Abstract
The molecular etiology of pseudoxanthoma elasticum (PXE), an autosomal recessive connective tissue disorder, has become increasingly complex as not only mutations in ATP-binding cassette family C member 6 (ABCC6) but also ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) and gamma-glutamyl carboxylase (GGCX) can cause resembling phenotypes. Identification of modifier genes, such as vascular endothelial growth factor A, has further contributed to the molecular heterogeneity of PXE. In such heterogeneous diseases, next-generation sequencing (NGS) allows to perform mutation screening of several genes in a single reaction. We explored whole-exome sequencing (WES) as an efficient diagnostic tool to identify the causal mutations in ABCC6, GGCX, ENPP1, and vitamin K epoxide reductase complex, subunit 1 (VKORC1) in 16 PXE patients. WES identified a causal ABCC6 mutation in 30 out of 32 alleles and one GGCX mutation, whereas no causal mutations in ENPP1 or VKORC1 were detected. Exomes with insufficient reads (⩽20 depth) for the four genes and patients with single mutations were further evaluated by Sanger sequencing (SS), but no additional mutations were found. The potential of WES compared with targeted NGS is the ease to examine target genes and the opportunity to search for novel genes when targeted analysis is negative. Together with low cost, rapid and less laborious workflow, we conclude that WES complemented with SS can provide a tiered approach to molecular diagnostics of PXE.
Introduction
Pseudoxanthoma elasticum (PXE; OMIM#264800) is an autosomal recessive connective tissue disorder, characterized by ectopic mineralization of elastic fibers in the extracellular matrix of the skin, eyes, and the cardiovascular system. The prevalence of PXE is estimated to be around 1 in 50,000 with a carrier frequency of ∼1:150–300 (Uitto et al., 2011). PXE is caused by mutations in the ABCC6 gene, which encodes a putative efflux transporter, ATP-binding cassette family C member 6 (ABCC6), primarily expressed in liver and kidney, and mediates ATP release from the liver (Jansen et al., 2014). Over 1000 mutant alleles have been identified in patients with PXE from varied ancestral and ethnic backgrounds (Chassaing et al., 2005; Miksch et al., 2005; Pfendner et al., 2007). Besides the presence of two pseudogenes (ABCC6-ψ1 and ABCC6-ψ2) containing sequences highly homologous to the 5′-end of ABCC6, the molecular diagnosis of PXE is becoming increasingly challenging as the molecular etiology of the disease becomes more complex (Pulkkinen et al., 2001). Indeed, PXE-related disorders such as PXE-like disease with coagulation deficiency, linked to mutations in the gamma-glutamyl carboxylase (GGCX; Vanakker et al., 2007) as well as overlapping phenotypes due to co-segregating ABCC6 and GGCX mutations (digenic inheritance) have been described (Li et al., 2009a) or encountered for vitamin K epoxide reductase complex, subunit 1 (VKORC1) genes (Li et al., 2009b). More recently, mutations in another gene, ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1), associated with the soft tissue mineralization disease generalized arterial calcification of infancy, were shown to be associated with a PXE phenotype, difficult to distinguish clinically from classic ABCC6-related PXE (Nitschke et al., 2012). In case of the acquired forms of PXE, associated with e.g., haemoglobinopathies and which shows similar clinical manifestations, no underlying genetic susceptibility has been identified yet (Baccarani-Contri et al., 2001; Hamlin et al., 2003).
Although fully penetrant, PXE is rarely present at birth and an accurate diagnosis can often only be made several years after birth when ocular complications such as subretinal neovascularization and hemorrhaging suddenly develop. In addition, the considerable intra- and interfamilial heterogeneity in clinical severity as well as the lack of reliable phenotype–genotype correlations pose a diagnostic challenge, particularly in young, mildly affected patients. The variability in clinical severity suggests the involvement of modifier genes; recently, it was suggested that variants in the vascular endothelial growth factor A gene were associated with a more severe PXE retinopathy (Hendig et al., 2013). It can be anticipated that, in the near future, other disease-modifying genes will be described.
The challenge to achieve a clinical diagnosis of PXE especially at younger age, combined with the genetic heterogeneity, stresses the importance of a molecular diagnosis. Sanger sequencing (SS) is the current golden standard for the detection of pathogenic variants (Tucker et al., 2009). However, it is well known that this approach is labor intensive, expensive, and time consuming, especially when large genes need to be analyzed (Vanakker and De Paepe, 2013). In PXE, the reported mutation detection rates obtained by SS are variable, ranging from 66% to 80–90%, probably reflecting the size and molecular variability of the cohorts examined (Pfendner et al., 2007; Hendig et al., 2013). Because larger insertions or deletions can be missed by direct sequencing, multiplex ligation-dependent probe amplification (MLPA) analysis to detect larger deletions/insertions is highly recommended (Costrop et al., 2010). The recent introduction of the next-generation sequencing (NGS) technology allows to perform a more efficient and cost-effective screening, particularly for disorders with several genes involved (Diamandis, 2009). Targeted enrichment of specific genes of interest or whole-exome sequencing (WES), sequencing all 20,805 coding genes in a single workflow, is now becoming available to efficiently diagnose such heterogeneous disorders.
