Significance
Accumulation of toxic lipids in macrophages or human plaques leads to endoplasmic reticulum (ER) stress and induction of autophagy. However, the mechanism by which ER stress and autophagy help in fine tuning cellular lipid homeostasis to counter free-cholesterol toxicity is not clear. Our studies demonstrate that cholesterol induces the translocation of ORMDL orosomucoid-like proteins out of the ER and targets them to autophagosomes, thereby relieving their negative regulation on cellular de novo sphingolipid synthesis. In addition, ORMDL3 has been recently implicated in childhood asthma and in eosinophil trafficking and activation. Thus, our finding of increased turnover of ORMDL proteins by free cholesterol is relevant not only in context to atherosclerotic disease progression but may also shed new insights on lipid homeostasis in other human diseases.
Keywords: ORMDL1, autophagy, free cholesterol, sphingomyelin, serine palmitoyl-CoA transferase
Abstract
Eukaryotic cells have evolved robust mechanisms to counter excess cholesterol including redistribution of lipids into different compartments and compensatory up-regulation of phospholipid biosynthesis. We demonstrate here that excess cellular cholesterol increased the activity of the endoplasmic reticulum (ER) enzyme serine palmitoyl-CoA transferase (SPT), the rate-limiting enzyme in sphingomyelin synthesis. This increased SPT activity was not due to altered levels of SPTLC1 or SPTLC2, the major subunits of SPT. Instead, cholesterol loading decreased the levels of ORMDL1, a negative regulator of SPT activity, due to its increased turnover. Several lines of evidence demonstrated that free-cholesterol–induced autophagy, which led to increased turnover of ORMDL1. Cholesterol loading induced ORMDL1 redistribution from the ER to cytoplasmic p62 positive autophagosomes. Coimmunoprecipitation analysis of cholesterol-loaded cells showed increased association between ORMDL1 and p62. The lysosomal inhibitor chloroquine or siRNA knockdown of Atg7 inhibited ORMDL1 degradation by cholesterol, whereas proteasome inhibitors showed no effect. ORMDL1 degradation was specific to free-cholesterol loading as autophagy induced by serum starvation or general ER stress did not lead to ORMDL1 degradation. ORMDL proteins are thus previously unidentified responders to excess cholesterol, exiting the ER to activate SPT and increase sphingomyelin biosynthesis, which may buffer excess cellular cholesterol.
Human atheroma macrophages accumulate large amounts of free cholesterol (FC), cholesterol esters (CEs), oxidized lipids, and oxysterols (1). Recent studies have established that accumulation of FC, oxidized phospholipids (PLs), and oxysterols in atherosclerotic plaques leads to endoplasmic reticulum (ER) stress, which in turn triggers the induction of autophagy to clear these toxic lipids (2–6). In addition, autophagy has been implicated as a key player in the clearance of cholesterol esters from foam cells and the regression of atherosclerotic lesions (2, 7, 8).
Sphingomyelin (SM) levels are inversely correlated with HDL function and are higher in atherosclerotic aorta compared with healthy aorta (9–13). Myriocin, a potent inhibitor of SM biosynthesis, decreases atherosclerosis in mouse models, which may be accounted by its effects on increasing reverse cholesterol transport (RCT) and decreasing cholesterol absorption (14–21).
The rate-limiting step in sphingomyelin biosynthesis is catalyzed by the ER resident serine palmitoyl-CoA transferase (SPT) enzyme complex, a heterodimer of serine palmitoyltransferase long-chain subunit (SPTLC)1 with either SPTLC2 or SPTLC3 (22). In yeast, the activity of SPT complex is posttranslationally inhibited by the orosomucoid-like (ORM) proteins. Under conditions of high sphingolipids, ORM proteins directly bind to the SPT complex and inhibit its activity, thus preventing further synthesis of sphingolipids (23, 24). When sphingolipid levels are low, ORM proteins are phosphorylated in N-terminal serine residues and dissociate from SPT complex (25), leading to increased sphingolipid biosynthesis (24, 25). However, no information is available about the mechanism by which mammalian ORM-like proteins (ORMDL1–3) regulate SPT activity. Recently, all three mammalian ORMDL proteins were shown to be involved in mediating a negative feedback response of ceramide biosynthesis upon addition of sphingosine (26). ORMDL3 has also been implicated in childhood asthma (27) and in promoting eosinophil trafficking and activation (28). Because mammalian ORMDL proteins have a truncated N-terminal domain and lack the phosphorylated serine residues found in yeast (29), the regulated binding to and inhibition of SPT by these ORMDL proteins must occur by a different mechanism than characterized in yeast.
Here, we show that FC loading in macrophages induced SPT activity without altering the levels of SPT complex enzymes. The molecular mechanism of this up-regulation was increased turnover of ORMDL proteins via autophagy upon FC loading. Our finding that ORMDL proteins are degraded by FC-induced autophagy provides insights into molecular pathways and mechanisms regulating lipid homeostasis upon the accumulation of excess cholesterol. We propose that ORMDL proteins play a previously unidentified role in mediating cross-talk between cholesterol and sphingolipid biosynthesis.
Results
FC Loading Increases SM Levels and Induces SPT Activity.
