Abstract
To examine the genomic reprogrammability of trophoblast stem (TS) cells using a nuclear transfer technique, we produced TS cloned embryos using five TS cell lines from three strains of mice (ICR, B6D2F1, and B6CBF1) as donors and observed developmental ability during preimplantation development. The developmental rates of the TS cloned embryos that developed to the two-cell, four- to eight-cell, morula, and blastocyst stages were 58–83%, 0–38.6%, 0–21.3%, and 0–15.9%, respectively, indicating that more than 50% of TS cloned embryos arrested at the two-cell stage. These TS cloned two-cell embryos were expressed low level of Dappa3 (also known as PGC7/Stella), indicating that zygotic gene activation (ZGA) was disrupted in these embryos. However, a small portion of the TS cloned embryos (0–15.9%) reached the blastocyst stage. In these TS cloned blastocysts, the numbers of trophectoderm (TE) and inner cell mass (ICM) cells were 31.9±4.6 and 12.1±3.0, respectively, which were not significantly different from those in the fertilized embryos. In addition, the gene expression analysis showed that Oct3/4, and Cdx2, which are ICM- and TE-specific marker genes, respectively, and Dppa3, and Hdac1, which are zygotic gene activation-related genes, were expressed in TS cloned blastocysts at the same levels as in the fertilized blastocysts. These results indicate that although TS cloned embryos are able to differentiate into ICM cells, the genomic reprogrammability of TS cells is very low following nuclear transfer.
Introduction
Mammalian development commences with the totipotent zygote, which is capable of developing into all of the specialized cells that make up the adult animal. The first cell lineage during mammalian embryogenesis is observed at the blastocyst stage prior to implantation. A blastocyst is composed of an inner cell mass (ICM) and trophectoderm (TE) (Watson and Cross, 2005). After the implantation, ICM cells differentiate into the embryo proper, whereas TE cells differentiate into the extraembryonic tissues, including the placenta.
In mice, embryonic stem (ES cells) and trophoblast stem (TS) cells are derived from the ICM and TE cells, respectively (Evans and Kaufman, 1981; Tanaka et al., 1998). In chimeras, ES cells contribute to embryonic tissues but not to extraembryonic tissues (Bradley et al., 1984). In contrast, TS cells differentiate only into extraembryonic tissues (Tanaka et al., 1998). These findings indicate that the developmental abilities of ICM and ES cells are restricted to embryonic tissues and that the developmental abilities of TE and TS cells are restricted to extraembryonic tissues. Moreover, it is generally thought that these developmental potentials do not extend beyond the boundaries between ICM and TE cells.
The ICM cells and ES cells have pluripotency and are able to differentiate into somatic and germ cells in vivo and in vitro. Somatic cells that have differentiated terminally cannot regain pluripotency. However, it was reported that somatic cells in which the expression of four genes (Oct3/4, Sox2, c-Myc, and Klf4) was transduced were able to revert to their epigenetic state to form ES-like cells. These cells, termed induced pluripotent stem (iPS) cells, have properties that are similar to those of ES cells (Takahashi and Yamanaka, 2006). This finding indicates that the differential potentials of somatic cells can be reinstated to the pluripotent state by direct reprogramming.
Moreover, when somatic cells were transferred into metaphase II (MII)-stage oocytes by a nuclear transfer technique, the nuclear-transferred embryos were able to develop to full term. To date, the cloned animals that have been produced by nuclear-transferred embryos in mammals include sheep (Wilmut et al., 1997), mice (Wakayama et al., 1998), cattle (Kato et al., 1998), pigs (Onishi et al., 2000), goats (Baguisi et al., 1999), rabbits (Chesne et al., 2002), cats (Shin et al., 2002), mules (Woods et al., 2003), horses (Galli et al., 2003), rats (Zhou et al., 2003), and dogs (Lee et al., 2005), suggesting that somatic cells are able to regain the totipotent state by nuclear transfer. These results confirm that somatic cells that are the derivatives of ICM cells can be reprogrammed to regain the pluripotent or totipotent states. Moreover, the derivatives from ICM cells are able to differentiate into extraembryonic tissues when the genomic reprogramming occurs after nuclear transfer. In addition, ES cells can be induced to differentiate into the TE lineage by the forced repression of Oct3/4 or the overexpression of Cdx2 (Niwa et al., 2000, 2005). Thus, the derivatives of ICM cells can also be directly reprogrammed to differentiate into the TE lineage.
Blastocysts comprise the mural and polar TE cells. The mural TE cells that are not in contact with the ICM generate the primary trophoblast giant cells. In contrast, the polar TE cells that are in contact with the ICM continue to divide (Watson and Cross, 2005). In 1998, Tsunoda and Kato showed that live mouse pups could be derived from mural TE nuclear-transferred embryos (Tsunoda and Kato, 1998). That was the first report that TE cells also have the ability to reacquire totipotency by nuclear transfer in mice. Moreover, the mural TE cells are able to differentiate into embryonic tissues when the genomic reprogramming occurs by nuclear transfer. These findings evoked the possibility that extraembryonic tissues are also useful for cloned animal production. However, it is difficult to produce TE nuclear-transferred embryos, because the preparation of mural TE cells as donors requires skilled techniques. Futhermore, it is difficult to prepare enough TE cells for nuclear transfer, because the mural TE cells have stopped mitotic cell division and easily differentiate into trophoblast giant cells in vitro.
In the present study, we focused on TS cells as donors for nuclear transfer. TS cells established from the polar TE cells in a blastocyst have the pluripotency to differentiate into placental tissues (Tanaka et al., 1998). TS cells continue to mitotic cell division with fibroblast growth factor 4 (FGF4). Moreover, as the TS cell culture system is established, it is easy to prepare enough donor cells for nuclear transfer. Thus, we examined the developmental ability of TS cloned embryos to evaluate the genomic reprogrammability of polar TE cells by nuclear transfer.
Materials and Methods
Animal ethics
All mice were maintained and used in accordance with the Guidelines for the Care and Use of Laboratory Animals, as specified by the Japanese Association for Laboratory Animal Science and by the Tokyo University of Agriculture.
