Eckwahl et al. used high-throughput sequencing to obtain a comprehensive description of the RNAs packaged by a model retrovirus, murine leukemia virus. The major encapsidated host RNAs are noncoding RNAs (ncRNAs) and members of the VL30 class of endogenous retroviruses. Packaging of both pre-tRNAs and U6 snRNA requires the nuclear export receptor Exportin-5. Adenylated and uridylated forms of these RNAs accumulate in cells and virions when the cytoplasmic exoribonuclease DIS3L2 and subunits of the RNA exosome are depleted.
Keywords: retrovirus, noncoding RNAs, RNA surveillance, exoribonucleases
Abstract
Although all retroviruses recruit host cell RNAs into virions, both the spectrum of RNAs encapsidated and the mechanisms by which they are recruited remain largely unknown. Here, we used high-throughput sequencing to obtain a comprehensive description of the RNAs packaged by a model retrovirus, murine leukemia virus. The major encapsidated host RNAs are noncoding RNAs (ncRNAs) and members of the VL30 class of endogenous retroviruses. Remarkably, although Moloney leukemia virus (MLV) assembles in the cytoplasm, precursors to specific tRNAs, small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs) are all enriched in virions. Consistent with their cytoplasmic recruitment, packaging of both pre-tRNAs and U6 snRNA requires the nuclear export receptor Exportin-5. Adenylated and uridylated forms of these RNAs accumulate in cells and virions when the cytoplasmic exoribonuclease DIS3L2 and subunits of the RNA exosome are depleted. Together, our data reveal that MLV recruits RNAs from a novel host cell surveillance pathway in which unprocessed and unneeded nuclear ncRNAs are exported to the cytoplasm for degradation.
A remarkable but poorly understood feature of retroviruses is their propensity to selectively encapsidate host cell RNAs. Up to 50% of virion RNA derives from the host (Berkowitz et al. 1996). An abundant component of all known retroviruses is the 7SL RNA subunit of the signal recognition particle (SRP) (Bishop et al. 1970; Onafuwa-Nuga et al. 2005, 2006). Each retrovirus also packages several tRNAs, including the particular tRNA needed to prime reverse transcription of its genome (Cen et al. 2002). For Moloney leukemia virus (MLV), probing for candidate RNAs revealed that the spliceosomal U6 small nuclear RNA (snRNA) and noncoding RNAs (ncRNAs) called Y RNAs are enriched within virions, while for human immunodeficiency virus type I (HIV-1), U6 snRNA and some nonprimer tRNAs are present (Jiang et al. 1993; Zaitseva et al. 2006; Houzet et al. 2007; Pavon-Eternod et al. 2010). Deep sequencing of short RNAs (<40 nucleotides [nt]) from HIV-1 and simian immunodeficiency virions suggests that some cellular microRNAs (miRNAs) are also packaged selectively (Schopman et al. 2012; Brameier et al. 2013). One function of host RNAs may be to promote virion assembly, since virus-like particles (VLPs) can form in the absence of viral genomic RNA (gRNA) (Levin et al. 1974), but RNA is needed to promote oligomerization of Gag, the major retrovirus structural protein (Johnson et al. 2002).
At least some host ncRNAs are selectively packaged. Recruitment of the primer tRNA requires interactions with reverse transcriptase and can involve binding of Gag to the respective tRNA synthetase (Levin and Seidman 1981; Cen et al. 2002). Additionally, some nonprimer tRNAs are abundant in virions, while others are poorly packaged (Onafuwa-Nuga et al. 2005; Pavon-Eternod et al. 2010). Consistent with specific recruitment, 7SL RNA is 250-fold more enriched in HIV-1 virions than actin mRNA (Onafuwa-Nuga et al. 2006). However, the full complement of packaged RNAs has not been reported for any retrovirus, and apart from the primer tRNAs, the mechanisms by which these RNAs are selected and the extent to which they contribute to virus replication are unknown.
Although both MLV and HIV assemble at the plasma membrane, the site at which host RNA packaging occurs has not been elucidated. At least one encapsidated RNA, U6 snRNA, is believed to reside entirely within nuclei (Hopper 2006). Consistent with nuclear packaging, some ncRNAs appear to be recruited immediately after synthesis (Onafuwa-Nuga et al. 2006; Wang et al. 2007; Garcia et al. 2009). For example, 7SL packaging by HIV-1 is reduced when SRP19, a protein that binds nascent 7SL and nucleates SRP assembly (Akopian et al. 2013), is overexpressed, suggesting that 7SL is packaged before SRP19 binds (Wang et al. 2007). Similarly, although Y RNAs are unstable in the absence of Ro60 protein, the RNAs are packaged efficiently in MLV-infected cells lacking Ro60, suggesting that packaging occurs before Ro60 binding (Garcia et al. 2009). However, while both HIV-1 and MLV Gag proteins have been detected in nuclei, it is controversial whether they normally traffic through nuclei (Stake et al. 2013). Moreover, as neither 7SL RNA nor Y RNAs are made as obligate 5′ or 3′ extended precursors, evidence that they are packaged as immature RNAs remains indirect.
Here, we used whole-transcriptome sequencing to characterize the RNAs encapsidated by MLV, a model retrovirus. In addition to previously described RNAs such as 7SL, Y RNAs, and U6, we identified several other ncRNAs that are packaged by MLV with high selectivity. Surprisingly, we discovered that precursors to specific tRNAs, snRNAs, and small nucleolar RNAs (snoRNAs) are all highly enriched in retroviruses. We show that MLV packages at least some of these normally nuclear RNAs from a novel host pathway in which newly synthesized ncRNAs are exported to the cytoplasm for degradation.
Results
Host ncRNAs are major components of the MLV ‘RNA packageome’
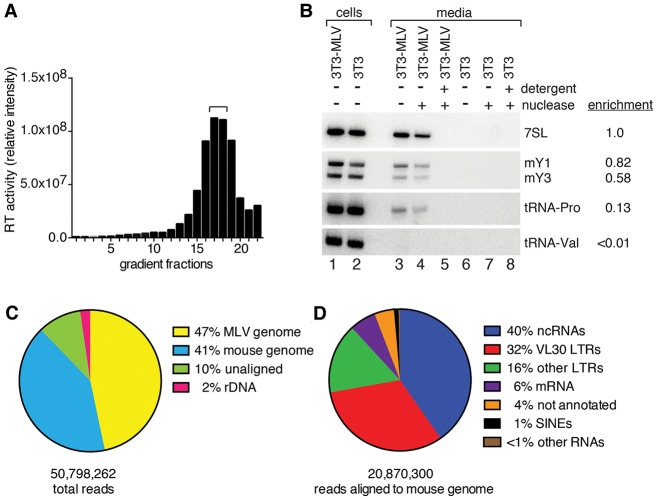
To identify packaged RNAs, we prepared highly purified virus and devised a cDNA synthesis protocol that would recover a wide range of RNAs in a single library. After harvesting culture supernatants from chronically infected NIH3T3 cells (NIH3T3-MLV) (Onafuwa-Nuga et al. 2005), MLV was purified on iodixanol gradients (Fig. 1A). To ensure that all RNA was within membranes, peak fractions were treated with micrococcal nuclease with or without Triton X-100 prior to RNA extraction. Using Northern blotting, we confirmed that known virion-enriched RNAs such as 7SL, Y RNAs, and the primer tRNAPro were present, and that a poorly encapsidated RNA, tRNAVal (Onafuwa-Nuga et al. 2005), was not detected (Fig. 1B). To capture a variety of RNAs, including mRNAs and RNAs with end modifications that preclude linker ligation (such as spliceosomal snRNAs), we fragmented the virion RNA prior to cDNA synthesis. Afterward, 50- 160-base-pair (bp) cDNAs were gel-purified, ligated to adapters, amplified, and sequenced. This strategy allowed recovery of diverse RNAs (see below) but omitted RNAs <50 nt, such as miRNAs. To identify contaminants, cDNA was prepared from uninfected cell medium that was subjected to identical manipulations (Supplemental Fig. 1).
Figure 1.
