Abstract
Induced pluripotent stem cells (iPSCs) are a seminal breakthrough in stem cell research and are promising tools for advanced regenerative therapies in humans and reproductive biotechnology in farm animals. iPSCs are particularly valuable in species in which authentic embryonic stem cell (ESC) lines are yet not available. Here, we describe a nonviral method for the derivation of bovine iPSCs employing Sleeping Beauty (SB) and piggyBac (PB) transposon systems encoding different combinations of reprogramming factors, each separated by self-cleaving peptide sequences and driven by the chimeric CAGGS promoter. One bovine iPSC line (biPS-1) generated by a PB vector containing six reprogramming genes was analyzed in detail, including morphology, alkaline phosphatase expression, and typical hallmarks of pluripotency, such as expression of pluripotency markers and formation of mature teratomas in immunodeficient mice. Moreover, the biPS-1 line allowed a second round of SB transposon-mediated gene transfer. These results are promising for derivation of germ line–competent bovine iPSCs and will facilitate genetic modification of the bovine genome.
Introduction
The recent reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka, 2006; Takahashi et al., 2007) was a seminal breakthrough in stem cell research and developmental biology. iPSCs were first derived from mouse fibroblasts by overexpression of four reprogramming factors—Oct4, Sox2, Klf4, and c-Myc—and subsequently from human fibroblasts by overexpression of OCT4, SOX2, LIN28, and NANOG (Takahashi and Yamanaka, 2006; Yu et al., 2007) after retroviral transfection. Retroviral and lentiviral delivery of core reprogramming factors allowed the derivation of iPSCs from several other mammals, including rat (Liao et al., 2009), dog (Shimada et al., 2010), pig (Esteban et al., 2009), rhesus monkey (Liu et al., 2008), sheep (Bao et al., 2011), goat (Ren et al., 2011), horse (Nagy et al., 2011), cattle (Han et al., 2011), buffalo (Deng et al., 2012), and of iPS-like cells from nonmammalian vertebrates (Rossello et al., 2013). iPSCs derived from somatic tissue are particularly promising in farm animals in which true, germline-competent embryonic stem cells (ESCs) could not yet be derived (Kumar et al., 2015). Previous approaches to derive bovine iPSCs employed retroviral (Han et al., 2011; Sumer et al., 2011), or lentiviral transduction (Cao et al., 2012) or plasmids (Huang et al., 2011; Wang et al. 2013).
However, the commonly employed method of viral gene delivery of the reprogramming factors is associated with considerable risk of insertional mutagenesis and genotoxicity (Wu and Dunbar, 2011). To overcome these risks, alternative methods, such as nonintegrating adenoviral vectors (Stadtfeld et al., 2008), plasmids (Yu et al., 2009), recombinant proteins (Zhou et al., 2009), modified mRNAs (Warren et al., 2010), and small molecules (Shi et al., 2008) were successfully used for iPSC derivation. However, the efficiency of reprogramming using these methods is significantly lower than that of retroviral or lentiviral vectors, and the alternative methods may require repetitive treatments to maintain pluripotency (Kumar et al., 2014).
Domestic cattle are an economically valuable livestock species raised both for meat and milk production (Malaver-Ortega et al., 2012). The availability of bovine pluripotent stem cells could facilitate the application of advanced reproductive technologies in this species. Numerous attempts to establish bovine ESC lines have been undertaken; however, they have had limited success (Gong et al., 2010; Jin et al., 2012; Maruotti et al., 2012; Saito et al., 1992; Saito et al., 2003; Solter et al., 2000). Most ESC-like cultures proliferated slowly, lost pluripotency markers, and ceased to divide at early passages (Cao et al., 2009; Malaver-Ortega et al., 2012; Maruotti et al., 2012; Munoz et al., 2008; Saito et al., 1992).
Genetic modification in cattle is currently being accomplished by somatic cloning using mainly fibroblasts. Gene targeting in somatic cells is difficult and relatively inefficient (Clark and Whitelaw, 2003; Garrels et al., 2012a; Kues and Niemann, 2011; Velazquez et al., 2014). Recent advances in the development of programmable DNA nucleases allow the targeted introduction of frameshift mutations of livestock genomes (Carlson et al., 2013; Gün and Kues, 2014; Hauschild-Quintern et al., 2013; Tan et al., 2013). Germ line–competent bovine iPSCs could be valuable for improving reproduction traits and biotechnological applications, including gene targeting and genetic modifications (Cao et al., 2012; Koh and Piedrahita, 2014). Bovine iPSCs may enhance the ability to develop transgenic cattle for the production of therapeutic proteins in milk and to introduce disease resistance and other valuable traits (Plews et al., 2012; Sumer et al., 2011).
Binary DNA transposon systems have a number of advantages over other vector systems used for iPSC derivation. They can carry large cargo and show no or minimal bias for integration into translated regions. A number of hyperactive transposase variants are available that approach the efficiency of retroviral or lentiviral gene transfer; the production costs are low and transposons are safe because they are noninfective and the removal of integrated transposons is possible. Transposons, specifically Sleeping Beauty (SB) and piggyBac (PB), have emerged as useful alternatives to virus-mediated reprogramming of somatic cells from different species, including human (Davis et al., 2013; Woltjen et al., 2009), murine (Grabundzija et al., 2013; Muenthaisong et al., 2012; Talluri et al., 2014; Tsukiyama et al., 2014), porcine (Kues et al., 2013), and equine somatic cells (Nagy et al., 2011). Until now, there are no reports on transposon-mediated reprogramming of bovine somatic cells to pluripotent iPSCs.
Recently, we have reported the successful derivation of iPSC lines from inbred and outbred mice, employing the SB and PB transposon systems (Talluri et al., 2014). Here, we assessed the PB transposon approach for the derivation of iPSC lines from bovine fetal fibroblasts (BFFs).
Materials and Methods
Ethics statement
Animals were maintained and handled according to the German laws for animal welfare and genetically modified organisms. The experiments were approved by an external ethics committee (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, AZ 33.9-42502-04-09/1718).
Transposon and helper plasmid constructs
The construct used in the present study was described recently (Talluri et al., 2014). Briefly, the PB-reprogramming transposon contains a CAGGS promoter-driven cassette containing SOX2, OCT4, KLF4, c-MYC, NANOG, and LIN28 cDNAs, each separated by human sequences coding for self-cleaving 2A peptides and flanked by PB-inverted terminal repeats (ITR), i.e., PB-6F. A cDNA coding for a hyperactive PB is driven by the cytomegalovirus (CMV) promoter on a helper plasmid (Li et al., 2013). The transposon and transposase plasmids were mixed in a 5:1 molar ratio and used for electroporation of BFFs. From the PB-6F reprogramming cassette, deletion clones were generated to produce PB-4F (SOX2, OCT4, KLF4, c-MYC) and PB-3F (SOX2, OCT4, KLF4). The SB reprogramming transposon carried the murine cDNAs of Oct4, Sox2, Klf4, and cMyc, each separated by sequences coding for self-cleaving 2A peptide. The SB transposase (SB100X) helper plasmid has been described before (Grabundzija et al., 2013; Ivics et al., 2014; Kues et al., 2013; Talluri et al., 2014).
