Significance
More than a thousand proteins are thought to contribute to mammalian chromatin and its regulation, but our understanding of the genomic occupancy and function of most of these proteins is limited. We have used a chromatin proteomic profiling approach to produce a catalogue of proteins associated with genomic regions whose chromatin is marked by specific modified histones. A substantial number of the newly identified proteins are associated with human disease. Future chromatin proteomic profiling studies should prove valuable for identifying additional chromatin-associated proteins in a broad spectrum of cell types.
Keywords: chromatin, genomics, proteomics
Abstract
More than a thousand proteins are thought to contribute to mammalian chromatin and its regulation, but our understanding of the genomic occupancy and function of most of these proteins is limited. Here we describe an approach, which we call “chromatin proteomic profiling,” to identify proteins associated with genomic regions marked by specifically modified histones. We used ChIP-MS to identify proteins associated with genomic regions marked by histones modified at specific lysine residues, including H3K27ac, H3K4me3, H3K79me2, H3K36me3, H3K9me3, and H4K20me3, in ES cells. We identified 332 known and 114 novel proteins associated with these histone-marked genomic segments. Many of the novel candidates have been implicated in various diseases, and their chromatin association may provide clues to disease mechanisms. More than 100 histone modifications have been described, so similar chromatin proteomic profiling studies should prove to be valuable for identifying many additional chromatin-associated proteins in a broad spectrum of cell types.
There are more than 1,000 transcription factors, cofactors, and chromatin regulators encoded in the mammalian genome, but we have limited understanding of the genomic occupancy and function of most of these (1–4). Understanding how these proteins interact with specific active and repressed portions of the genome would provide clues to their functions in global gene control, but limitations inherent in widely used genomic and proteomic technologies make acquiring this information laborious and expensive. Chromatin immunoprecipitation coupled to sequencing (ChIP-seq) can reveal the sites that a specific protein occupies in the genome (5–7) but is laborious and is limited by the availability of antibodies specific to candidate genome-binding proteins. Mass spectrometry (MS) can identify large populations of proteins present in specific preparations (8–10) but does not reveal how these proteins occupy specific portions of the genome. A recently developed approach combined ChIP with MS (ChIP-MS) to identify protein complexes that are associated with other proteins known to occupy sites in the genome (11–13). Here we adapt this approach to profile the proteins associated with chromatin containing specific histone modifications across the genome of mouse embryonic stem cells (mESCs). These chromatin modifications mark regions of the genome where specific transcriptional activities occur, thus implicating the associated proteins in these activities.
Results
Chromatin Proteomic Profiling Identifies Proteins Associated with Histone-Marked Regions of Chromatin.
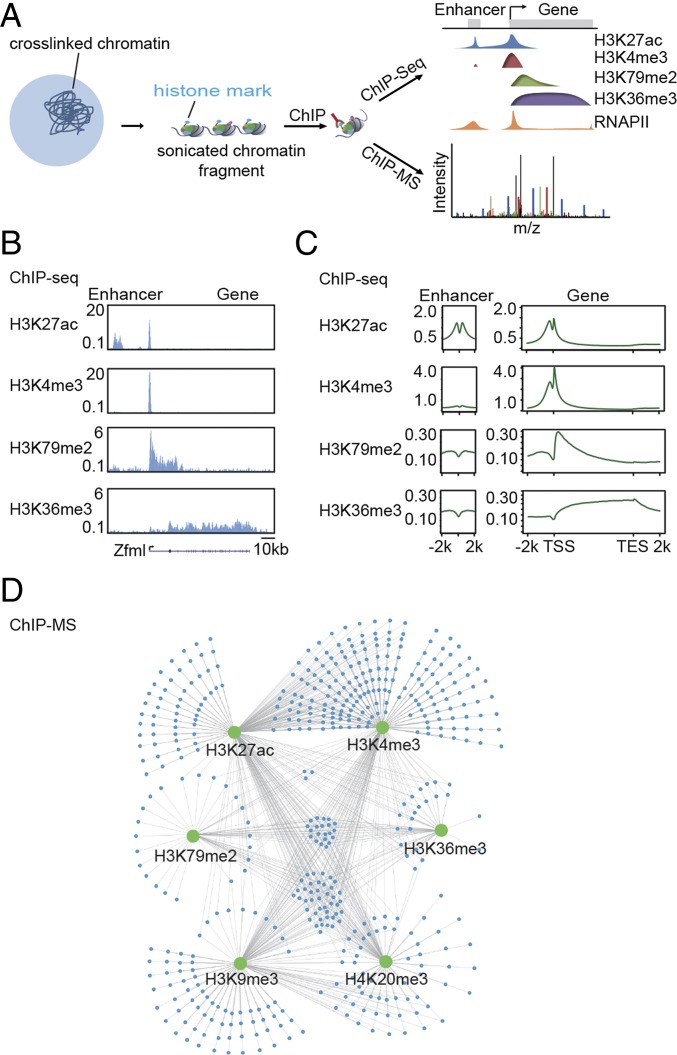