Elucidating the MLV RNA packageome. (A) Following iodixanol gradient fractionation, MLV was detected by assaying RT. Fractions 17 and 18 (bracket) were used to prepare cDNA. (B) To confirm that detected RNAs were within virions, virus-containing fractions were either untreated (lanes 3,6) or incubated with micrococcal nuclease in the absence (lanes 4,7) or presence (lanes 5,8) of Triton X-100. Extracted RNA was subjected to Northern blotting to detect the indicated ncRNAs. (Lanes 1,2) RNA from MLV-infected and uninfected cells. The level of each RNA in virions (lane 4) compared with its level in an arbitrary amount of total cell RNA (lane 1) was used to determine its enrichment in virions relative to 7SL RNA. (C) After aligning reads sequentially to the MLV genome, the rDNA, and the mouse genome (which lacks an intact 45S rDNA), the fraction of reads in each category is shown. (D) Summary of the alignment of reads to the mouse genome. RNA classes accounting for <1% of reads are not shown.
Bioinformatic analyses of the resulting data revealed that ∼47% of reads in the MLV data set aligned to the MLV genome, while ∼41% aligned to the mouse genome (Fig. 1C). Most reads that mapped to the mouse genome were derived from ncRNAs and VL30 long terminal repeats (LTRs), a class of retrotransposons that are abundantly packaged by MLV (French and Norton 1997). Consistent with previous reports (Onafuwa-Nuga et al. 2005; Rulli et al. 2007), mRNAs were not major components (Fig. 1D).
Using RPKM (reads per kilobase of transcript per million mapped reads) to normalize for RNA length and sequencing depth (Mortazavi et al. 2008), the most abundant RNAs included known packaged RNAs such as 7SL, Y RNAs, U6, and B1 short interspersed elements (SINEs) (Onafuwa-Nuga et al. 2005) as well as RNAs not detected previously (Supplemental Table S1). New RNAs included the RNA subunit of large cytoplasmic ribonucleoproteins (RNPs) known as vaults (Kickhoefer et al. 2001), the rodent-specific 4.5S(I) RNA (Ro-Choi et al. 1972), B2 SINEs (Krayev et al. 1982), and several nonprimer tRNAs (Supplemental Tables S1, S2). Consistent with estimates that each MLV virion contains six to eight copies of 7SL and four to five copies of mY1 RNA (Onafuwa-Nuga et al. 2005; Garcia et al. 2009), both RNAs had higher RPKM values than gRNA, which is present in two copies per virion. Moreover, since our cDNA protocol could underrepresent short RNAs (which are more likely to be fragmented to <50 nt), and some ncRNAs (such as tRNAs) are highly structured and/or contain modifications that impede reverse transcription, a cDNA-based metric such as RPKM may underestimate ncRNA levels.
Specific ncRNAs are highly enriched in MLV virions
To determine which RNAs are most selectively encapsidated, we used Northern blotting to compare the levels of each newly identified RNA in virions with its levels in an arbitrary amount of cell RNA. To allow comparisons, the enrichment of each RNA in virions was normalized to 7SL, the RNA reported previously to be most selectively packaged (Onafuwa-Nuga et al. 2005). Using this approach, vault RNA appeared more than twice as enriched in virions as 7SL, while scB1, a rodent-specific ncRNA related to the Alu family of human SINEs (Maraia 1991), was slightly more enriched (Fig. 2A). Transcripts from B2 SINEs were also enriched, although quantitation was complicated by their size heterogeneity and the finding that the encapsidated form was more discrete than the cellular transcripts (Fig. 2B). Other RNAs were present in virions but were less enriched than either 7SL or the primer tRNAPro. For example, two rodent-specific 4.5S RNAs, 4.5S(H) (Harada and Kato 1980) and 4.5S(I) (Ro-Choi et al. 1972), were detected in virions but were 16-fold to 33-fold less enriched than 7SL (Fig. 2A,B). As observed for B2 SINEs, the packaged 4.5S(I) RNAs were shorter than the majority of cellular transcripts. Whether the shorter RNAs are preferentially encapsidated or are processed after encapsidation is unknown.
Figure 2.
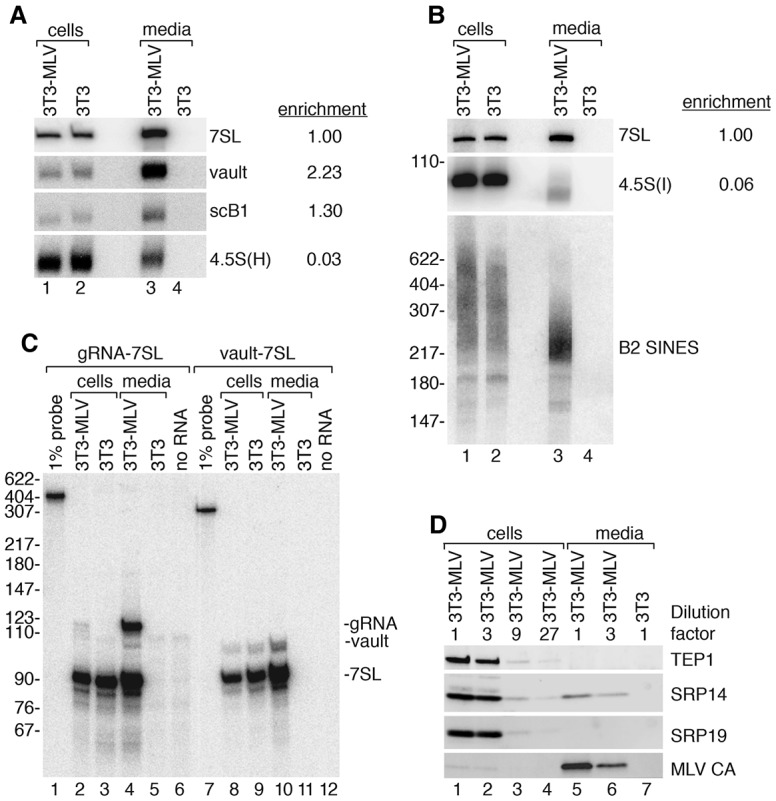
Host ncRNAs packaged by MLV. (A,B) To assess enrichment of the indicated RNAs in virions relative to 7SL, Northern blotting was used to compare their levels in virions (lane 3) and in an arbitrary amount of cell RNA (lane 1). RNA from uninfected cells (lane 2) and the media (lane 4) was also assayed. Since B2 SINEs were not present as discrete RNAs, their enrichment was not calculated. (C) RNase protection was used to determine the stoichiometry of vault RNA in virions. In lanes 1–6, a probe that detects both 7SL and MLV gRNA was used, while in lanes 7–12, the probe is complementary to 7SL and vault RNA. (Lanes 1,7) Undigested probe. (D) Immunoblotting was used to assess packaging of TEP1, SRP14, and SRP19. Serial dilutions were used to compare protein levels. Virus and cell lysates labeled “1” contain equal amounts of 7SL RNA, as measured by Northern blotting of RNAs in parallel samples. Capsid protein (CA) was detected as a control.
Vault RNA, but not its protein partner, is a stoichiometric component of virions
Although both vault and scB1 RNAs are enriched in virions, scB1 RNA is ∼100-fold less abundant in cells than 7SL (Chang and Maraia 1993) and thus is unlikely to be packaged in stoichiometric quantities. Since vault RNA levels have not been reported, we used RNase protection to determine its abundance relative to 7SL in cells and its copy number in virions. Using a probe complementary to 7SL and vault RNA followed by quantitation of the protected fragments, we determined that vault RNA is ∼20-fold less abundant than 7SL in cells and eightfold less abundant in virions (Fig. 2C). Using a similar approach, we confirmed that 7SL RNA is packaged in threefold to fourfold molar excess of gRNA or approximately six to eight copies per virion (Fig. 2C; Onafuwa-Nuga et al. 2005). Thus, approximately one copy of vault RNA is present per virion.
Since vault RNA is normally bound by TEP1 (Kickhoefer et al. 2001), we examined whether TEP1 was also packaged. To this end, we first normalized cell and virus lysates by the amounts of 7SL RNA detected in Northern blots of parallel samples. Since vault RNA is approximately twice as enriched in virions as 7SL (Fig. 2A), virus lysates contain twice as much vault RNA as cell lysates. Upon immunoblotting, TEP1 was detected in cell lysates even when the lysate was diluted 27-fold but was not detected in the undiluted virus lysate (Fig. 2D, cf. lanes 1–4 and 5). Thus, as observed for Y RNAs (Garcia et al. 2009), vault RNA may be recruited into virions without its major protein partner.
