Significance
c-Myc is ubiquitinated by SCFFbw7 and appears to be degraded mainly in the nucleolus, where it acts as a master regulator of ribosomal biogenesis. However, deubiquitination regulation of c-Myc in the nucleolus is previously unknown. Here, we found that ubiquitin-specific protease 36 (USP36), a nucleolar ubiquitin-specific protease, interacts with and deubiquitinates c-Myc in the nucleolus. It also interacts with Fbw7γ but not Fbw7α, yet it abolishes c-Myc degradation mediated by either Fbw7γ or Fbw7α. Ablation of USP36 reduced c-Myc levels and severely inhibited cancer cell proliferation. USP36 is overexpressed in a subset of human breast and lung cancers. Thus, our study reveals USP36 as a novel deubiquitinating enzyme controlling c-Myc’s nucleolar degradation, adding a missing and end-point regulator into the c-Myc degradation pathway.
Keywords: c-Myc, USP36, ubiquitination, deubiquitination, nucleolus
Abstract
c-Myc protein stability and activity are tightly regulated by the ubiquitin-proteasome system. Aberrant stabilization of c-Myc contributes to many human cancers. c-Myc is ubiquitinated by SCFFbw7 (a SKP1-cullin-1-F-box complex that contains the F-box and WD repeat domain-containing 7, Fbw7, as the F-box protein) and several other ubiquitin ligases, whereas it is deubiquitinated and stabilized by ubiquitin-specific protease (USP) 28. The bulk of c-Myc degradation appears to occur in the nucleolus. However, whether c-Myc is regulated by deubiquitination in the nucleolus is not known. Here, we report that the nucleolar deubiquitinating enzyme USP36 is a novel c-Myc deubiquitinase. USP36 interacts with and deubiquitinates c-Myc in cells and in vitro, leading to the stabilization of c-Myc. This USP36 regulation of c-Myc occurs in the nucleolus. Interestingly, USP36 interacts with the nucleolar Fbw7γ but not the nucleoplasmic Fbw7α. However, it abolished c-Myc degradation mediated both by Fbw7γ and by Fbw7α. Consistently, knockdown of USP36 reduces the levels of c-Myc and suppresses cell proliferation. We further show that USP36 itself is a c-Myc target gene, suggesting that USP36 and c-Myc form a positive feedback regulatory loop. High expression levels of USP36 are found in a subset of human breast and lung cancers. Altogether, these results identified USP36 as a crucial and bono fide deubiquitinating enzyme controlling c-Myc’s nucleolar degradation pathway.
The c-Myc oncoprotein is a pleiotropic transcription factor that regulates the expression of a large number of genes involved in the control of cell growth, proliferation, apoptosis, differentiation, angiogenesis, metabolism, and ribosomal biogenesis (1–3). Proper levels of c-Myc are essential for regulated cell growth and proliferation. However, aberrant overexpression of c-Myc is associated with most human cancers and overexpression of a c-myc transgene induces tumorigenesis in various tissues in mice (2, 4). Thus, the level and activity of c-Myc must be precisely controlled during normal cell homeostasis. One of the key mechanisms for this control is the regulation of c-Myc protein stability (2, 5, 6).
c-Myc is a short-lived protein in nontransformed cells that is degraded by the ubiquitin (Ub)- proteasome system (5, 6). It is transiently stabilized upon stimulation of cell growth. The growth-regulated turnover of c-Myc involves two conserved phosphorylation sites within Myc Box I, Threonine 58 (T58), and Serine 62 (S62) (6–8). Phosphorylation of c-Myc at S62 triggered by ERK and/or CDK kinases in response to growth signals increases c-Myc stability. With the cessation of growth signals, T58 is phosphorylated by GSK3β, whose activity is held in check by PI(3)K/Akt signaling in the presence of growth factor and receptor tyrosine kinase signaling (6–8). T58 phosphorylation then destabilizes c-Myc by facilitating S62 dephosphorylation and recruiting the T58 phosphorylation-dependent Ub ligase (E3) complex SCFFbw7 (a SKP1-cullin-1-F-box complex that contains the F-box and WD repeat domain-containing 7, Fbw7, as the F-box protein) to mediate c-Myc ubiquitination and degradation through the proteasome system (9–12). Supporting the importance of this phosphorylation-dependent c-Myc degradation pathway, a subset of Burkitt’s lymphomas harbors mutations at or around T58 that prevents its phosphorylation (13–16) and mutations and deletions of FBW7 are found in multiple human cancers (17). There are also other Ub ligases that regulate c-Myc protein stability and activity in a T58 phosphorylation-independent manner, including HectH9, PirH2, TRUSS, CHIP, SCFskp2, SCFβTRCP, and SCFFBXO28 E3 complexes (5, 18–21). Interestingly, SCFskp2-, SCFFBXO28-, and HectH9-mediated ubiquitination increases c-Myc activity (18–21) and SCFβTRCP antagonizes SCFFbw7 to ubiquitinate the N terminus of c-Myc and stabilizes c-Myc (20). Thus, ubiquitination plays a key and complex role in regulating both c-Myc protein turnover and its activity.
Like other posttranslational modifications, ubiquitination of c-Myc can be reversed by deubiquitinating enzymes (DUBs). The Ub-specific protease (USP) family member USP28 is so far the only known DUB for c-Myc (22). USP28 deubiquitinates and stabilizes c-Myc, whereas its knockdown reduces the levels and activity of c-Myc. There are three Fbw7 isoforms located in distinct subcellular compartments: Fbw7β and Fbw7α are localized in the cytoplasm and the nucleoplasm, respectively, whereas Fbw7γ is in the nucleolus (12, 17). Interestingly, USP28 binds to c-Myc through Fbw7α in the nucleoplasm, but not Fbw7γ in the nucleolus, and reverses Fbw7α- but not Fbw7γ- mediated c-Myc ubiquitination and degradation (22), suggesting that USP28 regulates c-Myc stability only in the nucleoplasm. Studies have indicated that c-Myc is mainly degraded in the nucleolus and proteasome inhibition leads to accumulation of c-Myc in the nucleolus (12, 23, 24). Also, c-Myc–mediated cell growth and tumorigenesis is well integrated with its role in enhancing ribosome biogenesis in the nucleolus (3, 25). Thus, the nucleolus plays a key role in regulating c-Myc levels and in mediating its oncogenic activity. However, whether c-Myc is deubiquitinated in the nucleolus is previously unknown.