In this study, we evaluated the use of WES as an efficient diagnostic tool in 16 clinically confirmed PXE patients. The advantages and the disadvantages of this approach are discussed.
Results
Clinical findings
The clinical characteristics of the PXE patients in this study are summarized in Table 1. All patients presented the typical skin and eye manifestations. The occurrence of cardiovascular disease was variable, which is a common observation in PXE. Only one patient suffered a gastrointestinal bleeding, whereas none had coagulation problems (abnormalities of the vitamin K–dependent coagulation factors II, VII, IX, and X).
Table 1. Overview of phenotypic characterization of 16 patients.
| Patient | Age (yrs) | Sex | Skin | Eyes | Cardiovascular | GI hemorrhage | Coagulation |
|---|---|---|---|---|---|---|---|
| 1 | 35 | M | 1 | 1 | 1 | − | − |
| 2 | 54 | F | 2 | 2 | 2 | − | − |
| 3 | 33 | M | 1 | 1 | 1 | − | − |
| 4 | 56 | M | 1 | 2 | 1 | − | − |
| 5 | 49 | F | 2 | 2 | 1 | − | − |
| 6 | 52 | M | 2 | 2 | 2 | + | − |
| 7 | 24 | F | 1 | 1 | 2 | − | − |
| 8 | 63 | F | 2 | 2 | 2 | − | − |
| 9 | 40 | F | 2 | 2 | 2 | − | − |
| 10 | 61 | F | 2 | 2 | 1 | − | − |
| 11 | 59 | M | 1 | 2 | 1 | − | − |
| 12 | 35 | F | 1 | 1–2 | 1 | − | − |
| 13 | 28 | F | 1 | 2 | 2 | − | − |
| 14 | 34 | M | 1 | 2 | 2 | − | − |
| 15 | 48 | M | 1 | 1 | 1 | − | − |
| 16 | 43 | F | 1 | 1 | 1 | − | − |
GI, gastrointestinal. Coagulation: deficiency of the vitamin K–dependent coagulation factors (factor II, VII, IX, and X).
WES analysis
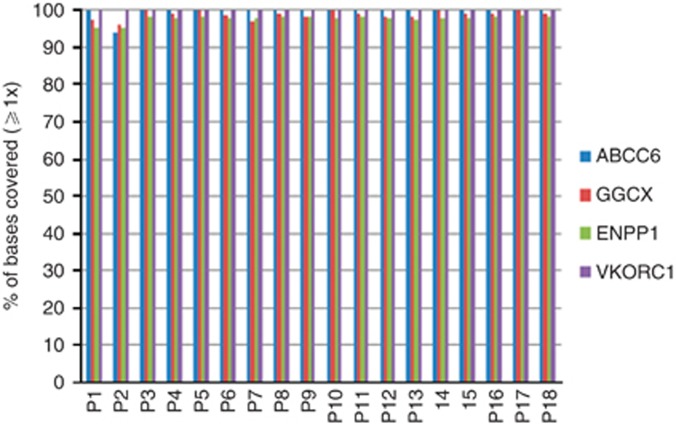
First, the quality of the exome capturing and sequencing in terms of the number of mapped reads, the coverage of the targeted exons, and the amount of detected single-nucleotide variants for all 16 patients were evaluated. On average 90% of the raw reads mapped on the human hg19 reference genome. We obtained a mean sequencing depth of 12 × to 53 × on the targeted exome bases across the samples. Averaging all samples, the mean depth on the targeted bases was 37x. Approximately, 98% of the targeted exome bases were covered across the samples. In relationship to the PXE phenotype, the quality of the coding regions of the ABCC6, GGCX, ENPP1, and VKORC1 genes was investigated in more detail. For all samples the overall sequencing depth was within the expected range (⩾97%,), except for patients 1 and 2 who revealed low quality data (Supplementary Tables S1—S8 online).