We also examined SRP protein packaging. SRP54, the last subunit to assemble into SRP (Akopian et al. 2013), is not detected in HIV-1 virions (Onafuwa-Nuga et al. 2006). We also failed to detect SRP19 (Fig. 2D), the subunit whose binding to 7SL is required for SRP54 association (Akopian et al. 2013). However, we detected low levels of SRP14, which binds with SRP9 as a heterodimer to the 7SL RNA Alu domain. Quantitation revealed that although the undiluted cell and virus lysates contained equal amounts of 7SL RNA, the amount of SRP14 in virions was 4.2-fold less than that detected in cell lysates (Fig. 2D, cf. lanes 1 and 5). Experiments to examine SRP9 packaging were unsuccessful, as the available antibodies do not detect this protein in NIH3T3 cells (Bovia et al. 1995). The finding that some SRP14 is encapsidated is consistent with the proposal that Gag recognizes 7SL RNA after SRP9/14 binding but before binding of the remaining subunits (Keene and Telesnitsky 2012). An alternative but not exclusive possibility is that scB1, which also binds SRP9/14 (Chang and Maraia 1993), is packaged with these proteins.
Precursors to tRNAs, snoRNAs, and snRNAs are preferentially encapsidated
Surprisingly, although precursors to tRNAs, snRNAs, and snoRNAs are rare in cells, these RNAs appeared enriched in virions. For example, many sequencing reads derived from unspliced forms of tRNA-Ile-UAU and tRNA-Tyr-GUA, two intron-containing tRNAs (Fig. 3A,B). Northern blotting confirmed that the unspliced pre-tRNAs, but not the mature tRNAs, were highly enriched in virions (Fig. 3C,D). Moreover, although cleaved tRNA-Ile-UAU halves were not detected in total cell RNA, they were packaged by MLV (Fig. 3C). We also detected reads from these and other tRNAs that included 5′ leader and/or 3′ trailer sequences (Fig. 3A,B,E). Notably, for at least one locus, a tRNA-His-GUG on chromosome 2, some reads extended through the first termination signal, stopping at the second site (TTTT) (Fig. 3E [arrowheads], F [underlined]). Northern blotting confirmed that pre-tRNAs corresponding to termination at both sites were enriched in virions (Fig. 3G). Remarkably, although pre-tRNAs generated by termination at the second site (with an ∼175-nt trailer) are 18-fold less abundant in cellular RNA than pre-tRNAs terminating at the first site, they were more selectively packaged. Both pre-tRNAs were more enriched in virions than the mature tRNA-His (Fig. 3G).
Figure 3.

MLV selectively packages pre-tRNAs. (A,B) Reads aligned to a tRNA-Ile-UAU (tRNA999) and a tRNA-Tyr-GUA (tRNA109) were visualized using the University of California at Santa Cruz (UCSC) genome browser. (Arrows) Transcription direction. Leader, intron, and trailer sequences are indicated by lines, while exons are denoted by boxes. The Y-axis indicates read depth. (C,D) Northern blotting was used to compare levels of precursor and mature forms of tRNA-Ile-UAU (C) and tRNA-Tyr-GUA (D) in cells and virions. The tRNA-Ile probe detected 3′ exon sequences. For tRNA-Tyr, the probes detected the intron of the most highly packaged locus, tRNA109 (D, top), or the spliced mature tRNA (middle). Because each of the 10 tRNA-Tyr loci has a different intron but the same mature sequence, the intron probe detected only tRNA109 transcripts, while the mature probe detected all spliced tRNA-Tyr-GUA. (Bottom panels) Blots were reprobed to detect 7SL. (E) UCSC genome browser visualization of reads aligned to a tRNA-His-GUG (tRNA587). (Arrowheads) Termination signals (≥TTTT). (F) The sequence of tRNA587 (underlined) and flanking sequences are shown. (Arrow) Possible transcription initiation site based on aligned reads. Termination signals are also underlined. (G) Northern blotting was performed to detect 3′ extended forms of tRNA-His-GUG (top), the mature tRNA (middle), and 7SL (bottom). The pre-tRNA probe detected tRNAs from three loci that have 26 nt of similar sequence in the 3′ trailers (tRNA587, tRNA1431, and tRNA1432). For tRNA1431 and tRNA1432, only transcripts terminating at the first site were detected in virions. The mature probe detected tRNAs from eight tRNA-His-GUG loci.
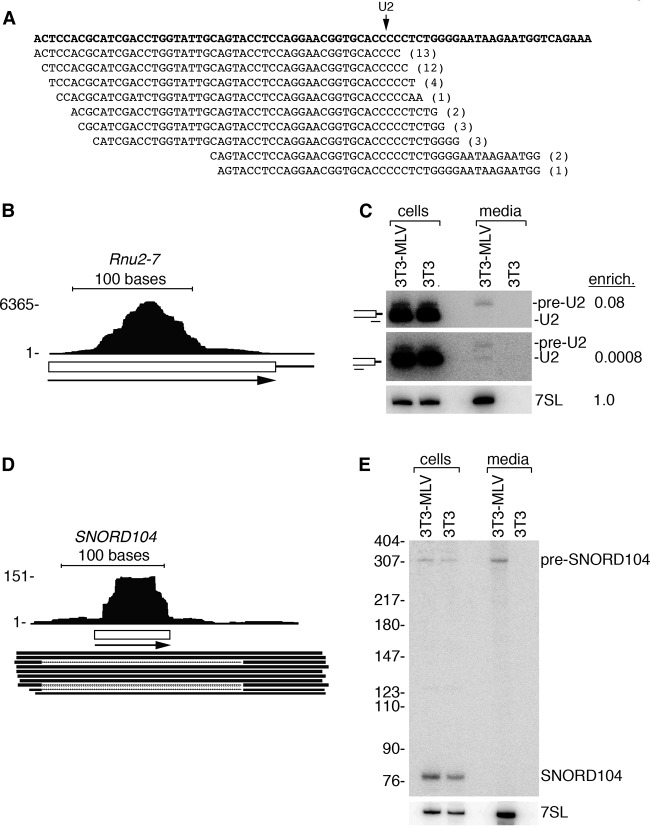
We also obtained evidence that MLV packages precursors to spliceosomal snRNAs and snoRNAs. For example, many reads derived from 3′ extended U2 spliceosomal snRNAs (Fig. 4A,B). This snRNA is cleaved cotranscriptionally to form a precursor with an ∼10-nt 3′ extension that is removed after the snRNA assembles with its core proteins (Matera and Wang 2014). By performing Northern blotting with a probe that preferentially detects the 3′ extended U2 (Fig. 4C, top panel) as well as a probe that detects the precursor and mature U2 snRNAs equally (middle panel), we confirmed that pre-U2 snRNAs are preferentially packaged compared with the far more abundant mature U2 snRNAs. Similarly, some reads mapped 5′ and 3′ to SNORD104, which encodes a member of the box C/D class of snoRNAs predicted to guide 2′-O- ribose methylation of 23S rRNA (Fig. 4D; Vitali et al. 2003). Northern blotting revealed that the form of SNORD104 in virions was an ∼320-nt RNA corresponding to the long ncRNA that is the snoRNA precursor (Fig. 4E). Our discovery that specific pre-tRNAs, pre-snRNAs, and pre-snoRNAs are highly enriched in virions allows us to conclude that MLV selectively packages newly synthesized ncRNAs.
Figure 4.
Selective packaging of snRNA and snoRNA precursors. (A) Reads aligning to the U2 snRNA 3′ end. The Rnu2-7 genomic sequence is at the top. (Arrow) Mature end; (parentheses) number of identical reads. (B) Reads aligned to the Rnu2-7 locus (chr11: 101,641,748–101,641,934). (C) Northern blot to detect pre-U2 (top), mature U2 (middle), and 7SL (bottom). The pre-U2 probe contains 8 nt complementary to the 3′ extension and 12 nt complementary to mature U2. (Top) Thus, pre-U2 snRNAs are preferentially detected. (Middle) Reprobing with a probe that is entirely complementary to mature U2 reveals that approximately half the encapsidated RNA is the pre-snRNA. Cartoons showing positions of these probes are at the left. (D) Reads aligned to SNORD104. ESTs spanning U104 snoRNA (box) are also shown. (E) Northern blot to examine SNORD104 packaging.
snRNA and tRNA precursors are packaged following nuclear export
Although MLV assembly takes place at the plasma membrane, some packaged ncRNAs, such as pre-U2 snRNAs, are present in both the nucleus and cytoplasm, while others, such as pre-tRNAs and U6 snRNAs, are believed to be confined to mammalian nuclei (Hopper 2006). To test whether these or other packaged RNAs access the cytoplasm by interacting with gRNA, we examined cells expressing a provirus lacking the Ψ sequence required for gRNA packaging. Although gRNA packaging was reduced 16.3-fold, packaging of most ncRNAs was similar to control Ψ+ cells (Supplemental Fig. 2). One exception, 4.5S(H) RNA, was reduced in virions, consistent with a proposal that this RNA base-pairs with gRNA (Supplemental Fig. 2B; Harada and Kato 1980).
