Abstract
Development of the metanephric kidney depends on tightly regulated interplay between self-renewal and differentiation of a nephron progenitor cell (NPC) pool. Several key factors required for the survival of NPCs have been identified, including fibroblast growth factor (FGF) signaling and the transcription factor Wilms' tumor suppressor 1 (WT1). Here, we present evidence that WT1 modulates FGF signaling by activating the expression of growth arrest-specific 1 (Gas1), a novel WT1 target gene and novel modulator of FGF signaling. We show that WT1 directly binds to a conserved DNA binding motif within the Gas1 promoter and activates Gas1 mRNA transcription in NPCs. We confirm that WT1 is required for Gas1 expression in kidneys in vivo. Loss of function of GAS1 in vivo results in hypoplastic kidneys with reduced nephron mass due to premature depletion of NPCs. Although kidney development in Gas1 knockout mice progresses normally until E15.5, NPCs show decreased rates of proliferation at this stage and are depleted as of E17.5. Lastly, we show that Gas1 is selectively required for FGF-stimulated AKT signaling in vitro. In summary, our data suggest a model in which WT1 modulates receptor tyrosine kinase signaling in NPCs by directing the expression of Gas1.
Keywords: Kidney development, Fibroblast growth factor signaling, Nephron progenitor cell, Mouse
Highlighted article: Wilms' tumor suppressor 1 (WT1) controls nephron numbers by regulating fibroblast growth factor signaling in nephron progenitors via Gas1, a novel WT1 target gene.
INTRODUCTION
The mammalian kidney is essential to the maintenance of body homeostasis by controlling water and salt balance and excreting waste products throughout life. This excretory function depends on its number of available kidney filtration units, nephrons, which is determined during kidney development. In mice, nephrogenesis ceases shortly after birth when the nephron progenitor cell (NPC) pool is completely exhausted (Short et al., 2014), whereas in humans the induction of new nephrons occurs entirely in the fetus. Crucially, in all mammals, it is not possible to induce new nephrons after the disappearance of NPCs. As such, the number of nephrons endowed during fetal kidney development in humans has been identified as a key risk factor for various diseases occurring later in life, including diabetes, hypertension and chronic kidney disease (Luyckx and Brenner, 2010). Therefore, pathways influencing nephron endowment during kidney development are now considered to be crucial to the pathogenesis of many diseases throughout life.
The development of the metanephric kidney depends on a tightly regulated and complex interplay of several cell lineages and structures derived from the intermediate mesoderm (Dressler, 2009). Metanephric development is initiated when the metanephric mesenchyme induces the outgrowth of the ureteric bud (UB) from the Wolffian duct. At the same time, the metanephric mesenchyme establishes a population of NPCs that give rise to nephrons. Signaling in both directions between the UB and NPCs results in an iterative process during which the UB undergoes branching morphogenesis to form ureteric tips and collecting ducts, while NPCs maintain a balance between self-renewal and differentiation. Signals from the UB induce NPCs to condense into pretubular aggregates (PTA) before undergoing mesenchymal-to-epithelial transition to form renal vesicles (RVs). RVs are further patterned and differentiated into primordial nephrons within S-shaped bodies, before a new round of branching and nephrogenesis is triggered (Dressler, 2009). Although the factors that affect branching morphogenesis and nephron induction were previously thought to be quantitatively constant throughout the period of nephrogenesis, recent studies have shown evidence of temporal and functional discontinuity in both processes (Short et al., 2014). As such, the NPC pool as a SIX2-positive (SIX2+) cell population can be divided into CITED1+, self-maintaining and CITED1-negative (CITED1−), induced compartments (Brown et al., 2013). Interestingly, proliferation rates in these compartments differ between early and late stages of renal development (Brown et al., 2013; Short et al., 2014). To date, the mechanisms controlling proliferation in these compartments remain unclear.
Research into nephrogenesis and NPCs has led to the identification of a large number of signaling pathways and molecules involved in regulating the balance between NPC self-renewal and differentiation, including the transcription factor Wilms' tumor suppressor protein 1 (WT1). In the absence of WT1, the metanephric mesenchyme undergoes apoptosis, resulting in renal agenesis (Kreidberg et al., 1993). Within NPCs, WT1 maintains the expression of a large number of genes required to promote the growth and differentiation of NPCs, including components and ligands of two major signaling pathways in NPCs – bone morphogenetic protein (BMP) and fibroblast growth factor (FGF) signaling, respectively (Hartwig et al., 2010; Motamedi et al., 2014). Both pathways are required for adequate proliferation rates within the NPC pool (Barak et al., 2012; Blank et al., 2009; Brown et al., 2011) and for an adequate NPC response to differentiation inductive signals (Blank et al., 2009; Brown et al., 2011; Motamedi et al., 2014). Interestingly, intracellular signaling cascades utilized by FGF and BMP signals overlap in various contexts. However, in NPCs, BMP has been shown to predominantly activate the mitogen-activated protein kinases (MAPKs) JNK and p38 (Blank et al., 2009; Di Giovanni et al., 2011; Motamedi et al., 2014), whereas FGF predominantly activates phosphoinositide 3-kinase (PI3K)-AKT pathways (Brown et al., 2011). Alteration of the activity of MAPK and PI3K signaling cascades has been linked to severe NPC defects, underscoring a need for tight control of signaling cascade levels and balances (Blank et al., 2009; Brown et al., 2011). To date, it is unclear how the balance between these intracellular cascades is regulated and how the correct levels of these signals are controlled in NPCs.
Based on our previous analysis of WT1 target genes in embryonic kidneys (Hartwig et al., 2010), the aim of this study is to identify the glycosylphosphatidylinositol (GPI)-anchored membrane protein growth-arrest-specific 1 (GAS1) as a novel and direct target gene of WT1 and characterize its function in NPCs. GAS1 is structurally related to glial-derived neurotrophic factor (GDNF) receptor alpha 1 (GFRA1) and has been shown to modulate intracellular signaling cascades in response to GDNF-Ret receptor tyrosine kinase (RTK) signaling (Cabrera et al., 2006). Furthermore, GAS1 has been identified as a co-receptor that binds to sonic hedgehog (SHH), resulting in amplification of SHH signals (Allen et al., 2007; Martinelli and Fan, 2007). We show that a WT1-responsive element in the Gas1 promoter is sufficient and required for Gas1 expression in embryonic kidneys. Loss of function of GAS1 results in hypoplastic kidneys due to decreased proliferation rates and premature depletion of NPCs. However, this phenotype does not manifest until later stages of kidney development, supporting the hypothesis that NPCs at early and late stages in development are governed by distinct though overlapping regulatory mechanisms. Finally, we link GAS1 to the activation of AKT downstream of FGF signals, providing a mechanistic basis for understanding the role of GAS1 in the control of NPC proliferation and differentiation. Taken together, these findings demonstrate how transcriptional regulation of gene expression can affect the level of RTK signaling in progenitor cells.
RESULTS
Identification of Gas1 as a WT1 target gene
WT1 is a transcription factor that is crucial to the maintenance and differentiation of NPCs in developing kidneys. Among the WT1 target genes identified in our previous study (Hartwig et al., 2010), Gas1 was of interest as a potential modifier of signal transduction pathways, the study of which would offer insight into how transcriptional regulation by WT1 would affect signaling in NPCs. Furthermore, the GUDMAP database had identified high level expression of Gas1 in NPCs.
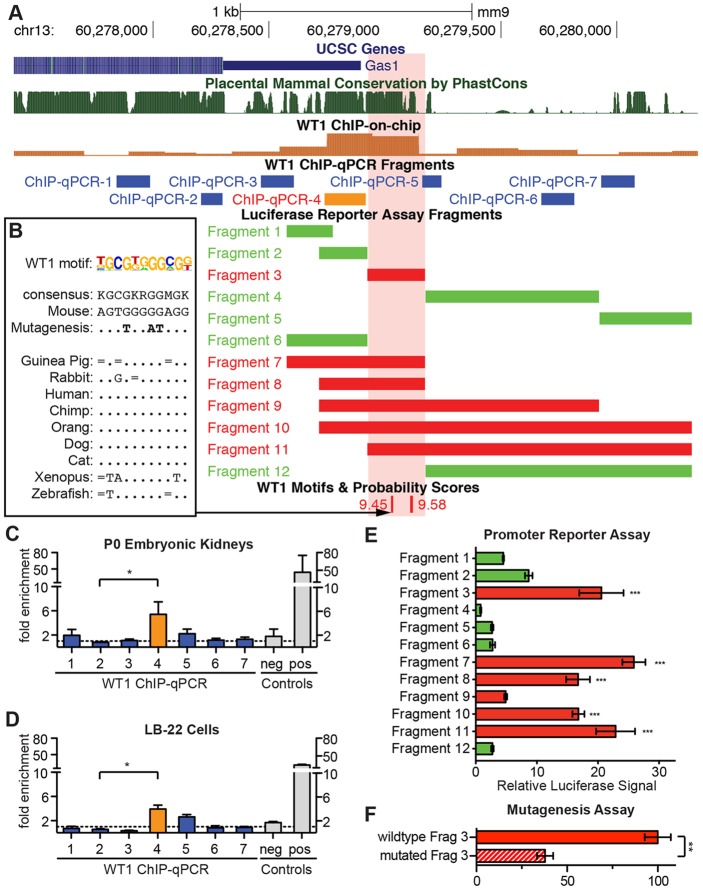
The Gas1 promoter was previously found to be bound by WT1 in a chromatin immunoprecipitation followed by array hybridization (ChIP-on-chip) screen in embryonic kidneys (Hartwig et al., 2010) with a 4.5-fold enrichment over background (adjusted P-value<0.0001, Fig. 1A, brown bars; supplementary material Fig. S1A). In order to confirm this interaction, we extracted chromatin from stage postnatal day (P) 0 wild-type kidneys and performed ChIP followed by qPCR using seven primer pairs tiled along the Gas1 promoter, 5′UTR and open reading frame (Fig. 1A,C). These experiments replicated the finding of WT1 binding to a conserved site, which was located close to the Gas1 transcription start site (TSS) in the same position as the two ChIP-on-chip probes with the highest enrichment. The specificity of these experiments was controlled by using an intronic site lacking a WT1 binding motif within the Gapdhs gene as negative control and the promoter of Kdm3a, which is tightly bound by WT1 (Hartwig et al., 2010), as a positive control (Fig. 1C, n=3). WT1 is thus bound to the Gas1 promoter in vivo.
