Significance
The search for new antifungal compounds is struggling to keep pace with emerging fungicide resistance. Through chemoprospecting of an untapped reservoir of inhibitory compounds, lignocellulosic hydrolysates, we have identified a previously undescribed antifungal agent, poacic acid. Using both chemical genomics and morphological analysis together for the first time, to our knowledge, we identified the cellular target of poacic acid as β-1,3-glucan. Through its action on the glucan layer of fungal cell walls, poacic acid is a natural antifungal agent against economically significant fungi and oomycete plant pathogens. This work highlights the chemical diversity within lignocellulosic hydrolysates as a source of potentially valuable chemicals.
Keywords: chemical genomics, high-dimensional morphometrics, lignocellulosic hydrolysates, fungal cell wall, Saccharomyces cerevisiae
Abstract
A rise in resistance to current antifungals necessitates strategies to identify alternative sources of effective fungicides. We report the discovery of poacic acid, a potent antifungal compound found in lignocellulosic hydrolysates of grasses. Chemical genomics using Saccharomyces cerevisiae showed that loss of cell wall synthesis and maintenance genes conferred increased sensitivity to poacic acid. Morphological analysis revealed that cells treated with poacic acid behaved similarly to cells treated with other cell wall-targeting drugs and mutants with deletions in genes involved in processes related to cell wall biogenesis. Poacic acid causes rapid cell lysis and is synergistic with caspofungin and fluconazole. The cellular target was identified; poacic acid localized to the cell wall and inhibited β-1,3-glucan synthesis in vivo and in vitro, apparently by directly binding β-1,3-glucan. Through its activity on the glucan layer, poacic acid inhibits growth of the fungi Sclerotinia sclerotiorum and Alternaria solani as well as the oomycete Phytophthora sojae. A single application of poacic acid to leaves infected with the broad-range fungal pathogen S. sclerotiorum substantially reduced lesion development. The discovery of poacic acid as a natural antifungal agent targeting β-1,3-glucan highlights the potential side use of products generated in the processing of renewable biomass toward biofuels as a source of valuable bioactive compounds and further clarifies the nature and mechanism of fermentation inhibitors found in lignocellulosic hydrolysates.
Lignocellulosics are a potential sugar feedstock for biofuels and bio-based chemicals. Before plant materials can be converted to biofuels by fermentation, their cell wall polysaccharides must be hydrolyzed to sugar monomers for microbial conversion (1). The hydrolysis processes generates, in addition to the sugars, small acids, furans, and other compounds that affect microbial growth and inhibit fermentation (2–5). The inhibitory effects of these compounds represent a challenge to efficient microbial bioconversion. The primary focus of lignocellulosic-derived inhibitor research has been to understand, evolve, and engineer tolerance in fermentative microbes (2). However, as natural antimicrobial agents, lignocellulosic fermentation inhibitors offer an untapped reservoir of bioactive compounds.
One increasingly important potential use of these inhibitors is as antifungal agents. Worldwide, fungicide-resistant pathogens pose a threat to agricultural sustainability. Pathogen resistance to conventional fungicides affects multiple crops (6, 7). Copper-based fungicides are effective in organic agriculture but facing restrictions because of copper accumulation in soils (8, 9). Furthermore, climate change is altering the global distribution of fungal pathogens (10, 11). New sources of fungicides are a necessity to keep pace with the evolution of resistant strains and emerging pathogens (12).
The antifungal activities of many of the inhibitors (e.g., ferulic acid and furfural) in hydrolysates have been described (13, 14), but new compounds continue to be discovered (15). One understudied class of compounds found in grasses and their hydrolysates is the dehydrodiferulates and compounds derived from them (hereafter all simply termed diferulates) (16, 17). In grasses, diferulates are produced by radical dimerization of ferulates that acylate arabinoxylan polysaccharides and function as powerful cell wall cross-linkers (16); derivatives of diferulates are released during the hydrolysis of biomass (16, 18, 19). At present, the structures of a range of diferulates have been described (16, 18), but the biological activities of isolated diferulates (beyond their function in the plant cell wall) have not been explored. Diferulates may be expected to have effects on organisms other than plants. One study found a negative correlation between diferulate concentration and colonization by corn-boring insects (20), but a direct effect of diferulates is unknown. Despite the well-documented antifungal activity of ferulic acid and its derivatives (13, 21, 22), no studies on the antifungal properties of diferulates have been described.
We screened a collection of diferulates found in lignocellulosic hydrolysates for antifungal activity using the yeast Saccharomyces cerevisiae as a discovery system for antifungal agents. We focused on the diferulate 8–5-DC (16) derived during hydrolysis from a major diferulate in grasses; we name this compound here as poacic acid, because it is found primarily in grasses (Poaceae). By applying both chemical genomics and morphological analysis, we predicted and confirmed that poacic acid binds to cell wall β-1,3-glucan. We showed its biological activity against not only yeast but also, the economically important fungal and oomycete pathogens Sclerotinia sclerotiorum, Alternaria solani, and Phytophthora sojae.
Results
Diferulates with Antifungal Activity.
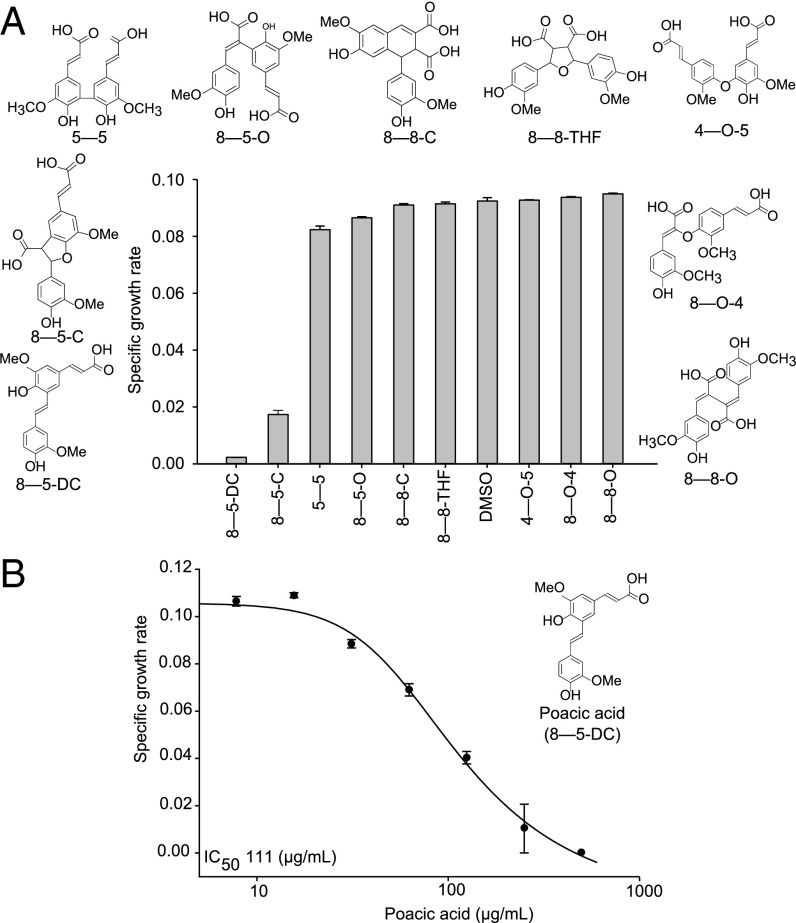
We tested a collection of nine diferulates known to occur in hydrolysates from corn stover for their effects on S. cerevisiae growth (Fig. 1A and Table S1, detailed nomenclature). Of these, only two had detectable bioactivity at the tested concentration of 1 mg/mL. In particular, poacic acid had the greatest antifungal activity, with an IC50 of 111 µg/mL (324 µM) against our control yeast (Fig. 1B). This inhibition is comparable with that of the widely used fungicides picoxystrobin (IC50 of 308 µM) and polyoxin D (IC50 of 340 µM) and substantially lower than that of the primary fungicide used in organic agriculture, copper sulfate (IC50 of 2.4 mM) (23–25).