To determine the effects of FC on cellular sphingomyelin levels, the levels of cellular cholesterol in RAW264.7 macrophages were modulated by addition of exogenous cholesterol. Treatments with either cyclodextrin cholesterol (Chol-CD) or acetylated LDL plus an inhibitor of acyl-CoA:cholesterol acyltransferase (AcLDL + ACATi) led to 3.5-and 3.2-fold increases in FC content, respectively, compared with unloaded cells (Fig. 1A), whereas the levels of SM were 3.3-fold higher in FC-loaded cells in comparison with unloaded cells (Fig. 1B). The protein levels of sphingomyelin phosphodiesterase 2 (SMPD2), the neutral sphingomyelinase that breaks SM into ceramide and phosphocholine, were unchanged (Fig. S1A), whereas lipid-raft levels were increased by FC loading (Fig. S1 B and C). To investigate the mechanism of FC-mediated increase in SM biosynthesis, we determined the activity of the rate-limiting SM biosynthetic enzyme complex, SPTLC. As shown in Fig. 1C, cells loaded with AcLDL have significantly higher SPT activity than control samples (∼30% increase vs. unloaded cells), which was further increased upon treatment with the ACATi (∼89% increase vs. unloaded cells). To ensure the specificity of the assay, the SPT inhibitor myriocin was added to the reaction, which strongly decreased SPT activity. The SPT activity was higher in cells treated with ACATi, suggesting that increased FC levels were influencing SPT activity. The effect of FC on SPT1 activity was confirmed by loading of RAW264.7 cells with 50 µg/mL cholesterol-CD, which led to significantly increased SPT activity (102% increase vs. unloaded cells, Fig. 1D).
Fig. 1.
FC loading increases SPT activity. (A) Cellular FC levels and (B) cellular SM levels, in RAW264.7 cells unloaded or loaded with 100 µg/mL AcLDL, 100 µg/mL AcLDL + 2 µg/mL ACATi, or 50 µg/mL cholesterol-cyclodextrin (Chol-CD) for 16 h (n = 3, mean ± SD, ***P < 0.005 vs. control, by ANOVA posttest). (C) SPT activity of RAW264.7 cells unloaded or loaded with 100 µg/mL AcLDL, in the absence or presence of 2 µg/mL ACATi for 16 h at 37 °C. Myriocin was used as negative control (n = 3, mean ± SD, different numbers above the bars show P < 0.05, by ANOVA posttest). (D) SPT activity of RAW264.7 cells after a 16-h incubation with or without 50 µg/mL cholesterol-CD (n = 3, mean ± SD, ***P < 0.005, by t test).
FC Accumulation Diminishes ORMDL1 Expression.
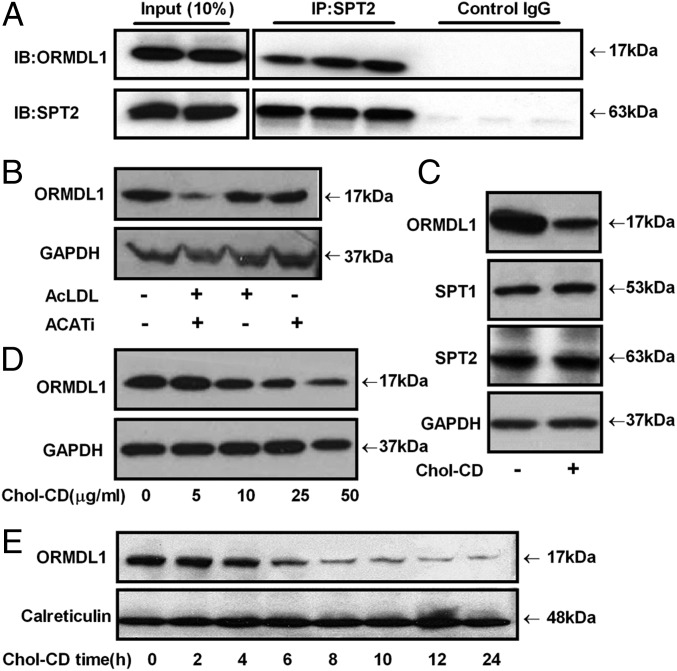
Because the SPT activity was induced upon FC loading, we tested whether FC affects the function of ORMDL proteins, the only known negative regulators of SPT activity. A coimmunoprecipitation (co-IP) assay was performed to determine whether there was a direct interaction between the SPT complex and ORMDL proteins in RAW264.7 cells. As shown in Fig. 2A, ORMDL1 directly binds to the SPTLC2 protein, the catalytic subunit of the SPT complex. We also determined the extent of association between ORMDL1 and SPTLC2 by performing Western blot analysis of bound (pellet) and unbound (supernatant) fractions and found that a significant fraction of ORMDL1 shifted from the unbound to the pellet fraction by pull down with SPTLC2 antibody (Fig. S2A). Next, the levels of ORMDL1 were determined in RAW264.7 cells loaded with FC either by treatment with AcLDL + ACATi or cholesterol-CD. As shown in Fig. 2 B and C, ORMDL1 levels were reduced upon FC loading. FC loading did not change the levels of two SPT subunits, SPTLC1 or SPTLC2, or control GAPDH levels (Fig. 2C), indicating that ORMDL1 was specifically reduced by FC loading. The effect of cholesterol-CD on ORMDL1 expression was dose dependent, with 25–50 µg/mL cholesterol-CD conferring robust decreases in ORMDL1 levels with no significant changes in GAPDH levels (Fig. 2D). We also performed a time-course study to determine when the cholesterol-mediated decrease of ORMDL1 was observed. As shown in Fig. 2E, the decrease in ORMDL1 levels was clearly evident by the 6-h time point, and this decrease was more prominent at later time points. We probed another ER resident protein calreticulin as a control and found no difference in its levels upon cholesterol loading (Fig. 2E). These data indicate that the decrease in ORMDL1 levels is a specific and coordinated response to cholesterol loading and is not due to extensive changes in the cellular contents. Cell-viability and apoptosis assays were performed to determine whether cholesterol loading induced cell death or apoptosis. As shown in Fig. S2 B and C, cholesterol loading under similar conditions did not cause apoptosis or affect cellular integrity. These data indicate that FC accumulation led to a specific reduction of the SPT inhibitor protein ORMDL1, which resulted in increased SPT activity without any changes in the levels of the SPTLC subunits.
Fig. 2.