Isolation of BFFs and iPSC generation
Primary BFFs were prepared from a 2- to 3-month-old fetus obtained from a local slaughterhouse. Fetal tissues were trypsinized and seeded on tissue culture plates. BFFs were cultured in high glucose Dulbecco's Modified Eagle Medium (DMEM), supplemented with 10% heat-inactivated fetal calf serum (PAA), 2 mM l-glutamine, 1 mM sodium pyruvate, 1% nonessential amino acids, 0.05 mM mercaptoethanol, 100 U/mL penicillin, and 100 μg/mL streptomycin. The cells were passaged with 0.25% trypsin/0.1% EDTA. Cells at passage 2 were electroporated with the transposon system using single pulses at 250V (single pulse/10 msec; BioRad Gene PulserXcell electroporator). Electroporated BFFs were cultured in fibroblast medium, which was later replaced by DMEM/nutrient mixture F-12, supplemented with 20% knockout serum replacement (Millipore), 1 mM l-glutamine, 0.1 mM nonessential amino acids, 0.1 mM mercaptoethanol, 100 U/mL penicillin, 100 μg/mL streptomycin, and supplemented with basic fibroblast growth factor (bFGF; 4 ng/mL or 8 ng/mL) and 1000 U/mL human leukemia inhibitory factor (hLIF). Presumptive bovine iPSC colonies were picked with a pipette under microscopic control and plated into individual wells of 96-well tissue culture plates containing trypsin. Trypsin was neutralized with DMEM and 10% FBS, and the cells within each well were then transferred to individual wells of 96-well tissue culture plates. Presumptive bovine iPSCs were maintained on gelatinized plates or plates seeded with inactivated MEFs feeders and enzymatically (trypsin/EDTA) subpassaged every second or third day. For gelatinization, the intended culture dishes were covered with sterile 1% gelatin in PBS and allowed to dry before cell seeding. BFFs and bovine iPSCs were cultured in a humidified atmosphere consisting of 5% CO2 in air at 37°C. An Olympus BX60 (Olympus) microscope equipped with epifluorescence and a 12-bit digital camera (Olympus DP 71) was used for fluorescence microscopic analysis of the cells.
Clonal expansion of cell colonies, alkaline phosphatase staining, and gene transfer
Putative iPSC colonies were picked around days 14–18 with the help of a pulled glass pipette. Subsequently, these colonies were treated with trypsin to split the colony and were subcultured until they reached confluency in 24-well plates on mitomycin C–inactivated mouse fetal fibroblasts feeders. For alkaline phosphatase (AP) staining, the cells were washed twice with phosphate-buffered saline (PBS) and fixed with 4% formaldehyde for 5 minutes. Then 2 mL of AP staining solution (0.4 mg/mL sodium–α-napthylphosphate and 1 mg/mL of Fast Red dissolved in AP buffer [25 mM TrisHCl pH 9.0, 0.15 M NaCl, 4 mM MgCl2] were transferred to the wells, and images were made with an Olympus BX 60 (Olympus, Hamburg, Germany) fluorescence microscope equipped with a high-resolution digital camera (Olympus DP71).
Trypsinized biPS-1 cells (300,000 cells) were electroporated (300 V square wave pulse, 10 msec) with a mixture of 1500 ng of pT2RMCE-Venus and 500 ng of pCMV-SB100X (Garrels et al., 2011). Five to 7 days after electroporation, individual colonies that showed specific Venus fluorescence were subcloned to a 96-well plate and then expanded.
Reverse transcription PCR
Total RNA was prepared from iPSC lines and tissues with TriReagent (Ambion), according to the manufacturer's instructions (Kues et al., 2013). Isolated total RNA was treated with RNase-free DNase (1 U/μg RNA) (Epicentre Biotechnologies) and 0.5 μg of RNA was used for cDNA synthesis. Reverse transcription (RT) was performed in a 20-μL volume consisting of 4 μL of 10× RT buffer (Invitrogen), 4 μL of 50 mM MgCl2 (Invitrogen), 4 μL of 10 mM dNTP solution (Bioline), 2 μL (20 Units) of RNasin (Applied Biosystems), 2 μL (50 units) of Moloney murine leukemia virus (MMLV) reverse transcriptase (Applied Biosystems), and 2 μL hexamers (50 μM) (Applied Biosystems). The samples were maintained at 25°C for 10 min for primer annealing and then incubated at 42°C for 1 h. Finally, the samples were heated to 95°C for 5 min. The cDNA was diluted 1:5, and 2 μL (10 ng) were used for PCR amplification. The cDNA samples were subjected to PCR amplification with the primer pairs listed in Table 1. The PCR reaction included an initial denaturation at 94°C for 5 min followed by 30–35 cycles of denaturation at 94°C for 1 min, annealing at 58°C to 62°C for 30 sec, and extension at 72°C for 30 sec. Amplification of the transcript of the housekeeping glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used for normalization.
Table 1.
