Pancreatic Ductal Adenocarcinoma (PDA) develops predominantly through pancreatic intraepithelial neoplasia (PanIN) and intraductal papillary mucinous neoplasm (IPMN) precursor lesions. Roy et al. identify critical antagonistic roles for Brg1, a catalytic subunit of the SWI/SNF complexes, during IPMN-PDA development. In mature duct cells Brg1 inhibits the dedifferentiation that precedes neoplastic transformation. In contrast, Brg1 promotes tumorigenesis in full-blown PDA by supporting a mesenchymal-like transcriptional landscape. JQ1 impairs PDA tumorigenesis by both mimicking some and inhibiting other Brg1-mediated functions.
Keywords: Brg1, IPMN, dedifferentiation, pancreatic cancer, Kras, EMT
Abstract
Pancreatic ductal adenocarcinoma (PDA) develops predominantly through pancreatic intraepithelial neoplasia (PanIN) and intraductal papillary mucinous neoplasm (IPMN) precursor lesions. Pancreatic acinar cells are reprogrammed to a “ductal-like” state during PanIN-PDA formation. Here, we demonstrate a parallel mechanism operative in mature duct cells during which functional cells undergo “ductal retrogression” to form IPMN-PDA. We further identify critical antagonistic roles for Brahma-related gene 1 (Brg1), a catalytic subunit of the SWI/SNF complexes, during IPMN-PDA development. In mature duct cells, Brg1 inhibits the dedifferentiation that precedes neoplastic transformation, thus attenuating tumor initiation. In contrast, Brg1 promotes tumorigenesis in full-blown PDA by supporting a mesenchymal-like transcriptional landscape. We further show that JQ1, a drug that is currently being tested in clinical trials for hematological malignancies, impairs PDA tumorigenesis by both mimicking some and inhibiting other Brg1-mediated functions. In summary, our study demonstrates the context-dependent roles of Brg1 and points to potential therapeutic treatment options based on epigenetic regulation in PDA.
Pancreatic ductal adenocarcinoma (PDA) is a deadly malignancy with a 5-year survival rate of 6% (Siegel et al. 2014). Untargeted polychemotherapy is the current standard of care but is too toxic for many patients to tolerate (Conroy et al. 2011; Ryan et al. 2014). Several novel chemotherapy and immunotherapy clinical trials are currently in progress, but conclusive evidence regarding their effectiveness has not been established. A better characterization of the molecular pathogenesis of this disease is thus essential to identify novel therapeutic approaches and the patients most likely to benefit from them.
Oncogenic KRAS drives PDA development, which progresses through histologically and prognostically distinct precursor lesions (Hezel et al. 2006). These lesions also appear to have distinct cells of origin. Pancreatic intraepithelial neoplasia (PanIN) lesions likely derive from exocrine acinar cells (Kopp et al. 2012), whereas pancreatic duct cells (PDCs) serve as the progenitor cells for intraductal papillary mucinous neoplasia (IPMN) (von Figura et al. 2014). Prognostically, PanIN-derived PDA is an aggressive disease associated with an average survival of 6 mo after diagnosis, usually due to late diagnosis. In contrast, IPMN-derived PDA is more indolent, with a 5-year survival after surgical resection approaching 50% (Poultsides et al. 2010; Mino-Kenudson et al. 2011; Matthaei et al. 2012). Thus, understanding the key differences in signals that regulate PanIN and IPMN formation is essential for prognostic and predictive biomarker development schemes that will in turn lead toward personalized therapy.
Loss of cellular identity, defined by decreased differentiation markers and inappropriate expression of progenitor markers, is a characteristic of PDA (Stanger and Hebrok 2013). Acinar–ductal metaplasia (ADM), a process in which acinar cells undergo dedifferentiation toward a duct-like state, precedes PanIN and PDA formation (Guerra et al. 2007; Zhu et al. 2007; De La O et al. 2008; Habbe et al. 2008). In contrast, IPMN lesions appear to arise from pancreatic duct but not acinar cells. We showed previously that the Brahma-related gene 1 (Brg1; human gene name SMARCA4), encoding the catalytic subunit of the SWI/SNF complex in the context of oncogenic Kras, leads to IPMN lesions and PDA (von Figura et al. 2014). Interestingly, Brg1 expression is higher in IPMN-associated PDA compared with IPMN in matched patient samples, suggesting that Brg1 may serve distinct, stage-specific roles in the pathogenesis of PDA (von Figura et al. 2014). To address this possibility, we compared and characterized the role of Brg1 at the “early” preinvasive stage of the cancer (IPMN) and the “late” PDA stage emanating from the very same precursor lesion.
We found that PDCs, like their acinar counterparts, also undergo a dedifferentiation step to form IPMN. We further show that Brg1 has antithetical functions during progression of IPMN to PDA. While the protein functions as a tumor suppressor early in the disease by inhibiting dedifferentiation of PDCs, it promotes late stage PDA progression at least in part by induction of epithelial-to-mesenchymal transition (EMT). Thus, Brg1 has bipolar contextual roles both preventing and promoting pancreatic tumorigenesis in a stage-dependent manner. Several of these malignant attributes can be successfully therapeutically addressed by epigenomic intervention with the BRD4 inhibitor JQ1, opening a new treatment paradigm in PDA. Therefore, the contextual functions of Brg1 in PDA of both preventing and promoting tumorigenesis can be therapeutically exploited in select patient subsets with this deadly disease.
Results
Loss of Brg1 promotes dedifferentiation of PDCs in the context of oncogenic Kras
Distinct PanIN and IPMN lesions can both progress to PDA (Hezel et al. 2006). We showed previously that Brg1 differentially regulates the development of these precancerous lesions (von Figura et al. 2014). Loss of Brg1 in conjunction with oncogenic Kras inhibits formation of PanIN from pancreatic acinar cells. In contrast, depletion of Brg1 in PDCs expressing oncogenic Kras promotes IPMN formation. To investigate why PDCs are sensitive to IPMN formation upon Brg1 elimination, we developed an in vitro model system using primary PDCs. PDCs were isolated from genetically modified mouse models (GEMM) carrying Lox-stop-Lox KrasG12D (LSLKrasG12D) and floxed alleles of Brg1 (Brg1f/f) following a previously published protocol (Reichert et al. 2013a,b). PDCs were isolated from 6- to 8-wk-old mice by DBA lectin sorting and cultured for 1 wk (Supplemental Fig. 1A,B). This protocol allows for the isolation and cultivation of a pure population of duct cells, as demonstrated by high expression of duct marker cytokeratin 19 and the absence of the acinar cell marker amylase or endocrine marker insulin (Supplemental Fig. 1C). One week after isolation, cells were infected with Cre recombinase-expressing adenovirus to initiate genetic recombination of the loxP sites at the Kras (activation of oncogenic Kras expression through elimination of the floxed stop allele) and Brg1 (elimination through recombination of Brg1 exons 2 and 3, Brg1f/f) loci (Supplemental Fig. 1D,E). Adenoviruses expressing RFP were used as a control to monitor transfection efficiency.
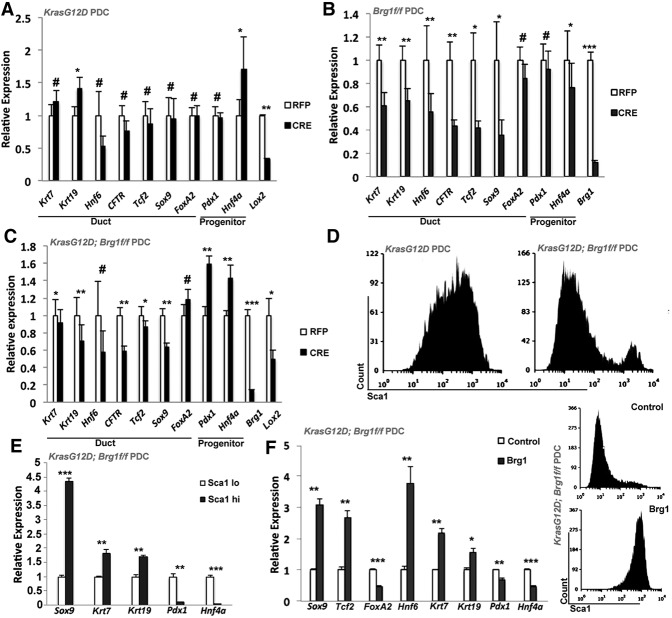
To determine whether duct cells lose their cellular identity while progressing toward IPMN, we first investigated the effects of oncogenic Kras on duct cell differentiation. With the exception of Krt19 (cytokeratin 19), oncogenic Kras does not alter the expression of mature duct cell markers, including Krt7 (cytokeratin 7), Hnf6 (ONECUT homeobox 1), Cftr (cystic fibrosis transmembrane conductance regulator), Tcf2 (HNF1 homeobox B), Sox9 (SRY sex-determining region box 9), and FoxA2 (Forkhead box A2) (Fig. 1A). While the expression of Pdx1 (pancreatic and duodenal homeobox 1), a marker normally expressed in duct progenitors and only at a low level in mature PDCs, was unaltered, another progenitor marker, Hnf4a (hepatocyte nuclear factor 4α), was up-regulated. As expected, Lox2 (lysyl oxidase 2), a gene known to be transcriptionally repressed by oncogenic Kras signaling, was significantly down-regulated (Gazin et al. 2007). In contrast, Brg1 deletion in the presence of wild-type Kras led to a dramatic decrease in the majority of the mature duct cell markers (FoxA2 expression appeared unaffected) (Fig. 1B), while the expression of the progenitor marker Pdx1 or Hnf4a was unchanged or even reduced. These findings suggest that loss of Brg1 degrades mature duct cell identity, as evidenced by attenuation of mature duct cell markers. Interestingly, concomitant activation of oncogenic Kras together with Brg1 elimination leads to a more pronounced dedifferentiated state. The transcriptional profile of KrasG12D; Brg1f/f PDCs revealed down-regulation of mature duct cell markers accompanied by enhanced expression of progenitor markers (Fig. 1C). Thus, simultaneous loss of Brg1 and activation of KrasG12D collaborates to erode the mature ductal state and promote the inappropriate activation of progenitor factors.