Fig. 1.
Bioactivity of diferulates. The bioactivity of nine diferulates against S. cerevisiae at 1 mg/mL was tested. Poacic acid (8–5-DC) had (A) the highest bioactivity and (B) an IC50 of 111 µg/mL (mean ± SE).
Chemical Genomics Predict That Poacic Acid Targets the Fungal Cell Wall.
To gain insight into the mode of action and the cellular target of poacic acid, we conducted chemical genomic analysis, a method that uses genome-wide collections of viable gene-deletion mutants to identify genes with deletions that confer sensitivity or resistance to bioactive compounds (26, 27). The resulting set of sensitive and resistant gene-deletion mutants associated with a response yields functional insight into the mode of action (26). We challenged a pooled mixture of ∼4,000 different yeast gene-deletion mutants with either poacic acid or a solvent control (DMSO). Sequencing of strain-specific DNA barcodes enabled us to decipher the relative fitness of each yeast mutant in the presence of the drug relative to the solvent control (28).
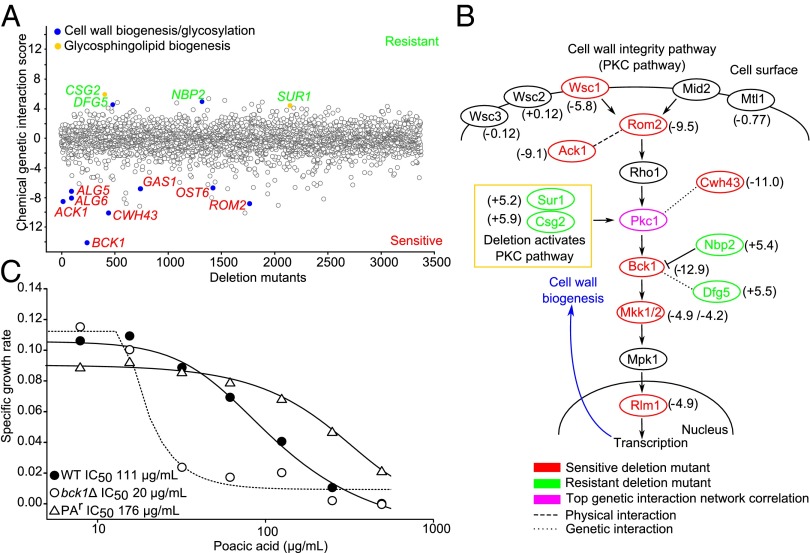
Deletion mutants for genes involved in cell wall and glycosylation-related processes were present in the top significantly sensitive and resistant strains (Fig. 2A, blue circles and Table S2). Among the top 10 deletion mutants sensitive to poacic acid, we detected enrichment for genes involved in the gene ontology (GO) category fungal-type cell wall organization (P < 0.01). These mutants included deletion alleles of BCK1 (bypass of C kinase), which encodes an MAPKKK in the Pkc1p (protein kinase C) cell wall integrity signaling pathway (PKC pathway); CWH43, which encodes a membrane protein involved in cell wall biogenesis and its null mutation is synthetically lethal with PKC1 mutants (29); ROM2 (Rho1 multicopy suppressor), a GDP/GTP exchange factor for Rho1p and another component of the PKC pathway; and ACK1, which seems to encode an upstream activator of Pkc1p and has a physical interaction with Rom2p. Overall, the PKC pathway was the most sensitive pathway to poacic acid (Pathway z score = −7.85) (Fig. 2B). This profile is similar to the chemical genomic profiles of other agents that target the cell wall and related processes (e.g., caspofungin) (26). Deletion mutants of BCK1 are hypersensitive to agents that compromise glycosylation (tunicamycin) and cell wall β-1,3-glucan biosynthesis (caspofungin) (26). We confirmed the sensitivity of the individual bck1∆ mutant and found a sixfold reduction in the IC50 against poacic acid compared with the control strain (WT) (Fig. 2C).
Fig. 2.
Chemical genomics of poacic acid. (A) Treatment of the yeast deletion collection with poacic acid revealed that mutants involved in cell wall biosynthesis and glycosylation were sensitive and resistant to the compound (blue). Mutants in genes involved in glycosphingolipid biogenesis were among the most resistant (yellow). (B) The PKC pathway, which governs cell wall integrity signaling, was the most sensitive pathway (Pathway z score = −7.85), with many members and interacting genes showing sensitivity to poacic acid (chemical genetic interaction score in parentheses). Comparison with the yeast genetic interaction network indicated that the genetic interaction profile of the essential gene PKC1 was most significantly correlated to the chemical genomic profile of poacic acid (P < 0.001). (C) Mutants of the gene encoding the cell wall signaling kinase BCK1 were sixfold more sensitive to poacic acid, whereas a poacic acid-resistant mutant (PAr) with an SNP in SUR1 had increased resistance compared with the control strain (mean and SE bars were removed for clarity).
Among the top significantly resistant strains, we detected significant enrichment for deletions of genes involved in the GO category glycosphingolipid biosynthetic process (P < 0.01) driven by csg2∆ (calcium sensitive growth) and sur1∆ (suppressor of Rvs161 and rvs167 mutations) (Fig. 2A, yellow circles and Table S2). Deletion of glycosphingolipid genes has been shown to activate the PKC pathway and cell wall biogenesis (Fig. 2B) (30). Involvement of SUR1 in poacic acid sensitivity was confirmed when we isolated a spontaneous chain-termination mutant in SUR1 able to form colonies on agar with 500 µg poacic acid/mL (Fig. 2C). Additionally, some other cell wall-related gene mutants were resistant to poacic acid. A deletion mutant of NBP2 (Nap1 binding protein) was resistant to poacic acid (Fig. 2A); Nbp2p down-regulates cell wall biogenesis through an interaction with Bck1p, which is activated by Pkc1p (Fig. 2B) (31). Thus, it seems that defects in the PKC pathway confer sensitivity to poacic acid, whereas activation of the PKC pathway confers resistance. A deletion mutant of DFG5 (defective for filamentous growth) also was resistant to poacic acid; DFG5 encodes a GPI-anchored protein involved in cell wall biogenesis that also has a genetic interaction with Bck1p (32, 33).
Because the chemical inhibitor of a gene product tends to mimic the loss-of-function phenotype of a mutant that inactivates the gene, the chemical–genomic profile for a bioactive compound can resemble the genetic interaction profile for the target (34). PKC1 has a genetic interaction profile that is most significantly correlated to the chemical genomic profile of poacic acid (Fig. 2B) (P < 0.0001). PKC1 is an essential gene required for growth and response to cell wall stress, and it has been implicated as a key mediator of cell wall-targeting drugs, such as the echinocandins (35). Together, these data narrowed our target search to the fungal cell wall. Poacic acid could directly damage the cell wall, inhibit a key cell wall synthesis enzyme, or disrupt the PKC pathway.
Morphological Analysis Revealed That Poacic Acid Affects the Fungal Cell Wall.