Fig. 1.
WT1 binds to the Gas1 promoter to control Gas1 expression. (A) Representation of the Gas1 promoter in the UCSC genome browser, including WT1 ChIP-on-chip signals, location of PCR products for ChIP-qPCR, promoter fragments used for luciferase assays and WT1 binding motifs abolished by mutagenesis. The red box indicates the WT1-sensitive element. (B) Cross-species conservation at the conserved WT1 motif abolished by mutagenesis. The WT1 motif logo, consensus sequence, bases changed by mutagenesis and genomic sequence from various species is shown. Dots indicate identical bases, bars represent bases that match the consensus. (C,D) Results of WT1 ChIP-qPCR in embryonic kidneys and LB-22 cells using PCR products as indicated in A. An intron of Gapdhs and the Kdm3b promoter are used as negative (neg) and positive (pos) controls, respectively. (E) Dual luciferase promoter reporter assays in LB-22 cells for fragments shown in A and fragments with mutated WT1 binding sites. (F) Relative luciferase reporter signals for the wild type and mutated (see B) WT1-responsive element. For C-F, data show the mean±s.e.m., n=3; *P<0.05; **P<0.01; ***P<0.001.
To then assess the functional significance of WT1 binding to the Gas1 gene, we determined whether loss of WT1 affected levels of Gas1 mRNA. Initial evidence that WT1 indeed controls Gas1 transcription was acquired through characterization of a novel WT1-expressing cell line, LB-22. LB-22 is an immortalized mesenchymal cell line derived from the nephrogenic zone of mouse embryonic kidneys that constitutively expresses abundant WT1. Characterization of the cell line by RNA expression microarray experiments revealed expression of several genes that are bona fide WT1 target genes in NPCs in vivo, such as Cxxc5, Pbx2, Sox4 and Rps6ka3 (Hartwig et al., 2010). However, the cell line lacks expression of most established marker genes of NPCs, such as Six2, Cited1 and Sall1. Hence, LB-22 does not accurately model NPCs in cell culture but, based on its constitutive WT1 expression, it is a useful tool to address WT1-mediated regulation of gene expression. In order to show that the binding of WT1 to the Gas1 promoter as described in embryonic kidneys is present in LB-22 cells, we repeated the ChIP-qPCR experiments, revealing the same binding site of WT1 in LB-22 cells (Fig. 1D). LB-22 cells can therefore serve as an accurate model of WT1-mediated regulation of Gas1 in cell culture.
Furthermore, we established siRNA-mediated knockdown of WT1 in LB-22 cells (supplementary material Fig. S1B) and used mRNA expression arrays (n=4) to analyze the effects of WT1 depletion on mRNA levels (supplementary material Fig. S1C,D). Unbiased analysis of summarized array datasets identified Gas1 as one of the most significantly downregulated genes upon treatment with Wt1 siRNA (supplementary material Fig. S1D). These results were validated by RT-qPCR and western blotting (supplementary material Fig. S1B). In conclusion, WT1 binds the Gas1 promoter and affects Gas1 mRNA and protein levels in LB-22 cells.
A WT1-responsive DNA element within the Gas1 promoter is required and sufficient for Gas1 expression
We used promoter-reporter and mutagenesis studies to establish direct regulation of Gas1 by WT1. Direct target genes contain functional transcription factor binding DNA motifs within their cis-regulatory domain (CRD). We therefore screened the conserved CRD surrounding the Gas1 TSS within −1.5 kb to 0.5 kb for instances of WT1 binding motifs using a published WT1 positional weight matrix (Hartwig et al., 2010). Based on the location of WT1 motifs, conservation, ChIP-qPCR and ChIP-on-chip results, we divided the Gas1 CRD into five sections that were subsequently cloned in several combinations (Fig. 1A, green and red bars) into a luciferase reporter vector containing a minimal promoter. When measuring relative luciferase signals generated by these fragments upon transfection into LB-22 cells, exclusively fragments containing a 200 bp region close to the Gas1 TSS produced a significant increase in reporter signals, indicating that a functional cis-regulatory element was located in this region (Fig. 1E). This promoter element overlapped with the WT1-bound region as identified by ChIP (Fig. 1A, red shaded box). To confirm that WT1 was indeed responsible for eliciting reporter signals, we repeated the reporter assays in LB-22 cells treated with WT1 and control siRNAs. Reporter assays in the presence of WT1 siRNA showed significantly reduced signals exclusively in fragments containing the WT1-responsive element (supplementary material Fig. S2A), indicating that our signals were specific to WT1 and that WT1 binding to these promoter fragments was required for efficient downstream mRNA expression. As experiments were performed in the presence of a minimal promoter within the reporter vectors, we next analyzed whether the endogenous promoter was sufficient to induce expression by using a promoterless luciferase reporter vector (supplementary material Fig. S2C). Again, we identified the same WT1-responsive element as sufficient to elicit significant reporter activity. In summary, we characterized a WT1-responsive element in the Gas1 promoter that is both required and sufficient to induce transcription in a WT1-dependent manner.
We next analyzed WT1 binding motifs within the WT1-responsive element and identified two sequences with increased motif probability scores (Fig. 1A,B), one of which showed substantial cross-species conservation (Fig. 1B). Site-directed mutagenesis was carried out on the luciferase reporter constructs to abolish both motifs (Fig. 1B). Transfection of these mutated WT1-reporter constructs resulted in a 60% decrease in luciferase signals when compared with those of wild type (Fig. 1F). Gas1 was thus characterized as a direct WT1 target gene in LB-22 cells.
WT1 is required for efficient Gas1 mRNA and protein expression in embryonic kidneys ex vivo
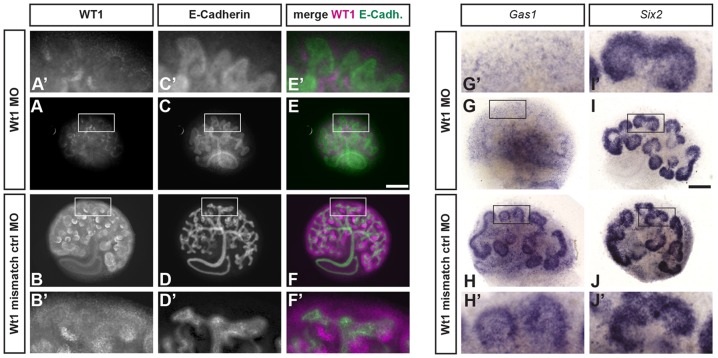
Having established that WT1 could regulate Gas1 expression in an in vitro model system, we next sought to determine whether our findings could be validated in embryonic kidneys. In a first approach, WT1 expression in embryonic kidney organ explants was inhibited using a previously described morpholino oligonucleotide (MO) technique (Fig. 2A-F) (Hartwig et al., 2010) and assayed for Gas1 mRNA levels. Staining for Gas1 mRNA by in situ hybridization (ISH) revealed diminished Gas1 expression in WT1 morphant cultures when compared with controls (Fig. 2G-J). Importantly, within the time frame of this experiment, signals for SIX2+ NPCs did not change between morphant cultures and controls, confirming that the loss of Gas1 signals was not due to loss of NPCs. Furthermore, the expression pattern of Gas1 in developing kidneys was identical to that of Six2, identifying the NPC as the key cell within embryonic kidneys for a WT1-directed regulation of Gas1. This ex vivo model therefore further supported our previous findings of the requirement of WT1 to activate Gas1 transcription in NPCs.
Fig. 2.
Knockdown of WT1 in kidney organ cultures results in decreased Gas1 mRNA. (A-F) Morpholino oligonucleotide (MO)-based knockdown of WT1 in kidney organ cultures results in decreased levels of WT1 protein. (G-J) In situ hybridization for Gas1 mRNA shows decreased signals in WT1 morphant cultures while Six2+ nephron progenitor cells are maintained. Scale bars: 100 µm.