Figure 1.
PDCs expressing oncogenic Kras and loss of Brg1 undergo dedifferentiation. Quantitative PCR analysis of duct cell differentiation markers in PDCs isolated from LSL- KrasG12D mice (A), Brg1f/f mice (B), and LSL-KrasG12D; Brg1f/f mice (C) infected with adenovirus expressing either RFP or Cre recombinase. (D) FACS analysis of KrasG12D and KrasG12D; Brg1f/f PDCs using the Sca1 antibody. (E) Quantitative PCR analysis of duct cell differentiation markers in PDCs sorted from KrasG12D; Brg1f/f PDCs based on Sca1 expression. (F, left) Quantitative PCR analysis of duct cell differentiation markers in KrasG12D; Brg1f/f PDCs with ectopic Brg1 expression. (Right) FACS analysis of Sca1 in KrasG12D; Brg1f/f PDCs expressing empty vector or vector expressing wild-type Brg1.
Pdx1 serves as an essential transcription factor in normal pancreas development during embryogenesis (Reichert and Rustgi 2011; Stanger and Hebrok 2013). It is ubiquitously expressed in the early epithelial cells of the developing pancreas, but its high expression is restricted to the pancreatic islet cells in the adult organ. Up-regulation of Pdx1 expression is observed during dedifferentiation of pancreatic exocrine cells upon injury as well as in pancreatic preinvasive lesions (Sharma et al. 1999; Park et al. 2011). Since Pdx1 was the unique progenitor marker up-regulated in KrasG12D; Brg1f/f PDCs, we questioned whether Pdx1high cells were those to have undergone dedifferentiation. A previous study has shown an inverse correlation between expression of Pdx1 and the cell surface marker Sca1 (also known as Ly6a [lymphocyte antigen 6 complex, locus A]) in pancreatic adenocarcinoma cells (Ischenko et al. 2014), providing a means to sort and compare PDCs based on their Pdx1 expression levels by using antibodies directed against Sca1 (Fig. 1D; Ischenko et al. 2014). KrasG12D PDCs showed a bell-shaped curve when assayed for Sca1 expression, with the majority of the cells marked by high levels of Sca1/lower levels of Pdx1 expression (Fig. 1D; Supplemental Fig. 2A). In contrast, depletion of Brg1 in the context of oncogenic Kras resulted in the vast majority of the cells assuming a Sca1low/Pdx1high phenotype (Fig. 1D,E). Differential expression of Sca1 and, by extrapolation, Pdx1 in Brg1 intact and depleted PDC lines expressing oncogenic Kras reflects the differentiation status of these cells and supports our prior observations (Fig. 1A–C). For example, we detected only a very small number of Sca1low/Pdx1high cells in KrasG12D PDCs, and these cells did not demonstrate any decrease in the expression of matured duct markers (Supplemental Fig. 2A,B). In contrast, Sca1low/Pdx1high cells from KrasG12D; Brg1-depleted PDCs displayed not only an increase in the progenitor markers Pdx1 and Hnf4a but also reduced expression of the mature duct markers Krt7, Krt19, and Sox9 (Fig. 1E). Thus, Sca1low cells marked by loss of Brg1 in the context of oncogenic Kras cannot sustain mature duct cell identity. The critical role for Brg1 in maintaining this duct differentiation state is further highlighted by the observation that the small population of Sca1high/Pdx1low KrasG12D; Brg1f/f PDCs have escaped Cre recombination of the Brg1 locus and therefore continued Brg1 expression (Supplemental Fig. 2C). Furthermore, forced re-expression of Brg1 (Supplemental Fig. 2D) in KrasG12D; Brg1f/f PDCs reduced progenitor markers but increased expression of duct markers and Sca1 (Fig. 1F), reaffirming the concept that reactivation of Brg1 in PDCs restores their mature identity and differentiation state. Collectively, these data show that Brg1 actively maintains ductal identity even in the presence of KrasG12D expression, thus playing a tumor-suppressive role (Shain et al. 2012).
Immortalized PDCs give rise to IPMN lesions in vivo
Based on our previously published work on Ptf1aCre; KrasG12D; Brg1f/f (PKB) mice (von Figura et al. 2014) and our above-described findings in duct cells, we hypothesized that the KrasG12D; Brg1f/f PDCs serve as the progenitors for IPMN lesions. To test the tumorigenic potential of PDCs, we established immortalized PDC lines from both KrasG12D and KrasG12D; Brg1f/f mice by adenoviral infection of Cre recombinase followed by serial passaging. These cell lines exhibit extremely low population doubling times compared with wild-type PDCs (Supplemental Fig. 3A). When grown in three-dimensional (3D) cultures, wild-type PDCs form small spheres. The appearance of these spheres is similar to those from KrasG12D PDCs, although the latter are more proliferative and thus significantly larger than their wild-type counterparts (Supplemental Fig. 3B). In contrast, KrasG12D; Brg1f/f PDCs form Krt19-positive irregular cystic structures displaying prominent protrusions that are morphologically distinct from those generated by both the wild-type and KrasG12D PDCs (Supplemental Fig. 3B,C). Orthotopic transplantation into the pancreata of immune-deficient mice was used to assess their tumor-forming capacities in vivo. Six weeks after transplantation, KrasG12D; Brg1f/f PDCs did form Krt19- and mucin-positive cysts harboring high levels of Ki67-positive cells reminiscent of IPMN lesions (Supplemental Fig. 3D,E). In contrast, KrasG12D PDCs developed either cystic structures that were mostly Ki67- and mucin-negative (Supplemental Fig. 3E) or Ki67-positive sporadic PanIN lesions but no IPMN. Thus, Brg1 loss in the context of oncogenic Kras is required for PDCs to develop into IPMN lesions upon transplantation.
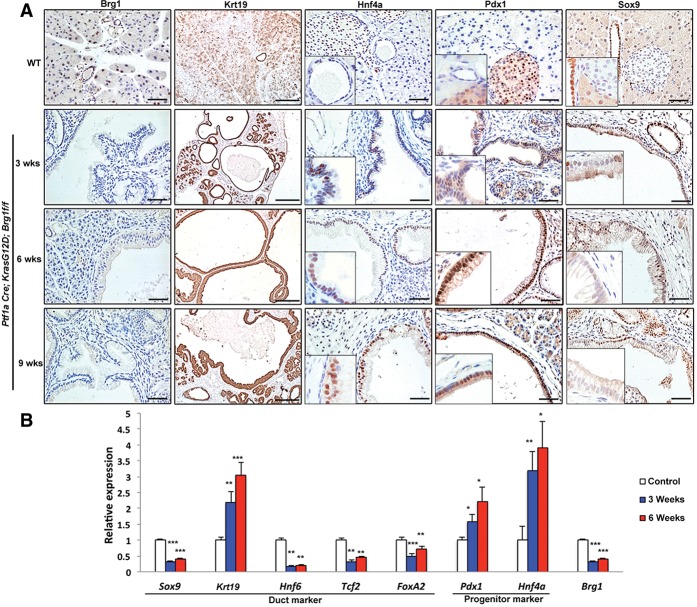
We next used the previously characterized PKB mice (von Figura et al. 2014) to evaluate the phenomenon of ductal dedifferentiation in vivo. PKB mice express Cre recombinase shortly after the onset of pancreas formation. PKB mice harbor IPMN lesions at 3 wk of age that progress to PDA by 9 wk. In order to better characterize the early stages of IPMN-derived PDA, we analyzed mice at 3, 6, and 9 wk of age. PKB mice displayed the expected loss of Brg1 expression in the IPMN structures that developed by 3 wk. The lesions were Krt19-positive but had low expression levels of the critical duct marker Sox9. Consistent with our observations in PDCs, these lesions express the progenitor markers Pdx1 and Hnf4a (Fig. 2A). Duct cells were isolated via DBA lectin staining from IPMN mice (Supplemental Fig. 4), and expression of mature duct and progenitor markers was compared with those in control mice. Quantitative PCR of transcripts isolated from IPMN lesions confirmed the “dedifferentiation” gene expression profile in 3- and 6-wk-old PKB mice (Fig. 2B). Thus, our data strongly suggest that duct-to-IPMN progression requires an intermediary step during which functional duct cells undergo a transcriptional shift toward a more plastic state susceptible for neoplastic transformation.