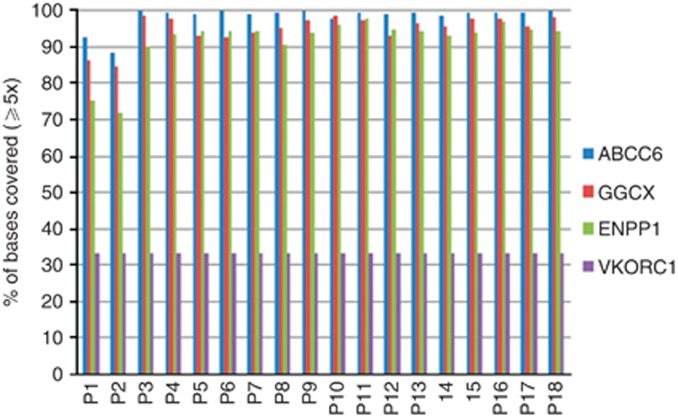
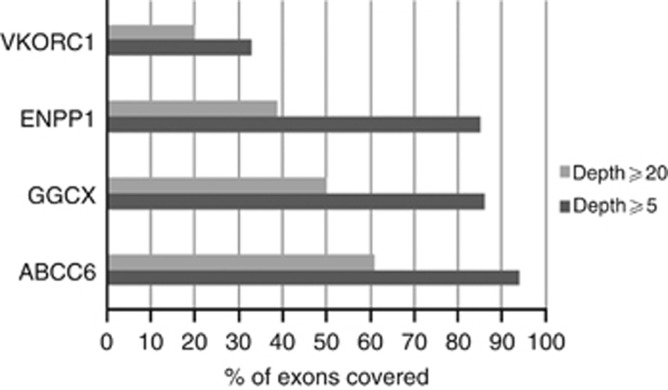
In order to identify the disease-causing mutations in all four genes, we analyzed all the exons that were 100% covered with ⩾1 and ⩾5 reads (Figures 1 and 2) and found that 93, 84, 84, and 33% of the exons for ABCC6, GGCX, ENPP1, and VKORC1, respectively, met this criteria for the overall sequencing depth of ⩾5 reads (Figure 2). Our further strategy was to investigate all exons with a depth lower compared with 20 by SS. Previously, we demonstrated that variants with a coverage <20 might be missed in 0.1% of the cases (Leeneer et al., 2011; Figure 3 and Supplementary Tables S5—S8 online). The latter analysis was performed for patients with only a single mutation (Table 2). For patients 1 and 2, coverage was poor for all four genes, which are in correlation with the poor overall sequencing depth of their exomes.
Figure 1.
Percentage (%) of bases covered of four targeted genes at ⩾1x depth.
Figure 2.
Percentage (%) of bases covered of four targeted genes at ⩾5x depth.
Figure 3.
Percentage (%) of exons covered in four genes using ⩾5 and ⩾20 depth as a filter.
Table 2. List of mutations found by WES and SS.
| Gene | Nucleotide change | Protein change | Patient ID | Hom/Het | WES | SS | Known/PUR | Reference |
|---|---|---|---|---|---|---|---|---|
| ABCC6 | c.C118T | p.(P40S) | P10 | Het | √ | √ | PUR | |
| ABCC6 | c.998+2 | 998+3del TG | P8 | Het | √ | √ | PUR | |
| ABCC6 | c.T1484A | p.(L495H) | P7 | Het | √ | √ | Known | Miksch et al., 2005 |
| ABCC6 | c.G1553A | p.(R518Q) | P11 | Hom | √ | √ | Known | Uitto et al., 2001 |
| ABCC6 | c.G1553A | p.(R518Q) | P12, P13, P14 | Het | √ | √ | Known | Uitto et al., 2001 |
| ABCC6 | c.G2263A | p.(G755R) | P11 | Het | √ | √ | Known | Pfendner et al., 2007 |
| ABCC6 | c.G2294A | p.(R765Q) | P3 | Het | √ | √ | Known | Le Saux et al., 2001 |
| ABCC6 | del2860_2865 | P12, P13,14 | Het | √ | √ | PUR | ||
| ABCC6 | c.T2911C | p.(W971R) | P11 | Het | √ | √ | PUR | |
| ABCC6 | Ex23_24del | P2 | Hom | √ | √ | Known | Ringpfeil et al., 2001 | |
| ABCC6 | c.T3032C | p.(L1011P) | P9 | Hom | √ | √ | PUR | |
| ABCC6 | c.C3190T | p.(A1064T) | P7 | Het | √ | √ | Known | Miksch et al., 2005 |
| ABCC6 | c.G3413A | p.(R1138Q) | P11 | Het | √ | √ | Known | Le Saux O, 2011 |
| ABCC6 | c.C3421T | p.(R1141X) | P4 | Hom | √ | √ | Known | Bergen et al., 2000 |
| ABCC6 | c.C3421T | p.(R1141X) | P52, P8, P162 | Het | √ | √ | Known | Bergen et al., 2000 |
| ABCC6 | c.C3490T | p.(R1164X) | P6, P15 | Hom | √ | √ | Known | Struk et al., 2000 |
| ABCC6 | c.G4198A | p.(E1400K) | P10 | Het | √ | √ | Known | Chassaing et al., 2004 |
| ABCC6 | c.C4216A | p.(Q1406K) | P3 | Het | √ | √ | PUR | |
| GGCX | c.C1321T | p.(R441C) | P7 | Het | √ | √ | PUR |
Het, heterozygous; Hom, homozygous; PUR, previously unreported; SS, Sanger sequencing; WES, whole-exome sequencing.