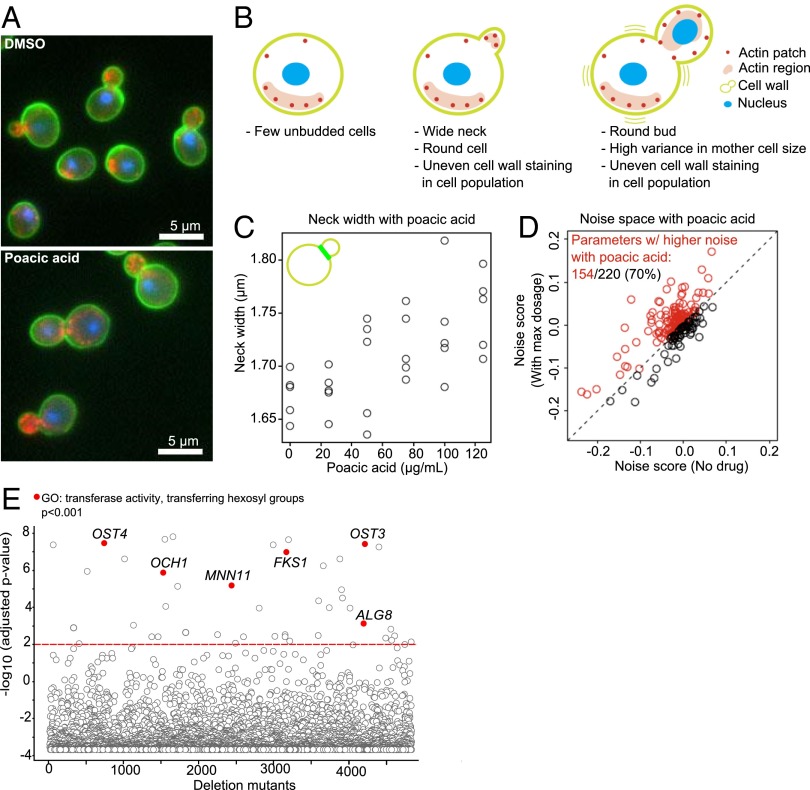
We investigated the morphological changes induced by poacic acid using high-dimensional microscopy (36, 37). Recently, two morphological features (a wide neck and morphological heterogeneity) were reported as common phenotypes in cells treated with agents known to affect the cell wall (38). The morphologies of cells exposed to poacic acid had both features (Fig. 3 A and B); they displayed dose-dependent increase in bud neck size (Fig. 3C) and heterogeneous morphologies (Fig. 3D). Because mutants displaying a high correlation with a drug phenotype can help identify targeted processes, we next compared the morphology of poacic acid-treated cells with the individual morphologies of 4,718 yeast deletion mutants (37, 39, 40). Forty-three deletion mutants had morphological profiles statistically similar (P < 0.01) to those of poacic acid (Fig. 3E and Table S3). Within the top correlations, we found significant enrichment of genes in the GO category transferase activity, transferring hexosyl groups (P < 0.001). This GO category contained genes responsible for key processes in the cell wall biogenesis, such as OCH1, which encodes a mannosyltransferase that initiates polymannose outer chain elongation, and FKS1 (FK506 sensitivity), which encodes a catalytic subunit of β-1,3-glucan synthase. Taken together, these data further indicate that poacic acid affects the yeast cell wall, consistent with the chemical genomic analysis.
Fig. 3.
Morphological characteristics of poacic acid-treated cells. Poacic acid treatment caused (A and B) abnormal cell morphology and (C and D) morphological characteristics similar to those caused by other cell wall-targeting agents. Poacic acid-treated cells had (C) a dose-dependent increased bud neck size and (D) heterogeneity of cell morphology. (E) The phenotype of poacic acid-treated cells was highly correlated with the phenotypes of mutants in genes involved in transferase activity transferring hexosyl groups (P < 0.001). The red line indicates an adjusted P value of <0.01.
Poacic Acid Is Synergistic with Drugs That Target the Cell Wall and Membrane Integrity.
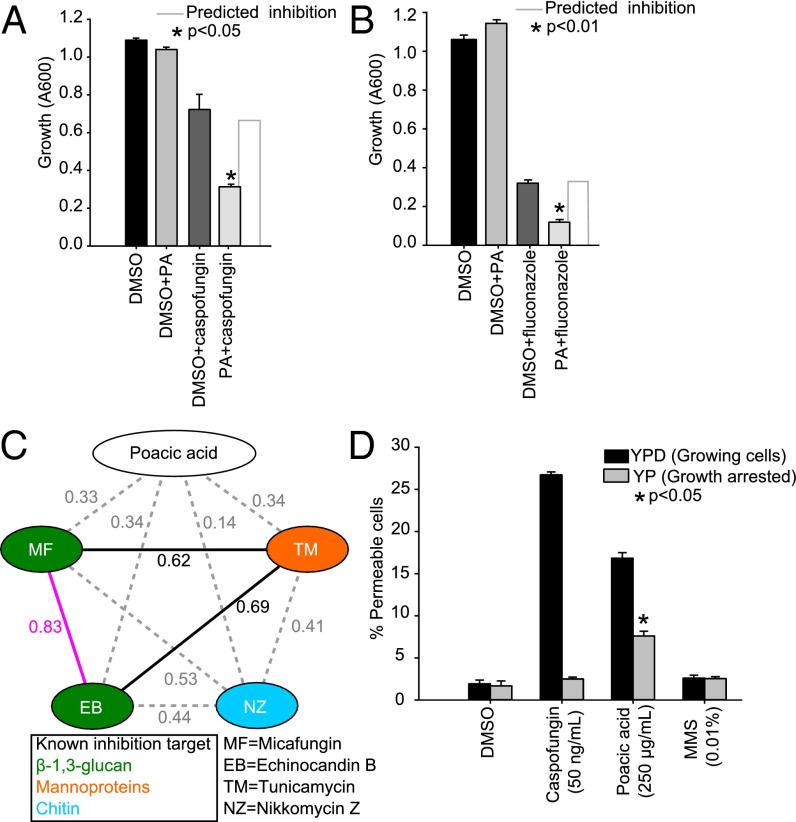
Given that poacic acid may directly target the cell wall or the integrity signaling pathway, we tested whether the mode of action of poacic acid differed from that of the echinocandin caspofungin. Echinocandins damage the yeast cell wall by noncompetitive binding of the β-1,3-glucan synthase complex at the Fks1p subunit (41, 42). Synergistic interactions occur with drugs targeting the same or a functionally related pathway but through different targets (43). We found significant synergistic effects (Fig. 4A) between poacic acid and caspofungin (P < 0.05). This interaction suggests that poacic acid targets the cell wall but does so through a mechanism distinct from that of caspofungin. Because echinocandins are also synergistic with antifungal azoles that target ergosterol biosynthesis and compromise membrane integrity (44), we tested and determined that poacic acid also displayed significant synergy with fluconazole (Fig. 4B) (P < 0.01).
Fig. 4.
Synergisms and the mode of action of poacic acid. (A) Poacic acid (125 µg/mL) is significantly synergistic with caspofungin (12.5 ng/mL). (B) Poacic acid (125 µg/mL) is also synergistic with fluconazole (3.8 µg/mL). (C) Morphological similarity between poacic acid and other cell wall-affecting agents was measured based on the correlation coefficient value (R) of their morphological profiles. (D) Poacic acid causes cell leakage within 4 h of treatment, similar to the cell wall-targeting compound caspofungin. The leakage is most apparent in actively growing cells [yeast extract peptone dextrose (YPD)] compared with cells arrested without a carbon source [yeast extract peptone (YP)]. DMSO and MMS were included as control agents that do not directly affect cell wall integrity. In arrested cells, poacic acid had significantly greater cell leakage than other treatments. One-way ANOVA and Tukey’s test were used to calculate the differences between treatments (mean ± SE). PA, poacic acid.
Poacic Acid-Induced Morphologies Are Unique Compared with Those from Other Cell Wall-Targeting Agents.
Compounds that induce similar morphological responses can be indicative of similar modes of action. To determine how similar the morphology induced by poacic acid is to that induced by other cell wall-affecting drugs, we compared their morphological profiles. Two echinocandins (micafungin and echinocandin B), both of which bind Fks1p, had morphological profiles that were highly correlated with each other, whereas poacic acid-treated cells had lower morphological correlations with these and other cell wall-affecting compounds (Fig. 4C). Thus, although there is some morphological similarity with other cell wall agents, the morphological response to poacic acid suggests that it may have a mode of action that is different from that of other cell wall-targeting agents.
