Significance
Plus-sense RNA viruses cause diverse pathologies in humans. Viral RNA genomes are selected to encode information both in their primary sequences and in their higher-order tertiary structures required to replicate and to evade host immune responses. We interrogated the physical structures of three evolutionarily divergent hepatitis C virus (HCV) RNA genomes using high-throughput chemical probing and found, along with all previously known RNA-structure–based regulatory elements, diverse previously uncharacterized structures that impact viral replication. We also characterized strategies by which the HCV genomic RNA structure masks detection by innate immune sensors. This structure-first strategy for comparative analysis of genome-wide RNA structure can be broadly applied to understand the contributions of higher-order genome structure to viral replication and pathogenicity.
Keywords: RNA structure, evolution, motif discovery, functional validation
Abstract
Hepatitis C virus (HCV) infects over 170 million people worldwide and is a leading cause of liver disease and cancer. The virus has a 9,650-nt, single-stranded, messenger-sense RNA genome that is infectious as an independent entity. The RNA genome has evolved in response to complex selection pressures, including the need to maintain structures that facilitate replication and to avoid clearance by cell-intrinsic immune processes. Here we used high-throughput, single-nucleotide resolution information to generate and functionally test data-driven structural models for three diverse HCV RNA genomes. We identified, de novo, multiple regions of conserved RNA structure, including all previously characterized cis-acting regulatory elements and also multiple novel structures required for optimal viral fitness. Well-defined RNA structures in the central regions of HCV genomes appear to facilitate persistent infection by masking the genome from RNase L and double-stranded RNA-induced innate immune sensors. This work shows how structure-first comparative analysis of entire genomes of a pathogenic RNA virus enables comprehensive and concise identification of regulatory elements and emphasizes the extensive interrelationships among RNA genome structure, viral biology, and innate immune responses.
Hepatitis C virus (HCV) currently infects over 170 million people. There is no vaccine, and therapy, generally involving treatment with IFN and ribavirin, is often ineffective (1). Efficacious anti-HCV therapeutics are becoming available (2), but the extent to which they will mitigate the hepatitis C disease burden remains to be seen. Roughly 70% of acutely infected individuals fail to clear the virus and become lifelong HCV carriers, at risk for progressive hepatic fibrosis, cirrhosis, and hepatocellular carcinoma (3).
HCV genomes are single-stranded, ∼9,650-nt, messenger-sense RNA molecules (4). The naked RNA initiates autonomous replication when transfected into cells and establishes chronic HCV infection in chimpanzees (5, 6). The HCV genomic RNA carries genetic information at two levels: a single large ORF encodes viral proteins and complex RNA structural elements regulate the viral replication cycle (4). Viral replication begins when conserved RNA elements in the 5′ UTR bind the 40S ribosome subunit and recruit essential translation factors (7). Translation produces a viral polyprotein that is cleaved by cellular and viral proteases to generate 10 viral proteins (4). The HCV genome is replicated through a negative-strand RNA intermediate by a viral RNA-dependent RNA polymerase (NS5B) in a process controlled by conserved RNA elements (8–17).
The HCV genomic RNA is physically compact (18) and highly structured (19). These features likely facilitate persistent HCV infections in humans by protecting the genome from degradation by innate antiviral defenses (20, 21). Two elements of this defense are RNase L, which cleaves in single-stranded regions (22), and diverse double-stranded RNA-induced antiviral immune responses (23). Selection of RNase L-resistant structures has likely led to the stable and compact HCV genome structure (24); in contrast, concurrent selection imposed by the requirement to evade double-stranded RNA-triggered innate immune sensors likely constrains the lengths of internal helices. Defining structural conformations of HCV RNA genomes is thus a critical step in understanding why human innate immune systems frequently fail to recognize and clear the virus.
The structure of 80% of the HCV genome is unexplored. Most existing structural data have been obtained using short RNA transcripts that may not fully recapitulate structures in the intact genome. Here we used selective 2′-hydroxyl acylation analyzed by primer extension, read out by mutational profiling (SHAPE-MaP) using massively parallel sequencing (25, 26), to model the structures of infectious HCV RNA genomes from genotypes 1a, 1b, and 2a. We developed a concise structure-first approach, based on SHAPE reactivity information, to identify structural features conserved across HCV genotypes. We discovered conserved structural elements that influence viral replication and identified genotype-specific structures that likely facilitate evasion of cellular innate immune responses.
Results
Interrogation of Three Divergent HCV RNA Genome Structures.
We examined genome-length synthetic RNA transcripts from three molecular HCV clones H77c, Con1, and JFH1. H77 and Con1 correspond to HCV genotypes 1a and 1b, respectively, share 80% sequence identity, and represent HCV strains accounting for the majority of infections worldwide (27). JFH1 represents genotype 2a (27) and shares only 70% sequence identity with genotype 1 viruses. SHAPE-MaP data were obtained at read depths that allow full recovery of the underlying structure information. SHAPE reagents react with RNA nucleotides to report local nucleotide flexibility (28) such that unconstrained single-stranded regions are reactive and base-paired elements are generally unreactive (29).
