Significance
HIV infection is associated with elevated inflammation and aberrant cellular immune activation. Indeed, the activation status of an HIV-infected individual is often more predictive of disease trajectory than viral load. Here, we highlight the importance of the replicative fitness of the transmitted viral variant in driving an early inflammatory state, characterized by T-cell activation and immune dysfunction. This impact on T-cell homeostasis is independent of protective host immune response genes and viral load. Highly replicating transmitted variants were also significantly more efficient at infecting memory CD4+ T cells, a population important for maintaining the latent viral reservoir. Together, these data provide a mechanism whereby viral replicative fitness acts as a major determinant of disease progression and persistence.
Keywords: HIV-1, Gag, replicative capacity, immune activation, pathogenesis
Abstract
HIV-1 infection is characterized by varying degrees of chronic immune activation and disruption of T-cell homeostasis, which impact the rate of disease progression. A deeper understanding of the factors that influence HIV-1–induced immunopathology and subsequent CD4+ T-cell decline is critical to strategies aimed at controlling or eliminating the virus. In an analysis of 127 acutely infected Zambians, we demonstrate a dramatic and early impact of viral replicative capacity (vRC) on HIV-1 immunopathogenesis that is independent of viral load (VL). Individuals infected with high-RC viruses exhibit a distinct inflammatory cytokine profile as well as significantly elevated T-cell activation, proliferation, and CD8+ T-cell exhaustion, during the earliest months of infection. Moreover, the vRC of the transmitted virus is positively correlated with the magnitude of viral burden in naive and central memory CD4+ T-cell populations, raising the possibility that transmitted viral phenotypes may influence the size of the initial latent viral reservoir. Taken together, these findings support an unprecedented role for the replicative fitness of the founder virus, independent of host protective genes and VL, in influencing multiple facets of HIV-1–related immunopathology, and that a greater focus on this parameter could provide novel approaches to clinical interventions.
From the start of the AIDS epidemic, HIV-1 infection has been characterized by a steady decline in CD4+ T cells that results in a state of overt immunodeficiency marked by an increased susceptibility to opportunistic infections and malignancies (1). Although a majority of HIV-1–infected individuals eventually progress to AIDS during their lifetime, they do so at drastically different rates (2). Several immunological abnormalities during HIV-1 infection have been identified that correlate with disease progression, such as a rapid and robust expression of proinflammatory cytokines, chronic immune activation, cellular exhaustion, and infection of vulnerable memory CD4+ T-cell subsets important for maintaining T-cell homeostasis (3). The degree to which these pathogenic mechanisms are triggered early in infection may explain the varying rates of disease progression among individuals.
It has been shown that the magnitude of immune activation during HIV-1 infection is established early, is relatively stable over time, and predicts the rate of disease progression better than viral load (4, 5). Successful antiretroviral treatment (ART) of HIV-1–infected individuals reduces viremia to undetectable levels, restores CD4+ T-cell counts to some degree, and significantly prolongs life (6). Despite this, ART does not fully restore immune function, and levels of residual immune activation are associated with an increased risk of morbidity and mortality (7, 8). Moreover, even with successful ART, the virus is not fully eradicated and viral rebound occurs upon treatment interruption (9). Strategies aimed at mitigating persistent immune activation and eradicating the latent viral reservoir will contribute immensely toward improving the quality of life of HIV-1–infected individuals and will help to curb the epidemic. Thus, a better understanding of the mechanisms driving HIV-1–induced immunological abnormalities and the processes by which they ultimately cause disease is crucial for unveiling novel avenues for pursuing these more advanced therapeutic interventions.
To date, research has primarily focused on identifying host factors that contribute to viral control and favorable disease outcomes, whereas viral characteristics have received less scrutiny (10, 11). HLA class I alleles such as HLA-B*57 and B*5801 have been shown to influence viral load and CD4+ T-cell decline through the induction of a strong CD8+ T-cell response that is able to target functionally vulnerable regions of the genome such as the structural protein Gag (10). The observation that not all individuals harboring such protective HLA class I alleles go on to become long-term nonprogressors suggests that other factors outside of host immunogenetics play a role in defining disease progression (12). Transmitted viral characteristics have been shown to impact viral load within heterosexual transmission pairs, suggesting that viral characteristics are heritable and can impact disease severity (11, 13, 14). Moreover, we recently showed that attenuated viral replicative capacity (vRC) of the transmitted virus, defined in vitro by the Gag sequence, was associated with a significant delay in CD4+ T-cell decline in individuals recently infected with HIV-1 subtype C (15). Because this clinical benefit appeared to be partially independent of set point viral load (SPVL), we hypothesized that high levels of transmitted/founder virus replication might initiate irreversible pathogenic events early in infection. Specifically, we hypothesized that high vRC might lead to exacerbated immune activation, elevated cellular dysfunction, and increased infection of memory CD4+ T-cell subsets, which in total might dictate the kinetics of subsequent disease progression (15).
To test this hypothesis, we have studied a unique cohort of 127 Zambian seroconvertors acutely infected (median 46 d postinfection) with HIV-1 subtype C that have up to 6 y of longitudinal follow-up. We show here that transmission of high-vRC HIV-1 is associated with a distinct inflammatory profile marked by significantly higher levels of proinflammatory cytokines, increased cellular immune activation and exhaustion, and higher levels of proviral burden in naive and central memory CD4+ T-cell subsets at early time points after infection. Thus, the replicative capacity of transmitted HIV-1, defined by the structural protein Gag, is a critical factor in defining early immune activation, the preservation or loss of CD4+ T-cell homeostasis, and the subsequent trajectory of disease progression. Interventions including early antiretroviral therapy or vaccine-induced immunity that impact these early events and that attenuate early viral replication will have a significant effect on the development of clinical disease.
Results
Viral Characteristics Determine HIV-1 Pathogenesis.
Previously, our work, and that of others, showed that transmitted viral characteristics significantly correlate with early SPVL (11, 13, 14) as well as CD4+ T-cell decline up to 3 y postinfection (15). Here, we sought to determine the underlying mechanisms by which vRC of transmitted HIV-1 impacts the trajectory of CD4 decline even in the context of viral control by previously identified host factors, such as protective HLA alleles, that also impact disease progression.
To assess the impact of the transmitted gag sequence on replicative capacity, we amplified the gag gene from plasma virus during acute infection time points (median 46 d after estimated date of infection), generated replication-competent virus by cloning the gag gene into a common proviral backbone (MJ4), and measured vRC in an in vitro cell culture assay as described previously (15, 16). In comparing six Gag-MJ4 chimeric viruses to transmitted/founder full-length infectious molecular clones derived from the same individuals, we find a strong positive correlation between the vRC of the chimeric viruses and the vRC of the full-length infectious molecular clones (Fig. S1). This indicates that although other genes undoubtedly play a role in defining in vitro HIV-1 replicative capacity, the contributions of gag are a significant component of the replicative capacity of the full-length virus.
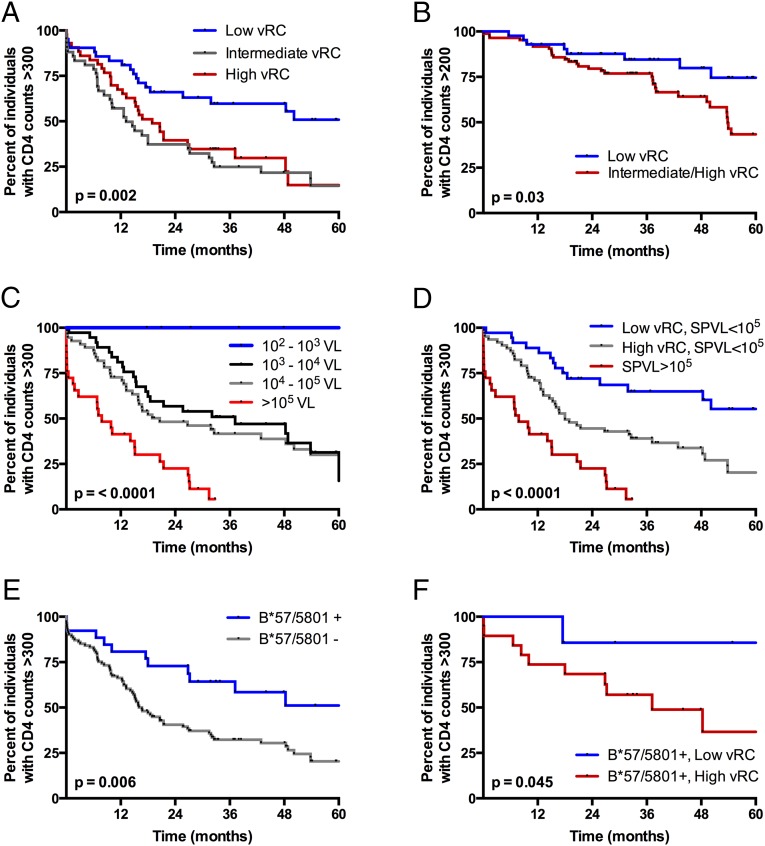
In this cohort of 127 acutely infected individuals from Zambia, low vRC significantly delayed the time to CD4+ T-cell counts <300 for up to 5 y postinfection (Fig. 1A, P = 0.002). The clearest benefit is observed with the lowest vRC tercile compared with the middle and highest tercile. A significant benefit remained even down to CD4+ T-cell counts of <200, the clinical definition of AIDS, when individuals infected with intermediate and highly replicating viruses were combined into one group (Fig. 1B, P = 0.03).