Here, we report that USP36 is a novel nucleolar c-Myc deubiquitinase. USP36 binds to and deubiquitinates c-Myc in cells and in vitro. Overexpression of wild-type (WT) USP36, but not its catalytic-inactive mutant (C131A), stabilizes c-Myc, whereas knockdown of USP36 reduces c-Myc levels and markedly suppresses cell proliferation. Interestingly, USP36 interacts with Fbw7γ but not Fbw7α. However, it abolished c-Myc degradation mediated both by Fbw7γ and by Fbw7α. Furthermore, we found that USP36 itself is a c-Myc target gene, suggesting that USP36 and c-Myc form a positive feedback regulatory loop. Altogether, these results identified USP36 as a crucial DUB controlling c-Myc’s nucleolar degradation pathway and impacting its oncogenic activity.
Results
USP36 Stabilizes c-Myc Protein and Stimulates Its Activity.
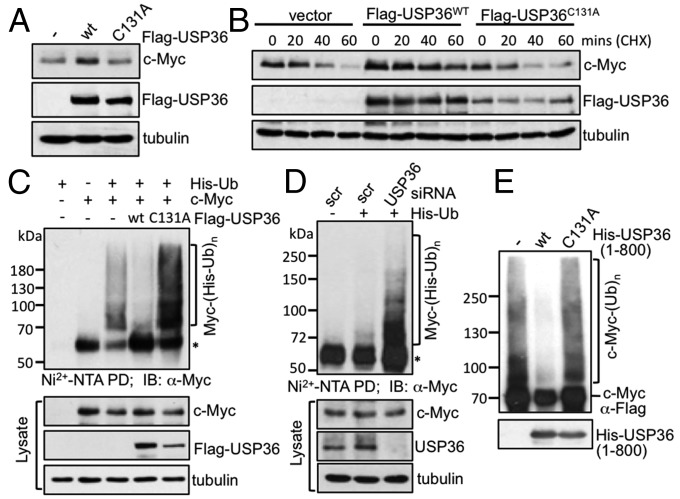
To examine whether nucleolar DUBs regulate c-Myc, we focused on the USP family member USP36. We found that overexpression of WT USP36 induced the levels of endogenous and ectopically expressed c-Myc in HeLa cells (Fig. 1A and Fig. S1A). This induction requires the DUB activity of USP36, as the catalytic-inactive C131A mutant of USP36 failed to induce c-Myc levels (Fig. 1A and Fig. S1A). The induction was dose-dependent (Fig. S1B) and is due to the stabilization of c-Myc, as overexpression of WT USP36, but not the C131A mutant, markedly prolonged the half-life of endogenous and ectopic c-Myc protein (Fig. 1B and Fig. S1C) without affecting the levels of c-myc mRNA (Fig. S1G). Overexpression of WT USP36, but not the C131A mutant, also increased the levels of endogenous c-Myc in HEK293 (Fig. S1D) and the immortalized mammary epithelial MCF10A cells (Fig. S1E), suggesting that c-Myc induction by USP36 is not a cell-type–specific effect. Of note, the inability of the C131A mutant (USP36C131A) to stabilize c-Myc is not due to its lower expression levels compared with WT USP36 (Fig. 1 A and B and Fig. S1 A–E), as at similar expression levels by transfecting cells with WT USP36 or USP36C131A plasmid at 1:3 ratio, WT USP36, but not USP36C131A, still markedly stabilized c-Myc (Fig. S1F). Together, these results demonstrate that USP36 stabilizes c-Myc and increases its levels. Consistently, overexpression of WT USP36, but not the C131A mutant, significantly increased the levels of c-Myc target genes, including prerRNA, E2F2 mRNA, and 5S rRNA catalyzed by RNA polymerase (Pol) I, II, and III, respectively (Fig. S1G), indicating that USP36 stimulates c-Myc transactivation activity in cells.
Fig. 1.
USP36 deubiquitinates and stabilizes c-Myc. (A) Overexpression of WT USP36, but not the C131A mutant, induces c-Myc levels. HeLa cells transfected with control or Flag-USP36 (WT or the C131A mutant) were assayed by IB. (B) USP36 stabilizes endogenous c-Myc. HeLa cells transfected with the indicated plasmids for 48 h were treated with 50 μg/mL cycloheximide (CHX). The cells were harvested at different time points and assayed by IB. (C) USP36 deubiquitinates c-Myc in cells. HEK293 cells transfected with the indicated plasmids were treated with MG132 for 6 h before harvesting. The cells were subjected to pulldown (PD) using the Ni2+-NTA bead under denaturing conditions, followed by IB. Asterisks indicate nonspecific PD of native c-Myc. (D) Knockdown of USP36 increases c-Myc ubiquitination. HeLa cells were transfected with scrambled or USP36 siRNA together with or without His-Ub for 48 h and treated with MG132 for 6 h. The cells were subjected to PD using the Ni2+-NTA bead, followed by IB. (E) USP36 deubiquitinates c-Myc in vitro. Ubiquitinated c-Myc was purified from 293 cells transfected with His-Ub and Flag–c-Myc using anti-Flag affinity purification. The ubiquitinated c-Myc was incubated with recombinant WT His-USP361-800 or its C131A mutant protein, followed by IB with anti-Flag antibody.