Figure 2.
PDCs undergo dedifferentiation en route to IPMN formation. (A) Brg1, Krt19, Hnf4a, Pdx1, and Sox9 immunohistochemistry analysis of either control or 3-, 6-, and 9-wk-old PKB animals. Bars: Brg1, Hnf4a, Pdx1, and Sox9, 100 μm; Krt19, 250 μm. (B) Quantitative PCR analysis of duct cell differentiation markers in DBA lectin-sorted PDCs from either control or 3- and 6-wk-old PKB mice.
Ectopic expression of Sox9 in PDCs stabilizes the mature ductal cell identity and reduces IPMN occurrence
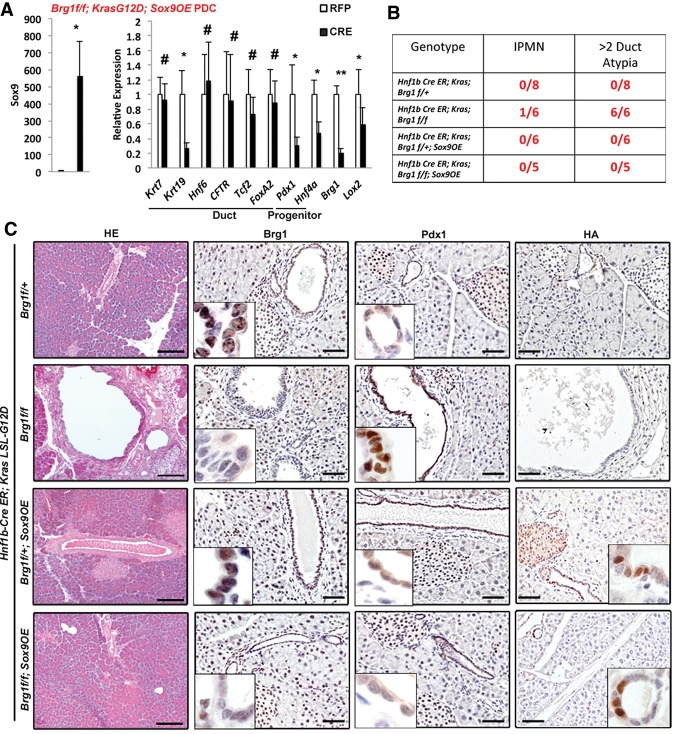
We next investigated how loss of Brg1 contributes to ductal dedifferentiation. Brg1 is an essential component of the SWI/SNF chromatin remodeling complexes known to regulate expression of target genes (Trotter and Archer 2008), and our prior work has demonstrated direct binding of Brg1 to Sox9 regulatory elements (von Figura et al. 2014). Overexpression of Brg1 increased Sox9 expression, raising the possibility that Sox9 might be involved in regulating the expression of mature duct and progenitor markers. To test this hypothesis, we isolated PDCs from KrasG12D; Brg1f/f; Sox9OE mice that permit simultaneous, adenoviral Cre-dependent expression of both oncogenic Kras and a Sox9-HA fusion protein in the context of Brg1 elimination. Notably, activation of Sox9 not only reduced the expression of progenitor markers Pdx1 and Hnf4a but, with the exception of Krt19, also restored the levels of mature duct cell markers (Fig. 3A). Thus, overexpression of Sox9 not only blocked duct dedifferentiation but also inhibited up-regulation of progenitor markers caused by Brg1 loss/oncogenic Kras, effectively rescuing duct cells from the dedifferentiation process that would otherwise follow KrasG12D activation with Brg1 loss.
Figure 3.
Ectopic expression of Sox9 in PDCs leads to changes in cellular identity and reduced IPMN occurrence. (A) Quantitative PCR analysis of duct cell differentiation markers in PDCs isolated from LSL-KrasG12D; Brg1f/f; Sox9OE mice infected with adenovirus expressing either RFP or Cre recombinase. (B) Analysis of IPMN or duct atypia occurrence for the indicated genotypes. (C) H&E, Brg1, Pdx1, and HA immunohistochemistry analysis of Hnf1bCreER; LSL- KrasG12D; Brg1f/+, Hnf1bCreER; LSL- KrasG12D; Brg1f/f, Hnf1bCreER; LSL- KrasG12D; Brg1f/+; Sox9OE, or Hnf1bCreER; LSL- KrasG12D; Brg1f/f; Sox9OE mice 4 wk after tamoxifen administration. Bars: H&E, 250 μm; HA, Brg1, and Pdx1, 50 μm.
To test whether ectopic Sox9 expression has similar effects on duct cell identity in vivo, we used the Hnf1bCre-ER mouse model system to manipulate Sox9, Brg1, and Kras specifically in adult PDCs. Tamoxifen treatment in Hnf1bCre-ER; KrasG12D; Brg1f/+ control mice did not result in the formation of any detectable duct abnormalities (Fig. 3B). As described previously (von Figura et al. 2014), elimination of Brg1 in the context of oncogenic Kras in Hnf1bCre-ER; KrasG12D; Brg1f/f mice promotes duct atypia, a precursor lesion for IPMN lesions, and occasional occurrence of IPMN. In contrast, constitutive up-regulation of Sox9 in Hnf1bCre-ER; KrasG12D; Brg1f/f; Sox9OE mice blocked increased Pdx1 expression and prevented any duct atypia or IPMN in these animals (Fig. 3C). Thus, overexpression of Sox9 can compensate for the loss of ductal Brg1 and interferes with the dedifferentiation process and subsequent IPMN formation even in the context of oncogenic Kras.
KrasG12D Brg1f/f PDCs display a transcriptome signature distinct from KrasG12D PDCs
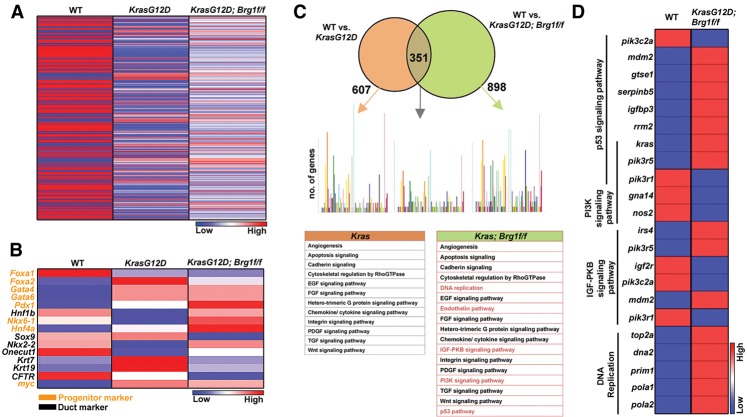
To gain mechanistic insights into how oncogenic Kras and loss of Brg1 function compromise the duct differentiation state, we sequenced RNA isolated from wild-type, KrasG12D, and KrasG12D; Brg1f/f PDCs 7 d after adenoviral infection (Fig. 4A). By initially focusing on the expression of genes involved in pancreatic organogenesis, we observed significant up-regulation of progenitor and down-regulation of ductal markers in KrasG12D; Brg1f/f PDCs as compared with wild-type PDCs (Fig. 4B). Next, we performed a global analysis of gene expression changes in KrasG12D PDCs in the presence and absence of Brg1 compared with wild-type PDCs. Using a false discovery rate (FDR) set at 0.05, we found 607 genes to be differentially expressed in KrasG12D PDCs when compared with wild-type PDCs and 898 genes to be differentially expressed in KrasG12D; Brg1f/f PDCs compared with controls (Fig. 4C). A total of 351 genes were common among the differentially expressed genes between KrasG12D and KrasG12D; Brg1f/f PDCs. Pathway analysis with the software program Panther 9.0 (Mi et al. 2013) revealed that the majority of signaling cascades in which at least five or more genes were differentially expressed was shared between KrasG12D and KrasG12D; Brg1f/f PDCs, including the Wnt, Tgf, Egf, Fgf, and apoptosis pathways. However, we identified at least five pathways that were uniquely activated in KrasG12D; Brg1f/f PDCs—DNA replication, PI3K signaling, p53 signaling, and IGF-PKB prominent among them (Fig. 4C). A detailed analysis of the expression levels of all of the genes belonging to these pathways revealed several intriguing features (Fig. 4D). Mdm2, a known inhibitor of p53, was significantly increased in KrasG12D; Brg1f/f PDCs, suggesting defects in cell cycle regulation. In support of this notion, deletion of Brg1 in the pancreas epithelium of transgenic mouse models expressing oncogenic Kras exhibited reduced p53 expression (von Figura et al. 2014). In addition, KrasG12D; Brg1f/f PDCs presented with up-regulation of genes that promote DNA replication. This finding is interesting given the prior observation that increased DNA replication may promote duct cell dedifferentiation into a progenitor-like state (Inada et al. 2008). Last, amplification of the PI3K signaling pathway in the setting of oncogenic Kras, an expression profile that we observed in KrasG12D; Brg1f/f PDCs, has been shown previously to induce dedifferentiation of duct cells (Watanabe et al. 2008). Thus, in summary, our RNA sequencing results indicate that loss of Brg1 in PDCs expressing oncogenic Kras provide a permissive environment through regulation of several critical pathways that may promote cellular dedifferentiation as an initial neoplastic event.