Patient with three mutations in ABCC6.
Patients with a single mutation in ABCC6.
Variants detected by exome sequencing
Exome sequencing of 16 patients revealed 30 mutant alleles in the ABCC6 gene, which results in an overall mutation detection rate of 93.75% (30/32). We did not take into account the occurrence of a third mutation when determining the mutation detection rate. Of these, five homozygous (P2, 4, 6, 9, 15), nine compound heterozygous (P1, 3, 7, 8, 10, 11, 12, 13, and 14), and two (P5 and 16) heterozygous mutations were identified (Table 2). In one patient (P2) homozygous deletion of exons 23 and 24 in ABCC6 was detected. In total, six mutations–– four amino acid substitutions, a 5 bp exonic deletion, and one intronic deletion–were previously unreported in the ABCC6 gene. The pathological nature of these previously unreported mutations was considered because the variants were not or very rarely present in dbSNP131 or in the 1,000 genomes database. In addition, all these previously unreported mutations were located in highly conserved regions (high Polyphen score) and could also be predicted by SIFT (Sorting Intolerant From Tolerant), Polyphen, and Mutation taster, suggesting their pathogenic nature. One patient (P7) shows a compound heterozygous ABCC6 mutation in combination with a missense mutation in the GGCX gene (Table 2). No mutations were found in the ENPP1 or VKORC1 gene in this cohort. In addition, exome sequencing resulted in 24 distinct exonic single-nucleotide polymorphisms (SNPs), 11 distinct SNPs in the intronic boundary, and one SNP in the 3′ UTR region in the four genes (Supplementary Table S9 online). Most of these SNPs have a minor allele frequency higher than 1% and are therefore not considered as disease causing. Four of these SNPs have a minor allele frequency <1% (Supplementary Table S10 online) and are difficult to exclude from the list of disease-causing variants. However, splice prediction programs did not reveal evidence for the presence of a splice site mutation of these rare SNPs.
Confirmation by SS and MLPA and analyzing the exons with insufficient reads
Mutations identified by WES could all be confirmed by SS. No additional mutation in all exons with a sequence depth coverage <20 for the two patients (P5, P16) with only one ABCC6 mutation was detected by SS (Table 2). In addition, MLPA analysis of these 2 patients for the ABCC6 gene revealed no deletion or insertion. In order to verify the missing allele, which is responsible for the PXE phenotype in the patients with only one mutation, whole-exome data analysis was performed. We used two approaches: first, we looked at whether homozygous or compound heterozygous mutations could be found in genes other than ABCC6, GGCX, ENPP1, and VKORC1. Supplementary Table S11 online summarizes the three homozygous variants, which were considered as possibly damaging and/or deleterious by in silico analysis. However, none of these candidate genes could be confidently linked with the pathomechanism of PXE. Using the same filtering criteria, no compound heterozygous variants could be retained. In a second approach, we looked at whether there could be a digenic inheritance present in P5 and P16. We therefore analyzed the heterozygous variants in the respective exome data sets. Nearly 1,500 variants (including missense, nonsense, frame-shift, in-frame indel, stop codon, splice donor/acceptor) were obtained, which were limited by using filtering criteria based on a population frequency ⩽1% and in silico analysis (variants designated as deleterious or probably damaging). This resulted in 17 missense variants (no other variant types fulfilled the filtering criteria) in 13 candidate genes, three of which overlapped in the two patients (Supplementary Table S12 online). We further looked into the function of these 13 genes and any relation with the pathomechanisms of PXE. No indisputable functional associations could be delineated, particularly for those three shared by both patients. At this point it remains difficult to make definite conclusions on the basis of the available data of these two patients. For both approaches, it will be necessary to evaluate a larger cohort of patients with incomplete genotypes using NGS to further evaluate these potential candidate genes and/or identify novel ones; this is, however, beyond the scope of this paper.
To rule out a whole exon deletion on one allele for the p.(R518Q) mutation in ABCC6, MLPA was also performed for patients P1 (with 3 ABCC6 mutation), which revealed no additional deletion. Unfortunately, no MLPA analysis is available for the three other genes (ENPP1, GGCX, and ENPP1).