We used ChIP-MS to identify proteins associated with specific euchromatic histone modifications, including H3K27ac, H3K4me3, H3K79me2, and H3K36me3, and heterochromatic histone modifications, including H3K9me3 and H4K20me3, in mESCs (Fig. 1). Active enhancers are occupied by nucleosomes with H3K27ac; active promoters are occupied by nucleosomes with H3K27ac and H3K4me3; and portions of transcribed gene bodies are occupied by nucleosomes with H3K79me2 and H3K36me3 (14, 15). In contrast, relatively silent genes are occupied by nucleosomes with H3K9me3 and H4K20me3 (16–18). We hypothesized that ChIP-MS, when used with antibodies specific to these chromatin modifications, would create a catalog of candidate proteins associated with specifically marked chromatin in vivo and that ChIP-seq analysis of the same material could confirm that the correct portions of the genome had been enriched by the procedure (Fig. 1A).
Fig. 1.
Chromatin proteomic profiling identifies proteins associated with histone-marked regions of chromatin. (A) Schematic overview of the chromatin proteomic profiling approach. (B) Gene track illustration of H3K27ac, H3K4me3, H3K79me2, and H3K36me3 ChIP-seq signals (in reads per million) at the Zfml gene locus. (C) Metagene analysis of H3K27ac, H3K4me3, H3K79me2, and H3K36me3 ChIP-seq showing the average signal (in reads per million) for these modifications at enhancers and genes. (D) Distribution of proteins that were enriched in the H3K27ac, H3K4me3, H3K79me2, H3K36me3, H3K9me3, and H4K20me3 ChIP-MS preparations relative to the IgG control and input samples. Green nodes represent different histone mark preparations. Blue nodes represent identified proteins.
We first carried out ChIP of crosslinked euchromatic fractions using antibodies that recognize histone H3K27ac, H3K4me3, H3K79me2, and H3K36me3 and subjected part of this material to DNA sequence analysis to confirm that previously established patterns of genome occupancy were obtained. The results, when analyzed for individual genes (Fig. 1B) or the population of genes (Fig. 1C), were consistent with the well-established locations of nucleosomes with these modifications (14, 15). We then subjected the rest of this material to MS to identify proteins that were enriched in these ChIP preparations relative to proteins found in the initial chromatin preparation (input) and in a control preparation (IgG). Four lines of evidence suggest that selective enrichment of specific chromatin-associated proteins was achieved: (i) ChIP-Western blot analysis indicated that modified histones were immunoprecipitated with specific antibodies (Fig. S1A); (ii) ChIP-MS preparations of modified histones contained proteins that were present at low abundance in the initial chromatin preparation (input), indicating that the identified proteins were not simply reflective of cellular abundance (Fig. S1B); (iii) modified histone ChIP-MS preparations contained proteins whose functions were enriched in relevant Gene Ontology categories, including transcription regulation and chromatin organization, which differed considerably from the categories enriched among the proteins in the IgG ChIP-MS preparation (Fig. S1C); and (iv) many of the proteins identified in these preparations have been shown previously to associate with chromatin containing nucleosomes with these modified histones, as described below.