Poacic Acid Causes Rapid Cell Leakage.
Cell wall-targeting agents, such as echinocandins, can lead to compromised cell integrity and ultimately, cell lysis from turgor pressure (45). We investigated whether poacic acid caused cell lysis in a similar way. We tested the extent of cell permeability after 4 h of treatment with poacic acid, caspofungin, methyl methanesulfonate (MMS), or DMSO using a propidium iodide dye that is taken up only by cells with compromised cell integrity. MMS, an agent that does not cause rapid cell wall damage, was included as a negative control. We found that both caspofungin and poacic acid caused rapid cell leakage, whereas MMS and DMSO did not (Fig. 4D).
When growth was arrested by depriving cells of a carbon source, the effect of caspofungin was diminished, supporting the known mode of action of echinocandins, which inhibit glucan synthesis. The effects of poacic acid were reduced in nonactively growing cells, but leakage was still significantly greater (P < 0.05) than in all other treatments (Fig. 4D). The mechanism by which poacic acid causes leakage is lessened without active growth, showing that the compound can still cause leakage in nonactively growing cells, unlike with echinocandins. This result could indicate a general disruption of cell wall integrity rather than an enzymatic target and thus, a different mode of action.
Poacic Acid Localizes to the Cell Surface and Targets β-1,3-Glucan.
We next sought to determine to which cell wall component poacic acid binds by localizing the compound in treated cells. As a ferulate derivative, poacic acid is fluorescent, enabling us to visualize its accumulation at the cell surface (Fig. 5A). Based on this result together with poacic acid’s chemical genomic profile, morphological profile, phenotypic similarity to fks1∆, and ability to cause cell leakage, we hypothesized that poacic acid targets the β-1,3-glucan layer and thus, rapidly compromises cell integrity, leading to cell lysis when turgor pressure bursts the weakened cell wall. The absence of chitin-related genes in both the chemical genomic and morphological profiles and the low correlation between the morphological profile of poacic acid and the chitin targeting compound nikkomycin Z led us to believe that the chitin layer is not the cell wall target of poacic acid. Furthermore, the uniform staining pattern with poacic acid is different from the calcofluor white staining of chitin, which preferentially binds the bud neck and bud scar.
Fig. 5.
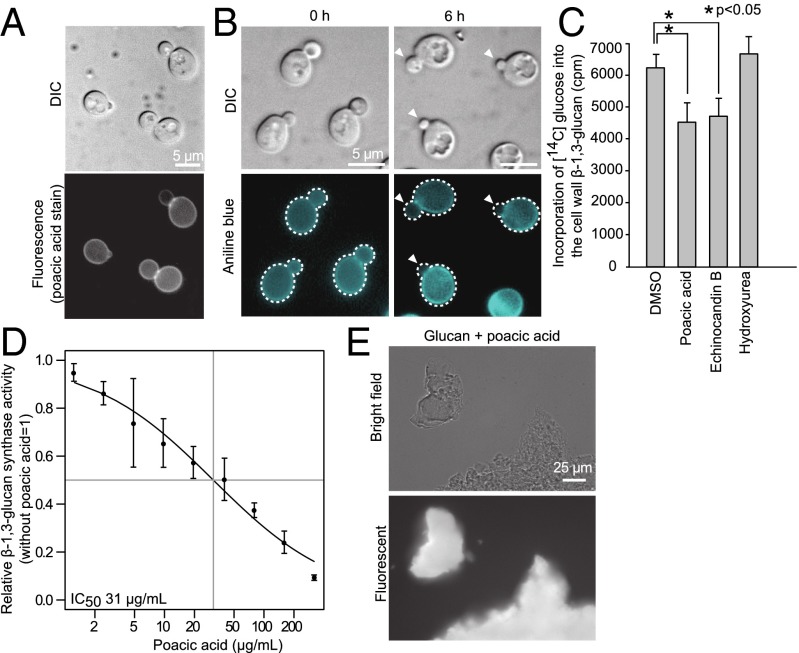
Poacic acid targets β-1,3-glucan. (A) Poacic acid is fluorescent and accumulates on the cell wall. Poacic acid inhibits β-1,3-glucan in vivo as shown by (B) the decrease in signal from aniline blue staining (arrowheads) and (C) the incorporation of 14C-labeled glucose into the β-1,3-glucan layer of the cell wall (P < 0.05). Concentrations of poacic acid, echinocandin B, and hydroxyurea were 250 µg/mL, 4 µg/mL, and 30 mM, respectively. (D) Poacic acid inhibits β-1,3-glucan synthase activity in vitro with an IC50 of 31 µg/mL. (E) Poacic acid directly binds purified yeast glucan. Student’s t test was used to determine significant differences (mean ± SD). DIC, differential interference contrast.
We suggest that the mode of poacic acid action is distinct from that of the echinocandins, acting through direct binding to the glucan fibrils rather than inhibition of glucan synthase. This hypothesis is supported by observations that poacic acid does not localize specifically to the site of bud growth, like Fks1p (46), but rather, binds across the entire cell surface (Fig. 5A). Poacic acid can inhibit β-1,3-glucan synthesis in vivo as shown by significantly decreased glucan staining in buds (Fig. 5B and Fig. S1) and significantly less 14C-glucose incorporation into the β-1,3-glucan layer after poacic acid treatment (Fig. 5C). We also observed an in vitro inhibition of β-1,3-glucan synthesis after poacic acid treatment (Fig. 5D). By incubating purified yeast glucan with poacic acid and observing fluorescence, we found that poacic acid directly binds β-1,3-glucan (Fig. 5E). Furthermore, although poacic acid can reduce aniline blue staining of β-1,3-glucan in buds, it does not change mannoprotein staining with fluorescent dye-conjugated Con A (Fig. S1), which suggests that poacic acid acts primarily on the formation of the glucan fibrils rather than by inhibiting mannoprotein assembly in the cell wall.
Poacic Acid Is an Inhibitor of Fungal and Oomycete Plant Pathogens.
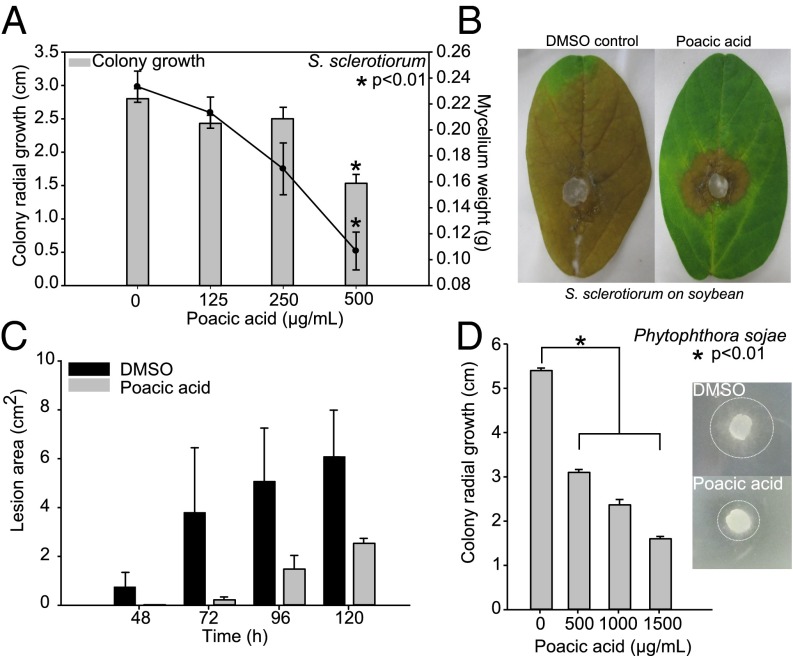
As a plant-derived natural product, poacic acid may have a potential use in organic agriculture, which is presently lacking in fungicide diversity beyond copper sulfate mixtures. We initially tested the effects of poacic acid on S. sclerotiorum, an ascomycete fungal pathogen with an extremely broad host plant range (>400 species) and worldwide distribution. In soybeans, S. sclerotiorum causes Sclerotinia stem rot or white mold of soybean. The incorporation of poacic acid into culture media caused a significant (P < 0.01) dose-dependent decrease in fungal growth both on agar plates and in liquid cultures, which was evidenced by decreases in colony radial growth and fungal mass (Fig. 6A). We further investigated whether poacic acid could inhibit lesion development in planta on detached soybean leaves. Solvent (DMSO) control or poacic acid (500 µg/mL) solutions were applied to detached leaves before inoculation with agar plugs containing actively growing mycelia of S. sclerotiorum. Lesion development was monitored daily up to 120 h postinoculation. Poacic acid treatment markedly reduced lesion development over this time course compared with the control (Fig. 6 B and C). We also found poacic acid to be similarly effective against the ascomycete A. solani, which causes early blight in tomato and potato crops (Fig. S2). These data show that poacic acid inhibits fungal growth in vitro and in planta, with promising agricultural applications.
