Significance
Both genetically altered and naturally long-lived mammals are more resistant to toxic compounds that may cause cancer and age-associated diseases than their shorter-lived counterparts. The mechanisms by which this stress resistance occurs remain elusive. We found that longer-lived rodent species had markedly higher levels of signaling activity of the multifunctional regulator nuclear factor erythroid 2-related factor (Nrf2) and that this increase in cytoprotective signaling appeared to be due to species differences in Kelch-like ECH-Associated Protein 1 (Keap1) and β-transducin repeat-containing protein (βTrCP) regulation of Nrf2 activity. Both of these negative regulators of Nrf2-signaling activity are significantly lower in longer-lived species. By targeting the proteins that regulate Nrf2 rather than Nrf2 itself, we may be able to identify new therapies that impact aging and age-associated diseases such as cancer.
Keywords: stress resistance, Nrf2, rodent longevity, naked mole-rat
Abstract
The preternaturally long-lived naked mole-rat, like other long-lived species and experimental models of extended longevity, is resistant to both endogenous (e.g., reactive oxygen species) and environmental stressors and also resists age-related diseases such as cancer, cardiovascular disease, and neurodegeneration. The mechanisms behind the universal resilience of longer-lived organisms to stress, however, remain elusive. We hypothesize that this resilience is linked to the activity of a highly conserved transcription factor, nuclear factor erythroid 2-related factor (Nrf2). Nrf2 regulates the transcription of several hundred cytoprotective molecules, including antioxidants, detoxicants, and molecular chaperones (heat shock proteins). Nrf2 itself is tightly regulated by mechanisms that either promote its activity or increase its degradation. We used a comparative approach and examined Nrf2-signaling activity in naked mole-rats and nine other rodent species with varying maximum lifespan potential (MLSP). We found that constitutive Nrf2-signaling activity was positively correlated (P = 0.0285) with MLSP and that this activity was also manifested in high levels of downstream gene expression and activity. Surprisingly, we found that species longevity was not linked to the protein levels of Nrf2 itself, but rather showed a significant (P < 0.01) negative relationship with the regulators Kelch-like ECH-Associated Protein 1 (Keap1) and β-transducin repeat-containing protein (βTrCP), which target Nrf2 for degradation. These findings highlight the use of a comparative biology approach for the identification of evolved mechanisms that contribute to health span, aging, and longevity.
The quest for molecular mechanisms that may stave off the aging process and extend good health remains elusive. Extended longevity in fungi, plants, and animals is strongly associated with enhanced stress resistance (1). Moreover, the degree of stress resistance is proportional to observed lifespan extension in long-lived mutants of yeasts (2), worms (3), flies (4), and laboratory mouse strains (5). Similarly, data based upon cell lines derived from long-lived mammals support this premise (6, 7). Clearly, long-lived species have evolved ways to shield their cells from both chronic [e.g., reactive oxygen species (ROS)] and unpredictable stressors. Surprisingly little is known about the mechanism(s) responsible. Lack of progress in this regard could reflect the predominant choice of short-lived animal models that are likely to have an inadequate molecular arsenal to naturally protect against the vagaries of aging (8). Comparative biology provides a powerful tool to evaluate whether nature, through millions of years of “evolutionary experimentation,” has evolved cellular mechanisms that may abrogate physiological declines during aging in long-lived species. In this study, we exploit the large variation in mammalian maximum species lifespan potential (MLSP) to determine whether species longevity may be linked to specific cytoprotective mechanisms.
Species MLSP is strongly dependent on body size, such that for every doubling of species body mass, there is a 16% increase in MLSP (9). However, even among rodents of similar body size, MLSP varies eightfold. At one end of the spectrum are mice that live approximately half as long as allometrically predicted (4 y), and at the other is the naked mole-rat, with a MLSP of ∼31 y in captivity, fivefold greater than its predicted MLSP. Throughout their long lives, these rodents experience a prolonged health span with attenuated declines in many physiological characteristics—e.g., cardiac function—and age-associated diseases that typically change with age (10). Unlike other small mammals that are particularly susceptible to cancer, spontaneous and experimentally induced neoplasia are extremely rare in naked mole-rats (10, 11). Naked mole-rats also tolerate oxidative damage, as well as other stressors, including DNA-damaging compounds and heat (7, 12, 13). We hypothesize that activity of a conserved cytoprotective signaling pathway, namely that of nuclear factor erythroid 2-related factor 2 (Nrf2), plays a pivotal role in the degree of stress resistance and may be directly linked to species longevity and health span.
Nrf2 is expressed in all tissues and is active under both homeostatic and stressful conditions (14, 15). It initiates the transcription of several hundred molecules that possess an antioxidant response element (ARE) (16, 17). These include the detoxicant glutathione S-transferase (GST) and the redox-regulator, NAD(P)H:quinone oxidoreductase 1 (NQO1), in addition to antioxidants, molecular chaperones [heat shock proteins (HSPs)], and proteasome subunits (16, 17). These multiple layers of protection play a pivotal role in toxin eradication and removal of oxidative stress, and they contribute to proteostasis, using both HSPs and the removal of damaged proteins via proteasomal degradation or autophagy (18). Nrf2 also protects against inflammation, cancer induction, and neurodegeneration, and it likely positively influences both the health span and lifespan of an organism (reviewed in ref. 18).
Surprisingly, little is known about Nrf2 signaling and its divergent regulation in species with disparate MLSP. Nrf2 regulation is thought to be largely driven by Kelch-like ECH-Associated Protein 1 (Keap1), which targets Nrf2 for proteasomal degradation (19). Recently, it has become clear that regulation of Nrf2 activity is more complex than previously reported, and numerous intracellular molecules modulate Nrf2 expression, activation, and half-life (19). We posit that modulation of Nrf2 activity and its regulatory proteins differs among rodents with disparate longevity. We test this hypothesis using 10 species, ranging in MLSP from 4 to 31 y. We focus our study specifically on rodents because this mammalian order is arguably the most successful and contains similarly sized short-lived and extremely long-lived species, thereby eliminating confounding allometric relationships. We hypothesize that those species expressing high constitutive activities of Nrf2 are primed to rapidly respond and neutralize stress, maintain good health during aging, and attain extraordinary longevity.