Primer Pairs Used in the Present Study
| Gene | Forward primer | Reverse primer | Size (bp) |
|---|---|---|---|
| POU5F1 | GTTCTCTTTGGAAAGGTGTTC | ACACTCGGACCACGTCTTTC | 313 |
| SOX2 | CATCCACAGCAAATGACAGC | TTTCTGCAAAGCTCCTACCG | 251 |
| c-MYC | CGCGGTCGCCTCCTTCTCGCCCAGG | GTCCGGGGAAGCGCAGGGC | 418 |
| KLF4 | GCCCCTAGAGGCCCACTT | CACAACCATCCCAGTCACAG | 433 |
| ALP1 | TTCAAACCGAAACACAAGCAC | GGTAAAGACGTGGGAGTGGTC | 378 |
| NOG | GTGTTTGGTGAACTCTCCTG | GGGAATTGAAATACTTGACAG | 307 |
| REX1 | GCAGAATGTGGGAAAGCCT | GACTGAATAAACTTCTTGC | 220 |
| AFP | AAGGCACCCTGTCCTGTATG | AGACACTCCAGCACGTTTCC | 330 |
| NESTIN | TCTGTCCTGAGCCCTACTCC | TTCTTTCACGTCCAGACCCG | 558 |
| β-TUBULIN | GGAGGGCGAGATGTACGAAG | GGGGTAAGATTGGGGGTGTG | 150 |
| GATA 4 | TGGAAGAAAGACGACGGGTG | TAGAGATAGCGACGCGGAGA | 104 |
| PAX 6 | GTCAGATCTGCCACTTCCCC | CATCTGCGCGCTCCTAGTTA | 305 |
| VIMENTIN | CCAGTCCGTGCTACCGC | ACGAGAAGTCCACCGAGTCC | 311 |
| pOSMKLN | CCAGCACTACCAGAGCGG | CCATCACCTCCACCACCTG | 201 |
| AMELEX | CCTGGCCTCTCTGACTCAG | AGGGAGATACACAGAGTGTG | 216 |
| GAPDH | CAAGGTCATCCATGACAACTTTG | CTACAGCAACAGGGTGGTGGAC | 496 |
| CDH1 | GACACTGGAGGTATCAGCGCAC | TGATCTGGACCAGCGACTTAGG | 194 |
| DPPA3 | TGC AAGTTGCCACTCAACTC | TCTTACCCCTCTCCGCCTAT | 158 |
| SALL4 | CGGGTGCTCCAA TGAACTAT | TGTCCTTCAAGATGA GCACG | 195 |
| STAT3 | GTGCATTGACAAAGACTCCG | AATCAGGGAGGCATCACAAT | 200 |
| SOCS3 | CCAGCCTGCGCCTCAAGACC | AAAGTGGCGCTGGTCCGAGC | 185 |
| FGF5 | GCGACTTCCTCTTCTTCCC | GCAGATGGAAACCGATGC | 151 |
| T | CCAGTACCCCAGCCTGTGGTCC | TGATGCCAGAGGCATCTCC | 595 |
Western blotting
Total protein isolation and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) have been described before (Kues et al., 2013). In brief, cells were extracted in RIPA buffer, and 10 μg of protein per slot was separated on a 12% SDS-PAGE gel, blotted to polyvinylidene difluoride (PVDF) membrane, blocked in 5% nonfat milk powder, and probed with a mouse polyclonal antibody against Oct4. The primary antibodies anti-Oct4 (Santa Cruz, sc-5279 (1:500)), anti-tubulin [Developmental Studies Hybridoma Bank (DHSB), E7 (1:500)] were used, respectively. Proteins were detected via horseradish peroxidase–coupled secondary antibody (1:10 000; Sigma) and an enhanced chemoluminescence reagent (ECL Plus, GE Healthcare).
Immunohistochemistry
Cells were seeded on gelatinized coverslips and fixed the next day in 80% methanol. The coverslips were washed three times with PBS/0.01% Triton-X100 and incubated with the anti-SSEA-1, anti SSEA-3, SSEA-4 (DHSB), and anti Oct-4 (1:200) antibodies (Santa Cruz), followed by incubation with secondary antibodies (1:2000, Invitrogen Molecular Probes, Darmstadt, Germany). Fibroblasts were used as controls, and samples without the first or without the secondary antibody were run in parallel.
Bioinformatic analysis of SB- and PB-homologous sequences in the bovine genome
To assess the possibility of whether bovine endogenous elements may interfere with the transposon-mediated gene transfer, the bovine genome BLAST analysis was performed against the cDNAs of SB and PB, and against the sequences of the ITRs of SB and PB. For the BLAST (BLASTN and BLAT) analyses, the www.ensembl.org resource, containing the Bos taurus genome (version 4.0), was employed.
Teratoma assay
To assess the teratoma potential, 1×106 cells of clonal bovine iPS-1 cultures were subcutaneously injected into the flank per CD-1 nude mice. Mice were analyzed for signs of tumor formation twice per week and sacrificed when tumor size exceeded a diameter of 1 cm. Tumors were fixed in 4% formaldehyde, embedded in paraffin, and sections were stained with hematoxylin and eosin for histological analyses.
Results
Reprogramming of bovine fetal fibroblasts to iPSCs by SB or PB transposons
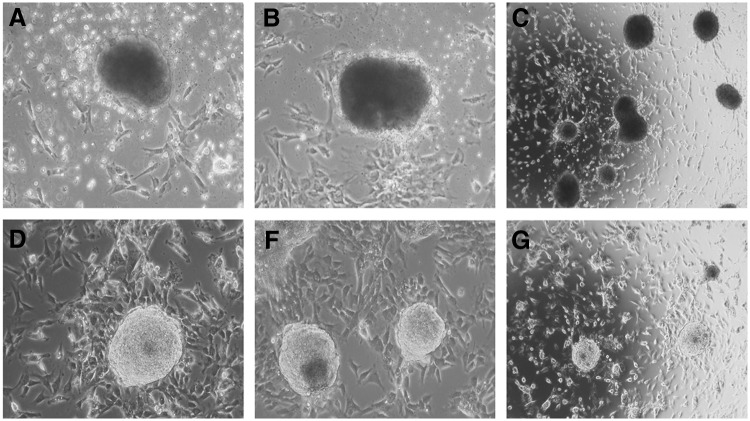
We first assessed two nonautonomous transposon systems, i.e., PB, carrying three, four, and six reprogramming factors, and SB, with four reprogramming factors (see Materials and Methods for the combinations of reprogramming factors), plus their respective helper plasmids (Fig. 1) for their ability to reprogram bovine somatic cells (Table 2). After 14–17 days, the electroporated BFFs started to form round and compact colonies. Two types of colonies were observed; one type appeared as large opaque colonies (Fig. 2A–C) and the other type of colonies was small and transparent (Fig. 2D–F). Colonies with the large opaque morphology were observed in treatment groups and the controls. The large, opaque colonies did not proliferate upon subpassaging and were not considered for further expansion. The non-proliferating colonies were not analyzed for senescence or apoptosis. Between days 10 and 20 postelectroporation, similar ratios of nonproliferating and presumptive iPSC colonies were found in the transposon groups. The small transparent colonies were only observed in transposon-treated cultures and were manually picked with a pulled pipette, then subcultured and expanded clonally (Table 2).