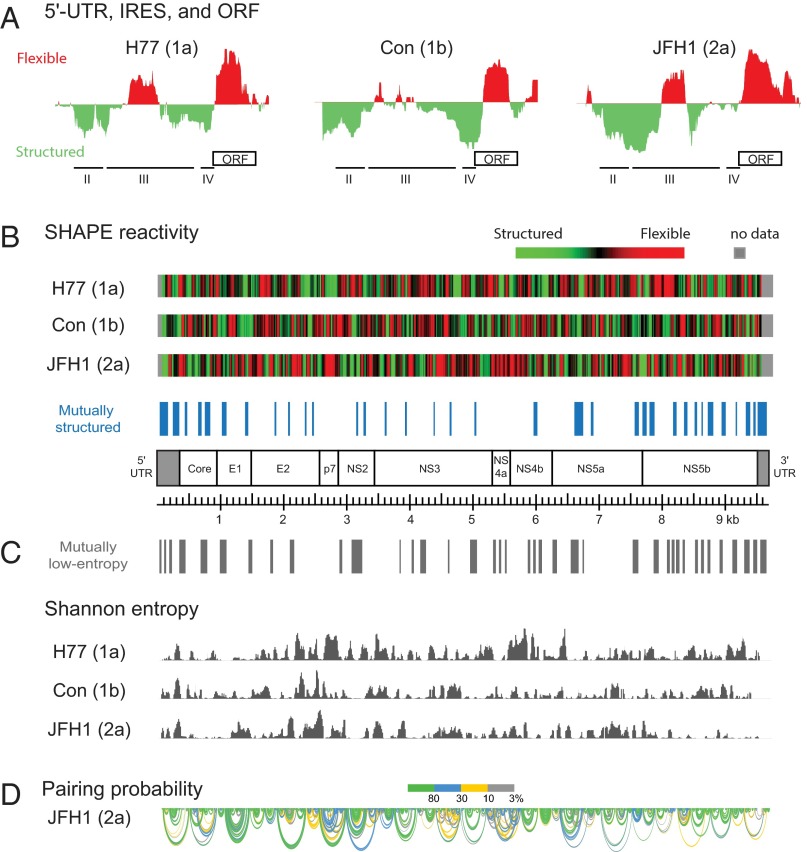
Regions known to have conserved structural features, such as the internal ribosome entry site (IRES) (7) and the unstructured region at the beginning of the ORF (30), had similar SHAPE profiles in each genome (Fig. 1A). Many regions had low SHAPE reactivities in all three genomes (Fig. 1B, blue bars). These structured regions included known regulatory elements, including the IRES (7) and NS5B cis-regulatory element (CRE) (8, 9, 14, 31), and also many regions with uncharacterized functions. The central regions of the individual genomes (encoding proteins E2 through NS4b) contained highly structured elements; however, the majority of these structured elements did not occur in the same regions in all three genomes (Fig. 1B, green regions in heat maps).
Fig. 1.
Global structural analysis of three HCV genomes. (A) SHAPE reactivities for each genome over the first 450 nt. IRES subdomains II–IV and the ORF are highlighted. Reactivities are shown relative to the global median. (B) Heat maps showing median SHAPE reactivities (51-nt windows) for each of three genomes. Regions identified as mutually structured across all three genomes are emphasized with blue bars. (C) Median Shannon entropies (51-nt windows) for each of three HCV genomes. Regions with low Shannon entropies in all three genomes are indicated above the histograms with gray bars. (D) Representative structural model for the JFH1 RNA genome. Helices are shown as arcs, colored according to base-pairing probabilities as calculated from the SHAPE-directed partition function (26). Regions with green arcs represent well-defined structures; regions with overlapping blue, yellow, and gray arcs likely sample multiple conformations. Full arc models for all three RNA genomes are provided in Figs. S1–S3.
Modeling HCV Genome Secondary Structures.
We identified regions within the HCV genomes most likely to form well-determined, stable secondary structures based on analysis of Shannon entropy and SHAPE reactivities (Fig. 1C). Shannon entropies are derived from a SHAPE-directed partition function (32) and report a measure of confidence in the predicted base-pairing pattern at each nucleotide (26, 33). Low entropy also corresponds to high confidence in structure modeling. Multiple regions with low Shannon entropy were shared by all three genomes, and these often overlapped in whole or in part with conserved regions of low SHAPE reactivity (Fig. 1 B and C).
Using SHAPE data as constraints (34), we generated experimentally informed secondary structure models for each genome (Fig. 1D and Figs. S1–S3). This approach has been validated for RNAs of known structure (34) and for identification of novel functional elements in large RNAs (26). The secondary structure models are illustrated using arc plots, which capture both predicted base pairing and the degree of variability in the structural models. Well-defined structures corresponding to highly probable helices are green. Alternative structures appear as overlapping blue, yellow, and gray arcs in the figures. In HCV genome regions where functional RNA structures have been previously characterized—for example, the IRES in the 5′ UTR (Fig. S4) and stem-loop elements within NS5B (15)—our genome-wide structural models corresponded closely with previously validated secondary structures.
Conservation of Structured Elements Across HCV Genotypes.
