Significance
Bone morphogenetic protein (BMP) activity is regulated by prodomains. Here, structures of BMP procomplexes reveal an open-armed conformation. In contrast, the evolutionarily related, latent TGF-β1 procomplex is cross-armed. We propose that in the TGF-β and BMP family, conversion between cross-armed and open-armed conformations may regulate release and activity of the growth factor.
Abstract
Bone morphogenetic proteins (BMPs) belong to the TGF-β family, whose 33 members regulate multiple aspects of morphogenesis. TGF-β family members are secreted as procomplexes containing a small growth factor dimer associated with two larger prodomains. As isolated procomplexes, some members are latent, whereas most are active; what determines these differences is unknown. Here, studies on pro-BMP structures and binding to receptors lead to insights into mechanisms that regulate latency in the TGF-β family and into the functions of their highly divergent prodomains. The observed open-armed, nonlatent conformation of pro-BMP9 and pro-BMP7 contrasts with the cross-armed, latent conformation of pro-TGF-β1. Despite markedly different arm orientations in pro-BMP and pro-TGF-β, the arm domain of the prodomain can similarly associate with the growth factor, whereas prodomain elements N- and C-terminal to the arm associate differently with the growth factor and may compete with one another to regulate latency and stepwise displacement by type I and II receptors. Sequence conservation suggests that pro-BMP9 can adopt both cross-armed and open-armed conformations. We propose that interactors in the matrix stabilize a cross-armed pro-BMP conformation and regulate transition between cross-armed, latent and open-armed, nonlatent pro-BMP conformations.
Members of the TGF-β family including bone morphogenetic proteins (BMPs) are biosynthesized and processed into complexes between large prodomains and smaller, C-terminal mature growth factor (GF) domains that are separated by proprotein convertase (PC) (furin) cleavage sites. In the original isolation of proteins responsible for BMP activity, bone was first demineralized with 0.5 M HCl. The resulting residual matrix was extracted with 6 M urea or 4 M guanidine HCl (1–3). During subsequent purification under largely denaturing conditions, the GF domains were separated from their prodomains. Therefore, little attention was paid to the potential existence of BMP procomplexes. However, evidence exists that BMP prodomains contribute to maintaining BMP GF domains inactive or latent in vivo. For example, early studies showed a 60-fold increase in total BMP activity during the first two purification steps following extraction of the BMP, which was interpreted as purification of BMP away from an inhibitor (2). This finding is consistent with the presence of largely latent complexes between BMPs, their prodomains, and extracellular matrix components in the insoluble residual matrix from which BMPs were purified. In agreement with a regulatory role for the prodomain, mutations of secondary PC sites within the prodomain perturb embryonic development in insects and vertebrates, suggesting that prodomains of several BMPs remain associated with GFs after secretion and regulate the distance over which BMPs signal (4–7). An important role for the prodomain in development is also illustrated by prodomain mutations, including in secondary PC cleavage sites, that cause human diseases (5, 7).
Pro-TGF-β is latent; however, when overexpressed as recombinant proteins, most BMPs are active. Although noncovalently associated with their GF after secretion, the prodomains of most BMPs do not bind strongly enough to prevent GF from binding to receptors and signaling (8, 9). To better understand such differences among members of the TGF-β family, we examine the structure of pro-BMP9 and compare it to the previously described, cross-armed conformation of pro-TGF-β1 (10). Although a member of the BMP subfamily and possessing chondrogenic and osteogenic activity, BMP9 is expressed in liver and is required for properly organized blood and lymphatic vascular development (11, 12). Mutations in the prodomain of BMP9, in its receptor Alk1, and in its coreceptor endoglin cause phenotypically overlapping hereditary hemorrhagic telangiectasias (13–15).
Here, we reveal surprising open-armed conformations of pro-BMPs 7 and 9. We propose that binding to interactors in the matrix may regulate transition between open-armed and cross-armed conformations in the TGF-β family and that these conformations regulate GF latency.
Results
Structures of BMP Procomplexes.