Since pre-U2 snRNAs undergo nuclear export followed by reimport as part of their biogenesis, we tested whether depleting components of this transport pathway affected packaging. We used siRNAs to deplete either PHAX, which adapts the pre-snRNAs for CRM1-dependent export, or snurportin (SPN), which binds the assembled snRNPs and adapts them for importinβ-mediated reimport (Matera and Wang 2014). Successful depletion was confirmed using RT followed by quantitative PCR (RT-qPCR) (Supplemental Fig. 3A). Strikingly, packaging of pre-U2 relative to mature U2 snRNA increased 2.8-fold when SPN was depleted, blocking nuclear reimport (Fig. 5A). Conversely, packaging of pre-U2 relative to mature U2 decreased twofold when nuclear export was impaired by depleting PHAX. Thus, pre-U2 may be packaged from the cytoplasm through a process that occurs in competition with the normal snRNP biogenesis pathway.
Figure 5.
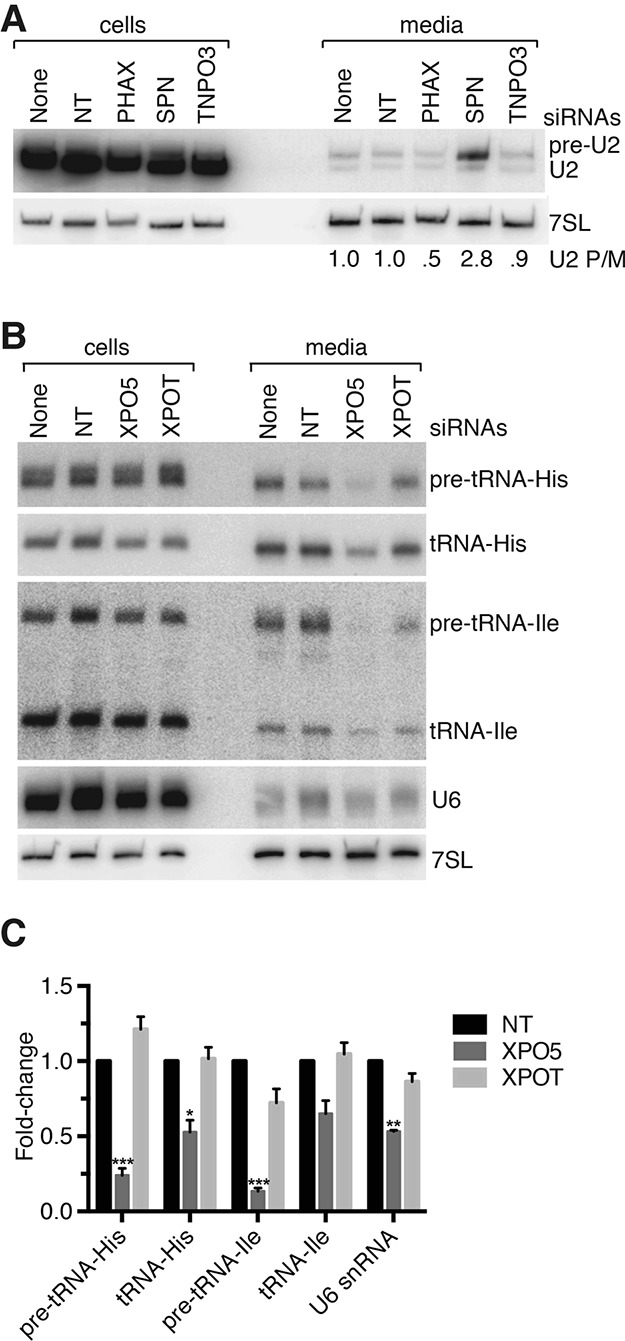
Nuclear export is required for pre-tRNA and snRNA packaging. (A) After transfecting cells with the indicated siRNAs, virus was harvested between 24 and 48 h, and cells were harvested after 48 h. After confirming that depletions were effective (Supplemental Fig. 3), Northern blotting was performed to detect U2 (top) and 7SL (bottom). The ratio of pre-U2 to mature U2 (P/M) in each virus sample is shown below. (B) After transfecting cells with the indicated siRNAs, virus was harvested between 48 and 72 h, and cells were harvested after 72 h. (C) Quantitation of results from B. Data are mean values from four biological replicates ± S.E.M. Results are relative to packaging when nontargeting siRNAs were transfected. Loading was normalized to 7SL. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001 (Student's t-test).
We also tested whether nuclear export was required for pre-tRNA packaging. Depleting either the major tRNA export receptor, Exportin-T, or Transportin-3, which may export tRNAs with damaged 3′ ends (Zhou et al. 2011), did not affect packaging of any examined RNAs (Fig. 5B; Supplemental Fig. 3). We also depleted Exportin-5 (XPO5), which exports both mature tRNAs and ncRNAs such as pre-miRNAs that possess a short 3′ overhang (Lee et al. 2011). Although 7SL packaging was unaffected (as measured by comparing 7SL with gRNA incorporation using RT-qPCR) (Supplemental Fig. 3D), pre-tRNA incorporation decreased. Quantitation revealed that pre-tRNA-His-GUG packaging decreased 4.2-fold, and pre-tRNA-Ile packaging decreased 7.4-fold (Fig. 5B,C). There was also a nearly twofold reduction in encapsidation of mature tRNA-His-GUG and U6 snRNA (Fig. 5B,C). Thus, packaging of at least some pre-tRNAs and U6 snRNAs requires XPO5-mediated nuclear export.
MLV packages ncRNAs from a host cell surveillance pathway
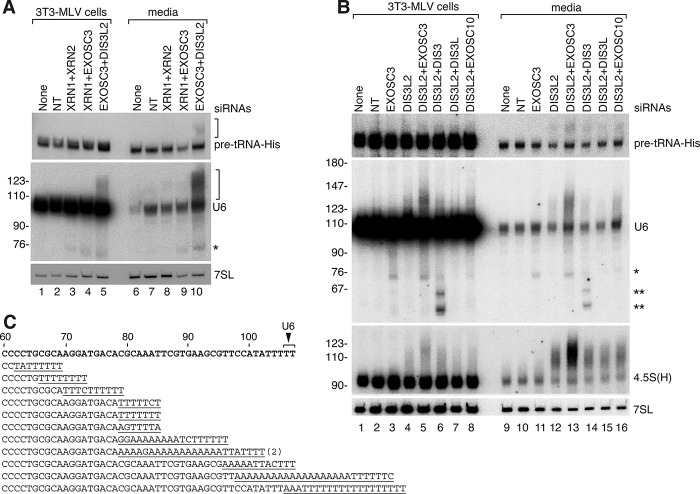
Our finding that XPO5 depletion impairs pre-tRNA and U6 snRNA packaging implies that these RNAs undergo nuclear export. However, in mammalian cells, tRNA end maturation and splicing occur in nuclei, and unspliced pre-tRNAs have not been detected in the cytoplasm (Hopper 2006). U6 is also believed to be confined to nuclei (Hopper 2006). As cytoplasmic forms of these RNAs could escape detection if they were degraded after export, we tested whether depleting one or more exoribonuclease increased packaging, as expected if MLV recruits the RNAs from a decay pathway. No changes in packaging were observed when two 5′-to-3′ exoribonucleases—cytoplasmic XRN1 and nuclear XRN2—were codepleted (Fig. 6A). However, upon depleting EXOSC3, a core subunit of the nuclease complex known as the RNA exosome, a truncated U6 RNA accumulated in host cells and was packaged (Fig. 6B, asterisk). Moreover, when DIS3L2, a cytoplasmic 3′-to-5′ exoribonuclease, was depleted, longer forms of U6 snRNA and 4.5S(H) RNA were detected in both cells and virions, and extended forms of pre-tRNA-His-GUG were detected in virions (Fig. 6B). Increased levels of the longer RNAs were present in both cells and virions when EXOSC3 and DIS3L2 were codepleted (Fig. 6A,B).