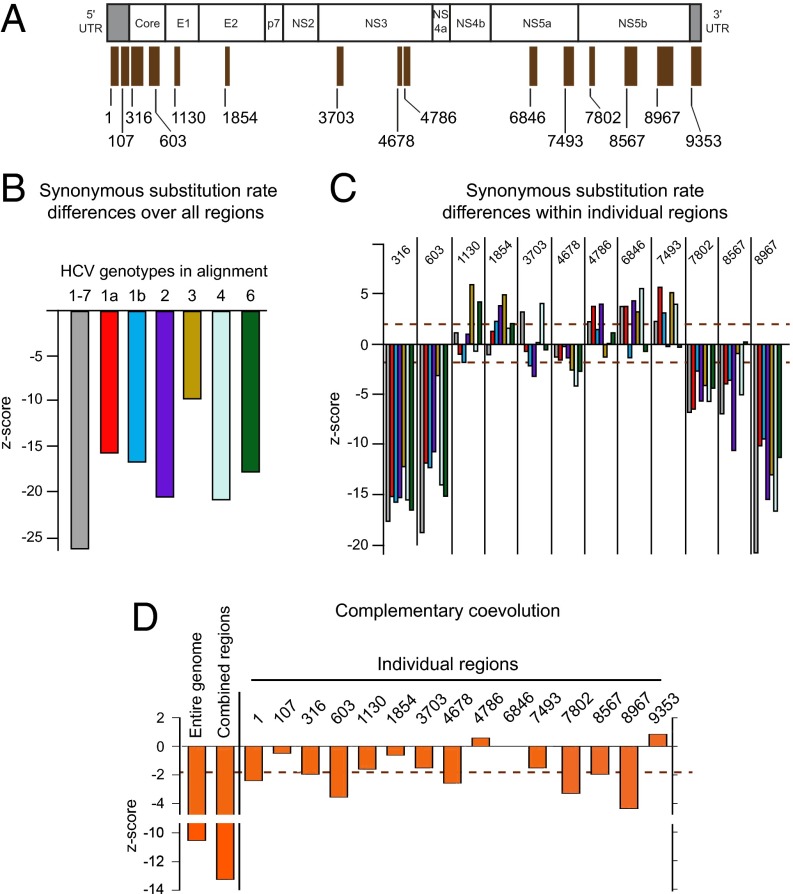
Using the SHAPE-directed RNA structural models, we identified 15 regions of 75 nt or more in which at least 75% of modeled base pairs occur at homologous positions in all three genomes (Fig. 2A and Table S1). We used two independent analyses to examine evolutionary pressures on these 15 structurally conserved regions. The presence of selection favoring the maintenance of base pairing within functional RNA elements is expected to drive: (i) reduced synonymous nucleotide substitution frequencies in codons and (ii) complementary coevolution between base-paired sites.
Fig. 2.
Selective pressures on higher-order HCV RNA structures. (A) Locations of 15 regions that have conserved base pairing in the SHAPE-directed models across all three HCV genotypes. Regions are numbered relative to their position in the H77c genome. (B) Synonymous substitution rates for each of seven individual genomic alignments over all regions of conserved base pairing combined. Larger (negative) values indicate lower synonymous substitution rates, consistent with evolutionary conservation of RNA structure. (C) Differences in synonymous substitution rates for individual regions. The z-scores above 1.96 and below −1.96 (dashed lines) are significant at the P < 0.05 level. The lower synonymous substitution rates in regions 316 and 603 may reflect evolutionary constraints imposed by an alternative ORF (48). (D) Complementary coevolution between base-paired sites. Values for predicted base pairs across the entire genome and over regions with high degrees of conserved base pairing are shown at left. Values below −1.96 (dashed line) are significant at the P < 0.05 level.
Twelve of the structurally conserved regions fall in the coding region. The relative synonymous substitution rates within these regions were examined across seven HCV sequence alignments, each including up to 100 sequences from clinical samples. Strong selection pressures can generally be detected with datasets of this size; however, currently available sequences are likely too few to confirm subtle selection of particular structures. The ORF elements had statistically significant low synonymous substitution rates across each of the seven alignments (Fig. 2B). Six regions displayed statistically significant decreases in the synonymous substitution rates in three or more HCV alignments (Fig. 2C). Of these, four regions (316, 603, 7802, and 8967) had significantly reduced synonymous substitution rates in all genotypes examined, indicating that selective forces acting at the RNA level have impacted these regions throughout the entirety of HCV evolution. Two regions (6846 and 7493) showed statistically significant increases in synonymous substitution rate in at least five HCV genotypes. These sequences are apparently subject to evolutionary pressures beyond those that maintain specific base pairs or coding potential.
There was strong evidence for coevolution of complementary base pairs in the complete full-genome SHAPE-informed structures of the HCV genomes (Fig. 2D and Figs. S1D, S2D, S3D, and S5B) (z-score = –10.5, P = 8.6 × 10−26). When only the 15 regions with conserved SHAPE-informed structures were considered, base pair coevolution was even more significant (z-score = –13.2, P = 8.8 × 10−40); associations were statistically significant for seven of the 15 structured regions (alignment positions 1, 316, 603, 4678, 7802, 8567, and 8967) (Fig. 2D). In sum, conservation of base pairing strongly supports the existence of multiple regions in HCV genomes that contain functionally conserved higher-order structures.
Importance of SHAPE Data.