Culture of TS cell lines
TS cells were established from blastocysts using three mouse strains (ICR, BDF1 (C57BL/6×DBA2), and BCF1 (C57BL/6×CBA) as described (Ogawa et al., 2009; Tanaka et al., 1998). TS cells were cultured in TS medium (RPMI-1640; Gibco Invitrogen, Carlsbad, CA) supplemented with 20% fetal bovine serum (FBS), 1 mM sodium pyruvate (Gibco), 100 μM β-mercaptoethanol (Sigma, St. Louis, MO, USA), 2 mM l-glutamine (Gibco), 100 U/mL penicillin, and 100 mg/mL streptomycin (Gibco). Mitomycin-treated mouse embryonic fibroblast conditioned medium (MEF-CM) was prepared as described previously (Tanaka et al., 1998). TS cell lines were cultured on mitomycin-treated MEFs with TS medium containing 25 ng/mL FGF4 (PeproTech EC, London, UK) and 1 mg/mL heparin (Sigma) (TS+F4H medium), or cultured without mitomycin-treated MEFs by using 70% MEF-CM and 30% TS medium, containing 25 ng/mL FGF4 and 1 mg/mL heparin (70 Cond F4H medium) to maintain the cells in an undifferentiated condition. The differentiation of TS cells was induced by removing FGF4, heparin, and MEF-CM from the culture medium.
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNAs were extracted from TS and ES cells by using ISOGEN (Nippon Gene, Tokyo). They were then treated with DNase (Promega, Madison, WI, USA) to eliminate genomic DNA. The genomic DNA-free total RNA was reverse-transcribed to cDNA by using SuperScript III Reverse Transcriptase (Invitrogen). The PCR was carried out with Ex Taq polymerase (Takara Bio, Shiga, Japan). The PCR conditions are listed in Table 1.
Table 1.
PCR Conditions
| Primer sequences | |||||
|---|---|---|---|---|---|
| Genes | Forward | Reverse | Annealing Temp (°C) | Cycles | Size (bp) |
| RT-PCR | |||||
| Eomes | CCACTGGATGAGGCAGGAGATTTCC | AGTCTTGGAAGGTTCATTCAAGTCC | 65 | 30 | 178 |
| Cdx2 | CGAAACCTGTGCGAGTGGATG | TGCTGTTCGTGGGTAGGAGGAG | 63 | 35 | 568 |
| Tpbpa | AAGTTAGGCAACGAGCGAAA | AGTGCAGGATCCCACTTGTC | 65 | 35 | 167 |
| Pl1 | CTCACTTGGAGCCTACATTGT | GGGGAAAGCATTACAAGTC | 65 | 30 | 773 |
| Pl2 | CCAGACAACATCGGAAGACC | GACCATGCAGACCAGAAAGT | 60 | 30 | 240 |
| Plpa | GGCAATCCAGTTATCCCAGA | GATCCCAGCCTTTCACACAT | 60 | 30 | 257 |
| Oct3/4 | GGCGTTCTCTTTGCAAAGGTGTTC | CTCGAACCACATCCTTCTCT | 60 | 30 | 313 |
| GAPDH | CCATCAACGACCCCTTCATT | GCCTGCTTCACCACCTTCTT | 60 | 30 | 680 |
| qPCR | |||||
| Dppa2 | TACCAAGCACCTGTCCTCCGC | CCCTGCTTCATTCTGGCCTCC | 60 | 198 | |
| Dppa3 | ATCGGGAAGAATTAGGAGCT | TTTACTTTATTCCTCAGCCCT | 60 | 222 | |
| Dppa4 | AGGAAGTCAGCACCACCGTAGT | AAGCTGGAAGAGGGCAATGGCT | 60 | 263 | |
| HDAC1 | GACCGGTTAGGTTGCTTCAA | GTCATGTTGGAAGGGCTGAT | 60 | 247 | |
| Cdx2 | AGGCTGAGCCATGAGGAGTA | GAGGTCCATAATTCCACTCA | 60 | 123 | |
| Oct3/4 | GGCGTTCTCTTTGCAAAGGTGTTC | CTCGAACCACATCCTTCTCT | 60 | 313 | |
| GAPDH | GTCGTGGAGTCTACTGGTGTC | GAGCCCTTCCACAATGCCAAA | 60 | 240 | |
TS cells as donors for nuclear transfer
TS cells differentiated into the trophoblast giant cells by DNA replication without cell division, even if they were cultured in the undifferentiation condition (70 Cond F4H medium). To avoid the differentiation into the trophoblast giant cells, we cultured TS cells on feeder cells (mitomycin-treated MEF). To synchronize the cells at metaphase, we cultured TS cells for 3 h in TS+T4H medium containing nocodazole (1 μg/mL) which is a microtubule polymerization inhibitor (Jincho et al., 2008).
ESCs as donors for nuclear transfer
ESCs (TT2 line) were cultured on feeder cells in Dulbecco's modified Eagle's medium (DMEM) medium containing 100 μM β-mercaptoethanol, 100 μM nonessential amino acid (Gibco), 1000 units/mL ESGRO (Millipore, Billerica, MA, USA), and 15% (vol/vol) FBS. ESCs were cultured with nocodazole (1 μg/mL) for 3 h to collect M-phase cells as donors for nuclear transfer.
Production of nuclear-transferred embryos
We collected superovulated oocytes from C57BL/6×DBA2 (BDF1) female mice at 14 h after an administration of 7.5 IU of human chorionic gonadotropin (hCG) (Teikokokuzoki, Tokyo). After the enucleation of the MII chromosomes, we transferred a TS cell arrested at M phase into an enucleated oocyte, using a piezo-driven system (Prime Tech, Ibaraki, Japan) in the M2 medium containing 5 μg/mL cytochalasin B. The nuclear-transferred embryos were cultured in KSOM medium for 2 h, and then they were activated artificially in Ca2+-free M16 medium containing SrCl2 (8 mM) for 6 h. The activated embryos that had a pseudo-prenucleus were cultured in KSOM medium to blastocysts.