A novel Wt1 hypomorphic mouse model confirms regulation of Gas1 by WT1
As RNA interference (RNAi)-based and ex vivo studies can be compromised by off-target effects and confounders introduced within the organ culture system, we sought to extend our ex vivo findings in an independent in vivo model where expression of Wt1 was reduced in embryonic kidneys. To this end, we generated Wt1ckd mice as a tool to investigate WT1 target genes. Wt1ckd mice conditionally express a short hairpin (sh)RNA (Coumoul et al., 2004; Shukla et al., 2007) directed against Wt1 in NPCs and derivatives by Six2:Cre-driven recombination. Wt1ckd mice were born in Mendelian ratios and were viable for up to 5 weeks. Although kidney development appeared to progress normally until stage embryonic day (E)14.5 (data not shown), at stage E16.5 WT1 hypomorphic phenotypes became evident, with premature loss of condensing mesenchyme surrounding the ureteric tips and lack of comma- and S-shaped bodies (Fig. 3A,B). Kidneys of newborn Wt1ckd mice were hypoplastic (Fig. 3C,D) and had reduced nephron endowment. We subsequently assayed the efficacy of our RNAi approach by confirmation of decreased Wt1 mRNA and protein levels in Wt1ckd kidneys using ISH at stage E14.5 (Fig. 3E,F) and immunofluorescence staining at stage E15.5 (Fig. 3K,L), respectively. Importantly, the SIX2+ NPC pool was preserved at stage E14.5 (Fig. 3I,J) and only moderately depleted at stage E15.5 (Fig. 3Q,R), rendering these stages suitable for WT1 target gene analysis.
Fig. 3.
Conditional knockdown of WT1 in nephron progenitors in vivo results in decreased Gas1 expression. (A,B) High-power histology of the nephrogenic zone of WT1 knockdown (Wt1ckd) kidneys and controls at stage E16.5. Loss of condensed mesenchyme and pretubular aggregates/induced nephrons at a preserved ureteric bud branch. (C,D) Low-power H&E stain of Wt1ckd kidneys at stage P0 shows hypoplasia with cystic malformations and increased stroma. (E-J) In situ hybridization for Wt1, Gas1 and Six2 mRNA at stage E14.5. Decreased signals for Wt1 and Gas1 are evident in Wt1ckd kidneys. NPCs are preserved at this stage. (K-R) Decreased WT1 and GAS1 in Wt1ckd are visualized by immunofluorescence at stage E15.5. NPCs are decreased in number in Wt1ckd kidneys. Scale bars: 10 µm in A,E,K; 50 µm in C.
The subsequent analysis of Gas1 expression in WT1ckd kidneys identified decreased Gas1 mRNA in NPCs at stage E14.5 (Fig. 3G,H) and decreased Gas1 protein at stage E15.5 (Fig. 3M,N) when compared with Cre-negative controls. Of note, although Wt1 mRNA and protein expression is increased in pretubular aggregates and renal vesicles (Fig. 3F,L), Gas1 expression in these structures was similar to that of Six2. Gas1 mRNA expression was abolished, whereas protein was retained at low levels until the renal vesicle stage before vanishing completely at the S-shaped body stage (Fig. 3H,N), suggesting that Gas1 function and gene regulation are specific to the NPC pool in embryonic kidneys. In summary, we established Gas1 as a bona fide direct WT1 target gene in embryonic kidneys in vivo.
GAS1 loss of function results in hypoplastic kidneys with decreased nephron endowment
As high expression levels of Gas1 in NPCs and the sharp decline of Gas1 levels upon nephron induction had already suggested a relevant function of Gas1 in kidney development, we next sought to determine the effects of Gas1 loss of function in embryonic kidneys. To this end, we made use of the previously published Gas1 knockout mouse (Martinelli and Fan, 2007).
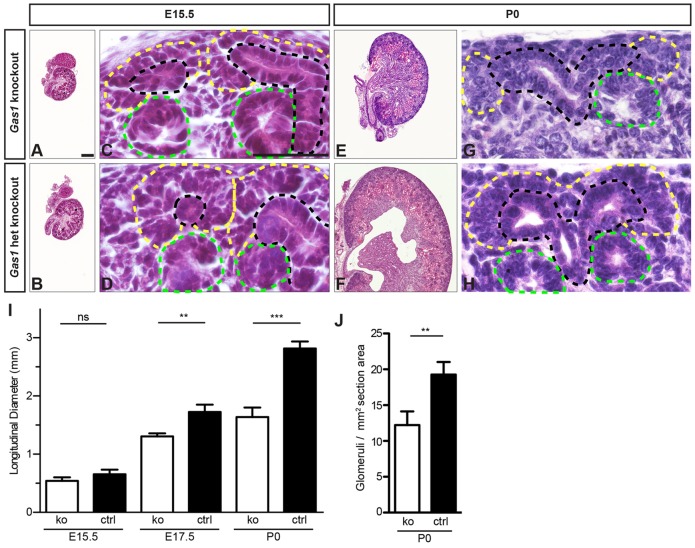
Initial histologic analysis of Gas1 null kidneys at stage E15.5 did not show significant differences when compared with heterozygous null controls (Fig. 4A,B,I). The overall size of Gas1 null kidneys at E15.5 was similar to that of control counterparts; condensing mesenchyme was clearly visible surrounding the ureteric tips and nephron induction appeared to be occurring normally, with evidence of one to two induced nephron structures (renal vesicles, comma- or S-shaped bodies) per ureteric branch (Fig. 4C,D). Similar numbers of fully differentiated glomeruli with positive nephrin expression were present in Gas1 null and wild-type kidneys (data not shown). However, analysis of kidney size at stage E17.5 and later showed significant hypoplasia of Gas1 null kidneys, with knockout kidneys at stage P0 being about half the size of heterozygous knockout control kidneys (Fig. 4E,F,I). High-power histologic analysis at stage P0 suggested perturbation of the NPC pool and nephrogenesis in Gas1 knockout kidneys with premature depletion of condensing mesenchyme surrounding the ureteric tips and a reduced number of induced nephrons (Fig. 4G,H). Decreased nephron endowment as measured by counting glomeruli was also evident (Fig. 4J). Therefore, loss of function of Gas1 did not appear to affect early renal development and establishment of the nephrogenic zone; however, a quantitative effect on nephrogenesis at stages in renal development where the bulk of nephrons is formed was clearly evident.
Fig. 4.
Gas1 knockout kidneys are hypoplastic and show disorganization of the nephrogenic zone. (A-D) Histology of Gas1 knockout kidneys at stage E15.5. Kidneys are normal and adequately sized with condensing mesenchyme (yellow line) and two renal vesicles (green line) surrounding a ureteric bud branch (black line). het, heterozygous. Scale bars: 50 µm (low power), 10 µm (high power). (E-H) Histology at stage P0. Gas1 knockout kidneys are hypoplastic. Condensed mesenchyme is reduced and only a single renal vesicle is present. (I) The longitudinal diameters of kidney sections including the papilla at various stages. ko, knockout; ctrl, heterozygous knockout. Data show the mean±s.d., n≥4; ns, not significant; **P<0.01; ***P<0.001. (J) The numbers of glomeruli in Gas1 knockout and control kidneys at P0. Data show the mean±s.d.; **P<0.01.
GAS1 controls nephron endowment by ensuring adequate nephron progenitor cell proliferation
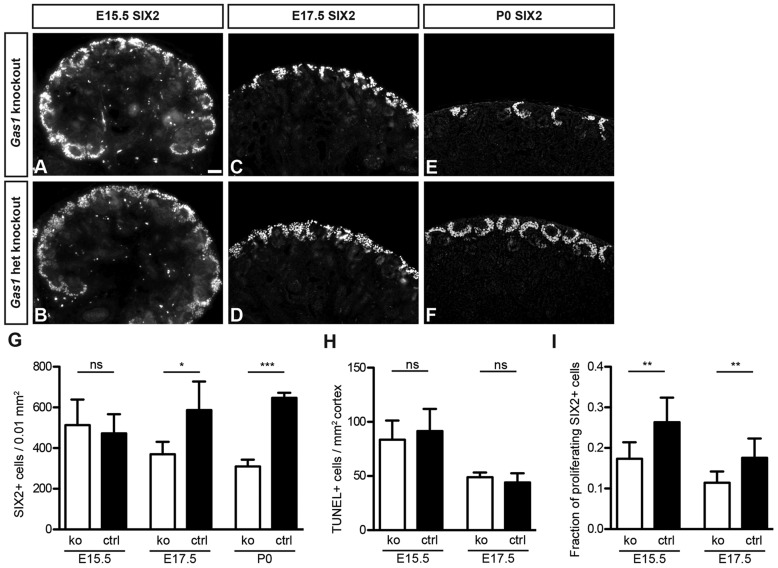
We continued to investigate the SIX2+ NPC pool in Gas1 knockout kidneys in order to elucidate the role for GAS1 in NPC self-renewal and differentiation. The expression pattern of Gas1 and the depletion of condensed mesenchyme in Gas1 knockout kidneys suggested a role for GAS1 in the maintenance of NPCs. Similar to the histological findings, at stage E15.5 no differences in numbers of SIX2+ NPCs or their distribution were evident upon examination of SIX2 expression and quantitative analyses of SIX2-expressing cells (Fig. 5A,B,G). Beginning at stage E17.5, however, premature depletion of the NPC pool became visible, with fewer SIX2+ cells surrounding ureteric tips and a significant reduction in NPC numbers (Fig. 5C,D,G). NPC depletion became significantly more pronounced by P0 (Fig. 5E-G).