We then carried out ChIP of crosslinked heterochromatic fractions using antibodies that recognize histone H3K9me3 and H4K20me3 and subjected this material to MS to identify proteins that were enriched in these ChIP preparations, which were expected to be substantially different from the euchromatic preparations. Indeed, the majority of the proteins identified in the heterochromatic histone ChIP-MS preparations were not found in the euchromatic histone H3K27ac, H3K4me3, H3K79me2, and H3K36me3 ChIP-MS preparations (Fig. S1D). The extent of overlap of proteins identified in all the preparations is shown in Fig. 1D and is summarized in Table S1, which largely reflects the overlap of histone modifications across the genome (Fig. 1A).
The proteins identified by chromatin proteomic profiling were grouped into three categories, “known,” “implicated,” and “novel,” based on prior evidence for their association with regions of the genome marked by these histone modifications (Fig. 2A). The known category contained proteins reported to associate with chromatin in the UniProt, GeneCards, or PubMed databases. The implicated group contained proteins that have not been reported to associate with chromatin but contain DNA-binding domains, chromatin-binding domains, or belong to a protein complex known to associate with chromatin. The novel category contained proteins not previously reported or implicated in chromatin association. We found 332 proteins known to be associated with chromatin, 46 proteins that have been implicated, and 114 novel candidates (Fig. 2A).
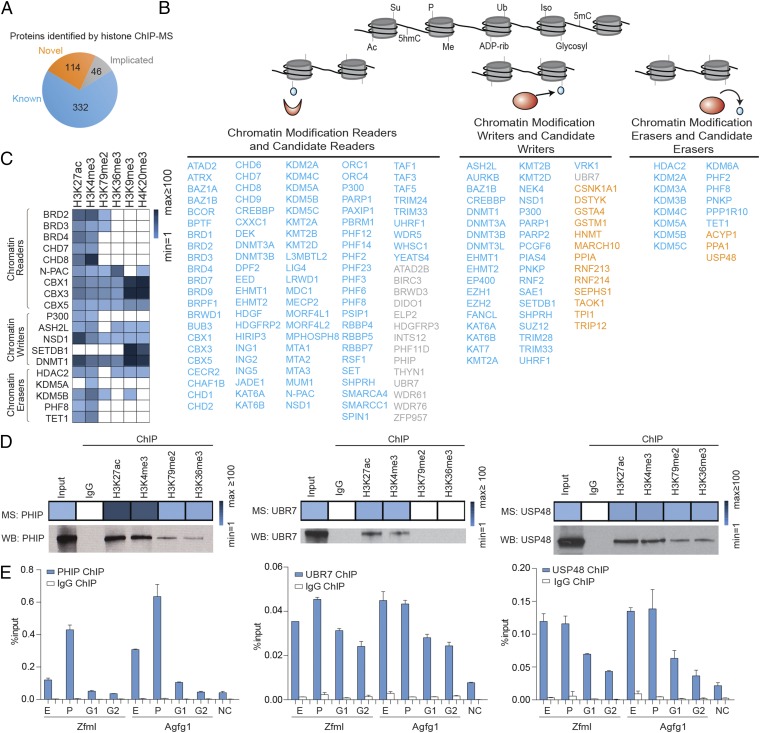
Fig. 2.