Karyotype analysis
The nuclear-transferred embryos were washed in phosphate-buffered saline (PBS) and mounted on glass slides. The embryos were fixed in freshly prepared chilled Carnoy's solution (glacial acetic acid:methanol=1:3). After the embryos were washed in ethanol, the chromosomes were stained with acetic orcein and washed with aceto-glycerol solution (glycerol:glacial acetic acid:water=1:1:3). Spindle formation was observed under an inverted microscope.
Cell counting
Blastocysts were incubated with 0.5% acid tyrode to remove the zona pellucidae. Zona-free blastocysts were reacted with rabbit anti-mouse serum at 37°C for 40 min, and then were treated with guinea pig complement at 37°C for 40 min. Samples were stained with Hoechst 33342 and propidium iodide (PI) for 40 min. The nuclei were counted under a fluorescence microscope.
Immunostaining
Blastocysts were incubated with 0.5% acid tyrode to remove the zona pellucidae and fixed with 4% paraformaldehyde for 1 h. Samples were treated with 0.25% Triton X-100 in PBS (−) for 1 h to induce the membrane permeability, and incubated with blocking solution [0.1% TritonX-100, 1% skim milk, and 5% FBS in PBS (−)] for 1 h. After blocking, the samples were treated with mouse anti-CDX2 monoclonal antibody (CDX2-88; BioGenex, Fremont, CA) or mouse anti-OCT3/4 monoclonal antibody (sc-5279; Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 2 h. After washing, they were cultured with Alexa 594–labeled mouse immunoglobulin G (IgG; Invitrogen, Carlsbad, CA, USA) for 30 min, and the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Image acquisition was performed using an Olympus IX71 fluorescence microscope (Olympus, Tokyo) with a Cool Snap camera (Nippon Roper, Tokyo) and MetaMorph software (Molecular Devices, Downingtown, PA).
Quantitative gene expression analysis of donor cells and cloned two-cell embryos and blastocyst
The two-cell-stage nuclear-transferred embryos were collected at about 24 h after activation. Five two-cell embryos were lysed in 75 μL of Buffer RLT (Qiagen; Valencia, CA) containing 1% β-mercaptoethanol, and total RNA was extracted using an RNeasy Micro Kit (Qiagen). Blastocysts from nuclear-transferred embryos were collected at 4 days after activation. One blastocyst embryo was lysed in 75 μL of Buffer RLT (Qiagen) containing 1% β-mercaptoethanol, and total RNA was extracted using an RNeasy Micro Kit (Qiagen). Total RNA was reverse-transcribed to cDNA using SuperScript III Transcriptase (Invitrogen). cDNA samples synthesized from TS cells and ESCs as described above were used for the quantitative analysis.
The quantitative gene expression analysis was performed by real-time PCR using a 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). PCR products were detected with SYBR Green using Power SYBR Green master mix (Applied Biosystems).
For the quantitative analysis, the PCR products amplified with primers were electrophoresed on agarose gels and purified with the Wizard SV Gel and PCR Clean-Up System (Promega). Ten-fold serially diluted PCR products were used as the external standards for the real-time PCR. The expression levels of target mRNAs were determined from the appropriate standard curve and normalized relative to the amount of mRNA.
Statistical analysis
We compared the developmental rates of embryos cloned from TS and ESCs using the chi-squared test and Fisher's exact probability test. The relative transcription levels of blastocysts and donor cells determined by quantitative RT-PCR were analyzed by one-way analysis of variance (ANOVA) followed by Tukey's honest significant difference (HSD) test using JMP software (SAS Institute. Cary, NC, USA). Differences at p<0.05 were considered significant.
Results
Establishment of TS cell lines
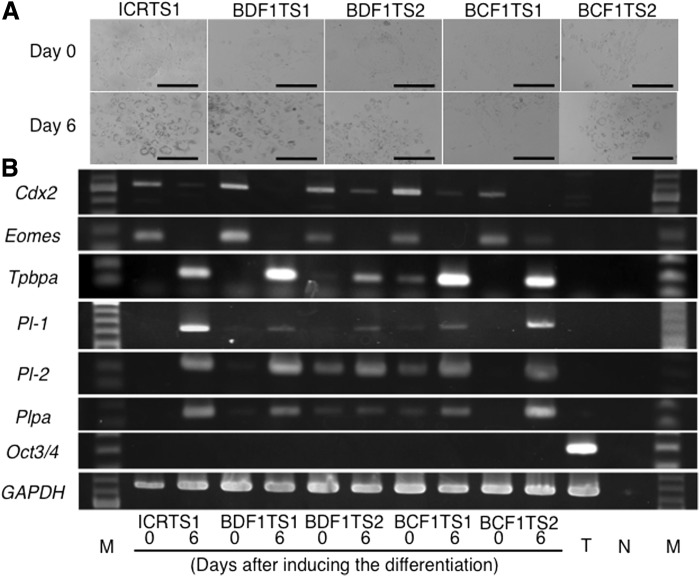
We established seven (ICR), six (BDF1), and nine (BCF1) TS cell lines from 20, 10, and 34 blastocysts, respectively (Table 2). These established cell lines (one ICR line, two BDF1 lines, and two BCF1 lines) showed the epithelial cell–like appearance and the distinct colony boundary at day 0, and at day 6 after inducing the differentiation they showed a giant cell–specific morphology with a large cytoplasm and nucleus and numerous granules around the nucleus (Fig. 1A).
Table 2.
Establishment of Trophoblast Stem Cells from Three Strains of Mice
| Strains | Blastocysts | Outgrowth (%) | Cell lines (%) |
|---|---|---|---|
| ICR | 20 | 20 (100.0) | 7 (35.0) |
| B6D2F1 | 10 | 9 (90.0) | 6 (60.0) |
| B6CBF1 | 34 | 33 (97.1) | 9 (26.5) |
FIG. 1.
(A) Morphology of five TS cell lines (one ICR line, two BDF1 lines, and two BCF1 lines) at days 0 and 6 after inducing the differentiation. (B) Expression of two TS marker genes (Cdx2 and Eomes), one spongiotrophoblast marker gene (Tpbpa), three giant cell marker genes (Pl-1, Pl-2, and Plpa), and one ES marker gene (Oct3/4) days 0 and 6 after inducing the differentiation. T, TT2; N, Negative control; M, 100-bp ladder marker.