Figure 4.
KrasG12D; Brg1f/f PDCs have a distinct transcriptome signature. (A) Heat map showing differential expression signature between wild-type, KrasG12D, and KrasG12D; Brg1f/f PDCs. (B) Heat map showing expression of genes implicated in pancreas development between wild-type, KrasG12D, and KrasG12D; Brg1f/f PDCs. (C) Pathway enrichment analysis for differentially expressed genes between wild-type PDCs and KrasG12D or KrasG12D; Brg1f/f PDCs. Uniquely activated pathways are marked in red. Each colored bar represents the number of genes that belong to a particular pathway. (D) Heat map demonstrating expression of genes that belong to the indicated pathways between wild-type and KrasG12D; Brg1f/f PDCs.
Re-expression of Brg1 in Brg1-deficient cancer cell lines enhances tumorigenicity
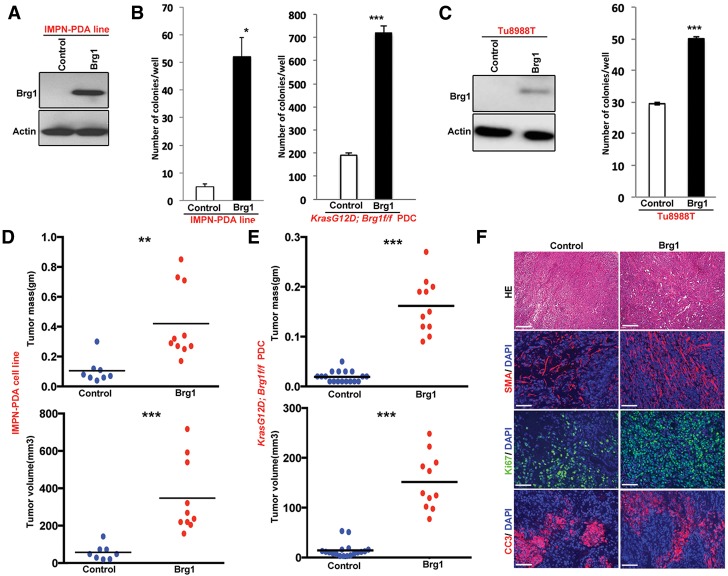
As Brg1 re-expression reverses the dedifferentiation state in KrasG12D; Brg1f/f PDCs (Fig. 1F) via redirecting cells toward a mature duct state, we reasoned that re-expression of Brg1 may reduce tumorigenicity in established cancer cells. To test this hypothesis, we ectopically re-expressed Brg1 (“Brg1 put back”) in PDA cell lines derived from the KrasG12D; Brg1f/f mice using a retroviral Brg1-Flag construct (Fig. 5A; Sif et al. 2001). Somewhat unexpectedly, re-expression of Brg1 increased anchorage-independent growth in both the IPMN-PDA-derived tumor cell lines and immortalized KrasG12D; Brg1f/f PDCs (Fig. 5B). Similar results were obtained when Brg1 was ectopically expressed in a human PDA line, Tu8988T, a cell line lacking Brg1 protein (Fig. 5C; Elsasser et al. 1992; Hoffman et al. 2014). Thus, reinstatement of Brg1 expression in pancreatic duct and cancer cells carrying oncogenic Kras is not growth inhibitory and appears to paradoxically support tumor maintenance.
Figure 5.
Re-expression of Brg1 in KrasG12D; Brg1f/f PDCs and IPMN-derived PDA tumor cells enhances their tumorigenic properties. (A) Western blot showing ectopic expression of Brg1 in IPMN-derived PDA. (B) Brg1 put backs in KrasG12D; Brg1f/f PDCs and IPMN-derived PDA show increased anchorage-independent growth compared with Brg1-null cells. (C, left) Western blot depicting ectopic expression of Brg1 in the Brg1-null human PDA cell line Tu8988T. (Right) Soft agar colony formation assay showing that Brg1-null human PDA cell line Tu8988T forms more colonies on soft agar upon ectopic expression of Brg1. Brg1 put backs in IPMN-derived PDA lines (D) and KrasG12D; Brg1f/f PDCs (E) were subcutaneously injected into NOD scidγ (NSG) mice and monitored for tumor growth. Mice were sacrificed 4 wk after inoculation, and tumors were excised. Tumor mass and volume are significantly higher in Brg1 put backs. (F) Costaining of Ki67, smooth muscle actin (SMA), cleaved caspase 3, and DAPI of tumors obtained in D. Bars: H&E, 250 μm; Ki67, cleaved caspase 3, and SMA, 100 μm.
To evaluate the consequences of Brg1 re-expression in vivo, we performed subcutaneous injection of the control and Brg1 “put back” cancer lines into immune-compromised NOD scidγ (NSG) mice. We again observed a significant increase in tumor burden in mice injected with Brg1 “put back” compared with control cells expressing empty vector (Fig. 5D,E). Brg1-overexpressing tumors retained high levels of Brg1 (Supplemental Fig. 5A) and expressed less Pdx1 and Hnf4a (Supplemental Fig. 5B,C). However, these tumors displayed higher proliferative indexes marked by Ki67-positive cells and contained more smooth muscle actin (SMA)-positive stroma, a negative prognostic factor for survival of pancreatic cancer patients (Fig. 5F; Supplemental Fig. 5D,E; Fujita et al. 2010; Sinn et al. 2014). No dramatic differences were found in cleaved caspase 3-positive cells undergoing apoptosis between the control and Brg1 “put back” tumors (Fig. 5F; Supplemental Fig. 5F). Also, Brg1-overexpressing tumors were poorly differentiated, further pointing toward their increased aggressiveness (Supplemental Fig. 5G). Thus, our cell culture and in vivo findings support the notion of Brg1 playing antagonistic roles during IPMN-PDA formation, changing from a tumor suppressor in mature duct cells to an oncogene during later stages of neoplasia. To correlate this observation with the human disease condition, we analyzed the Brg1 expression status in low-grade IPMN, high-grade IPMN, and IPMN-derived PDA patient samples. Notably, we found that Brg1 expression is higher in PDA compared with its precursor IPMN lesions (Supplemental Fig. 6).