Discussion
In this study, we applied WES in combination with SS as an efficient molecular tool for the molecular diagnosis of PXE. Sixteen patients with a clear PXE phenotype were investigated for mutations in the ABCC6, GGCX, ENPP1, and VKORC1 genes, which revealed an overall mutation yield rate of 93.75%. With the applied degree of coverage (⩾5), WES was able to detect all the mutations in the four genes. As mentioned earlier, consideration of sequence depth coverage of ⩾5 as a filter implicates a 5% risk of missing heterozygous mutations. We therefore considered, in a second step, sequence depth coverage of ⩾20 as a filter to achieve a mutation detection rate of ∼100% for those patients with only one mutation and performed SS for those exons; however, no additional mutations were identified in the four genes.
In this cohort, 14 patients with homozygous or compound heterozygous mutations were identified. Two patients present an interesting genotype. Patient P1 carries three known disease-causing mutations in ABCC6, a phenomenon that has been described before by Pfendner et al. (2007), who identified two subjects with three ABCC6 mutations. However, no remarkable phenotypic differences were found in these patients compared with the other PXE patients. In two patients only a single ABCC6 mutation was detected. In autosomal recessive diseases, the heterozygous state is most often indistinguishable from the healthy state. However, a partial PXE phenotype has been reported in some heterozygous carriers (Martin et al., 2008). In some of these, the p.(R1141X) mutation has been correlated with the presence of coronary artery disease (Trip et al., 2002; Sherer et al., 2001). In our cohort, two heterozygous patients, P5 and P16, carry the p.(R1141X) mutation (Table 2). Although these patients present occlusive vessel disease, the presence of ocular and skin manifestations rather suggests that a second mutation has been missed, owing to limitations of the current technology, in the regions of the ABCC6 gene, which were not analyzed––the 3′ and 5′ UTR, deep intronic sequences or promotor region––or in another gene. Further detailed examination of the complete exome of patient P5 and P16 did not reveal obvious mutations in other genes. In patient P7, we observed compound heterozygosity for two ABCC6 missense substitution (p.(L495H) and p.(A1064T)) mutations, as well as a co-inherited missense substitution (p.(R441C)) mutation in the GGCX gene. Vanakker et al. (2011) described a patient with biallelic ABCC6 mutation and a gain-of-function SNP in GGCX who displayed a phenotype partly reminiscent of both PXE and the PXE-like disease with coagulation deficiency (Vanakker et al., 2011). However, we remain cautious at this moment on the role of the co-inherited GGCX mutation, which may be acting as a disease modifier. Indeed, inter- as well as intra-familial clinical variability in PXE suggests that the contribution of factors other than ABCC6 variants also affects the phenotypic features (Bacchelli et al., 1999; Christen-Zach et al., 2006).
We showed that the WES approach in combination with SS is an effective strategy in a polygenic disease that PXE has become. It is well known that WES kits do not capture all genes because of limitations inherent in the processes of exome enrichment, library construction, and amplification. As most exons of the four genes have a read depth of >5, a range by which all mutations were detected, the number of exons that still need to be analyzed by SS is relatively low. Of course, this is gene dependent and might not be applicable for other sets of genes. We estimated that the reagent cost of screening by the method presented here is cheaper compared with the conventional methods. At the same time the analysis is much less time consuming and more efficient, whereas specificity and sensitivity are the same. The advantage of WES compared with targeted enrichment of specific genes of interest by array of PCR technology is that optimization is independent of the genes involved as a standard procedure can be used. The limited coverage compared with a targeted enrichment approach is compensated by SS.
Another important advantage of WES is that it also provides us the opportunity to screen for other PXE-related genes including GGCX, ENPP1, and VKORC1 in a single workflow. In addition, the availability of the complete exome sequence allows us to investigate other genes not yet related to PXE at this moment. Nevertheless, the combination with SS is essential to fill the gaps with insufficient coverage but also to adequately sequence the exons located in the pseudogenic region. The close sequence homology of the two ABCC6 pseudogenes at the 5' site hampers mutation detection, especially when a NGS approach is used, which is based on an alignment of sequences to a reference sequence. Nevertheless, we were able to identify the c.998+2_998+3 delTG in exon 8, indicating that despite the high similarity of the pseudogenic region, gene-specific mutations are identified.
Besides identification of the causal genes, exome sequencing in these patients will provide the opportunity to study the involvement of modifier genes, such as GGCX, to explain the variability in phenotypic expression between patients. The efficacy of such a strategy, particularly when studying extreme phenotypes, has previously been demonstrated in cystic fibrosis (Emond et al., 2012).
In summary, we demonstrate that WES could be a rapid, noninvasive, highly effective and economical method to screen mutations in PXE and PXE-associated phenotypes, as their molecular etiology becomes increasingly more complex, whereas their clinical resemblance often makes it difficult to predict the causal gene(s). Further, WES provides the opportunity to identify additional genes involved in PXE, both disease-causing and disease-modifying. Although the use of WES does currently not provide sufficient coverage of all exons for comprehensive variant calling in a clinical diagnostic setting, combined with SS and MLPA, it can be considered as a reliable, efficient, and cost-effective approach to molecular diagnostics of PXE.
