Significance
The transatlantic slave trade resulted in the forced movement of over 12 million Africans to the Americas. Although many coastal shipping points are known, they do not necessarily reflect the slaves’ actual ethnic or geographic origins. We obtained genome-wide data from 17th-century remains of three enslaved individuals who died on the Caribbean island of Saint Martin and use them to identify their genetic origins in Africa, with far greater precision than previously thought possible. The study demonstrates that genomic data can be used to trace the genetic ancestry of long-dead individuals, a finding that has important implications for archeology, especially in cases where historical information is missing.
Keywords: ancient DNA, genomics, slave trade
Abstract
Between 1500 and 1850, more than 12 million enslaved Africans were transported to the New World. The vast majority were shipped from West and West-Central Africa, but their precise origins are largely unknown. We used genome-wide ancient DNA analyses to investigate the genetic origins of three enslaved Africans whose remains were recovered on the Caribbean island of Saint Martin. We trace their origins to distinct subcontinental source populations within Africa, including Bantu-speaking groups from northern Cameroon and non-Bantu speakers living in present-day Nigeria and Ghana. To our knowledge, these findings provide the first direct evidence for the ethnic origins of enslaved Africans, at a time for which historical records are scarce, and demonstrate that genomic data provide another type of record that can shed new light on long-standing historical questions.
Historians have long been interested in the origins of the millions of enslaved Africans who were transported to the Americas, but there is little direct evidence to go on (1, 2). Although much is known about the volume and changing demographic trends of the Atlantic slave trade, the African origins of the enslaved remain largely unknown (3). Genome-wide analyses of SNPs provide a powerful tool for estimating individual ancestry, and several studies (e.g., refs. 4 and 5) have shown that they can be used to infer an individual’s geographic origin with great accuracy. In the present study, we used genome-wide data to trace the origins of three enslaved Africans whose remains were recovered in the Zoutsteeg area of Philipsburg on the Caribbean island of Saint Martin (Materials and Methods). Previous reports (6) suggest that the “Zoutsteeg Three,” as they became known locally, were likely born in Africa as opposed to the New World. But they did not reveal where in Africa they originated.
Bayesian analysis of individual calibrated radiocarbon dates suggests that the burials date between A.D. 1660 and 1688 (for more details, see SI Appendix, Section 2). During this period, Saint Martin—like other islands—relied to a large extent on African slave labor, but there are no records about the slaves’ origins in Africa. The Transatlantic Slave Trade Database (slavevoyages.org), a large online repository containing information on over 35,000 slaving voyages, lists only a single vessel that arrived in Saint Martin between A.D. 1650 and 1700, although there were undoubtedly more for which there are no records. Moreover, the lone entry does not give the African port of embarkation or any information on the slave cargo. Given the lack of documentation, we embarked on a genomic study with the goal of identifying the genetic origins of the Zoutsteeg Three in Africa.
Results
Initial shotgun sequencing revealed that the DNA in the samples was very poorly preserved (SI Appendix, Section 8). This result was expected because DNA preservation in the Caribbean is known to be poor (7). The fraction of nonredundant reads mapping to the human reference genome varied between 0.3% and 7.6%, and the sequences showed all features typical of ancient DNA, including short average read lengths (∼67 bp), characteristic fragmentation patterns, and an increased frequency of apparent C-to-T substitutions toward the 5′ ends of molecules (see Table 1 and SI Appendix, Fig. S7). To increase sequencing efficiency and lower cost, we enriched the ancient DNA libraries using two recently developed whole-genome capture methods (8, 9). Following enrichment, we generated between 0.1- and 0.5-fold genome-wide coverage for the three individuals, which proved to be sufficient to infer their likely origins within Africa.
Table 1.
Modeled radiocarbon dates and sequencing results for the Zoutsteeg Three
| Sample | Modeled 14C age* | Sex† | Nuclear coverage | Contamination (%)‡ | Damage (%)§ | Mt coverage | Mt haplogroup | Y haplogroup |
| STM1 | A.D. 1660–1688 | M | 0.3× | 0.63 | 16.8 | 641× | L3b1a | R1b1c-V88 |
| STM2 | A.D. 1660–1688 | M | 0.1× | 0.22 | 23.2 | 543× | L3d1b | — |
| STM3 | A.D. 1660–1688 | F | 0.5× | 0.15 | 14.9 | 651× | L2a1f | — |
Modeled age range of the samples based on Bayesian analysis of individual calibrated radiocarbon dates.