To confirm the differentiation ability of the five cell lines, we examined the expression of two TS marker genes (Cdx2 and Eomes), one spongiotrophoblast maker gene (Tpbpa), three trophoblast giant cell marker genes (Pl-1, Pl-2, and Plpa), and one ES marker gene (Oct3/4) in these TS cell lines (Fig. 1B). Cdx2 and Eomes were detected in undifferentiated (D0, day 0 after inducing the differentiation) TS cells, but were not detected in differentiated cells (D6, day 6 after inducing the differentiation). In contrast, Tpbpa, Pl-1, Pl-2, and Plpa were expressed in differentiated cells. Oct3/4 was not detected in either undifferentiated or differentiated TS cells.
These results indicate that these five cell lines showed the typical character of TS cells, and these TS cells were used in this study as donors for nuclear transfer.
Development of TS and ES cloned embryos
To investigate whether genomes of TS cells can be reprogrammed by transferring them into oocytes, we compared the in vitro development of reconstructed embryos that received the five lines of TS cells with the development of reconstructed embryos that received TT2 ES cells (Table 3). We found that 82.4% of the ES cloned embryos activated and excluded the polar body. The developmental rate of the ES cloned embryos to blastocyst stage was 64.8%. In contrast, 58.4–70.4% of the TS cloned embryos activated and excluded the polar body. Although 58.4–84.0% of the TS cloned embryos developed to the two-cell stage, the developmental rate to blastocyst stage was only 0–21.3%.
Table 3.
Development of Embryos Cloned from Embryonic Stem Cells and Trophoblast Stem Cells
| No. of embryos developed to | ||||||
|---|---|---|---|---|---|---|
| Donor cell | No. of oocytes injected | No. of oocytes activated (%) | Two cell (%) | Four to eight cell (%) | Morula (%) | Blastocyst (%) |
| ICRTS1 | 294 | 207 (70.4)b | 153 (73.9)b | 80 (38.6)b | 4 (21.3)b | 33 (15.9)b |
| BDF1TS1 | 115 | 69 (60.0)c | 41 (59.4)c | 0 (0.0)e | 0 (0.0)d | 0 (0.0)c |
| BDF1TS2 | 151 | 101 (58.4)c | 59 (58.4)c | 7 (6.9)d | 6 (5.9)cd | 3 (3.0)c |
| BCF1TS1 | 143 | 94 (65.7)bc | 79 (84.0)b | 33 (35.1)b | 19 (20.2)b | 4 (4.3)c |
| BCF1TS2 | 227 | 160 (70.4)b | 127 (79.4)b | 30 (18.8)c | 17 (10.6)c | 15 (9.4)b |
| ES (TT2) | 131 | 108 (82.4)a | 101 (93.5)a | 95 (88.0)a | 82 (75.9)a | 70 (64.8)a |
Significantly different at p<0.05.
These results indicated that the developmental rates of the TS cloned embryos during the preimplantation stage were very low. In particular, the developmental rates of the TS cloned embryos using BDF1 TS cells as donors were significantly low, and almost all of the TS cloned embryos were arrested at the two-cell stage. Consequently, the developmental rate of the TS cloned embryos to the blastocyst stage was less than 15.9%.
Metaphase spindle formation in cloned embryos
The rates of spindle formation in the TS and ES cloned embryos at 3 h after nuclear transfer are shown in Table 4 and Figure 2. Among the ES cloned embryos, 80% formed normal metaphase spindles that showed the alignment of condensed chromosomes on the metaphase-like spindles (Fig. 2a). In contrast, 66.7% of the TS cloned embryos formed normal metaphase spindles.
Table 4.
Spindle Formation in Embryos Cloned from Embryonic Stem Cells and Trophoblast Stem Cells
| Donor cell | No. of oocytes examined | No. of oocytes that formed metaphase spindles (%) | No. of oocytes that formed normal metaphase spindles (%) |
|---|---|---|---|
| ICRTS1 | 15 | 12 (80.0) | 10 (66.7) |
| ES (TT2) | 10 | 9 (90.0) | 8 (80.0) |
FIG. 2.
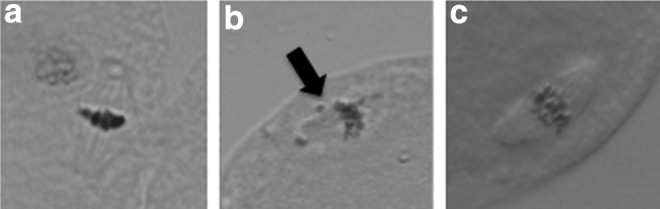
Metaphase spindle in cloned embryos at 3 h after nuclear transfer (a, b) and in an MII oocyte (c). A normal spindle was formed (a) and an abnormal spindle was formed (b). Chromosome misalignment is indicated by an arrow.
Number of TE and ICM cells in cloned blastocysts
To evaluate the quality of blastocysts derived from TS cloned embryos, we examined the numbers of TE and ICM cells in those blastocysts (Fig. 3). The mean numbers of TE and ICM cells in a blastocyst derived from a TS cloned embryo were 31.9±4.6 and 12.1±3.0, respectively. These numbers were not significantly different from those obtained with ES cloned and fertilized embryos (ES cloned embryos: TE, 29.2±7.9, ICM, 12.8±3.3; fertilized embryos: TE, 33.8±7.0, ICM, 15.1±5.2).
FIG. 3.
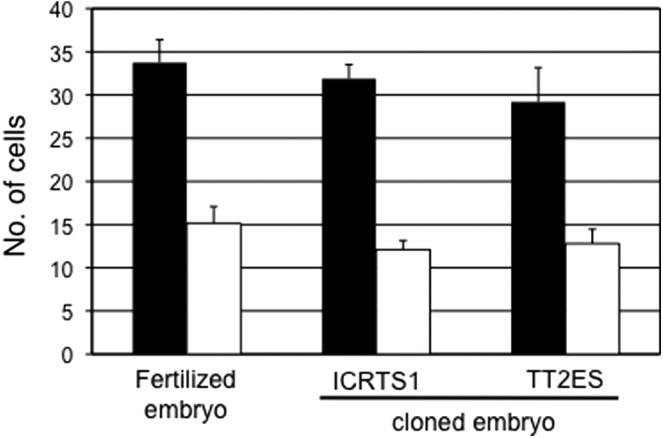
Numbers of TE and ICM cells in the blastocysts. Black bars, TE cells; white bars, ICM cells.