Materials and Methods
Approval and consent
Written, informed consent was obtained from all patients, and the Declaration of Helsinki protocols were followed. This study was approved by the local Ethical Committees (Ghent University Hospital and University Hospital of Angers, France).
Phenotypical characterization
All patients in the present study were diagnosed with PXE based on combination of skin pathology (the characteristic findings of elastic fiber mineralization and fragmentation in the middermis of lesional skin, using Verhoef-van Giesson and Von Kossa staining), and fundus finding. The phenotype of the patients was summarized as follows (Table 1): skin—S0: no macroscopic skin lesions, S1: papular lesions, S2; plaques of papules; eyes—E0: no retinopathy, E1: uncomplicated peau d'orange and/or angioid streaks, E2: subretinal neovascularisation, hemorrhaging and vision loss; cardiovascular—C0: no cardiovascular symptoms, C1: non-occlusive vasculopathy, C2: occlusive vessel disease. Further, the occurrence of gastrointestinal hemorrhaging and coagulation problems were evaluated.
Extraction of genomic DNA
Genomic DNA was extracted from the patient's peripheral whole blood using the High pure PCR Template Preparation Kit (Roche Applied Sciences, Mannheim, Germany) or the genomic DNA purification kit (Quick-gDNA MiniPrep-D3024, Zymo Research, Freiburg, Germany), or the QIAmp DNA Mini Kit on the QIAcube robot (Qiagen, Hilden, Germany) according to standard protocol.
Whole-Exome Sequencing
Exome capture was performed according to the manufacturer's protocol using the SeqCap EZ Human Exome Library v3.0 kit (Roche/Nimblegen, Madison, WI). After ligation of the barcoded Illumina (Eindhoven, The Netherlands) adapters, samples were pooled per 2 before capturing. Consequently, 6 samples were sequenced per Illumina HiSeq 2500 flow cell lane. On average 50E6 2x100 bp paired end reads were sequenced per sample.
The CLC Genomics Workbench v6.5.1 (CLC Bio, Qiagen, Aarhus, Denmark) software was used for read mapping against the human genome reference sequence (NCBI, GRCh37.p5/hg19), duplicate read removal, coverage analysis, and variant calling.
Variants were annotated with an in-house developed software package, which makes use of the Ensembl API and Alamut Batch. To reduce the number of variants, synonymous variants with no predicted splice site disruption, variants with an allelic frequency in the global 1,000G population >1%, and technical variants (i.e., coverage <3 and observation in only one sequence direction) were filtered out. The possible impact of variants on the protein level and on the correct splicing was further assessed by Alamut (http://www.interactive-biosoftware.com/software/alamut/overview), Polyphen 2 (Adzhubei et al., 2010), and SIFTs (Kumar et al., 2009) for conservation, physicochemical changes, and splice site effects.
Initially, data analysis was restricted to variants in ABCC6, GGCX, ENPP1, and VKORC1. In patients where only one mutation was found the whole exome was analyzed.
SS
SS was performed to confirm the variants found with WES in the ABCC6 and GGCX gene in all patients. Primers used for amplifying fragments harboring individual variants were designed by LightScanner primer design software (version: 1.0.R.84; Salt Lake City, UT) and the sequences of these primers are listed in Supplementary Table S13 online. Touch down PCR was used to amplify the genomic fragments followed by SS on an ABI3130XL or ABI3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA).
MLPA
MLPA analysis of the ABCC6 gene was performed to evaluate the presence of larger deletions or insertions by using commercially available SALSA MLPA kit PO92-B3 (MRC-Holland, Amsterdam, The Netherlands) and according to the manufacturer's recommendations (www.mlpa.com). The P092-B3 probemix contains one probe for each exon of the ABCC6 gene, with the exception of exon 1, 3, 6, 16, 19, 20, 29, 31. A total of 100 ng genomic DNA from each patient in total 5 μl reaction volume was used. Fragments generated by MLPA were detected by using an ABI3130XL or ABI3730XL capillary electrophoresis system (Applied Biosystems). Size, concentration, and quantification of fragments were analyzed using Coffalyser (MRC-Holland).
Acknowledgments
We thank the Biobank of Angers University Hospital France (CRB-CHU Angers France) for DNA extraction and blood cell freezing, as well as the PXE France patient support group for funding. We acknowledge all the patients for taking part in this study and Elke D'Haese and Inge Vereecke for technical support with SS and MLPA. This study was supported by a BOF research fellowship from Ghent University to OMV, a research grant from the Research Foundation—Flanders (Belgium; G.0241.11N) to ADP and OMV, and a Methusalem grant (BOF08/01M01108) to ADP. OMV is a Senior Clinical Investigator of the Research Foundation—Flanders (Belgium).