Chromatin regulators enriched in the histone mark ChIP-MS preparations in mESCs. (A) Number of known, implicated, and novel chromatin-associated proteins that were enriched in the histone mark preparations. (B) Known (blue), implicated (gray), and novel (orange) chromatin-associated proteins. (C) Relative enrichment of selected chromatin regulators in various ChIP preparations. (D) Relative enrichments of PHIP, UBR7, and USP48 in various ChIP preparations. Histone mark ChIP followed by Western blot analysis of PHIP, UBR7, and USP48; 0.25% of input was loaded for ChIP Western blot. (E) ChIP-qPCR analysis of PHIP, UBR7, and USP48 with primers targeting enhancers (E), promoters (P), and gene body regions (G1 and G2) of Zfml and Agfg1. NC, negative control region for qPCR. Error bars indicate SD based on two independent experiments.
Chromatin Regulators Enriched in ChIP-MS.
We focused our initial analysis on proteins that are known or are likely to be histone modification readers, writers, and erasers (Fig. 2B and Table S1). A large number of proteins that have well-established roles in these activities were enriched in the appropriate preparations (Fig. 2C and Table S1). For example, the bromodomain reader BRD4, the histone acetyltransferase writer CBP/P300, and the histone deacetylase eraser HDAC2 were especially enriched in the H3K27ac preparation, consistent with their functions at enhancers and promoters (19). The methyltransferase writer ASH2L and the demethylase erasers PHF8, KDM5A, and KDM5B were especially enriched in the H3K4me3 preparation, consistent with their functions in promoter regions (20–22). The PWWP domain reader N-PAC, an LSD2/KDM1B cofactor that stimulates H3K4 demethylation, was especially enriched in the H3K36me3 preparation (23). The chromobox readers CBX1, CBX3, and CBX5 and the histone methyltransferase writer SETDB1 were especially enriched in the H3K9me3 and H4K20me3 preparations, consistent with their functions in heterochromatic regions (24). There were many additional proteins identified in these preparations that have established enzymatic activities (25) but that have not been reported to be associated with chromatin (Fig. 2B and Table S1); these candidate histone readers, writers, and erasers thus may contribute to chromatin regulation.
To confirm that a subset of the candidate proteins was in fact associated with specific histone-marked genomic regions suggested by the ChIP-MS data, we further studied PHIP, UBR7, and USP48, proteins for which specific and ChIP-capable antibodies were available. We anticipate that PHIP, UBR7, and USP48 may function as a chromatin reader, writer, and eraser, respectively; this possibility warrants future investigation. ChIP followed by Western blot analysis confirmed that PHIP, UBR7, and USP48 were associated with the corresponding histone mark ChIP-MS preparations (Fig. 2D). ChIP-quantitative PCR (qPCR) then was performed for PHIP, UBR7, and USP48 with probes targeting enhancers, promoters, and gene bodies. The ChIP-qPCR signals for PHIP, UBR7, and USP48 were relatively enriched in enhancers and promoters, consistent with the enrichment of PHIP, UBR7, and USP48 in the H3K27ac and H3K4me3 preparations (Fig. 2E, Fig. S2, and Table S2). Small differences in the relative enrichments at different regions are seen with ChIP-qPCR and ChIP-MS, likely reflecting the difference between a gene-specific view using ChIP-qPCR and the global average view provided by ChIP-MS, as well as technical differences in the two approaches.
Transcription Factors and Cofactors Enriched in ChIP-MS.