Relative expression levels of mRNA transcripts in donor cells and cloned two-cell embryos and blastocysts
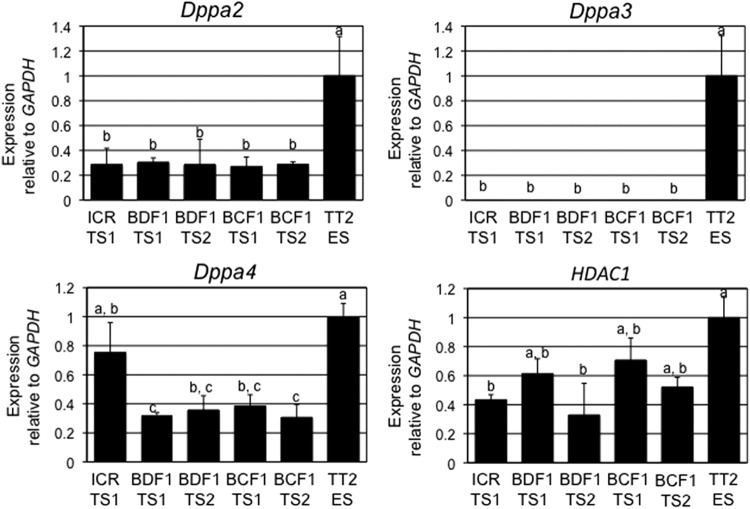
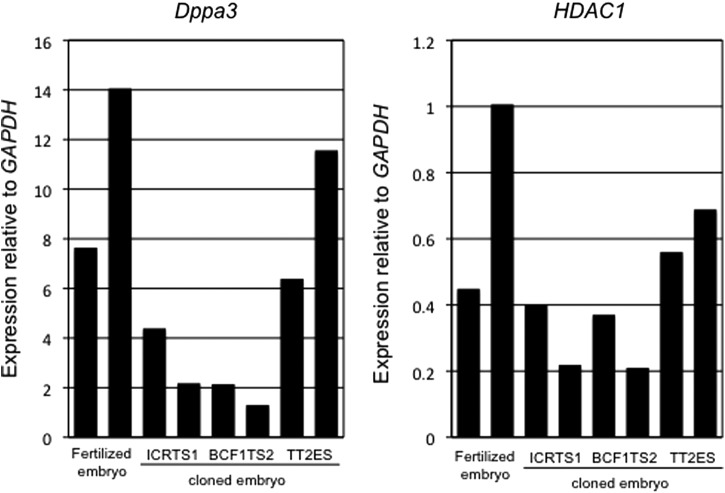
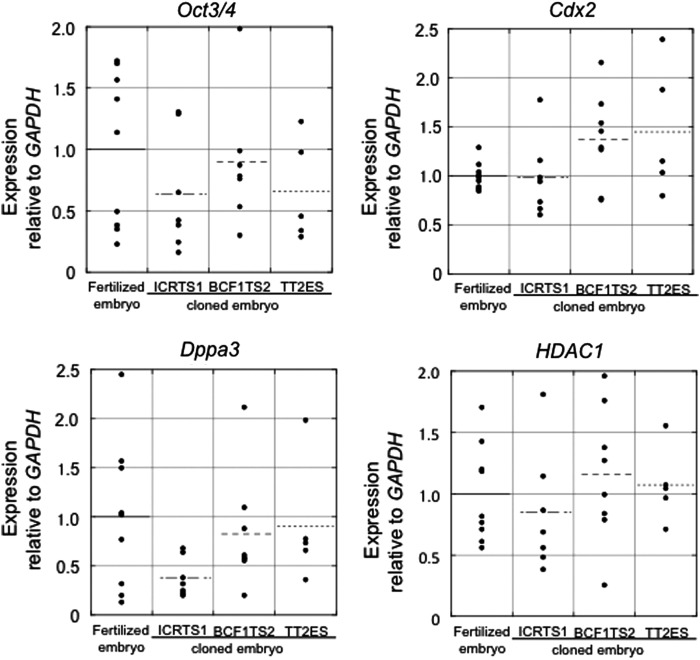
The developmental arrest of the TS clones might have been caused by a failure to activate genes of the major zygotic gene activation (ZGA) at the two-cell stage. To evaluate the characteristics of blastocysts from TS cloned embryos, we conducted an expression analysis of four ZGA-related genes (Dppa2, Dppa3, Dppa4, and Hdac1) (Inoue et al., 2006) and Oct3/4 and Cdx2, the pluripotent marker genes of ES and TS cells, respectively, in TS and ES cells and cloned two-cell embryos and blastocysts by performing a real-time quantitative RT-PCR. We found that the expression levels of two genes (Dppa2 and Dppa4) in the TS cells were less than one-half of those in the ES cells (except for the expression of Dppa4 in ICRTS1) (Fig. 4). The expression level of HDAC1 in the TS cells was 30–70% of that in the ES cells. In contrast, the expression of Dppa3 was repressed completely in the TS cells. The expression level of HDAC1 in TS cloned two-cell embryos was the same in fertilized and ES cloned embryos. However, the expression of Dppa3 in TS cloned embryos was lower than fertilized and ES cloned embryos (Fig. 5). In contrast, the expression levels of four genes (Cdx2, Oct3/4, Dppa3, and HDAC1) in blastocysts were the same in the TS and ES cloned embryos compared to the fertilized embryos (Fig. 6).
FIG. 4.