Biological sex inferred from the ratio of reads mapping to X and Y chromosomes (44).
Likelihood-based contamination estimate based on mtDNA reads (10).
Frequency of C-to-T misincorporations at 5' ends of sequencing reads.
Although the presence of characteristic damage patterns and short average read lengths suggest that authentic ancient molecules were sequenced, it is possible that some degree of modern contamination could be present in the data. Therefore, we used a previously published Bayesian method (10) to detect contamination and found very low (i.e., <1%) levels overall (see Table 1 and SI Appendix, Section 9). We then merged our ancient sequence data with genotype data from the Human Genome Diversity Cell Line Panel (HGDP) reference panel (11) and used principal component analysis (PCA) (12) to confirm the individuals’ African ancestry (for more details, see SI Appendix, Section 12). Because of low depth, we randomly sampled a single allele from both ancient and modern individuals, as done in ref. 13. As expected, all three individuals fell within African variation as defined by PC1 and PC2 (SI Appendix, Fig. S17). This clustering was retained upon restricting the analysis to sequences showing signs of ancient DNA damage (14) (SI Appendix, Fig. S18), indicating that the signal was not driven by modern DNA contamination.
D-Statistic Test.
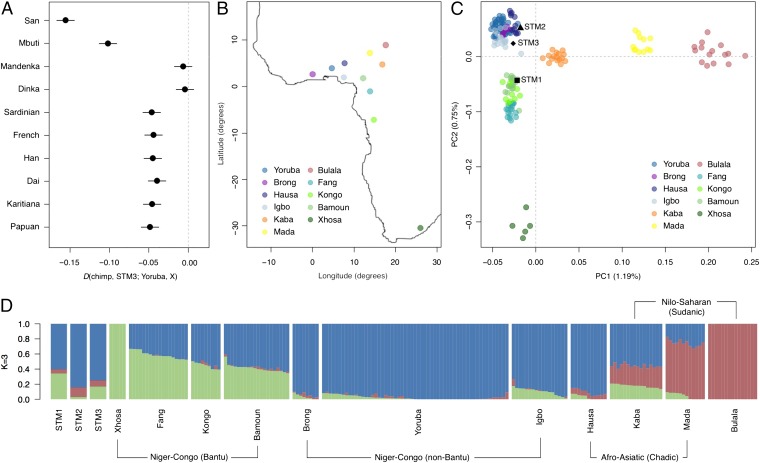
We then tested the relationships between each sample (henceforth referred to as STM1, STM2, and STM3) and 11 populations from across the world for which whole genome data were available (15), using a D-test of the form D (chimpanzee, STM; Yoruba, X), where X stands for a population other than Yoruba (for more details, see SI Appendix, Section 13). We found that the STMs were significantly more closely related to the Yoruba than to any non-African population (Fig. 1A), again confirming their African origins. Within Africa, the STMs appeared significantly more closely related to the Yoruba than to hunter-gatherer populations (San, Mbuti Pygmies). This was not surprising, as the San and the Mbuti were not represented in the transatlantic slave trade. For the Mandenka and Dinka, the D-test results were not significant, suggesting that these populations are equally closely related to the STMs as are the Yoruba. The lack of rejection for the Dinka was surprising, as this population—from southern Sudan—is not known to have been involved in the Atlantic slave trade (16).
Fig. 1.
Genetic affinities of the Zoutsteeg individuals. (A) D-statistic test results for STM3. Error bars correspond to 3 SEs of the D-statistic. Results for STM1 and STM2 are plotted in SI Appendix, Fig. S18. (B) Sampling locations for the 11 African populations in our reference panel (17). (C) Procrustes-transformed PCA plot of the Zoutsteeg individuals with African reference panel samples. (D) Ancestry proportions for the Zoutsteeg individuals and those of 188 African individuals in the reference panel, as inferred by ADMIXTURE analysis (19).
Principal Component Analysis.
To refine our assignment within Africa, we compared our samples to another reference panel, consisting of genotype data from 11 West African populations (Fig. 1B) (17). We intersected 294,651 sites from this reference panel with our sequence data (SI Appendix, Section 14) and conducted PCA to determine whether the individuals showed close affinity to a particular population within the panel. For each of the three individuals, we merged the sequence data with the reference panel genotypes and calculated PC1 and PC2 based on the overlapping sites (SI Appendix, Fig. S19). We then combined the three analyses using Procrustes transformation, as done in ref. 13. Interestingly, the samples clustered with different populations: Bantu-speaking groups in the case of STM1 (specifically, Bamoun) and non-Bantu–speaking groups for STM2 and STM3 (Fig. 1C). We observed similar patterns using the probabilistic model of population splits and divergence implemented in TreeMix (18) (SI Appendix, Fig. S20).
