Significance
Cancer is a leading cause of mortality worldwide, with the identification of novel drug targets and chemotherapeutic agents being a high priority in the fight against it. The NEET proteins mitoNEET (mNT) and nutrient-deprivation autophagy factor-1 (NAF-1) were recently shown to be required for cancer cell proliferation. Utilizing a combination of experimental and computational techniques, we identified a derivative of the mitocan cluvenone that binds to NEET proteins at the vicinity of their 2Fe-2S clusters and facilitates their destabilization. The new drug displays a high specificity in the selective killing of human epithelial breast cancer cells, without any apparent effects on normal breast cells. Our results identify the 2Fe-2S clusters of NEET proteins as a novel target in the chemotherapeutic treatment of breast cancer.
Keywords: NEET proteins, mitocan, iron-sulfur proteins, mitochondria, rational drug design
Abstract
Identification of novel drug targets and chemotherapeutic agents is a high priority in the fight against cancer. Here, we report that MAD-28, a designed cluvenone (CLV) derivative, binds to and destabilizes two members of a unique class of mitochondrial and endoplasmic reticulum (ER) 2Fe-2S proteins, mitoNEET (mNT) and nutrient-deprivation autophagy factor-1 (NAF-1), recently implicated in cancer cell proliferation. Docking analysis of MAD-28 to mNT/NAF-1 revealed that in contrast to CLV, which formed a hydrogen bond network that stabilized the 2Fe-2S clusters of these proteins, MAD-28 broke the coordinative bond between the His ligand and the cluster’s Fe of mNT/NAF-1. Analysis of MAD-28 performed with control (Michigan Cancer Foundation; MCF-10A) and malignant (M.D. Anderson–metastatic breast; MDA-MB-231 or MCF-7) human epithelial breast cells revealed that MAD-28 had a high specificity in the selective killing of cancer cells, without any apparent effects on normal breast cells. MAD-28 was found to target the mitochondria of cancer cells and displayed a surprising similarity in its effects to the effects of mNT/NAF-1 shRNA suppression in cancer cells, causing a decrease in respiration and mitochondrial membrane potential, as well as an increase in mitochondrial iron content and glycolysis. As expected, if the NEET proteins are targets of MAD-28, cancer cells with suppressed levels of NAF-1 or mNT were less susceptible to the drug. Taken together, our results suggest that NEET proteins are a novel class of drug targets in the chemotherapeutic treatment of breast cancer, and that MAD-28 can now be used as a template for rational drug design for NEET Fe-S cluster-destabilizing anticancer drugs.
Mitochondria function as a key point of convergence for both the intrinsic and extrinsic apoptotic pathways, as well as play an essential role in regulating cellular metabolism and energy production (1). As such, they are emerging as important pharmacological targets for cancer therapy (2, 3). Small molecules that exert their bioactivity by selectively localizing and targeting mitochondria are often referred to as mitocans (4). The Garcinia genus of tropical plants has yielded a structurally intriguing family of xanthone-derived natural compounds collectively referred to as caged Garcinia xanthones (CGXs) (5). Gambogic acid, the archetype of this family, inhibits tumor growth in various animal models with minimal side effects and little toxicity on immune and hemopoietic systems (6, 7). Its anticancer activity has been associated with mitochondrial membrane polarization, inhibition of the B-cell lymphoma 2 (Bcl-2) family of proteins, accumulation of reactive oxygen species (ROS), suppression the NF-κB signaling pathway, and inhibition of proteasome activity (4–10). In previous studies, we determined that cluvenone (CLV) is the pharmacophoric structure of CGX (8, 9). CLV displayed good tumor selectivity in the NCI60 cell panel, as well as high differential cytotoxicity in cancer vs. normal tissue studies (10). We also demonstrated that CLV preferentially localizes to the mitochondria and induces cell death (11, 12). To improve further on the tumor selectivity of CLV, we designed and synthesized the hydroxylated derivatives MAD-28 and MAD-44 (13).
Localized in the outer mitochondrial membrane (14, 15), a potential site of action of mitocans, the NEET proteins nutrient-deprivation autophagy factor-1 (NAF-1) and mitoNEET (mNT) are essential in tumor growth regulation. These proteins constitute a novel family of iron-sulfur (2Fe-2S) proteins defined by a unique CDGSH amino acid sequence in their Fe-S cluster-binding domain (16). They are involved in several human pathologies, including diabetes, cystic fibrosis, Wolfram syndrome 2, neurodegeneration, and muscle atrophy (15, 17, 18). NAF-1 was found to interact with Bcl-2 and Beclin 1 and was proposed to regulate autophagy and apoptosis (19). Deficiency in mNT or NAF-1 causes an alteration in iron and ROS homeostasis in animal and plant cells, and deficiency in NAF-1 results in decreased mitochondrial function and stability, as well as activation of autophagy in mouse and human cells (14, 15, 17, 20, 21). We recently demonstrated that suppression of NAF-1 or mNT expression via shRNA in human breast cancer cells resulted in the accumulation of iron and ROS in mitochondria, a shift from respiration to glycolysis, the activation of autophagy, and the suppression of cell proliferation and tumor growth (21). Many of these phenotypical effects are analogous to the effects induced by mitocans, leading us to the hypothesis that small molecules that bind NEET proteins and alter their molecular features could have a significant pharmacological potential in cancer treatment.