Results
Increased Nrf2 Signaling Activity in Long-Lived Naked Mole-Rats.
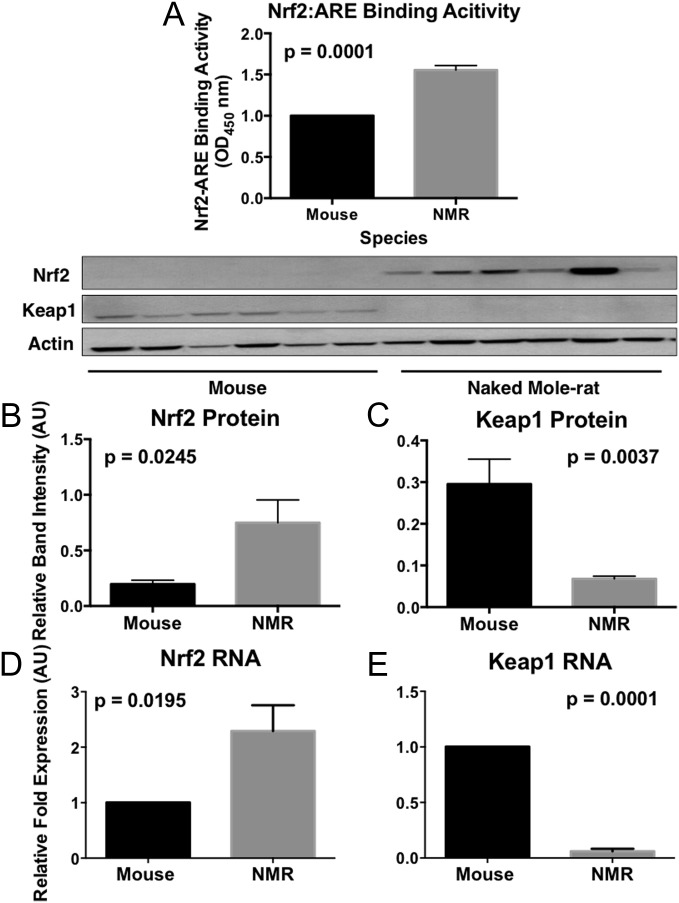
We examined Nrf2 signaling by measuring the rates at which Nrf2 binds to the ARE in liver tissue of young, physiologically age-matched long-lived naked mole-rats and short-lived mice. We chose to use liver tissue because this tissue is histologically indistinguishable in these two species and is relatively homogenous in cellular composition compared with other tissue types. Also, liver is the primary detoxification organ for xenobiotic compounds. Nrf2:ARE binding was 50% (P = 0.0001) higher in naked mole-rats compared with mice (Fig. 1A). Western blot analyses in liver tissues revealed that total Nrf2 protein levels were almost fivefold higher (n = 6, P = 0.0245) in naked mole-rats than in mice (Fig. 1B). Also, the key negative regulator of Nrf2, Keap1, was threefold lower (P = 0.0037) in naked mole-rats (Fig. 1C), resulting in increased levels of free Nrf2 and presumably an extended half-life. Similarly, mRNA levels of Nrf2 were 2.5-fold higher (P = 0.0195) in naked mole-rats (Fig. 1D), whereas Keap1 was lower (Fig. 1E; P = 0.0001) than observed in mice. Thus, the higher Nrf2:ARE binding activity in the naked mole-rat is most likely due to constitutively high levels of total Nrf2 protein paired with very low levels of the inhibitory protein Keap1.
Fig. 1.
Preternaturally long-lived naked mole-rats have higher Nrf2-signaling activity than short-lived mice. (A) Liver tissue of naked mole-rats have higher Nrf2–ARE-binding (n = 4) activity than that of mice (P = 0.0001). (B and C) Nrf2 total protein levels are higher (B; n = 6; P = 0.0245) and Keap1 levels lower (C; P = 0.0037) in naked mole-rats. (D and E) Quantitative PCR similarly shows that Nrf2 transcript levels are markedly higher (D; P = 0.0195) and Keap1 levels strikingly lower (E; P = 0.0001) in the longer-lived species. These data indicate that the higher Nrf2:ARE binding activity is due to increased Nrf2 protein availability as a result of low levels of Keap1 and concomitant decreased Nrf2 ubiquitination and proteasomal degradation. Error bars represent SEM.
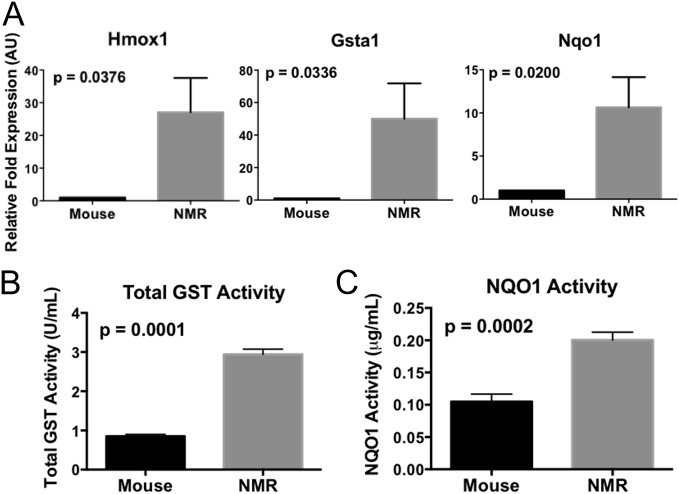
We further measured mRNA levels of some Nrf2–ARE signaling targets, including heme oxygenase 1 (Hmox1), Glutathione-S-transferase alpha 1 isoform (Gsta1), and Nqo1 (n = 5 or 6; Fig. 2A). All of these transcripts were significantly higher in naked mole-rats (P < 0.05). Similarly, the activities of the enzymes GST and NQO1, determined by using colorimetric assays (Fig. 2 B and C), were significantly higher in naked mole-rat liver tissue than those of mice, indicating that the high total Nrf2 protein levels and elevated Nrf2:ARE binding activity in the naked mole-rat facilitate higher levels of transcriptional activation of down-stream cytoprotective molecules, even under nonstressed basal conditions.