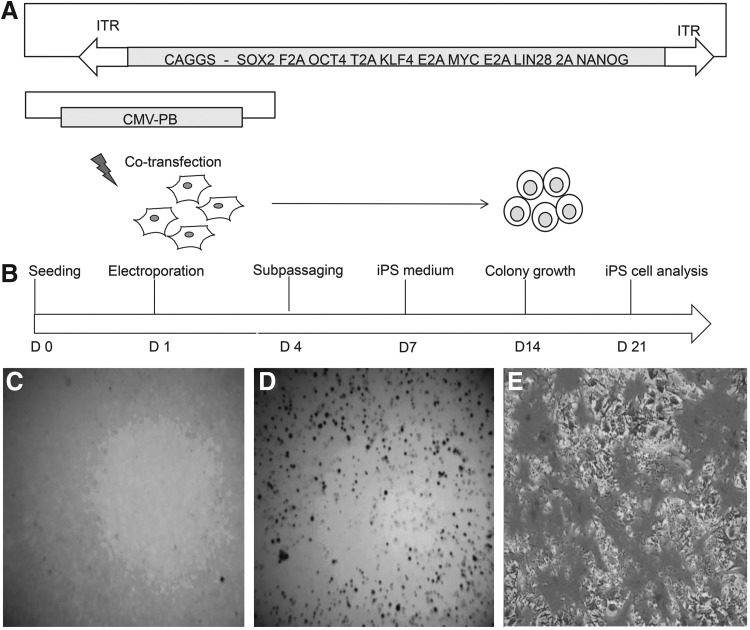
FIG. 1.
Experimental outline followed for reprogramming of bovine fetal fibroblasts. (A) Reprogramming PB-6F transposon system. (B) Time schedule followed for transposon mediated reprogramming (C) AP activity of bovine fetal fibroblasts. (D and E) AP activity of biPS-1 cells at P25 and P30 [low (4×) and high magnification (20×)].
Table 2.
Sleeping Beauty and piggyBac Transposon Reprogramming of Bovine Fibroblasts
| bFGF (4 ng/mL) | bFGF (4 ng/mL)+2i | hLIF (1000 U/mL)+2i | bFGF (8 ng/mL)+hLIF (1000 U/mL) | bFGF+hLIF | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transposase | PB | SB | PB | SB | PB | SB | PB | SB | —– | —– | —– | ||||||||
| Transposon type | 3F | 4F | 6F | 4F | 3F | 4F | 6F | 4F | 3F | 4F | 6F | 4F | 3F | 4F | 6F | 4F | 3F | 4F | 6F |
| No. of replicates | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 4 | 4 | 4 |
| Colony formation rate (%) | 0.006 | 0.005 | 0.014 | 0.003 | 0.003 | 0.010 | 0.021 | 0.012 | 0.003 | 0.018 | 0.006 | 0.009 | 0.005 | 0.012 | 0.024 | 0.011 | 0 | 0 | 0 |
| No. of colonies picked | 14 | 16 | 19 | 12 | 15 | 19 | 20 | 16 | 11 | 16 | 10 | 18 | 18 | 14 | 21 | 17 | 0 | 0 | 0 |
| No. of opaque/nonproliferating colonies found | 42 | 37 | 28 | 54 | 51 | 48 | 33 | 36 | 28 | 47 | 39 | 34 | 56 | 52 | 46 | 57 | 54 | 26 | 40 |
| Stable iPSC line/maximal passage | 0 | 0 | 1/P12a | 0 | 0 | 0 | 1/P18a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1/P40b | 0 | 0 | 0 | 0 |
| AP expression | — | — | No | — | — | — | Yes | — | — | — | Yes | — | — | — | Yes | — | — | — | — |
| Pluripotency gene expression (RT-PCR) | — | — | No | — | — | — | No | — | — | — | No | — | — | — | Yes | — | — | — | — |
| Teratoma formation | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 4/6 | NA | NA | NA | NA |
Cells ceased proliferation.
Culture with ongoing proliferation.
bFGF, basic fibroblast growth factor; hLIF, human leukemia inhibitory factor; PB, piggyBac transposon system; SB, Sleeping Beauty transposon system; iPSC, induced pluripotent stem cell; AP, alkaline phosphatase; NA, not assessed.
FIG. 2.
Morphology of bovine colonies after transposon electroporation. (A–C) Opaque, non-proliferative colonies (magnifications, 20× and 4×). (D–F) Presumptive bovine iPSC colonies (magnifications, 20× and 4×).
A stable iPSC line (biPS-1) could be established using the PB-6F transposon. Cultures obtained from SB-4F, PB-3F, and PB-4F could not be maintained for longer passages and showed no or weak AP activity. The biPS-1 line could be maintained for more than 40 passages, was successfully stored frozen, and analyzed in detail. Histochemical staining of these colonies showed strong expression of AP (Fig. 1C–E). The biPS-1 cell line displayed typical ESC-like colonies with a dome-shaped morphology and with distinct round, clear-cut borders (Fig. 2D–F).
Culture conditions
To support bovine iPSC proliferation, different medium combinations were tested (Table 1). The basic iPSC medium contains DMEM with low glucose, knockout serum replacement, l-glutamine, penicillin and streptomycin, β-mercaptoethanol, and nonessential amino acids. This medium was supplemented with bFGF, hLIF, and kinase inhibitors PD and CHIR, either alone or in combinations thereof (Table 2). Basic iPSC medium supplemented with bFGF (8 ng/mL) and hLIF (1000 U/mL) provided adequate culture conditions for maintaining the pluripotent status of the biPS-1 cells.
Characterization of biPS-1 cells
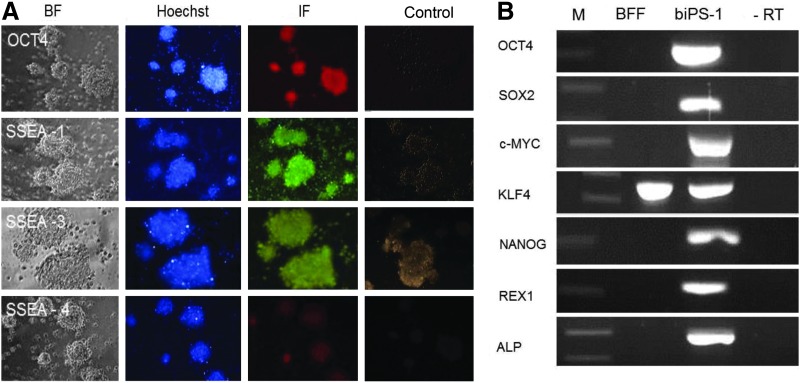
Expression of the endogenous pluripotency markers OCT4, SSEA-1, and SSEA-3 was detected by immunofluorescence (Fig. 3A), whereas the biPS-1 cells stained only faintly for SSEA-4 (Fig. 3A). Gene expression analysis by species-specific RT-PCR revealed that the biPS-1 cells expressed the endogenous pluripotency genes OCT4, SOX2, c-MYC, KLF4, NANOG, REX1, ALP (Fig. 3B), CDH1, STAT3, SALL, DPPA, and SOCS (Fig. S1; Supplementary Data are available at www.liebertpub.com/cell/), whereas brachyury (T) and FGF5, markers of primed epiblast stem cells (EpiSCs), were not detected (Fig. S1). Total protein from the biPS-1 cells was used for immunodetection of the OCT-4 protein (Fig. S2). The biPS-1 morphology is presented in Fig. S3. The in vitro differentiation potential of PB-6F transposon-derived iPSCs was evaluated by derivation of embryoid bodies by the hanging drop method (Fig. S3). The biPS-1 cell line had a normal karyotype, and no abnormalities were observed during chromosomal spreading (Fig. S3).