Fig. 6.
Poacic acid inhibits the growth of fungal and oomycete plant pathogens. (A) Colony growth on plates and mycelia weight of S. sclerotiorum (strain 1980) in liquid culture were significantly inhibited by poacic acid in a dose-dependent manner. (B and C) A single aerosol treatment of poacic acid (500 µg/mL) before inoculation inhibited white mold lesion development on soybean leaves in planta. (D) Representative photographs were taken 96 h postinoculation. Poacic acid significantly inhibited colony growth of P. sojae (field isolate 7 d of growth). Dashed circles in Inset indicate the mycelium front after 2 d of growth. One-way ANOVA and Tukey’s test were used to calculate the difference between drug treatments among treatments (mean ± SE).
Fungi generally have 30–80% glucan in their cell walls (47); similarly, oomycetes have a cell wall containing β-1,3-glucan and β-1,6-glucan, but unlike fungi, oomycete walls contain a cellulose layer rather than chitin. Oomycetes are broadly distributed, economically significant pathogens, and a natural fungicide that could affect both true fungi and oomycetes by disruption of the glucan layer could be of high value. We found that poacic acid significantly reduces colony growth of the oomycete P. sojae (Fig. 6D) (P < 0.01), a widespread pathogen that causes root and stem rot of soybeans. Given its effectiveness against both fungi and oomycetes, poacic acid may have potential as a plant-derived fungicide with broad action.
Discussion
Through chemoprospecting of lignocellulosic hydrolysates, we have identified a promising antifungal agent. Combining chemical genomic and morphological analyses, we determined that poacic acid targets β-1,3-glucan within fungal cell walls. Inhibition of glucan synthesis in vivo and in vitro and cell-wide localization and direct binding of purified glucan indicate that the compound can bind to β-1,3-glucans in the growing glucan fibrils as well as the mature wall. The cell wall dye Congo red may also bind growing glucan fibrils (48), but it also binds to chitin, and biochemical evidence indicates that the primary target of Congo red is chitin (49). Poacic acid targets the β-1,3-glucan layer of fungal cell walls in a manner distinct from that of other cell wall-affecting agents (e.g., caspofungin and nikkomycin Z) and therefore, represents a previously undescribed compound targeting β-1,3-glucan. Although we found no effects of poacic acid on mannoprotein assembly, direct binding of glucan fibrils outside the plasma membrane may also result in inhibition of cell wall assemblages, such as the glucan-transglycolase Gas1p and the chitin transglycosylases Crh1p and Utr2p, that connect chitin chains to glucans, which require glucan as a substrate or cosubstrate.
In nature, diferulates strengthen monocot cell walls by cross-linking polysaccharides (arabinoxylans) to each other and polysaccharides to lignin—in both cases by radical coupling mechanisms (16, 17). It remains unknown if poacic acid has a similar interaction with the fungal cell wall polysaccharides. However, this work raises the question about whether diferulates may have a dual role as antifungal secondary metabolites that confer protection to certain plants. Other diferulates may have the same potential to bind glucan, but the size/physical properties of poacic acid may contribute to its more optimal bioactivity.
Against yeast, the bioactivity of poacic acid is similar to the widely used fungicide picoxystrobin (IC50 of 308 µM), lower than thiabendazole (IC50 of 607 µM), and considerably more toxic than copper sulfate (IC50 of 2.4 mM) (25). Poacic acid may also have potential to be used combined with agricultural azoles through its documented synergism to slow the development of azole resistance. Although there are conventional fungicides effective at lower doses (e.g., captan at IC50 of 19 µM and prochloraz at IC50 of 132 µM) (25), most conventional agents are specific to either the Eumycota or Oomycota, whereas poacic acid affects both. Options for organic agriculture are limited to copper-based fungicides, which are facing increasing restrictions because of copper accumulation in soil ecosystems (8, 9). Furthermore, as a natural plant-based phenolic acid, poacic acid would likely be rapidly broken down in the soil and would not accumulate (50). Field trials, application strategies (e.g., seed treatment vs. foliar spray), and more diverse pathogen tests will be necessary to determine its performance as an agricultural fungicide and evaluate its persistence in the environment, but it may also provide an important and abundant lead compound that could be modified for increased efficacy.
Although it was identified from lignocellulosic hydrolysates for bioethanol fermentation, poacic acid alone is not likely to be a primary inhibitor affecting fermentation given its relatively low concentration (0.1 µM) (Table S4). However, it could be synergistic with other inhibitory agents (e.g., furfural and phenolics) or other diferulates (e.g., 8–5-C) (2, 16). The combined effects of diferulates with other inhibitors could significantly affect lignocellulosic biofuel synthesis by fungi, but such combinatorial effects have yet to be tested.
The complementary profiling methodologies that we applied to the analysis of poacic acid’s effects, including chemical genomic profiling and morphological profiling, are powerful and can provide high-resolution predictions of targeted processes; this work highlights the power of the combined approach. Given the increasing throughput of both techniques thanks to advances in next generation sequencing and automated microscopy, the use of both genetic and morphological approaches in large-scale screening of drug libraries may allow unbiased whole-cell target identification with less reliance on target-centric high-throughput screening methods.
This study was designed to identify novel bioactive compounds from lignocellulosic hydrolysates. Given the goal of cellulosic ethanol production [60 billion L/y by 2022, requiring 0.6–1.2 trillion L hydrolysate/y assuming 5–10% (vol/vol) ethanol before distillation] (51, 52), even low-abundance compounds within hydrolysates could be available in significant quantities. We have detected monomeric ferulate in hydrolysates at markedly higher levels (up to 1.7 mM in alkaline H2O2-treated corn stover). If synthesized from the recovered ferulate component posthydrolysis, poacic acid may confer greater value to lignocellulosic conversion. Our results highlight the chemical diversity within lignocellulosic hydrolysates as a source of potentially valuable chemicals.
Methods
Detailed methods are provided in SI Methods.
Chemical Genomic Analysis.