Fig. 1.
HIV-1 replicative capacity, when defined by the transmitted Gag sequence, is an independent predictor of CD4+ T-cell decline in ART-naive, HIV-1–infected individuals. Kaplan–Meier (KM) survival analysis was performed to evaluate the effects of viral and host factors on HIV-1 pathogenesis. Endpoints were defined as CD4+ T-cell counts <300 (A and C–F) and <200 (B). (A) KM analysis demonstrating the effect of in vitro vRC on CD4+ T-cell decline. Low vRC, middle vRC, and high vRC represent the lowest, middle, and upper third of vRC scores, respectively. (B) KM analysis demonstrating the effect of low vRC on the time to end-stage disease or CD4+ T-cell counts <200. (C) KM analysis demonstrating the effect of log10 increases in set point VL on CD4+ T-cell decline. (D) KM analysis demonstrating the additive effect of vRC and set point VL on CD4+ T-cell decline. (E) KM analysis demonstrating the effect of canonical protective HLA class I alleles, B*57 and B*5801, on CD4+ T-cell decline. (F) KM analysis demonstrating the effect of vRC in individuals expressing protective HLA-B*57 or B*5801. All statistics were generated from the log-rank test.
We have previously shown an association between early SPVL and vRC (15). Thus, we sought to more definitively determine if the replication capacity defined by the gag gene affected CD4 decline in a manner linked to, or independent of, the well-documented effect of early SPVL on subsequent disease progression. In this cohort, we found SPVLs >105 RNA copies/mL to be associated with poor outcomes for all volunteers (Fig. 1C, P < 0.0001); however, vRC significantly dichotomized the trajectory of CD4 decline (P < 0.0001) in individuals with SPVLs <105 (Fig. 1D), a majority (77%) of the cohort. This suggested independent, but additive effects, of both vRC and early SPVL on HIV disease progression. Moreover, whereas carriage of B*57/B*5801 alleles protects against CD4 decline in this cohort (Fig. 1E, P = 0.006), vRC significantly dichotomizes disease trajectories of those with these protective HLA alleles (Fig. 1F, P = 0.04). This is confirmed in a multivariable Cox proportional hazards model assessing the relative risk of vRC in the context of other well-established predictors of HIV disease progression. We find that low vRC, early SPVL, and canonical protective HLA class I alleles (B*57, B*5801) were each highly significant independent predictors of CD4 decline (Table 1). Of note, the protective effect of being infected with low-RC viruses as opposed to high-RC viruses was similar to that of HLA-B*57 or B*5801 alleles, as evidenced by their similar hazard ratios (Table 1).
Table 1.
Host and viral characteristics independently predict CD4+ T-cell decline
| Cox proportional hazards model,* time to CD4 <300 | |||
| Factors tested | HR | 95% CI | P value |
| Female | 1.10 | 0.67–1.70 | 0.78 |
| Low vRC, lowest tercile | 0.48 | 0.28–0.80 | 0.004 |
| B*57/5801 | 0.45 | 0.23–0.81 | 0.006 |
| Set point VL† | 10.00 | 2.86–44.1 | 0.0004 |
CI, confidence interval; HR, hazard ratio.
Multivariable Cox proportional hazards model with an endpoint defined as a single CD4+ T-cell count reading below 300 (n = 127).
Set point VL was defined as the earliest nadir viral load, between 3 mo and 9 mo postinfection, which remained stable for subsequent viral load readings.
Taken together, these data firmly establish vRC as a distinct contributor to HIV disease progression. Moreover, they suggest that vRC may modulate innate immune events very early after infection, which could alter both the establishment of an inflammatory state and the development of an effective adaptive immune response capable of controlling viremia. To further test this hypothesis, we assessed early levels of circulating inflammatory cytokines, immune activation, and exhaustion in T-cell compartments, as well as viral burden in different CD4+ T-cell subsets.
Viral Replicative Capacity Alters Early Inflammatory Cytokine Profiles.
Acute HIV infection is characterized by a rapid and robust expression of type I interferons (IFN-I), IFN-I–stimulated genes, and inflammatory cytokines (17). Disruption of the gut-associated lymphoid tissue (GALT) and subsequent microbial translocation have also been shown to contribute significantly to this inflammatory state, possibly through a positive feedback loop (18). This inflammatory response, particularly during chronic infection, contributes to disease progression (19, 20). Therefore, we analyzed the levels of 16 inflammatory cytokines, chemokines, and markers of gut damage and microbial translocation at or before seroconversion to assess the effect of vRC on the early inflammatory milieu [n = 33; previously dichotomized into low- and high-vRC phenotypes (15)]. We found that vRC was positively correlated with a number of inflammatory cytokines (Table 2), most notably IL-6 and IL-1β, two proinflammatory cytokines previously implicated in driving aberrant CD4+ T-cell turnover and impairing homeostatic proliferation (21). Of note, vRC was also strongly correlated with elevated levels of IL-10, an important antiinflammatory cytokine linked to T-cell dysfunction in HIV infection (22, 23).
Table 2.
High vRC is associated with significantly increased levels of inflammatory cytokines at seroconversion
| Analyte | Low vRC,* mean pg/mL | High vRC,† mean pg/mL | P value‡ |
| IL-6 | 1.88 | 3.94 | 0.004 |
| IL-10 | 5.5 | 10.73 | 0.004 |
| IL-1β | 0.21 | 0.57 | 0.008 |
| IFNγ | 4.38 | 10.26 | 0.014 |
| IP-10 | 639.56 | 1,108.43 | 0.018 |
| TNFα | 10.76 | 13.87 | 0.028 |
| IL-7 | 1.81 | 2.65 | 0.046 |
| IFNα2 | 21.09 | 31.48 | 0.048 |
Low vRC was defined as the lowest third of all replicative capacity scores in the larger cohort of 127 seroconvertors. Of this lowest third, 12 were randomly selected and analyzed for the presence of 16 analytes associated with inflammation.
High vRC was defined as the upper two-thirds of all replicative capacity scores in the larger cohort of 127 seroconvertors. Of this upper two-thirds, 21 were randomly selected and analyzed for the presence of 16 analytes associated with inflammation.
P values were generated using Student’s t test and reported P values are one tailed.