Fig. 2.
Transcript expression of downstream molecules regulated by Nrf2 is significantly higher in naked mole-rats than in mice. Naked mole-rats have higher levels of several gene transcripts that are dominantly regulated by Nrf2 (n = 5 or 6). (A) These include Hmox1 (Left; P = 0.0376), Gsta1 (Center; P = 0.0336), and Nqo1 (Right; P = 0.0200). (B and C) Elevated Nrf2-signaling activity in the naked mole-rat was further confirmed via higher total GST (B; P = 0.0001) and NQO1 activities (C; P = 0.0002) than observed in mice. Error bars represent SEM.
Nrf2-Signaling Activity Is Positively Correlated with Maximum Lifespan.
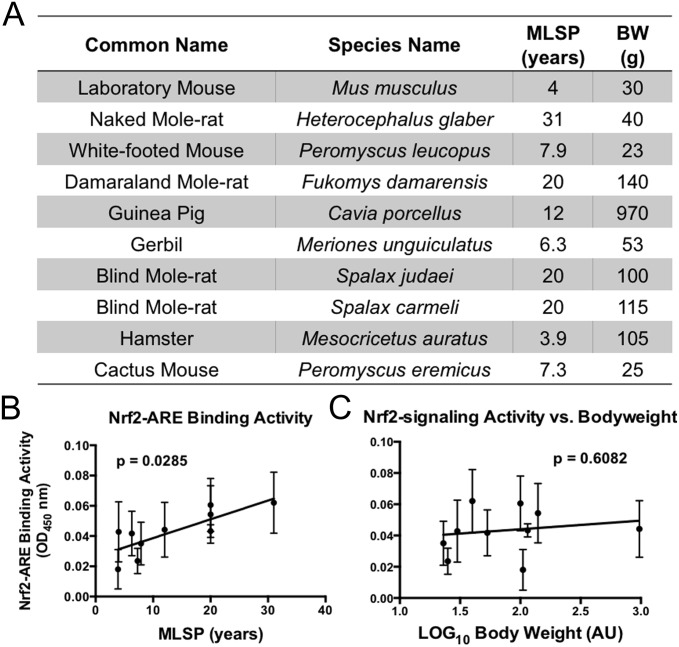
To establish whether high basal levels of Nrf2 activity in the naked mole-rat reflect an idiosyncratic property of the naked mole-rat or a common mechanism directly linked to species longevity, we used a multispecies comparative approach including eight additional rodent species representing eight distinct rodent families with widely divergent longevities (Fig. 3A). We found a striking correlation between Nrf2:ARE binding activity and MLSP in these 10 species (Fig. 3B; P = 0.0285, n = 3 or 4 for each species).
Fig. 3.
Nrf2–ARE binding activity is significantly correlated with the lifespan of rodent species. (A) Ten different rodent species with varying MLSPs were used to asses basal Nrf2 signaling activities. (B) Liver Nrf2–ARE binding activity was positively correlated with MLSP in the 10 rodent species (n = 4 per species; P = 0.0285), whose MLSP varied between 4 and 31 y and whose body weights ranged from 20 g to almost 1 kg. (C) Nrf2 signaling did not correlate with bodyweight (P = 0.6082). Error bars represent SEM.
In contrast, we found no significant correlation between species average body weight and Nrf2:ARE activity levels in these 10 species (Fig. 3C; P = 0.6082); this elevation in Nrf2–ARE signaling activity is not related to body size. When data were further adjusted for phylogenetic lineage, there was no additional effect on the relationship between Nrf2-signaling activity and MLSP, indicating that levels of Nrf2-signaling activity was linked to MLSP, independent of body weight and phylogenetic relatedness.
Surprisingly, despite the strong correlation between Nrf2 activity and species MLSP under nonstressed, basal conditions, no such relationship was evident between total protein levels of Nrf2 and MLSP (Table 1 and Fig. S1). These data suggest that regulatory mechanisms controlling levels of free Nrf2 rather than total Nrf2 protein levels are responsible for this relationship.
Table 1.
Multiple proteins that regulate Nrf2-signaling activity correlated with MLSP in rodents
| Protein | Regulation of Nrf2 signaling | Relationship with MLSP | R2 | P value |
| Nrf2 | Increased levels of total Nrf2 protein may lead to increased Nrf2-signaling activity | y = 0.4253x + 4.812 | 0.07832 | 0.1035 |
| p21 | Binds to Nrf2, preventing Keap1 binding and degradation | y = −0.03173x + 1.129 | 0.08131 | 0.0871 |
| p38MAPK | Phosphorylated p38MAPK potentially phosphorylates and activates Nrf2 signaling | y = 0.02161x + 1.306 | 0.009601 | 0.5475 |
| P62/SQSTM1 | Competitively binds to Keap1, preventing Nrf2 degradation | y = −0.01620x + 0.8494 | 0.1696 | 0.0102* |
| PALB2 | Binds to Keap1 to inhibit Nrf2 degradation and promote Nrf2 activity | y = 0.1446x + 2.292 | 0.04845 | 0.1093 |
| WTX | Competitively binds to Keap1 to prevent Nrf2 binding and ubiquitination | y = 0.2896x + 0.2954 | 0.07801 | 0.2330 |
| DPP3 | Competitively binds to Keap1 to prevent Nrf2 binding and ubiquitination | y = 0.1621x + 1.685 | 0.09239 | 0.0674 |
| Keap1 | Targets Nrf2 for ubiquitination and degradation via the proteasome | y = −0.04825x + 2.065 | 0.2396 | 0.0013* |
| βTrCP | Binds to Nrf2 to promote ubiquitination and degradation via the proteasome | y = −0.01547x + 0.6708 | 0.2010 | 0.0037* |
| SIAH2 | Suppresses and contributes to the degradation of Nrf2 | y = 0.02748x + 1.630 | 0.01734 | 0.4180 |
| CRIF1 | Increases proteasome degradation of Nrf2; Redox independent | y = 0.07633x + 2.348 | 0.02128 | 0.3691 |
We measured 11 different proteins that have been shown to regulate cytoprotective Nrf2-signaling activity to determine whether there was a correlation between protein expression in liver tissue and MLSP in 10 different rodent species (*P < 0.05). Surprising, total Nrf2 protein level did not correlate with MLSP (P = 0.1035); however, its negative regulators Keap1 (P = 0.0013) and βTrCP (P = 0.0037) did negatively correlate with MLSP. Notably, p62/SQSTM1, a player in autophagy regulation, also negatively correlated with MLSP (P = 0.0102).