The global minimum free-energy secondary structures generated with and without experimental data shared only 73% of predicted base pairs. This level of dissimilarity leads to substantial, nontrivial differences in modeled structures (35). Only 7 of the 15 regions with mutually conserved structures were present in some form in the structures predicted without use of SHAPE data (Fig. S5). When the Shannon entropy was calculated omitting SHAPE data, overall levels of entropy were approximately twofold higher, consistent with an important role for the experimental constraints in distinguishing between otherwise similarly probable helices. Moreover, the fivefold fewer mutual low-entropy regions identified in the absence of SHAPE information only partially overlap with those detected with SHAPE data (Fig. S5). SHAPE data were also critical for detecting evolutionary pressures favoring the maintenance of genome structures (Fig. S6). Without prior categorization of sites into structured and nonstructured groups it would be extremely difficult (if not impossible) to use these metrics alone to identify sites of conserved base pairing. In sum, experimental SHAPE constraints had a significant impact on, and were essential for, success of the structure-first approach explored in this work.
Functional Characterization of Conserved RNA Elements.
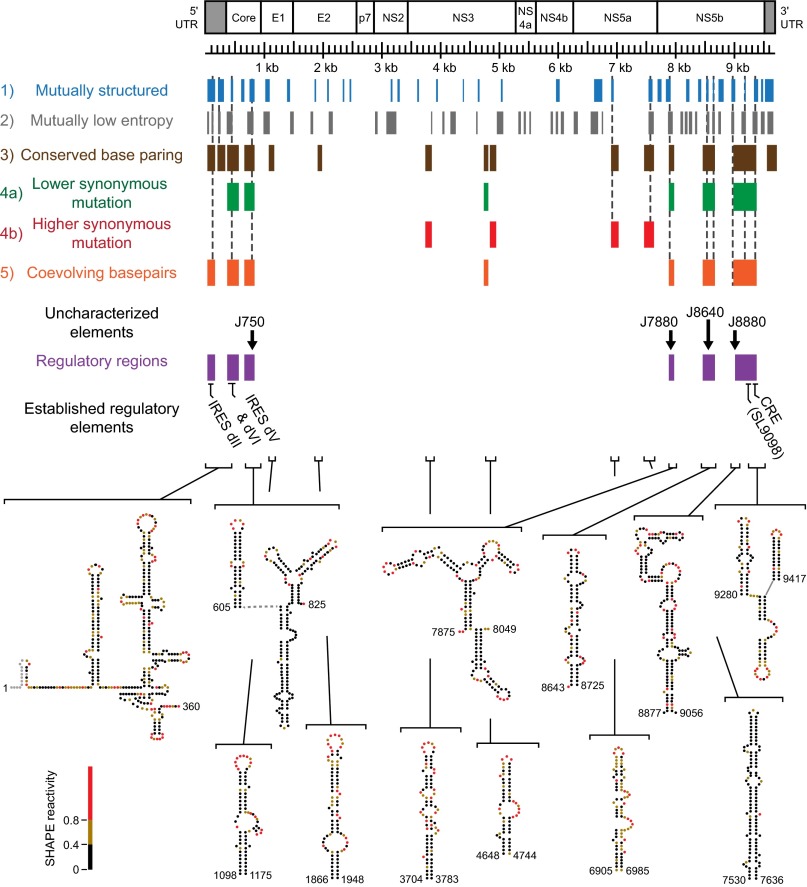
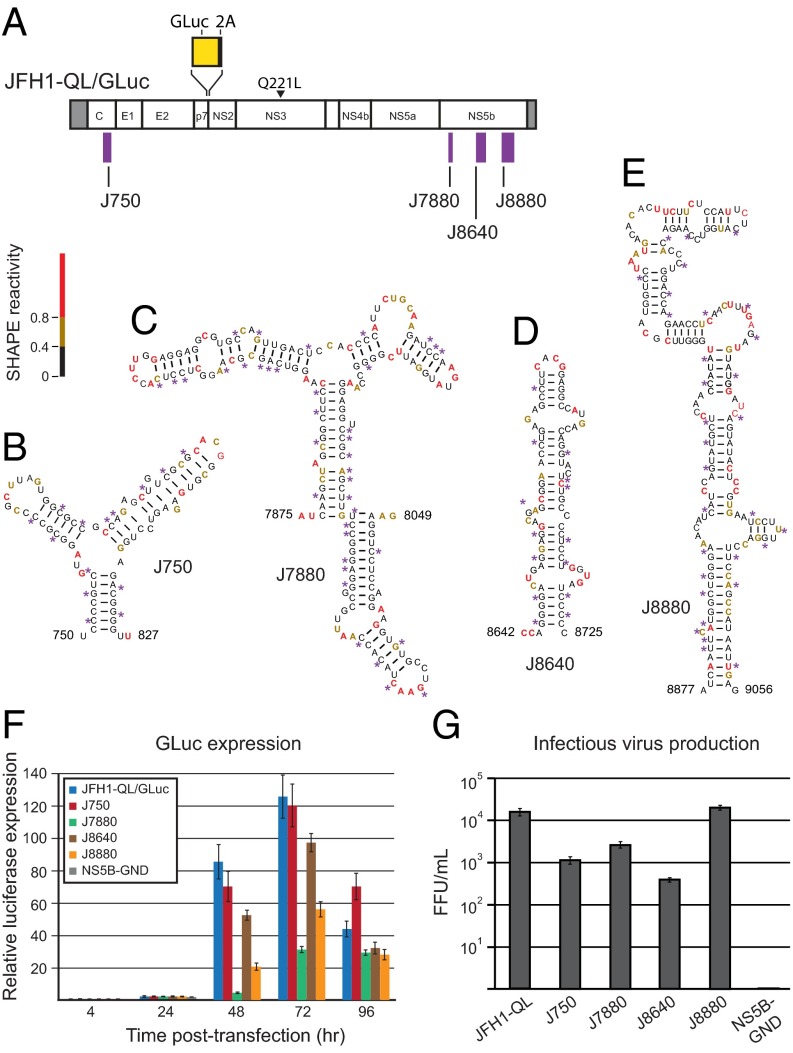
The structure-first analysis identified six regions, comprising at least nine individual structural elements, with especially strong evidence of natural selection (Fig. 3, purple boxes). Five elements correspond to regulatory motifs that have been well characterized: the IRES domains II–IV, SL9098, and CRE (13–17). We focused on the four uncharacterized RNA elements (Fig. 4A). We introduced mutations that disrupted the structures of each of these RNA elements in a cell culture-adapted strain (JFH1-QL, genotype 2a) (36). Mutations maintained amino acid sequence and avoided rare codons (Fig. 4 B–E and Table S2). Replication was detected by expression of a Gaussia princeps (GLuc) luciferase gene inserted in the HCV ORF distant from the structures of interest (Fig. 4A) following transfection of synthetic transcripts into Huh-7.5 cells (37). GLuc activity reflects viral polyprotein synthesis and is a good surrogate measure of genome replication. Structure-disrupting mutations within elements J7880 and J8880, both located in NS5B, reduced replication by four- and twofold, respectively, but both RNAs were replication competent, producing substantially more luciferase than a nonviable mutant (NS5B-GND) (Fig. 4F).