Chemical genomic analysis of poacic acid was performed as described previously (26, 53). The tested yeast deletion collection had ∼4,000 strains using the genetic background described by Andrusiak (54). The optimal inhibitory concentration of poacic acid for chemical genomic profiling [70–80% growth vs. solvent control in yeast extract-peptone-galactose medium after 24 h of growth] was determined using an eight-point dose curve. A concentration of 88 µg/mL inhibited growth within this range; 200-µL cultures of the pooled deletion collection of S. cerevisiae were grown with 88 µg/mL poacic acid (n = 3) or a DMSO control in triplicate for 48 h at 30 °C. Genomic DNA was extracted using the Epicentre MasterPure Yeast DNA Purification Kit. Mutant-specific molecular barcodes were amplified with specially designed multiplex primers (28). The barcodes were sequenced using an Illumina MiSeq. Replicates of each condition, poacic acid (n = 3) or DMSO (n = 2), were sequenced. The barcode counts for each yeast deletion mutant in the presence of poacic acid were compared to the DMSO control conditions to determine sensitivity or resistance of individual strains (the chemical genetic interaction score) (26). To determine a P value for each top sensitive and resistant mutant, we used the EdgeR package (55, 56). A Bonferroni-corrected hypergeometric distribution test was used to search for significant enrichment of GO terms among the top 10 sensitive and resistant deletion mutants (57). To understand the pathways that were most affected by poacic acid, we developed a protein complex/pathway score based on the summation of the z scores for each complex/pathway (Pathway z score). Correlation of the chemical genomic profile of poacic acid with the yeast genetic interaction network was performed as previously described (34).
Multivariate Morphological Analysis.
Cells of budding yeast S. cerevisiae (BY4741 his3∆::KanMX; hereafter, his3∆) were cultured in 2 mL 1% Bacto yeast extract (BD Biosciences), 2% Bacto peptone (BD Biosciences), and 2% glucose (YPD) with 0, 25, 50, 75, 100, or 125 μg/mL poacic acid or a DMSO control at 25 °C for 16 h until the early log phase. The maximum concentration of the drug (125 μg/mL) was determined based on the growth inhibition rates (10%). Cell fixation, staining, and observation were performed (n = 5) as described previously (37). Images of cell shape, actin, and nuclear DNA were analyzed using the image processing software CalMorph (version 1.2), which extracted a total of 501 morphological quantitative values from at least 200 individual cells in each experiment (37). Images were processed using Photoshop CS2 (Adobe Systems) for illustrative purposes.
To assess the morphological similarity between the cells treated with poacic acid and nonessential deletion mutants or cells treated with other cell wall-affecting drugs, their morphological profiles were compared as described previously (36). To identify functional gene clusters, the most significant similar mutants (43 genes, P < 0.01 after Bonferroni correction, t test) were selected as a query for GO term analysis (GO term finder in the Saccharomyces genome database).
To extract independent and characteristic features of morphology induced by poacic acid, a two-step principal component analysis was performed as described previously (39). To compare phenotypic noise in the yeast population (poacic acid vs. DMSO), the noise score was calculated as described previously (58).
Measurement of in Vivo β-1,3-Glucan Synthesis.
Inhibition of in vivo β-1,3-glucan synthesis was measured as described previously with slight modification (n = 3) (59). Yeast cells (his3∆) were grown in YPD to early log phase at 25 °C. The cultured cells were diluted to 1 × 107 cells per 1 mL with 1 mL of YPD medium containing one-tenth the glucose containing 23.125 kBq [14C]glucose (ARC0122; American Radiolabeled Chemicals) and test compounds [250 µg/mL for poacic acid, 4 µg/mL for echinocandin B, 30 mM for hydroxyurea (negative control), or 0.4% (vol/vol) DMSO as a solvent control]. The cells were radiolabeled by culturing at 25 °C for 2 h. The labeled cells were harvested and incubated with 1 N NaOH at 80 °C for 30 min. The insoluble pellets were resuspended in 10 mM Tris⋅HCl, pH 7.5, containing 5 mg/mL zymolyase 100T (Seikagaku) and incubated at 37 °C for 18 h. After digestion, the zymolyase-resistant material was removed by centrifugation (15,000 × g for 15 min), and the zymolyase-degradation product (mostly β-1,3-glucan) was purified by ultrafiltration with a centrifugal filter membrane (Amicon Ultra 0.5 mL; molecular weight cutoff is 10,000; Millipore). The flow-through fraction was mixed with scintillation mixture (Ultima Gold; PerkinElmer), and radioactivity was measured by a scintillation counter (LSC-6100; Aloka). The differences in the incorporation rates in samples were normalized by ∆OD600 measured before and after the labeling period.
Measurement of β-1,3-Glucan Synthase Activity in the Membrane Fraction.
After the membrane fraction was prepared from S. cerevisiae BY4741 as described previously (n = 3) (60), β-1,3-glucan synthase activity was measured as described previously (n = 3) (61) with slight modification. Briefly, 20 µL membrane fraction (∼70 µg total protein) was added to a reaction mixture (final volume of 100 µL) containing 50 mM Tris⋅HCl, pH 7.5, 10 mM potassium fluoride, 1 mM EDTA, 0.2 mM UDP-Glc (with 89 Bq UDP-[Glucose-14C]; NEC403; PerkinElmer), and different concentrations of poacic acid (1.25, 2.5, 5, 10, 20, 40, 80, 160, or 320 µg/mL). The reaction mixture was incubated at 25 °C for 30 min and stopped by the addition of ethanol. To trap reaction product (β-1,3-glucan polymer), the reaction mixture was filtered through the membrane filter (mixed cellulose esters; 0.2 µm in pore size; ADVANTEC), washed one time with 2 mL distilled water, and dried at room temperature. After addition of scintillation mixture (Econofluor-2; PerkinElmer), radioactivity was measured by a scintillation counter (LSC-6100; Aloka). Inhibition curves and IC50 values were determined using R software (ver. 3.0.1) by sigmoidal curve fitting with the glm function.
Growth Inhibition of Plant Pathogens.
To test inhibition in liquid culture, a dose curve of 0, 125, 250, and 500 µg/mL poacic acid in 100 mL potato dextrose broth (n = 3) was used. Cultures of S. sclerotiorum strain 1980 were inoculated with 100 µL homogenized mycelia and grown at 25 °C for 48 h. The mycelia in the liquid media were dried and weighed. The growth inhibition of poacic acid on solid agar cultures (potato dextrose agar) was assessed by generating replicate plates (n = 3) containing 0, 125, 250, and 500 µg/mL poacic acid. Plates were inoculated with an actively growing plug of S. sclerotiorum and grown at 25 °C. The mycelial radial growth after 48 h was measured. Inhibition of S. sclerotiorum in planta was tested by inoculating detached soybean leaves of the commercial variety Williams 82 with an agar plug of actively growing S. sclerotiorum mycelia. Leaves were treated one time before inoculation with either an aerosol spray of water with DMSO (control) or a 500 µg/mL solution of poacic acid. Leaves were incubated in a moist environment, and lesion development was monitored up to 120 h postinoculation. Field strains of P. sojae and A. solani were grown on cornmeal and potato dextrose agar plates, respectively, at room temperature for 7 and 5 d, respectively, before measurement. The growth inhibition of poacic acid was assessed at 0, 500, 1,000, and 1,500 µg/mL in replicate plates (n = 3). Agar plugs from actively growing cultures were placed at the center of the plates and allowed to grow at room temperature. Colony diameter was monitored for each treatment in a time-course experiment. One-way ANOVA and Tukey’s test were used to calculate the differences between drug treatments among treatments.
Supplementary Material
Acknowledgments
We thank T. K. Sato, E. Hendel, S. Morford, and N. Keller for critical discussions of the manuscript. J.S.P., F.L., L.H., A.J.H., A.U., J.J.C., S.M., I.M.O., and J.R. are funded by Department of Energy (DOE) Great Lakes Bioenergy Research Center DOE Biological and Environmental Research Office of Science Grant DE-FC02-07ER64494. J.S.P., F.L., and M.K. are supported by Wisconsin Alumni Research Foundation Award MSN178899. H.O. is a research fellow of the Japan Society for the Promotion of Science. S.C.L. is supported by a RIKEN Foreign Postdoctoral Fellowship. A.R., D.L.S., and M.K. are supported by Wisconsin Soybean Marketing Board Grant MSN172403. R.D. and C.L.M. are supported by National Institutes of Health Grants 1R01HG005084-01A1, 1R01GM104975-01, and R01HG005853 and National Science Foundation Grant DBI 0953881. C.L.M. and C.B. are supported by the Canadian Institute for Advanced Research Genetic Networks Program. M.K. is supported by United Soybean Board Grant MSN143317. Y.O. is supported by Ministry of Education, Culture, Sports, Science and Technology, Japan Grant for Scientific Research 24370002.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410400112/-/DCSupplemental.