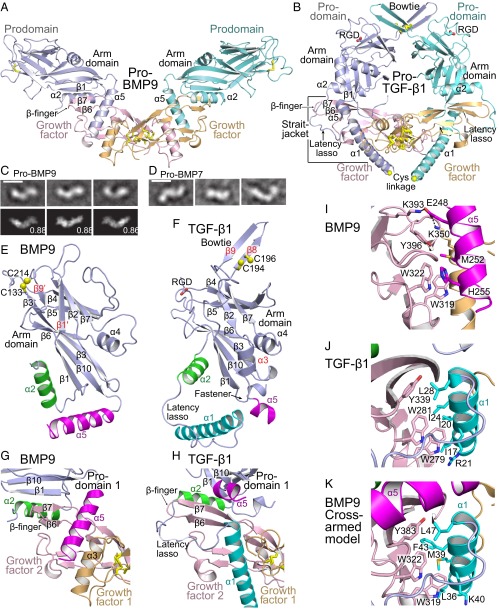
In marked contrast to the cross-armed, ring-like conformation of pro-TGF-β1 (10), crystal structures of natively glycosylated pro-BMP9 reveal an unexpected, open-armed conformation (Fig. 1 A and B and Table S1). All negative stain EM class averages show an open-armed conformation for pro-BMP9 (Fig. 1C and Fig. S1) and a similar, although less homogenous, open-armed conformation for pro-BMP7 (Fig. 1D and Fig. S2). Crystal structure experimental electron density is excellent (Fig. S3) and allows us to trace the complete structure of each pro-BMP9 arm domain (residues 63–258; Fig. 1E). As in pro-TGF-β1, the arm domain has two β-sheets that only partially overlap. Hydrophobic, nonoverlapping portions of the β-sheets are covered by meandering loops and the α4-helix (Fig. 1 E and F). Comparison of pro-BMP9 and pro-TGF-β1 arm domains defines a conserved core containing two four-stranded β-sheets and the α4-helix (labeled in black in Fig. 1 E and F).
Fig. 1.
Structures. (A and B) Cartoon diagrams of pro-BMP9 (A) and pro-TGF-β1 (10) (B) with superimposition on GF dimers. Disulfides (yellow) are shown in stick. (C and D) Representative negative-stain EM class averages of pro-BMP9 (C) and pro-BMP7 (D). Best correlating projections of the pro-BMP9 crystal structure with their normalized cross-correlation coefficients are shown below class averages. (Scale bars, 100 Å.) (E and F) BMP9 and TGF-β1 prodomains shown in cartoon after superimposition. Core arm domain secondary structural elements are labeled in black and others in red. Helices that vary in position between cross- and open-armed conformations are color-coded. Spheres show Cys S atoms. (G–K) Prodomain–GF interactions in pro-BMP9 (G and I), pro-TGF-β1 (H and J), and a model of cross-armed pro-BMP9 (K). Structures are superimposed on the GF monomer. Colors are as in A, B, E, and F. Key residues are shown in stick.
One of the BMP9 arm domain β-sheets joins a finger-like β-sheet in the GF to form a super β-sheet (Fig. 1 A and G). Each GF monomer has a hand-like shape. The two BMP9 GF hands dimerize in their palm regions (Fig. 1A and Fig. S4) in an interface that buries 1,280 Å2 of solvent-accessible surface.
The interface between each BMP9 prodomain and GF monomer buries 1,440 Å2. The larger size of the prodomain–GF interface than the GF–GF interface emphasizes its significance, as does the super β-sheet interface between the prodomain and GF and the burial of hydrophobic residues by this interface and by the prodomain α2-helix (Fig. 1A). A specialization in pro-BMP9 not present in pro-TGF-β1 is a long α5-helix (Fig. 1 A, B, E, and F) that is a C-terminal appendage to the arm domain and that separately interacts with the GF dimer to bury 750 Å2 (Fig. 1A).
Despite markedly different arm domain orientations, topologically identical secondary structure elements form the interface between the prodomain and GF in pro-BMP9 and pro-TGF-β1: the β1-strand and α2-helix in the prodomain and the β6- and β7-strands in the GF (Fig. 1 A, B, G, and H). The outward-pointing, open arms of pro-BMP9 have no contacts with one another, which results in a monomeric prodomain–GF interaction. In contrast, the inward pointing arms of pro-TGF-β1 dimerize through disulfides in their bowtie motif, resulting in a dimeric, and more avid, prodomain-GF interaction (Fig. 1 A and B).
Twists at two different regions of the interface result in the remarkable difference in arm orientation between BMP9 and TGF-β1 procomplexes. The arm domain β1-strand is much more twisted in pro-TGF-β1 than in pro-BMP9, enabling the β1-β10-β3-β6 sheets to orient vertically in pro-TGF-β and horizontally in pro-BMP9 in the view of Fig. 1 A and B. In addition, if we imagine the GF β7- and β6-strands as forefinger and middle finger, respectively, in BMP9, the two fingers bend inward toward the palm, with the β7 forefinger bent more, resulting in cupping of the fingers (Fig. 1 G and H and Fig. S4). In contrast, in TGF-β1, the palm is pushed open by the prodomain amphipathic α1-helix, which has an extensive hydrophobic interface with the GF fingers and inserts between the two GF monomers (Fig. 1B) in a region that is remodeled in the mature GF dimer and replaced by GF monomer–monomer interactions (10).
Role of Elements N and C Terminal to the Arm Domain in Cross- and Open-Armed Conformations.