Quantitative mRNA expression of Dppa2, Dppa3, Dppa4, and HDAC1 genes in TS cells (ICRTS1, BDF1TS1, BDF1TS2, BCF1TS1, and BCF1TS2) and ES cells (TT2). The relative amounts of transcripts for Dppa2, Dppa3, Dppa4, and HDAC1 genes are expressed relative to GAPDH values. Data were normalized to TT2 ES cell levels. The expression level of each line indicates the mean±standard error of the mean (SEM) of three trials. Bars with different letters above them differ significantly (p<0.05).
FIG. 5.
Quantitative mRNA expression of Dppa3 and HDAC1 genes in two-cell embryos derived from TS (ICRTS1 and BCF1TS) and ES (TT2) cloned embryos collected at about 24 h after activation. The relative amounts of transcripts for Dppa3 and HDAC1 genes are expressed relative to GAPDH values. The expression level of each lane means mRNA expression of five two-cell embryos.
FIG. 6.
Quantitative mRNA expression of Oct3/4, Cdx2, Dppa3, and HDAC1 genes in single blastocysts derived from TS (ICRTS1 and BCF1TS2) and ES (TT2) cloned embryo. The relative amounts of transcripts for Oct3/4, Cdx2, Dppa3, and HDAC1 genes are expressed relative to GAPDH values. Data were normalized to control blastocyst levels. Median values are indicated by dot bars.
Localization of OCT3/4 in cloned blastocysts
An immunostaining study revealed that OCT3/4 was localized in the nuclei of ICM cells in blastocysts derived from fertilized embryos (Fig. 7). In the TS cloned blastocysts, the localization of OCT3/4 was restricted to the nuclei of ICM cells.
FIG. 7.
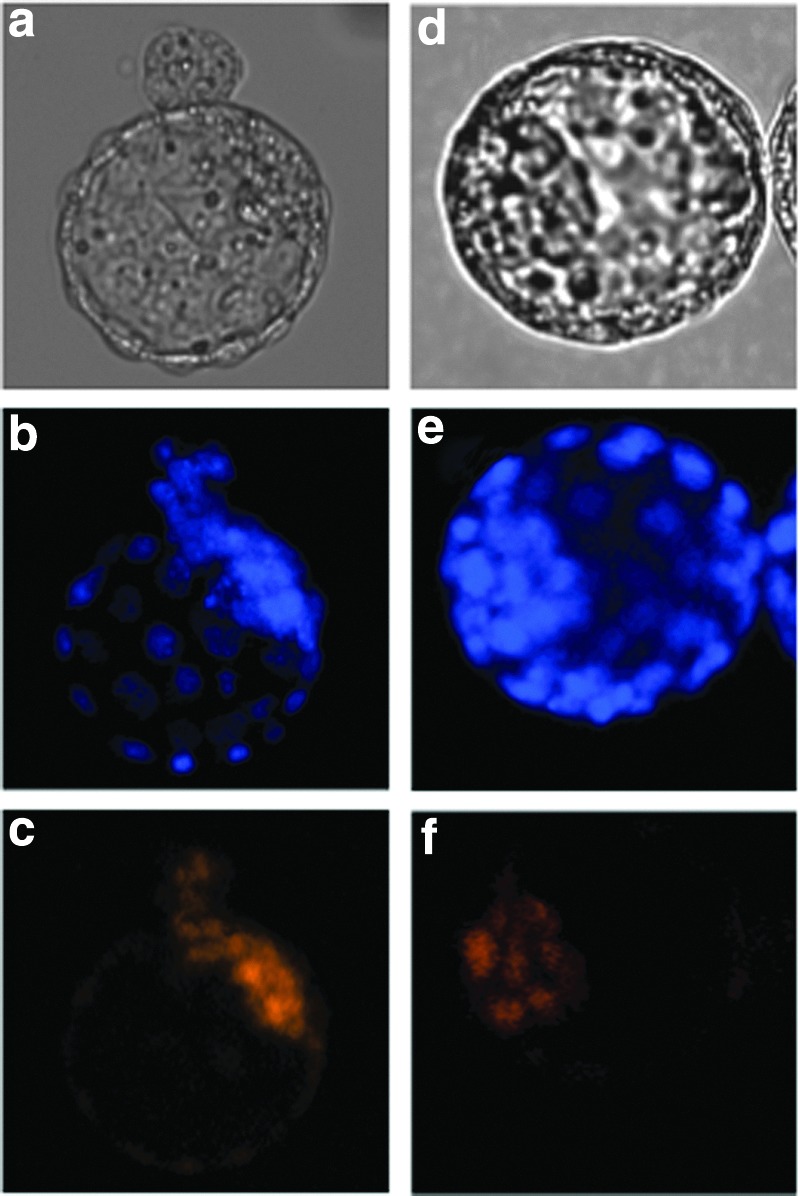
Localization of OCT3/4 in a blastocyst. (a–c) TS cloned embryo; (d–f) fertilized embryo. (a and d) Bright field; (b and e) DAPI staining; (c and f) OCT3/4 staining.
Discussion
In the present study, we examined the genomic reprogrammability of TS cells by evaluating the developmental ability of TS cloned embryos. In TS cloned embryos, more than 50% of them were arrested at the two-cell stage and few embryos reached to the blastocyst stage. Moreover, the expression level of the ZGA-related gene, such as Dppa3, in TS cloned two-cell embryos was very low. These results indicate that the deficiency of the genomic reprogammability of TS cells is due to the lack of ZGA.
Although most somatic or ES clones fail to develop soon after implantation, more than 50% of them are able to develop to the blastocyst stage. In contrast, cloned embryos derived from hematopoietic stem cells (HSCs) were arrested at the two-cell stage, and only 6% of them reached the blastocyst stage (Inoue et al., 2006). The poor development of HSCs cloned embryos was consistent with the failure of expression of Hdac1, a key regulator of ZGA, at the two-cell stage (Zeng and Schultz, 2005). This low expression level of Hdac1 may be inherited from the expression pattern in HSCs (Inoue et al., 2006), indicating that the lack of ZGA-related gene expression in donor cells affects the development of cloned embryos beyond the two-cell stage. The developmental ability of TS cloned embryos was similar to HSCs clones; therefore, we examined the expression of ZGA-regulated genes in TS cells and two-cell embryos and blastocysts from TS cloned embryos.