Re-expression of Brg1 induces an EMT-like phenotype
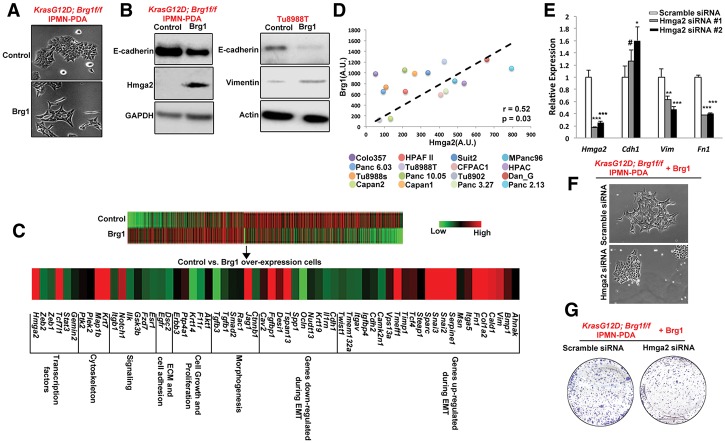
We next analyzed how re-expression of Brg1 contributed to the enhanced tumorigenicity of established tumor lines. Of note, in addition to the increase in SMA expression (Fig. 5), we observed that re-expressing Brg1 in a KrasG12D; Brg1f/f IPMN-derived PDA cell line changed the cellular appearance toward a more elongated and spindle-shaped form, a characteristic of EMT (Fig. 6A). In order to confirm whether Brg1 re-expression conferred more mesenchymal features, we investigated the protein levels of epithelial (E-cadherin) and mesenchymal (vimentin) markers in the control and Brg1 “put back” cells. Consistent with the morphological changes, Brg1-expressing mouse and human cancer cells showed decreased E-cadherin protein levels and increased vimentin expression (Fig. 6B). RNA sequencing of control and Brg1 “put back” lines also revealed that the Brg1-re-expressing cell line adopted an overall transcriptional profile consistent with increased EMT, including enhanced gene expression of several mesenchymal genes and down-regulation of epithelial genes (Fig. 6C). Notably, this transcriptional analysis revealed Hmga2 (high mobility group AT-hook 2) as a significantly up-regulated gene in Brg1 “put back” (Fig. 6B,C). Hmga2, an architectural protein that maintains chromatin structure, has previously been shown to induce EMT in pancreatic cancer, and our prior work demonstrated that Brg1 binds to the Hmga2 promoter and regulates the histone H3K4 methylation levels to activate its transcription (von Figura et al. 2014). Furthermore, tumors derived from Brg1 “put back” IPMN-PDA cells exhibit increased Hmga2 level (Supplemental Fig. 7). We further looked at SMARCA4 and HMGA2 expression in a previously published human data set (Collisson et al. 2011) consisting of several human pancreatic cancer cell lines (Fig. 6D). In agreement with the results obtained from murine cells, there was a strong positive correlation between SMARCA4 and HMGA2 expression (Pearson correlation = 0.52, P = 0.03) across 16 out of 20 cell lines in the data set, while four cell lines did not show a positive correlation (MiaPaCa2, HupT3, SW1990, and Panc1). To further confirm the role of Hmga2 as a critical mediator of the aggressive cancer phenotype, we reduced Hmga2 expression by two independent siRNAs in KrasG12D; Brg1f/f IPMN-PDA cell lines with ectopic Brg1 expression. Attenuated Hmga2 level resulted in decreased expression of mesenchymal markers vimentin/fibronectin and increased expression of the epithelial marker E-cadherin (Fig. 6E). Hmga2-depleted cells also presented with a more epithelial-like morphology (Fig. 6F) and impaired growth potential when compared with control cells expressing Hmga2 (Fig. 6G). Thus, our data indicate that re-expression of Brg1 in pancreatic cancer cell lines promotes EMT, likely through activation of Hmga2 expression.
Figure 6.
Re-expression of Brg1 induces a mesenchymal phenotype in mouse and human Brg1-deficient pancreatic cancer cell lines. (A) Bright-field images of IPMN-derived KrasG12D; Brg1f/f PDA cells expressing a control empty vector and Brg1 put back. (B) Brg1 put backs in KrasG12D; Brg1f/f in IPMN-derived PDA lines and Tu8988T were probed for epithelial marker E-cadherin, EMT regulator Hmga2, and mesenchymal marker vimentin. (C) Heat map showing an induction of a pronounced mesenchymal gene signature in Brg1-overexpressing IPMN-derived KrasG12D; Brg1f/f PDA cells. (D) Correlation between expression of Brg1 and Hmga2 in human pancreatic cancer cell lines. Plots show signal intensities as measured by Affymetrix chip. Spearman correlation coefficient is shown. (E) Quantitative PCR analysis of Hmga2, Cdh1 (E-cadherin), Vimentin, and Fn1 (fibronectin). (F,G) Bright-field images (F) and crystal violet staining (G) in scrambled siRNA or Hmga2 siRNA transfected IPMN-derived KrasG12D; Brg1f/f PDA cells expressing wild-type Brg1. Lower levels of crystal violet in Hmga2 siRNA transfected cells indicate reduced proliferative capacity.
An epigenomic approach to PDA treatment
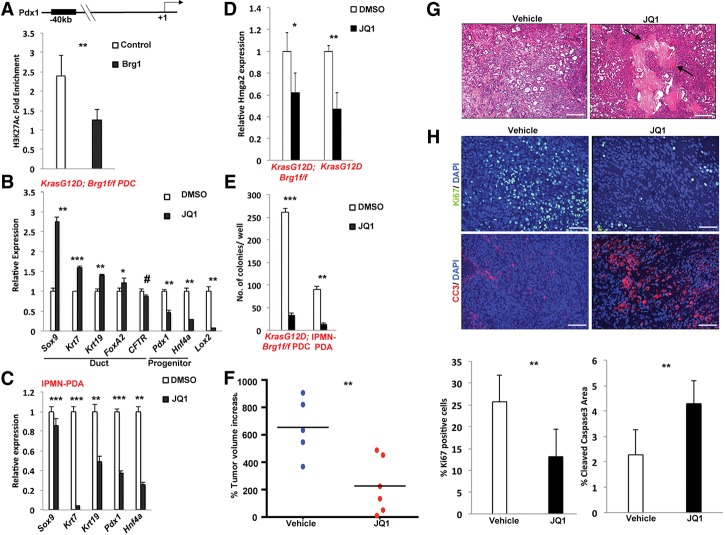
Whereas current targeted therapeutic approaches focus on directly targeting oncogenes or their downstream effectors, our findings suggest that an alternative or complementary approach might be to interfere with the increase of progenitor markers in PDA tumors. Our data point to up-regulation of Pdx1 and Hnf4a as the critical markers of the ductal dedifferentiation step during the transition of duct cells towards neoplasia. However, while reintroduction of Brg1 did block expression of these progenitor markers in PDA cells, ectopic expression of Brg1 also induced an EMT phenotype, which may be in part due to up-regulation of Hmga2. Therefore, we reasoned that simultaneously reversing the dedifferentiation process and interfering with HMGA2-mediated EMT might provide a unique therapeutic opportunity. In other words, our aim was to explore whether combined down-regulation of Pdx1 and Hnf4a coupled with Hmga2 repression could be used to reduce tumor progression. Fortunately, JQ1, a drug in clinical trial for hematological malignancies (Filippakopoulos et al. 2010; Delmore et al. 2011), has been shown recently to decrease tumorigenicity of pancreatic cancer cells through Hmga2 repression (Sahai et al. 2014). Furthermore, JQ1 targets the BET family of transcription factors and disrupts the interaction between them and acetylated H3K27 (H3K27ac) to favor a more repressive gene expression profile (De Raedt et al. 2014). This activity is similar to that of Brg1, which promotes a repressive gene expression pattern by reducing H3K27ac levels (Zhang et al. 2014) as shown here for a predicted distal enhancer element of Pdx1 (Fig. 7A; http://www.dcode.org). JQ1-treated KrasG12D; Brg1f/f PDCs displayed increased expression of mature duct markers and attenuated expression of progenitor markers (Fig. 7B), suggesting a return to a more mature state. However, in IPMN-derived PDA cancer cells, JQ1 was able to reduce the expression of Pdx1 and Hnf4a but unable to increase the expression of mature duct markers such as Sox9 (Fig. 7C). Nonetheless and as expected, JQ1 treatment did reduce expression of Hmga2 in PDCs expressing oncogenic Kras independently of Brg1 (Fig. 7D). Furthermore, JQ1 resulted in reduced soft agar colony formation (Fig. 7E) and slowed the growth of IPMN-PDA tumor cell lines implanted subcutaneously into immune-deficient mice (Fig. 7F). The reduction in tumor progression was associated with abundant necrosis (Fig. 7G), reduced proliferation, and increased apoptosis (Fig. 7H). Thus, our data implicate JQ1 as a promising therapeutic agent for PDA through its dual activities on inhibiting Hmga2-mediated EMT and the expression of progenitor markers.
Figure 7.
JQ1 can be exploited for the treatment of duct-derived pancreatic cancer. (A) Chromatin immunoprecipitation (ChIP) analysis of H3K27ac enrichment on the indicated promoter regions. Values are expressed as fold enrichment over IgG control. (B) Quantitative PCR analysis of duct cell differentiation markers in KrasG12D; Brg1f/f PDCs treated with DMSO or JQ1. (C) Quantitative PCR analysis of duct cell differentiation markers in IPMN-derived PDA cell line treated with DMSO or JQ1. (D) Quantitative PCR analysis of Hmga2 expression in DMSO- or JQ1-treated KrasG12D or KrasG12D; Brg1f/f PDCs. (E) Quantitation of soft agar assay of KrasG12D; Brg1f/f PDCs or IPMN-derived PDA cell lines treated with DMSO or JQ1. (F) NSG mice were subcutaneously injected with an IPMN-PDA cell line. Once the tumor reached 100 mm3, mice were randomized and treated with JQ1 or vehicle. The graph shows the percent change in tumor volume at the study end point. (G) H&E stainings of vehicle- or JQ1-treated tumors. Arrows point to necrotic lesions. Bar, 250 μm. (H, top) Costaining of Ki67, cleaved caspase 3, and DAPI of tumors treated with vehicle or JQ1. Bar, 100 μm. (Bottom) Percentage of Ki67-positive or cleaved caspase 3-positive cells in vehicle- or JQ1-treated tumors. Ten random fields from three mice in each group were chosen for quantification.