Glossary
- ABCC6
ATP-binding cassette family C member 6
- ENPP1
ectonucleotide pyrophosphatase/phosphodiesterase 1
- GGCX
gamma-glutamyl carboxylase
- MLPA
multiplex ligation-dependent probe amplification
- NGS
next-generation sequencing
- PXE
pseudoxanthoma elasticum
- SNP
single-nucleotide polymorphism
- SS
Sanger sequencing
- VKORC1
vitamin K epoxide reductase complex, subunit 1
- WES
whole-exome sequencing
The authors state no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/jid
Supplementary Material
References
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarani-Contri M, Bacchelli B, Boraldi F, et al. Characterization of pseudoxanthoma elasticum-like lesions in the skin of patients with beta-thalassemia. J Am Acad Dermatol. 2001;44:33–39. doi: 10.1067/mjd.2001.110045. [DOI] [PubMed] [Google Scholar]
- Bacchelli B, Quaglino D, Gheduzzi D, et al. Identification of heterozygote carriers in families with a recessive form of pseudoxanthoma elasticum. Mol Pathol. 1999;12:1112–1123. [PubMed] [Google Scholar]
- Bergen AA, Plomp AS, Schuurman EJ, et al. Mutations in ABCC6 cause pseudoxanthoma elasticum. Nat Genet. 2000;25:228–231. doi: 10.1038/76109. [DOI] [PubMed] [Google Scholar]
- Chassaing N, Martin L, Mazereeuw J, et al. Novel ABCC6 mutations in pseudoxanthoma elasticum. J Invest Dermatol. 2004;122:608–613. doi: 10.1111/j.0022-202X.2004.22312.x. [DOI] [PubMed] [Google Scholar]
- Chassaing N, Martin L, Calvas P, et al. Pseudoxanthoma elasticum: a clinical, pathophysiological and genetic update including 11 novel ABCC6 mutations. J Med Genet. 2005;42:881–892. doi: 10.1136/jmg.2004.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen-Zach S, Huber M, Struk B, et al. Pseudoxanthoma elasticum: evaluation of diagnostic criteria based on molecular data. Br J Dermatol. 2006;155:89–93. doi: 10.1111/j.1365-2133.2006.07278.x. [DOI] [PubMed] [Google Scholar]
- Costrop LM, Vanakker O, van Laer L, et al. Novel deletions causing pseudoxanthoma elasticum underscore the genomic instability of the ABCC6 region. J Hum Genet. 2010;55:112–117. doi: 10.1038/jhg.2009.132. [DOI] [PubMed] [Google Scholar]
- Diamandis EP. Next-generation sequencing: a new revolution in molecular diagnostics. Clin Chem. 2009;55:2088–2092. doi: 10.1373/clinchem.2009.133389. [DOI] [PubMed] [Google Scholar]
- Emond MJ, Louie T, Emerson J, et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet. 2012;44:886–889. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin N, Beck K, Bacchelli B, et al. Acquired Pseudoxanthoma elasticum-like syndrome in beta-thalassaemia patients. Br J Haematol. 2003;122:852–854. doi: 10.1046/j.1365-2141.2003.04484.x. [DOI] [PubMed] [Google Scholar]
- Hendig D, Knabbe C, Gotting C. New insights into the pathogenesis of pseudoxanthoma elasticum and related soft tissue calcification disorders by identifying genetic interactions and modifiers. Front Genet. 2013;4:114. doi: 10.3389/fgene.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RS, Duijst S, Mahakena S, et al. ATP-binding cassette subfamily C member6-mediated ATP secretion by the liver is the main source of the mineralization inhibitor inorganic pyrophosphate in the systemic circulation. Arterioscler Thromb Vasc Biol. 2014;34:1985–1989. doi: 10.1161/ATVBAHA.114.304017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;47:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Le Saux O, Beck K, Sachsinger C, et al. A spectrum of ABCC6 mutations is responsible for pseudoxanthoma elasticum. Am J Hum Genet. 2001;69:749–764. doi: 10.1086/323704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Saux O, Fulop K, Yamaguchi Y, et al. Expression and in vivo rescue of human ABCC6 disease-causing mutants in mouse liver. PLoS One. 2011;6:e24738. doi: 10.1371/journal.pone.0024738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeneer KD, Schrijver JD, Clement L, et al. Practical tools to implement massive parallel pyrosequencing of PCR products in next generation molecular diagnostics. PLoS ONE. 2011;6:1–7. doi: 10.1371/journal.pone.0025531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Grange DK, Armstrong NL, et al. Mutations in the GGCX and ABCC6 genes in a family with pseudoxanthoma elasticum-like phenotypes. J Invest Dermatol. 2009;129:553–563. doi: 10.1038/jid.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Schurgers LJ, Smith AC, et al. Co-existent pseudoxanthoma elasticum and vitamin K-dependent coagulation factor deficiency: compound heterozygosity for mutations in the GGCX gene. Am J Pathol. 2009;174:534–540. doi: 10.2353/ajpath.2009.080865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L, Bonicel P, Daudon P, et al. Heterozygosity for a single mutation in the ABCC6 gene may closely mimic PXE. Arch Dermatol. 2008;144:301–306. doi: 10.1001/archderm.144.3.301. [DOI] [PubMed] [Google Scholar]