References
- 1.Sun Y, Cheng J. Hydrolysis of lignocellulosic materials for ethanol production: A review. Bioresour Technol. 2002;83(1):1–11. doi: 10.1016/s0960-8524(01)00212-7. [DOI] [PubMed] [Google Scholar]
- 2.Piotrowski JS, et al. Death by a thousand cuts: The challenges and diverse landscape of lignocellulosic hydrolysate inhibitors. Front Microbiol. 2014;5(2014):90. doi: 10.3389/fmicb.2014.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmqvist E, Hahn-Hägerdal B. Fermentation of lignocellulosic hydrolysates. II: Inhibitors and mechanisms of inhibition. Bioresour Technol. 2000;74(1):25–33. [Google Scholar]
- 4.Skerker JM, et al. Dissecting a complex chemical stress: Chemogenomic profiling of plant hydrolysates. Mol Syst Biol. 2013;9:674. doi: 10.1038/msb.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.FitzPatrick M, Champagne P, Cunningham MF, Whitney RA. A biorefinery processing perspective: Treatment of lignocellulosic materials for the production of value-added products. Bioresour Technol. 2010;101(23):8915–8922. doi: 10.1016/j.biortech.2010.06.125. [DOI] [PubMed] [Google Scholar]
- 6.Avenot HF, Sellam A, Karaoglanidis G, Michailides TJ. Characterization of mutations in the iron-sulphur subunit of succinate dehydrogenase correlating with Boscalid resistance in Alternaria alternata from California pistachio. Phytopathology. 2008;98(6):736–742. doi: 10.1094/PHYTO-98-6-0736. [DOI] [PubMed] [Google Scholar]
- 7.Leroch M, Kretschmer M, Hahn M. Fungicide resistance phenotypes of Botrytis cinerea isolates from commercial vineyards in south west Germany. J Phytopathol. 2011;159(1):63–65. [Google Scholar]
- 8.Wightwick AM, Salzman SA, Reichman SM, Allinson G, Menzies NW. Effects of copper fungicide residues on the microbial function of vineyard soils. Environ Sci Pollut Res Int. 2013;20(3):1574–1585. doi: 10.1007/s11356-012-1114-7. [DOI] [PubMed] [Google Scholar]
- 9.Mackie KA, Müller T, Zikeli S, Kandeler E. Long-term copper application in an organic vineyard modifies spatial distribution of soil micro-organisms. Soil Biol Biochem. 2013;65(2013):245–253. [Google Scholar]
- 10.Altizer S, Ostfeld RS, Johnson PTJ, Kutz S, Harvell CD. Climate change and infectious diseases: From evidence to a predictive framework. Science. 2013;341(6145):514–519. doi: 10.1126/science.1239401. [DOI] [PubMed] [Google Scholar]
- 11.Garrett KA, Dendy SP, Frank EE, Rouse MN, Travers SE. Climate change effects on plant disease: Genomes to ecosystems. Annu Rev Phytopathol. 2006;44(1):489–509. doi: 10.1146/annurev.phyto.44.070505.143420. [DOI] [PubMed] [Google Scholar]
- 12.Alexander BD, Perfect JR. Antifungal resistance trends towards the year 2000. Implications for therapy and new approaches. Drugs. 1997;54(5):657–678. doi: 10.2165/00003495-199754050-00002. [DOI] [PubMed] [Google Scholar]
- 13.Sarma BK, Singh UP. Ferulic acid may prevent infection of Cicer arietinum by Sclerotium rolfsii. World J Microbiol Biotechnol. 2003;19(2):123–127. [Google Scholar]
- 14.Heer D, Sauer U. Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb Biotechnol. 2008;1(6):497–506. doi: 10.1111/j.1751-7915.2008.00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayakody LN, Hayashi N, Kitagaki H. Identification of glycolaldehyde as the key inhibitor of bioethanol fermentation by yeast and genome-wide analysis of its toxicity. Biotechnol Lett. 2011;33(2):285–292. doi: 10.1007/s10529-010-0437-z. [DOI] [PubMed] [Google Scholar]
- 16.Ralph J, Quideau S, Grabber JH, Hatfield RD. Identification and synthesis of new ferulic acid dehydrodimers present in grass cell walls. J Chem Soc. 1994;23:3485–3498. [Google Scholar]
- 17.Ralph J, et al. Cell wall cross-linking in grasses by ferulates and diferulates. In: Lewis NG, Sarkanen S, editors. Lignin and Lignan Biosynthesis, ACS Symposium Series. American Chemical Society; Washington, DC: 1998. pp. 209–236. [Google Scholar]
- 18.Vismeh R, et al. Profiling of diferulates (plant cell wall cross-linkers) using ultrahigh-performance liquid chromatography-tandem mass spectrometry. Analyst (Lond) 2013;138(21):6683–6692. doi: 10.1039/c3an36709f. [DOI] [PubMed] [Google Scholar]
- 19.Bunzel M, Ralph J, Marita JM, Hatfield RD, Steinhart H. Diferulates as structural components in soluble and insoluble cereal dietary fibre. J Sci Food Agric. 2001;81(7):653–660. [Google Scholar]
- 20.Santiago R, et al. Diferulate content of maize sheaths is associated with resistance to the Mediterranean corn borer Sesamia nonagrioides (Lepidoptera: Noctuidae) J Agric Food Chem. 2006;54(24):9140–9144. doi: 10.1021/jf061830k. [DOI] [PubMed] [Google Scholar]
- 21.Baranowski JD, Davidson PM, Nagel CW, Branen AL. Inhibition of Saccharomyces cerevisiae by naturally occurring hydroxycinnamates. J Food Sci. 1980;45(3):592–594. [Google Scholar]
- 22.Piotrowski JS, Morford SL, Rillig MC. Inhibition of colonization by a native arbuscular mycorrhizal fungal community via Populus trichocarpa litter, litter extract, and soluble phenolic compounds. Soil Biol Biochem. 2008;40(3):709–717. [Google Scholar]
- 23.Jo WJ, et al. Identification of genes involved in the toxic response of Saccharomyces cerevisiae against iron and copper overload by parallel analysis of deletion mutants. Toxicol Sci. 2008;101(1):140–151. doi: 10.1093/toxsci/kfm226. [DOI] [PubMed] [Google Scholar]
- 24.Rogers B, et al. The pleitropic drug ABC transporters from Saccharomyces cerevisiae. J Mol Microbiol Biotechnol. 2001;3(2):207–214. [PubMed] [Google Scholar]
- 25.Fai PB, Grant A. A rapid resazurin bioassay for assessing the toxicity of fungicides. Chemosphere. 2009;74(9):1165–1170. doi: 10.1016/j.chemosphere.2008.11.078. [DOI] [PubMed] [Google Scholar]
- 26.Parsons AB, et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126(3):611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 27.Ho CH, et al. Combining functional genomics and chemical biology to identify targets of bioactive compounds. Curr Opin Chem Biol. 2011;15(1):66–78. doi: 10.1016/j.cbpa.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 28.Smith AM, et al. Quantitative phenotyping via deep barcode sequencing. Genome Res. 2009;19(10):1836–1842. doi: 10.1101/gr.093955.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Yken H, et al. Saccharomyces cerevisiae YCRO17c/CWH43 encodes a putative sensor/transporter protein upstream of the BCK2 branch of the PKC1-dependent cell wall integrity pathway. Yeast. 2001;18(9):827–840. doi: 10.1002/yea.731. [DOI] [PubMed] [Google Scholar]