Fig. 3.
Comparative HCV RNA genome structure analysis. Summary of notable regions based on each analysis class: structured regions (blue), regions of low Shannon entropy (gray), regions of conserved base pairing (brown), regions with low synonymous substitution rates (green), regions with high synonymous substitution rates (red), and regions with evidence of complementary coevolution of base pairs (orange) for all three HCV genomes. Six regions (purple) have conserved base paring in the SHAPE-directed structural models and phylogenetic support for broad conservation. These regions are annotated with the four structural elements (J750, J7880, J8640, and J8880) evaluated in HCV replication assays. SHAPE-informed secondary structure models of the JFH1 genome are shown for regions with notable features conserved across all three HCV RNA genomes. Nucleotides are colored by SHAPE reactivity.
Fig. 4.
Functional regulatory structures within the JFH1 RNA genome. (A) JFH1-QL/GLuc expression construct. The four structural elements tested in functional assays are shown. RNA secondary structure models for (B) J750, (C) J7880, (D) J8640, and (E) J8880. The positions of structure-disrupting, silent mutations are shown with asterisks. Nucleotides are colored by SHAPE reactivity (see scale). (F) Relative levels of HCV-encoded G. princeps luciferase protein, normalized to the 4-h time point, secreted by cells transfected with JFH1-QL/GLuc RNA or structure disrupting mutants. The bars show the means of triplicate measurements; error bars report SDs. NS5B-GND is a lethal (negative) control. (G) Titers of infectious virus generated 72 h posttransfection by JFH1-QL (no GLuc2A insertion), the lethal mutant NS5B-GND, and structure-disrupting mutants. Histograms show the mean of triplicate measurements; error bars report SDs.
We also measured production of infectious virus in assays that depend on competency at every stage of the viral replication cycle (37, 38). Mutant J750 (Fig. 4B), with changes in the Core protein coding region, and J8640 (Fig. 4D), located within the NS5B coding region, had 25-fold and 60-fold reductions in infectious virus yields compared with wild-type viral RNA, respectively. This large effect contrasts with the GLuc assays that showed that these mutants replicate at levels close to that of the parent (Fig. 4 F and G). This finding suggests these two structures play important roles in viral assembly. Mutant J7880 (Fig. 4C), with substitutions in the NS5B coding region, produced fourfold less infectious virus than the parental RNA (Fig. 4G), consistent with the observed reduction in GLuc expression (Fig. 4F). Virus yields from mutant J8880 (Fig. 4E) were similar to yields from the parent RNA, consistent with the twofold effect of these substitutions on GLuc expression (compare Fig. 4 F and G). In sum, all four of these structural mutants were deficient in HCV genome replication or infectious virus production.
Role of RNA Structure in Immune Evasion.