FC loading diminishes ORMDL1 expression. (A) RAW264.7 cell lysates were immunoprecipitated (IP) with anti-SPT2 and immunoblotted (IB) with anti-ORMDL1 antibody, demonstrating that ORMDL1 and SPT2 are in a protein complex. (B) Levels of ORMDL1 in RAW264.7 cells unloaded or loaded with 100 µg/mL AcLDL, in the absence or presence of 2 µg/mL ACATi for 16 h. GAPDH levels were used as a loading control. (C) Levels of ORMDL1, SPT1, and SPT2 in RAW264.7 cells after a 16-h incubation with or without 50 µg/mL Chol-CD. (D) ORMDL1 levels in RAW264.7 cells in the presence of the indicated dose of Chol-CD for 16 h. (E) ORMDL1 levels in RAW264.7 cells in the presence of 50 µg/mL Chol-CD for indicated time points. Calreticulin, an ER resident protein, was used as control.
ORMDL1 Is Destabilized by FC Accumulation.
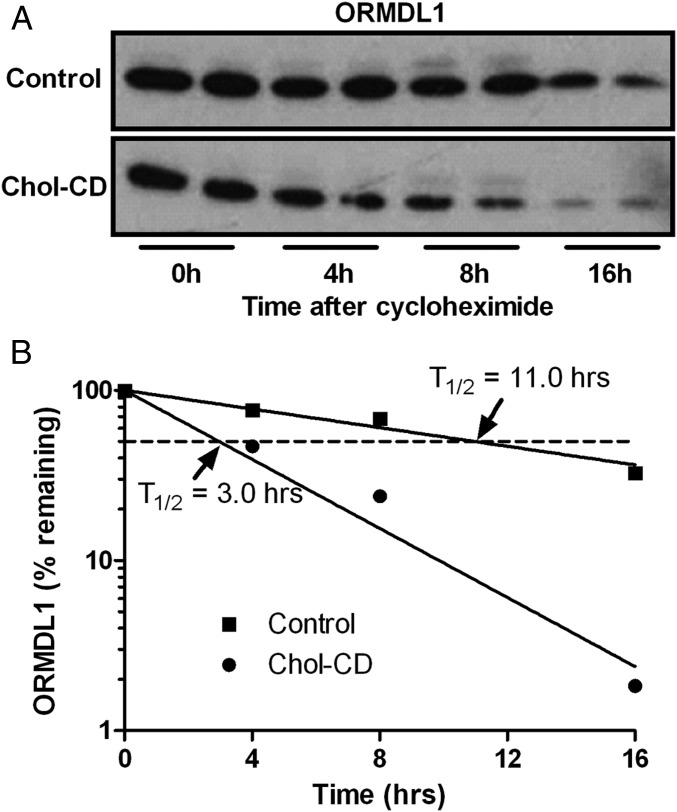
The decreased levels of ORMDL1 upon FC accumulation may be due to decreased synthesis or increased degradation. We performed a cycloheximide-chase experiment to determine the turnover of ORMDL1 protein in unloaded and FC-loaded RAW264.7 cells. FC-loaded cells exhibited a marked increase in the turnover rate of ORMDL1 vs. unloaded cells (Fig. 3). The calculated half-life (T1/2) of ORMDL1 was 3.0 h and 11.0 h in FC-loaded and control cells, respectively (Fig. 3B). We confirmed that cholesterol loading increased the turnover of ORMDL1 through a [14C] leucine pulse-chase immunoprecipitation study. Similar to the cycloheximide results, we observed increased turnover of ORMDL1 upon cholesterol loading (Fig. S3 A and B). Because FC loading has been shown to induce ER stress (30), we tested and found that ORMDL1 levels were not affected by general ER stress induced by tunicamycin (Fig. S3C). Next, we tested whether FC-induced degradation of ORMDL1 could be blocked by treatment with either proteasomal or lysosomal inhibitors. FC-mediated ORMDL1 degradation was not blocked by two proteasomal inhibitors (Fig. S3 D and E), but was efficiently rescued by the lysosomal inhibitor chloroquine (Fig. S3E), indicating that an active lysosomal degradation pathway was required for FC-induced ORMDL1 degradation.
Fig. 3.
FC loading increases ORMDL1 degradation. (A) RAW264.7 cells were incubated in the presence of 20 µg/mL cycloheximide, with or without 50 µg/mL cholesterol-CD for the indicated time and were analyzed for ORMDL1 levels by Western blot. (B) Semilog plot of ORMDL1 levels normalized to the levels before the cyclohexamide chase in FC-loaded and unloaded cells.
ORMDL Proteins Are Translocated out of the ER upon FC Loading.
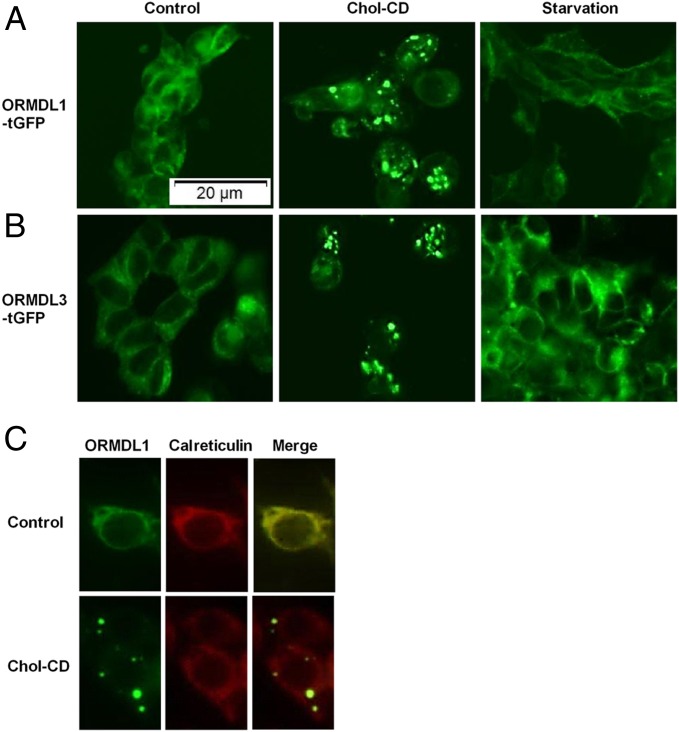
ORMDL protein localization studies were performed in stably transfected HEK293 cells carrying ORMDL1 or ORDML3–turbo GFP (tGFP) constructs. Under control growth conditions, both ORMDL1 and ORMDL3–tGFP fusion proteins in HEK293 cells were localized at the ER (Fig. 4 A and B), consistent with a previous report (29). In FC-loaded HEK293 cells, ORMDL1 and ORMDL3–tGFP fusion proteins were exported out of the ER and localized in puncta like structures in the cytoplasm (Fig. 4). These data indicated that FC-loading effects were shared by ORMDL1/3 proteins. In cholesterol-loaded RAW macrophages, endogenous ORMDL1 protein, observed by indirect immunofluorescence, was also translocated from the ER to cytoplasmic puncta structures, whereas endogenous calreticulin remained in the ER with no cytoplasmic puncta localization (Fig. 4C). These data indicated a specific exit of ORMDL1 from the ER upon cholesterol loading.