Figure 6.
Truncated and 3′ extended ncRNAs accumulate in cells and virions when DIS3L2 and/or the RNA exosome is depleted. (A,B) After transfecting MLV-infected cells with the indicated siRNAs, virus was harvested between 48 and 72 h, and cells were harvested at 72 h. After normalizing virus by RT activity, Northern blots were performed to detect the indicated RNAs. (*) Truncated U6 that accumulates when EXOSC3 is depleted; (**) truncated U6 RNAs that accumulate when DIS3 is depleted. (C) Sequences of 12 independent clones obtained from performing 3′ rapid amplification of cDNA ends (RACE) on virion RNA from cells depleted of EXOSC3 and DIS3L2. Nontemplated nucleotides are underlined. One sequence was obtained twice. The wild-type U6 sequence is at the top, with the 3′ end indicated by the arrow. A total of 25 cDNAs were sequenced. The remaining clones consisted of 10 sequences terminating between U6 nucleotides 60–78 and three full-length U6 sequences.
Since the exosome contains three distinct catalytic subunits that differ in their subcellular location, we determined whether inactivation of specific subunits together with DIS3L2 resulted in increased packaging of the longer RNAs. Consistent with reports that the catalytic subunits EXOSC10, DIS3, and DIS3L have some overlapping functions (Tomecki et al. 2010), depleting individual subunits together with DIS3L2 did not strongly enhance packaging of longer forms of U6, 4.5S(H) RNA, and pre-tRNA-His-GUG compared with depleting DIS3L2 alone (Fig. 6B, cf. lanes 14–16 and 12). However, upon depleting DIS3, which has both endonucleolytic and exonucleolytic activities (Tomecki et al. 2010), two U6 fragments accumulated in both cells and virions (Fig. 6B, lane 14, asterisks). DIS3 is both nucleoplasmic and cytoplasmic (Tomecki et al. 2010), making the location where these fragments are generated unclear. However, the cytoplasmic location of DIS3L2 (Lubas et al. 2013) together with our finding that XPO5 is important for packaging these RNAs support a model in which MLV packages U6 snRNAs and unprocessed pre-tRNAs that are exported to the cytoplasm for decay.
Since 3′ tails are often added to RNAs destined for decay (Houseley and Tollervey 2009), we determined whether the longer U6 RNAs contained additional 3′ sequences. Because most U6 snRNA terminates in a 2′,3′ cyclic phosphate (Lund and Dahlberg 1992), we removed this modification using HCl and shrimp alkaline phosphatase prior to carrying out 3′ rapid amplification of cDNA ends (3′ RACE). This revealed that a heterogeneous mixture of degradation intermediates, many of which contained oligo-U tails, accumulated when EXOSC3 and DIS3L2 were depleted (Fig. 6C). Moreover, for some RNAs, the U-tail was preceded by an oligo(A) tail (Fig. 6C). If, as in yeast, these RNAs undergo oligoadenylation by the Trf4/5–Air1/2–Mtr4 polyadenylation (TRAMP) complex, which targets RNAs for degradation by the nuclear exosome (Houseley and Tollervey 2009), one possibility is that those oligo(A)-containing RNAs that escape exosome-mediated decay undergo subsequent oligo(U) addition by terminal U transferases (TUTs).
As similar truncated and extended U6 snRNAs were detected in uninfected cells when DIS3L2 and exosome subunits were depleted (Fig. 7A; Supplemental Fig. 4), we carried out cell fractionation to determine their subcellular location. Because some nuclear RNAs, such as pre-tRNAs, leak into the cytoplasm when mammalian cells are fractionated using standard procedures (Hashimoto and Steitz 1983), we prepared cytosol with digitonin, which in low concentrations preferentially permeabilizes the plasma membrane, leaving the nuclear membrane and endoplasmic reticulum intact (Adam et al. 1990). To minimize leakage, permeabilization was carried out in the presence of wheat germ agglutinin, which binds glycosylated nuclear pore proteins and inhibits transport (Finlay et al. 1987). Using this approach, 96%–98% of 7SK RNA, which is confined to nuclei (Prasanth et al. 2010), was retained within the permeabilized cells, as was most pre-tRNA-Ile (Fig. 7B). Moreover, since mature SRP is largely localized to the endoplasmic reticulum, ∼65%–70% of 7SL RNA remained within permeabilized cells. Consistent with the hypothesis that some U6 snRNAs undergo nuclear export, 7%–11% of the mature U6 snRNA and 33% of the extended U6 snRNAs that accumulate when DIS3L2 and EXOSC3 are depleted were present in cytosol (Fig. 7B). As our fractionation was designed to obtain cytosol free of nuclear contaminants rather than recover all cytoplasmic RNAs, these numbers represent minimal estimates of the amount of U6 that accesses the cytoplasm. These data support a model in which at least some nuclear ncRNAs undergo nuclear export and cytoplasmic decay.
Figure 7.
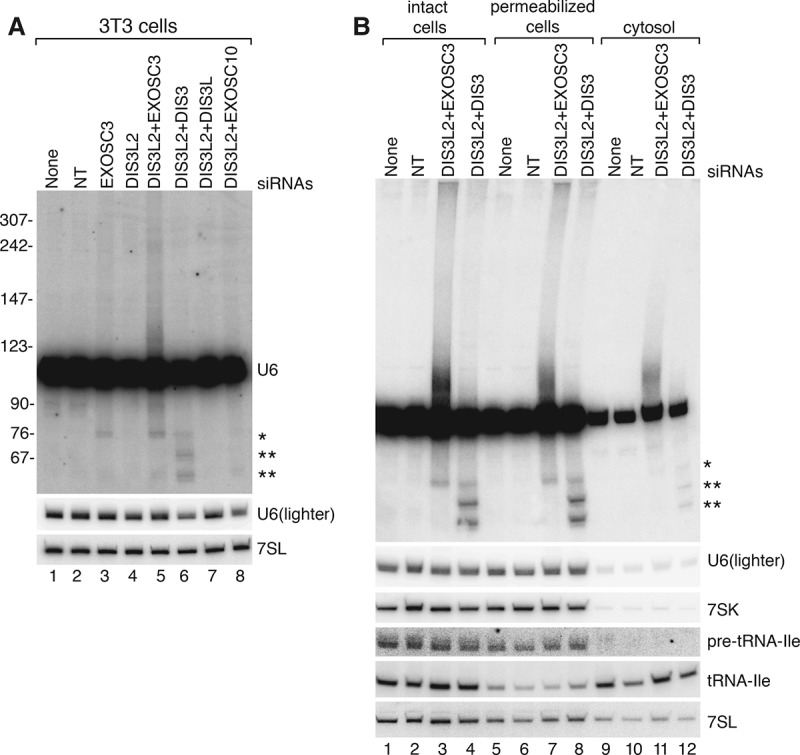
Aberrant forms of U6 snRNA also accumulate in uninfected cells when DIS3L2 and the exosome are depleted. (A) Uninfected 3T3 cells were transfected with the indicated siRNAs and harvested at 72 h. Northern blotting was performed to detect U6 and 7SL RNAs. (*) Truncated U6 RNAs that accumulate when exosome subunits are depleted (see Fig. 6). (B). After transfecting with the indicated siRNAs, uninfected cells were either untreated (lanes 1–4) or permeabilized with digitonin (lanes 5–8) to release cytosol (lanes 9–12). RNA was extracted and subjected to Northern blotting to detect the indicated RNAs.
Discussion
Although packaging of host ncRNAs by retroviruses was described >40 years ago, both the extent to which specific ncRNAs are encapsidated and the RNA features that confer selective recruitment have been unknown. Using deep sequencing, we discovered that host ncRNA biogenesis intermediates, which exist only transiently within cells, are strikingly enriched within MLV virions. At least some of these ncRNAs are packaged from a previously undetected pathway in which unprocessed and unneeded nuclear RNAs are exported to the cytoplasm for degradation.
Implications for retroviral replication
Our data support a model in which retroviruses package newly synthesized ncRNAs in kinetic competition with their normal maturation, RNP assembly, and quality control pathways. This model is consistent with reports that 7SL packaging by HIV-1 is reduced when SRP19 is overexpressed (Wang et al. 2007) and that protein components of the mature RNPs, such as SRP54 and Ro60, are not detected in virions (Onafuwa-Nuga et al. 2006; Garcia et al. 2009). Since nascent RNAs are more likely to contain protein-free regions that would be accessible for Gag binding, sequence-specific interactions may not be necessary for encapsidation of many of these RNAs. However, similar to the well-studied interactions between the primer tRNA, the MLV genome, and nucleocapsid (NC) protein (Miller et al. 2014), some packaged ncRNAs may be selected through binding of Gag or other proteins to specific RNA sequence or structural features. Consistent with specific interactions, truncated 7SL RNAs containing only the Alu domain compete with full-length 7SL for HIV-1 packaging (Keene and Telesnitsky 2012).