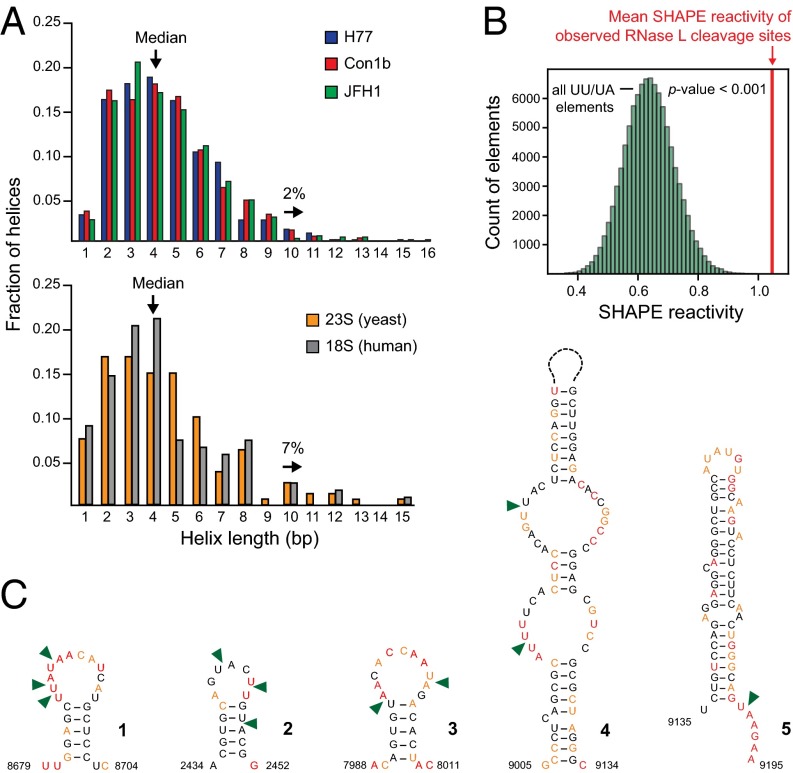
HCV RNA genomes have evolved under selective pressures exerted by innate immune sensors that recognize long double-stranded RNA helices (23). Based on SHAPE-informed secondary structure models, we generated global profiles of lengths of RNA helices in the three HCV genomes (Fig. 5A, Upper). Because many helices in the minimum free-energy structures have high Shannon entropies (Figs. S1–S3), these profiles represent an upper limit on the extent of stable base pairing. The median helix length is four consecutive canonical base pairs, 90% of helices contain seven base pairs or fewer, and only 2% of helices are longer than nine base pairs. The longest modeled individual helix in each genome is 15 or 16 base pairs, and each is located at a different position in the genome. In a randomly generated RNA, containing the same dinucleotide distribution as the HCV RNA, the median helix length is 3 base pairs and the maximum helix size is 10–11 base pairs. The overall helix distribution for HCV is similar to that of ribosomal RNAs (Fig. 5A, Lower). In rRNAs the median helix length is also four base pairs, but 7% of helices are longer than nine base pairs. The helix lengths observed in HCV are substantially below the 16-base pair helix length recognized by the immune sensor, PKR (39). Although highly structured overall (18), HCV appears to have evolved to minimize its visibility to innate immune sensors.
Fig. 5.
Relationships between HCV genome structure and innate immune factor recognition features. (A) RNA helix lengths in the SHAPE-directed folding models for each of three HCV genomes. For comparison, helix lengths for models of the human 18S and yeast 28S ribosomal RNAs (49) are shown. (B) Comparison of measured RNase L cleavage sites (22, 24) with mean SHAPE reactivities randomly sampled from UU/UA dinucleotides in the H77c genome. Green bars show a bootstrap analysis of mean SHAPE reactivities for 10,000 populations of UU/UA motifs chosen at random from the H77 genome. The mean SHAPE reactivity for efficient RNase L cleavage sites (red line) lies well outside the distribution expected by chance. (C) RNA structure models for the five strongest RNase L cleavage sites (triangles) in the H77 genome. Nucleotides are colored by SHAPE reactivity.
HCV RNA is also targeted by the RNase L system (40), which cleaves RNA at single-stranded, UU/UA (22, 24), or UNN (41) motifs. RNase L cleaves the H77 HCV genome with high-efficiency at 10 sites (22, 24). These RNase L cleavage sites occur in highly flexible, unconstrained regions of the SHAPE-informed genome model with a mean SHAPE reactivity of 1.05, much greater than the average for UU/UA motifs (Fig. 5B); most are in flexible loop regions 5′ of a stable helix of at least five base pairs (Fig. 5C). Critically, the vast majority of RNase L motifs in the H77 HCV genome occur in structural contexts unfavorable for RNase L cleavage.
Discussion
Using a structure-first approach, we characterized the evolutionary forces shaping genome-wide structure in HCV. SHAPE probing revealed 15 highly structured RNA elements that were conserved in three diverse HCV genomes (Fig. 1B, green regions and blue bars). Every validated regulatory element discovered over the past two decades of HCV research—including the IRES, J8647, SL9098, and CRE elements (8, 9, 14, 17, 31)—were located within these elements (Fig. 3). The importance of experimental data for defining accurate structural models is clear based on analysis of the IRES region: inclusion of SHAPE data improved the accuracy of de novo single-sequence structure modeling from 42 to 96% for recovery of accepted base pairs (Figs. S1–S4).
Within the UTRs and in the Core and NS5B protein coding sequences, we identified four previously uncharacterized structural elements (Fig. 3, purple regions). Reduced synonymous substitution rates and complementary coevolution of base-paired nucleotides (Fig. 2 C and D) suggest that evolution has specifically maintained RNA structures in these regions since the last common ancestor of the contemporary HCV lineages. In the newly identified conserved structures, destabilization caused substantial effects on HCV replication or infectious virus production (Fig. 4). The initial SHAPE analysis that identified these structures was performed on synthetic RNA transcripts. The reverse molecular genetics analyses involved transfecting these RNAs into cells where they initiate a complete viral replication cycle. Thus, the structures identified in synthetic RNA transcripts substantially reflect the structure of replicating viral RNA.