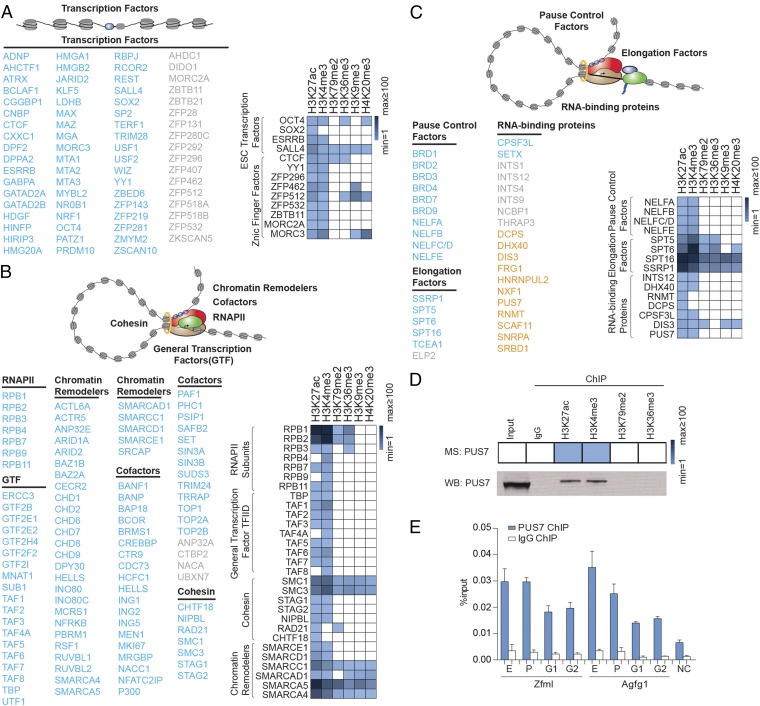
We extended our analysis of proteins in the ChIP-MS preparations to transcription factors (Fig. 3A), proteins associated with transcription initiation (Fig. 3B), and proteins associated with the elongation apparatus (Fig. 3C). The well-studied ES cell pluripotency transcription factors OCT4, SOX2, ESRRB, and SALL4, as well as many additional transcription factors, were found to be enriched mostly in the H3K27ac and H3K4me3 preparations (Fig. 3A), as is consistent with their established presence at enhancers and promoter-proximal regions (19, 26, 27). Many transcription factors that previously have not been described as playing roles in ES cell gene expression also were found in these regions, e.g., ZFP131, which previously was reported to regulate Oct4 gene expression in an shRNA screen in mESCs (28). ZFP296 and ZFP532 also were found in our preparations. These transcription factors are specifically expressed in ES cells, suggesting potential roles in ES cell-specific transcriptional regulation (Fig. 3A) (29). Many known components of the transcription initiation apparatus were found in the ChIP-MS preparations (Fig. 3B), including RNA polymerase II, transcriptional cofactors (such as the ATP-dependent chromatin remodeling complexes with SMARC subunits), and the general transcription factor TFIID (containing TBP and TAF subunits) (1, 30, 31). TFIID was found enriched only in the promoter-associated H3K27ac and H3K4me3 preparations, as is consistent with its known promoter-binding properties (Fig. 3B). Some proteins associated with the transcription-initiation apparatus (RNA polymerase II, cohesin) are thought to traverse actively transcribed genes and were found in all the euchromatin preparations (Fig. 3B). Proteins associated with the elongation apparatus included pause-control factors, elongation factors, and several RNA-binding proteins (Fig. 3C) (32, 33). Recent studies indicate that RNA-binding proteins can be associated with specific portions of chromatin, in part because of their interaction with RNA species associated with the transcriptional machinery (34, 35). For example, the integrator complex, an snRNA-processing machinery, can be detected at the gene promoters, where it contributes to the regulation of transcriptional initiation and pause release (36). The RNA-binding proteins observed in these chromatin preparations included the RNA helicase DHX40, the RNA-capping enzyme RNMT, the RNA-decapping enzyme DCPS, the RNA cleavage factor CPSF3L, and many others.
Fig. 3.
Transcriptional regulators enriched in the active histone mark ChIP-MS preparations in mESCs. (A–C) Transcription factors (A), proteins associated with the transcription-initiation apparatus (B), and proteins associated with the elongation apparatus (C) were enriched in the histone mark preparations. (D, Upper) Relative enrichment of PUS7 in various ChIP preparations. (Lower) Histone mark ChIP followed by Western blot analysis of PUS7. Of the input, 0.25% was loaded for ChIP-Western blotting. (E) ChIP-qPCR analysis of PUS7 with primers targeting enhancers (E), promoters (P), and gene body regions (G1 and G2) of Zfml and Agfg1. NC, negative control region for PCR. Error bars indicate SD based on two independent experiments.