USP36 Deubiquitinates c-Myc.
USP36 is a cysteine protease (26); thus, we then examined whether USP36 regulates c-Myc stability through deubiquitination. Using in vivo ubiquitination assays (27), we found that WT USP36, but not the C131A mutant, significantly reduced the ubiquitinated species of c-Myc (Fig. 1C). Of note, the C131A mutant increased the levels of the ubiquitinated species of c-Myc, indicating a dominant-negative effect of this mutant. Conversely, knockdown of endogenous USP36 significantly increased the ubiquitinated species of c-Myc (Fig. 1D). To determine whether USP36 directly deubiquitinates c-Myc in vitro, we purified ubiquitinated c-Myc from 293 cells transfected with Flag–c-Myc and His-Ub using affinity purification with anti-Flag (M2) agarose beads and Flag-peptide elution. The ubiquitinated c-Myc was incubated with purified recombinant USP domain-containing His-USP361–800 (WT or C131A) or control buffer followed by immunoblot (IB) with anti-Flag antibody. As shown in Fig. 1E, WT USP36, but not the C131A mutant, markedly reduced the ubiquitinated species of c-Myc. These results demonstrate that USP36 is a bono fide c-Myc deubiquitinase and the N-terminal fragment containing 800 amino acids of USP36 is sufficient to deubiquitinate c-Myc.
USP36 Interacts with c-Myc in Cells and in Vitro.
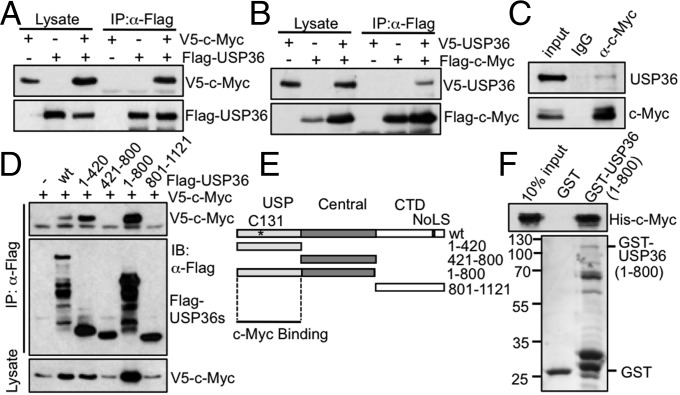
Next, we asked whether we can detect USP36 interaction with c-Myc. As shown in Fig. 2A, ectopic V5–c-Myc was coimmunoprecipitated with Flag-USP36 using anti-Flag antibody only when both plasmids were transfected. Also, V5-USP36 was coimmunoprecipitated with Flag–c-Myc using anti-Flag antibody when both plasmids were transfected (Fig. 2B). Further, endogenous USP36 was specifically immunoprecipitated by anti–c-Myc antibodies but not control IgG (Fig. 2C). These results suggest that USP36 interacts with c-Myc in cells. To determine which domain of USP36 interacts with c-Myc, we constructed a panel of Flag-tagged USP36 deletion mutants, including the N-terminal USP domain (1–420), central domain (421–800), and the C-terminal nucleolar localization signal (NoLS)-containing domain (801–1121) (Fig. 2E). We cointroduced WT USP36 or its deletion mutants with V5–c-Myc into 293 cells and performed coimmunoprecipitation (co-IP) assays. As shown in Fig. 2D, c-Myc specifically coimmunoprecipitated with the mutants containing the N-terminal USP domain, but not the mutants lacking this domain. The N-terminal USP domain containing mutants USP361–420 and USP361–800 also induced the levels of c-Myc (Fig. 2D, Bottom). This is consistent with the His-USP361–800 fragment deubiquitinating c-Myc in vitro (Fig. 1E). Co-IP assays using cells treated with MG132 to normalize the c-Myc protein levels showed the similar results (Fig. S2A). Together, these results indicate that the N-terminal USP domain of USP36 is necessary for binding to c-Myc. In addition, His–c-Myc purified from bacteria was specifically bound by purified GST-USP361–800 protein, but not GST alone, indicating that USP36 directly interacts with c-Myc in vitro (Fig. 2F). Further, USP36C131A also binds to c-Myc in cells (Fig. S2B), indicating that USP36 stabilization of c-Myc requires its catalytic activity in addition to binding to c-Myc.
Fig. 2.
USP36 interacts with c-Myc in cells and in vitro. (A and B) Co-IP of ectopic USP36 with ectopic c-Myc. H1299 cells transfected with V5–c-Myc and Flag-USP36 (A) or V5-USP36 and Flag–c-Myc (B) individually or together were assayed by co-IP. (C) Co-IP between endogenous USP36 and c-Myc. Lysates from 293 cells were assayed by co-IP with anti–c-Myc (Y69) or control IgG. (D and E) The N-terminal USP domain of USP36 is required for binding to c-Myc. HEK293 cells transfected with V5–c-Myc together with control or Flag-USP36 (WT or deletion mutants) were assayed by co-IP with anti-Flag antibody (D). The diagram of the Flag-USP36 and its deletion mutants are shown in E. (F) USP36 interacts with c-Myc in vitro. Purified GST or GST-USP36 immobilized on glutathione beads was incubated with purified His–c-Myc. Bound proteins were assayed by IB with anti-Myc (Top) and anti-GST (Bottom).
USP36 Interacts with Fbw7γ, but Not Fbw7α, and Inhibits c-Myc Degradation Mediated by Either Fbw7γ or Fbw7α.