A straitjacket in pro-TGF-β1 composed of the prodomain α1-helix and latency lasso encircles the GF on the side opposite the arm domain (Fig. 1B). Sequence for putative α1-helix and latency lasso regions is present in pro-BMP9 (Fig. 2A); however, we do not observe electron density corresponding to this sequence in the open-armed pro-BMP9 map. Furthermore, in the open-armed pro-BMP9 conformation, the prodomain α5-helix occupies a position that overlaps with the position of the α1-helix in the cross-armed pro-TGF-β1 conformation (Fig. 1 A, B, G, and H). The differing twists between the arm domain and GF domains in open-armed and cross-armed conformations relate to the distinct ways in which the prodomain α5-helix in pro-BMP9 and the α1-helix in pro-TGF-β1 bind to the GF (Fig. 1 A and B). The strong sequence signature for the α1-helix in pro-BMP9, which is essential for the cross-armed conformation in pro-TGF-β, suggests that pro-BMP9 can also adopt a cross-armed conformation (Discussion).
Fig. 2.
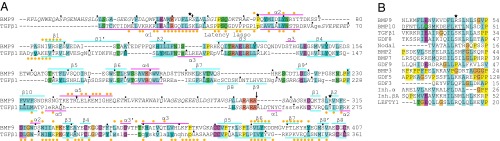
Sequence alignments. (A) Sequences aligned structurally with SSM (26). Structurally aligned residues are in uppercase; lowercase residues are not structurally aligned. Residues with missing density are in italics. Decadal residues are marked with black dots. Residues with more than 50% solvent accessible surface area buried in prodomain–GF interfaces are marked with gold dots. Secondary structural elements or named regions are shown with lines. Long black arrow marks cleavage sites between the prodomain and GF; short black arrows mark putative PC cleavage sites after the α1-helix and in the α5-helix. Stars mark HHT-like mutation sites. (B) Sequence alignment of representative TGF-β family members in the prodomain α1-helix (overlined for TGF-β1).
In absence of interaction with a prodomain α1-helix, the GF dimer in pro-BMP9 is much more like the mature GF (1.6-Å RMSD for all Cα atoms) than in pro-TGF-β1 (6.6-Å RMSD; Fig. S4). Moreover, burial between the GF and prodomain dimers is less in pro-BMP9 (2,870 Å2) than in pro-TGF-β1 (4,320 Å2). In the language of allostery, GF conformation is tensed in cross-armed pro-TGF-β1 and relaxed in open-armed pro-BMP9.
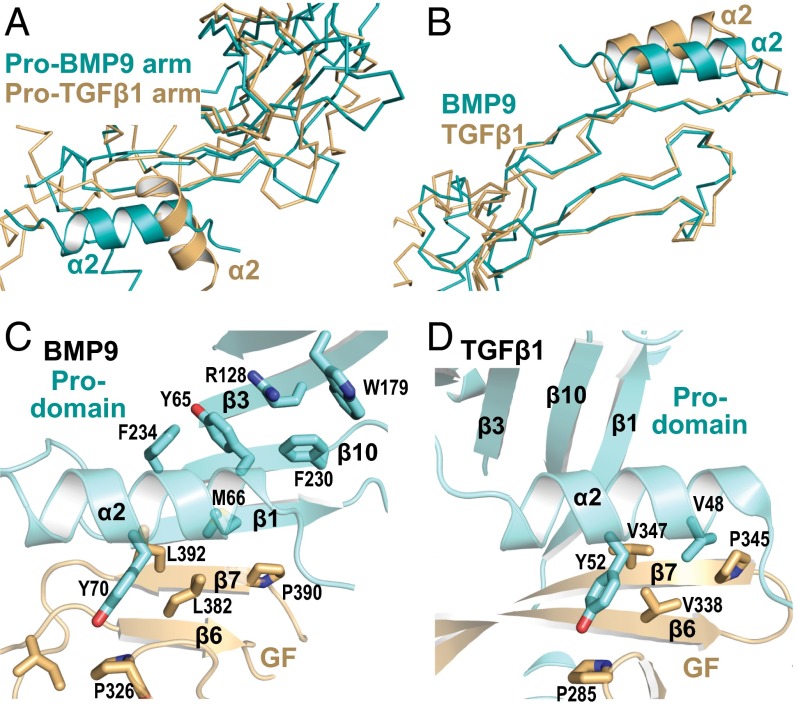
The prodomain α2-helix covers the interface between the arm and GF domains and acts as a buffer between the open-armed and cross-armed conformations. With superimposition based on arm domains, α2-helix orientation differs by ∼90° in pro-BMP9 and pro-TGF-β1 (Figs. 1 E and F and 3A), whereas with superimposition based on GF monomers, α2-helix orientation is similar in pro-BMP9 and pro-TGF-β1 (Figs. 1 G and H and 3B). Thus, if we imagine shape-shifting between open- and cross-armed conformations of pro-BMP9 (Discussion), the α2-helix moves in harmony with the GF rather than the arm domain.
Fig. 3.