FIG. 3.
Immunostaining and RT-PCR of pluripotency genes for biPS-1. (A) Immunostaining of OCT4, SSEA-1, SSEA-3, and SSEA-4 (brightfield (BF); blue, Hoechst33342 nuclear counterstain; red and yellow, antibody staining); controls, without first antibody, Bars, 100 μm. (B) RT-PCR analysis of biPS-1 cells. M, marker; BFF, bovine fetal fibroblasts; biPS-1, bovine iPS line; -RT, no RT.
To assess the suitability of biPS-1 cells for genetic modifications, trypsinized cells were electroporated with a SB-Venus transposon and a SB helper plasmid. Venus is a yellow-shifted derivative of enhanced green fluorescent protein (EGFP) and can be specifically excited with EGFP filter blocks (Garrels et al., 2012b). Colonies expressing Venus could be detected 3 days after electroporation. Subcloning allowed enrichment and expansion of a Venus-positive biPS subline Fig. S3).
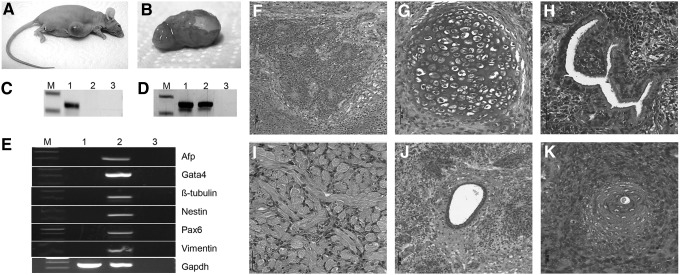
To analyze the in vivo differentiation potential, biPS-1 cells were injected subcutaneously into the flank of six immunodeficient nude mice. After 4 weeks, four mice had developed solid tumors (Fig. 4A, B). Samples from teratoma tissue were genotyped with bovine-specific primers of the AMELEX gene to confirm the bovine origin of the tumor tissue (Fig. 4C, D). Gene expression analysis of teratoma samples confirmed expression of differentiation markers such as vimentin, α-fetoprotein (AFP), nestin, PAX6, GATA, and tubulin (Fig. 4E). Histological examination revealed that the teratoma tissue contained cell types indicative of the three germ lineages, including muscle and cartilage tissue (mesoderm), neural rosettes (ectoderm), and keratinized and mucosa-producing epithelium (endoderm) (Fig. 4F–K, Fig. S4).
FIG. 4.
Teratoma development. (A) Immunodeficient mouse, 4 weeks after injection with biPS-1 cells. (B) Isolated tumor tissue. (C) Confirmation of bovine origin of teratoma by species-specific PCR of AMELEX gene. (D) Cross-species reactive PCR (lane M, marker; lane 1, teratoma tissue; lane 2, murine gDNA; and lane 3, no template). (E) Gene expression profile of teratoma tissue for differentiation markers (lane 1, biPS-1 cells; lane 2, teratoma; lane 3, no RT; lane M, marker). (F–K) Histological analysis of bovine teratoma. (F) Neuronal rosettes. (G) Bone and cartilage tissue. (H) Ciliated epithelium. (I) Striated musculature. (J) Mucosa-producing epithelium. (K) Keratinized epithelium.
Discussion
Here, we have reported the successful reprogramming of BFFs to pluripotent iPSCs by means of transposon-mediated gene transfer. The transposition of six reprogramming factors seems to be advantageous over the other approaches used in the present study, because only with PB-6F could we establish a stable iPSC line, called biPS-1. The biPS-1 cells exhibited typical features of pluripotent stem cells, including in vivo differentiation with the formation of mature teratomas.
Previously, bovine iPSCs have been derived from bovine fetal and adult fibroblasts by retroviral or lentiviral transduction (Cao et al., 2012; Han et al., 2011; Sumer et al., 2011) or nonintegrating plasmids (Huang et al., 2011; Wang et al., 2013). Retroviral and lentiviral vectors are the most commonly used gene delivery systems because they have high transduction rates; lentiviral vectors even allow transduction of nonproliferating cells (Coroadinha et al., 2010; Segura et al., 2013). However, the drawbacks of virus-mediated reprogramming are manifold and include the potential immune response (Manno et al., 2006; Thomas et al., 2003), their expensive production (Grimm et al., 1998; Tiscornia et al., 2006), the preferential integration in 5′-regions (UTRs) of genes that in turn may be associated with insertional mutagenesis (Thomas et al., 2003; Wu et al., 2003; Wu et al., 2006), the limited cargo capacity (Thomas et al., 2003), and integrated “pro-viruses” that cannot easily be removed after reprogramming. Bacterial plasmid approaches are nonviral alternatives for the derivation of bovine iPSCs (Huang et al, 2011; Wang et al., 2013); however, they may integrate randomly at sites of spontaneous double-strand breaks, and they also cannot easily removed after reprogramming.
This has prompted research into suitable alternatives to viral and plasmid reprogramming (Li et al., 2011; Okita et al., 2008; Talluri et al., 2014; Woltjen et al., 2009; Worsdorfer et al., 2013; Yu et al., 2009; Zhou et al., 2009). The SB and PB transposon systems share many advantages for mediating epigenetic reprogramming and possess a high cargo capacity of >100 kb (Rostovskaya et al., 2012).
Here, PB and SB transposon systems with a variable number of reprogramming factors were applied to derive pluripotent iPSC lines from bovine fibroblasts. We identified the PB-6F construct to be compatible with reprogramming, provided the culture medium had been enriched with bFGF and hLIF. It has been shown that virus-mediated reprogramming of bovine somatic cells required the addition of ectopic NANOG cDNA to the Oct4, Klf4, Sox2, and c-Myc (OKSM) factors for successful epigenetic reprogramming (Sumer et al., 2011). These data are in accordance with a recent study in the mouse (Buganim et al., 2014) showing that the quality of murine iPSC lines correlated positively with the number of reprogramming factors used. The biPS-1 cell line in the present experiments was established with the aid of the PB-6F transposon, which contains NANOG and LIN28. We also observed that culture conditions play a pivotal role in establishing and maintaining the biPS-1 line.