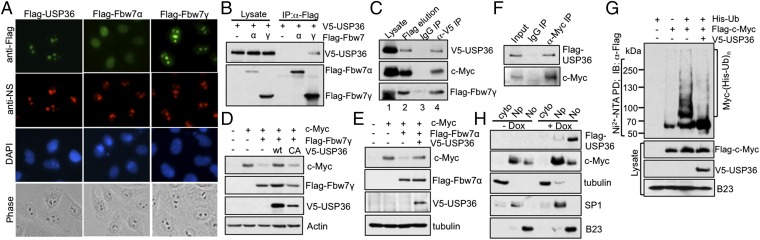
c-Myc can be ubiquitinated and degraded by expression of all three isoforms of Fbw7 in cotransfection assays (11), but it appears to be degraded mainly in the nucleolus (12). Fbw7γ is localized in the nucleolus, whereas Fbw7α and Fbw7β are localized in the nucleoplasm and cytoplasm, respectively (12) (Fig. 3A). USP36 is mainly localized in the nucleolus (28, 29) (Fig. 3A and Fig. S3 E and F). Therefore, we tested whether USP36 interacts with specific Fbw7 isoforms in a complex with c-Myc. As shown in Fig. 3B, USP36 was specifically coimmunoprecipitated with Fbw7γ, but not Fbw7α, in cells. Conversely, Flag-Fbw7γ, but not Flag-Fbw7α, was specifically coimmunoprecipitated with V5-USP36 by anti-V5, but not control IgG, in cells cotransfected with both V5-USP36 and Flag-Fbw7γ (Fig. S3A) or Fbw7α (Fig. S3B). Knocking down of Fbw7γ neither resulted in the relocalization of Fbw7α from the nucleoplasm to the nucleolus (Fig. S3C) nor its binding to USP36 (Fig. S3D), suggesting that Fbw7α does not compensate for Fbw7γ in the regulation of c-Myc by USP36. To determine whether USP36 and Fbw7γ form a complex with c-Myc, we performed sequential co-IP assays. Cell lysates from H1299 cells cotransfected with V5-USP36, Flag-Fbw7γ, and c-Myc were first immunoprecipitated with anti-Flag antibody followed by elution with Flag-peptide (Fig. 3C, lane 2). The elution containing Flag-Fbw7γ–associated proteins was then immunoprecipitated with control IgG (Fig. 3C, lane 3) or anti-V5 antibody (Fig. 3C, lane 4). As shown in Fig. 3C, c-Myc was coimmunoprecipitated in the first anti-Flag IP as well as in the secondary anti-V5 IP, suggesting that USP36 forms a tertiary complex with c-Myc and Fbw7γ in cells. Interestingly, overexpression of USP36 abolished the reduction of c-Myc mediated by either Fbw7γ (Fig. 3D) or Fbw7α (Fig. 3E). These results suggest that in addition to USP36 deubiquitinating c-Myc ubiquitinated by Fbw7γ in the nucleolus, it may also deubiquitinate c-Myc following Fbw7α-mediated ubiquitination in the nucleoplasm. Alternatively, Fbw7α may act sequentially with Fbw7γ to regulate c-Myc as in the case of cyclin E, and thus, nucleolar USP36 can still impact Fbw7α-mediated c-Myc ubiquitination (30).
Fig. 3.
USP36 interacts with Fbw7γ, but not Fbw7α; inhibits c-Myc degradation mediated by either Fbw7γ or Fbw7α; and deubiquitinates c-Myc in the nucleolus. (A) Immunofluorescence (IF) staining. HeLa cells transfected with Flag-USP36, Flag-Fbw7γ, or Flag-Fbw7α were immunostained with anti-Flag (green) and anti-nucleostemin (NS) (red). (B) USP36 interacts with Fbw7γ, but not Fbw7α. HeLa cells transfected with the indicated plasmids were assayed by co-IP. (C) USP36 forms a complex with c-Myc and Fbw7γ. HeLa cells transfected with Flag-Fbw7γ, V5-USP36, and c-Myc were subjected to IP with anti-Flag, followed by elution with the Flag peptide. The elution was then subjected to co-IP with control IgG or anti-V5. (D and E) USP36 inhibits c-Myc degradation mediated by Fbw7γ or Fbw7α. HeLa cells transfected with the indicated plasmids were assayed by IB. (F) USP36 interacts with c-Myc in the nucleolus. The nucleolar fraction isolated from HeLa cells expressing Flag-USP36 as in Fig. S3E was assayed by co-IP. (G) USP36 deubiquitinates c-Myc in the nucleolus. The nucleoli isolated from HeLa cells transfected with the indicated plasmids were assayed by Ni2+-NTA PD. (H) Overexpression of USP36 increases the levels of c-Myc in both the nucleoplasm and the nucleolus. HeLa–TO (tetracycline operator)–Flag-USP36 cells were induced without or with doxycycline (dox) for 12 h and assayed by cell fractionation and IB.
USP36 Interacts with and Deubiquitinates c-Myc in the Nucleolus.
The nucleolar localization of USP36 and its interaction with Fbw7γ indicates that it interacts with c-Myc in the nucleolus. To test this hypothesis, we performed cell fractionation followed by co-IP using nucleolar lysates. As shown in Fig. S3E, ectopically expressed USP36 is predominantly detected in the nucleolar fraction, whereas c-Myc is mainly localized in the nucleoplasm, with substantial presence in the nucleolar fraction as well. Co-IP using the isolated nucleolar lysates showed that USP36 interacted with c-Myc in the nucleolus (Fig. 3F). Further in vivo ubiquitination assays showed that USP36 was able to deubiquitinate c-Myc in the nucleolus (Fig. 3G) but not in the nucleoplasm (Fig. S3 G and H). However, c-Myc levels are increased in both the nucleoplasm and nucleolar fractions upon overexpression of USP36 (Fig. 3H). Thus, USP36 can deubiquitinate and stabilize c-Myc in the nucleolus, and the deubiquitinated c-Myc could shuttle from the nucleolus back to the nucleoplasm.