Fig. 4.
ORMDL proteins are translocated out of the ER upon FC loading. (A and B) Localization of ORMDL1–turbo green fluorescent protein (tGFP), and ORMDL3–tGFP fusion proteins, respectively, in stably transfected HEK293 cell lines after a 16-h incubation with or without 50 µg/mL Chol-CD, or 4 h serum starvation as a positive control for autophagy induction. (C) Indirect immunofluorescence microscopy showing localization of endogenous ORMDL1 and calreticulin with or without 50 µg/mL Chol-CD, using ORMDL1 and calreticulin antibodies.
Because cholesterol loading has been shown to induce membrane whorls or aggregates mainly composed of phospholipids due to increased PC synthesis (31), we performed filipin staining and found that the cholesterol-loaded cells showed stronger filipin positive signal at the plasma membrane and increased number of filipin-positive intracellular vesicles (Fig. S4A). Overall, no dramatic differences were found in cholesterol localization in loaded or unloaded RAW macrophages (Fig. S4A). To determine whether free cholesterol was found in ORMDL1-containing puncta, transiently transfected RAW–ORMDL1–tGFP macrophages were loaded with cholesterol for 16 h. The ORMDL1-containing puncta were not specifically stained by filipin (Fig. S4B). We conclude that ORMDL1-positive puncta were not particularly enriched with cholesterol but there is a possibility that these structures may contain low amounts of cholesterol, which were not readily stained by filipin.
Based on the effect of chloroquine in blocking the FC-induced turnover of ORMDL1 and the FC-mediated transfer of ORMDL1 to cytoplasmic puncta, we investigated the role of autophagy as the mechanism for FC-induced degradation of ORMDL1. First we tested whether induction of general autophagy can induce ORMDL1 translocation from the ER to the cytoplasm. Serum starvation, a known inducer of autophagy, failed to mediate ORMDL proteins’ export from the ER (Fig. 4 A and B).
FC Loading Induces Autophagy.
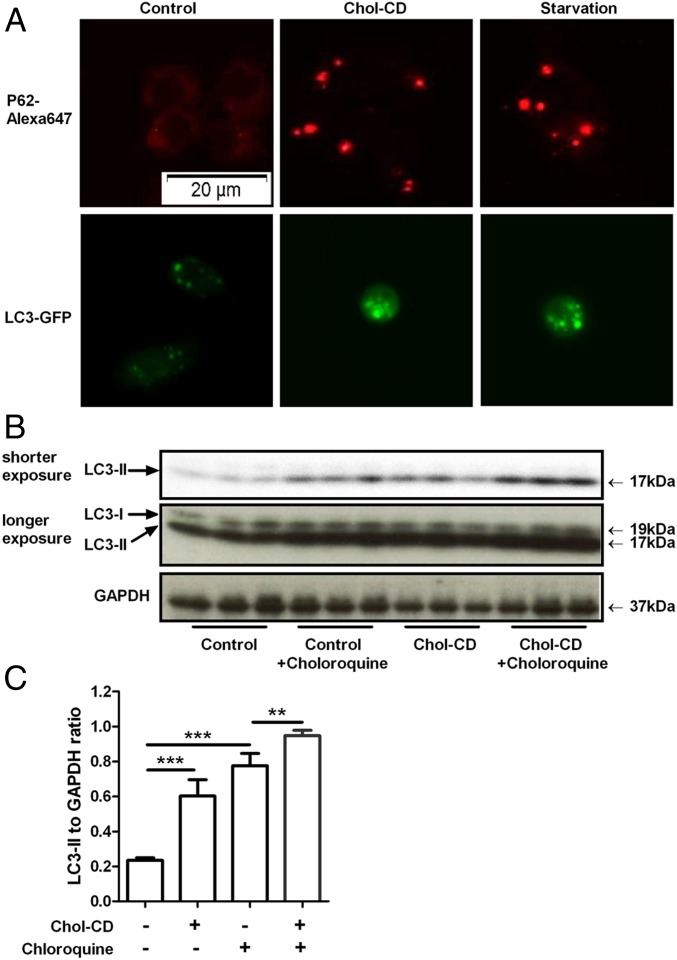
We then tested whether FC loading induced autophagy, which may be responsible for ORMDL1 turnover. RAW264.7 cells loaded with FC or subjected to serum starvation were probed for p62 and LC3. The adaptor protein p62 binds to unfolded proteins and damaged membranes to chaperone them to the autophagophore, and LC3 is a protein required for autophagophore formation. The unloaded cells expressed p62, but almost no p62 signal was found in puncta, whereas FC-loaded and serum-starved cells showed multiple p62 puncta per cell (Fig. 5A). Similar to p62, LC3–GFP localization in puncta was increased by FC loading as well as by serum starvation (Fig. 5A), both findings indicative of the induction of autophagy. To evaluate whether the increased LC3 puncta was due to increased autophagic initiation or due to decreased autophagic flux leading to an accumulation of autophagic markers, we determined the level of LC3-II (lipidated form) by Western blotting in the presence or absence of chloroquine. If chloroquine further increases LC3-II levels, then autophagic flux is active. RAW264.7 cells were treated with Chol-CD for 16 h and chloroquine was added in the indicated wells during the last 4 h of treatment. LC3-II protein levels were increased by 3.3-fold in FC-loaded cells in comparison with unloaded cells (Fig. 5 B and C). When cells were treated with chloroquine, LC3-II protein levels were significantly increased compared with the no-chloroquine condition. These results suggest that FC loading increased autophagy initiation and maintained autophagic flux.