- Miksch S, Lumsden A, Guenther UP, et al. Molecular genetics of pseudoxanthoma elasticum: type and frequency of mutations in ABCC6. Hum Mutat. 2005;26:235–248. doi: 10.1002/humu.20206. [DOI] [PubMed] [Google Scholar]
- Nitschke Y, Baujat G, Botschen U, et al. Generalized arterial calcification of infancy and pseudoxanthoma elasticum can be caused by mutations in either ENPP1 or ABCC6. Am J Hum Genet. 2012;90:25–39. doi: 10.1016/j.ajhg.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfendner EG, Vanakker OM, Terry SF, et al. Mutation detection in the ABCC6 gene and genotype-phenotype analysis in a large international case series affected by pseudoxanthoma elasticum. J Med Genet. 2007;44:621–628. doi: 10.1136/jmg.2007.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen L, Nakano A, Ringpfeil F, et al. Identification of ABCC6 pseudogenes on human chromosome 16p: implications for mutation detection in pseudoxanthoma elasticum. Hum Genet. 2001;109:356–365. doi: 10.1007/s004390100582. [DOI] [PubMed] [Google Scholar]
- Ringpfeil F, Nakano A, Uitto J, et al. Compound heterozygosity for a recurrent 16.5-kb Alu-mediated deletion mutation and single-base-pair substitutions in the ABCC6 gene results in pseudoxanthoma elasticum. Am J Hum Genet. 2001;68:642–652. doi: 10.1086/318807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer DW, Bercovitch L, Lebwohl M. Pseudoxanthoma elasticum: significane of limited phenotype expression in parents of affected offspring. J Am Acad Dermatol. 2001;44:534–537. doi: 10.1067/mjd.2001.112401. [DOI] [PubMed] [Google Scholar]
- Struk B, Cai L, Zach S, et al. Mutations of the gene encoding the transmembrane transpoter protein ABC-C6 cause pseudoxanthoma elasticum. J Mol Med. 2000;78:282–286. doi: 10.1007/s001090000114. [DOI] [PubMed] [Google Scholar]
- Trip MD, Smulders YM, Wegman JJ. Frequent mutation in the ABCC6 (R1141X) is associated with a strong increase in the prevalence of coronary artery disease. Circulation. 2002;106:773–775. doi: 10.1161/01.cir.0000028420.27813.c0. [DOI] [PubMed] [Google Scholar]
- Tucker T, Marra M, Friedman JM. Massively parallel sequencing: the next big thing in genetic medicine. Am J Hum Genet. 2009;85:142–154. doi: 10.1016/j.ajhg.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto J, Bercovitch L, Terry SF, et al. Pseudoxanthoma Elasticum: Progress in diagnostics and research towards treatment summary of the 2010 PXE international research meeting. Am J Med Genet A. 2011;155:1517–1526. doi: 10.1002/ajmg.a.34067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto J, Pulkkinen L, Ringpfeil F. Molecular genetics of pseudoxanthoma elasticum: a metabolic disorder at the environment-genome interface. Trends Mol Med. 2001;7:13–17. doi: 10.1016/s1471-4914(00)01869-4. [DOI] [PubMed] [Google Scholar]
- Vanakker OM, Leroy BP, Schurgers LJ, et al. Atypical presentation of pseudoxanthoma elasticum with abdominal cutis laxa: evidence for a spectrum of ectopic calcification disorders. Am J Med Genet A. 2011;155A:2855–2859. doi: 10.1002/ajmg.a.34264. [DOI] [PubMed] [Google Scholar]
- Vanakker OM, Martin L, Gheduzzi D, et al. Pseudoxanthoma elasticum-like phenotype with cutis laxa and multiple coagulation factor deficiency represents a separate genetic entity. J Invest Dermatol. 2007;127:581–587. doi: 10.1038/sj.jid.5700610. [DOI] [PubMed] [Google Scholar]
- Vanakker OM, De Paepe A. Pharmacogenomics in children: advantages and challenges of next generation sequencing applications. Int J Pediatr. 2013;13:136524. doi: 10.1155/2013/136524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.