To confirm that an RNA-binding protein was associated with specific histone-marked genomic regions, we further studied a pseudouridine synthase, PUS7, for which a specific and ChIP-capable antibody was available. PUS7 recently was found to be required for mRNA pseudouridylation in yeast and human cells (37, 38); however, its chromatin association is unknown. We used ChIP followed by Western blot analysis to confirm that PUS7 was associated with the corresponding histone mark ChIP-MS preparations (Fig. 3D). We then performed ChIP-qPCR for PUS7 with probes targeting enhancers, promoters, and gene bodies. The ChIP-qPCR signals for PUS7 were relatively enriched in enhancers and promoters, consistent with the enrichment of PUS7 in the H3K27ac and H3K4me3 preparations (Fig. 3E). This evidence suggests that PUS7 may play a role at the cotranscriptional level.
Discussion
The chromatin proteomic profiling approach described here combines ChIP-MS and ChIP-seq both to identify proteins associated with specific types of chromatin and to map that type of chromatin genome-wide. The mESC chromatin proteomic profiling data produced a catalog of chromatin-associated proteins, which includes a large number of known and candidate chromatin regulators and transcriptional regulators. Our results overlap and confirm chromatin-associated proteins identified in other MS studies (8, 12) but also provide a larger catalog of chromatin proteins associated with a larger variety of chromatin types. We confirmed that several of the candidates (PHIP, UBR7, USP48, and PUS7) were indeed associated with appropriate histone-marked segments of the genome as detected by chromatin proteomic profiling. These results indicate that chromatin proteomic profiling is a powerful means to identify proteins associated with chromatin containing specific histone modifications.
Although many well-studied proteins were found in the expected ChIP-MS preparations, we noted two limitations of the ChIP-MS chromatin proteomic profiling data. Proteins that are known to be present in these regions of chromatin were not detected, showing that the method did not produce a complete catalog. For example, five of the smaller subunits of the 12-subunit RNA polymerase II enzyme were not detected (39). The transcription factors NANOG and KLF4 co-occupy many promoters together with OCT4, SOX2, ESRRB, and SALL4 (19), but these were not detected. The failure to detect these and other proteins may result from their relatively small size, low abundance, loss during preparation, or poor signal detection by MS. Another potential limitation of the ChIP-MS chromatin proteomic profiling data is that some proteins are found both in the expected preparations and in others. For example, the H3K4me3 writer ASH2L was found most abundantly in the H3K4me3 preparation but also was found in the H3K27ac and H3K36me3 preparations. The presence of ASH2L in the H3K27ac preparation was expected, because this modification occurs in the same region as H3K4me3 (Fig. 1A), but it was not expected to be present in the H3K36me3 preparation. In the future, ChIP-MS of single nucleosomes prepared through nuclease digestion or ChIP-MS using sequential chromatin immunoprecipitations of multiple histone marks might help increase the specificity of chromatin proteomic profiling.