Fig. 5.
FC loading induces autophagy. (A) RAW264.7 cells or LC3–GFP transfected RAW264.7 cells were either loaded with 50 µg/mL Chol-CD for 16 h or serum starved for 4 h as a positive control for autophagy induction. p62 was visualized by indirect immunofluorescence (red, Top), and LC3–GFP protein (green, Lower) is shown. (B) RAW264.7 cells were treated with 50 µg/mL Chol-CD for 16 h. To assess autophagic flux, each condition was repeated in the presence of chloroquine for the last 4 h of incubation. The amount of LC3-II was assessed by Western blot and densitometry analysis. (C) LC3-II levels were presented relative to GAPDH levels (n = 3, mean ± SD, **P < 0.01, ***P < 0.001 by ANOVA posttest).
FC Loading Leads to ORMDL1 Localization in Autophagosomes.
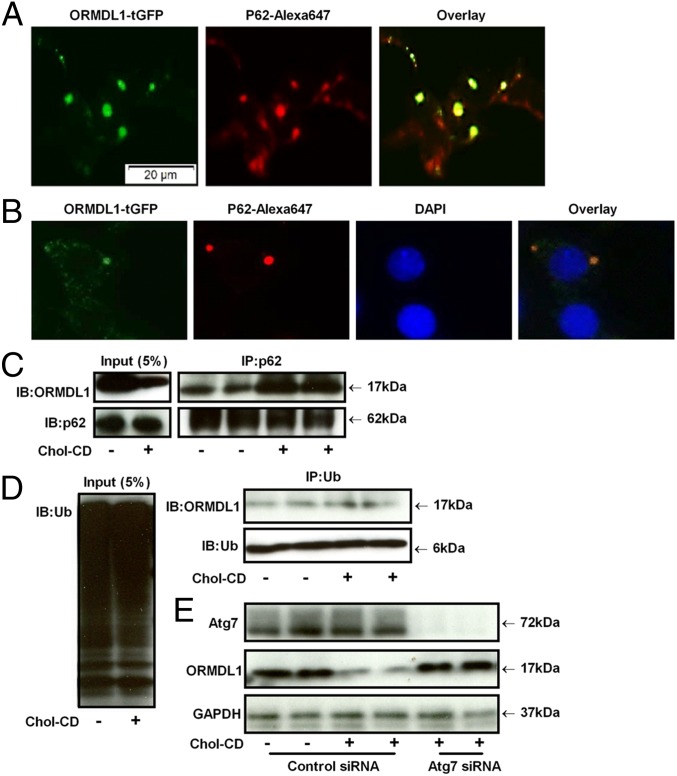
Because we found that FC induced autophagy, we tested whether ORMDL1 was exported to autophagosomes in FC-loaded cells. The ORMDL1–GFP fusion protein was colocalized in cytoplasmic puncta with p62 in FC-loaded ORMDL1–tGFP stably transfected HEK293 cells (Fig. 6A), indicating transport of ORMDL1 from the ER to autophagosomes.
Fig. 6.
Autophagosome formation in degradation of ORMDL1 upon FC loading. (A) ORMDL1–tGFP fusion proteins (green) stably transfected HEK293 cells were loaded with 50 µg/mL Chol-CD for 16 h and further stained with p62 antibody (red). (B) RAW macrophages were loaded with 50 µg/mL Chol-CD for 16 h and further stained with p62 (red) and ORMDL1 (green) antibodies, respectively. (C) RAW264.7 cells were loaded with 50 µg/mL Chol-CD for 16 h, and the cell lysates were immunoprecipitated with anti-p62 antibody and immunoblotted with ORMDL1 antibody. (D) Cell lysates were immunoprecipitated with antiubiquintin (Ub) antibody, followed by immunoblotting using ORMDL1 antibody. (E) RAW264.7 cells were transfected with siRNA against Atg7 for 48 h and then incubated with or without 50 µg/mL Chol-CD for 16 h. The levels of Atg7, ORMDL1, and GAPDH were assessed by Western blot.
Next, we tested FC-loaded RAW macrophages to determine whether endogenous ORMDL1 was translocated to autophagosomes, by staining these cells with p62 and ORMDL1 antibodies. As shown in Fig. 6B, ORMDL1 and p62 were found to be localized in the same cytoplasmic puncta, confirming the trafficking of endogenous ORMDL1 to autophagosomes upon FC loading in RAW macrophages.
To further confirm the interaction between p62 and ORMDL1, we performed a co-IP assay in control and FC-loaded RAW macrophages. As shown in Fig. 6C, the FC-loaded cells showed increased association between p62 and ORMDL1 compared with control cells, despite overall lower levels of ORMDL1. Interestingly, a co-IP experiment using antiubiquitin antibody did not show significant increase in association between ubiquitin and ORMDL1 (Fig. 6D). These data indicate the role of autophagy in removal of ORMDL1 from cytoplasm to lysosomes. We then inhibited autophagy initiation by knockdown of endogenous Atg7 using siRNA in RAW264.7 cells (32, 33). ORMDL1 Western blot analysis of control siRNA-treated cells showed the FC-mediated reduction of ORMDL1 as expected, whereas the Atg7 siRNA-treated cells were resistant for FC-mediated reduction of ORMDL1 (Fig. 6E). These data demonstrate that the FC effect on ORMDL1 turnover is mediated by autophagy. However, we already observed that autophagy induction by serum starvation did not lead to ORMDL1 mislocalization to cytoplasmic puncta (Fig. 4A), suggesting that ORMDL1 autophagy is specific for FC loading.