Intriguingly, mutations that disrupted the four newly identified elements had distinct phenotypes when examined in the JFH1 virus. Mutations within elements J750 and J8640 had dramatic effects on infectious virus production but only modest impact on expression of a luciferase reporter protein embedded in the HCV polyprotein. Our analysis of the J8640 element revealed a phenotype distinct from that reported recently (17); we observed a much larger effect on virus production than on genome replication (Fig. 4). In contrast, mutations in the J7880 and J8880 elements had comparable significant effects on HCV-driven luciferase expression and production of infectious virus. Elements J750 and J8640 may therefore selectively impact late-stage events, like assembly or packaging. We did not observe phenotypes when the J750 and J8640 elements were mutated in the context of the genotype 1a virus, H77S.3 (Fig. S7), even though structure-based analyses provided strong evidence that these RNA elements are under evolutionary selection in both genotypes 1a and 2. H77S.3 replicates less efficiently in cell culture than JFH1-QL (Fig. 4G and Fig. S7E) and, under our cell-culture conditions, these elements presumably do not affect rate limiting viral replication processes.
In the central part of the genome, we identified many well-defined (low Shannon entropy) structures that do not appear to involve evolutionarily conserved base-paired configurations (Figs. 1 and 2C). These regions may be evolving under a complex mix of pressures related to maintaining highly compacted physical configurations (18, 19) while evading immune recognition. HCV genomes replicate in the presence of innate immune response sensors that detect single-stranded (RNase L) and double-stranded (PKR and others) viral RNAs. This work supports a model in which HCV RNA genome structure balances these opposing pressures by evolving extensive double-stranded structures consisting predominantly of short helices (Fig. 5).
The models presented here also provide a framework for understanding RNA genome structure interactions that govern HCV pathogenesis. For example, two recent chimpanzee studies monitored chronic infection by H77c viruses and independently detected a silent mutation, A7586G, that arose early and was subsequently maintained during infection (42, 43). In the context of our H77c structure model, this mutation yields an impressive 6.6 kcal/mol stabilization of a structural element in the H7430 region (Fig. S8), close to the upper limit attainable by converting any mismatch to a canonical base pair (44). These results suggest that, in the presence of an active immune system, stabilizing the H7420 structure confers increased viral fitness.
In sum, these genome-scale, structure-first comparative analyses and structural models provide a critical foundation for understanding how the HCV genome interacts with both viral and host proteins during replication, functions in viral gene expression, and contributes to evasion of innate cellular immune responses. This concise analytical strategy is broadly applicable to studying diverse single-stranded RNA viruses that pose serious current and emerging threats to public health.
Methods
SHAPE Modification of Genomic RNAs.
Full-length genomic RNAs for HCV-H77c, HCV-JFH1, and HCV-Con1 were synthesized in vitro (5, 6). Purified genomes were folded in 50 mM Hepes (pH 8.0), 5 mM MgCl2, 200 mM potassium acetate (pH 7.5) by heating to 65 °C and slowly cooling to 37 °C. RNA was modified with 1M7 and SHAPE-MaP libraries prepared (26). Full SHAPE data and details regarding genomic subclones, SHAPE-MaP massively parallel sequencing, and RNA structure analysis are available in SI Methods and Datasets S1 and S2. SHAPE reactivities are not reported for the poly-U region and X-tail in the 3′ UTR because of low sequencing depth.
HCV Bioinformatic Analyses.
We assembled clinically derived HCV sequences (SI Methods, Figs. S7 and S8, and Dataset S3) and used two parametric maximum-likelihood approaches to examine selective maintenance of structural elements. The FUBAR method (45) was applied to seven HCV datasets to test for statistically significant fluctuations in synonymous substitution rates between codons containing base-paired versus unpaired nucleotides (46). A modification of the Spidermonkey approach (47) was used to examine complementary coevolution at base-paired sites (46) using an alignment of 250 representative sequences drawn from six HCV genotypes.
HCV Replication Assays.
Structure-disrupting mutations were created in the JFH1-QL and H77S.3 genomes and their GLuc2A counterparts (Table S2), and synthetic RNAs produced from these clones were then assayed for their replication competence by measuring luciferase expression and infectious virus yields (36–38).
Supplementary Material
Acknowledgments
This work was supported by NIH Grants GM064803 (to K.M.W.) and AI095690, AI109965, and CA164029 (to S.M.L.); and the South African National Research Foundation NBIG UID 86935 (to D.P.M.). D.M.M. was a Lineberger Postdoctoral Fellow in the Basic Sciences (T32-CA009156) and a Fellow of the American Cancer Society (PF-11-172-01-RMC). S.W. was supported as an infectious disease fellow (T32-AI007151).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416266112/-/DCSupplemental.