Fig. 5.
Progressive loss of SIX2+ nephron progenitor cells in Gas1 knockout kidneys due to decreased proliferation. (A-F) Immunofluorescence staining for SIX2 on Gas1 knockout and control kidneys at various stages. A decreased number of SIX2+ nuclei is evident with progression of renal development. het, heterozygous. Scale bar: 10 µm. (G) Quantification of SIX2+ nuclei normalized to cortex surface at various embryonic stages. Data show the mean±s.d., n≥4; ns, not significant; *P<0.05; ***P<0.001. (H) Quantification of TUNEL+ signals normalized to cortical area. Increased cell death is excluded as a reason for NPC loss. Data show the mean±s.d., n=3; ns, not significant. (I) Quantification of SIX2+/BrdU+ cells at E15.5 and E17.5. The proliferative fraction of SIX2+ cells in Gas1 knockout kidneys is decreased at E15.5 and E17.5. ko, knockout; ctrl, heterozygous knockout. Data show the mean±s.d., n=3; **P<0.01.
NPCs can be depleted either by differentiation, cell death or inadequate self-renewal. GAS1 had previously been implicated in apoptosis and cell-cycle control (Del Sal et al., 1992; Liu et al., 2001; Mellstrom et al., 2002). In order to identify the defect responsible for premature depletion of the NPC pool, we first examined NPC proliferation and cell death in Gas1 knockout kidneys. A quantitative analysis of TUNEL+ cells in the nephrogenic zone did not reveal a difference between Gas1 knockout and control kidneys at stages E15.5 and E17.5, suggesting that NPC death was not a primary factor in their depletion (Fig. 5H). However, quantitative analysis of NPC proliferation demonstrated fewer BrdU+/SIX2+ NPCs as a fraction of the overall SIX2+ NPCs at both E15.5 and E17.5 (Fig. 5I). As numbers of SIX2+ cells did not yet differ at E15.5 between Gas1 knockout and control kidneys, the mechanism by which GAS1 affects proliferation of NPCs appears to be particularly relevant to the maintenance of NPCs at later stages in kidney development.
A role for GAS1 in the maintenance of NPCs was corroborated in a second model using two overlapping MOs directed against Gas1 mRNA to block translation in E12.5 kidney organ cultures. In this model, an efficient knockdown of GAS1 was achieved within 18 h of cultivation in the presence of a low MO concentration (supplementary material Fig. S4A-G). Increased MO concentrations and longer cultivation periods resulted in rapid depletion and finally abolishment of NPCs in Gas1 morphant cultures in a time- and dose-dependent manner (supplementary material Fig. S4H-M). As opposed to the NPC depletion phenotype occurring at later stages in kidney development of Gas1 knockout kidneys, within the context of this ex vivo model, this phenotype was exacerbated and already evident within 24 h of cultivation in the presence of MOs.
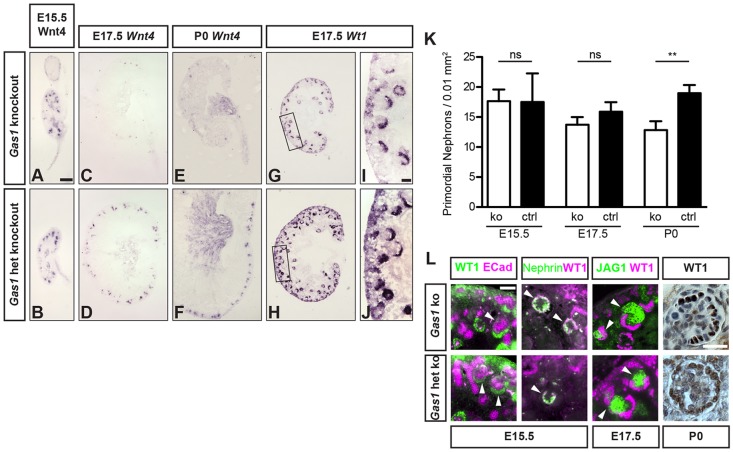
Gas1 knockout kidneys were also examined for abnormalities in nephron differentiation. Consistent with the histology, at stage E15.5, Gas1 knockout kidneys did not show any defects in the induction of Wnt4+ PTAs or RVs (Fig. 6A,B). With declining numbers of NPCs at E17.5, however, very few Wnt4+ structures were present in Gas1 knockout kidneys, progressing to their complete absence at stage P0 (Fig. 6C-F). Similar findings were obtained when analyzing induced nephrons using increased Wt1 levels in PTAs and RVs as a marker (Fig. 6G-J). Of note, S-shaped bodies that were induced in Gas1 knockout kidneys at any stage, including E17.5 and later, showed normal patterning in terms of glomerular WT1+, proximal tubular JAG1+ and distal tubular E-cadherin+ domains, and were capable of forming morphologically normal glomeruli (Fig. 6L), consistent with a role for GAS1 in maintaining NPCs, rather than affecting their differentiation into nephrons. Indeed, the number of NCAM+ primordial nephrons normalized to section area was significantly reduced in Gas1 knockout kidneys at stage P0 (Fig. 6K; supplementary material Fig. S3). Furthermore, there was no indication of ectopic or precocious differentiation of nephrons in Gas1 knockout kidneys. In summary, loss of GAS1 results in decreased proliferation of NPCs and establishes GAS1 as a novel factor required for the maintenance of NPCs during mid and later stages of kidney development in vivo.
Fig. 6.
Nephrogenesis decreases prematurely in Gas1 knockout kidneys. (A-J) In situ hybridization for Wnt4 and Wt1. Normal signal distribution at E15.5 and near loss of Wnt4+ structures in Gas1 knockout beginning at stage E17.5. Wt1+ S-shaped bodies and glomeruli are visible at E17.5, PTAs and RVs are lacking. Scale bars: 50 µm (low power), 10 µm (high power). (K) Quantification of NCAM+ primordial nephrons normalized to cortical surface. ko, knockout; ctrl, heterozygous knockout. Data show the mean±s.d.; ns, not significant; **P<0.01. (L) Immunofluorescence staining of induced nephrons and glomeruli (arrowheads) for JAG1, WT1, E-cadherin and nephrin show normal signal patterning. Immunohistochemistry for WT1 in glomeruli shows normal distribution of podocytes at P0. het, heterozygous. Scale bars: 5 µm.
GAS1 directs the intracellular response to FGF9 and FGF20 stimulation towards activation of AKT
GAS1 has previously been shown to function in two major signaling pathways with relevance to kidney development; namely, SHH signaling and the GDNF-Ret RTK axis (Allen et al., 2007; Cabrera et al., 2006; Izzi et al., 2011; Martinelli and Fan, 2007). With regard to SHH signaling, GAS1 functions as a SHH co-receptor facilitating SHH signal transmission to GLI1 and full-length GLI3 (Allen et al., 2007; Martinelli and Fan, 2007). However, the kidney cortical mesenchyme and NPCs require signaling of repressive, cleaved GLI3-R isoforms that are generated in the absence of SHH ligand (Cain et al., 2009), inconsistent with a possible role for GAS1 in potentiating SHH signaling in NPCs. We therefore investigated whether GAS1 plays a role in RTK signaling in NPCs.
As GAS1 has been shown to modulate signaling downstream of RET (Cabrera et al., 2006), a key pathway in ureteric branching that also feeds back to NPCs through WNT11 (Costantini and Kopan, 2010; Majumdar et al., 2003), we investigated whether expression patterns and levels of ureteric tip genes dependent on RET were altered in Gas1 knockout kidneys (Fig. 7A-G). No changes in expression patterns by ISH and mRNA levels by RT-qPCR were identified for Wnt11, Crlf1, Etv4 and Cxcr4. These data indicated that GAS1 does not exert a non-cell-autonomous effect on RET signals in ureteric tip cells, and further suggest that decreased NPC proliferation in Gas1 knockout kidneys is not a consequence of decreased RET-dependent pro-proliferative signals from ureteric tips.
Fig. 7.

Loss of function of Gas1 does not affect RET signaling in vivo. (A-F) In situ hybridization on stage E15.5 kidney sections for Wnt11, Etv5 and Crlf1 mRNAs as genes depending on RET signaling shows no difference in expression patterns. het, heterozygous. Scale bar: 50 µm. (G) Real-time qPCR results for RET-dependent genes on total RNA extracted from E13.5 kidneys. ko, knockout; ctrl, heterozygous knockout. Data show the mean±s.e.m., n=3; ns, not significant.