Keap1 and βTrCP Regulate Nrf2-Signaling Activity in Long-Lived Species.
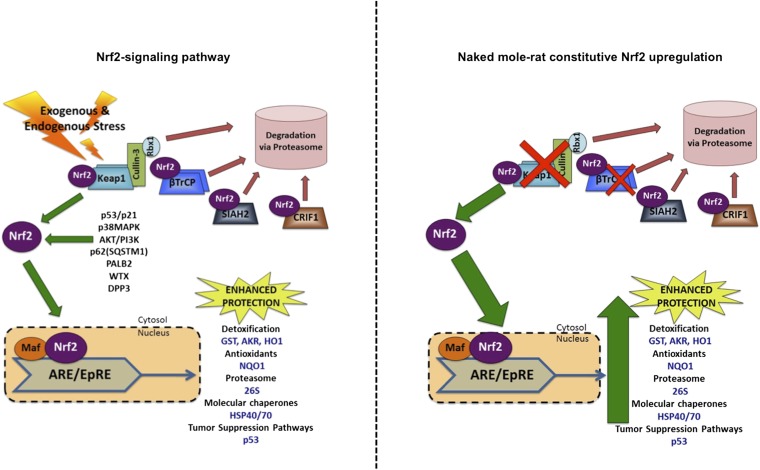
Because levels of Nrf2 total protein did not correlate with MLSP, we evaluated whether levels of Keap1, as well as 11 other proteins known to influence Nrf2 signaling (Table 1 and Fig. S1), were correlated with MLSP. Cyclin-dependent kinase inhibitor p21 positively influences Nrf2 signaling by binding to Nrf2 and thereby preventing it from binding to Keap1 and thwarting Nrf2 ubiquitination and degradation (20). Other molecules do not directly interact with Nrf2 but rather with its regulators (e.g., Keap1) and indirectly modulate Nrf2 degradation and signaling activity. These include sequestosome 1 (SQSTM1/p62) (21), the partner and localizer of BRCA2 (PALB2), Wilms tumor gene on X chromosome (WTX), and dipeptidyl-peptidase 3 (DPP3) (22–24). Also, β-transducin repeat-containing protein (βTrCP), seven in absentia homolog 2 (SIAH2), and CR6-interacting factor 1 (CRIF1) act in a similar fashion to Keap1, promoting Nrf2 degradation, although the specific mechanisms by which they act have yet to be fully elucidated (25–27). Because Nrf2 phosphorylation is required for Nrf2 nuclear migration and its binding to the ARE (28), phosphorylating kinases such as p38 mitogen-activated protein kinase (p38MAPK) may directly phosphorylate Nrf2 or impact other proteins that carry out this function (29), thereby influencing Nrf2 signaling.
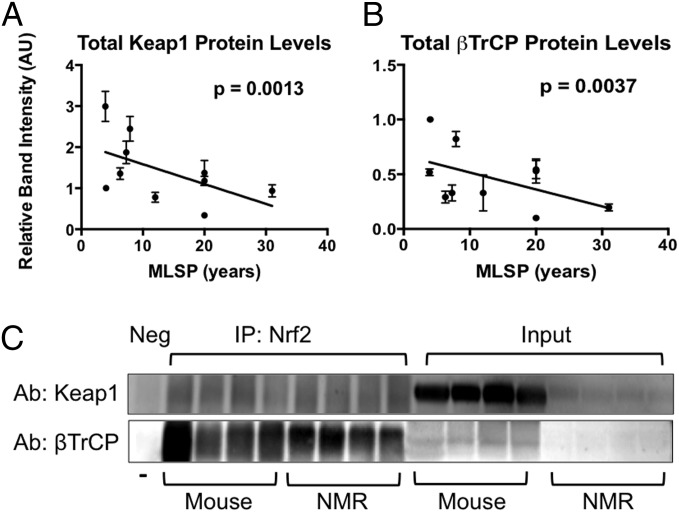
Of the 11 proteins we tested (Table 1 and Fig. S1), we observed significant relationships with only 3 of these proteins with MLSP (Table 1)—namely, Keap1 (Fig. 4A; P = 0.0013), p62/SQSTM1 (P = 0.0102), and βTrCP (Fig. 4B; P = 0.0037). These molecules were negatively correlated with Nrf2, suggesting that the lower their levels, the greater the Nrf2 activity and the longer the MLSP. For Keap1 and βTrCP to impact Nrf2 activity, Nrf2 must bind to either protein for subsequent ubiquitination and degradation. We confirmed that these proteins were bound to Nrf2 via immunoprecipitation (Fig. 4C). Additionally, the βTrCP mRNA levels were significantly lower in naked mole-rats compared with mice (Fig. S2; P = 0.0001). Thus, it is highly likely that both Keap1 and βTrCP contribute to the regulation of Nrf2 signaling and that their levels are inversely linked to MLSP and longevity (Fig. 5).
Fig. 4.