The prodomain α2-helix. (A and B) Superimpositions based on the arm domain (A) or on the GF (B), which are shown as Cα traces with the α2-helix in cartoon. (C and D) Prodomain α2-helix environments in pro-BMP9 (C) and pro-TGF-β1 (D) shown after superimposition on the prodomain α2-helix and GF β6 and β7-strands. Key interacting residues are shown in stick.
Prodomain α2-helix association with the GF is stabilized by conserved interactions. Homologous α2-helix residues Tyr-70 in pro-BMP9 and Tyr-52 in pro-TGF-β1 stack against homologous Pro-326 and Pro-285 residues in their GFs (Fig. 3 C and D). Moreover, homologous α2-helix residues Met-66 in pro-BMP9 and Val-48 in pro-TGFβ1 are buried in a hydrophobic cavity on their GFs (Fig. 3 C and D).
The α2-helix in open-armed pro-BMP9 interacts with the arm domain in a way not seen in cross-armed pro-TGF-β1. Tyr-65 from the α2-helix together with Trp-179 and Phe-230 from the arm domain form an aromatic cage (Fig. 3C). Arm residue Arg-128 at the center of this cage forms π–cation interactions with Tyr-65 and Trp-179 (Fig. 3C). Residues for the π-cation cage are well conserved in BMP4, 5, 6, 7, 8, and 10, GDF5, 6, and 7, and GDF15 (Fig. S5). However, in BMP2 and BMP15, Arg-128 is replaced by Gln, potentially weakening association of the prodomain with the GF in the open-armed conformation.
The similar arm domain cores and α2-helices in the prodomains of BMP9 and TGF-β1 are remarkable, given that the prodomains have only 11% identity in sequence and have 12 insertions/deletions (Fig. 2A). This contrasts with the 25% identity between their GF domains (Fig. 2A). Among notable differences, pro-BMP9 lacks the 14-residue bowtie in pro-TGF-β1 that disulfide links the two arm domains together and has in its place a β7-β9' loop (Fig. 2A). The two cysteine residues in the TGF-β1 arm domain, Cys-194 and Cys-196 (Fig. 1F), form reciprocal interchain disulfide bonds (10). In contrast, our pro-BMP9 structure shows that the two arm domain cysteines, Cys-133 and Cys-214, form an intrachain disulfide that links the β3 strand to the β7-β9' loop (Fig. 1E). The disulfide helps stabilize an extension of the β3-strand in BMP9 and the formation of the β1'- and β9'-strands unique to pro-BMP9 that add onto the β2-β7-β5-β4 sheet (Fig. 1 E and F).
The α5-helix in pro-BMP9 is its most surprising specialization. It is much longer than in pro-TGF-β1, orients differently (Fig. 1 E and F), and binds to a similar region of the GF domain as the α1-helix in pro-TGF-β1. However, the prodomain α1 and α5-helices orient differently on the GF domain (Fig. 1 A, B, G, and H). The BMP9 prodomain α5-helix inserts into the hydrophobic groove formed by the fingers of one GF monomer and the α3-helix of the other monomer (Fig. 1A). This association is stabilized by a cluster of specific interactions (Fig. 1I). Glu-248, at the N terminus of the α5-helix, forms salt bridges with GF residues Lys-393 and Lys-350. In the middle of the α5-helix, Met-252 plunges into a hydrophobic cavity. At the C terminus, His-255 stacks against GF residue Trp-322 (Fig. 1I). However, GF burial by the pro-BMP9 α5-helix (750 Å2) is less than by the pro-TGF-β1 α1-helix (1,120 Å2) or α1-helix plus latency lasso (1,490 Å2). Furthermore, when crystals were cryo-protected with a 10% higher concentration of ethanol (3.25-Å dataset; Table S1), density for the α5-helix was present in one monomer but not the other (Fig. S6).
Prodomain Functions.
We next asked if interactions of the two BMP9 prodomains with the GF dimer are independent or cooperative. Isothermal calorimetry (ITC) showed that, irrespective of whether increasing amounts of prodomain were added to GF or vice versa, heat production showed a single sigmoidal profile (Fig. 4 A and B). Curves fit well to a model in which the two binding sites are independent, and yielded KD values of 0.8–1.0 μM at pH 4.5, which maintains BMP9 solubility.
Fig. 4.