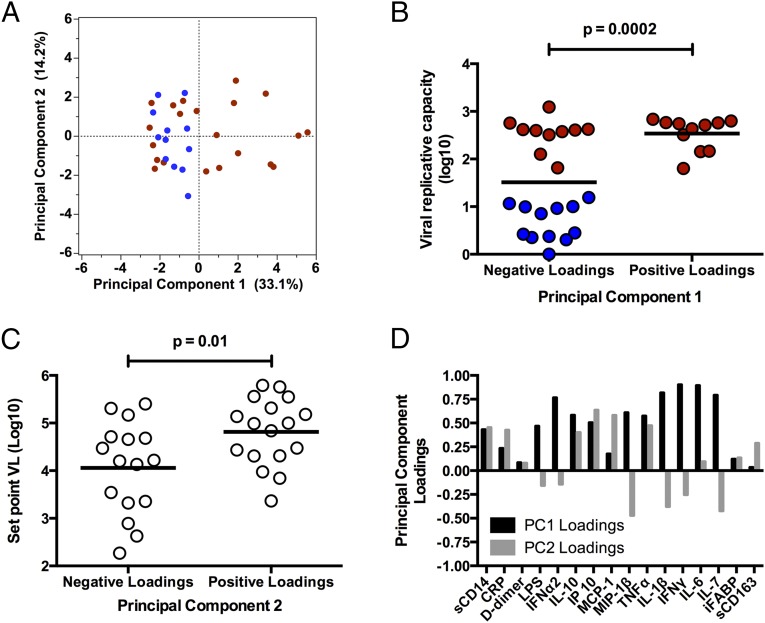
Because acute HIV-1 infection is associated with a complex cytokine storm (17), we aimed to define distinct inflammatory “profiles” that could explain the impact of vRC, by using an unsupervised data reduction tool, principal component analysis (PCA), which groups linear variables into combinations, termed principal components (PCs) (Fig. 2A). Strikingly, principal component 1 (PC1), which describes the greatest variation in the dataset, significantly correlates with vRC, in that individuals with positive loadings for PC1 (elevated levels of inflammation) tend to have higher vRC, whereas those with negative loadings (low levels of inflammation) are significantly enriched for poorly replicating viruses (Fig. 2B, P = 0.0002). Moreover, principal component 2 (PC2), which by definition is uncorrelated with PC1, describes the second-greatest variation in the data and is significantly correlated with SPVL (Fig. 2C, P = 0.01) but not with vRC. The differences in analyte loadings between PC1 and PC2 are shown in Fig. 2D, and the inflammatory cytokines that substantially contribute to PC1 and -2 are depicted schematically in Fig. S2. This result further highlights the independence of factors associated with initial viral replication and subsequent adaptive immune control of SPVL. Moreover, it demonstrates that viruses with high vRC are correlated with a distinct inflammatory cytokine profile characterized by a heightened type I and type II IFN response and elevated levels of key inflammatory cytokines such as IL-6 and IL-1β.
Fig. 2.
High vRC is associated with a distinct cytokine profile early in infection, characterized by elevated levels of inflammatory cytokines. Principal component analysis (PCA) was used to reduce dimensionality and extract principal components composed of linear combinations of 16 different analytes measured at the seroconversion time point (n = 33). (A) Principal component 1 (PC1) and principal component 2 (PC2) scores are depicted in a two-dimensional scatter plot for each individual. The first principal component (x axis), which describes the greatest variation in the dataset, has positive loadings associated with an increased production of key inflammatory cytokines. Red, individuals with high vRC; blue, individuals with low vRC. (B and C) Associations between vRC and PC1 loadings (B) and early set point VL and PC2 loadings (C). (D) Bar graph depicting analyte loadings for PC1 (black) and PC2 (gray). Loadings were extracted from the PCA correlation matrix and represent the degree of contribution of each analyte to both principal components. Positive loadings for PC1 are characterized by increased levels of key inflammatory cytokines, such as IL-6 and IL-1β, as well as increased production of type I (IFNα2) and type II (IFNγ) interferons. Positive loadings for PC2 are characterized by increased levels of IP-10 and MCP-1 and decreased levels of MIP-1β. Statistical comparisons were made using Student’s t test. P values are one tailed.
Replicative Capacity Is Associated with Levels of CD8+ T-Cell Activation and Exhaustion.
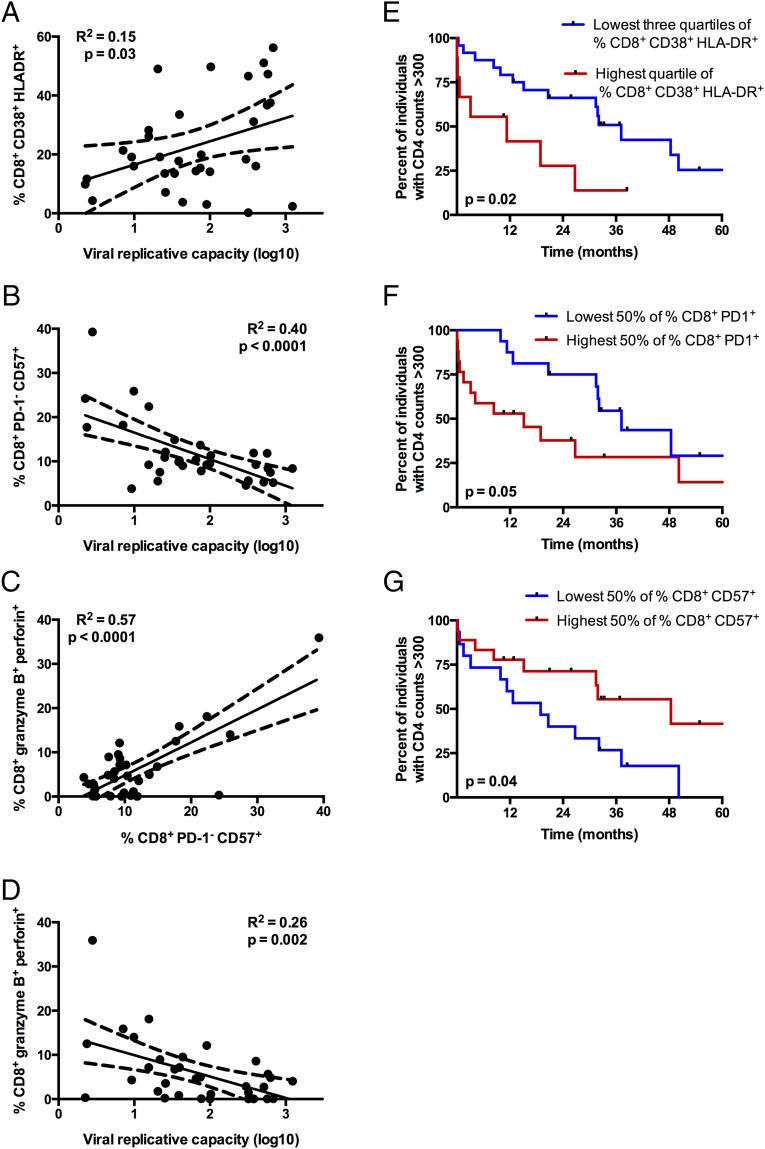
Chronic immune activation is a hallmark of HIV-1 infection; it often persists following ART and is a more reliable predictor of disease progression than viral load (4, 5). Therefore, we assessed the impact of vRC on levels of cellular immune activation by measuring the coexpression of CD38 and HLA-DR on CD8+ T cells isolated within 3 mo postinfection for a subset of individuals under study (n = 33). We found that vRC is positively correlated with the expression of CD38 and HLA-DR on CD8+ T cells (Fig. 3A, P = 0.03). Consistent with previous studies (4, 24), we observe that higher CD8+ T-cell activation in this cohort is associated with faster CD4+ T-cell decline (Fig. 3E, P = 0.02), thus positioning T-cell activation as a link between vRC and subsequent CD4+ T-cell decline.
Fig. 3.
High vRC is associated with increased CD8+ T-cell activation and lower cytotoxic potential. (A–D) Cryopreserved PBMCs were collected less than 3 mo after the estimated date of infection (n = 33) and stained with two multicolor flow cytometry panels evaluating either memory subsets and markers of cellular activation and exhaustion (A and B) or markers of T-cell cytotoxicity (C and D). (A) Correlation between vRC and the percentage of total CD8+ T cells coexpressing CD38 and HLA-DR. (B) Correlation between vRC and the percentage of CD8+ T cells that are CD57+ but remain PD-1−. (C) Association between the percentages of PD-1−/CD57+and granzyme B+/perforin+ CD8+ T cells. (D) Correlation between cytotoxic potential in CD8+ T cells, as measured by coexpression of granzyme B and perforin, and vRC. Correlation statistics were generated using linear regression. Solid lines indicate trend lines, and dashed lines represent 95% confidence bands. (E–G) Kaplan–Meier survival curves depict associations between markers of cellular activation and risk for CD4+ T-cell decline, defined as the time to CD4+ T-cell counts <300. Kaplan–Meier survival statistics were generated from the log-rank test.