Signaling of FGF9 and FGF20 to NPCs is required for their maintenance and proliferation (Barak et al., 2012). Interestingly, the modulating effect of Gas1 on RET signaling has been shown to be dependent on the interaction of RET with FRS2A (Cabrera et al., 2006), a phospho-tyrosine-binding RTK adaptor molecule that also plays key roles in mediating FGF signaling. Therefore, we next investigated whether GAS1 modulates FGF signaling by knocking down Gas1 in LB-22 cells. LB-22 cells endogenously express FGF receptor 2 (FGFR2), the receptor for FGF8, FGF9 and FGF20, key FGF ligands expressed in NPCs and RVs (Barak et al., 2012; Grieshammer et al., 2005; Perantoni et al., 2005). We stimulated LB-22 cells with these FGF ligands after transfection with Gas1 siRNA or scramble control siRNA and assayed for the phosphorylation of p42/p44 MAPK (ERK; MAPK3/MAPK1, respectively – Mouse Genome Informatics) and AKT (Fig. 8A,B). Interestingly, the phosphorylation of AKT upon FGF stimulation was reduced to baseline levels in the Gas1 knockdown condition, whereas ERK phosphorylation was not affected. These findings indicate a novel function for GAS1, in potentiating signaling from FGF receptors that is selective to the AKT pathway as opposed to affecting activation of ERK. Therefore, we assayed AKT phosphorylation in NPCs of E15.5 Gas1 knockout kidneys and controls by immunohistochemistry (Fig. 8C-G). Indeed, phospho-AKT signals in NPCs were considerably reduced upon loss of GAS1, whereas signals in ureteric tips, the kidney capsule and developing nephrons appeared to be unchanged. Furthermore, we aimed to rescue the FGF signaling defect by overstimulating Gas1 morphant kidney explants with excess FGF9 (data not shown). However, in line with GAS1 modulating RTK signals on the intracellular level, neither the dynamics of NPC depletion nor the extent of the NPC pool appeared to be different between overstimulation and control conditions. In summary, we identify a modulating effect of GAS1 on FGF signaling in vitro as well as in vivo and suggest that GAS1 drives the NPC response to FGF signals towards intracellular activation of AKT to promote NPC proliferation.
Fig. 8.
Depletion of Gas1 modulates FGF signaling through AKT in vitro and in vivo. (A) Western blots of whole-cell lysates from LB-22 cells treated with Gas1 siRNA or scramble control and stimulated with 50 ng/ml FGF as indicated. (B) Densitometry of repeat western blots as in A. Results are normalized to the non-stimulated, vehicle-treated control and to total AKT, total ERK or tubulin, respectively. Data show the mean±s.e.m., n≥3; ns, not significant. (C-G) Immunohistochemistry for phospho-Akt at E15.5 shows diminished phospho-Akt signal in condensed mesenchyme upon Gas1 knockout. Black boxes in C,D correspond to high-power panels. Ureteric tips are labeled with asterisks. Arrowheads indicate condensed mesenchyme. Scale bars: 200 µm (low power), 25 µm (high power), 75 µm (mid power).
DISCUSSION
The data provided in this study establish the GPI-anchored membrane molecule GAS1 as a novel and direct WT1 target gene in NPCs. Furthermore, we show in embryonic kidneys that GAS1 controls nephron endowment by ensuring NPC proliferation is sufficient to maintain the SIX2+ NPC population. In the absence of GAS1, early metanephric development, establishment of a nephrogenic niche and initial nephrogenesis proceed normally. However, loss of GAS1 results in premature depletion of the NPC pool at later stages in kidney development with subsequent decreased nephrogenesis and kidney hypoplasia. Mechanistically, our data suggest that GAS1 affects signaling downstream of FGF9 and FGF20 to phosphorylate AKT but not ERK, allowing for the activation of pro-proliferative pathways downstream of AKT in NPCs. These data establish a link between WT1 and FGF signaling in NPCs through transcriptional control of Gas1 and provide novel insight into how WT1 maintains NPCs.
WT1 directly activates Gas1 transcription
Homozygous deletion of WT1 has long been known to result in kidney agenesis due to premature loss of NPCs (Kreidberg et al., 1993). Recent studies employing ChIP techniques to identify WT1 binding sites in a genome-wide fashion provided further insight into this phenotype, identifying a large number of bona fide and candidate WT1 target genes (Hartwig et al., 2010; Motamedi et al., 2014). Importantly, the WT1 binding site close to the Gas1 TSS was replicated independently in one of these studies (Motamedi et al., 2014). Genome-wide WT1 ChIP experiments have provided new insight into the mechanisms by which WT1 regulates its target genes. Interestingly, both promoter (Hartwig et al., 2010; Motamedi et al., 2014) and distal WT1 binding events (Essafi et al., 2011; Martinez-Estrada et al., 2010; Motamedi et al., 2014) in putative enhancers appear to be involved in WT1 target gene regulation. In the present study, we dissect the Gas1 promoter using luciferase reporter assays and show that a short 200 bp WT1-responsive promoter fragment is sufficient and required to endogenously activate Gas1 transcription. These data indicate that, in NPCs, at least a subset of WT1 target genes in NPCs depends on promoter WT1 binding events for efficient transcription. These results are in agreement with previous findings that WT1 binding at target promoters in NPCs activates transcription of a large number of key genes in renal development (Hartwig et al., 2010; Motamedi et al., 2014). A puzzling aspect of the transcriptional regulation of Gas1 and other WT1 target genes in NPCs is the fact that their expression in NPCs depends on WT1, whereas they are silenced in RVs despite increased WT1 levels in these structures. Consistent with these findings, Gas1 mRNA has been shown to be downregulated upon induction of canonical WNT signaling in FACS-sorted NPCs, an effect that was attributed to a co-operative SIX2 and β-catenin binding site downstream of the Gas1 locus (Park et al., 2012). Therefore, Gas1 transcription in NPCs might be dependent on co-operative SIX2 and WT1 effects, with SIX2 being bound at a distal regulatory element and WT1 at the promoter. Once the differentiation of NPCs is induced by canonical WNT signals (Carroll et al., 2005; Karner et al., 2011), β-catenin binding at the SIX2-bound regulatory element might disrupt this co-operation (Park et al., 2012) and result in Gas1 silencing in RVs. Another possible explanation for the incongruence of Gas1 and Wt1 expression patterns might be a differential utilization of the WT1-dependent co-activator CBP/p300 (Wang et al., 2001) and co-repressor BASP1 (Carpenter et al., 2004) between NPCs and RVs, resulting in different epigenetic regulation on the chromatin level at the Gas1 locus as has been described for the Wnt4, Snai1 and Cdh1 loci in various tissues (Essafi et al., 2011; Martinez-Estrada et al., 2010). In the light of our data that implicate GAS1 in modulating FGF signals during kidney development, differential transcriptional regulation of Gas1 between NPCs and RVs might, in part, account for differences in FGF activity within NPCs versus nephrons. For example, FGF8 signals derived from RVs are required for Wnt4 and Lhx1 expression within the vesicles (Grieshammer et al., 2005; Perantoni et al., 2005). However, in contrast to NPC-derived FGF20, FGF8 signals are not sufficient to maintain the progenitor status in NPCs and might direct FGF signals towards responses other than AKT phosphorylation (Barak et al., 2012). In summary, our results suggest that WT1 binding to promoters is a key event in WT1 target gene regulation in NPCs, which is in contrast to the more prevalent enhancer-based transcriptional regulation as observed in SIX2 targets in NPCs (Park et al., 2012).
GAS1 maintains NPC expansion at late stages in kidney development
Decreased nephron endowment during renal development has been shown to be a key risk factor for hypertension and chronic kidney disease later in life (Luyckx et al., 2013), highlighting the importance of identifying genetic modifiers of nephron number during kidney development. Reduced nephron endowment can result from perturbations within several cell pools and in different morphogenetic processes during kidney development, including the progenitor cell niche, the kidney stroma, branching morphogenesis and nephrogenesis (Kopan et al., 2014). The majority of loss-of-function phenotypes affecting genes expressed in NPCs result in renal agenesis, failed establishment of the NPC niche or in severe early impairment of nephrogenesis, featuring kidney rudiments with few if any induced nephrons (Kopan et al., 2014). Loss-of-function phenotypes associated with reduced NPC proliferation have been reported, including Bmp7 (Blank et al., 2009; Dudley et al., 1995), c-myc (Couillard and Trudel, 2009), Dlg1 and Cask (Ahn et al., 2013), Mi-2-NurD (Denner and Rauchman, 2013) and the miR-17-92 cluster (Marrone et al., 2014). However, with the exception of the miR-17-92 knockout, these phenotypes are characterized by early growth deficits and reduced nephron numbers evident by E13.5 (Ahn et al., 2013) or E14.5 (Denner and Rauchman, 2013; Dudley et al., 1995). By contrast, establishment of the niche and the nephrogenic zone in early metanephric development appear to be unaffected in Gas1 knockout kidneys. A possible explanation for these divergent phenotypes all associated with NPC proliferation deficits is differential regulation of proliferation in early (E13.5) as compared to late (E17.5) NPC pools. In fact, temporal discontinuity with respect to NPC proliferation rates and cell cycle lengths in both CITED1+ and CITED1− NPC compartments has recently been described (Short et al., 2014), with the majority of proliferating NPCs being contributed by a slow-cycling compartment at E17.5, whereas at E13.5 proliferation was predominantly in a fast-cycling NPC compartment. Given that Gas1 knockout kidneys are phenotypically normal until E15.5, our data suggest that GAS1 might primarily regulate the slow-cycling NPC population.