Knockdown of USP36 Reduces the Levels of c-Myc and Suppresses Cell Proliferation.
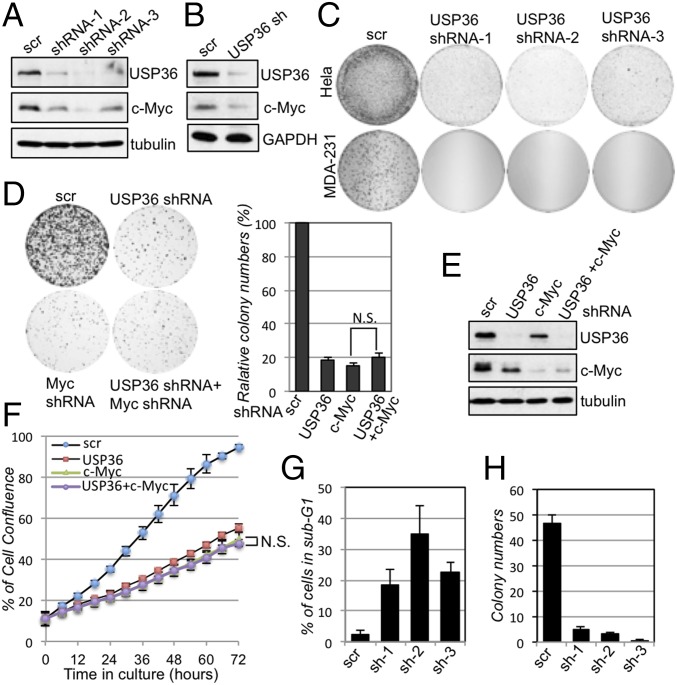
To determine the role of endogenous USP36 in regulating c-Myc, we examined whether knockdown of USP36 affects c-Myc levels and activity. As shown in Fig. 4A, lentiviral-mediated knockdown of USP36 by three individual shRNAs significantly reduced the levels of endogenous c-Myc protein in HeLa cells compared with the scrambled RNA control. Knockdown of USP36 also reduced the levels of endogenous c-Myc in MCF10A cells (Fig. 4B) and breast cancer MDA-MB-231 cells (Fig. S4A). In addition, knockdown of USP36 reduced the levels of doxycycline (dox)-induced ectopic V5–c-MycWT (Fig. S4B), but not the Fbw7-resistant mutant V5–c-MycT58A (Fig. S4C) in MCF10A cells, compared with scrambled control. Similarly, knockdown of USP36 also reduced the levels of stably transfected c-MycWT, but not c-MycT58A, in HeLa cells (Fig. S4D). The reduction of c-Myc by USP36 knockdown can be rescued by concomitant knockdown of Fbw7 (Fig. S4E), suggesting the opposite coregulation of c-Myc by Fbw7 and USP36 in the nucleolus. Consistently, knockdown of USP36 by three individual shRNAs drastically suppressed the proliferation of HeLa and breast cancer MDA-MB-231 cells (Fig. 4C) as well as SKBR3 cells (Fig. S4F) as determined by colony formation assays and cell confluence over time in culture using an IncuCyte live cell imaging system (Fig. S4G). These effects are not off-target effects, as overexpression of shRNA-3–resistant USP36 in HeLa cells rescued the c-Myc reduction by knockdown of endogenous USP36 (Fig. S4H) and abolished the inhibitory effect of USP36 knockdown on cell proliferation (Fig. S4I). Knockdown of USP36 also significantly induced apoptosis in cells as evident by the increases in sub-G1 cells infected with USP36 shRNA (Fig. 4G). Of note, overexpression of c-MycT58A did not rescue the inhibitory effect of USP36 knockdown on cell proliferation, suggesting that other targets in addition to c-Myc are involved in cell growth inhibition upon USP36 knockdown. One possibility is that cells with c-MycT58A overexpression depend on elevated levels of ribosomal biogenesis for cell growth and proliferation, and this may require USP36 regulation of additional targets such as B23 and RNA Pol I (29, 30). Thus, knockdown of USP36 in these cells is synthetic lethal. Nevertheless, upon c-Myc knockdown, additional growth inhibitory effects of USP36 knockdown were not observed (Fig. 4 D–F). Together, these results suggest that endogenous USP36 regulates c-Myc levels and this correlates with its role in promoting cell proliferation and cell survival.
Fig. 4.
Knockdown of USP36 reduces c-Myc levels and suppresses cell proliferation. (A and B) Knockdown of USP36 reduces c-Myc levels. HeLa (A) and MCF10A (B) cells were infected with scrambled or USP36 shRNA-encoding lentiviruses. The cell lysates were assayed for the expression of c-Myc and USP36 by IB. (C) Knockdown of USP36 suppresses cell proliferation. HeLa or MDA-MB-231 cells were infected with control or USP36 shRNA-encoding lentiviruses. The cells were cultured for up to 3 wk. The colonies were visualized by staining with crystal violet. (D–F) Knockdown of c-Myc does not further increase the inhibitory effects of knockdown of USP36 on cell proliferation. HeLa cells infected with control, USP36 shRNA, or c-Myc shRNA-encoding lentiviruses alone or together were subjected to colony formation assay (D) or measurement of cell confluence over time using IncuCyte System (F). A representative IB detection of the expression of USP36 and c-Myc is shown in E. (G) Knockdown of USP36 induces apoptosis. HeLa cells infected with control or USP36 shRNA-encoding lentiviruses were stained with propidium iodide (PI) followed by flow cytometry analysis. The average percentages of cells in sub-G1 are shown. (H) Soft agar colony formation assays. MDA-MB-231 breast cancer cell lines were infected with control or USP36 shRNA-encoding lentiviruses, followed by colony formation assay in soft agar.