ADMIXTURE Analysis.
To further explore the genetic ancestry of the STMs, we used the maximum-likelihood–based clustering algorithm ADMIXTURE (19). When assuming three ancestral populations (K = 3), the clusters in the reference panel mirror the grouping of individuals in the space defined by PC1 and PC2: a cluster predominating in Bantu-speaking populations, a cluster for non-Bantu West African populations, and a third restricted mostly to Kaba, Mada, and Bulala (Fig. 1D). The distribution of these components in our samples indicates that STM1 has a higher proportion of Bantu-specific ancestry, whereas STM2 and STM3 carry higher proportions of the component prevalent among the non-Bantu–speaking Yoruba, Brong, and Igbo. Notably, STM2 also shows a slightly higher proportion of the component prevalent among the Kaba, Mada, and Bulala, perhaps suggesting closer affinity with Chadic or Sudanic speakers (Fig. 1D).
Uniparental Markers.
Furthermore, we determined the individuals’ mitochondrial DNA (mtDNA) haplogroups and the Y-chromosome haplogroup of STM1 (SI Appendix, Section 11). The mtDNAs were assigned to haplogroups L3b1a, L3d1b2, and L2a1f, respectively. Tracing these lineages to particular regions in Africa is challenging because of their pan-continental distribution, which is the result of thousands of years of population movements (e.g., the Bantu migrations) and continued gene flow (20–22). Nevertheless, we note that haplogroup L3b1a is one of the most common lineages found in the Lake Chad Basin (23). This finding is noteworthy, because the Y-chromosome lineage of this individual (STM1) was identified as belonging to haplogroup R1b1c-V88, which—although quite rare in Africa on the whole—occurs at extremely high frequency in the Lake Chad Basin, rising to 95% in one population of northern Cameroon (24).
Discussion
Taken together, the genetic data suggest that STM1 may have originated among Bantu-speaking groups in northern Cameroon, whereas STM2 and STM3 more likely originated among non-Bantu speakers living in present-day Nigeria and Ghana. To our knowledge, these findings provide the first direct evidence for the ethnic origins of enslaved Africans, with the important caveat that the modern reference populations might not be the same as the historical populations who lived in the same locations at the time of the Atlantic slave trade. Nevertheless, the data suggest that the Africans who reached Saint Martin were drawn from diverse cultures and societies. This finding highlights interesting questions regarding the formation of Creole communities and the survival of African cultures in the Americas. Chief among these is to what extent Africans were able to maintain African cultural beliefs and practices following their arrival in the New World (see, for example, refs. 25–27).
Genome-wide analyses clearly bear great potential to predict a person’s place of origin, but we caution that there are also limitations. First, accuracy is limited by the number of markers used, although new methods (e.g., ref. 28) are constantly being developed, claiming to achieve greater accuracy using relatively small sets of makers. Unfortunately, however, many of these methods rely on called genotypes, which makes them unsuited to low-coverage ancient genome studies. Second, accuracy also depends on the appropriate samples being represented in our reference panels, highlighting the need for more comprehensive sampling and sequencing of human populations across Africa. Third, tracing the ancestry of admixed individuals is more complicated, but advances are also being made in the study of the ancestral composition of admixed genomes (see, for example, refs. 29 and 30). Many of these limitations will be overcome, as more data are being generated and new analytical methods are being developed.
Conclusion
Our study has several major implications. First, it demonstrates that it is possible to obtain genome-wide data from poorly preserved archeological remains found in tropical settings like the Caribbean. This has important implications for future ancient DNA studies in the region, including those addressing pre-Columbian population movements (7). Second, our study underscores the value of whole-genome capture methods (e.g., ref. 8) for ancient DNA research. These new methods enable us to use samples that were previously thought to be beyond our reach because of their low endogenous DNA contents. Third, our study highlights the power of genome-wide analyses for tracing the genetic origins of long-dead individuals, in this case victims of the transatlantic slave trade. As our study shows, genomic data can provide an alternative kind of record that can help shed light on long-standing historical questions, in cases where documentary records are scarce or unavailable.
Materials and Methods
Samples.