Although the TS cloned embryos were also arrested at the two-cell stage, the expression level of Hdac1 in the TS cells was not so different from that in the ES cells. In contrast, Dppa3 (which is also a regulator of ZGA) was not expressed in the TS cells. Moreover, the expression level of Dppa3 in TS cloned two-cell embryos was low. The expression of Dppa3 persisted during the implantation after fertilization (Iqbal et al., 2011). Dppa3 was required to protect the DNA methylation of the maternal genome after fertilization (Nakamura et al., 2007). Dppa3-null embryos exhibited abnormal cleavage at the two- to four-cell stage, and only 3% of them reached the blastocyst stage (Nakamura et al., 2007). Therefore, TS cloned embryos might be arrested at the two-cell stage due to the deficiency of Dppa3 expression at this stage.
Treatment with class IIb histone deacetylase inhibitors such as trichostatin A (TSA), Scriptaid, suberoylanilide hydroxamic acid, and oxamflatin improved the developmental rate of mouse nuclear-transferred embryos (Kishigami et al., 2006, 2007; Ono et al., 2010; Rybouchkin et al., 2006). TSA enhanced the histone acetylation and decondensation of chromosomes and the levels of newly synthesized RNA in two-cell embryos, indicating that genomic reprogramming in nuclear-transferred embryos was improved by TSA treatment (Bui et al., 2010). These findings suggest that genomic reprogramming such as histone modification is necessary for the achievement of ZGA in nuclear-transferred embryos.
The rates of development to blastocyst and to term were higher in ICM cloned embryos than in ES cloned embryos, although developmental regression and developmental disorders of the placenta have been observed in ICM clones as well as ES clones (Ono and Kono, 2006). Both ICM cells and ES cells have pluripotency, but the global gene expression patterns of ICM and ES cells are significantly different (Nakanishi et al., 2012; Reijo Pera et al., 2009; Tang et al., 2010). Moreover, the differentially methylated regions that were hypermethylated in ES cells were hypomethylated in ICM cells (Nakanishi et al., 2012). The methylation statuses were not so different between the ICM and TE cells. Together these findings indicate that the reprogrammability of methylation status in ES cells and TS cells is lower than that in ICM and TE cells, and that this is the reason for the decline in the developmental ability of ES and TS cloned embryos compared to ICM and TE cloned embryos.
The iPS cell experiment revealed that reprogramming could be induced in somatic cells. TS cells were also converted to iPS cells by the overexpression of Oct3/4, Sox2, Klf4, and cMyc (Kuckenberg et al., 2011). Moreover, Wu et al. showed that the overexpression of a single transcription factor, Oct3/4, in TS cells is sufficient to reprogram TS cells into a pluripotent state (Wu et al., 2011). Although the rate of TS cells converted to iPS cells was lower than those of somatic cells, these TS cells had acquired the potential to differentiate into somatic and germ cells. These results revealed that TS cells are also able to regain the pluripotency to differentiate into embryonic cells.
In the present study, blastocysts derived from TS cloned embryos formed ICM cells that expressed OCT3/4, and the numbers of ICM cells were not significantly different from those of the fertilized embryos. Moreover, the expression level of Oct3/4 in the TS cloned blastocysts was the same as that in the fertilized blastocysts. These results indicated that TS cells also have the reprogrammability to differentiate into ICM cells by nuclear transfer. In addition, although the expression level of Dppa3 in TS cloned two-cell embryos was lower, the expression level of Dppa3 in the TS cloned blastocysts was also the same as that in the fertilized blastocysts. These results indicate that the TS cloned embryos developed into blastocysts due to the recovery of the expression of ZGA-related genes. In the future, the differentiation ability of TS cloned embryos could be established by embryo transfer or the establishment of ES cells.
Acknowledgments
This work was supported by a grant from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (No. 22658084).
Author Disclosure Statement
The authors declare that there are no conflicts of interest.
References
- Baguisi A., Behboodi E., Melican D.T., Pollock J.S., Destrempes M.M., Cammuso C., Williams J.L., Nims S.D., Porter C.A., Midura P., Palacios M.J., Ayres S.L., Denniston R.S., Hayes M.L., Ziomek C.A., Meade H.M., Godke R.A, Gavin W.G., Overström E.W., and Echelard Y. (1999). Production of goats by somatic cell nuclear transfer. Nat. Biotechnol. 17, 456–461 [DOI] [PubMed] [Google Scholar]
- Bradley A., Evans M., Kaufman M.H., and Robertson E. (1984). Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309, 255–256 [DOI] [PubMed] [Google Scholar]
- Bui H.T., Wakayama S., Kishigami S., Park K.K., Kim J.H., Thuan N.V., and Wakayama T. (2010). Effect of trichostatin A on chromatin remodeling, histone modifications, DNA replication, and transcriptional activity in cloned mouse embryos. Biol. Reprod. 83, 454–463 [DOI] [PubMed] [Google Scholar]
- Chesne P., Adenot P.G., Viglietta C., Baratte M., Boulanger L., and Renard J.P. (2002). Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat. Biotechnol. 20, 366–369 [DOI] [PubMed] [Google Scholar]
- Evans M.J., and Kaufman M.H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- Galli C., Lagutina I., Crotti G., Colleoni S., Turini P., Ponderato N., Duchi R., and Lazzari G. (2003). Pregnancy: A cloned horse born to its dam twin. Nature 424, 635. [DOI] [PubMed] [Google Scholar]
- Inoue K., Ogonuki N., Miki H., Hirose M., Noda S., Kim J.M., Aoki F., Miyoshi H., and Ogura A. (2006). Inefficient reprogramming of the hematopoietic stem cell genome following nuclear transfer. J. Cell Sci. 119, 1985–1991 [DOI] [PubMed] [Google Scholar]