Knockdown of USP36 Suppresses Anchorage-Independent Growth of Breast Cancer Cell Lines.
To determine whether USP36 plays a role in cell transformation, we performed anchorage-independent growth of breast cancer MDA-MB-231 cells. As shown in Fig. 4H and Fig. S4J, knockdown of USP36 by shRNA significantly reduced the colony formation in soft agar for MDA-MB-231 cells. These results suggest that USP36 may play a key role in cell proliferation and tumor cell growth.
USP36 Is a c-Myc Target Gene.
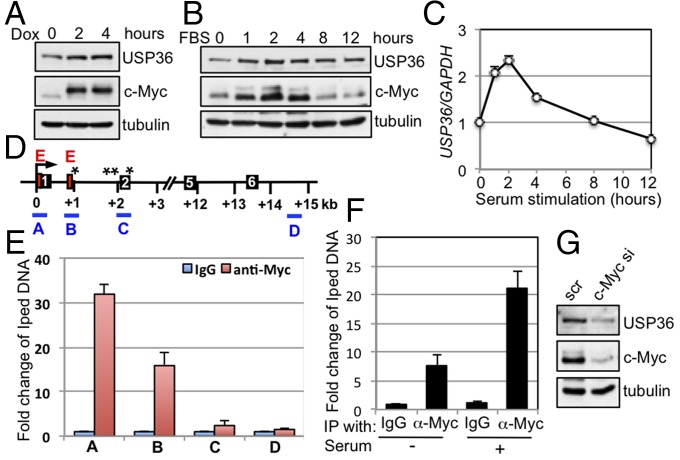
During our observation using tet-inducible expression of c-Myc (e.g., Fig. S4B), we observed that USP36 is induced by c-Myc expression. To confirm this observation, we also induced the expression of c-Myc in 293-TO-HA-Myc (Fig. 5A) and U2OS-TO-Flag-Myc (Fig. S5A) cells with dox. The results clearly showed that the levels of USP36 protein were increased by induced expression of c-Myc in both cells in a time-dependent manner (Fig. 5A and Fig. S5A). This correlates with the increased expression of USP36 mRNA (Fig. S5 B and C). We also observed that USP36 protein was rapidly increased along with c-Myc following serum stimulation, peaking at 2 h after stimulation (Fig. 5B), and USP36 mRNA was also rapidly increased and peaked at 2 h (Fig. 5C). These results indicate that USP36 may be a c-Myc target gene. By analyzing the promoter and exon–intron sequences of the human USP36 gene, we found two canonical c-Myc–binding E box motifs and a number of noncanonical E box sites in a region spanning exon 1, intron 1, and exon 2 (Fig. 5D). Chromatin IP (ChIP)–qPCR assays (Fig. 5E) showed that c-Myc specifically bound to the exon 1 (A in Fig. 5E) and intron 1 (B in Fig. 5E) E box-containing regions and to a lesser extent to exon 2 (noncanonical E boxes, C in Fig. 5E), but not the intron 6 region (D in Fig. 5E). Further, the binding of c-Myc to the USP36 gene was increased following serum stimulation (Fig. 5F). Consistently, knockdown of c-Myc reduced the levels of USP36 (Fig. 5G). Together, these results demonstrate that USP36 is a c-Myc target gene. Interestingly, knockdown of USP36 significantly abolished the c-Myc induction following serum stimulation (Fig. S5D), demonstrating that USP36 plays a critical role in c-Myc stabilization in response to growth signals. Together, our results suggest that USP36 may form a positive feedback regulatory loop with c-Myc.
Fig. 5.
USP36 is a target gene of c-Myc. (A) c-Myc induces the expression of USP36. 293-TO-Myc cells were cultured in the presence of dox for different times as indicated, followed by IB. (B and C) Dynamic expression of USP36 and c-Myc in cells following serum stimulation. HeLa cells cultured in 0.2% FBS containing medium for 48 h were stimulated with 20% FBS and harvested at the indicated time points. The cells were assayed for USP36 expression by IB (B) or RT-qPCR assays (C). (D) Diagram of the USP36 gene exon–intron regions. Bars indicate the four regions amplified by ChIP–qPCR assays. Red boxes and asterisks indicate canonical and noncanonical E boxes, respectively. Filled boxes indicate exons. The numbers of exons are also indicated. Arrows indicate the transcription start. (E) c-Myc specifically binds to the E box-containing exon 1 and intron 1, but not intron 6, regions of the USP36 gene. HeLa cells were subjected to ChIP using control IgG or anti–c-Myc antibodies followed by qPCR amplification of the indicated genomic regions of the USP36 gene. (F) Serum stimulation increases the c-Myc binding to the USP36 gene. HeLa cells were serum starved for 48 h, followed by stimulation with 20% FBS for 2 h, as in B. The serum-starved and restimulated cells were subjected to ChIP–qPCR to detect the USP36 promoter DNA (exon 1). (G) Knockdown of endogenous c-Myc reduces USP36 levels. HeLa cells transfected with c-Myc or scrambled siRNA were assayed by IB.
USP36 Is Deregulated in Breast Cancers.