How might these RNAs contribute to the retrovirus life cycle? Speculatively, since newly synthesized ncRNAs are more likely to contain protein-free regions than components of mature RNPs, they are ideally suited to contribute to Gag dimerization and/or oligomerization, a process that is enhanced by RNA. Even the shortest RNAs found here to be virion components, such as pre-tRNAs and the ∼95-nt 4.5S(H) and 4.5S(I) RNAs, are longer than the 15–16 nt needed to support efficient assembly of HIV-1 and Rous sarcoma virus in vitro (Campbell and Rein 1999; Ma and Vogt 2002) In this scenario, the RNAs selected for packaging would be those with sufficient stretches of protein-free RNA to allow Gag dimerization. Such a role would allow a large variety of nascent RNAs, most of which are encapsidated in substoichiometic amounts, to collectively contribute to virion assembly.
The packaged ncRNAs could also affect retrovirus replication by interacting with host cell sensors that detect incoming virus. For example, toll-like receptor 7 (TLR7) restricts endocytosed MLV and HIV-1 virions (Beignon et al. 2005; Kane et al. 2011). Because HIV-1 containing NC mutations that impair gRNA binding failed to activate TLRs, the responsible nucleic acid was assumed to be gRNA (Beignon et al. 2005). However, these studies do not rule out contributions from host RNAs, particularly since many packaged RNAs, such as 7SL and Y RNAs, should be excellent TLR7 ligands, as they lack nucleotide modifications and contain polyuridine-rich tails, two criteria of strong activators (Kariko et al. 2005). Moreover, under conditions where retroviruses undergo accelerated uncoating, such as during Trim5α restriction (Sanz-Ramos and Stoye 2013), RNAs that contain 5′ triphosphates, such as 7SL, Y RNAs, and pre-tRNAs, could potentially interact with cytosolic sensors such as RIG-I.
Do those RNAs—such as 7SL RNA, Y RNAs, and vault RNAs—that are present in one or more copy per virion have additional functions? Although studies in which 7SL packaging by HIV-1 was reduced by 90% failed to identify defects in virus assembly (Wang et al. 2007), this RNA could contribute to retrovirus replication in specific cell types or during some stress conditions. Moreover, the contributions of the other abundant packaged ncRNAs remain to be examined. As was demonstrated for 7SL RNA (Wang et al. 2007), it may be possible to prevent packaging of these RNAs by overexpressing core proteins of their respective mature RNPs. Given the increasing number of functions that ncRNAs are now known to play within host cells, it would not be surprising if host ncRNAs also contributed to retrovirus life cycles. Our identification of the MLV “RNA packageome” will facilitate these future analyses.
A new host cell ncRNA surveillance pathway
Our studies of RNA packaging by MLV revealed a novel ncRNA surveillance pathway in mammalian cells. In budding yeast, both U6 and pre-tRNAs are polyadenylated by TRAMP and degraded by the nuclear exosome (Houseley and Tollervey 2009). Moreover, the few yeast pre-tRNAs with 5′ and 3′ extensions that access the cytoplasm are reimported into the nucleus (Kramer and Hopper 2013). Our finding that tailed versions of pre-tRNAs, U6, and other ncRNAs accumulate when DIS3L2 and EXOSC3 are depleted both supports a role for the exosome in decay of these RNAs in mammalian cells and reveals the existence of a new ncRNA decay pathway involving DIS3L2. Although DIS3L2 was known to contribute to decay of pre-let-7 miRNAs and at least some mRNAs (Gallouzi and Wilusz 2013; Lim et al. 2014), our work reveals that newly synthesized ncRNAs are major targets of this exoribonuclease. Since Saccharomyces cerevisiae differs from most eukaryotes in lacking DIS3L2 and enzymes that uridylate RNA 3′ ends, the role of DIS3L2 in degrading newly synthesized ncRNAs may be widespread.
Although some pre-tRNAs and U6 snRNAs undergo nuclear export, we favor a model in which degradation of these RNAs occurs in both the nucleus and cytoplasm. Consistent with nuclear decay, only a fraction of the tailed and truncated U6 RNAs that accumulate when exosome subunits and DIS3L2 are depleted are present in cytosol (Fig. 7B). Although this could reflect their localization to other cell structures, such as the nuclear pore or cytoplasmic organelles, a role for the nuclear exosome is supported by our finding that some U6 RNAs that accumulate when EXOSC3 and DIS3L2 are codepleted contain nontemplated poly(A) tracts, as expected if they were targets of a TRAMP polymerase. Since all of the characterized U6 tails terminate in oligo(U), a modification that enhances DIS3L2 activity (Faehnle et al. 2014), newly synthesized ncRNAs that escape degradation by the nuclear exosome may be exported to the cytoplasm, where they undergo oligo(U) addition and degradation by DIS3L2. Although these exosome and DIS3L2 pathways are another example of redundancy in RNA decay pathways (Houseley and Tollervey 2009), the finding that truncated U6 RNAs accumulate upon EXOSC3 or DIS3 depletion (Fig. 6B) indicates that the exosome also plays a unique role in U6 decay. Since DIS3, but not the other exonucleases, is also an endonuclease (Tomecki et al. 2010), endonucleolytic cleavage may be required for effective decay of some structured ncRNAs.
We do not know whether other “nuclear” RNAs packaged by MLV, such as the SNORD104 precursor and newly synthesized 7SL, also access the cytoplasm. Experiments in which we depleted CRM1/XPO1, which exports partly assembled SRP in yeast and mammalian cells (Hutten and Kehlenbach 2007), did not impair 7SL packaging (MJ Eckwahl and SL Wolin, unpubl.). However, as pre-U2 snRNAs, pre-tRNAs, and U6 are all exported prior to encapsidation, we consider it likely that at least some other “nuclear” RNAs also access the cytoplasm. In this scenario, our failure to block packaging of these RNAs by depleting a small number of export receptors may indicate that the RNAs can be exported via multiple receptors or that receptors not yet tested are involved.
What might be the role of this new ncRNA surveillance pathway? Since at least some targeted RNAs, such as pre-tRNAs and 4.5S(H) RNAs, should contain 5′ triphosphates and base-paired regions, degradation of these RNAs could prevent activation of cytosolic sensors such as RIG-I. Consistent with this idea, it was reported that the cytoplasmic exosome-associated SKIV2L RNA helicase is important for preventing activation of these sensors during the unfolded protein response (UPR), a stress response that includes exosome-mediated decay of specific mRNA fragments (Eckard et al. 2014). Given much evidence that inappropriate activation of cytosolic sensors by endogenous nucleic acids contributes to the development of autoimmune diseases such as systemic lupus erythematosus (Crow 2014), an exciting possibility is that degradation of unprocessed and unassembled ncRNAs by DIS3L2 and the RNA exosome is another mechanism for limiting activation of these sensors and preventing autoimmunity.
Materials and methods
Cells and virus
NIH3T3 cell lines were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum, 100 U/mL penicillin-streptomycin, and 2 mM L-glutamine. Virus was harvested from the media of cells chronically infected with MLV (NIH3T3-MLV) (Onafuwa-Nuga et al. 2005). Virus-containing media were passed through 0.2-μm filters and stored at −80°C until use. RT was assayed as described (Telesnitsky et al. 1995) using poly(rA) as template, oligo(dT) as a primer, and [α-32P]TTP to measure incorporation. Reactions were spotted on filters and quantified using a Typhoon phosphorimager and ImageQuant software. To examine the role of gRNA in ncRNA packaging, ET cells, which are HEK293T cells that produce murine Env (Pfeiffer et al. 1999), were transfected with either the MLV provirus plasmid pMLV GPP (which has env replaced by a puromycin resistance cassette) (Pfeiffer et al. 1999) or a Ψ− pMLV GPP derivative (Miyazaki et al. 2010). The resulting virus was used to infect NIH3T3 cells in the presence of 8 μg/mL hexadimethrine bromide (Polybrene, Sigma). Infected cells were selected with 6 μg/mL puromycin.