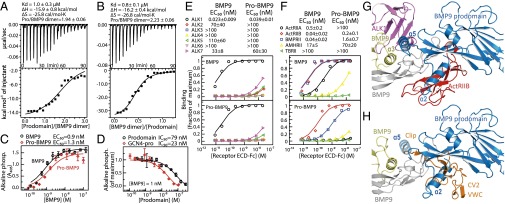
Binding of BMP9 to the prodomain and receptors. (A and B) Representative ITC data at 15 °C adding material either to the BMP9 dimer (A) or prodomain (B) in the calorimetry cell. (Upper) Baseline-corrected raw data. (Lower) Integrated heats fit to the independent-binding site model. (C and D) Differentiation of C2C12 cells measured by alkaline phosphatase production. (C) Comparison of BMP9 and pro-BMP9. (D) Inhibition of 1 nM BMP9 by GCN4-linked, oligomeric BMP9 prodomain and native prodomain. (E and F) EC50 values of BMP9 and pro-BMP9 for Fc-fused type I (E) or type II (F) receptor ectodomains measured using quantitative ELISA. Data are plotted as the fraction of maximal bound in each experiment and fit to the equation of fractional saturation. Graphs show average of triplicates in one experiment; numerical values show mean EC50 and sd from three such experiments. (G and H) Superimpositions on pro-BMP9 of the BMP9–receptor complex (17) (G) and BMP2-crossveinless2 complex (H) in identical orientations. For clarity, BMP in receptor and inhibitor complexes is omitted, and the α5-helix is transparent in H.
A critical question concerning BMP prodomains is whether the BMP9 prodomain inhibits GF signaling and whether making the BMP9 prodomain dimeric as in pro-TGF-β1 would provide sufficient avidity to keep the GF latent. Consistent with previous studies (16), pro-BMP9 and BMP9 showed little difference in inducing C2C12 cell differentiation, with EC50 values of 1.3 and 0.8 nM, respectively (Fig. 4C). However, by doing the experiment a different way, i.e., by titrating the prodomains into assays with 1 nM BMP9, we found that the prodomain is inhibitory, with IC50 = 79 nM (Fig. 4D). Thus, a large excess of the BMP9 prodomain over the GF is required to inhibit signaling, whereas at equimolar 1-nM concentrations of prodomain and GF monomers, the BMP9 prodomain has little effect on signaling. We added a cysteine-bearing GCN4 coil at the N terminus of the BMP9 prodomain to disulfide-link BMP9 prodomains into dimers or oligomers (Fig. S7). Multimerization modestly lowered the IC50 to 23 nM (Fig. 4D) but did not induce latency at equimolar concentrations.
To determine the basis for the partial inhibition of BMP9 signaling by the prodomain, we compared binding of receptor Fc fusions to pro-BMP9 and BMP9 in quantitative ELISA (Fig. 4 E and F). Among the seven type I receptors, ALK1 bound by far the strongest, and there was little difference in affinity and no difference in selectivity between BMP9 and pro-BMP9 (Fig. 4E). In contrast, the prodomain altered both EC50 values and selectivity of type II receptors (Fig. 4F). Whereas BMP9 bound ActRIIB and BMPRII similarly and 10-fold better than ActRIIA, pro-BMP9 bound ActRIIB 10-fold better than BMPRII and did not detectably bind ActRIIA (Fig. 4F). The EC50 values for type I and II receptors agree with previous affinity measurements with BMP9 (16, 17). Interestingly, the BMP7 prodomain also competes binding of BMP7 to type II but not type I receptors (18), consistent with the open-armed conformation of pro-BMP7 (Fig. 1D).
Discussion
BMPRII and ActRIIA are coexpressed with ALK1 and mediate BMP9 signaling in endothelial cells (12, 19). Furthermore, ALK1, which primarily functions as a receptor for BMP9 and BMP10, has recently been implicated as an important target of antiangiogenic tumor therapy (17). Together with the finding that BMP9 complexed with its prodomain circulates in the bloodstream at physiologically relevant concentrations (11), our finding that the BMP9 prodomain blocks binding to ActRIIA and alters selectivity for ActRIIB compared with BMPRII has important implications for BMP9 signaling in vivo.
The pro-BMP9 structure explains the selective effect of BMP prodomains on type II receptor binding (18) (Fig. 4G). The prodomain arm domain and α2-helix occupy the type II receptor binding site. In contrast, only the prodomain α5-helix blocks the type I receptor binding site (Fig. 4G). Selective displacement of the α5-helix by the type I receptor with retention of arm domain association is consistent with the relatively weak α5-helix interface described above. In contrast, arm domain binding to the type II receptor site is strong enough to alter receptor affinity and selectivity.
Antagonists including noggin, gremlins, and chordins bind to BMP GFs and regulate activity. However, little attention has been paid to whether BMP prodomains might prevent antagonist binding. Interestingly, the BMP-inhibiting fragment of the chordin family member crossveinless-2 binds to interfaces on BMP2 (20) similar to those on BMP9 to which the prodomain binds (Fig. 4H). The von Willebrand factor C (VWC) domain binds to a similar site on the GF fingers as the arm domain, whereas an N-terminal appendage called clip binds to the same site as the prodomain C-terminal appendage, the α5-helix (Fig. 4H). Whether prodomains can protect GF from inhibitors, as well as prevent GF binding to receptors, deserves study.