Discussion
Pioneering studies from the laboratory of Tabas and coworkers have shown that the ER is the site of cholesterol-induced cell cytotoxicity (30), and that FC loading induces de novo PC biosynthesis in macrophages to buffer excess FC and prevent cell death (31, 34). Here we demonstrate that FC loading also leads to increased SM synthesis and the previously unidentified mechanism by which this occurs, FC-induced autophagy of ORM proteins, which negatively regulate SPT activity. Autophagy in macrophages has previously been shown to play a protective role against atherosclerosis progression; and, it is a major pathway for the lysosomal hydrolysis of CE stored in lipid droplets, producing FC that can be effluxed by ATP-binding cassette family members ABCA1 or ABCG1 (2, 7).
Recently, three pools of plasma membrane cholesterol were defined in an elegant study (35) and sphingomyelin was shown to sequester a specific pool of cholesterol in plasma membrane. Interestingly, the overall cholesterol distribution pattern, observed by filipin staining, remained unchanged by depletion of sphingomyelin (36, 37). Previous studies have highlighted the intricate relationship between sphingomyelin and cholesterol levels (14, 18, 35, 38–42), indicating that modulation of biosynthetic as well as metabolic pathways mediating cross-talk between cholesterol and sphingolipids is essential for cellular lipid homeostasis. Sphingomyelin biosynthesis is regulated by the rate-limiting SPT enzyme complex, which is under negative regulation by ORMDL proteins, which reside in the ER and were initially identified as ER stress response pathway proteins (29). The role of ORM proteins in sphingolipid homeostasis was first established by novel yeast studies from the laboratories of Weissman and Chang and coworkers, showing both direct and negative interactions between ORM proteins and the SPT complex (23, 24). Our study identifies ORMDL1 as a missing link between sterol and sphingolipid pathways in mammalian cells.
Previous studies have also shown that 25-OH cholesterol stimulates SM synthesis, and that this was mainly mediated by the induction of ceramide transport from the ER to Golgi via enhanced recruitment of ceramide transfer protein (CERT) to Golgi membranes (31, 43). The association of SPT activity with cholesterol loading is controversial, with various studies showing effects ranging from negative, to positive, to no correlation (43–46). These studies used different cell types, including human fibroblasts, CHO cells, and human intestinal cells. These anomalies may be explained based on different cholesterol-loading conditions and different cell types, which may have different capacities to convert FC to the inert CE.
We found that FC loading in RAW264.7 cells led to a dose-dependent decrease in ORMDL1 levels due to its increased turnover. Our data indicate that a threshold level of FC loading is essential for FC-induced ORMDL1 degradation. FC loading led to export of ORMDL1 from the ER to cytoplasmic puncta, which we identified as autophagosomes based on their colocalization with p62. Also, FC loading was sufficient to induce autophagy in RAW264.7 cells. We believe that once a threshold level of FC in the ER is reached, autophagy is initiated and ORMDL1 exits from the ER to the autophagosomes, thus leading to degradation of ORMDL1 and relieving its repression of SPT activity. Under normal growth conditions, we demonstrated the direct interaction between ORMDL1 and the SPT complex by co-IP, implying that SPT activity is repressed in the basal state. Just the export of ORMDL1/3 from the ER would be sufficient to separate them from the SPT complex and derepress SPT activity. Although FC loading led to the selective redistribution of ORMDL1 from the ER to autophagosomes, there is no evidence for bulk ER autophagy (ER-phagy), as SPTLC1/2 and calreticulin levels were not decreased and SPT activity was increased. Thus, ORMDL proteins may be exported from the ER before recognition for autophagosomal targeting. The mechanism for the FC-induced ORMDL protein exit from the ER and its targeting to autophagosomes is still not clear. We speculate that ORMDL proteins may interact with protein or lipid molecules that are specifically ejected from the ER upon FC loading. ORMDL proteins may also interact with other proteins that are partitioned between the ER and Golgi [for example, oxysterol binding proteins (OSBPs) and CERT] to facilitate ORMDL exit from the ER, or they may be transported to the autophagosome via direct membrane contact between the ER and the autophagophore or autophagosome membrane. Alternatively, ORMDLs may be translocated to the cytoplasm first and then engulfed by the autophagic machinery, or they may be released in vesicles that are recognized by p62 and directed toward the autophagic route for degradation.
Interestingly, ER stress caused by tunicamycin failed to degrade ORMDL1, indicating that FC-mediated ER stress is specifically required for the translocation of ORMDL1 from the ER. Autophagy induction by serum starvation also led to robust autophagosome generation but failed to translocate ORMDL1 from the ER. Thus, two independent events are needed for the FC-induced degradation of ORMDL1: (i) ORMDL1 translocation from the ER and (ii) induction of autophagy. ORMDL1 is only selectively targeted for degradation when both of these events occur simultaneously, as shown in our model in Fig. S5.
Our finding that ORMDL proteins are degraded by FC-induced autophagy in macrophages provides insights into mechanisms regulating lipid homeostasis upon excess accumulation of cholesterol and is thus relevant in the context of atherosclerotic plaque formation and progression.
Materials and Methods
A detailed description of the materials and methods used in this study is provided in SI Materials and Methods. Basic methods are summarized below.
Cell Culture and Reagents.
All cell culture incubations were performed at 37 оC, unless otherwise indicated, in a humidified 5% CO2 incubator. Cells were grown in DMEM with 10% FBS (vol/vol) with antibiotics. Other materials are described in SI Materials and Methods.
SM/FC Measurements.
Cellular SM and FC levels were measured as described earlier (47, 48). RAW264.7 cells grown in 12-well plates were loaded with 50 µg/mL Chol-CD or with 100 µg/mL AcLDL ± 2 µg/mL ACATi for 16 h at 37 °C.