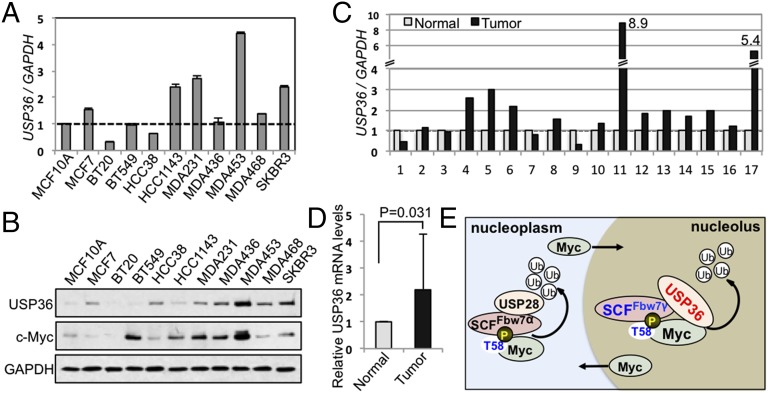
The above results suggest that USP36 might possess an oncogenic role. To determine whether the expression of USP36 is altered in human cancers, we searched publicly available databases such as the cBiol Cancer Genomics Portal (cBioPortal), where the TCGA data suggest that USP36 is frequently amplified or overexpressed in various human cancers, including breast invasive carcinomas (Fig. S6A). We therefore analyzed the USP36 expression in a series of human breast cancer cell lines. We found that USP36 mRNA is overexpressed in several cell lines, including SKBR3, MDA-MB-453, MDA-MB-231, and HCC1143 cells (Fig. 6A). Elevated protein expression is also detected in these cell lines as well as others, such as MDA-MB-468, MDA-MB-436, HCC38, and MCF7 cells (Fig. 6B). In most of these USP36-overexpressing breast cancer cell lines, c-Myc levels are also increased (Fig. 6B). Consistently, we detected higher USP36 mRNA levels (equal to or more than a twofold increase) in primary tumors than the adjacent normal tissues in 6 out of 17 (35.3%) breast cancer patients (Fig. 6C) (average increase, 2.19-fold; P = 0.031; Fig. 6D). Further, USP36 expression is drastically increased in 64% (7 out of 11 patients) of lung cancer patients compared with normal adjacent controls (Fig. S6B). Thus, USP36 is frequently overexpressed in tested human breast and lung cancers.
Fig. 6.
USP36 is overexpressed in cancer cells. (A and B) USP36 is overexpressed in breast cancer cell lines. The expression of USP36 was examined in a total of 10 breast cancer cell lines by RT-qPCR (A) and IB (B) compared with the immortalized mammary epithelia MCF10A cells. (C and D) USP36 is overexpressed in a subset of primary breast cancers. Tissues from breast cancer patients as well as patient-matched normal adjacent tissues were assays for USP36 mRNA expression by RT-qPCR (C). The average 2.19-fold increase of USP36 expression is shown in D. (E) A schematic diagram illustrating the role of USP36 in regulating c-Myc in the nucleolus (see Discussion for details).
Discussion
Ubiquitination plays a key and complex role in the regulation of c-Myc protein turnover and activity. c-Myc is ubiquitinated by Fbw7γ in the nucleolus, where c-Myc is mainly degraded (12, 23, 24), but the nucleolus is also the site where c-Myc contributes to ribosomal biogenesis (3). Thus, the nucleolus plays a central role in regulating c-Myc levels and oncogenic activity. However, whether c-Myc stability can be regulated by deubiquitination in the nucleolus was not known. In this study, we identified USP36 as a novel nucleolar DUB for c-Myc, counteracting c-Myc ubiquitination and degradation in the nucleolus.
USP36 and the c-Myc Nucleolar Degradation Pathway.
SCFFbw7-mediated ubiquitination and proteasomal degradation play key roles in linking c-Myc turnover to growth factor-dependent signaling. It is known that both Fbw7α and Fbw7γ, when overexpressed, can promote c-Myc ubiquitination and degradation, although the bulk of c-Myc degradation appears to occur in the nucleolus (12, 23, 24). However, how c-Myc stability is controlled by the Fbw7 isoforms in specific cell compartments under physiological conditions is still not clear. Recently, it has been shown that Fbw7α and Fbw7γ function sequentially to regulate the turnover of cyclin E, another Fbw7 analog substrate, instead of targeting cyclin E independently in their respective compartments (31, 32). Fbw7α promotes initial isomerization of cyclin E by PIN1 without ubiquitinating cyclin E, possibly coupled with nucleolar translocation, and allows successive ubiquitination of cyclin E by Fbw7γ in the nucleolus (31, 32). Thus, it will be interesting in future studies to examine whether Fbw7α may also promote isomerization of c-Myc by PIN1 that facilitates c-Myc translocation into the nucleolus, where it is polyubiquitinated by Fbw7γ. Our cell fractionation assays showed that USP36 interacts with c-Myc and deubiquitinates c-Myc in the nucleolus. Also, USP36 interacts with Fbw7γ, but not Fbw7α, and forms a tertiary complex with c-Myc and Fbw7γ, indicating that this tertiary complex assembles in the nucleolus, where c-Myc stability is dynamically controlled by the Fbw7γ–USP36 axis. This specific interaction between USP36 and Fbw7γ in the nucleolus may contribute to the specific USP36 regulation of c-Myc in the nucleolus. It is interesting to examine whether other mechanisms such as other posttranslational modifications of USP36 and c-Myc may involve this regulation as well.
Previously, USP28 has been shown to deubiquitinate c-Myc in the nucleus and suppress c-Myc degradation mediated by Fbw7α, but not Fbw7γ, suggesting that USP28 controls c-Myc stability primarily in the nucleoplasm (22). Together, we propose that c-Myc may be controlled dynamically and cooperatively by the USP28-–Fbw7α and USP36–Fbw7γ axes at the nucleoplasm and the nucleolus, respectively. USP28 may suppress the action of Fbw7α in the nucleoplasm, whereas USP36 ultimately counteracts Fbw7γ and controls c-Myc degradation in the nucleolus, acting as an end-point regulator of the c-Myc degradation pathway (Fig. 6E). This finding will therefore add a missing and key regulator into the c-Myc degradation pathway. Future studies are warranted to test how USP36–Fbw7γ and USP28–Fbw7α axes coordinately regulate c-Myc stability and function and how their deregulation contributes to human cancers.
USP36 Regulation of c-Myc and Ribosomal Biogenesis.