Discussion
Brg1 is historically known for its tumor-suppressive functions, and, recently, the role of SWI/SNF complex mutations has come into the spotlight for numerous malignancies (Kadoch et al. 2013; Shain and Pollack 2013; Helming et al. 2014). However, a number of recent reports have associated Brg1 with oncogenic properties. For example, Brg1 has been shown to be required for leukemia maintenance (Buscarlet et al. 2014), and elevated Brg1 expression is observed in gastric carcinoma, melanoma, and prostate cancer (Sentani et al. 2001; Sun et al. 2007; Lin et al. 2010). Our previous work also demonstrated that there are cellular context-dependent roles of Brg1. For example, ablation of Brg1 in pancreatic acinar cells inhibits Kras-driven PanIN formation (von Figura et al. 2014), likely because Brg1 plays a critical role in oncogenic Kras-driven acinar ductal dedifferentiation (Supplemental Fig. 8). Here, we reconcile these apparently opposing roles of Brg1 in tumor formation in a Kras-driven PDA model. We demonstrate that duct cells, like acinar cells, undergo a previously unappreciated dedifferentiation event en route to IPMN formation. In mature duct cells, Brg1 inhibits this dedifferentiation event and thus IPMN formation through regulation of genes that sustain duct cell identity, including Sox9. In marked contrast, reactivation of Brg1 promotes cancer properties in established tumor cells both in vitro and in vivo. These discrepant functions emphasize the contextual dependence of Brg1 activities in PDA. It should be noted that such context-dependent regulation of PDA is not uncommon. For example, TGF-β acts as a tumor suppressor during initiation of PDA but promotes tumorigenesis in an established state of the tumor (Bardeesy et al. 2006; Ijichi et al. 2006; Izeradjene et al. 2007; Melisi et al. 2008; Yang et al. 2010; Ostapoff et al. 2014). Similarly, while low levels of Wnt aids in oncogenic Kras-driven PanIN formation, high levels of the signaling pathway prevent KrasG12D-driven ADM and PanIN formation (Morris et al. 2010; Zhang et al. 2013). Thus, Brg1 joins a growing list of factors whose effects on pancreatic cancer formation depends on the timing and differentiation state of the cells in which they are active.
We showed previously that acinar cells undergo a dedifferentiation step to assume a duct-like state during PanIN-to-PDA progression (Kopp et al. 2012). Here we observed that duct cells similarly and transiently pass through a dedifferentiation step to occupy a less mature state, a process that is inhibited by Brg1. Duct dedifferentiation appears to require two distinct processes. On the one hand, the expression of factors maintaining duct identity (e.g., Sox9 and Krt7) needs to be reduced. On the other hand, progenitor factors normally expressed at a very low level in mature duct cells (including Pdx1 and Hnf4a) are reactivated. Our studies reveal important synergistic functions between oncogenic Kras and the loss of Brg1 in permitting duct dedifferentiation and thus initiation of IPMN. Neither oncogenic Kras nor Brg1 elimination by itself appears to be sufficient to drive IPMN formation from duct cells. In contrast, simultaneous loss of Brg1 with ectopic expression of oncogenic Kras promotes a dramatic change in duct identity, resulting in neoplastic transformation. Pdx1 re-expression caused by Brg1 loss/expression of oncogenic Kras emerges as a critical factor for this process. Thus, changes in the epigenetic state in the context of the Kras oncogene activation initiate duct-derived IPMN lesions. Of note, our results do show that a very small population of Pdx1high cells exists in KrasG12D PDCs, indicating that the oncogene by itself is able to compromise cellular identity even in the presence of Brg1. Single-cell analysis would have to be used in future studies to determine whether oncogenic Kras compromises the epigenetic state in a subset of duct cells.
At the onset of our studies, we hypothesized that redifferentiation of cancer cells toward the cell of origin would interfere with tumor progression. Considering our data that Brg1 sustains the identity of primary duct cells, we reasoned that Brg1 re-expression in PDA cells would have beneficial effects. Somewhat surprisingly, Brg1 reactivation promoted tumor formation, pointing to a switch from a tumor suppressor to an oncogene in cells that have undergone neoplastic transformation. Brg1 interacts with different coactivators (such as histone acetyltransferase CBP/p300) and corepressors (e.g., mSin3a histone deacetylase complex) to alter gene expression (Pal et al. 2003; Naidu et al. 2009). This plasticity in Brg1 interaction might promote distinct roles commensurate with a particular developmental or disease state. Additionally, SWI/SNF complexes can use either Brg1 or Brm as the ATPase catalytic subunit, further diversifying its regulatory role in transcription. Interestingly, Brg1- or Brm-containing complexes exhibit antagonistic function during osteoblast differentiation (Flowers et al. 2009), raising the possibility that these distinct complexes regulate diverse processes during disease progression. To determine the mechanism underlying the disparate roles of Brg1 in initiation and maintenance of PDA, we performed deep sequencing analysis of IPMN-derived PDA cells expressing ectopic Brg1. Brg1 overexpression did reactivate expression of mature duct markers, but the expression of the progenitor markers Pdx1 and Hnf4a, while reduced, remained significantly above what is found in wild-type duct cells. Thus, Brg1 put back initiates but does not complete redifferentiation of cancer cells to duct cells.
Our gene expression analysis further revealed a transition to a mesenchymal-like state in duct-derived cancer cells. One of the up-regulated genes was Hmga2, a transcription factor known to induce EMT and metastasis in gastric, mammary, and pancreatic cancer (Thuault et al. 2008; Watanabe et al. 2009; Zha et al. 2013). In a recent study, Munshi and colleagues (Sahai et al. 2014) showed that the BET bromodomain inhibitor JQ1 suppresses Hmga2 expression in pancreatic cancer cell lines, and we observed the same response in KrasG12D; Brg1f/f PDC lines. Notably, reactivation of Brg1 up-regulates Hmga2 expression, raising the question of whether combining redifferentiation toward a mature duct state with inhibition of the mesenchymal phenotype would be efficient in reducing tumor progression. JQ1, a drug currently tested in clinical trials for hematologic malignancies, has previously been shown to block Hmga2 expression. In addition, our data demonstrate that JQ1 reduces the expression of Pdx1 and Hnf4a, progenitor markers that become up-regulated during IPMN development. Moreover, JQ1 treatment reduced the tumorigenicity of KrasG12D; Brg1f/f PDCs and IPMN-derived murine cancer cell lines both in vitro and in vivo. Therefore, it is tempting to speculate that blocking EMT through Hmga2 inhibition and reversing inappropriate duct dedifferentiation through reduction of Pdx1/Hnf4a expression work in tandem to elicit these effects. Notably, Pdx1 and Hnff4a are also up-regulated during PanIN-derived PDA. Future experiments should address the specific contributions of Brd4 and other JQ1 targets on the progression of both PanIN- and IPMN-derived PDA.
There are conflicting reports regarding the correlation of Brg1 expression and overall survival in pancreatic cancer. Several studies, including ours, have shown that low Brg1 expression is correlated with poor prognosis (Dal Molin et al. 2012; von Figura et al. 2014). However, contrasting reports also exist demonstrating high Brg1 association with reduced overall survival (Liu et al. 2014). It is important to consider that the above-mentioned clinical specimens are all PDA samples without any knowledge of the preinvasive lesions from which these tumors have originated; for example, duct or acinar cells. It is highly plausible that the cellular origin of the PDA plays a decisive role in the outcome of Brg1 association with prognosis. Brg1 has dichotomous function for PanIN- and IPMN-derived PDA (von Figura et al. 2014). Therefore, the prognostic implications of Brg1 expression levels may actually depend on the preinvasive lesion that gave rise to the PDA being evaluated. Indeed, for PanIN-derived PDA, low Brg1 expression is associated with poor prognosis (von Figura et al. 2014). In contrast, high Brg1 expression is found in IPMN-PDA. Furthermore, the stage of the disease is important for the outcome of these analyses. For example, one study showed that high Brg1 expression was only associated with stage IV PDA samples (Numata et al. 2013). Recently, SWI/SNF targeted therapeutic strategies for cancer have been gaining momentum (Helming et al. 2014). However, our results show that it may be critical to define the stage and origin of the PDA when exploring this chemotherapeutic route. A recent report further demonstrated that high Brg1 expression is associated with gemcitabine resistance, a property also exhibited by increased expression of Hmga2 (Liu et al. 2014). Therefore, specific targeting of the Brg1–Hmga2 axis may be beneficial to reduce EMT-related tumor aggressiveness and also sensitize resistant tumors to chemotherapy. As JQ1 treatment had only a cytostatic effect in our tumor model, combining JQ1 with a cytotoxic chemotherapy regimen might be a promising approach for PDA patients.