Keap1 and BTrCP levels are significantly linked to rodent MLSP and likely contribute to longevity-associated up-regulation in Nrf2-signaling activity. (A and B) Both Keap1 (A; P = 0.0013) and βTrCP (B; P = 0.0037) were negatively correlated with MLSP in rodents (n = 4 for each of 10 species). Both of these proteins negatively regulate Nrf2 by binding to Nrf2 and targeting it for degradation. (C) Using immunoprecipitation, we confirmed that Keap1 and Nrf2 were bound to Nrf2 in liver tissues of both mice and naked mole-rats (n = 6). Error bars represent SEM.
Fig. 5.
Nrf2 signaling is regulated by several distinct mechanisms. Nrf2 activity is augmented by exogenous (i.e., xenobiotic compounds) and/or endogenous (i.e., ROS) stressors. Free Nrf2 can bind to the ARE in the nucleus and activates hundreds of cytoprotective molecules, including detoxification enzymes like GST, antioxidant enzymes (e.g., NQO1), proteasome subunits, and molecular chaperones. Nrf2 activity is largely negatively regulated by Keap1, which binds to Nrf2 and targets it for ubiquitination and proteasomal degradation. Recently, other proteins have been shown to influence Nrf2 activity either by interacting directly with Nrf2 or with Keap1. Positive regulators, including p21 and p38MAPK, directly activate Nrf2 signaling, whereas P62/SQSTM1, PALB2, WTX, and DPP3 interact with Keap1 and thereby activate Nrf2 signaling. Other negative regulators of Nrf2, including βTrCP, SIAH2, and CRIF1, directly target it for degradation independent of Keap1. CRIF1 regulates Nrf2 signaling independent of the cell’s redox status. All of these molecules may be potential targets for cancer prevention or longevity therapeutics.
Discussion
Given the importance of Nrf2 in cytoprotection and xenobiotic metabolism, we confirmed our hypothesis that the degree of protection afforded by Nrf2 activity was positively linked to species longevity. Clearly, long-lived species are primed to rapidly and effectively respond to stressors before these damaging agents can compromise somatic integrity, impair cell function, or induce cell death. High constitutive levels of Nrf2 activity support our hypothesis that Nrf2 is the “guardian of health span and the gate-keeper of species longevity” (18). To our knowledge, we also show for the first time that increased levels of Nrf2 activity in long-lived species are not due to increased expression of Nrf2 protein, but rather are due to reduced expression of its negative regulators—namely, Keap1 and βTrCP.
Insights from the Naked Mole-Rat.
Constitutive Nrf2 levels and activity are markedly higher in the longest-lived rodent than in short-lived mice (Figs. 1 and 2). Given the pivotal role of Nrf2 in cytoprotection, proteostasis, and xenobiotic metabolism, such high levels of Nrf2 activity likely contribute to the extraordinary resilience of naked mole-rats to a diverse suite of stressors. In vitro studies using primary cultures of skin fibroblasts from mice and naked mole-rats treated with heavy metals, chemotherapeutics, and oxidative stressors revealed that naked mole-rats require 2- to 100-fold higher concentrations of toxins to kill 50% of cells 24 h after exposure (7, 13). Similar resistance to beta amyloid is observed in neurons in culture and in brains ex vivo (30). Enhanced cytoprotection afforded by the twofold to fourfold higher levels of Nrf2 signaling in the brain may also contribute to sustained neuronal number and integrity (30), well-maintained cardiac function (31), unchanged levels of oxidative damage during aging (32), and resistance to spontaneous and experimentally induced cancers (11).
Nrf2 also regulates the transcription of antioxidants, molecular chaperones, and proteasome subunits. These Nrf2-regulated genes are constitutively expressed at high levels up to 80-fold higher than observed in mice (Fig. 2A). Such high levels of expression and activity provide a diverse and at-the-ready arsenal to combat both inevitable endogenous stressors (e.g., ROS) and the barrage of unpredictable xenobiotics (e.g., food toxins or chemical pollutants). Moreover, they also enable the removal and repair of any damage that may have been induced by these stressors (33, 34). As such, these primed protective mechanisms likely contribute to better maintenance of both proteostasis and somatic integrity, thus contributing to extended health span and delayed aging.
Nrf2-dependent downstream molecules, GST and NQO1, show twofold to fivefold higher activities in naked mole-rats (Fig. 2 B and C). Similarly, long-lived dwarf mice have elevated basal GST levels and activity (35), and a similar GST profile is associated with increased lifespan and toxin resistance in Caenorhabditis elegans (36). NQO1 activity also confers multiple layers of protection by combating oxidative stress, stabilizing membranes, and inhibiting apoptosis. Furthermore, NQO1 facilitates the complete reduction and detoxification of highly reactive quinones. Like longer-lived dwarf and caloric-restricted mice, naked mole-rats have higher basal levels than do wild-type mice (Fig. 2C) (37, 38) facilitating augmented protection.
Nrf2 Regulation in Species with Divergent Longevity.
To our knowledge, we report for the first time a significant, positive relationship between Nrf2 activity and species longevity (Fig. 3). These 10 different rodent species represent five phylogenetically distinct families, a 500-fold range in body mass (20 g to 1 kg), and MLSPs ranging from 4 to 31 y. Only two of the species used in this study are in the same rodent family (Bathyergidae). This strictly subterranean family had very similar Nrf2-dependent traits to those of another long-living subterranean rodent, Spalax judaei (Spalacidae), which is more closely related to mice and rats than to the Bathyergidae. High levels of Nrf2 activity in underground-dwelling species may reflect convergent evolution of similar protective properties in response to a habitat where heavy metals and other soil toxins prevail and where variable oxygen atmospheres and concomitant oxidative stress are commonly encountered.