Lipid-Raft Quantification.
A flow-cytometry–based CTB Alexa-647 GM1 binding assay was used to determine the presence of lipid rafts in cells loaded with free cholesterol as described previously (49).
SPT Activity Assay.
SPT activity was measured in cell membrane homogenate using a radiochemical assay as described earlier (50).
Western Blotting/Co-IP.
For ORMDL1 Western blot analysis, RAW264.7 cells were loaded with 50 µg/mL cholesterol-CD, or with 100 µg/mL AcLDL with or without 2 µg/mL ACATi for 16 h. For LC3 Western blot, RAW264.7 cells were incubated with or without 50 µg/mL cholesterol-CD for 16 h; indicated wells received 30 µM chloroquine during the last 4 h of incubation.
Cycloheximide-Chase Analysis.
RAW264.7 cells were cultured in the presence of 20 µg/mL cycloheximide and 50 µg/mL cholesterol-CD for 0–16 h. Cells were lysed and blotted for ORMDL1 expression. To quantify the rate of degradation of ORMDL1, the bands were analyzed by using densitometry and plotted against time.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant P01 HL098055 (to J.D.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422455112/-/DCSupplemental.
References
- 1.Katz SS, Shipley GG, Small DM. Physical chemistry of the lipids of human atherosclerotic lesions. Demonstration of a lesion intermediate between fatty streaks and advanced plaques. J Clin Invest. 1976;58(1):200–211. doi: 10.1172/JCI108450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao X, et al. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab. 2012;15(4):545–553. doi: 10.1016/j.cmet.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011;145(3):341–355. doi: 10.1016/j.cell.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lusis AJ. Atherosclerosis. Nature. 2000;407(6801):233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sergin I, Razani B. Self-eating in the plaque: What macrophage autophagy reveals about atherosclerosis. Trends Endocrinol Metab. 2014;25(5):225–234. doi: 10.1016/j.tem.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Razani B, et al. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 2012;15(4):534–544. doi: 10.1016/j.cmet.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouimet M, et al. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13(6):655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinet P, Ritchey B, Smith JD. Physiological difference in autophagic flux in macrophages from 2 mouse strains regulates cholesterol ester metabolism. Arterioscler Thromb Vasc Biol. 2013;33(5):903–910. doi: 10.1161/ATVBAHA.112.301041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hornemann T, Worgall TS. Sphingolipids and atherosclerosis. Atherosclerosis. 2013;226(1):16–28. doi: 10.1016/j.atherosclerosis.2012.08.041. [DOI] [PubMed] [Google Scholar]
- 10.Smith EB. Intimal and medial lipids in human aortas. Lancet. 1960;1(7128):799–803. doi: 10.1016/s0140-6736(60)90680-2. [DOI] [PubMed] [Google Scholar]
- 11.Portman OW, Alexander M. Metabolism of sphingolipids by normal and atherosclerotic aorta of squirrel monkeys. J Lipid Res. 1970;11(1):23–30. [PubMed] [Google Scholar]
- 12.Jiang XC, et al. Plasma sphingomyelin level as a risk factor for coronary artery disease. Arterioscler Thromb Vasc Biol. 2000;20(12):2614–2618. doi: 10.1161/01.atv.20.12.2614. [DOI] [PubMed] [Google Scholar]
- 13.Jeong TS, et al. Increased sphingomyelin content of plasma lipoproteins in apolipoprotein E knockout mice reflects combined production and catabolic defects and enhances reactivity with mammalian sphingomyelinase. J Clin Invest. 1998;101(4):905–912. doi: 10.1172/JCI870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park TS, Rosebury W, Kindt EK, Kowala MC, Panek RL. Serine palmitoyltransferase inhibitor myriocin induces the regression of atherosclerotic plaques in hyperlipidemic ApoE-deficient mice. Pharmacol Res. 2008;58(1):45–51. doi: 10.1016/j.phrs.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Glaros EN, et al. Myriocin slows the progression of established atherosclerotic lesions in apolipoprotein E gene knockout mice. J Lipid Res. 2008;49(2):324–331. doi: 10.1194/jlr.M700261-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Glaros EN, Kim WS, Garner B. Myriocin-mediated up-regulation of hepatocyte apoA-I synthesis is associated with ERK inhibition. Clin Sci (Lond) 2010;118(12):727–736. doi: 10.1042/CS20090452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park TS, et al. Inhibition of sphingomyelin synthesis reduces atherogenesis in apolipoprotein E-knockout mice. Circulation. 2004;110(22):3465–3471. doi: 10.1161/01.CIR.0000148370.60535.22. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, et al. Serine palmitoyltransferase (SPT) deficient mice absorb less cholesterol. Biochim Biophys Acta. 2009;1791(4):297–306. doi: 10.1016/j.bbalip.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasumov T, et al. 2H2O-based high-density lipoprotein turnover method for the assessment of dynamic high-density lipoprotein function in mice. Arterioscler Thromb Vasc Biol. 2013;33(8):1994–2003. doi: 10.1161/ATVBAHA.113.301700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hojjati MR, et al. Effect of myriocin on plasma sphingolipid metabolism and atherosclerosis in apoE-deficient mice. J Biol Chem. 2005;280(11):10284–10289. doi: 10.1074/jbc.M412348200. [DOI] [PubMed] [Google Scholar]
- 21.Glaros EN, et al. Inhibition of atherosclerosis by the serine palmitoyl transferase inhibitor myriocin is associated with reduced plasma glycosphingolipid concentration. Biochem Pharmacol. 2007;73(9):1340–1346. doi: 10.1016/j.bcp.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 22.Hornemann T, Richard S, Rütti MF, Wei Y, von Eckardstein A. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J Biol Chem. 2006;281(49):37275–37281. doi: 10.1074/jbc.M608066200. [DOI] [PubMed] [Google Scholar]