In summary, our findings point to Brg1 as a context-dependent mediator of Kras-driven pancreatic tumorigenesis. Support for this concept comes from the opposing roles of Brg1 during the initiation of IPMN and the progression of IPMN-derived PDA. Future studies are needed to clarify how Brg1-regulated epigenetic and genetic changes contribute to such stage-specific effects. Deciphering the temporal activities of the SWI/SNF complex should provide important insights into their potential as therapeutic targets in numerous human malignancies.
Materials and methods
Mouse lines
The following mice strains were used: Ptf1a-Cre (gift of Christopher Wright, Vanderbilt University, Nashville, TN), Hnf1b-CreERT2 (gift of Jorge Ferrer, Imperial College, London, UK), KrasG12D (gift of Dave Tuveson, Cold Spring Harbor Laboratory), Brg1flox (gift of David Reisman, University of Florida, with permission of Pierre Chambon), and Sox9OE mice. Mice were crossed on a mixed background. Tamoxifen citrate (TEVA Pharmaceuticals) was administered by oral gavage at a dose of 10 mg per gavage three times a week for 1 wk. The University of California at San Francisco Institutional Animal Care and Use Committee (IACUC) approved all mouse experiments.
Immunohistochemistry and immunofluorescence
Mouse tissue was fixed overnight in Z-FIX (Anatech, catalog no. 174) at 4°C with gentle shaking, washed with PBS for 30 min at room temperature, embedded in paraffin, and cut into sections. Antigen retrieval was performed by boiling in antigen retrieval Citra solution (Biogenex, catalog no. HK086-9K). Blocking was performed using either 5% BSA in PBS or by blocking solution provided in MOM kit (Vector Laboratories, catalog no. BMK-2202). For primary antibodies, incubation was performed overnight at 4°C in a humidified chamber; secondary antibody incubation was for 1 h at room temperature. For immunohistochemistry, slides were developed using the ABC kit (Vector Laboratories, catalog no. PK-6100) and DAB kit (Vector Laboratories, catalog no. SK-4100) or the Envision kit (DAKO, catalog no. K-4011) or MOM kit (Vector Laboratories) and counterstained with hematoxylin. For immunofluorescence, slides were mounted with DAPI hard set (Vector Laboratories, catalog no. H-1500). Primary antibodies used in this study were rabbit anti-Brg1 (Santa Cruz Biotechnology, catalog no. sc-10768), rabbit anti-Krt19 (Epitomics, catalog no. AC-0073), rabbit anti-Sox9 (Sigma, catalog no. HPA001758), mouse anti-Ki-67 (BD Pharmingen, catalog no. 550609), Armenian hamster anti-Mucin1 (Neomarkers, catalog no. HM-1630), rabbit anti-Mucin2 (Santa Cruz Biotechnology, catalog no. sc-15334), rabbit anti-cleaved caspase 3 (Cell Signaling, catalog no. 9661), rabbit anti-Pdx1 (Millipore, catalog no. 07-696), rat HA (Roche, catalog no. 1867423), rabbit anti-Hnf4a (Santa Cruz Biotechnology, catalog no. sc-8987), and mouse anti-SMA (Sigma, catalog no. A5228).
RNA deep sequencing
Eight-hundred nanograms of total RNA isolated from the control and Brg1-expressing IPMN-derived PDA lines was used to generate libraries with the Illumina TruSeq RNA sample version 2 preparation kit (catalog no. RS-122-2001). Each library was diluted to ∼10 nM prior to loading and sequenced using a HiSeq 2000 instrument generating paired 100-base-pair reads. Reads were then mapped to the mouse genome (NCBI37/mm9) using TopHat version 2 (Trapnell et al. 2009) with default parameters. Transcript assembly and differential expression analysis was performed using Cufflinks and Cuffdiff using default parameters.
3D duct cell culture
3D culture of PDCs was performed using a previously published protocol (Reichert et al. 2013a). Briefly, chamber slides were coated with collagen type I. Cells were mixed with collagen and placed on the top layer. PDC full medium was replaced every 2 d.
Drug treatment
JQ1 was a kind gift from Dr. Mike Kagey and Dr. Steven Landau, Tensha Therapeutics. JQ1 dissolved in DMSO was used to treat cells at a concentration of 1 μM. DMSO vehicle was used as a control.
Once tumors were ∼100–150 mm3 in volume, mice were randomized and treated with either vehicle (10% hydroxypropyl β cyclodextrin; Sigma, catalog no. C0926) or JQ1 dissolved in vehicle. Fifty micrograms of JQ1 per kilogram of body weight was intraperitoneally injected 5 d a week (Monday–Friday).
Isolation and culture of duct cells
PDCs were isolated following a previously published protocol (Reichert et al. 2013a). Briefly, pancreas was harvested in ice-cold HBSS buffer. Next, pancreas was digested with collagenase type V and trypsinized for 5 min, and trypsinization was stopped using soy trypsin inhibitor. Cells were filtered using a 40-μm cell strainer. Next, cells were centrifuged, stained with DBA lectin FITC, and pulled down with anti-FITC microbeads using a miniMACS separation unit. Cells were grown on collagen-coated plates in PDC medium.
Statistical analysis
To determine the P-values for all the experiments performed, Student's t-test was performed. A P-value of <0.05 was assumed as statistically significant. P < 0.05, P < 0.01, and P < 0.001 are represented with single, double, and triple asterisks. P > 0.05 is represented by a number sign. Population doubling times for cell lines were calculated by the exponential growth function in GraphPad Prism. All statistical analyses were performed with either GraphPad Prism (version 4.0c) or Microsoft Excel.
Cell culture
Human cells (Tu8988T) were cultured in DMEM + 10% FBS. Mouse cells were isolated from PDA mice as described previously (von Figura et al. 2014) and cultured in DMEM + 10% FBS.
Supplementary Material
Acknowledgments
We thank Dr. Christopher Wright for sharing the Ptf1a-Cre mice, Dr. David Tuveson for the KrasG12D mice, and Dr. Pierre Chambon and Dr. David Reisman for the Brg1f/f mice. We also thank Dr. Sapna Puri and Dr. Julia Carnevale for critical reading of the manuscript, Debbie Ngow for tissue processing, and all of the Hebrok/Collisson laboratory members for their helpful discussion. This work was supported by National Institutes of Health (NIH)/National Cancer Institute (NCI) grants R01CA172045 and R01CA112537 to M.H., and NIH/NCI grants R01CA178015 and K08CA137153 to E.A.C. E.S.S. is funded by the 2011 Pancreatic Cancer Action Network-American Association for Cancer Research Pathway to Leadership Award. N.R. is supported by a Basic Science Post-doctoral Fellowship from the American Association of Cancer Research. A.U. is supported by a fellowship from Daichi Sankyo.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.256628.114.