This association of Nrf2 activity with MLSP further reveals that, as species independently evolved longer lifespans, they also augmented Nrf2 activity, such that for a 10-y increase in lifespan, Nrf2 activity increases 1.4-fold (Fig. 3B). It is likely that increased Nrf2 signaling enhances species fitness and contributes to delayed rates of aging and ultimately to prolonged MLSP and health span. In short-lived mice, both Nrf2 levels and downstream signaling components in the nervous system decrease with age (39) and likely contribute to their observed age-related cognitive and physiological declines. Mouse models of extended longevity [i.e., methionine-restricted (40), little (37), and dwarf (35) mice] all show signs of enhanced Nrf2 activity with elevated activities of Nrf2-dependent enzymes—e.g., GST. Conversely, when the homolog of Nrf2 is knocked out in invertebrate models, lifespan extension associated with caloric restriction is abrogated (41), further supporting the premise that Nrf2 is a conserved mechanism linked to lifespan regulation. Also, given that changes in Nrf2 signaling are closely associated with cancer, toxin resistance, good health, and prolonged longevity, understanding how it is regulated is of critical importance and may lead to potential therapeutics that extend longevity and health span as well as abrogate age-associated diseases.
Unlike Nrf2 activity, total Nrf2 protein levels did not show a significant relationship with MLSP (Table 1 and Fig. S1). We therefore focused on other regulators of Nrf2 activity, including its negative regulator, Keap1 (14). Keap1 tethers most Nrf2 in the cytoplasm and regulates its proteasomal degradation via direct binding with an E3 ligase. Although Keap1 knockout mice are not viable and die within the first 3 weeks of life (42), down-regulation of Keap1 expression in mice increases resistance to toxins (43). Experimental manipulation of Keap1 levels alters both fruit fly toxin resistance and modulates their lifespan (44). Our multispecies comparative study similarly shows a significant inverse relationship between MLSP and Keap1 levels (Table 1, Fig. 4A, and Fig. S1), suggesting that, commensurate with their lifespan, this negative regulator is instrumental in modulating Nrf2 activity. Two other regulators of Nrf2 activity, p62/SQSTM1 and βTrCP, also showed significant relationships with MLSP and highlight the importance of multiple regulatory pathways and longevity.
The negative relationship with p62 and MLSP was surprising in that p62 binds to Keap1, preventing Nrf2 degradation and thus promoting Nrf2 activity (Table 1 and Fig. S1). However, because p62 is also known to modulate autophagy, its effects here on lifespan may reflect elevated levels of autophagy in long-lived species, rather than Nrf2 activity. Accumulation of p62 is reportedly linked to a rapid aging phenotype and increased cancer risk (45); thus, it is not a surprise that longer-lived rodents have lower levels of p62 than their shorter-lived counterparts. Lower levels of p62 may also explain the increase in autophagy observed in naked mole-rat cells and tissues (12).
Recent reports revealed that Nrf2-signaling pathways are negatively regulated by βTrCP, an E3 ubiquitin ligase that binds to Nrf2, promoting proteasomal degradation (27). We found that βTrCP negatively correlates with MLSP (Table 1 and Fig. 4B). βTrCP is associated with several intracellular processes, including replication checkpoints and responses to both DNA damage and xenobiotic agents. It is likely that these actions are linked to MLSP. Oxidative stress, Akt/Gsk3β, and Sirt1 pathways modulate βTrCP expression, implying that βTrCP may also be critical in the life-extending effects of caloric restriction (46–48). βTrCP may be an exciting therapeutic target for longevity-associated increases in Nrf2 signaling because it is highly conserved among mice, naked mole-rats, and even humans, with homology >97% among these three evolutionarily distinct species. Moreover, low levels of this protein appear to have fewer harmful side effects than those of Keap1 (42).
This multispecies comparative study exploits the untapped resource of natural variation in longevity among similar-sized rodents and, to our knowledge, reveals for the first time that Nrf2 activity is an essential modulator of species longevity. The significant positive relationship between Nrf2 activity and rodent MLSP is not due to total Nrf2 protein levels, but rather is associated with its regulation by Keap1 and βTrCP. These findings highlight previously unidentified directions and overlooked mechanisms to target for extending good health well into old age.
Materials and Methods
Animal Care.
C57bl/6 mice, naked mole-rats, white-footed mice, deer mice, guinea pigs, gerbils, and hamsters were raised in-house under Association for Assessment and Accreditation of Laboratory Animal Care International standards of care. All animal protocols were approved by the University of Texas Health Science Center at San Antonio Institutional Animal Care and Use Committee. Spalax ehrenbergi carmeli and judaei were housed individually in Haifa University, and harvested tissues were shipped to our laboratory on dry ice. All animals used in these studies were young, healthy individuals from both sexes. Immediately after euthanasia, tissues were flash-frozen in liquid nitrogen and stored at −80 °C until use.
Immunoblotting.
Sections of liver tissue with homogenized in radioimmunoprecipitation assay buffer (RIPA), and electrophoresis was performed on a 4–20% gradient gel (Bio-Rad) and transferred to a PVDF membrane. Membranes were probed with the following antibodies per manufacturer recommendation: Nrf2 (sc-13032; Santa Cruz), Keap1 (sc-33569; Santa Cruz), p21 (sc-397; Santa Cruz), p38MAPK (no. 9215; Cell Signaling), p62/SQSTM1 (sc-10117; Santa Cruz), PALB2 (sc-160647; Santa Cruz), WTX (sc-138078; Santa Cruz), DPP3 (sc-55640; Santa Cruz), βTrCP (sc-8863; Santa Cruz), SIAH2 (sc-5508; Santa Cruz), and CRIF1 (sc-103443; Santa Cruz). Bands were quantified by using ImageJ software and normalized to GAPDH (G8795; Sigma). Antibody epitopes were aligned with known rodent sequences to evaluate species specificity.
Nrf2:ARE Binding Activity.
Nrf2:ARE binding activity was assessed by using a TransAm Binding Assay (Active Motif). Briefly, nuclear extracts from liver lysates were incubated on the ARE–oligonucleotide-coated plate and incubated with Nrf2 antibody. Binding was detected via a colorimetric reaction at a wavelength of 450 nm (n = 3–4 per species).