Previously, bovine iPSCs were cultured in medium supplemented with growth factors including FGF and LIF (Cao et al., 2012; Sumer et al., 2011) and kinase inhibitors (PD0325901 and CHIR99021) or with N2/B27 medium with LIF (Huang et al., 2011). Here, we tested bFGF, hLIF, and kinase inhibitors alone or in combination (Table 2). The biPS-1 cell line was established in medium supplemented with bFGF (8 ng/mL) and hLIF (1000 U/mL), suggesting that the bovine iPSCs use different pluripotency signaling pathways and require different culture conditions than human or murine iPSCs. Speculatively, bovine iPSCs may need NANOG on top of OSKM, and FGF and knockout serum to maintain the pluripotency status (Han et al., 2011). The exogenous genes used for reprogramming in the current study were not silenced as reported in previous studies in which viral vectors had been used for reprogramming (Cao et al., 2012; Han et al., 2011; Sumer et al., 2011).
The biPS-1 line could be successfully employed for further transposon-based genetic modification and was compatible with expanding a Venus-positive biPS-1 subculture. This shows that biPS-1 is suitable for additive and potentially for subtractive genetic modification and may be used for the production of genetically modified cattle via SCNT (Cao et al., 2012; Saito et al., 2003; Verma et al., 2012). Recently, CRISPR/Cas genome editing was shown in bovine iPSCs (Heo et al., 2014).
In conclusion, we show for the first time that transposon technology is a useful nonviral method suitable for reprogramming BFFs to iPSCs. Our results emphasize that the selected reprogramming factors and culture conditions play a crucial role in the efficiency of deriving and maintaining pluripotent bovine iPSCs. An attractive and advantageous feature of the PB system is that the transposon can be excised seamlessly, and the current development of excision-competent/integration-defective PB variants will facilitate the derivation of footprint-free iPSCs without permanent alterations in their genome (Li et al., 2013; Meir et al., 2013).
Supplementary Material
Acknowledgments
The authors thank S. Holler, N. Cleve, D. Herrmann, M. Ziegler, and K. Klingemann for excellent technical assistance. This work was supported by an Indian Council of Agricultural Research (ICAR) International Fellowship to T.R.T., and a DBT CREST Fellowship to D.K., and a grant from the Bundesministerium für Bildung und Forschung to Z.I. (ReGene, grant number 01GN1003A). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. T.R.T is enrolled at the Hannover Graduate School for Veterinary Pathobiology, Neuroinfectiology and Translational Medicine (HGNI), and this work represents a partial fulfilment of the requirements for the degree of a Ph.D.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Bao L., He L., Chen J., Wu Z., Liao J., Rao L., Ren J., Li H., Zhu H., Qian L., Gu Y., Dai H., Xu X., Zhou J., Wang W., Cui C., and Xiao L. (2011). Reprogramming of ovine adult fibroblasts to pluripotency via drug-inducible expression of defined factors. Cell Res. 21, 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Markoulaki S., van Wietmarschen N., Hoke H., Wu T., Ganz K., Akhtar-Zaidi B., He Y., Abraham B.J., Porubsky D., Kulenkampff E., Faddah D.A., Shi L., Gao Q., Sarkar S., Cohen M., Goldmann J., Nery J.R., Schultz M.D., Ecker J.R., Xiao A., Young R.A., Lansdorp P.M., and Jaenisch R. (2014). The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell 15, 295–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Yang P., Pu Y., Sun X., Yin H., Zhang Y., Zhang Y., Li Y., Liu Y., Fang F., Zhang Z., Tao Y., and Zhang X. (2012). Characterization of bovine induced pluripotent stem cells by lentiviral transduction of reprogramming factor fusion proteins. Int. J. Biol. Sci. 8, 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Wang F., Chen Z., Liu Z., Mei C., Wu H., Huang J., Li C., Zhou L., and Liu L. (2009). Isolation and culture of primary bovine embryonic stem cell colonies by a novel method. J. Exp. Zool. A Ecol. Genet. Physiol. 311, 368–376 [DOI] [PubMed] [Google Scholar]
- Carlson D.F., Tan W., Hackett P.B., and Fahrenkrug S.C. (2013). Editing livestock genomes with site-specific nucleases. Reprod. Fertil. Dev. 26, 74–82 [DOI] [PubMed] [Google Scholar]
- Clark J., and Whitelaw B. (2003). A future for transgenic livestock. Nat. Rev. Genet. 4, 825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coroadinha A.S., Gama-Norton L., Amaral A.I., Hauser H., Alves P.M., and Cruz P.E. (2010). Production of retroviral vectors: Review. Curr. Gene Ther. 10, 456–473 [DOI] [PubMed] [Google Scholar]
- Davis R.P., Nemes C., Varga E., Freund C., Kosmidis G., Gkatzis K., de Jong D., Szuhai K., Dinnyes A., and Mummery C.L. (2013). Generation of induced pluripotent stem cells from human foetal fibroblasts using the Sleeping Beauty transposon gene delivery system. Differentiation 86, 30–37 [DOI] [PubMed] [Google Scholar]
- Deng Y., Liu Q., Luo C., Chen S., Li X., Wang C., Liu Z., Lei X., Zhang H., Sun H., Lu F., Jiang J., and Shi D. (2012). Generation of induced pluripotent stem cells from buffalo (Bubalus bubalis) fetal fibroblasts with buffalo defined factors. Stem Cells Dev. 21, 2485–2494 [DOI] [PubMed] [Google Scholar]
- Esteban M.A., Xu J., Yang J., Peng M., Qin D., Li W., Jiang Z., Chen J., Deng K., Zhong M., Cai J., Lai L., and Pei D. (2009). Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J. Biol. Chem. 284, 17634–17640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrels W., Mates L., Holler S., Dalda A., Taylor U., Petersen B., Niemann H., Izsvak Z., Ivics Z., and Kues W.A. (2011). Germline transgenic pigs by Sleeping Beauty transposition in porcine zygotes and targeted integration in the pig genome. PloS One 6, e23573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrels W., Ivics Z., and Kues W.A., (2012a). Precision genetic engineering in large mammals. Trends Biotechnol. 30, 386–393 [DOI] [PubMed] [Google Scholar]
- Garrels W., Holler S., Cleve N., Niemann H., Ivics Z., and Kues W.A. (2012b). Assessment of fecundity and germ line transmission in two transgenic pig lines produced by sleeping beauty transposition. Genes (Basel) 3, 615–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G., Roach M.L., Jiang L., Yang X., and Tian X.C. (2010). Culture conditions and enzymatic passaging of bovine ESC-like cells. Cell. Reprogram. 12, 151–160 [DOI] [PubMed] [Google Scholar]