GAS1 selectively modulates the AKT branch of the FGF signaling pathway
Multiple signaling pathways have been reported to regulate the balance between self-maintenance and differentiation in NPCs, including FGF, BMP and canonical WNT signals (Barak et al., 2012; Blank et al., 2009; Brown et al., 2011, 2013; Carroll et al., 2005; Park et al., 2012), all of which interact with or are activated by WT1 (Akpa et al., 2014; Hartwig et al., 2010; Kim et al., 2010; Motamedi et al., 2014). The FGF ligand FGF20 is transcriptionally activated by WT1 in NPCs (Motamedi et al., 2014) and is connected to NPC proliferation through activation of PI3K-AKT (Barak et al., 2012; Brown et al., 2011), whereas Bmp7 is a WT1 target gene (Hartwig et al., 2010) that promotes NPC proliferation by activation of JNK MAPK (Blank et al., 2009; Motamedi et al., 2014). Both MAPK and PI3K-AKT cascades have well-established roles in cell cycle control; however, the mechanisms by which these signaling cascades are balanced and regulated in response to BMP and FGF signals is not well understood. Our results suggest that WT1-dependent transcriptional control of Gas1 might promote FGF-induced intracellular signals to activation of PI3K-AKT. Interestingly miR-17-92, whose loss of function results in a late NPC proliferation defect as well (Marrone et al., 2014), has also been shown to modulate the PI3K-AKT pathway (Olive et al., 2009), underlining the importance of this signaling cascade in NPCs. Taken together, we expand the findings from WT1 ChIP-on-chip and ChIP-seq studies in NPCs (Hartwig et al., 2010; Motamedi et al., 2014) to show that WT1 can affect signaling in NPCs by transcriptional regulation on multiple levels, including ligands, receptors and RTK-interacting cofactors such as GAS1.
GAS1 has previously been shown to complex with GFRA1, GDNF and RET to modulate PI3K-AKT RTK signaling in the GDNF-RET pathway (Cabrera et al., 2006). Importantly, direct GDNF-GAS1 interactions were not required for modulation of intracellular signals, suggesting a GAS1-dependent mechanism downstream of ligand binding. Accordingly, our results that implicate a role for GAS1 in the FGF signaling pathway suggest that GAS1 modulates signaling downstream of FGF ligands, as well. Altered FGF expression in response to GAS1 loss of function has been previously described in the developing limb (Liu et al., 2002). Given the widespread expression of GAS1 in various anlagen (Lee and Fan, 2001) and the critical role of FGF signaling in a large number of developmental processes, GAS1 might play an important regulatory role in FGF signaling in other developing organs.
In summary, we identify a novel mechanism by which WT1 regulates the progenitor population by transcriptional activation of Gas1. GAS1 adjusts the gain on RTK signaling in NPCs to regulate NPC proliferation through the PI3K-AKT pathway in a manner that results in defects late in kidney development. Eventually, these mechanisms governed by GAS1 in embryonic kidneys play an important role in determining nephron mass and are therefore relevant to the risk of various diseases later in life.
MATERIALS AND METHODS
LB-22 cells
LB-22 cells were derived from metanephric organ cultures (see below) of E12.5 embryos obtained from ‘immortomice’ (Charles River Laboratories) (Jat et al., 1991). The nephrogenic zone was dissected from the organoid after cultivation for 48 h and triturated into a single-cell suspension. Clones from individual cells were expanded in DMEM-F12/50:50 medium with L-glutamine (Cellgro) supplemented with 10% fetal bovine serum (FBS, Hyclone), 25 ng/ml prostaglandin E1 (Calbiochem), 2 ng/ml heparan-stabilized basic FGF, 100 nM hydrocortisone, 2 nM triiodothyronine, 5 µg/ml insulin (all from Sigma) and 5 µg/ml transferrin (Roche), and subsequently screened for WT1 expression by western blotting. The WT1-positive clone LB-22 was maintained at 33°C in the presence of 10 U/ml interferon γ (R&D) and transferred to interferon-free medium at 37°C upon transfection.
RNAi experiments
Stealth siRNA duplexes (Invitrogen) were used to knock down Wt1 and Gas1 in LB-22 cells (for oligo sequences see supplementary material Table S1). Cells were transfected with 25 pmol gene-specific or control siRNA in 25 µl Lipofectamine 2000 (Invitrogen) per 5×105 cells. After transfection, cells were maintained at 70-80% confluency. RNA and protein were harvested at 48 and 72 h after transfection, respectively.
Stimulation of LB-22 cells with FGFs was carried out at 70% confluency after 6 h of serum starvation. FGF8, FGF9 or FGF20 (all from Peprotech) were added at concentrations as indicated together with 5 µg/ml heparin (Sigma).
Western blotting
Whole-cell lysates from LB-22 cells and kidney organ cultures were prepared either in high-salt RIPA (500 mM NaCl, 50 mM Tris-HCl pH 7.4, 5 mM EDTA, 1 mM EGTA, 0.1% SDS, 1% Igepal, 0.5% sodium deoxycholate, 1× Roche protease inhibitor mix) for experiments involving WT1 or in RIPA (same as above, except 300 mM NaCl, 0.4 mM Na3VO4, 4 mM NaF) for all other experiments (all reagents from Sigma). Equal amounts of protein lysates were resolved on 10% polyacrylamide gels and transferred to PVDF membranes. Standard western blotting was performed with antibodies against WT1 (C19, Santa Cruz sc-192, rabbit polyclonal, 1:500), β-tubulin (Santa Cruz sc-9104, rabbit polyclonal, 1:1000), GAS1 (R&D AF2644, goat polyclonal, 1:500), SIX2 (Proteintech 11562-1-AP, rabbit polyclonal, 1:500), phospho-AKT, AKT, phospho-ERK and ERK (4695S, 4370, 4060, 4691, all from Cell Signaling, rabbit monoclonals, 1:1000).
RNA isolation, RT-qPCR and microarrays
RNA was isolated from LB-22 cells and embryonic kidneys by means of the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RNA was quantified on a Nanodrop-2000 instrument and equal amounts per sample were reverse transcribed into cDNA with the SuperScript III RT Kit (Invitrogen) with oligo-dT primers. Real-time quantitative PCR was carried out in triplicate using the QuantiTect SYBR Green HotStart Kit (Qiagen) on a Cepheid SmartCycler. Primer sequences are provided in supplementary material Table S1. Data analysis was carried out as described previously (Pfaffl, 2001).
For microarray studies, RNA samples were hybridized to Mouse 430.2 expression microarrays (Affymetrix) at the Boston Children's Hospital Microarray Core Facility after processing according to the manufacturer's instructions. Microarray data have been deposited at Gene Expression Omnibus (GEO) under accession number GSE66356.
Chromatin immunoprecipitation
Chromatin immunoprecipitation on P0 kidneys was carried out as described previously (Hartwig et al., 2010). LB-22 cells were crosslinked with 1% formaldehyde in PBS for 5 min, washed in PBS and sonicated in RIPA buffer (300 mM NaCl, 1 mM EDTA, 10 mM Tris-HCl pH 7.4, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 0.1% sodium deoxycholate, 0.1 M DTT, 0.25% N-lauroyl-sarcosine) using a Misonix S-4000 sonicator (all reagents from Sigma). ChIP was carried out using a WT1 antibody (C19, Santa Cruz sc-192) and normal rabbit serum (both from Santa Cruz) as control. Real-time qPCR of ChIP and input DNA was carried out as described above. For primers see supplementary material Table S1.
Dual luciferase reporter assays and mutagenesis
Fragments of the Gas1 promoter were amplified from mouse genomic DNA and directionally cloned into the Gateway-compatible pENTR-dTOPO entry vectors (Invitrogen). The pGL4.14 and pGL4.26 firefly luciferase vectors (Promega) were rendered Gateway-compatible destination vectors by means of the Gateway Vector Conversion System (Invitrogen). Entry and destination vectors were then recombined using the LR Clonase II Kit (Invitrogen) to yield functional reporter plasmids. The pGL4.74 Renilla luciferase vector (Promega) was used as the transfection control.
LB-22 cells were transfected for 6 h with 1 µg reporter plasmid and 0.1 µg Renilla plasmid in Lipofectamine 2000 (Invitrogen) per well in 24-well plates. Luciferase signals were assayed 48 h after transfection using DualGlo chemistry (Promega) on a FLUOstar luminometer (Omega). For reporter assays in WT1 knockdown cells, LB-22 cells were first transfected with siRNAs as described above and re-transfected with luciferase plasmids at 24 h, and luciferase was assayed at 72 h.
For mutagenesis studies, the QuikChange2 Site Directed Mutagenesis Kit (Stratagene) was used according to the manufacturer's instructions. Primers used for mutagenesis can be found in supplementary material Table S1.
Mice and metanephric organ cultures
WT1 knockdown mice were established by cloning a WT1-specific shRNA (see supplementary material Table S1 for sequence) into the pBS/U6-loxPNeo vector (Shukla et al., 2007). Plasmids were microinjected into the pronuclei of single-cell mouse embryos, which were transferred into pseudopregnant foster mothers. Germline transmission was established and F1 offspring were genotyped for the presence of the transgene.
Six2:Cre mice and Gas1 knockout mice have been described previously (Kobayashi et al., 2008; Martinelli and Fan, 2007). Genotyping primers and conditions are given in supplementary material Table S1. Embryos for metanephric organ cultures were harvested from pregnant CD-1 wild-type mice at stage E12.5.