In addition to cellular immune activation, CD8+ T-cell exhaustion is characteristic of pathogenic HIV/simian immunodeficiency virus (HIV/SIV) infection. Exhaustion of CD8+ T cells is marked by the increased expression of the inhibitory receptor, programmed death 1 (PD-1), and levels of PD-1 expression predict the rate of disease progression (25, 26). PD-1hi CD8+ T cells are typically CD57low (25, 26); however, the contrasting PD-1lowCD57high CD8+ T cells are more resistant to apoptosis (27). We find that a greater percentage of CD8+ T cells isolated from individuals infected with low-RC viruses displayed high levels of CD57 while maintaining low levels of PD-1 (Fig. 3B, P < 0.0001) relative to individuals infected by high-RC viruses. In CD8+ T cells, markers of exhaustion are often associated with impaired cytotoxic function (28). Indeed, dual expression of granzyme B and perforin was positively correlated with the frequency of PD-1−/CD57+ CD8+ T cells (Fig. 3C, P < 0.0001) and was inversely correlated with vRC (Fig. 3D, P = 0.002). Consequently, we find that PD-1 expression is associated with faster disease progression, whereas CD57 expression is protective in terms of CD4+ T-cell decline, in this cohort (Fig. 3 F and G; P = 0.05 and 0.04, respectively).
It is important to point out that although CD8+ T-cell activation was also significantly correlated with VL measured at the time the peripheral blood mononuclear cells (PBMCs) were collected (P = 0.01, R2 = 0.18; Fig. S3A), which is consistent with previous reports, the impact of VL on the reduction of either CD8+ PD-1−/CD57+ cells (P = 0.03, R2 = 0.14; Fig. S3B) or CD8+ granzyme B+/perforin+ cells (P = 0.02, R2 = 0.17; Fig. S3C) was much less significant than the observed impact of vRC on these parameters. Moreover, in a multivariate partial least-squares regression analysis, in which both vRC and plasma VL were important predictors of these CD8+ T-cell phenotypes, vRC consistently had the greater influence (Fig. S3D). Thus, reduced percentages of these potentially protective CD8+ cells are not simply a function of antigen load as reflected by VL in the plasma.
Taken together, these data suggest that individuals infected with low-RC viruses mount a more functional and less exhausted cytotoxic T-lymphocyte (CTL) response early in infection that may provide an extended clinical benefit for the individual.
Replicative Capacity Predicts Levels of CD4+ T-Cell Activation and Proliferation.
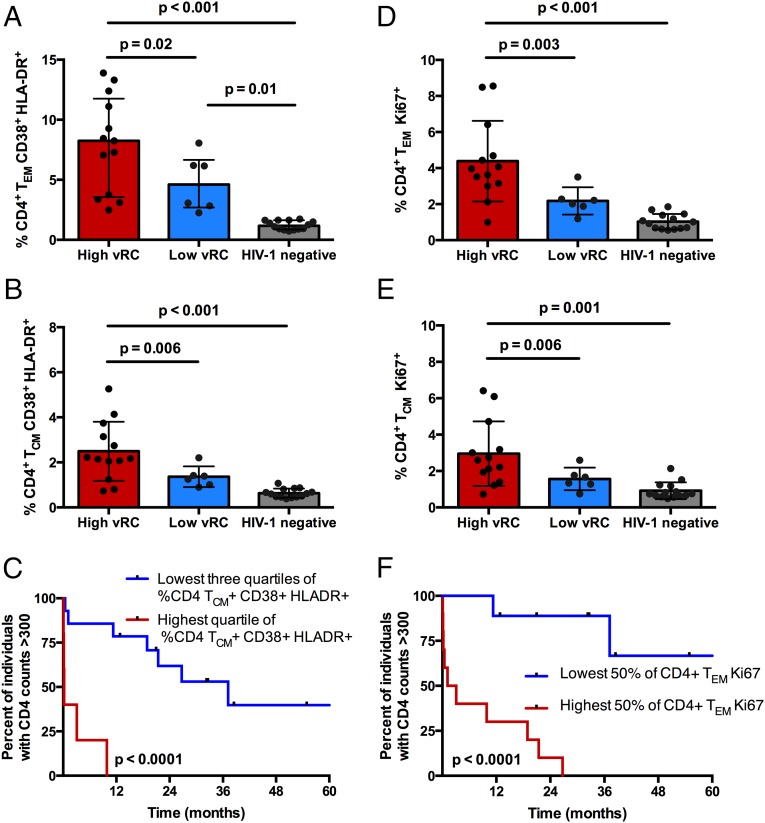
Immune activation in HIV-1 infection is associated with global immunological dysfunction, characterized by increased cellular turnover and ultimately the disruption of critical CD4+ T-cell homeostasis, indicative of progressive disease (29, 30). The percentages of CD4+ T cells expressing CD38/HLA-DR and Ki67 were measured in PBMCs isolated less than 3 mo postinfection for a subset of individuals (n = 19, see Materials and Methods for sample selection). Expression of these markers was assessed on total CD4+ T cells as well as in the context of different CD4+ T-cell subsets: naive (TN) (CD27+, CD45RO−), central memory (TCM) (CD27+/−, CD45RO+, CCR7+), and effector memory (TEM) (CD27+/−, CD45RO+, CCR7−).
High vRC was associated with increased expression of CD38 and HLA-DR on total (Fig. S4A, P = 0.02) and TEM (Fig. 4A, P = 0.02) CD4+ T cells; however, this association was most pronounced in the TCM (Fig. 4B, P = 0.006) compartment. Consistent with previous reports (4, 31), high levels of CD4+ T-cell activation (CD38+/HLA-DR+) are also associated with faster CD4+ T-cell decline (Fig. 4C, P < 0.0001). Furthermore, individuals infected with high-RC viruses exhibited a significantly greater percentage of CD4+ T cells expressing Ki67. This association was most striking in the effector memory T-cell subset (Fig. 4D, P = 0.003), but was also significant for total (Fig. S4B, P = 0.008) and central memory CD4+ T cells (Fig. 4E, P = 0.006). Increased CD4+ T-cell proliferation, demonstrated by elevated expression of Ki-67 on CD4+ T-cell subsets, was also associated with faster CD4+ T-cell decline, as has been reported previously (Fig. 4F, P < 0.0001) (30, 32). Furthermore, individuals infected with poorly replicating viruses displayed expression levels of CD38/HLA-DR and Ki67 on their CD4+ T cells, which, although somewhat elevated, for the most part did not differ significantly compared with those of HIV-uninfected Zambians (Fig. 4 B, D, and E). Thus, these results suggest that, shortly after transmission, individuals infected with low-vRC viruses preserve an immune system more similar to that of a healthy individual.
Fig. 4.
Viral RC is associated with increased cellular activation and proliferation in CD4+ T-cell memory subsets. Cryopreserved PBMCs isolated at 3 mo after the estimated date of infection from individuals with high vRC (n = 13) and low vRC (n = 6) as well as cryopreserved PBMCs from HIV-negative Zambians (n = 14) were stained for markers of activation and cellular turnover and for delineation of memory T-cell subsets. Statistical comparisons were made using the Student’s t test. (A and B) Comparison of the percentage of CD4+ TEM (A) and TCM (B) coexpressing the activation markers CD38 and HLA-DR in HIV-negative Zambians and individuals infected with either low-RC or high-RC viruses. (D and E) Comparison of the percentage of CD4+ TEM (D) and TCM (E) positive for Ki67 in HIV-negative Zambians and individuals infected with either low-RC or high-RC viruses. (C and F) Kaplan–Meier survival analyses (with an endpoint defined as CD4+ T-cell counts <300) depict associations between the percentage of memory CD4+ T cells expressing markers of activation (C) and proliferation (F) and the decline in total CD4+ T cells. Kaplan–Meier survival statistics were generated from the log-rank test.
Despite the stratification of these individuals into those infected with low- and high-RC viruses, an analysis of VL at the time these PBMC samples were collected did not reveal any significant difference in mean VL between the two groups (mean VL for low vRC = 4.5, mean VL for high vRC = 4.8; P = 0.26; Student’s t test).
Early Inflammatory Cytokine Profiles Associated with vRC Are Linked to Activated T-Cell Phenotypes.