References
- Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, et al. 2006. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev 20: 3130–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscarlet M, Krasteva V, Ho L, Simon C, Hebert J, Wilhelm B, Crabtree GR, Sauvageau G, Thibault P, Lessard JA. 2014. Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood 123: 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, et al. 2011. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 17: 500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardiere C, et al. 2011. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 364: 1817–1825. [DOI] [PubMed] [Google Scholar]
- Dal Molin M, Hong SM, Hebbar S, Sharma R, Scrimieri F, de Wilde RF, Mayo SC, Goggins M, Wolfgang CL, Schulick RD, et al. 2012. Loss of expression of the SWI/SNF chromatin remodeling subunit BRG1/SMARCA4 is frequently observed in intraductal papillary mucinous neoplasms of the pancreas. Hum Pathol 43: 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La O JP, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, Murtaugh LC. 2008. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci 105: 18907–18912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, Helin K, Hornick JL, Mautner V, Kehrer-Sawatzki H, et al. 2014. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 514: 247–251. [DOI] [PubMed] [Google Scholar]
- Elsasser HP, Lehr U, Agricola B, Kern HF. 1992. Establishment and characterisation of two cell lines with different grade of differentiation derived from one primary human pancreatic adenocarcinoma. Virchows Arch B Cell Pathol Incl Mol Pathol 61: 295–306. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers S, Nagl NG Jr, Beck GR Jr, Moran E. 2009. Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. J Biol Chem 284: 10067–10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H, Ohuchida K, Mizumoto K, Nakata K, Yu J, Kayashima T, Cui L, Manabe T, Ohtsuka T, Tanaka M. 2010. α-Smooth muscle actin expressing stroma promotes an aggressive tumor biology in pancreatic ductal adenocarcinoma. Pancreas 39: 1254–1262. [DOI] [PubMed] [Google Scholar]
- Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. 2007. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature 449: 1073–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, Dubus P, Sandgren EP, Barbacid M. 2007. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11: 291–302. [DOI] [PubMed] [Google Scholar]
- Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, Feldmann G, Stoffers DA, Konieczny SF, Leach SD, et al. 2008. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci 105: 18913–18918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helming KC, Wang X, Roberts CW. 2014. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell 26: 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. 2006. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 20: 1218–1249. [DOI] [PubMed] [Google Scholar]
- Hoffman GR, Rahal R, Buxton F, Xiang K, McAllister G, Frias E, Bagdasarian L, Huber J, Lindeman A, Chen D, et al. 2014. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci 111: 3128–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, Wright CV, Moses HL. 2006. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-β signaling in cooperation with active Kras expression. Genes Dev 20: 3147–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada A, Nienaber C, Katsuta H, Fujitani Y, Levine J, Morita R, Sharma A, Bonner-Weir S. 2008. Carbonic anhydrase II-positive pancreatic cells are progenitors for both endocrine and exocrine pancreas after birth. Proc Natl Acad Sci 105: 19915–19919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischenko I, Petrenko O, Hayman MJ. 2014. Analysis of the tumor-initiating and metastatic capacity of PDX1-positive cells from the adult pancreas. Proc Natl Acad Sci 111: 3466–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeradjene K, Combs C, Best M, Gopinathan A, Wagner A, Grady WM, Deng CX, Hruban RH, Adsay NV, Tuveson DA, et al. 2007. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell 11: 229–243. [DOI] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. 2013. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, von Figura G, Mayes E, Liu FF, Dubois CL, Morris JP, Pan FC, Akiyama H, Wright CVE, Jensen K, et al. 2012. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22: 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Wong RP, Martinka M, Li G. 2010. BRG1 expression is increased in human cutaneous melanoma. Br J Dermatol 163: 502–510. [DOI] [PubMed] [Google Scholar]
- Liu X, Tian X, Wang F, Ma Y, Kornmann M, Yang Y. 2014. BRG1 promotes chemoresistance of pancreatic cancer cells through crosstalking with Akt signalling. Eur J Cancer 50: 2251–2262. [DOI] [PubMed] [Google Scholar]
- Matthaei H, Norris AL, Tsiatis AC, Olino K, Hong SM, Dal Molin M, Goggins MG, Canto M, Horton KM, Jackson KD, et al. 2012. Clinicopathological characteristics and molecular analyses of multifocal intraductal papillary mucinous neoplasms of the pancreas. Ann Surg 255: 326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melisi D, Ishiyama S, Sclabas GM, Fleming JB, Xia Q, Tortora G, Abbruzzese JL, Chiao PJ. 2008. LY2109761, a novel transforming growth factor β receptor type I and type II dual inhibitor, as a therapeutic approach to suppressing pancreatic cancer metastasis. Mol Cancer Ther 7: 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41: D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino-Kenudson M, Fernandez-del Castillo C, Baba Y, Valsangkar NP, Liss AS, Hsu M, Correa-Gallego C, Ingkakul T, Perez Johnston R, Turner BG, et al. 2011. Prognosis of invasive intraductal papillary mucinous neoplasm depends on histological and precursor epithelial subtypes. Gut 60: 1712–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JP IV, Wang SC, Hebrok M. 2010. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer 10: 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu SR, Love IM, Imbalzano AN, Grossman SR, Androphy EJ. 2009. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene 28: 2492–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata M, Morinaga S, Watanabe T, Tamagawa H, Yamamoto N, Shiozawa M, Nakamura Y, Kameda Y, Okawa S, Rino Y, et al. 2013. The clinical significance of SWI/SNF complex in pancreatic cancer. Int J Oncol 42: 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapoff KT, Cenik BK, Wang M, Ye R, Xu X, Nugent D, Hagopian MM, Topalovski M, Rivera LB, Carroll KD, et al. 2014. Neutralizing murine TGFβR2 promotes a differentiated tumor cell phenotype and inhibits pancreatic cancer metastasis. Cancer Res 74: 4996–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S. 2003. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol 23: 7475–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Hong SM, Klimstra DS, Goggins MG, Maitra A, Hruban RH. 2011. Pdx1 expression in pancreatic precursor lesions and neoplasms. Appl Immunohistochem Mol Morphol 19: 444–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poultsides GA, Reddy S, Cameron JL, Hruban RH, Pawlik TM, Ahuja N, Jain A, Edil BH, Iacobuzio-Donahue CA, Schulick RD, et al. 2010. Histopathologic basis for the favorable survival after resection of intraductal papillary mucinous neoplasm-associated invasive adenocarcinoma of the pancreas. Ann Surg 251: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert M, Rustgi AK. 2011. Pancreatic ductal cells in development, regeneration, and neoplasia. J Clin Invest 121: 4572–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert M, Takano S, Heeg S, Bakir B, Botta GP, Rustgi AK. 2013a. Isolation, culture and genetic manipulation of mouse pancreatic ductal cells. Nat Protoc 8: 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert M, Takano S, von Burstin J, Kim SB, Lee JS, Ihida-Stansbury K, Hahn C, Heeg S, Schneider G, Rhim AD, et al. 2013b. The Prrx1 homeodomain transcription factor plays a central role in pancreatic regeneration and carcinogenesis. Genes Dev 27: 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DP, Hong TS, Bardeesy N. 2014. Pancreatic adenocarcinoma. N Engl J Med 371: 1039–1049. [DOI] [PubMed] [Google Scholar]
- Sahai V, Kumar K, Knab LM, Chow CR, Raza SS, Bentrem DJ, Ebine K, Munshi HG. 2014. BET bromodomain inhibitors block growth of pancreatic cancer cells in three-dimensional collagen. Mol Cancer Ther 13: 1907–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentani K, Oue N, Kondo H, Kuraoka K, Motoshita J, Ito R, Yokozaki H, Yasui W. 2001. Increased expression but not genetic alteration of BRG1, a component of the SWI/SNF complex, is associated with the advanced stage of human gastric carcinomas. Pathobiology 69: 315–320. [DOI] [PubMed] [Google Scholar]
- Shain AH, Pollack JR. 2013. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 8: e55119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, Maitra A, Pollack JR. 2012. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci 109: E252–E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Zangen DH, Reitz P, Taneja M, Lissauer ME, Miller CP, Weir GC, Habener JF, Bonner-Weir S. 1999. The homeodomain protein IDX-1 increases after an early burst of proliferation during pancreatic regeneration. Diabetes 48: 507–513. [DOI] [PubMed] [Google Scholar]
- Siegel R, Ma J, Zou Z, Jemal A. 2014. Cancer statistics, 2014. CA Cancer J Clin 64: 9–29. [DOI] [PubMed] [Google Scholar]
- Sif S, Saurin AJ, Imbalzano AN, Kingston RE. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev 15: 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn M, Denkert C, Striefler JK, Pelzer U, Stieler JM, Bahra M, Lohneis P, Dorken B, Oettle H, Riess H, et al. 2014. α-Smooth muscle actin expression and desmoplastic stromal reaction in pancreatic cancer: results from the CONKO-001 study. Br J Cancer 111: 1917–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger BZ, Hebrok M. 2013. Control of cell identity in pancreas development and regeneration. Gastroenterology 144: 1170–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A, Tawfik O, Gayed B, Thrasher JB, Hoestje S, Li C, Li B. 2007. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate 67: 203–213. [DOI] [PubMed] [Google Scholar]
- Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. 2008. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem 283: 33437–33446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter KW, Archer TK. 2008. The BRG1 transcriptional coregulator. Nucl Recept Signal 6: e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Figura G, Fukuda A, Roy N, Liku ME, Morris Iv JP, Kim GE, Russ HA, Firpo MA, Mulvihill SJ, Dawson DW, et al. 2014. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol 16: 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Saito H, Ueda J, Evers BM. 2008. Regulation of pancreatic duct cell differentiation by phosphatidylinositol-3 kinase. Biochem Biophys Res Commun 370: 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Ueda Y, Akaboshi S, Hino Y, Sekita Y, Nakao M. 2009. HMGA2 maintains oncogenic RAS-induced epithelial–mesenchymal transition in human pancreatic cancer cells. Am J Pathol 174: 854–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Pang Y, Moses HL. 2010. TGF-β and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol 31: 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha L, Zhang J, Tang W, Zhang N, He M, Guo Y, Wang Z. 2013. HMGA2 elicits EMT by activating the Wnt/β-catenin pathway in gastric cancer. Dig Dis Sci 58: 724–733. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Morris JP IV, Yan W, Schofield HK, Gurney A, Simeone DM, Millar SE, Hoey T, Hebrok M, Pasca di Magliano M. 2013. Canonical wnt signaling is required for pancreatic carcinogenesis. Cancer Res 73: 4909–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li B, Li W, Ma L, Zheng D, Li L, Yang W, Chu M, Chen W, Mailman RB, et al. 2014. Transcriptional repression by the BRG1–SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports 3: 460–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Shi G, Schmidt CM, Hruban RH, Konieczny SF. 2007. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. Am J Pathol 171: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.