The samples used in this study stem from three 17th-century burials that were recovered during construction work in the Zoutsteeg area of Philipsburg, the capital of the Caribbean island of Saint Martin in 2010. Skeletal analysis suggested that the individuals—two males and one female—were of African ancestry and that they had been aged between 25 and 40 y at the time of death. The most striking feature of the skeletons was that all three had culturally modified teeth (SI Appendix, Section 1). Similar types of dental modification are known to have been practiced by different groups in Africa but a look at the ethnographic literature (e.g., refs. 31 and 32) suggests that they cannot be used to infer points of origin or specific “tribal” affiliations.
DNA Extraction and Library Preparation.
DNA was extracted from tooth roots using a silica-based method (33) and eluted in 60 μL EB. Thirty microliters of extract were then built into Illumina libraries using the NEBNext DNA Sample Prep Master Mix Set 2 (New England Biolabs) and Illumina-specific adapters (34) following the manufacturer’s instructions, with some minor changes to the protocol (SI Appendix, Section 3). The remaining 30 μL of DNA extract were built into Illumina libraries using a single-stranded library preparation protocol, as described in ref. 35 but without first removing deoxyuracils. Both sets of libraries were amplified and indexed in 50-μL PCR reactions, purified, quantified, and pooled for sequencing (for more details, see SI Appendix, Section 3).
Whole-Genome Capture.
We used two whole-genome capture methods to enrich two sets of aDNA libraries in their human DNA content. Both methods make use of biotinylated RNA probes transcribed from genomic DNA libraries to capture the human DNA in the aDNA libraries. The first method, which we refer to as WISC (Whole Genome In-Solution Capture), was carried out as described in ref. 8, using home-made biotinylated RNA probes. For the second capture experiment, we used the MYbait Human Whole Genome Capture Kit (MYcroarray), following the manufacturer’s instructions (9). Following the capture experiments, the enriched libraries were amplified again, purified, quantified, and sequenced on an Illumina HiSeq 2000 (for more details, see SI Appendix, Section 4).
Data Processing.
We used AdapterRemoval (36) to trim adapter sequences and to remove adapter dimers and low quality reads (SI Appendix, Section 5). Filtered fastq files were mapped to the human reference genome version hg18 and hg19, but replacing the mitochondria with the revised Cambridge Reference Sequence (37). Mapping was done using BWA v0.7.5a-r405 (38), keeping only reads with mapping quality 30 and above. Duplicate reads were removed using SAMtools’ (39) rmdup function. BAM files from different runs were merged using SAMtools merge. Sequencing error rates were estimated to be on the order of 0.3% (SI Appendix, Section 6). MapDamage2 (40) was used to rescale the quality of bases that had a mismatch to the reference likely derived from damage (for more details, see SI Appendix, Section 7).
mtDNA and Y-Chromosome Haplogroups.
mtDNA haplogroups were determined by recovering reads mapping to the revised Cambridge Reference Sequence (37) from the BAM files and generating a consensus sequence and list of variants using SAMtools/BCFtools v0.1.19 (38). Indels and hotspot mutations were excluded from analysis. Haplogroups were determined using HaploGrep (41). The maximum parsimony tree (SI Appendix, Fig. S12) was built using mt-Phyl (eltsov.org). The Y-chromosome haplogroup for STM1 was determined by assembling a panel of phylogenetically informative SNPs, with emphasis on those lineages previously reported to occur at appreciable frequencies within Africa. For more details, see SI Appendix, Section 11.
Reference Panels.
For the genome-wide analyses we used three previously published reference data sets, including: (i) a dataset of 11 modern genomes used in ref. 15, (ii) genotype data for 854 unrelated individuals from 52 worldwide populations from the HGDP reference panel described in ref. 11, and (iii) a reference panel consisting of 146 individuals from 11 sub-Saharan populations genotyped on the Affymetrix 500k array set (17). Before merging with the aDNA sequence data, we randomly sampled one allele at each site and for each individual in the reference panel and made such site homozygous for the drawn allele, as described in ref. 13. For more details, see SI Appendix, Section 12.
Principal Component Analysis.
For each file containing the genotypes of the sample and reference panels, we ran smartpca (EIGENSOFT v4.0) (42) to perform PCA. Eigenvectors were plotted independently for each dataset using RStudio (www.rstudio.com). To visualize the three samples in a single PCA plot we used Procrustes transformation as done in ref. 13. We transformed the first two PCs calculated for each intersected dataset to match the reference-only PC1 and PC2. When transforming the PCs, the ancient individual was excluded. The configuration of transformed PC1 and PC2 was then applied to the ancient individuals, and transformed coordinates were overlaid on the reference-only PC1 and PC2 plot (Fig. 1C). For more details, see SI Appendix, Section 14.