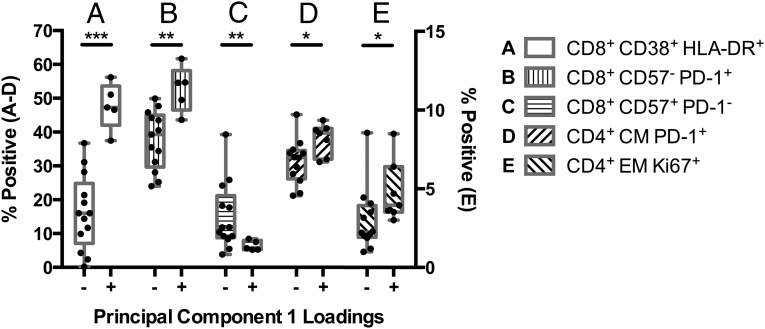
To determine whether changes in T-cell activation are associated with the distinct inflammatory cytokine profiles associated with vRC, we compared individuals with positive (increased inflammatory cytokine levels) and negative loadings for PC1 (Fig. 2). Individuals with positive loadings for PC1 presented with significantly increased CD8+ T-cell activation (Fig. 5A, P < 0.0001) and exhaustion (Fig. 5 B and C, P = 0.002 and 0.005, respectively). Positive loadings were further associated with increased frequency of PD-1+ CD4+ TCM cells (Fig. 5D, P = 0.01), as well as higher levels of CD4+ TEM cell proliferation, as measured by Ki67 expression (Fig. 5E, P = 0.04). Thus, higher levels of inflammatory cytokines are closely linked to the activated, exhausted T-cell phenotypes observed in individuals infected with high-RC viruses.
Fig. 5.
Inflammatory cytokine profiles associated with vRC correlate with T-cell activation. Inflammatory cytokines measured at seroconversion in 33 acutely infected individuals were used to define distinct cytokine profiles via principal Component Analysis (Fig. 2). In this group of 33 individuals, a subset of 18 and 19 individuals had also been immunophenotyped for CD8+ and CD4+ T-cell activation, respectively. Positive loadings (elevated inflammatory cytokines) were defined as a PC1 score >0 and negative loadings were defined as a PC1 score <0. (A–C) The differences in CD8+ T-cell activation in individuals with positive (elevated inflammatory cytokines) or negative loadings for PC1. (D and E) The differences in CD4+ T-cell activation in individuals with positive (elevated inflammatory cytokines) or negative loadings for PC1. The left-hand y axis displays the percentage of positive cells for A–D; the right-hand y axis displays the percentage of Ki67+ cells for E. (*P < 0.05, **P < 0.01, ***P < 0.001; statistical comparisons were made using Student’s t test, P values are one tailed.)
High vRC Is Associated with Increased Proviral Burden in CD4+ T-Cell Subsets.
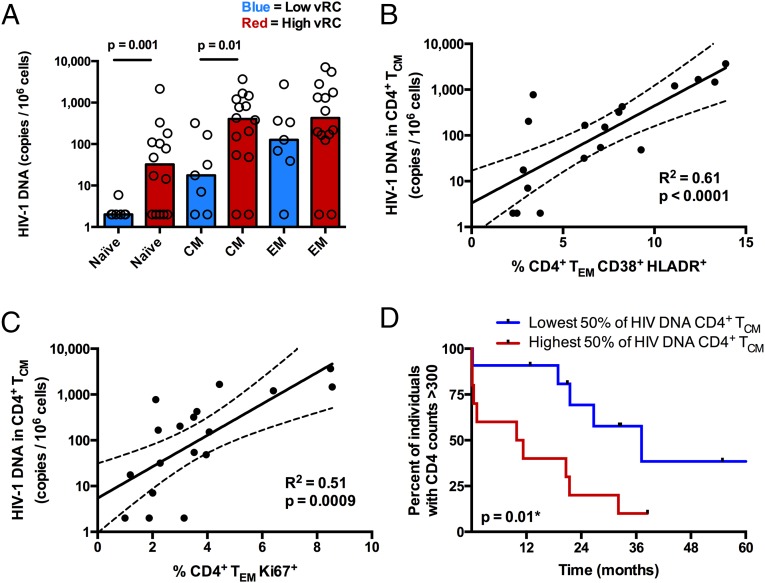
Establishment of the latent HIV reservoir occurs early during acute infection and sets the stage for viral persistence. Thus, even in the context of suppressive antiretroviral therapy, HIV-1 cannot be fully eradicated (9, 33, 34). To determine whether the replicative capacity of the transmitted/founder virus influences the proviral DNA burden during early infection, levels of cell-associated viral DNA were measured in naive, central memory, and effector memory CD4+ T-cell subsets sorted from PBMCs isolated 3 mo postinfection (n = 21). Despite no significant difference in mean VL between the two groups at this time point (mean VL for low vRC = 4.4, mean VL for high vRC = 4.8; P = 0.13; Student’s t test), we observed that high vRC was associated with a significant increase in the amount of cell-associated viral DNA in both TCM (P = 0.01) and TN (P = 0.001) CD4+ subsets (Fig. 6A). Because high vRC was associated with increased levels of immune activation and proliferation, we sought to determine whether a direct association exists between these markers and the magnitude of viral burden in TCM, a key population in CD4+ T-cell homeostasis. We find that HIV-1 DNA in TCM positively correlates with expression of CD38+/HLADR+ on TEM (Fig. 6B, P < 0.0001) and, to a lesser extent, on TCM (R2 = 0.3, P = 0.01). Similarly, HIV-1 DNA in TCM was positively correlated with Ki67 expression on TEM CD4+ cells (Fig. 6C, P = 0.0009). Moreover, higher levels of cell-associated HIV DNA in central memory CD4+ T cells were associated with an accelerated loss of CD4+ T cells (Fig. 6D, P = 0.01).
Fig. 6.
Viral RC correlates with the burden of HIV-1 viral DNA in CD4+ TCM and TN. (A) Cryopreserved PBMCs isolated 3 mo postinfection (n = 21) were stained for markers to delineate memory phenotypes, and CD4+ T cells were sorted into three distinct populations, TN (CD27+, CD45RO−), TEM (CD27+/−, CD45RO+, CCR7−), and TCM (CD27+/−, CD45RO+, CCR7+). Cell-associated HIV-1 DNA was quantified by real-time PCR amplification of the HIV-1 subtype C integrase gene. Cell copy number was normalized based on real-time PCR amplification of the human albumin gene. (A) Bar graphs depicting the magnitude of viral burden in TN, TCM, and TEM between individuals infected with either low-RC or high-RC viruses. (B and C) In a majority of the same individuals (n = 18), a fraction of unsorted PBMCs were stained for markers of cellular activation and proliferation. (B) Correlation between the magnitude of viral burden in CD4+ TCM and the percentage of activated (CD38+/HLA-DR+) CD4+ TEM. (C) Correlation between the magnitude of viral burden in CD4+ TCM and the percentage of proliferating (Ki67+) CD4+ TEM. Correlation statistics were generated using linear regression. Solid lines indicate trend lines, and dashed lines represent 95% confidence bands. (D) A Kaplan–Meier survival analysis (with an endpoint defined as CD4+ T-cell counts <300) depicts the effect of viral burden in CD4+ TCM on total CD4+ T-cell decline. Kaplan–Meier survival statistics were generated from the log-rank test.
Discussion
Recent studies have implicated transmitted viral characteristics in explaining the heritability of HIV-1 pathogenesis (11, 15, 35), and the results delineated above argue that a major component of this heritability is the replicative capacity of the transmitted/founder virus. Moreover, in this cohort of Zambian seroconvertors, acutely infected with HIV-1 subtype C, we demonstrate that vRC is a key contributor to CD4+ T-cell decline that can only be modulated but not reversed by the protective effects of HLA class I alleles such as B*57 and B*5801. This role in disease pathogenesis is consistent with studies of the lymphocytic choriomeningitis virus (LCMV) infection model, where the higher replicative capacity LCMV clone 13 strain goes on to induce chronic infection whereas the more attenuated Armstrong strain is quickly contained by the immune system (36). Although we were unable to accurately measure peak viral load in this cohort of HIV-1 acutely infected individuals, it seems likely that viruses with high replicative capacity have the potential to induce much higher levels of peak viremia, which then induces exacerbated immunopathology that cannot be completely reversed by the immune response. Thus, it is clear that early interactions between the virus and the host have the potential to set in motion a series of events that ultimately define the trajectory of disease progression.
The first interactions between HIV-1 and the host immune response occur at the interface of the virus and the innate immune system. Viral replication results in an initial cytokine storm that is thought to be largely deleterious (17). We show here that the replicative capacity of HIV-1 modulates the magnitude and composition of the early inflammatory milieu, perhaps through differential activation of innate sensing mechanisms that lead to multiple inflammatory consequences. Notably, the inflammatory state associated with high vRC is characterized by elevated levels of key mediators, such as IFNα, IL-1β, and IL-10, which are known to drive pathogenesis (37).