- 23.Han S, Lone MA, Schneiter R, Chang A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl Acad Sci USA. 2010;107(13):5851–5856. doi: 10.1073/pnas.0911617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breslow DK, et al. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463(7284):1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2011;108(48):19222–19227. doi: 10.1073/pnas.1116948108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siow DL, Wattenberg BW. Mammalian ORMDL proteins mediate the feedback response in ceramide biosynthesis. J Biol Chem. 2012;287(48):40198–40204. doi: 10.1074/jbc.C112.404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 28.Ha SG, et al. ORMDL3 promotes eosinophil trafficking and activation via regulation of integrins and CD48. Nat Commun. 2013;4:2479. doi: 10.1038/ncomms3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hjelmqvist L, et al. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 2002;3(6):RESEARCH0027. doi: 10.1186/gb-2002-3-6-research0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng B, et al. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5(9):781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 31.Shiratori Y, Okwu AK, Tabas I. Free cholesterol loading of macrophages stimulates phosphatidylcholine biosynthesis and up-regulation of CTP: Phosphocholine cytidylyltransferase. J Biol Chem. 1994;269(15):11337–11348. [PubMed] [Google Scholar]
- 32.Pattison JS, Osinska H, Robbins J. Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes. Circ Res. 2011;109(2):151–160. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabas I, Marathe S, Keesler GA, Beatini N, Shiratori Y. Evidence that the initial up-regulation of phosphatidylcholine biosynthesis in free cholesterol-loaded macrophages is an adaptive response that prevents cholesterol-induced cellular necrosis. Proposed role of an eventual failure of this response in foam cell necrosis in advanced atherosclerosis. J Biol Chem. 1996;271(37):22773–22781. doi: 10.1074/jbc.271.37.22773. [DOI] [PubMed] [Google Scholar]
- 35.Das A, Brown MS, Anderson DD, Goldstein JL, Radhakrishnan A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife. 2014;3:3. doi: 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slotte JP, Tenhunen J, Pörn I. Effects of sphingomyelin degradation on cholesterol mobilization and efflux to high-density lipoproteins in cultured fibroblasts. Biochim Biophys Acta. 1990;1025(2):152–156. doi: 10.1016/0005-2736(90)90092-3. [DOI] [PubMed] [Google Scholar]
- 37.Pörn MI, Slotte JP. Localization of cholesterol in sphingomyelinase-treated fibroblasts. Biochem J. 1995;308(Pt 1):269–274. doi: 10.1042/bj3080269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohvo H, Olsio C, Slotte JP. Effects of sphingomyelin and phosphatidylcholine degradation on cyclodextrin-mediated cholesterol efflux in cultured fibroblasts. Biochim Biophys Acta. 1997;1349(2):131–141. doi: 10.1016/s0005-2760(97)00126-4. [DOI] [PubMed] [Google Scholar]
- 39.Scheek S, Brown MS, Goldstein JL. Sphingomyelin depletion in cultured cells blocks proteolysis of sterol regulatory element binding proteins at site 1. Proc Natl Acad Sci USA. 1997;94(21):11179–11183. doi: 10.1073/pnas.94.21.11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu XX, Tabas I. Sphingomyelinase enhances low density lipoprotein uptake and ability to induce cholesteryl ester accumulation in macrophages. J Biol Chem. 1991;266(36):24849–24858. [PubMed] [Google Scholar]
- 41.Slotte JP, Bierman EL. Depletion of plasma-membrane sphingomyelin rapidly alters the distribution of cholesterol between plasma membranes and intracellular cholesterol pools in cultured fibroblasts. Biochem J. 1988;250(3):653–658. doi: 10.1042/bj2500653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bektas M, et al. Sphingosine 1-phosphate lyase deficiency disrupts lipid homeostasis in liver. J Biol Chem. 2010;285(14):10880–10889. doi: 10.1074/jbc.M109.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ridgway ND. 25-Hydroxycholesterol stimulates sphingomyelin synthesis in Chinese hamster ovary cells. J Lipid Res. 1995;36(6):1345–1358. [PubMed] [Google Scholar]
- 44.Leppimäki P, Kronqvist R, Slotte JP. The rate of sphingomyelin synthesis de novo is influenced by the level of cholesterol in cultured human skin fibroblasts. Biochem J. 1998;335(Pt 2):285–291. doi: 10.1042/bj3350285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris IR, et al. Parallel regulation of sterol regulatory element binding protein-2 and the enzymes of cholesterol and fatty acid synthesis but not ceramide synthesis in cultured human keratinocytes and murine epidermis. J Lipid Res. 1998;39(2):412–422. [PubMed] [Google Scholar]
- 46.Chen H, Born E, Mathur SN, Field FJ. Cholesterol and sphingomyelin syntheses are regulated independently in cultured human intestinal cells, CaCo-2: Role of membrane cholesterol and sphingomyelin content. J Lipid Res. 1993;34(12):2159–2167. [PubMed] [Google Scholar]
- 47.Hojjati MR, Jiang XC. Rapid, specific, and sensitive measurements of plasma sphingomyelin and phosphatidylcholine. J Lipid Res. 2006;47(3):673–676. doi: 10.1194/jlr.D500040-JLR200. [DOI] [PubMed] [Google Scholar]
- 48.Robinet P, Wang Z, Hazen SL, Smith JD. A simple and sensitive enzymatic method for cholesterol quantification in macrophages and foam cells. J Lipid Res. 2010;51(11):3364–3369. doi: 10.1194/jlr.D007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gulshan K, Brubaker G, Wang S, Hazen SL, Smith JD. Sphingomyelin depletion impairs anionic phospholipid inward translocation and induces cholesterol efflux. J Biol Chem. 2013;288(52):37166–37179. doi: 10.1074/jbc.M113.512244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rütti MF, Richard S, Penno A, von Eckardstein A, Hornemann T. An improved method to determine serine palmitoyltransferase activity. J Lipid Res. 2009;50(6):1237–1244. doi: 10.1194/jlr.D900001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.