Metanephric organ cultures and MO knockdown of WT1 were performed as described previously (Hartwig et al., 2010). For Gas1 MO knockdown in organ culture, kidneys of E12.5 CD-1 wild-type embryos were treated with MOs (Vivo-Morpholino, GeneTools) for 18 h or 24 h, with a morphant kidney being exposed to a mixture of two Gas1-blocking oligonucleotides at concentrations as indicated in the figure (supplementary material Fig. S4), while the opposite kidney of the same embryo was treated with either control MO only or a mixture of sub-efficient Gas1 MO1 and control MO at concentrations as indicated. All animal studies were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee at Boston Children's Hospital.
Immunofluorescence, immunohistochemistry, in situ hybridization and phenotypic quantification
Immunofluorescence was carried out using antibodies against WT1 (C19, Santa Cruz sc-192, 1:250), GAS1 (R&D AF2644, 1:250), SIX2 (Proteintech 11562-1-AP, 1:200), E-cadherin (BD Biosciences 610404, mouse monoclonal, 1:200), NCAM (Sigma C6680, mouse monoclonal, 1:200), JAG1 (Santa Cruz sc-6011, goat polyclonal, 1:200) and pan-cytokeratin (Sigma C5992, mouse monoclonal, 1:150). Whole-mount images of organ cultures were edited using the background subtraction function in NIKON NIS Elements software to remove autofluorescence of the polyethylene membrane. The Apoptag Fluorescein Direct In Situ Detection Kit (Millipore) was used for TUNEL staining as per the manufacturer's instructions. BrdU labeling of proliferating cells was achieved by injecting mice with 300 µg of BrdU (Roche) per gram of body weight at 1 h before sacrifice. BrdU was detected with the BrdU Labeling and Detection Kit (Roche) according to the manufacturer's instructions.
For immunohistochemistry, paraffin-embedded formalin-fixed tissue was de-waxed in xylenes and rehydrated. Epitope retrieval was carried out by boiling in TE (10 mM Tris pH 9, 1 mM EDTA) for 20 min. Slides were blocked in 3% H2O2, 5% (w/v) BSA and using the Avidin/Biotin Blocking Kit (Vector Labs) as per the manufacturer's instructions. The phospho-Akt antibody (1:100, Cell Signaling) was applied at 4°C over night, slides were washed and the signal was detected using a biotinylated anti-rabbit secondary antibody (1:400, Jackson) with the Vectastain ABC Kit (Vector Labs) and di-aminobenzidine, as per the manufacturer's instructions. Slides were counterstained in Hematoxylin, dehydrated and mounted with histomount.
Whole-mount and section in situ hybridization was carried out following the Genito-Urinary Molecular Anatomy Project protocols accessible at http://www.gudmap.org/Research/Protocols/McMahon.html. Probes were Six2, Wt1, Gas1 (a gift of Andrew P. McMahon, Keck School of Medicine of USC, Los Angeles, CA, USA), Wnt4, Wnt9b, Etv5, Crlf1 and Wnt11.
For phenotypic characterization, at least four kidneys per stage and genotype were evaluated. SIX2+ nuclei were counted on immunofluorescent sections and normalized to the product of section thickness and the length of the capsular circumference present on the section. For evaluation of TUNEL signals, the area between capsule and corticomedullary border was computed, and TUNEL+ nuclei within this area were counted and normalized to cortical area. The proliferative index was calculated by dividing the numbers of SIX2+/BrdU+ nuclei by numbers of all SIX2+ nuclei. Only nuclei located on the capsular face of adjacent ureteric tips were evaluated. Nephrogenesis was assessed by counting NCAM+ comma- and S-shaped bodies and normalizing this number to the product of capsular circumference and section thickness.
Statistics
All statistical analyses were carried out using GraphPad Prism 5. One-way ANOVA with Tukey's post hoc test was used to analyze ChIP-qPCR and luciferase reporter assay data. Significance was assumed at P<0.05 in Tukey's test. Two-way ANOVA with Bonferroni's post hoc test was used to analyze luciferase reporter assays in RNAi experiments, western blot densitometry results for AKT and ERK, and RT-qPCR experiments with significance requiring P<0.05 in Bonferroni's test. Student's t-test was used to test for differences between groups in BrdU incorporation assays, TUNEL assays, densitometry of Gas1 knockdown and phenotypic quantifications.
Microarray results were normalized to the trimmed mean (top and bottom 2% of values excluded for mean calculation) in each sample. Perseus framework software was used to compute q-values, and for principal component analysis (Cox and Mann, 2012).
Screening for WT1 motifs in the Gas1 promoter was done by adding log-odds scores for each base in the WT1 positional weight matrix (Hartwig et al., 2010) using the TRED algorithm. (Zhao et al., 2005).
Supplementary Material
Acknowledgements
We thank Martyna Bruetting for excellent technical assistance, Martin Hoehne for advice in image processing, Andrew P. McMahon for providing reagents, Shan Qin for valuable discussions and the Harvard Rodent Histopathology Core Facility for histology services.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
M.K. and J.A.K. developed the concept of the study. M.K., E.B., M.O.L., L.L., B.T., V.A.S., M.E.T., L.B. and C.-M.F. performed experiments. M.K., E.B., S.H. and M.M.R. analyzed data. M.K. and J.A.K. wrote the manuscript with input from S.H., C.-M.F., T.B., B.S. and all other co-authors.
Funding
M.K. received scholarships from the German Research Foundation [KA3217/2-1]; the German Hypertension Society; and CECAD Cologne. M.O.L. was a recipient of a KoelnFortune scholarship. M.M.R. was supported by a Fritz-Scheler scholarship of the KfH Foundation for Preventive Medicine; and by a UoC postdoctoral grant in the framework of the German Research Foundation Excellence Initiative. T.B. is supported by the German Research Foundation [BE2212 and SFB329]. J.A.K. received funding from the National Institutes of Health/the National Institute of Diabetes and Digestive and Kidney Diseases [NIDDK R01 DK087794-A1]. E.B. was supported by a fellowship grant from the National Kidney Foundation. B.S. was supported by the German Research Foundation [SCHE 1562/2-1]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.119735/-/DC1
References
- Ahn S.-Y., Kim Y., Kim S. T., Swat W. and Miner J. H. (2013). Scaffolding proteins DLG1 and CASK cooperate to maintain the nephron progenitor population during kidney development. J. Am. Soc. Nephrol. 24, 1127-1138 10.1681/ASN.2012111074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akpa M. M., Iglesias D. M., Chu L. L., Cybulsky M., Bravi C. and Goodyer P. (2014). Wilms tumour suppressor, WT1, suppresses epigenetic silencing of the beta-catenin gene. J. Biol. Chem. 290, 2279-2288 10.1074/jbc.M114.573576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen B. L., Tenzen T. and McMahon A. P. (2007). The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 21, 1244-1257 10.1101/gad.1543607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak H., Huh S.-H., Chen S., Jeanpierre C., Martinovic J., Parisot M., Bole-Feysot C., Nitschke P., Salomon R., Antignac C. et al. (2012). FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev. Cell 22, 1191-1207 10.1016/j.devcel.2012.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U., Brown A., Adams D. C., Karolak M. J. and Oxburgh L. (2009). BMP7 promotes proliferation of nephron progenitor cells via a JNK-dependent mechanism. Development 136, 3557-3566 10.1242/dev.036335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. C., Adams D., de Caestecker M., Yang X., Friesel R. and Oxburgh L. (2011). FGF/EGF signaling regulates the renewal of early nephron progenitors during embryonic development. Development 138, 5099-5112 10.1242/dev.065995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. C., Muthukrishnan S. D., Guay J. A., Adams D. C., Schafer D. A., Fetting J. L. and Oxburgh L. (2013). Role for compartmentalization in nephron progenitor differentiation. Proc. Natl. Acad. Sci. USA 110, 4640-4645 10.1073/pnas.1213971110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera J. R., Sanchez-Pulido L., Rojas A. M., Valencia A., Manes S., Naranjo J. R. and Mellstrom B. (2006). Gas1 is related to the glial cell-derived neurotrophic factor family receptors alpha and regulates Ret signaling. J. Biol. Chem. 281, 14330-14339 10.1074/jbc.M509572200 [DOI] [PubMed] [Google Scholar]
- Cain J. E., Islam E., Haxho F., Chen L., Bridgewater D., Nieuwenhuis E., Hui C.-C. and Rosenblum N. D. (2009). GLI3 repressor controls nephron number via regulation of Wnt11 and Ret in ureteric tip cells. PLoS ONE 4, e7313 10.1371/journal.pone.0007313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B., Hill K. J., Charalambous M., Wagner K. J., Lahiri D., James D. I., Andersen J. S., Schumacher V., Royer-Pokora B., Mann M. et al. (2004). BASP1 is a transcriptional cosuppressor for the Wilms’ tumor suppressor protein WT1. Mol. Cell. Biol. 24, 537-549 10.1128/MCB.24.2.537-549.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll T. J., Park J.-S., Hayashi S., Majumdar A. and McMahon A. P. (2005). Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev. Cell 9, 283-292 10.1016/j.devcel.2005.05.016 [DOI] [PubMed] [Google Scholar]