It has been shown that USP36 regulates the stability of the nucleolar proteins B23 and fibrillarin, which are critical for rRNA processing and ribosomal biogenesis (29). Yeast DUB ubp10, the human USP36 ortholog, also localizes in the nucleolus; controls the stability of Rpa190, the largest subunit of yeast RNA Pol I; and is required for proper ribosomal biogenesis and cell growth (30). These studies indicate that USP36 may play a role in ribosomal biogenesis. Our finding that USP36 controls c-Myc protein stability in the nucleolus further supports this notion, as c-Myc is a master regulator of ribosome biogenesis by controlling transcription catalyzed by all three RNA Pols, leading to enhanced production of rRNA, ribosomal proteins, and ribosomal biogenesis accessory factors (3, 25). By deubiquitinating and stabilizing c-Myc in the nucleolus, USP36 can directly promote c-Myc function in the nucleolus by enhancing RNA Pol I activity. Deubiquitinated c-Myc is then also translocated to the nucleoplasm (Fig. 3H), where it can enhance RNA Pol II and III activity as well. Furthermore, USP36 itself is a c-Myc target gene, suggesting that USP36 and c-Myc form a positive feedback regulatory loop. Altogether, these results reveal that USP36 is a novel positive c-Myc regulator controlling c-Myc’s nucleolar degradation pathway to enhance c-Myc’s oncogenic activity. Indeed, we observed that USP36 is overexpressed in a number of human cancers, including breast and lung cancers. Future studies would include the examination of USP36 regulation of c-Myc in mouse tumorigenesis models and whether USP36 could be a therapeutic target.
Materials and Methods
Cell lines were cultured and maintained as described in SI Materials and Methods. Flag-tagged USP36 (WT and the C131A mutant) plasmids and rabbit polyclonal anti-USP36 serum were previously described (28, 29). Flag–Fbw7 plasmids were provided by B. E. Clurman (Fred Hutchinson Cancer Research Center (Seattle, WA) (12). A detailed description of all other plasmids, in vivo and in vitro ubiquitination assays, RNA interference, IB, co-IP, ChIP, reverse transcriptase–quantitative PCR (RT-qPCR), cell proliferation, cell fractionation, flow cytometry, immunofluorescence staining, GST-fusion protein association, and soft agar assays can be found SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Bruce E. Clurman, from the Fred Hutchinson Cancer Research Center, for providing plasmids and reagents. This work was supported by National Institutes of Health/National Cancer Institute Grants R00 CA127134 and R01 CA160474, Department of Defense Grant W81XWH-10-1-1029 (to M.-S.D.), and a grant from the Medical Research Foundation of Oregon (to X.-X.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.S.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411713112/-/DCSupplemental.
References
- 1.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6(8):635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 2.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 3.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10(4):301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 4.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18(19):3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 5.Farrell AS, Sears RC. MYC degradation. Cold Spring Harb Perspect Med. 2014;4(3):a014365. doi: 10.1101/cshperspect.a014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol. 2006;16(4):288–302. doi: 10.1016/j.semcancer.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Lutterbach B, Hann SR. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol. 1994;14(8):5510–5522. doi: 10.1128/mcb.14.8.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sears R, et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14(19):2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moberg KH, Mukherjee A, Veraksa A, Artavanis-Tsakonas S, Hariharan IK. The Drosophila F box protein archipelago regulates dMyc protein levels in vivo. Curr Biol. 2004;14(11):965–974. doi: 10.1016/j.cub.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 10.Yada M, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23(10):2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101(24):9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. 2004;14(20):1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 13.Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95(6):2104–2110. [PubMed] [Google Scholar]
- 14.Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: Stabilization of c-Myc in Burkitt’s lymphoma cells. Mol Cell Biol. 2000;20(7):2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: Cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18(3):717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY Complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 17.Welcker M, Clurman BE. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8(2):83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 18.Cepeda D, et al. CDK-mediated activation of the SCF(FBXO) (28) ubiquitin ligase promotes MYC-driven transcription and tumourigenesis and predicts poor survival in breast cancer. EMBO Mol Med. 2013;5(7):999–1018. doi: 10.1002/emmm.201202341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11(5):1177–1188. doi: 10.1016/s1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 20.Popov N, Schülein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(β-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol. 2010;12(10):973–981. doi: 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- 21.von der Lehr N, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11(5):1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 22.Popov N, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9(7):765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 23.Arabi A, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7(3):303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 24.Grandori C, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7(3):311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 25.Dai MS, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J Cell Biochem. 2008;105(3):670–677. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quesada V, et al. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem Biophys Res Commun. 2004;314(1):54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- 27.Sun XX, Challagundla KB, Dai MS. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2012;31(3):576–592. doi: 10.1038/emboj.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Endo A, Kitamura N, Komada M. Nucleophosmin/B23 regulates ubiquitin dynamics in nucleoli by recruiting deubiquitylating enzyme USP36. J Biol Chem. 2009;284(41):27918–27923. doi: 10.1074/jbc.M109.037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endo A, et al. Nucleolar structure and function are regulated by the deubiquitylating enzyme USP36. J Cell Sci. 2009;122(Pt 5):678–686. doi: 10.1242/jcs.044461. [DOI] [PubMed] [Google Scholar]
- 30.Richardson LA, et al. A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell Reports. 2012;2(2):372–385. doi: 10.1016/j.celrep.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhaskaran N, et al. Fbw7α and Fbw7γ collaborate to shuttle cyclin E1 into the nucleolus for multiubiquitylation. Mol Cell Biol. 2013;33(1):85–97. doi: 10.1128/MCB.00288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Drogen F, et al. Ubiquitylation of cyclin E requires the sequential function of SCF complexes containing distinct hCdc4 isoforms. Mol Cell. 2006;23(1):37–48. doi: 10.1016/j.molcel.2006.05.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.