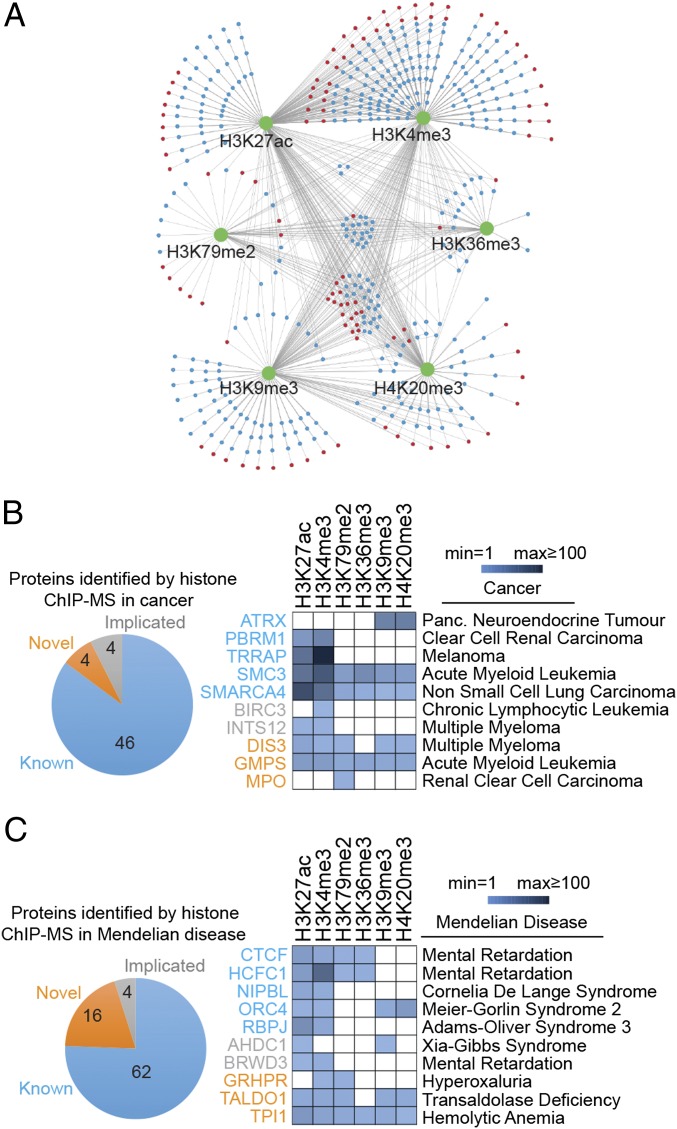
We note that many proteins identified by this chromatin proteomic profiling study have been implicated in various diseases (40–43) but have poorly understood functions (Fig. 4 and Table S3). Nearly 20% of the proteins in these chromatin preparations (110 proteins) have been implicated in cancer and/or Mendelian diseases. Future studies of these proteins, nearly one-fourth of which fell into the novel or implicated categories, may lead to additional insights into disease mechanisms.
Fig. 4.
Proteins identified by chromatin proteomic profiling that are associated with human diseases. (A) Distribution of proteins (blue or red nodes) that were enriched in each histone mark ChIP-MS preparation (green nodes) relative to the IgG control and input samples. Red nodes represent proteins associated with diseases. (B and C) The number and relative enrichment of chromatin-associated proteins identified in this study that are mutated in cancers (B) and Mendelian diseases (C).
The chromatin proteomic profiling study described here produced a catalog of known, implicated, and novel proteins associated with genomic regions whose chromatin is marked by specific modified histones. More than 100 additional histone modifications have been reported (44), so further chromatin proteomic profiling studies should prove valuable for identifying additional chromatin-associated proteins in ES cells and other cell types. Chromatin proteomic profiling also should prove useful in investigating changes in protein composition in dynamic settings such as drug exposure, metabolism, cell growth/cell cycle, stress response, and development.
Materials and Methods
Cell Culture.
V6.5 (C57BL/6–129) mESCs were grown on irradiated murine embryonic fibroblasts (MEFs) under standard ES cell culture conditions as described previously (45). mESCs were collected for specific experiments after being grown for two passages without MEFs.
Antibodies.
The following antibodies were used for Western blot analysis, ChIP-MS, and ChIP-seq/qPCR: H3K27ac (ab4729), H3K4me3 (ab8580), H3K79me2 (ab3594), H3K36me3 (ab9050), H3K9me3 (ab8898), and H4K20me3 (ab9053) (all from Abcam). The USP48 (A301-190A) and PHIP (A302-055A) antibodies were purchased from Bethyl Laboratories. The UBR7 (NBP1-88409) antibody was purchased from NOVUS. The PUS7 (HPA024116) antibody was purchased from Sigma.
ChIP-MS and ChIP-Seq.
We used 6–8 × 108 mESCs for each ChIP-MS and 6–8 × 107 mESCs for each ChIP-seq. See SI Materials and Methods for further details regarding the protocols and data processing.