- Costantini F. and Kopan R. (2010). Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev. Cell 18, 698-712 10.1016/j.devcel.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillard M. and Trudel M. (2009). C-myc as a modulator of renal stem/progenitor cell population. Dev. Dyn. 238, 405-414 10.1002/dvdy.21841 [DOI] [PubMed] [Google Scholar]
- Coumoul X., Li W., Wang R.-H. and Deng C. (2004). Inducible suppression of Fgfr2 and Survivin in ES cells using a combination of the RNA interference (RNAi) and the Cre-LoxP system. Nucleic Acids Res. 32, e85 10.1093/nar/gnh083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. and Mann M. (2012). 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13Suppl. 16, S12 10.1186/1471-2105-13-S16-S12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Sal G., Ruaro M. E., Philipson L. and Schneider C. (1992). The growth arrest-specific gene, gas1, is involved in growth suppression. Cell 70, 595-607 10.1016/0092-8674(92)90429-G [DOI] [PubMed] [Google Scholar]
- Denner D. R. and Rauchman M. (2013). Mi-2/NuRD is required in renal progenitor cells during embryonic kidney development. Dev. Biol. 375, 105-116 10.1016/j.ydbio.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanni V., Alday A., Chi L., Mishina Y. and Rosenblum N. D. (2011). Alk3 controls nephron number and androgen production via lineage-specific effects in intermediate mesoderm. Development 138, 2717-2727 10.1242/dev.059030 [DOI] [PubMed] [Google Scholar]
- Dressler G. R. (2009). Advances in early kidney specification, development and patterning. Development 136, 3863-3874 10.1242/dev.034876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley A. T., Lyons K. M. and Robertson E. J. (1995). A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev. 9, 2795-2807 10.1101/gad.9.22.2795 [DOI] [PubMed] [Google Scholar]
- Essafi A., Webb A., Berry R. L., Slight J., Burn S. F., Spraggon L., Velecela V., Martinez-Estrada O. M., Wiltshire J. H., Roberts S. G. E. et al. (2011). A wt1-controlled chromatin switching mechanism underpins tissue-specific wnt4 activation and repression. Dev. Cell 21, 559-574 10.1016/j.devcel.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshammer U., Cebrian C., Ilagan R., Meyers E., Herzlinger D. and Martin G. R. (2005). FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development 132, 3847-3857 10.1242/dev.01944 [DOI] [PubMed] [Google Scholar]
- Hartwig S., Ho J., Pandey P., MacIsaac K., Taglienti M., Xiang M., Alterovitz G., Ramoni M., Fraenkel E. and Kreidberg J. A. (2010). Genomic characterization of Wilms’ tumor suppressor 1 targets in nephron progenitor cells during kidney development. Development 137, 1189-1203 10.1242/dev.045732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzi L., Levesque M., Morin S., Laniel D., Wilkes B. C., Mille F., Krauss R. S., McMahon A. P., Allen B. L. and Charron F. (2011). Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Dev. Cell 20, 788-801 10.1016/j.devcel.2011.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jat P. S., Noble M. D., Ataliotis P., Tanaka Y., Yannoutsos N., Larsen L. and Kioussis D. (1991). Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proc. Natl. Acad. Sci. USA 88, 5096-5100 10.1073/pnas.88.12.5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner C. M., Das A., Ma Z., Self M., Chen C., Lum L., Oliver G. and Carroll T. J. (2011). Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development 138, 1247-1257 10.1242/dev.057646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S., Yoon S. K., Bollig F., Kitagaki J., Hur W., Whye N. J., Wu Y. P., Rivera M. N., Park J. Y., Kim H. S. et al. (2010). A novel Wilms tumor 1 (WT1) target gene negatively regulates the WNT signaling pathway. J. Biol. Chem. 285, 14585-14593 10.1074/jbc.M109.094334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A., Valerius M. T., Mugford J. W., Carroll T. J., Self M., Oliver G. and McMahon A. P. (2008). Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3, 169-181 10.1016/j.stem.2008.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Chen S. and Little M. (2014). Nephron progenitor cells: shifting the balance of self-renewal and differentiation. Curr. Top. Dev. Biol. 107, 293-331 10.1016/B978-0-12-416022-4.00011-1 [DOI] [PubMed] [Google Scholar]
- Kreidberg J. A., Sariola H., Loring J. M., Maeda M., Pelletier J., Housman D. and Jaenisch R. (1993). WT-1 is required for early kidney development. Cell 74, 679-691 10.1016/0092-8674(93)90515-R [DOI] [PubMed] [Google Scholar]
- Lee C. S. and Fan C.-M. (2001). Embryonic expression patterns of the mouse and chick Gas1 genes. Mech. Dev. 101, 293-297 10.1016/S0925-4773(01)00283-0 [DOI] [PubMed] [Google Scholar]
- Liu Y., May N. R. and Fan C.-M. (2001). Growth arrest specific gene 1 is a positive growth regulator for the cerebellum. Dev. Biol. 236, 30-45 10.1006/dbio.2000.0146 [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu C., Yamada Y. and Fan C. M. (2002). Growth arrest specific gene 1 acts as a region-specific mediator of the Fgf10/Fgf8 regulatory loop in the limb. Development 129, 5289-5300. [DOI] [PubMed] [Google Scholar]
- Luyckx V. A. and Brenner B. M. (2010). The clinical importance of nephron mass. J. Am. Soc. Nephrol. 21, 898-910 10.1681/ASN.2009121248 [DOI] [PubMed] [Google Scholar]
- Luyckx V. A., Bertram J. F., Brenner B. M., Fall C., Hoy W. E., Ozanne S. E. and Vikse B. E. (2013). Effect of fetal and child health on kidney development and long-term risk of hypertension and kidney disease. Lancet 382, 273-283 10.1016/S0140-6736(13)60311-6 [DOI] [PubMed] [Google Scholar]
- Majumdar A., Vainio S., Kispert A., McMahon J. and McMahon A. P. (2003). Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development 130, 3175-3185 10.1242/dev.00520 [DOI] [PubMed] [Google Scholar]
- Marrone A. K., Stolz D. B., Bastacky S. I., Kostka D., Bodnar A. J. and Ho J. (2014). MicroRNA-17∼92 is required for nephrogenesis and renal function. J. Am. Soc. Nephrol. 25, 1440-1452 10.1681/ASN.2013040390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli D. C. and Fan C.-M. (2007). Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev. 21, 1231-1243 10.1101/gad.1546307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Estrada O. M., Lettice L. A., Essafi A., Guadix J. A., Slight J., Velecela V., Hall E., Reichmann J., Devenney P. S., Hohenstein P. et al. (2010). Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat. Genet. 42, 89-93 10.1038/ng.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellstrom B., Cena V., Lamas M., Perales C., Gonzalez C. and Naranjo J. R. (2002). Gas1 is induced during and participates in excitotoxic neuronal death. Mol. Cell. Neurosci. 19, 417-429 10.1006/mcne.2001.1092 [DOI] [PubMed] [Google Scholar]
- Motamedi F. J., Badro D. A., Clarkson M., Rita Lecca M., Bradford S. T., Buske F. A., Saar K., Hubner N., Brandli A. W. and Schedl A. (2014). WT1 controls antagonistic FGF and BMP-pSMAD pathways in early renal progenitors. Nat. Commun. 5, 4444 10.1038/ncomms5444 [DOI] [PubMed] [Google Scholar]
- Olive V., Bennett M. J., Walker J. C., Ma C., Jiang I., Cordon-Cardo C., Li Q.-J., Lowe S. W., Hannon G. J. and He L. (2009). miR-19 is a key oncogenic component of mir-17–92. Genes Dev. 23, 2839-2849 10.1101/gad.1861409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-S., Ma W., O'Brien L. L., Chung E., Guo J.-J., Cheng J.-G., Valerius M. T., McMahon J. A., Wong W. H. and McMahon A. P. (2012). Six2 and Wnt regulate self-renewal and commitment of nephron progenitors through shared gene regulatory networks. Dev. Cell 23, 637-651 10.1016/j.devcel.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perantoni A. O., Timofeeva O., Naillat F., Richman C., Pajni-Underwood S., Wilson C., Vainio S., Dove L. F. and Lewandoski M. (2005). Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development 132, 3859-3871 10.1242/dev.01945 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short K. M., Combes A. N., Lefevre J., Ju A. L., Georgas K. M., Lamberton T., Cairncross O., Rumballe B. A., McMahon A. P., Hamilton N. A. et al. (2014). Global quantification of tissue dynamics in the developing mouse kidney. Dev. Cell 29, 188-202 10.1016/j.devcel.2014.02.017 [DOI] [PubMed] [Google Scholar]
- Shukla V., Coumoul X. and Deng C.-X. (2007). RNAi-based conditional gene knockdown in mice using a U6 promoter driven vector. Int. J. Biol. Sci. 3, 91-99 10.7150/ijbs.3.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lee S. B., Palmer R., Ellisen L. W. and Haber D. A. (2001). A functional interaction with CBP contributes to transcriptional activation by the Wilms tumor suppressor WT1. J. Biol. Chem. 276, 16810-16816 10.1074/jbc.M009687200 [DOI] [PubMed] [Google Scholar]
- Zhao F., Xuan Z., Liu L. and Zhang M. Q. (2005). TRED: a transcriptional regulatory element database and a platform for in silico gene regulation studies. Nucleic Acids Res. 33Suppl. 1, D103-D107 10.1093/nar/gki004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.