Elevated levels of IFNα can be detected in HIV-infected individuals as early as 6 d after the presence of detectable plasma viremia (17). As the first plasma samples available for analysis in this cohort were collected a median of 46 d postinfection, our measurements most likely reflect the contraction phase. It is reasonable to postulate that in HIV-1 infection the degree of this contraction could significantly affect severity of disease. This is supported by data from the SIV model, where robust expression of IFN-stimulated genes (ISGs) is observed in both natural SIV infection of sooty mangabeys and pathogenic infection of rhesus macaques, but where expression of these ISGs is quickly resolved in sooty mangabeys by an active down-regulation process (38). Our data show that individuals infected with highly replicating viruses are less able to resolve acute levels of IFNα (and by extension, ISGs) by the time of seroconversion, consistent with what is observed in pathogenic SIV infection.
Additionally, vRC was positively correlated with levels of IL-1β, a highly inflammatory cytokine associated with pyroptosis, an inflammatory form of programmed cell death recently linked to abortive HIV infection of quiescent CD4+ T cells (39). It is likely that increased viral production during the acute phase of HIV-1 infection increases the occurrence of abortive infection. This would escalate pyroptotic cell death, drive the production and release of IL-1β, and serve to foment immune activation and exacerbated immunopathology.
Enhanced production of IL-10 was also observed in individuals infected with high-RC viruses. Although IL-10 is traditionally seen as an antiinflammatory cytokine capable of reducing inflammation, in this setting it may be a surrogate of hyperactivation of the innate immune response. Previous studies demonstrated that increased PD-1 expression by monocytes, due to the presence of microbial products and inflammatory cytokines in HIV-1 infection, can lead to the production IL-10, which further disrupts T-cell function (23). Moreover, IL-10 blockade restored the antigen-induced proliferation of HIV-specific CD4+ and CD8+ T cells in vitro, as well as the production of effector cytokines by HIV-specific CD4+ T cells (22). Thus, in the present study, enhanced IL-10 production may reflect a dysregulated innate and adaptive immune response brought on by intensified viral replication during acute infection.
Previous studies showed that the level of immune activation is established very early after HIV-1 infection and predicts disease progression even when viral loads are suppressed either immunologically or by ART (4, 5, 8). For the first time to our knowledge, we demonstrate that levels of aberrant immune activation in CD8+ and CD4+ T cells can be significantly attributed to the replicative capacity of the transmitted virus. We further link the increased activation, exhaustion, and proliferation of T cells to inflammatory cytokine profiles, suggesting a mechanism by which vRC, through induction of an early innate immune response, influences levels of chronic cellular immune activation. This is supported by previously reported associations between CD8+ T-cell activation and the presence of inflammatory cytokines (40, 41).
Whether the elevated inflammatory response and exacerbated immunopathology associated with highly replicating viruses are the consequence of molecular signatures specific to Gag and TRIM5 sensing (42) or higher overall antigen load remains to be determined. Increased vRC likely results in elevated levels of HIV-1 epitope presentation and recognition by CTLs, which in turn could lead to aberrant T-cell activation via T-cell receptor (TCR) overstimulation (43). In support of this hypothesis, we found vRC to be positively correlated with PD-1 expression on CD8+ T cells. Because PD-1 expression in chronic HIV infection has previously been shown to be most pronounced on HIV-specific CD8+ T-cell populations (25, 26), this is consistent with increased circulating antigen being a factor linking vRC to increased markers of immune activation and exhaustion in the CD8+ T-cell compartment. It is important to note, however, that loss of protective CD8+ PD-1−/CD57+ cells at these early time points is strongly associated with increasing vRC and to a lesser degree with contemporaneous VL, suggesting that it is not simply antigen load that defines this pathogenic effect. A key factor governing this observation may be that vRC is a heritable characteristic of the virus that remains relatively stable at these early times after infection, whereas, in contrast, VL is in significant flux due to the onset of the adaptive immune response. Thus, vRC, as a measurable phenotypic trait, may provide a correlate of viral damage to the immune system in tissues or at even earlier times where and when VL is difficult to measure.
Finally, we demonstrated that the amount of HIV-1 DNA harbored by both TCM and naive (CD27+/CD45RO−) CD4+ T cells is highly associated with viral replicative capacity. It is possible that the increased levels of HIV-1 DNA found in the “naive” CD4+ T cells could be due in part to the presence of T memory stem cells (TSCM) that would be found within our sorted naive T-cell populations (44, 45). These long-lived memory T cells with stem cell-like properties are susceptible to HIV-1 or SIV in pathogenic infections but are spared in nonpathogenic SIV disease (46). Similarly, the long-lived TCM CD4+ cells are spared in nonpathogenic SIV infection, and infection of this subset has been linked to the immunopathology of HIV-1 infection (30, 47). Moreover, recent evidence demonstrates that reduced infection of TSCM and TCM CD4+ cells is associated with a nonprogression phenotype in viremic individuals (48). The results of the present study provide further evidence that HIV-1 infection of these cell types occurs early, is linked to increased CD4+ T-cell activation, and in TCM significantly predicts disease progression, perhaps by disrupting the capacity to renew the CD4+ TEM population (30). TCM CD4+ cells have also been highlighted as an integral population for the maintenance of latency and viral persistence (33). Our findings suggest that the extent to which HIV infects this population and establishes a latent reservoir might be influenced by characteristics of the transmitted/founder virus.
Taken together, these results support an unprecedented role for the viral replicative capacity of the transmitted/founder virus, in determining the early inflammatory state and general immune dysfunction of HIV-1–infected individuals as well as their subsequent disease trajectory. The data presented here suggest that viral characteristics can provide independent information about an individual’s risk for disease progression and point to a previously unidentified target for interventions to reduce immune activation and viral burden. Vaccine-induced immune responses or interventions that effectively attenuate vRC in the earliest stages of infection could not only have a dramatic impact on viral control and disease progression, but, based on our recent findings (35) also impact the efficiency of subsequent transmission to other partners. Moreover, reducing the size of the viral burden in key T-cell populations before antiretroviral treatment could augment cure strategies aimed at eliminating the latent reservoir.
Materials and Methods
Study Subjects.
All participants in the Zambia Emory HIV Research Project (ZEHRP) discordant couples cohort in Lusaka, Zambia were enrolled in human subjects protocols approved by both the University of Zambia Research Ethics Committee and the Emory University Institutional Review Board. Before enrollment, individuals received counseling and signed a written informed consent form agreeing to participate.
The subjects included in this study were selected from the ZEHRP cohort based on being recently infected with HIV-1. All subjects were initially seronegative partners within serodiscordant cohabitating heterosexual couples that subsequently seroconverted. All subjects were antiretroviral therapy naive and were identified a median of 46 (interquartile range = 33–49) d after the estimated date of infection (EDI). The algorithm used to determine the EDI has been previously described (49). All subjects were infected by HIV-1 subtype C viruses.
Generation of Gag-MJ4 Chimeras and Transmitted/Founder Full-Length Infectious Molecular Clones.
Gag-MJ4 chimeras were generated from frozen plasma isolated at the seroconversion time point for 127 subjects as previously described (15, 16). Briefly, nested PCR primer sets were used to amplify the gag gene from patient plasma and cloned into the MJ4 provirus, using a restriction enzyme-based approach. Replication-competent virus was generated by transfection of 293T cells with proviral plasmids, and infectious units were assessed on the TZM-bl indicator cell line. GXR25 cells were infected at a constant multiplicity of infection of 0.05, and viral production in the supernatant was evaluated using a radiolabeled reverse transcriptase assay at days 2, 4, and 6 postinfection. Replication capacity scores were generated by dividing the log10-transformed slope of replication from days 2–6 for each Gag-MJ4 chimera by the log10-transformed slope of replication of wild-type MJ4. The sequences of the cloned gag genes are available at GenBank under accession nos. KP715723–KP715849.
Full-length genome cDNA synthesis and near full-length single genome amplification were performed as described previously (50) on seroconversion plasma samples for six individuals where Gag-chimera vRC had been measured. Sequencing of multiple amplicons allowed for confirmation of single-variant transmission by star-like phylogeny and to infer the near full-length sequence of the transmitted/founder (TF) virus. Full-length genome infectious molecular clones of the entire TF sequence were constructed as described previously (50). Viral stocks were generated and infectious units assessed in the same way as Gag-MJ4 chimera viruses. Replication curves were generated by infecting PHA-stimulated PMBCs at a constant multiplicity of infection of 0.01. Viral output was assessed at days 0, 2, 4, 6, and 8 d postinfection via a radiolabeled reverse transcriptase assay, and replication scores were generated in the same way as for Gag-MJ4 chimeras.