To purify MLV for deep sequencing, virus was first concentrated by sedimenting filtered media from NIH3T3-MLV cells and NIH3T3 cells through 2-mL cushions of 20% sucrose in 1× Dulbecco's phosphate-buffered saline (PBS) (Gibco) in a Beckman SW28 rotor at 25,000 rpm for 2 h at 4°C. The pellets were resuspended in 0.5 mL of PBS and fractionated in a continuous gradient of 8%–22% iodixanol (Optiprep, Sigma-Aldrich) in PBS in a Beckman SW41 rotor at 27,500 rpm for 1 h at 4°C. Fractions with peak RT activity and corresponding fractions from uninfected cells were incubated for 30 min at 37°C with 40 U of micrococcal nuclease (Takara Biotech) in 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 2.5 mM CaCl2, and 0.5× PBS plus residual iodixanol. Aliquots were digested in the same buffer with 0.1% Triton X-100 (Sigma). After digestion was stopped by addition of EGTA to 5 µM, RNA was extracted with Trizol-LS (Invitrogen). For Northern analyses, after virus was concentrated by sedimenting through sucrose, pellets were extracted with Trizol. Total RNA was extracted from ∼85% confluent cells using Trizol.
Library preparation and sequencing
After ERCC (External RNA Controls Consortium) RNA spike-in mix (Ambion, Life Technologies) was added to the RNA, each sample was divided in three and fragmented (Ambion fragmentation buffer) for 2, 5, or 7 min at 70°C. After reactions were combined and RNA was precipitated, a modified Illumina TruSeq protocol was performed. Briefly, first strand cDNA synthesis was initiated with random hexamers. After second strand synthesis, 50- to 160-bp cDNAs were excised from agarose gels and purified using a Nucleospin gel purification kit (Machery-Nagel). After cDNA ends were repaired with the End-It DNA end-repair kit (Epicentre), a single dA was added to the 3′ ends with Klenow fragment (3′ → 5′ exo−) (Roche), and Illumina adapters were ligated. Libraries were amplified by 14 cycles of PCR and gel-purified. After quantitating (KAPA library quantification kit; Kapa Biosystems), libraries were loaded onto a HiSeq 2000 Illumina sequencing flow cell and sequenced for 50 bases at the Yale Stem Cell Center. After demultiplexing based on index sequences, adapters were removed (FASTX-toolkit), and reads were aligned using Bowtie (Langmead et al. 2009), allowing up to three mismatches. Reads were first mapped to the MLV genome (accession no. NC_001501), then to 45S rDNA (NR_046233), and finally to the mouse mm10 assembly. Many of the ∼10% of reads that did not align (Fig. 1C) were of bacterial origin. In contrast, ∼90% of reads from the uninfected data set failed to align (Supplemental Fig. 1). Sailfish (Patro et al. 2014) was used to determine RPKM, while HTSeq (Anders et al. 2015) was used to assign reads to genomic features.
Northern blotting and RNase protection analyses
RNAs were fractionated in 5% or 8% polyacrylamide–8.3 M urea gels and transferred to Hybond-N (Amersham) in 0.5× TBE at 150 mA for 16 h. Northern blots were hybridized with [32P]-labeled oligonucleotide probes as described (Sim et al. 2009). Probes are listed in Supplemental Table S3. Signals were quantified using a Typhoon imager system.
To detect 7SL and MLV gRNA by RNase protection, pEG467-10 (Garcia et al. 2009) was linearized with HindIII and transcribed with T3 polymerase. To detect vault and 7SL RNAs, DNA consisting of nucleotides 7–138 of vault RNA flanked by BamHI and XbaI sites was amplified and cloned into the same sites of pEG467-10, replacing gRNA and generating pVault-7SL. After linearization with HindIII, transcription with T3 RNA polymerase (Promega) in the presence of [α-32P]rCTP produced an ∼315-nt RNA complementary to 99 nt of 7SL and 132 nt of vault RNA. RNase protection assays were as described (Garcia et al. 2009). Molar ratios of the two RNAs were determined by normalizing for the number of radiolabeled C's in each protected fragment.
siRNA transfections, RT-qPCR, and 3′ RACE
Smartpool siRNAs for XRN1, XRN2, EXOSC3, EXOSC10, DIS3, DIS3L, DIS3L2, PHAX, SPN, TNPO3, and nontargeting control siRNA #3 were from Dharmacon. Transfections were as described (Sim et al. 2009) except that Lipofectamine RNAiMAX (Invitrogen) and 20 nM siRNAs were used. For PHAX, SPN, and TNPO3, virus was harvested between 24 and 48 h, and cells were harvested at 48 h after transfection. For other siRNAs, virus was harvested between 48 and 72 h, and cells were harvested at 72 h after transfection.
To measure mRNA depletion, RNA was treated with DNase I (Roche), primed with random hexamers, and reverse-transcribed using iScript cDNA synthesis kit (Bio-Rad). For experiments measuring ncRNAs, DNase-treated RNA was primed with random hexamers and extended with SuperScript III RT (Invitrogen) using an annealing temperature of 65°C. cDNAs were subjected to qPCR using a SYBR Green real-time PCR master mix (Bio-Rad). Primers used for RT-qPCR are listed in Supplemental Table S3.
For 3′ RACE, RNA was first incubated with 10 mM HCl for 4 h on ice followed by incubation with 5 U of shrimp alkaline phosphatase (Takara) to remove 3′ cyclic phosphates. After phenol-chloroform extraction, the RNA was ligated to RNA oligonucleotide L3, treated with DNase I, reverse-transcribed using SuperScript III and oligonucleotide P3, and amplified with oligonucleotides P3 and U6 (Supplemental Table S3). PCR products were cloned using Zero Blunt PCR cloning kit (Invitrogen).
Immunoblotting
Virus and cell pellets were resuspended in RIPA buffer (10 mM Tris Cl at pH 8.0, 140 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) with 1 mM PMSF and 1× protease inhibitor cocktail (Roche). After sedimenting at 16,000 g, cleared lysates were subjected to immunoblotting as described (Yoo and Wolin 1994). Antibodies were anti-CA (Invitrogen), anti-SRP19 and anti-SRP14-9 (Bovia et al. 1995), and anti-TEP1 (Kickhoefer et al. 2001).
Cell permeabilization
Untreated and siRNA-transfected 3T3 cells were permeabilized largely as described (Garcia et al. 2009). After washing twice with 1× PBS and once with RSB100 (10 mM Tris-HCl at pH 7.4, 2.5 mM MgCl2, 100 mM NaCl), adherent cells were incubated with 1 mL of RSB100 containing 25 μg/mL digitonin (Calbiochem) and 1 μg/μL wheat germ agglutinin (Sigma) for 5 min on ice while rotating on an orbital shaker. To obtain cytosol, the buffer–digitonin mixture was removed and spun at 2000 rpm for 5 min, and the supernatant was collected. Permeabilized cells were washed with PBS and collected in Trizol.
Accession number
The sequencing data associated with this project have been deposited in NCBI's Sequence Read Archive (SRA) under accession number SRP053582.
Supplementary Material
Acknowledgments
We thank N. Kolev for advice on library preparation; S. Johnson, S. Kharytonchyk, X. Chen, H. Arnion, C. Belair, and M. Boccitto for other technical advice; K. Strub and V. Kickhoefer for gifts of antibodies; and W. Mothes and J. Steitz for comments on the manuscript. This work was supported by a National Science Foundation Graduate Research Fellowship to M.J.E., National Institutes of Health R21 AI094059 and Collaborative Development Grant from the Center for HIV RNA Studies (P50 GM103297) to S.L.W., grant P50 GM103297 to A.T., and Training Grant T32GM007223 to M.J.E.. This paper is dedicated to the memory of our friend and colleague, Elisabetta Ullu.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.258731.115.