Real-Time PCR.
RNA was isolated from frozen liver tissue (n = 5–6 per species) by using the mirVana isolation kit (Life Technologies). cDNA was made by using the Quantitect Reverse Trasncriptase Kit (Qiagen). Real-time PCR was performed by using the SYBR Green Real-Time PCR Master Mix (Life Technologies) protocol. Efficiency and accuracy was determined by titration and visualization on gel. Primer details are provided in Tables S1 and S2.
Enzyme Activity Assays.
NQO1 activity assays were performed by using the protocol outlined in Prochaska et al. (49). Homogenized liver samples were incubated with a NADPH-generating reaction system, menadione, and MTT. Absorbance was measured at 595 nm after 5 min of reaction time. Total GST activity was determined by using liver samples homogenized in a cold potassium phosphate buffer, and activity was determined by using the total GST assay kit (no. 703302; Carman Chemical).
Immunoprecipitation.
Immunoprecipitations were performed in mouse and naked mole-rat liver homogenates by using a Pierce Crosslink Immunoprecipitation Kit. A total of 1,000 μg of protein in liver homogenates was incubated with Nrf2 (Santa Cruz) and Keap1 (Santa Cruz) after these antibodies were cross-linked to agarose beads. Protein interactions were analyzed via Immunoblot for βTrCP and Keap1 binding, respectively.
Statistical Analysis.
GraphPad Prism software was used for most statistical analyses. Relationships between protein expression, enzyme activities, and binding activity between mice and naked mole-rats were analyzed via unpaired T-test. Error bars for all graphs represent SEM, and all calculated P values are provided in figures. Multispecies comparisons were performed by using phyologenetic independent contrast analyses of the linear regressions of MLSP with Nrf2–ARE binding activity and total protein expression as described (50).
Supplementary Material
Acknowledgments
We thank Megan Smith for care of the naked mole-rat and Damaraland mole-rat colonies; and Dr. Tomas Pavlicek and Pavel Tovbin for assisting Y.H.E. in the collection of Spalax tissue. This work was supported by grants from American Federation for Aging Research, the Glenn Foundation for Medical Research, and the NIH/National Institute on Aging (NIA) (to R.B.). K.N.L. was supported by NIA Training Grant T32 AG021890. The Nathan Shock Center partially contributed to the cost of animal husbandry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417566112/-/DCSupplemental.
References
- 1.Martin GM, Austad SN, Johnson TE. Genetic analysis of ageing: Role of oxidative damage and environmental stresses. Nat Genet. 1996;13(1):25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- 2.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292(5515):288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 3.Larsen PL. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1993;90(19):8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin YJ, Seroude L, Benzer S. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science. 1998;282(5390):943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- 5.Harper JM, et al. Stress resistance and aging: Influence of genes and nutrition. Mech Ageing Dev. 2006;127(8):687–694. doi: 10.1016/j.mad.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapahi P, Boulton ME, Kirkwood TB. Positive correlation between mammalian life span and cellular resistance to stress. Free Radic Biol Med. 1999;26(5-6):495–500. doi: 10.1016/s0891-5849(98)00323-2. [DOI] [PubMed] [Google Scholar]
- 7.Lewis KN, Mele J, Hornsby PJ, Buffenstein R. Stress resistance in the naked mole-rat: The bare essentials - a mini-review. Gerontology. 2012;58(5):453–462. doi: 10.1159/000335966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buffenstein R, Nelson OL, Corbit KC. Questioning the preclinical paradigm: Natural, extreme biology as an alternative discovery platform. Aging (Albany, NY Online) 2014;6(11):913–920. doi: 10.18632/aging.100704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulbert AJ, Pamplona R, Buffenstein R, Buttemer WA. Life and death: Metabolic rate, membrane composition, and life span of animals. Physiol Rev. 2007;87(4):1175–1213. doi: 10.1152/physrev.00047.2006. [DOI] [PubMed] [Google Scholar]
- 10.Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: Insights from a successfully aging species. J Comp Physiol B. 2008;178(4):439–445. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- 11.Liang S, Mele J, Wu Y, Buffenstein R, Hornsby PJ. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber) Aging Cell. 2010;9(4):626–635. doi: 10.1111/j.1474-9726.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez VI, et al. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci USA. 2009;106(9):3059–3064. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salmon AB, Sadighi Akha AA, Buffenstein R, Miller RA. Fibroblasts from naked mole-rats are resistant to multiple forms of cell injury, but sensitive to peroxide, ultraviolet light, and endoplasmic reticulum stress. J Gerontol A Biol Sci Med Sci. 2008;63(3):232–241. doi: 10.1093/gerona/63.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh K, Ishii T, Wakabayashi N, Yamamoto M. Regulatory mechanisms of cellular response to oxidative stress. Free Radic Res. 1999;31(4):319–324. doi: 10.1080/10715769900300881. [DOI] [PubMed] [Google Scholar]
- 15.McMahon M, et al. The Cap’n’Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61(8):3299–3307. [PubMed] [Google Scholar]
- 16.Reddy NM, et al. Genetic dissection of the Nrf2-dependent redox signaling-regulated transcriptional programs of cell proliferation and cytoprotection. Physiol Genomics. 2007;32(1):74–81. doi: 10.1152/physiolgenomics.00126.2007. [DOI] [PubMed] [Google Scholar]
- 17.Jaiswal AK. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic Biol Med. 2000;29(3-4):254–262. doi: 10.1016/s0891-5849(00)00306-3. [DOI] [PubMed] [Google Scholar]
