN. 2006. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J Gen Virol 87: 1991–1995. [DOI] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. 2009. MENε/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci 106: 2525–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. 2009. MENε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics C25: 4.10.1–4.10.14. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. 2013. lincRNAs: genomics, evolution, and mechanisms. Cell 154: 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. 2014. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell 55: 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. 1999. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol 19: 2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. 2001. A transcriptional switch mediated by cofactor methylation. Science 294: 2507–2511. [DOI] [PubMed] [Google Scholar]
- Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, Bedford MT. 2008. CARM1 promotes adipocyte differentiation by coactivating PPARγ. EMBO Rep 9: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Hanke JH, Carayannopoulos L, Craft CM, Capra JD, Tucker PW. 1993. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol Cell Biol 13: 5593–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, Bedford MT. 2010. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell 40: 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi Kenneth A, Grinstein Jonathan D, Dorrestein Pieter C, Rosenfeld Michael G. 2011. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT. 2014. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell 53: 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. 2012. Long noncoding RNAs with snoRNA ends. Mol Cell 48: 219–230. [DOI] [PubMed] [Google Scholar]
- Yin QF, Hu SB, Xu YF, Yang L, Carmichael GG, Chen LL. 2015. snoVectos for nuclear expression of RNA. Nucleic Acids Res 43: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG. 2001. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106: 465–475. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Chen CY, Yedavalli VS, Jeang KT. 2013a. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio 4: e00596–00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. 2013b. Circular intronic long noncoding RNAs. Mol Cell 51: 792–806. [DOI] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL. 2009. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev 19: 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.