- Gün G., and Kues W.A. (2014). Current progress of genetically engineered pig models for biomedical research. Biores. Open Access 3, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabundzija I., Wang J., Sebe A., Erdei Z., Kajdi R., Devaraj A., Steinemann D., Szuhai K., Stein U., Cantz T., Schambach A., Baum C., Izsvak Z., Sarkadi B., and Ivics Z. (2013). Sleeping Beauty transposon-based system for cellular reprogramming and targeted gene insertion in induced pluripotent stem cells. Nucleic Acids Res. 41, 1829–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D., Kern A., Rittner K., and Kleinschmidt J.A. (1998). Novel tools for production and purification of recombinant adeno-associated virus vectors. Hum. Gene Ther. 9, 2745–2760 [DOI] [PubMed] [Google Scholar]
- Han X., Han J., Ding F., Cao S., Lim S.S., Dai Y., Zhang R., Zhang Y., Lim B., and Li N. (2011). Generation of induced pluripotent stem cells from bovine embryonic fibroblast cells. Cell Res. 21, 1509–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo Y.T., Quan X., Xu Y.N., Baek S., Choi H., Kim N.H., and Kim J. (2014). CRISPR/Cas9 nuclease-mediated gene knock-in in bovine-induced pluripotent cells. Stem Cells Dev. 24, November3, 2014. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Hauschild-Quintern J., Petersen B., Cost G.J., and Niemann H. (2013). Gene knockout and knockin by zinc-finger nucleases: current status and perspectives. Cell. Mol. Life Sci. 70, 2969–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Li T., Alonso-Gonzalez L., Gorre R., Keatley S., Green A., Turner P., Kallingappa P.K., Verma V., and Oback B. (2011). A virus-free poly-promoter vector induces pluripotency in quiescent bovine cells under chemically defined conditions of dual kinase inhibition. PloS One 6, e24501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z., Garrels W., Mátés L., Yau T.Y., Bashir S., Zidek V., Landa V., Geurts A., Pravenec M., Rülicke T., Kues W.A., and Izsvák Z. (2014). Germline transgenesis in pigs by cytoplasmic microinjection of Sleeping Beauty transposons. Nat. Protoc. 9, 810–827 [DOI] [PubMed] [Google Scholar]
- Jin M., Wu A., Dorzhin S., Yue Q., Ma Y., and Liu D. (2012). Culture conditions for bovine embryonic stem cell-like cells isolated from blastocysts after external fertilization. Cytotechnology 64, 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S., and Piedrahita J.A. (2014). From “ES-like” cells to induced pluripotent stem cells: A historical perspective in domestic animals. Theriogenology 81, 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kues W.A., and Niemann H. (2011). Advances in farm animal transgenesis. Prev. Vet. Med. 102, 146–156 [DOI] [PubMed] [Google Scholar]
- Kues W.A., Herrmann D., Barg-Kues B., Haridoss S., Nowak-Imialek M., Buchholz T., Streeck M., Grebe A., Grabundzija I., Merkert S., Martin U., Hall V.J., Rasmussen M.A., Ivics Z., Hyttel P., and Niemann H. (2013). Derivation and characterization of sleeping beauty transposon-mediated porcine induced pluripotent stem cells. Stem Cells Dev. 22, 124–135 [DOI] [PubMed] [Google Scholar]
- Kumar D., Talluri T.R., Anand T., and Kues W.A. (2015). Induced pluripotent stem cells: Mechanisms, achievements and perspectives in farm animals. World J. Stem Cells 26, 315–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Burnight E.R., Cooney A.L., Malani N., Brady T., Sander J.D., Staber J., Wheelan S.J., Joung J.K., McCray P.B., Jr, Bushman F.D., Sinn P.L., and Craig N.L. (2013). piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. USA 110, E2279–E2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Q., Yin X., Yang W., Du Y., Hou P., Ge J., Liu C., Zhang W., Zhang X., Wu Y., Li H., Liu K., Wu C., Song Z., Zhao Y., Shi Y., and Deng H. (2011). Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 21, 196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Cui C., Chen S., Ren J., Chen J., Gao Y., Li H., Jia N., Cheng L., Xiao H., and Xiao L. (2009). Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell 4, 11–15 [DOI] [PubMed] [Google Scholar]
- Liu H., Zhu F., Yong J., Zhang P., Hou P., Li H., Jiang W., Cai J., Liu M., Cui K., Qu X., Xiang T., Lu D., Chi X., Gao G., Ji W., Ding M., and Deng H. (2008). Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell 3, 587–590 [DOI] [PubMed] [Google Scholar]
- Malaver-Ortega L.F., Sumer H., Liu J., and Verma P.J. (2012). The state of the art for pluripotent stem cells derivation in domestic ungulates. Theriogenology 78, 1749–1762 [DOI] [PubMed] [Google Scholar]
- Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B., Dake M., Kaye R., Razavi M., Zajko A., Zehnder J., Rustagi P.K., Nakai H., Chew A., Leonard D., Wright J.F., Lessard R.R., Sommer J.M., Tigges M., Sabatino D., Luk A., Jiang H., Mingozzi F., Couto L., Ertl H.C., High K.A., and Kay M.A. (2006). Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 12, 342–347 [DOI] [PubMed] [Google Scholar]
- Maruotti J., Munoz M., Degrelle S.A., Gomez E., Louet C., Diez C., de Longchamp P.H., Brochard V., Hue I., Caamano J.N., and Jouneau A. (2012). Efficient derivation of bovine embryonic stem cells needs more than active core pluripotency factors. Mol. Reprod. Dev. 79, 461–477 [DOI] [PubMed] [Google Scholar]
- Meir Y.J., Lin A., Huang M.F., Lin J.R., Weirauch M.T., Chou H.C., Lin S.J., and Wu S.C. (2013). A versatile, highly efficient, and potentially safer piggyBac transposon system for mammalian genome manipulations. FASEB J. 27, 4429–4443 [DOI] [PubMed] [Google Scholar]
- Muenthaisong S., Ujhelly O., Polgar Z., Varga E., Ivics Z., Pirity M.K., and Dinnyes A. (2012). Generation of mouse induced pluripotent stem cells from different genetic backgrounds using Sleeping beauty transposon mediated gene transfer. Exp. Cell Res. 318, 2482–2489 [DOI] [PubMed] [Google Scholar]
- Munoz M., Rodriguez A., De Frutos C., Caamano J.N., Diez C., Facal N., and Gomez E. (2008). Conventional pluripotency markers are unspecific for bovine embryonic-derived cell-lines. Theriogenology 69, 1159–1164 [DOI] [PubMed] [Google Scholar]
- Nagy K., Sung H.K., Zhang P., Laflamme S., Vincent P., Agha-Mohammadi S., Woltjen K., Monetti C., Michael I.P., Smith L.C., and Nagy A. (2011). Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev. 7, 693–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Nakagawa M., Hyenjong H., Ichisaka T., and Yamanaka S. (2008). Generation of mouse induced pluripotent stem cells without viral vectors. Science 322, 949–953 [DOI] [PubMed] [Google Scholar]