The crystal structure of pro-BMP9 begins to reveal how prodomains contribute to the tremendous functional diversity among the 33 members of the TGF-β family. Many of these members have prodomains that differ even more than BMP9 and TGF-β, which have only 11% sequence identity. Prodomain divergence may increase the specificity of GF signaling in vivo by regulating procomplex localization, movement, release, and activation in the extracellular environment.
The open-armed pro-BMP7 and 9 and cross-armed pro-TGF-β1 conformations differ greatly. Overall learnings from protein families that can adopt multiple conformations, such as tyrosine kinases, integrins, G protein-coupled receptors, membrane channels, and membrane transporters, show that when markedly distinct conformations are glimpsed for individual members, most family members can visit each state, often in a manner that is regulated by other interactors. Thus, we hypothesize that most members of the TGF-β family can visit both cross-armed and open-armed conformations. TGF-β is a later evolving family member; whereas BMPs and activins are found in all metazoans, TGF-β is found only in deuterostomes. Furthermore, TGF-β is the only known member with disulfide-linked arm domains. Thus, trapping pro-TGF-β in a solely cross-armed conformation with disulfides may be a later evolutionary adaptation.
The amino acid sequence of a protein is constrained by its structure, and sequence conservation in evolution is a powerful predictor of protein structure and conformation. The prodomain α1-helix has an important function in stabilizing the cross-armed conformation but has no function in the open-armed conformation, as shown by lack of electron density and presence of the prodomain α5-helix in a position that prevents α1-helix binding. In support of the hypothesis that pro-BMP9 can adopt a cross-armed conformation, the amino acid sequence corresponding to the α1-helix is highly conserved (44–79% identity at residues 29–47) among human, mouse, zebrafish, and chicken BMP9s. Indeed, the sequence of the α1-helix is more conserved than the remainder of the prodomain (33–74% identity). Furthermore, the prodomain α1-helix sequence and its amphipathic signature are also conserved among diverse representatives of the 33-member TGF-β family including BMP7 (Fig. 2B). Importantly, the α1-helix and its amphipathic signature are highly conserved between pro-TGF-β1 and pro-BMP9 (Fig. 2). These results support the hypothesis that pro-BMP9 and other TGF-β family members can adopt an α1-helix-bound, cross-armed conformation similar to that of TGF-β1.
To more directly test evolutionary support for a cross-armed BMP9 conformation, we made a pro-TGF-β1–like model of pro-BMP9 that uses the BMP9 conformation of the arm domains, superimposed on the cross-armed orientation of the arm domains in pro-TGF-β1, and pro-TGF-β1–like conformations of prodomain α1- and α2-helices and GF domains (Movie S1). In addition to the α1-helix, pro-BMP9 also includes a latency lasso-like sequence, including an identical PSQ sequence (Fig. 2A). There are no clashes between the two pro-BMP9 arm domains in the crossed-arm conformation; notably, the arm domains come close together at their β4 and β5-strands, which are on the side of the arm domain conserved between pro-BMP9 and pro-TGF-β1 (Movie S1). The extensive, amphipathic α1-helix–GF interface in pro-TGF-β1 is recapitulated well in the cross-armed pro-BMP9 model, and the long α5-helix can adopt a conformation similar to the shorter α5-helix in pro-TGF-β1 without clashes (Fig. 1K). These results compellingly support a cross-armed conformation for pro-BMP9. A plausible pathway for structural interconversion between open-armed and cross-armed conformations of BMP9 can be described in which crossing of the arms is accompanied by dissociation of the α5-helix from the GF and its replacement by the α1-helix and latency lasso (Movie S1).
The strong evolutionary and 3D structural support for a cross-armed conformation of BMP9 (and also BMP7; Fig. 2B) contrasts with our lack of observation of cross-armed BMP7 and BMP9 conformations in EM (Fig. 1 C and D). However, this is easily explicable, because it is compatible with a lower energy of the open-armed conformation for the isolated procomplex, and on the other hand, with a lower energy of the cross-armed conformation for the procomplex bound to an interactor. For BMPs in bone, such interactors may be present in the residual matrix, and release from interactors may in part be responsible for the increase in BMP activity found after extraction by denaturants and purification (2).