TreeMix Analysis.
We used the probabilistic model of population splits and divergence implemented in TreeMix (18) to infer ancestry graphs (SI Appendix, Fig. S20). As input we used estimated allele frequencies for our samples and the populations in our reference panel (17). For each dataset we ran 100 bootstraps with random seeds and with the -noss and -global flags to disable sample size correction, and perform a round of global rearrangements of the graph, respectively. Additionally, the number of SNPs per block was calculated for each dataset to allow ∼1,000 blocks. Finally, the root of the tree was set to Xhosa. For more details, see SI Appendix, Section 15.
ADMIXTURE Analysis.
We used the maximum-likelihood–based clustering algorithm ADMIXTURE (19) to estimate the genetic structure in our merged dataset (for more details, see SI Appendix, Section 16). We first estimated the cross-validation error with the -cv flag for K values between 1 and 6. This analysis revealed that the CV error increased with K, probably reflecting the very low Fst between the populations in the reference panel. For K = 4 to K = 6 we ran 100 replicates using a random seed and kept the Q (ancestral cluster proportions) and P (inferred ancestral cluster allele frequencies) matrices from the run with the best log likelihood. We used the P matrix from each K to estimate the most likely cluster proportions in the ancient samples as was done in ref. 43. SI Appendix, Fig. S21 shows the converged runs from K = 2 to K = 6 for STM1, STM2, and STM3 and 11 sub-Saharan populations in our reference panel (17).
Supplementary Material
Acknowledgments
We thank the staff at the Danish National High-throughput Sequencing Centre for technical support; P. F. Campos, M. Rasmussen, T. Korneliussen, F. Sánchez-Quinto, P. Skoglund, A. Moreno-Estrada, F. Mendez, and P. Underhill for their various input and helpful discussions; and J.S. Handler for reading earlier drafts of the paper and providing valuable comments. The Centre for Geogenetics is funded by the Danish National Research Foundation (DNRF94). This work was supported in part by Marie Curie Fellowships from the Directorate General for Research and Innovation of the European Commission FP7/2007-2013/236435 and 317184 (to H.S. and M.E.A.); Grants FP7/2007-2013/269442 and 319209 from the European Research Council (to H.S. and M.E.A.); National Science Foundation Grants DMS-1201234 and DGE-1147470 (to M.C.A.A. and G.D.P.); Swiss National Science Foundation Fellowship PBSKP3-143529 (to A.S.M.); National Institute of Health Grants NRSA 5F32HG007342 and K99 GM104158 (to M.L.C. and P.L.F.J.); Leverhulme Early Career Fellowship ECF-2012-123 (to M.W.D.); Ministerio de Ciencia e Innovación Grant SAF2011-26983 (to A.S.); the “Plan Galego IDT” EM 2012/045 (to A.S.); “Sistema Universitario Gallego-Modalidad REDES” Grant 2012-PG226 from the Xunta de Galicia (to A.S.); Lundbeck Foundation Grant R52-A5062 (to M.T.P.G.); and Danish Council for Independent Research Grant 10-081390 (to M.T.P.G.).
Footnotes
Conflict of interest statement: C.D.B. is the founder of IdentifyGenomics, LLC, and is on the Scientific Advisory Boards of Personalis, Inc., and Ancestry.com as well as the Medical Advisory Board of InVitae. M.L.C. is now the Chief Scientific Officer at IdentifyGenomics, LLC. None of this played a role in the design, execution, or interpretation of experiments and results presented here.
This article is a PNAS Direct Submission. R.A.K. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the European Nucleotide Archive, www.ebi.ac.uk/ena (project accession no. PRJEB8269). The mapped data are available upon request.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421784112/-/DCSupplemental.