- Iqbal K., Jin S.G., Pfeifer G.P., and Szabo P.E. (2011). Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. USA 108, 3642–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jincho Y., Sotomaru Y., Kawahara M., Ono Y., Ogawa H., Obata Y., and Kono T. (2008). Identification of genes aberrantly expressed in mouse embryonic stem cell-cloned blastocysts. Biol. Reprod. 78, 568–576 [DOI] [PubMed] [Google Scholar]
- Kato Y., Tani T., Sotomaru Y., Kurokawa K., Kato J., Doguchi H., Yasue H., and Tsunoda Y. (1998). Eight calves cloned from somatic cells of a single adult. Science 282, 2095–2098 [DOI] [PubMed] [Google Scholar]
- Kishigami S., Mizutani E., Ohta H., Hikichi T., Thuan N.V., Wakayama S., Bui H.T., and Wakayama T. (2006). Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem. Biophys. Res. Commun. 340, 183–189 [DOI] [PubMed] [Google Scholar]
- Kishigami S., Bui H.T., Wakayama S., Tokunaga K., Van Thuan N., Hikichi T., Mizutani E., Ohta H., Suetsugu R., Sata T., and Wakayama T. (2007). Successful mouse cloning of an outbred strain by trichostatin A treatment after somatic nuclear transfer. J. Reprod. Dev. 53, 165–170 [DOI] [PubMed] [Google Scholar]
- Kuckenberg P., Peitz M., Kubaczka C., Becker A., Egert A., Wardelmann E., Zimmer A., Brustle O., and Schorle H. (2011). Lineage conversion of murine extraembryonic trophoblast stem cells to pluripotent stem cells. Mol. Cell. Biol. 31, 1748–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.C., Kim M.K., Jang G., Oh H.J., Yuda F., Kim H.J., Hossein M.S., Kim J.J., Kang S.K., Schatten G., and Hwang W.S. (2005). Dogs cloned from adult somatic cells. Nature 436, 641. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Tanaka S, Shiota K, Nakano T. (2007). PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 9, 64–71 [DOI] [PubMed] [Google Scholar]
- Nakanishi M.O., Hayakawa K., Nakabayashi K., Hata K., Shiota K., and Tanaka S. (2012). Trophoblast-specific DNA methylation occurs after the segregation of the trophectoderm and inner cell mass in the mouse periimplantation embryo. Epigenetics 7, 173–182 [DOI] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., and Smith A.G. (2000). Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- Niwa H., Toyooka T., Shimosato D., Strumpf D., Takahashi K., Yagi R., and Rossant J. (2005). Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929 [DOI] [PubMed] [Google Scholar]
- Ogawa H., Shindo N., Kumagai T., Usami Y., Shikanai M., Jonwn K., Fukuda A., Kawahara M., Sotomaru Y., Tanaka S., Arima T., and Kono T. (2009). Developmental ability of trophoblast stem cells in uniparental mouse embryos. Placenta 30, 448–456 [DOI] [PubMed] [Google Scholar]
- Onishi A., Iwamoto M., Akita T., Mikawa S., Takeda K., Awata T., Hanada H., and Perry A.C. (2000). Pig cloning by microinjection of fetal fibroblast nuclei. Science 289, 1188–1190 [DOI] [PubMed] [Google Scholar]
- Ono T., Li C., Mizutani E., Terashita Y., Yamagata K., and Wakayama T. (2010). Inhibition of class IIb histone deacetylase significantly improves cloning efficiency in mice. Biol. Reprod. 83, 929–937 [DOI] [PubMed] [Google Scholar]
- Ono Y., and Kono T. (2006). Irreversible barrier to the reprogramming of donor cells in cloning with mouse embryos and embryonic stem cells. Biol. Reprod. 75, 210–216 [DOI] [PubMed] [Google Scholar]
- Reijo Pera R.A., DeJonge C., Bossert N., Yao M., Hwa Yang J.Y., Asadi N.B., Wong W., Wong C., and Firpo M.T. (2009). Gene expression profiles of human inner cell mass cells and embryonic stem cells. Differentiation 78, 18–23 [DOI] [PubMed] [Google Scholar]
- Rybouchkin A., Kato Y., and Tsunoda Y. (2006). Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol. Reprod. 74, 1083–1089 [DOI] [PubMed] [Google Scholar]
- Shin T., Kraemer D., Pryor J., Liu L., Rugila J., Howe L., Buck S., Murphy K., Lyons L., and Westhusin M. (2002). A cat cloned by nuclear transplantation. Nature 415, 859. [DOI] [PubMed] [Google Scholar]
- Takahashi K., and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Kunath T., Hadjantonakis A.K., Nagy A., and Rossant J. (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 [DOI] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K., and Surani M.A. (2010). Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 6, 468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda Y., and Kato Y. (1998). Not only inner cell mass cell nuclei but also trophectoderm nuclei of mouse blastocysts have a developmental totipotency. J. Reprod. Fertil. 113, 181–184 [DOI] [PubMed] [Google Scholar]
- Wakayama T., Perry A.C., Zuccotti M., Johnson K.R., and Yanagimachi R. (1998). Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 394, 369–374 [DOI] [PubMed] [Google Scholar]
- Watson E.D., and Cross J.C. (2005). Development of structures and transport functions in the mouse placenta. Physiology (Bethesda) 20, 180–193 [DOI] [PubMed] [Google Scholar]
- Wilmut I., Schnieke A.E., McWhir J., Kind A.J., and Campbell K.H. (1997). Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813 [DOI] [PubMed] [Google Scholar]
- Woods G.L., White K.L., Vanderwall D.K., Li G.P., Aston K.I., Bunch T.D., Meerdo L.N., and Pate B.J. (2003). A mule cloned from fetal cells by nuclear transfer. Science 301, 1063. [DOI] [PubMed] [Google Scholar]
- Wu T., Wang H., He J., Kang L., Jiang Y., Liu J., Zhang Y., Kou Z., Liu L., Zhang X., and Gao S. (2011). Reprogramming of trophoblast stem cells into pluripotent stem cells by Oct4. Stem Cells 29, 755–763 [DOI] [PubMed] [Google Scholar]
- Zeng F., and Schultz R.M. (2005). RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev. Biol. 283, 40–57 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Renard J.P., Le Friec G., Brochard V., Beaujean N., Cherifi Y., Fraichard A., and Cozzi J. (2003). Generation of fertile cloned rats by regulating oocyte activation. Science 302, 1179. [DOI] [PubMed] [Google Scholar]