- 18.Lewis KN, Mele J, Hayes JD, Buffenstein R. Nrf2, a guardian of health span and gatekeeper of species longevity. Integr Comp Biol. 2010;50(5):829–843. doi: 10.1093/icb/icq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278(24):21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 20.Chen W, et al. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34(6):663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 22.Ma J, et al. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol Cell Biol. 2012;32(8):1506–1517. doi: 10.1128/MCB.06271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camp ND, et al. Wilms tumor gene on X chromosome (WTX) inhibits degradation of NRF2 protein through competitive binding to KEAP1 protein. J Biol Chem. 2012;287(9):6539–6550. doi: 10.1074/jbc.M111.316471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, et al. A genomic screen for activators of the antioxidant response element. Proc Natl Acad Sci USA. 2007;104(12):5205–5210. doi: 10.1073/pnas.0700898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baba K, Morimoto H, Imaoka S. Seven in absentia homolog 2 (Siah2) protein is a regulator of NF-E2-related factor 2 (Nrf2) J Biol Chem. 2013;288(25):18393–18405. doi: 10.1074/jbc.M112.438762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang HJ, Hong YB, Kim HJ, Bae I. CR6-interacting factor 1 (CRIF1) regulates NF-E2-related factor 2 (NRF2) protein stability by proteasome-mediated degradation. J Biol Chem. 2010;285(28):21258–21268. doi: 10.1074/jbc.M109.084590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rada P, et al. SCF/beta-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31(6):1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277(45):42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 29.Zipper LM, Mulcahy RT. Inhibition of ERK and p38 MAP kinases inhibits binding of Nrf2 and induction of GCS genes. Biochem Biophys Res Commun. 2000;278(2):484–492. doi: 10.1006/bbrc.2000.3830. [DOI] [PubMed] [Google Scholar]
- 30.Edrey YH, et al. Amyloid beta and the longest-lived rodent: The naked mole-rat as a model for natural protection from Alzheimer’s disease. Neurobiol Aging. 2013;34(10):2352–2360. doi: 10.1016/j.neurobiolaging.2013.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimes KM, Lindsey ML, Gelfond JA, Buffenstein R. Getting to the heart of the matter: Age-related changes in diastolic heart function in the longest-lived rodent, the naked mole rat. J Gerontol A Biol Sci Med Sci. 2012;67(4):384–394. doi: 10.1093/gerona/glr222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andziak B, et al. High oxidative damage levels in the longest-living rodent, the naked mole-rat. Aging Cell. 2006;5(6):463–471. doi: 10.1111/j.1474-9726.2006.00237.x. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez KA, Edrey YH, Osmulski P, Gaczynska M, Buffenstein R. Altered composition of liver proteasome assemblies contributes to enhanced proteasome activity in the exceptionally long-lived naked mole-rat. PLoS ONE. 2012;7(5):e35890. doi: 10.1371/journal.pone.0035890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez KA, et al. A cytosolic protein factor from the naked mole-rat activates proteasomes of other species and protects these from inhibition. Biochim Biophys Acta. 2014;1842(11):2060–2072. doi: 10.1016/j.bbadis.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown-Borg HM, Rakoczy SG. Glutathione metabolism in long-living Ames dwarf mice. Exp Gerontol. 2005;40(1-2):115–120. doi: 10.1016/j.exger.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Ayyadevara S, et al. Lifespan and stress resistance of Caenorhabditis elegans are increased by expression of glutathione transferases capable of metabolizing the lipid peroxidation product 4-hydroxynonenal. Aging Cell. 2005;4(5):257–271. doi: 10.1111/j.1474-9726.2005.00168.x. [DOI] [PubMed] [Google Scholar]
- 37.Amador-Noguez D, Yagi K, Venable S, Darlington G. Gene expression profile of long-lived Ames dwarf mice and Little mice. Aging Cell. 2004;3(6):423–441. doi: 10.1111/j.1474-9728.2004.00125.x. [DOI] [PubMed] [Google Scholar]
- 38.Hyun DH, Emerson SS, Jo DG, Mattson MP, de Cabo R. Calorie restriction up-regulates the plasma membrane redox system in brain cells and suppresses oxidative stress during aging. Proc Natl Acad Sci USA. 2006;103(52):19908–19912. doi: 10.1073/pnas.0608008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan W, et al. Nrf2 activity is lost in the spinal cord and its astrocytes of aged mice. In Vitro Cell Dev Biol Anim. 2009;45(7):388–397. doi: 10.1007/s11626-009-9194-5. [DOI] [PubMed] [Google Scholar]
- 40.Miller RA, et al. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4(3):119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447(7144):545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 42.Wakabayashi N, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35(3):238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 43.Reisman SA, Yeager RL, Yamamoto M, Klaassen CD. Increased Nrf2 activation in livers from Keap1-knockdown mice increases expression of cytoprotective genes that detoxify electrophiles more than those that detoxify reactive oxygen species. Toxicol Sci. 2009;108(1):35–47. doi: 10.1093/toxsci/kfn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14(1):76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137(6):1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, et al. SCFbeta-TRCP-mediated degradation of NEDD4 inhibits tumorigenesis through modulating the PTEN/Akt signaling pathway. Oncotarget. 2014;5(4):1026–1037. doi: 10.18632/oncotarget.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren H, et al. The E3 ubiquitin ligases β-TrCP and FBXW7 cooperatively mediates GSK3-dependent Mcl-1 degradation induced by the Akt inhibitor API-1, resulting in apoptosis. Mol Cancer. 2013;12:146. doi: 10.1186/1476-4598-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo SR, et al. SIRT1 suppresses cellular accumulation of β-TrCP E3 ligase via protein degradation. Biochem Biophys Res Commun. 2013;441(4):831–837. doi: 10.1016/j.bbrc.2013.10.146. [DOI] [PubMed] [Google Scholar]
- 49.Prochaska HJ, Santamaria AB. Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: A screening assay for anticarcinogenic enzyme inducers. Analytical Biochemistry. 1988;169:328–336. doi: 10.1016/0003-2697(88)90292-8. [DOI] [PubMed] [Google Scholar]
- 50.Edrey YH, et al. Sustained high levels of neuregulin-1 in the longest-lived rodents; a key determinant of rodent longevity. Aging Cell. 2012;11(2):213–222. doi: 10.1111/j.1474-9726.2011.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.