Evaluation of Plasma Cytokines.
Plasma levels of cytokines and chemokines were measured using MILLIPLEX Human Cytokine/Chemokine detection kits (Millipore). High sensitivity kits were used for measurement of IFNγ, IL-1β, IL-6, and IL-7 and regular sensitivity kits were used for IFNα2, IL-10, IP-10, MCP-1, MIP-1β, and TNFα and were used according to the manufacturer’s instructions. Samples were run in duplicate with all individuals on the same plate and wells with low bead count or coefficient of variance >30% were excluded from subsequent analysis. Plates were read on the Bio-Plex 3D Suspension Array System (Bio-Rad). Levels of sCD14 (R&D Systems), CRP (Millipore), sCD163 (Trillium Diagnostics), and D-dimer (American Diagnostica) were all measured using standard ELISA-based assays according to the manufacturer’s instructions. Measurement of LPS levels was performed using the LAL Chromogenic Endotoxin Quantification kit (American Diagnostica). Intestinal fatty acid binding protein (I-FABP) was measured using a commercially available ELISA DuoSet assay (R&D Systems) according to the manufacturer’s instructions with minor adjustments. Plasma samples were diluted to 10% (vol/vol) in diluent from the R&D Systems soluble CD14 ELISA kit (DC140) and plates were blocked with Sigma Blocking Buffer.
Flow Cytometry Analysis.
Cryopreserved PBMCs isolated from 33 HIV-1–infected Zambians at a median of 49 d after the estimated date of infection were analyzed by flow cytometry to immunophenotype CD8+ T cells. The 12-parameter cytometric staining panel is depicted in Table S1. Frozen PBMCs were thawed, washed once with R20 [RPMI 1640 (Sigma) containing 20% FCS (Sigma), 1% 1 M Hepes buffer (Sigma), 1% l-glutamine (Sigma), 1% penicillin–streptomycin (Sigma), and 1% sodium pyruvate (Sigma)], and then rested overnight in 4 mL of R20 at 37 °C, 5% CO2, 95% humidity.
For each individual, 106 PBMCs per well were washed once with PBS (Sigma) before staining. When evaluating markers of T-cell cytotoxicity, an 11-parameter cytometric panel, shown in Table S2, was used. For the intracellular markers granzyme B and perforin, Cytofix/Cytoperm (BD) was used for permeabilization and staining per the manufacturer’s protocol.
CD4+ T-cell memory populations and markers of activation and proliferation were analyzed in a separate flow analysis, using a 12-parameter cytometric panel shown in Table S3. Cryopreserved PBMCs isolated from a subset of 19 HIV-1–infected Zambians at 3 mo postseroconversion as well as 14 uninfected healthy Zambians were assessed for activation and proliferation, using 12-parameter flow cytometric analysis. The individuals were chosen based on replication capacity phenotype, overlap with other parameters measured, and the sample availability at this time point.
Approximately 20 million PBMCs from each individual were first thawed and then washed twice with 10 mL of complete RPMI supplemented with 2 μL of DNase. Once the cells were counted using an automated cell counter, ∼2 million cells were set aside for staining with the above 12-parameter panel, and the rest of the cells were used for sorting if they were derived from an HIV-1–infected individual (see below for further methods on sorting). Cells were then washed with 3 mL of Dulbeco’s PBS without Ca+/Mg+ (Invitrogen) and stained for 5 min at room temperature with Aqua Live/Dead amine dye-AmCyan (Invitrogen). Anti-CCR7-PE-CY7 was added to the cells and incubated at 37 °C for 15 min. The rest of the monoclonal antibodies were then added and incubated at room temperature for 30 min (without anti-Ki67). The cells were then permeabilized and stained for the intracellular marker Ki67 for 30 min at room temperature, using the BD perm/wash kit (Fisher) following the manufacturer’s directions.
All flow cytometry data were collected on an LSRII cytometer with FACSDiVa Version 6.1.3 software. Analyses of these data were performed using FlowJo Version 9.7.5 software (TreeStar).
Detection of Cell-Associated HIV-1 Viral DNA in CD4+ T-Cell Compartments.
Cryopreserved PBMCs isolated from a subset of 21 HIV-1–infected individuals at 3 mo postseroconversion were stained to delineate memory CD4+ T-cell subsets, using the seven-parameter cytometric panel displayed in Table S4. Samples were sorted into naive CD4 T cells (CD3+, CD4+, CD8−, CD27+, CD45RO−), central memory CD4 T cells (CD3+, CD4+, CD8−, CD27+/−, CD45RO+, CCR7+), and effector memory CD4 T cells (CD3+, CD4+, CD8−, CD27+/−, CD45RO+, CCR7−), using a FACSAria II flow cytometer (Becton Dickinson). Sorted populations were checked for purity and were found to be between 94.5% and 99.4% specific for the population desired.
Live, sorted CD4+ T-cell subsets were immediately lysed and genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen). Samples were eluted in nuclease-free water and analyzed for DNA concentration. DNA samples were analyzed by quantitative real-time PCR (qPCR) for total cell-associated HIV DNA. Approximately 10,000 cell equivalents of genomic DNA were loaded in a 50-µL qPCR reaction, using a custom-designed HIV clade C primer and probe set designed to detect HIV int sequences derived from the patient population. Albumin was used as an internal control to quantify the number of genomes present. Both HIV and albumin absolute copies were determined using an external standard curve. HIV clade C primer and probe sequences are Fwd 5′-GTTATYCCAGCAGARACAGG-3′, Rev 5′-TGACTTTGRGGATTGTAGGG-3′, and probe 5′-RGCAGCCTGYTGGTGGGC-3′. Human albumin primer and probe sequences are Fwd 5′-TGCATGAGAAAACGCCAGTAA-3′, Rev 5′-ATGGTCGCCTGTTCACCAA-3′, and probe 5′-FAM-TGACAGAGTCACCAAATGCTGCACAGAA-3′. qPCRs were performed using the Taqman Universal master mix (Life Technologies), 0.2 μM of each primer, and 0.125 μM of probe. All assays were performed on the ABI 7500 system (Life Technologies).
Statistical Analyses.
All statistical analysis was performed using JMP, version 11 (SAS Institute). All bivariate continuous correlations were performed using standard linear regression. One-way comparison of means was performed using Student’s t test, and one-tailed P values are reported. Kaplan–Meier survival curves and Cox proportional hazards models were performed using an endpoint defined as a single CD4+ T-cell count reading less than 300, unless otherwise specified, and statistics reported for survival analyses were generated from the log-rank test.
PCA was performed using the JMP version 11 statistical package. Extreme positive values for each of the 16 analytes measured in plasma were Winsorized to the 90th percentile. Missing values were imputed using multiple linear regression models, and individuals for whom more than three cytokine values were missing were excluded. All 16 analytes that were measured in 33 individuals were used to extract latent variables.
Depiction of cytokine loadings for PC1 and PC2 (Fig. S2) was generated using Cytoscape v3.0. The loading matrix (list of pairwise correlations between each latent variable and each of the 16 analytes tested) was extracted from the PCA and used to represent the strength of contribution of each analyte to either PC1 or PC2.
Multivariate partial least-squares regression portraying the independent contributions of vRC and VL to CD8+ T-cell activation phenotypes (Fig. S3D) was performed with JMP version 11, using the SIMPLS algorithm and K-fold cross validation, where K = 7.
Supplementary Material
Acknowledgments
We thank all the volunteers in Zambia who participated in this study and all the staff at the Zambia–Emory HIV Research Project in Lusaka who made this study possible. We thank Emmanuel Cormier, Jon Allen, Sheng Luo, and Paul Farmer for technical assistance, sample management, and database management. We also thank Rafick-Pierre Sékaly and Ali Filali for helpful discussions and important biostatistics support. We thank Kiran Gill and Barbara Cervasi at the Emory Vaccine Center/Emory Center for AIDS Research Flow Core (Grant P30 AI050409) for performing cell sorting and for assisting with flow cytometry experiments. This study was funded by Grants R01 AI64060 and R37 AI51231 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIAID, NIH) (to E.H.). This work was also supported in part by the Virology Core at the Emory Center for AIDS Research (Grant P30 AI050409 from the NIAID, NIH), by the Yerkes National Primate Research Center Base Grant (2P51RR000165-51) through the National Center for Research Resources P51RR165, and by the Office of Research Infrastructure Programs/OD P51OD11132. This study is supported in part by the International AIDS Vaccine Initiative (IAVI) (S.A.A.), whose work is made possible by generous support from many donors, including the Bill & Melinda Gates Foundation, the Ministry of Foreign Affairs of Denmark, Irish Aid, the Ministry of Finance of Japan, the Ministry of Foreign Affairs of the Netherlands, the Norwegian Agency for Development Cooperation, the UK Department for International Development, and the US Agency for International Development (USAID). The full list of IAVI donors is available www.iavi.org. D.T.C., M.J.D., and J.L.P. were supported in part by Action Cycling Fellowships. E.H. is a Georgia Eminent Scholar.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KP715723–KP715849).