Accession Codes.
The ChIP-seq data for H3K27ac, H3K4me3, H3K79me2, and H3K36me3 are available at the Gene Expression Omnibus under the accession number GSE62380.
Supplementary Material
Acknowledgments
We thank Eric Spooner at the Whitehead Proteomics Core for mass spectrometry; Tom Volkert and Sumeet Gupta at the Whitehead Genome Technology Core for sequencing; Zi Peng Fan and Violaine Saint-André for protein domain analysis; Jessica Reddy and Lyndon Zhang for helpful bioinformatics discussions; and Jill Dowen, Denes Hnisz, and Jurian Schuijers for comments on the manuscript. This work was supported by National Institutes of Health Grants HG002668 (to R.A.Y.), HG006046 (to R.A.Y.), and HD045022 (to R.J.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE62380).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502971112/-/DCSupplemental.
References
- 1.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: Transcription enters a new era. Cell. 2014;157(1):13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: Function, expression and evolution. Nat Rev Genet. 2009;10(4):252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 3.Dawson MA, Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Maston GA, Landt SG, Snyder M, Green MR. Characterization of enhancer function from genome-wide analyses. Annu Rev Genomics Hum Genet. 2012;13(13):29–57. doi: 10.1146/annurev-genom-090711-163723. [DOI] [PubMed] [Google Scholar]
- 5.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 6.Dunham I, et al. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155(1):39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vermeulen M, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142(6):967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444(7117):364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 10.Bensimon A, Heck AJR, Aebersold R. Mass spectrometry-based proteomics and network biology. Annu Rev Biochem. 2012;81(81):379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- 11.Wang CI, et al. Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat Struct Mol Biol. 2013;20(2):202–209. doi: 10.1038/nsmb.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soldi M, Bonaldi T. The proteomic investigation of chromatin functional domains reveals novel synergisms among distinct heterochromatin components. Mol Cell Proteomics. 2013;12(3):764–780. doi: 10.1074/mcp.M112.024307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soldi M, Bonaldi T. The ChroP approach combines ChIP and mass spectrometry to dissect locus-specific proteomic landscapes of chromatin. J Vis Exp. 2014;86 doi: 10.3791/51220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 18.Campos EI, Reinberg D. Histones: Annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 19.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen J, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128(6):1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466(7305):508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seward DJ, et al. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat Struct Mol Biol. 2007;14(3):240–242. doi: 10.1038/nsmb1200. [DOI] [PubMed] [Google Scholar]
- 23.Fang R, et al. LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Mol Cell. 2013;49(3):558–570. doi: 10.1016/j.molcel.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Safran M, et al. GeneCards version 3: The human gene integrator. Database. 2010 doi: 10.1093/database/baq020. baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng H-H, Surani MA. The transcriptional and signalling networks of pluripotency. Nat Cell Biol. 2011;13(5):490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- 27.Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C, Macleod I, Su AI. BioGPS and MyGene.info: Organizing online, gene-centric information. Nucleic Acids Res. 2013;41(Database issue) D1:D561–D565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21(9):327–335. [PubMed] [Google Scholar]
- 31.Lessard JA, Crabtree GR. Chromatin regulatory mechanisms in pluripotency. Ann Rev Cell ad Devl Biol. 2010;26:503–532. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81(81):119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47(47):483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144(1):16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji X, et al. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153(4):855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardini A, et al. Integrator Regulates Transcriptional Initiation and Pause Release following Activation. Mol Cell. 2014;56:1–12. doi: 10.1016/j.molcel.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz S, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlile TM, et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cramer P. Multisubunit RNA polymerases. Curr Opin Struct Biol. 2002;12(1):89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- 40.Forbes SA, et al. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152(6):1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Online Mendelian Inheritance in Man OM-NIoGM (2014) (Johns Hopkins University, Baltimore). Available at omim.org/. Accessed September 1, 2014.
- 44.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.