References
- 1.Palmer CA. From Africa to the Americas: Ethnicity in the early black communities of the Americas. J World Hist. 1995;6(2):223–236. [Google Scholar]
- 2.Morgan PD. The cultural implications of the Atlantic slave trade: African regional origins, American destinations and new world developments. Slavery Abol. 1997;18(1):122–145. [Google Scholar]
- 3.Northrup D. Igbo and Myth Igbo: Culture and ethnicity in the Atlantic world, 1600–1850. Slavery Abol. 2000;21(3):1–20. [Google Scholar]
- 4.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456(7218):98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lao O, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18(16):1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder H, et al. The Zoutsteeg Three: Three new cases of African types of dental modification from Saint Martin, Dutch Caribbean. Int J Osteoarchaeol. 2014;24(6):688–696. [Google Scholar]
- 7.Lalueza-Fox C, Gilbert MT, Martínez-Fuentes AJ, Calafell F, Bertranpetit J. Mitochondrial DNA from pre-Columbian Ciboneys from Cuba and the prehistoric colonization of the Caribbean. Am J Phys Anthropol. 2003;121(2):97–108. doi: 10.1002/ajpa.10236. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter ML, et al. Pulling out the 1%: Whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am J Hum Genet. 2013;93(5):852–864. doi: 10.1016/j.ajhg.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. MYcroarray (2013) MYbaits Manual. Available at www.mycroarray.com/pdf/MYbaits-manual.pdf. Accessed March 1, 2014.
- 10.Fu Q, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol. 2013;23(7):553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70(Pt 6):841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- 12.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skoglund P, et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336(6080):466–469. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- 14.Skoglund P, et al. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc Natl Acad Sci USA. 2014;111(6):2229–2234. doi: 10.1073/pnas.1318934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer M, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338(6104):222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diouf S. Servants of Allah: African Muslims Enslaved in the Americas. New York Univ Press; New York, NY: 2013. [Google Scholar]
- 17.Bryc K, et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci USA. 2010;107(2):786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8(11):e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salas A, et al. The African diaspora: mitochondrial DNA and the Atlantic slave trade. Am J Hum Genet. 2004;74(3):454–465. doi: 10.1086/382194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salas A, Carracedo A, Richards M, Macaulay V. Charting the ancestry of African Americans. Am J Hum Genet. 2005;77(4):676–680. doi: 10.1086/491675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ely B, Wilson JL, Jackson F, Jackson BA. African-American mitochondrial DNAs often match mtDNAs found in multiple African ethnic groups. BMC Biol. 2006;4:34. doi: 10.1186/1741-7007-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerezo M, Černý V, Carracedo Á, Salas A. New insights into the Lake Chad Basin population structure revealed by high-throughput genotyping of mitochondrial DNA coding SNPs. PLoS ONE. 2011;6(4):e18682. doi: 10.1371/journal.pone.0018682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cruciani F, et al. Human Y chromosome haplogroup R-V88: A paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages. Eur J Hum Genet. 2010;18(7):800–807. doi: 10.1038/ejhg.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mintz SW, Price R. The Birth of African-American Culture: An Anthropological Perspective. Beacon Press; Boston, MA: 1992. [Google Scholar]
- 26.Midlo Hall G. Slavery and African Ethnicities in the Americas: Restoring the Links. Univ of North Carolina Press; Chapel Hill, NC: 2007. [Google Scholar]
- 27.Lovejoy PE, editor. Identity in the Shadow of Slavery. Continuum; London, UK: 2009. [Google Scholar]
- 28.Elhaik E, et al. Genographic Consortium Geographic population structure analysis of worldwide human populations infers their biogeographical origins. Nat Commun. 2014;5:3513. doi: 10.1038/ncomms4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang WY, Novembre J, Eskin E, Halperin E. A model-based approach for analysis of spatial structure in genetic data. Nat Genet. 2012;44(6):725–731. doi: 10.1038/ng.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno-Estrada A, et al. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9(11):e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Jehring H. Die künstliche Deformierung der Zähne. Z Ethnol. 1882;14:213–262. [Google Scholar]
- 32.Lignitz H. Die künstlichen Zahnverstümmelungen in Afrika im Lichte der Kulturkreisforschung. Anthropos. 1919-1922;14-15:891–943. [Google Scholar]
- 33.Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat Protoc. 2007;2(7):1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- 34.Meyer M, Kircher M. Illumina Sequencing Library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010(6):pdb.prot5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 35.Gansauge MT, Meyer M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat Protoc. 2013;8(4):737–748. doi: 10.1038/nprot.2013.038. [DOI] [PubMed] [Google Scholar]
- 36.Lindgreen S. AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC Res Notes. 2012;5:337. doi: 10.1186/1756-0500-5-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrews RM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jónsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29(13):1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kloss-Brandstätter A, et al. HaploGrep: A fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat. 2011;32(1):25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- 42.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 43.Sikora M, et al. Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe. PLoS Genet. 2014;10(5):e1004353. doi: 10.1371/journal.pgen.1004353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skoglund P, et al. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J Arch Sci. 2013;40:4477–4482. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.