- 30.Jesch SA, Gaspar ML, Stefan CJ, Aregullin MA, Henry SA. Interruption of inositol sphingolipid synthesis triggers Stt4p-dependent protein kinase C signaling. J Biol Chem. 2010;285(53):41947–41960. doi: 10.1074/jbc.M110.188607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohkuni K, Okuda A, Kikuchi A. Yeast Nap1-binding protein Nbp2p is required for mitotic growth at high temperatures and for cell wall integrity. Genetics. 2003;165(2):517–529. doi: 10.1093/genetics/165.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitagaki H, Wu H, Shimoi H, Ito K. Two homologous genes, DCW1 (YKL046c) and DFG5, are essential for cell growth and encode glycosylphosphatidylinositol (GPI)-anchored membrane proteins required for cell wall biogenesis in Saccharomyces cerevisiae. Mol Microbiol. 2002;46(4):1011–1022. doi: 10.1046/j.1365-2958.2002.03244.x. [DOI] [PubMed] [Google Scholar]
- 33.Surma MA, et al. A lipid E-MAP identifies Ubx2 as a critical regulator of lipid saturation and lipid bilayer stress. Mol Cell. 2013;51(4):519–530. doi: 10.1016/j.molcel.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327(5964):425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinoso-Martín C, Schüller C, Schuetzer-Muehlbauer M, Kuchler K. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot Cell. 2003;2(6):1200–1210. doi: 10.1128/EC.2.6.1200-1210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohnuki S, Oka S, Nogami S, Ohya Y. High-content, image-based screening for drug targets in yeast. PLoS ONE. 2010;5(4):e10177. doi: 10.1371/journal.pone.0010177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohya Y, et al. High-dimensional and large-scale phenotyping of yeast mutants. Proc Natl Acad Sci USA. 2005;102(52):19015–19020. doi: 10.1073/pnas.0509436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada H, Ohnuki S, Roncero C, Konopka JB, Ohya Y. Distinct roles of cell wall biogenesis in yeast morphogenesis as revealed by multivariate analysis of high-dimensional morphometric data. Mol Biol Cell. 2014;25(2):222–233. doi: 10.1091/mbc.E13-07-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohnuki S, et al. Analysis of the biological activity of a novel 24-membered macrolide JBIR-19 in Saccharomyces cerevisiae by the morphological imaging program CalMorph. FEMS Yeast Res. 2012;12(3):293–304. doi: 10.1111/j.1567-1364.2011.00770.x. [DOI] [PubMed] [Google Scholar]
- 40.Iwaki A, Ohnuki S, Suga Y, Izawa S, Ohya Y. Vanillin inhibits translation and induces messenger ribonucleoprotein (mRNP) granule formation in saccharomyces cerevisiae: Application and validation of high-content, image-based profiling. PLoS ONE. 2013;8(4):e61748. doi: 10.1371/journal.pone.0061748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balashov SV, Park S, Perlin DS. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob Agents Chemother. 2006;50(6):2058–2063. doi: 10.1128/AAC.01653-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson ME, Edlind TD. Topological and mutational analysis of Saccharomyces cerevisiae Fks1. Eukaryot Cell. 2012;11(7):952–960. doi: 10.1128/EC.00082-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cokol M, et al. Systematic exploration of synergistic drug pairs. Mol Syst Biol. 2011;7(2011):544. doi: 10.1038/msb.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiraz N, et al. Synergistic activities of three triazoles with caspofungin against Candida glabrata isolates determined by time-kill, Etest, and disk diffusion methods. Antimicrob Agents Chemother. 2010;54(5):2244–2247. doi: 10.1128/AAC.01527-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cassone A, Mason RE, Kerridge D. Lysis of growing yeast-form cells of Candida albicans by echinocandin: A cytological study. Sabouraudia. 1981;19(2):97–110. [PubMed] [Google Scholar]
- 46.Utsugi T, et al. Movement of yeast 1,3-β-glucan synthase is essential for uniform cell wall synthesis. Genes Cells. 2002;7(1):1–9. doi: 10.1046/j.1356-9597.2001.00495.x. [DOI] [PubMed] [Google Scholar]
- 47.Free SJ. Fungal cell wall organization and biosynthesis. Adv Genet. 2013;81:33–82. doi: 10.1016/B978-0-12-407677-8.00002-6. [DOI] [PubMed] [Google Scholar]
- 48.Kopecká M, Gabriel M. The influence of congo red on the cell wall and (1----3)-beta-D-glucan microfibril biogenesis in Saccharomyces cerevisiae. Arch Microbiol. 1992;158(2):115–126. doi: 10.1007/BF00245214. [DOI] [PubMed] [Google Scholar]
- 49.Imai K, Noda Y, Adachi H, Yoda K. A novel endoplasmic reticulum membrane protein Rcr1 regulates chitin deposition in the cell wall of Saccharomyces cerevisiae. J Biol Chem. 2005;280(9):8275–8284. doi: 10.1074/jbc.M409428200. [DOI] [PubMed] [Google Scholar]
- 50.Chen L, et al. Trichoderma harzianum SQR-T037 rapidly degrades allelochemicals in rhizospheres of continuously cropped cucumbers. Appl Microbiol Biotechnol. 2011;89(5):1653–1663. doi: 10.1007/s00253-010-2948-x. [DOI] [PubMed] [Google Scholar]
- 51.Westbrook J, Barter GE, Manley DK, West TH. A parametric analysis of future ethanol use in the light-duty transportation sector: Can the US meet its Renewable Fuel Standard goals without an enforcement mechanism? Energy Policy. 2014;65(2014):419–431. [Google Scholar]
- 52.Lau MW, Dale BE. Cellulosic ethanol production from AFEX-treated corn stover using Saccharomyces cerevisiae 424A(LNH-ST) Proc Natl Acad Sci USA. 2009;106(5):1368–1373. doi: 10.1073/pnas.0812364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fung S-Y, et al. Unbiased screening of marine sponge extracts for anti-inflammatory agents combined with chemical genomics identifies girolline as an inhibitor of protein synthesis. ACS Chem Biol. 2014;9(1):247–257. doi: 10.1021/cb400740c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrusiak K. 2012. Adapting S. cerevisiae chemical genomics for identifying the modes of action of natural compounds. Masters thesis (University of Toronto, Toronto) [DOI] [PubMed]
- 55.Robinson DG, Chen W, Storey JD, Gresham D. Design and analysis of Bar-seq experiments. G3 (Bethesda) 2014;4(1):11–18. doi: 10.1534/g3.113.008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boyle EI, et al. GO:TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20(18):3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yvert G, et al. Single-cell phenomics reveals intra-species variation of phenotypic noise in yeast. BMC Syst Biol. 2013;7(1):54. doi: 10.1186/1752-0509-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abe M, Qadota H, Hirata A, Ohya Y. Lack of GTP-bound Rho1p in secretory vesicles of Saccharomyces cerevisiae. J Cell Biol. 2003;162(1):85–97. doi: 10.1083/jcb.200301022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abe M, et al. Yeast 1,3-β-glucan synthase activity is inhibited by phytosphingosine localized to the endoplasmic reticulum. J Biol Chem. 2001;276(29):26923–26930. doi: 10.1074/jbc.M102179200. [DOI] [PubMed] [Google Scholar]
- 61.Inoue SB, et al. Characterization and gene cloning of 1,3-β-D-glucan synthase from Saccharomyces cerevisiae. Eur J Biochem. 1995;231(3):845–854. doi: 10.1111/j.1432-1033.1995.tb20770.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.