References
- 1.Alter HJ. HCV natural history: The retrospective and prospective in perspective. J Hepatol. 2005;43(4):550–552. doi: 10.1016/j.jhep.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19(7):837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leone N, Rizzetto M. Natural history of hepatitis C virus infection: From chronic hepatitis to cirrhosis, to hepatocellular carcinoma. Minerva Gastroenterol Dietol. 2005;51(1):31–46. [PubMed] [Google Scholar]
- 4.Choo QL, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88(6):2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94(16):8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66(3):1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol. 2000;74(4):2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H, Shin H, Wimmer E, Paul AV. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J Virol. 2004;78(20):10865–10877. doi: 10.1128/JVI.78.20.10865-10877.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friebe P, Bartenschlager R. Role of RNA structures in genome terminal sequences of the hepatitis C virus for replication and assembly. J Virol. 2009;83(22):11989–11995. doi: 10.1128/JVI.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friebe P, Lohmann V, Krieger N, Bartenschlager R. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J Virol. 2001;75(24):12047–12057. doi: 10.1128/JVI.75.24.12047-12057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi M, Lemon SM. 3′ Nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol. 2003;77(6):3557–3568. doi: 10.1128/JVI.77.6.3557-3568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diviney S, et al. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J Virol. 2008;82(18):9008–9022. doi: 10.1128/JVI.02326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You S, Stump DD, Branch AD, Rice CM. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol. 2004;78(3):1352–1366. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuplin A, Evans DJ, Simmonds P. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J Gen Virol. 2004;85(Pt 10):3037–3047. doi: 10.1099/vir.0.80141-0. [DOI] [PubMed] [Google Scholar]
- 16.Vassilaki N, et al. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J Virol. 2008;82(23):11503–11515. doi: 10.1128/JVI.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu D, et al. Systematic analysis of enhancer and critical cis-acting RNA elements in the protein-encoding region of the hepatitis C virus genome. J Virol. 2013;87(10):5678–5696. doi: 10.1128/JVI.00840-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis M, Sagan SM, Pezacki JP, Evans DJ, Simmonds P. Bioinformatic and physical characterizations of genome-scale ordered RNA structure in mammalian RNA viruses. J Virol. 2008;82(23):11824–11836. doi: 10.1128/JVI.01078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmonds P, Tuplin A, Evans DJ. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: Implications for virus evolution and host persistence. RNA. 2004;10(9):1337–1351. doi: 10.1261/rna.7640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Washenberger CL, et al. Hepatitis C virus RNA: Dinucleotide frequencies and cleavage by RNase L. Virus Res. 2007;130(1-2):85–95. doi: 10.1016/j.virusres.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li K, Lemon SM. Innate immune responses in hepatitis C virus infection. Semin Immunopathol. 2013;35(1):53–72. doi: 10.1007/s00281-012-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate-dependent endonuclease. Science. 1981;212(4498):1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 23.Peisley A, Hur S. Multi-level regulation of cellular recognition of viral dsRNA. Cell Mol Life Sci. 2013;70(11):1949–1963. doi: 10.1007/s00018-012-1149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han JQ, Wroblewski G, Xu Z, Silverman RH, Barton DJ. Sensitivity of hepatitis C virus RNA to the antiviral enzyme ribonuclease L is determined by a subset of efficient cleavage sites. J Interferon Cytokine Res. 2004;24(11):664–676. doi: 10.1089/jir.2004.24.664. [DOI] [PubMed] [Google Scholar]
- 25.Weeks KM, Mauger DM. Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res. 2011;44(12):1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat Methods. 2014;11(9):959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornberg M, et al. A systematic review of hepatitis C virus epidemiology in Europe, Canada and Israel. Liver Int. 2011;31(Suppl 2):30–60. doi: 10.1111/j.1478-3231.2011.02539.x. [DOI] [PubMed] [Google Scholar]
- 28.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J Am Chem Soc. 2008;130(37):12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460(7256):711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rijnbrand R, et al. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA. 2001;7(4):585–597. doi: 10.1017/s1355838201000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol. 2000;7(12):1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 32.Reuter JS, Mathews DH. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathews DH. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10(8):1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hajdin CE, et al. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci USA. 2013;110(14):5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice GM, Leonard CW, Weeks KM. RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA. 2014;20(6):846–854. doi: 10.1261/rna.043323.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma Y, Yates J, Liang Y, Lemon SM, Yi M. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J Virol. 2008;82(15):7624–7639. doi: 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimakami T, et al. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology. 2011;140(2):667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci USA. 2006;103(7):2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng X, Bevilacqua PC. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10(12):1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han JQ, Barton DJ. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA. 2002;8(4):512–525. doi: 10.1017/s1355838202020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han Y, et al. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science. 2014;343(6176):1244–1248. doi: 10.1126/science.1249845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Callendret B, et al. Transmission of clonal hepatitis C virus genomes reveals the dominant but transitory role of CD8+ T cells in early viral evolution. J Virol. 2011;85(22):11833–11845. doi: 10.1128/JVI.02654-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi M, et al. Evolution of a cell culture-derived genotype 1a hepatitis C virus (H77S.2) during persistent infection with chronic hepatitis in a chimpanzee. J Virol. 2014;88(7):3678–3694. doi: 10.1128/JVI.03540-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner DH, Mathews DH. NNDB: The nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2010;38(Database issue):D280–D282. doi: 10.1093/nar/gkp892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murrell B, et al. FUBAR: A fast, unconstrained Bayesian approximation for inferring selection. Mol Biol Evol. 2013;30(5):1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muhire BM, et al. Evidence of pervasive biologically functional secondary structures within the genomes of eukaryotic single-stranded DNA viruses. J Virol. 2014;88(4):1972–1989. doi: 10.1128/JVI.03031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poon AF, Lewis FI, Frost SD, Kosakovsky Pond SL. Spidermonkey: Rapid detection of co-evolving sites using Bayesian graphical models. Bioinformatics. 2008;24(17):1949–1950. doi: 10.1093/bioinformatics/btn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McMullan LK, et al. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci USA. 2007;104(8):2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cannone JJ, et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.