- Plews J.R., Gu M., Longaker M.T., and Wu J.C. (2012). Large animal induced pluripotent stem cells as pre-clinical models for studying human disease. J. Cell. Mol. Med. 16, 1196–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Pak Y., He L., Qian L., Gu Y., Li H., Rao L., Liao J., Cui C., Xu X., Zhou J., Ri H., and Xiao L, (2011). Generation of hircine-induced pluripotent stem cells by somatic cell reprogramming. Cell Res. 21, 849–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossello R.A., Chen C.C., Dai R., Howard J.T., Hochgeschwender U., and Jarvis E.D. (2013). Mammalian genes induce partially reprogrammed pluripotent stem cells in non-mammalian vertebrate and invertebrate species. eLife 2, e00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovskaya M., Fu J., Obst M., Baer I., Weidlich S., Wang H., and Smith A.J., Anastassiadis K., and Stewart A.F. (2012). Transposon-mediated BAC transgenesis in human ES cells. Nucleic Acids Res. 40, e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Strelchenko N., and Niemann H. (1992). Bovine embryonic stem cell-like cell lines cultured over several passages. Roux's Arch. Dev. Biol. 201, 134–141 [DOI] [PubMed] [Google Scholar]
- Saito S., Sawai K., Ugai H., Moriyasu S., Minamihashi A., Yamamoto Y., Hirayama H., Kageyama S., Pan J., Murata T., Kobayashi Y., Obata Y., and Yokoyama K.K. (2003). Generation of cloned calves and transgenic chimeric embryos from bovine embryonic stem-like cells. Biochem. Biophys. Res. Commun. 309, 104–113 [DOI] [PubMed] [Google Scholar]
- Segura M.M., Mangion M., Gaillet B., and Garnier A. (2013). New developments in lentiviral vector design, production and purification. Expert Opin. Bioll. Ther. 13, 987–1011 [DOI] [PubMed] [Google Scholar]
- Shi Y., Desponts C., Do J.T., Hahm H.S., Scholer H.R., and Ding S. (2008). Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell 3, 568–574 [DOI] [PubMed] [Google Scholar]
- Shimada H., Nakada A., Hashimoto Y., Shigeno K., Shionoya Y., and Nakamura T. (2010). Generation of canine induced pluripotent stem cells by retroviral transduction and chemical inhibitors. Mol. Reprod. Dev. 77, 2. [DOI] [PubMed] [Google Scholar]
- Solter D. (2000). Mammalian cloning: Advances and limitations. Nat. Rev. Genet. 1, 199–207 [DOI] [PubMed] [Google Scholar]
- Stadtfeld M., Nagaya M., Utikal J., Weir G., and Hochedlinger K. (2008). Induced pluripotent stem cells generated without viral integration. Science 322, 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumer H., Liu J., Malaver-Ortega L.F., Lim M.L., Khodadadi K., and Verma P.J. (2011). NANOG is a key factor for induction of pluripotency in bovine adult fibroblasts. J. Anim. Sci. 89, 2708–2716 [DOI] [PubMed] [Google Scholar]
- Takahashi K., and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- Talluri T.R., Kumar D., Glage S., Garrels W., Ivics Z., Debowski K., Behr R., and Kues W.A. (2014). Non-viral reprogramming of fibroblasts into induced pluripotent stem cells by Sleeping Beauty and piggyBac transposons. Biochem. Biophys. Res. Commun. 450, 581–587 [DOI] [PubMed] [Google Scholar]
- Tan W., Carlson D.F., Lancto C.A., Garbe J.R., Webster D.A., Hackett P.B., and Fahrenkrug S.C. (2013). Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc. Natl. Acad. Sci. USA 110, 16526–16531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.E., Ehrhardt A., and Kay M.A. (2003). Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 4, 346–358 [DOI] [PubMed] [Google Scholar]
- Tiscornia G., Singer O., and Verma I.M. (2006). Production and purification of lentiviral vectors. Nat. Protoc. 1, 241–245 [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Kato-Itoh M., Nakauchi H., and Ohinata Y. (2014). A comprehensive system for generation and evaluation of induced pluripotent stem cells using piggyBac transposition. PloS One 9, e92973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez M.A., Kues W.A., and Niemann H. (2014). Biomedical applications of ovarian transvaginal ultrasonography in cattle. Anim. Biotechnol. 25, 266–293 [DOI] [PubMed] [Google Scholar]
- Verma R., Holland M.K., Temple-Smith P., and Verma P.J. (2012). Inducing pluripotency in somatic cells from the snow leopard (Panthera uncia), an endangered felid. Theriogenology 77, 220–228, 228.e221–222 [DOI] [PubMed] [Google Scholar]
- Wang S.W., Wang S.S., Wu D.C., Lin Y.C., Ku C.C., Wu C.C., Chai C.Y., Lee J.N., Tsai E.M., Lin C.L., Yang R.C., Ko Y.C., Yu H.S., Huo C., Chuu C.P., Murayama Y., Nakamura Y., Hashimoto S., Matsushima K., Jin C., Eckner R., Lin C.S., Saito S., and Yokoyama K.K. (2013). Androgen receptor-mediated apoptosis in bovine testicular induced pluripotent stem cells in response to phthalate esters. Cell Death Dis. 4, e907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A., Daley G.Q., Brack A.S., Collins J.J., Cowan C., Schlaeger T.M., and Rossi D.J. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K., Michael I.P., Mohseni P., Desai R., Mileikovsky M., Hamalainen R., Cowling R., Wang W., Liu P., Gertsenstein M., Kaji K., Sung H.K., and Nagy A. (2009). piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsdorfer P., Thier M., Kadari A., and Edenhofer F. (2013). Roadmap to cellular reprogramming—manipulating transcriptional networks with DNA, RNA, proteins and small molecules. Curr. Mol. Med. 13, 868–878 [DOI] [PubMed] [Google Scholar]
- Wu C., and Dunbar C.E. (2011). Stem cell gene therapy: The risks of insertional mutagenesis and approaches to minimize genotoxicity. Front. Med. 5, 356–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li Y., Crise B., and Burgess S.M. (2003). Transcription start regions in the human genome are favored targets for MLV integration. Science 300, 1749–1751 [DOI] [PubMed] [Google Scholar]
- Wu X., Luke B.T., and Burgess S.M. (2006). Redefining the common insertion site. Virology 344, 292–295 [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., Slukvin I.I., and Thomson J.A. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., and Thomson J.A. (2009). Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Wu S., Joo J.Y., Zhu S., Han D.W., Lin T., Trauger S., Bien G., Yao S., Zhu Y., Siuzdak G., Scholer H.R., Duan L., and Ding S. (2009). Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4, 381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.