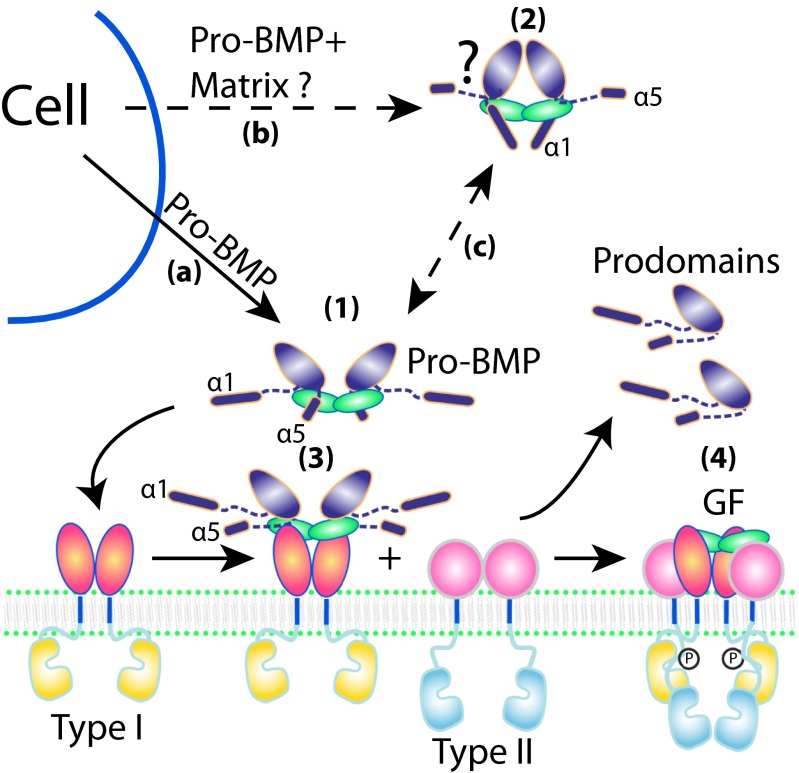
We hypothesize that cross-armed and open-armed conformations of TGF-β family members correspond to latent and nonlatent states, respectively, and propose a model for conformational regulation of release from storage and latency (Fig. 5). Some family members may be secreted as isolated procomplexes in signaling-competent, open-armed conformations (Fig. 5, pathway a and structure 1). Other family members may be secreted together with or immediately form complexes after secretion with extracellular matrix components including heparin, proteoglycans, fibrillin, and latent TGF-β binding proteins (8, 9, 21–23). These interactors may stabilize the cross-armed conformation (Fig. 5, pathway b and structure 2), and enable the GF domain, which is very short lived in vivo, to remain latent and reach storage concentrations as high as 100 ng/g in demineralized bone (24). Release from storage in vivo may then yield the open-armed conformation, which is ready for receptor or inhibitor binding (Fig. 5, pathway c).
Fig. 5.
Models for pro-BMP9 structures and binding to receptors. Models for the open-armed, nonlatent pro-BMP9 conformation characterized here (1), the proposed (dashed lines) pro-TGF-β1–like cross-armed conformation of pro-BMP9 (2), and stepwise binding (18) to type I (3) and type II receptors (4).
TGF-β family members with long sequences at the ends of their prodomains that may have α5-helix–like functions include BMP3, BMP10, BMP15, GDF5, 6, 7, and 9, anti-Müllerian hormone, and nodal (Fig. S5). A number of these, including BMP9 and BMP10, have basic sequences resembling PC cleavage sites (25) in or before the α5-helix (Fig. 2A and Fig. S5). Moreover, many TGF-β family members have PC or tolloid cleavage sites in or after the prodomain α1-helix that regulate activation or signaling (6, 7, 9, 10, 25) (Fig. S5). Indeed, recombinant pro-BMP9 preparations contain a minor component cleaved at a putative PC site in this region (Fig. 2A and Methods). Thus, potential mechanisms for regulating the switching between open-armed and cross-armed procomplex conformations include removal of the α1- or α5-helix by proteases in addition to binding to extracellular matrix components. Our results suggest that the open-armed conformation of pro-BMP9 can readily bind to type I receptors, with displacement of the α5-helix (Fig. 5, structure 3). The final step in signaling could then be binding to type II receptors, with complete prodomain dissociation, consistent with a previous model of stepwise receptor binding and prodomain displacement (18) (Fig. 5, structure 4).
Methods
Pro-BMP9 and pro-BMP7 were purified from supernatants of CHO and HEK293 cell transfectants, respectively. Crystals formed in 0.15 M zinc acetate, 0.1 M sodium cacodylate, pH 5.8, and 4% (vol/vol) isopropanol. Phases were solved using Zn anomalous diffraction. Complete methods are described in SI Methods.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4YCG and 4YCI).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501303112/-/DCSupplemental.