See Commentary on page 3591.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421607112/-/DCSupplemental.
References
- 1.Masur H, et al. CD4 counts as predictors of opportunistic pneumonias in human immunodeficiency virus (HIV) infection. Ann Intern Med. 1989;111(3):223–231. doi: 10.7326/0003-4819-111-3-223. [DOI] [PubMed] [Google Scholar]
- 2.Muñoz A, et al. Multicenter AIDS Cohort Study Group Acquired immunodeficiency syndrome (AIDS)-free time after human immunodeficiency virus type 1 (HIV-1) seroconversion in homosexual men. Am J Epidemiol. 1989;130(3):530–539. doi: 10.1093/oxfordjournals.aje.a115367. [DOI] [PubMed] [Google Scholar]
- 3.Sodora DL, Silvestri G. Immune activation and AIDS pathogenesis. AIDS. 2008;22(4):439–446. doi: 10.1097/QAD.0b013e3282f2dbe7. [DOI] [PubMed] [Google Scholar]
- 4.Deeks SG, et al. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104(4):942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- 5.Giorgi JV, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis. 1999;179(4):859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 6.Palella FJ, Jr, et al. HIV Outpatient Study Investigators Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med. 1998;338(13):853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 7.Baker JV, et al. Terry Beirn Community Programs for Clinical Research on AIDS (CPCRA) Poor initial CD4+ recovery with antiretroviral therapy prolongs immune depletion and increases risk for AIDS and non-AIDS diseases. J Acquir Immune Defic Syndr. 2008;48(5):541–546. doi: 10.1097/QAI.0b013e31817bebb3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunt PW, et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis. 2003;187(10):1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 9.Pierson T, McArthur J, Siliciano RF. Reservoirs for HIV-1: Mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu Rev Immunol. 2000;18:665–708. doi: 10.1146/annurev.immunol.18.1.665. [DOI] [PubMed] [Google Scholar]
- 10.Martin MP, Carrington M. Immunogenetics of HIV disease. Immunol Rev. 2013;254(1):245–264. doi: 10.1111/imr.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser C, et al. Virulence and pathogenesis of HIV-1 infection: An evolutionary perspective. Science. 2014;343(6177):1243727. doi: 10.1126/science.1243727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Migueles SA, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97(6):2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goepfert PA, et al. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med. 2008;205(5):1009–1017. doi: 10.1084/jem.20072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yue L, et al. Cumulative impact of host and viral factors on HIV-1 viral-load control during early infection. J Virol. 2013;87(2):708–715. doi: 10.1128/JVI.02118-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prince JL, et al. Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog. 2012;8(11):e1003041. doi: 10.1371/journal.ppat.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claiborne DT, Prince JL, Hunter E. A restriction enzyme based cloning method to assess the in vitro replication capacity of HIV-1 subtype C Gag-MJ4 chimeric viruses. J Vis Exp. 2014;90:e51506. doi: 10.3791/51506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stacey AR, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83(8):3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12(12):1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 19.Kuller LH, et al. INSIGHT SMART Study Group Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med. 2008;5(10):e203. doi: 10.1371/journal.pmed.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandler NG, et al. INSIGHT SMART Study Group Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203(6):780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shive CL, et al. Inflammatory cytokines drive CD4+ T-cell cycling and impaired responsiveness to interleukin 7: Implications for immune failure in HIV disease. J Infect Dis. 2014;210(4):619–629. doi: 10.1093/infdis/jiu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brockman MA, et al. IL-10 is up-regulated in multiple cell types during viremic HIV infection and reversibly inhibits virus-specific T cells. Blood. 2009;114(2):346–356. doi: 10.1182/blood-2008-12-191296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Said EA, et al. Programmed death-1-induced interleukin-10 production by monocytes impairs CD4+ T cell activation during HIV infection. Nat Med. 2010;16(4):452–459. doi: 10.1038/nm.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, et al. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16(2):83–92. doi: 10.1097/00042560-199710010-00003. [DOI] [PubMed] [Google Scholar]
- 25.Day CL, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443(7109):350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 26.Trautmann L, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med. 2006;12(10):1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- 27.Petrovas C, et al. Differential association of programmed death-1 and CD57 with ex vivo survival of CD8+ T cells in HIV infection. J Immunol. 2009;183(2):1120–1132. doi: 10.4049/jimmunol.0900182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang D, et al. Most antiviral CD8 T cells during chronic viral infection do not express high levels of perforin and are not directly cytotoxic. Blood. 2003;101(1):226–235. doi: 10.1182/blood-2002-03-0791. [DOI] [PubMed] [Google Scholar]
- 29.Douek DC. Disrupting T-cell homeostasis: How HIV-1 infection causes disease. AIDS Rev. 2003;5(3):172–177. [PubMed] [Google Scholar]
- 30.Okoye A, et al. Progressive CD4+ central memory T cell decline results in CD4+ effector memory insufficiency and overt disease in chronic SIV infection. J Exp Med. 2007;204(9):2171–2185. doi: 10.1084/jem.20070567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunt PW, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197(1):126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orendi JM, et al. Activation and cell cycle antigens in CD4+ and CD8+ T cells correlate with plasma human immunodeficiency virus (HIV-1) RNA level in HIV-1 infection. J Infect Dis. 1998;178(5):1279–1287. doi: 10.1086/314451. [DOI] [PubMed] [Google Scholar]
- 33.Chomont N, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 35.Carlson JM, et al. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345(6193):1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, et al. Visualizing antigen-specific and infected cells in situ predicts outcomes in early viral infection. Science. 2009;323(5922):1726–1729. doi: 10.1126/science.1168676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keating SM, Jacobs ES, Norris PJ. Soluble mediators of inflammation in HIV and their implications for therapeutics and vaccine development. Cytokine Growth Factor Rev. 2012;23(4-5):193–206. doi: 10.1016/j.cytogfr.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosinger SE, et al. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest. 2009;119(12):3556–3572. doi: 10.1172/JCI40115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doitsh G, et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505(7484):509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liovat AS, et al. Acute plasma biomarkers of T cell activation set-point levels and of disease progression in HIV-1 infection. PLoS ONE. 2012;7(10):e46143. doi: 10.1371/journal.pone.0046143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmons RP, et al. HIV-1 infection induces strong production of IP-10 through TLR7/9-dependent pathways. AIDS. 2013;27(16):2505–2517. doi: 10.1097/01.aids.0000432455.06476.bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472(7343):361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138(1):30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 44.Gattinoni L, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17(10):1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lugli E, et al. Superior T memory stem cell persistence supports long-lived T cell memory. J Clin Invest. 2013;123(2):594–599. doi: 10.1172/JCI66327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cartwright EK, et al. Divergent CD4+ T memory stem cell dynamics in pathogenic and nonpathogenic simian immunodeficiency virus infections. J Immunol. 2014;192(10):4666–4673. doi: 10.4049/jimmunol.1303193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paiardini M, et al. Low levels of SIV infection in sooty mangabey central memory CD4+ T cells are associated with limited CCR5 expression. Nat Med. 2011;17(7):830–836. doi: 10.1038/nm.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klatt NR, et al. Limited HIV infection of central memory and stem cell memory CD4+ T cells is associated with lack of progression in viremic individuals. PLoS Pathog. 2014;10(8):e1004345. doi: 10.1371/journal.ppat.1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haaland RE, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009;5(1):e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deymier MJ, et al. Particle infectivity of HIV-1 full-length genome infectious molecular clones in a subtype C heterosexual transmission pair following high fidelity amplification and unbiased cloning. Virology. 2014;468–470:454–461. doi: 10.1016/j.virol.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.