References
- Adam SA, Marr RS, Gerace L. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol 111: 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian D, Shen K, Zhang X, Shan SO. 2013. Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem 82: 693–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beignon AS, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, Bhardwaj N. 2005. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J Clin Invest 115: 3265–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz R, Fisher J, Goff SP. 1996. RNA packaging. Curr Top Microbiol Immunol 214: 177–218. [DOI] [PubMed] [Google Scholar]
- Bishop JM, Levinson WE, Quintrell N, Sullivan D, Fanshier L, Jackson J. 1970. The low molecular weight RNAs of Rous sarcoma virus. I. The 4 S RNA. Virology 42: 182–195. [DOI] [PubMed] [Google Scholar]
- Bovia F, Fornallaz M, Leffers H, Strub K. 1995. The SRP9/14 subunit of the signal recognition particle (SRP) is present in more than 20-fold excess over SRP in primate cells and exists primarily free but also in complex with small cytoplasmic Alu RNAs. Mol Biol Cell 6: 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameier M, Ibing W, Hofer K, Montag J, Stahl-Hennig C, Motzkus D. 2013. Mapping the small RNA content of simian immunodeficiency virions (SIV). PLoS One 8: e75063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Rein A. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol 73: 2270–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S, Javanbakht H, Kim S, Shiba K, Craven R, Rein A, Ewalt K, Schimmel P, Musier-Forsyth K, Kleiman L. 2002. Retrovirus-specific packaging of aminoacyl-tRNA synthetases with cognate primer tRNAs. J Virol 76: 13111–13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DY, Maraia RJ. 1993. A cellular protein binds B1 and Alu small cytoplasmic RNAs in vitro. J Biol Chem 268: 6423–6428. [PubMed] [Google Scholar]
- Crow MK. 2014. Type I interferon in the pathogenesis of lupus. J Immunol 192: 5459–5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckard SC, Rice GI, Fabre A, Badens C, Gray EE, Hartley JL, Crow YJ, Stetson DB. 2014. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat Immunol 15: 839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle CR, Walleshauser J, Joshua-Tor L. 2014. Mechanism of Dis3l2 substrate recognition in the Lin28–let-7 pathway. Nature 514: 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Newmeyer DD, Price TM, Forbes DJ. 1987. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J Cell Biol 104: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French NS, Norton JD. 1997. Structure and functional properties of mouse VL30 retrotransposons. Biochim Biophys Acta 1352: 33–47. [DOI] [PubMed] [Google Scholar]
- Gallouzi IE, Wilusz J. 2013. A DIStinctively novel exoribonuclease that really likes U. EMBO J 32: 1799–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia EL, Onafuwa-Nuga A, Sim S, King SR, Wolin SL, Telesnitsky A. 2009. Packaging of host mY RNAs by murine leukemia virus may occur early in Y RNA biogenesis. J Virol 83: 12526–12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada F, Kato N. 1980. Nucleotide sequences of 4.5S RNAs associated with poly(A)-containing RNAs of mouse and hamster cells. Nucleic Acids Res 8: 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto C, Steitz JA. 1983. Sequential association of nucleolar 7-2 RNA with two different autoantigens. J Biol Chem 258: 1379–1382. [PubMed] [Google Scholar]
- Hopper AK. 2006. Cellular dynamics of small RNAs. Crit Rev Biochem Mol Biol 41: 3–19. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. 2009. The many pathways of RNA degradation. Cell 136: 763–776. [DOI] [PubMed] [Google Scholar]
- Houzet L, Paillart JC, Smagulova F, Maurel S, Morichaud Z, Marquet R, Mougel M. 2007. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res 35: 2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S, Kehlenbach RH. 2007. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol 17: 193–201. [DOI] [PubMed] [Google Scholar]
- Jiang M, Mak J, Ladha A, Cohen E, Klein M, Rovinski B, Kleiman L. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol 67: 3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MC, Scobie HM, Ma YM, Vogt VM. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J Virol 76: 11177–11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Case LK, Wang C, Yurkovetskiy L, Dikiy S, Golovkina TV. 2011. Innate immune sensing of retroviral infection via Toll-like receptor 7 occurs upon viral entry. Immunity 35: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K, Buckstein M, Ni H, Weissman D. 2005. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23: 165–175. [DOI] [PubMed] [Google Scholar]
- Keene SE, Telesnitsky A. 2012. Cis-acting determinants of 7SL RNA packaging by HIV-1. J Virol 86: 7934–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Liu Y, Kong LB, Snow BE, Stewart PL, Harrington L, Rome LH. 2001. The telomerase/vault-associated protein TEP1 is required for vault RNA stability and its association with the vault particle. J Cell Biol 152: 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EB, Hopper AK. 2013. Retrograde transfer RNA nuclear import provides a new level of tRNA quality control in Saccharomyces cerevisiae. Proc Natl Acad Sci 110: 21042–21047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev AS, Markusheva TV, Kramerov DA, Ryskov AP, Skryabin KG, Bayev AA, Georgiev GP. 1982. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res 10: 7461–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Jiko C, Yamashita E, Tsukihara T. 2011. Selective nuclear export mechanism of small RNAs. Curr Opin Struct Biol 21: 101–108. [DOI] [PubMed] [Google Scholar]
- Levin JG, Seidman JG. 1981. Effect of polymerase mutations on packaging of primer tRNAPro during murine leukemia virus assembly. J Virol 38: 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JG, Grimley PM, Ramseur JM, Berezesky IK. 1974. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J Virol 14: 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159: 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. 2013. Exonuclease hDIS3L2 specifies an exosome-independent 3′–5′ degradation pathway of human cytoplasmic mRNA. EMBO J 32: 1855–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. 1992. Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA's. Science 255: 327–330. [DOI] [PubMed] [Google Scholar]
- Ma YM, Vogt VM. 2002. Rous sarcoma virus Gag protein–oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J Virol 76: 5452–5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ. 1991. The subset of mouse B1 (Alu-equivalent) sequences expressed as small processed cytoplasmic transcripts. Nucleic Acids Res 19: 5695–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Wang Z. 2014. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SB, Yildiz FZ, Lo JA, Wang B, D'Souza VM. 2014. A structure-based mechanism for tRNA and retroviral RNA remodelling during primer annealing. Nature 515: 591–595. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y, Irobalieva RN, Tolbert BS, Smalls-Mantey A, Iyalla K, Loeliger K, D'Souza V, Khant H, Schmid MF, Garcia EL, et al. 2010. Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J Mol Biol 404: 751–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Onafuwa-Nuga AA, King SR, Telesnitsky A. 2005. Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J Virol 79: 13528–13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onafuwa-Nuga AA, Telesnitsky A, King SR. 2006. 7SL RNA, but not the 54-kd signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. RNA 12: 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Mount SM, Kingsford C. 2014. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol 32: 462–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon-Eternod M, Wei M, Pan T, Kleiman L. 2010. Profiling non-lysyl tRNAs in HIV-1. RNA 16: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer JK, Topping RS, Shin NH, Telesnitsky A. 1999. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J Virol 73: 8441–8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Camiolo M, Chan G, Tripathi V, Denis L, Nakamura T, Hubner MR, Spector DL. 2010. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol Biol Cell 21: 4184–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro-Choi TS, Redy R, Henning D, Takano T, Taylor CW, Busch H. 1972. Nucleotide sequence of 4.5 S ribonucleic acid of Novikoff hepatoma cell nuclei. J Biol Chem 247: 3205–3222. [PubMed] [Google Scholar]
- Rulli SJ Jr, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. 2007. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J Virol 81: 6623–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Ramos M, Stoye JP. 2013. Capsid-binding retrovirus restriction factors: discovery, restriction specificity and implications for the development of novel therapeutics. J Gen Virol 94: 2587–2598. [DOI] [PubMed] [Google Scholar]
- Schopman NC, van Montfort T, Willemsen M, Knoepfel SA, Pollakis G, van Kampen A, Sanders RW, Haasnoot J, Berkhout B. 2012. Selective packaging of cellular miRNAs in HIV-1 particles. Virus Res 169: 438–447. [DOI] [PubMed] [Google Scholar]
- Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, Wolin SL. 2009. The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell 20: 1555–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stake MS, Bann DV, Kaddis RJ, Parent LJ. 2013. Nuclear trafficking of retroviral RNAs and Gag proteins during late steps of replication. Viruses 5: 2767–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A, Blain S, Goff SP. 1995. Assays for retroviral reverse transcriptase. Methods Enzymol 262: 347–362. [DOI] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. 2010. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J 29: 2342–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P, Royo H, Seitz H, Bachellerie JP, Huttenhofer A, Cavaille J. 2003. Identification of 13 novel human modification guide RNAs. Nucleic Acids Res 31: 6543–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 2007. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol 81: 13112–13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. 1994. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol 14: 5412–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva L, Myers R, Fassati A. 2006. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol 4: e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Sokolskaja E, Jolly C, James W, Cowley SA, Fassati A. 2011. Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS Pathog 7: e1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.