References
- 1.Wang EA, et al. Purification and characterization of other distinct bone-inducing factors. Proc Natl Acad Sci USA. 1988;85(24):9484–9488. doi: 10.1073/pnas.85.24.9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luyten FP, et al. Purification and partial amino acid sequence of osteogenin, a protein initiating bone differentiation. J Biol Chem. 1989;264(23):13377–13380. [PubMed] [Google Scholar]
- 3.Celeste AJ, et al. Identification of transforming growth factor beta family members present in bone-inductive protein purified from bovine bone. Proc Natl Acad Sci USA. 1990;87(24):9843–9847. doi: 10.1073/pnas.87.24.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui Y, et al. The activity and signaling range of mature BMP-4 is regulated by sequential cleavage at two sites within the prodomain of the precursor. Genes Dev. 2001;15(21):2797–2802. doi: 10.1101/gad.940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison CA, Al-Musawi SL, Walton KL. Prodomains regulate the synthesis, extracellular localisation and activity of TGF-β superfamily ligands. Growth Factors. 2011;29(5):174–186. doi: 10.3109/08977194.2011.608666. [DOI] [PubMed] [Google Scholar]
- 6.Constam DB. Regulation of TGFβ and related signals by precursor processing. Semin Cell Dev Biol. 2014;32:85–97. doi: 10.1016/j.semcdb.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama T, Marqués G, Wharton KA. A large bioactive BMP ligand with distinct signaling properties is produced by alternative proconvertase processing. Sci Signal. 2012;5(218):ra28. doi: 10.1126/scisignal.2002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory KE, et al. The prodomain of BMP-7 targets the BMP-7 complex to the extracellular matrix. J Biol Chem. 2005;280(30):27970–27980. doi: 10.1074/jbc.M504270200. [DOI] [PubMed] [Google Scholar]
- 9.Sengle G, Ono RN, Sasaki T, Sakai LY. Prodomains of transforming growth factor β (TGFβ) superfamily members specify different functions: Extracellular matrix interactions and growth factor bioavailability. J Biol Chem. 2011;286(7):5087–5099. doi: 10.1074/jbc.M110.188615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi M, et al. Latent TGF-β structure and activation. Nature. 2011;474(7351):343–349. doi: 10.1038/nature10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bidart M, et al. BMP9 is produced by hepatocytes and circulates mainly in an active mature form complexed to its prodomain. Cell Mol Life Sci. 2012;69(2):313–324. doi: 10.1007/s00018-011-0751-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, et al. Context-dependent signaling defines roles of BMP9 and BMP10 in embryonic and postnatal development. Proc Natl Acad Sci USA. 2013;110(29):11887–11892. doi: 10.1073/pnas.1306074110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castonguay R, et al. Soluble endoglin specifically binds bone morphogenetic proteins 9 and 10 via its orphan domain, inhibits blood vessel formation, and suppresses tumor growth. J Biol Chem. 2011;286(34):30034–30046. doi: 10.1074/jbc.M111.260133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David L, Feige JJ, Bailly S. Emerging role of bone morphogenetic proteins in angiogenesis. Cytokine Growth Factor Rev. 2009;20(3):203–212. doi: 10.1016/j.cytogfr.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Wooderchak-Donahue WL, et al. BMP9 mutations cause a vascular-anomaly syndrome with phenotypic overlap with hereditary hemorrhagic telangiectasia. Am J Hum Genet. 2013;93(3):530–537. doi: 10.1016/j.ajhg.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown MA, et al. Crystal structure of BMP-9 and functional interactions with pro-region and receptors. J Biol Chem. 2005;280(26):25111–25118. doi: 10.1074/jbc.M503328200. [DOI] [PubMed] [Google Scholar]
- 17.Townson SA, et al. Specificity and structure of a high affinity activin receptor-like kinase 1 (ALK1) signaling complex. J Biol Chem. 2012;287(33):27313–27325. doi: 10.1074/jbc.M112.377960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sengle G, Ono RN, Lyons KM, Bächinger HP, Sakai LY. A new model for growth factor activation: Type II receptors compete with the prodomain for BMP-7. J Mol Biol. 2008;381(4):1025–1039. doi: 10.1016/j.jmb.2008.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scharpfenecker M, et al. BMP-9 signals via ALK1 and inhibits bFGF-induced endothelial cell proliferation and VEGF-stimulated angiogenesis. J Cell Sci. 2007;120(Pt 6):964–972. doi: 10.1242/jcs.002949. [DOI] [PubMed] [Google Scholar]
- 20.Zhang JL, et al. Crystal structure analysis reveals how the Chordin family member crossveinless 2 blocks BMP-2 receptor binding. Dev Cell. 2008;14(5):739–750. doi: 10.1016/j.devcel.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 21.Sengle G, et al. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J Biol Chem. 2008;283(20):13874–13888. doi: 10.1074/jbc.M707820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S, et al. Activin A binds to perlecan through its pro-region that has heparin/heparan sulfate binding activity. J Biol Chem. 2010;285(47):36645–36655. doi: 10.1074/jbc.M110.177865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson SB, Goldberg AL, Whitman M. Identification of a novel pool of extracellular pro-myostatin in skeletal muscle. J Biol Chem. 2008;283(11):7027–7035. doi: 10.1074/jbc.M706678200. [DOI] [PubMed] [Google Scholar]
- 24.Pietrzak WS, Woodell-May J, McDonald N. Assay of bone morphogenetic protein-2, -4, and -7 in human demineralized bone matrix. J Craniofac Surg. 2006;17(1):84–90. doi: 10.1097/01.scs.0000179745.91165.73. [DOI] [PubMed] [Google Scholar]
- 25.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11(5):367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 26.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.