To test this hypothesis, we studied the potential of different derivatives of the mitocan (CLV, MAD-44, and MAD-28) to bind to the NEET proteins mNT and NAF-1 and to affect their cluster stability, and we correlated the biochemical and biophysical effects of these compounds on NEET proteins with their biological activity toward cancer cell metabolism, physiology, and viability. Here, we report that MAD-28, a derivative of the caged Garcinia mitocan CLV, binds to mNT and NAF-1 and facilitates the destabilization of their clusters. Docking analysis of MAD-28, CLV, and MAD-44 to mNT and NAF-1 revealed that in contrast to CLV, which formed a hydrogen bond network that stabilized the 2Fe-2S clusters of these proteins, or MAD-44, which did not affect the Fe-S cluster coordination of mNT and NAF-1, MAD-28 broke the coordination bond between H87/H114 and the cluster Fe of mNT/NAF-1, resulting in destabilization of their clusters. In vivo analysis of MAD-28 performed with control and malignant epithelial breast cells revealed that MAD-28 had a high specificity in the selective killing of cancer cells, without any apparent effects on normal breast cells. MAD-28 was less effective when applied to cancer cells with suppressed levels of NAF-1 or mNT. Taken together, our results suggest that the Fe-S clusters of the NEET proteins mNT and NAF-1 could be used as novel drug targets in the chemotherapeutic treatment of breast cancer cells.
Results
Biological Activity of CLV, MAD-28, and MAD-44 Toward Human Epithelial Breast Cancer Cells.
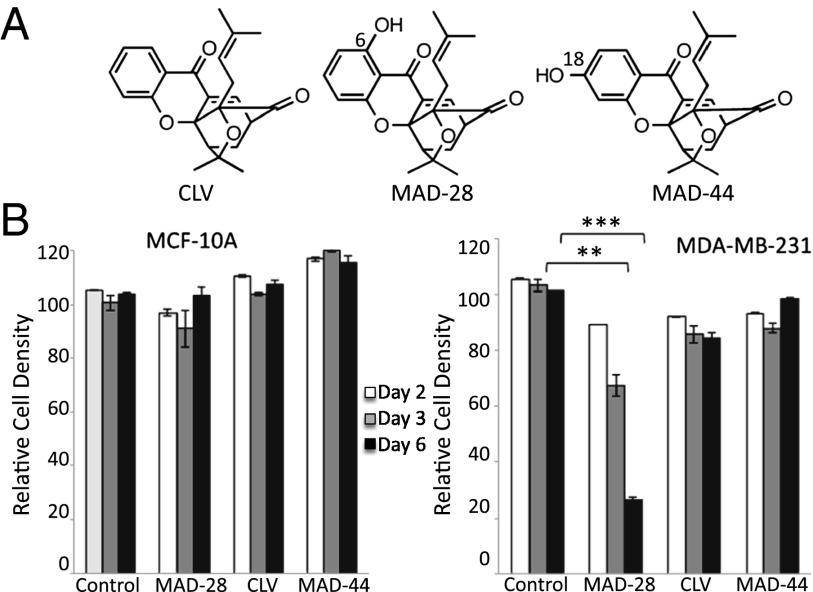
The mitocan CLV and its two derivatives MAD-28 and MAD-44 (Fig. 1A) were synthesized as described in Materials and Methods and SI Appendix (13) (SI Appendix, Fig. S1), and their ability to inhibit cancer cell growth was tested using epithelial breast cancer cells [M.D. Anderson–metastatic breast (MDA-MB)-231 and Michigan Cancer Foundation (MCF)-7], and control breast epithelial cells (MCF-10A) (Fig. 1B and SI Appendix, Fig. S2). Of the three CLV derivatives tested, only MAD-28 showed high potency in the selective killing of breast cancer cells, although it was nontoxic to normal breast cells (Fig. 1B). In contrast to CLV or MAD-44, which did not affect the proliferation of cancer cells within the time and concentration ranges tested, MAD-28 showed a selective killing rate of up to 70% within 6 d of treatment, with no apparent effect on the viability of control MCF-10A cells (Fig. 1B). Because MAD-28 was found to have the highest potency of the three compounds tested, it was selected for further analysis.
Fig. 1.
Structure and selective cytotoxicity of CLV, MAD-28, and MAD-44. (A) Structures of CLV, MAD-28, and MAD-44. The chemical synthesis of MAD-28 and MAD-44 is described in SI Appendix, Fig. S1B. (B) Effect of MAD-28, CLV, and MAD-44 on cell survival of control noncancerous human breast cells MCF-10A (Left) or human epithelial breast cancer cells MDA-MB-231 (Right). Alamar blue cell viability measurements were performed on cells at 2, 3, and 6 d following application of 1.25 μM MAD-28, CLV, or MAD-44. The results show that the three different compounds had no significant effect on the viability of control cells and that MDA-28 had a potent and selective killing effect on cancer cells. Percentage of cell survival was determined based on cells treated with DMSO. Error bars represent SD obtained from three individual experiments. **P < 0.01; ***P < 0.001.
MAD-28 Localizes to Mitochondria and Causes a Decrease in Mitochondrial Function in Breast Cancer Cells.
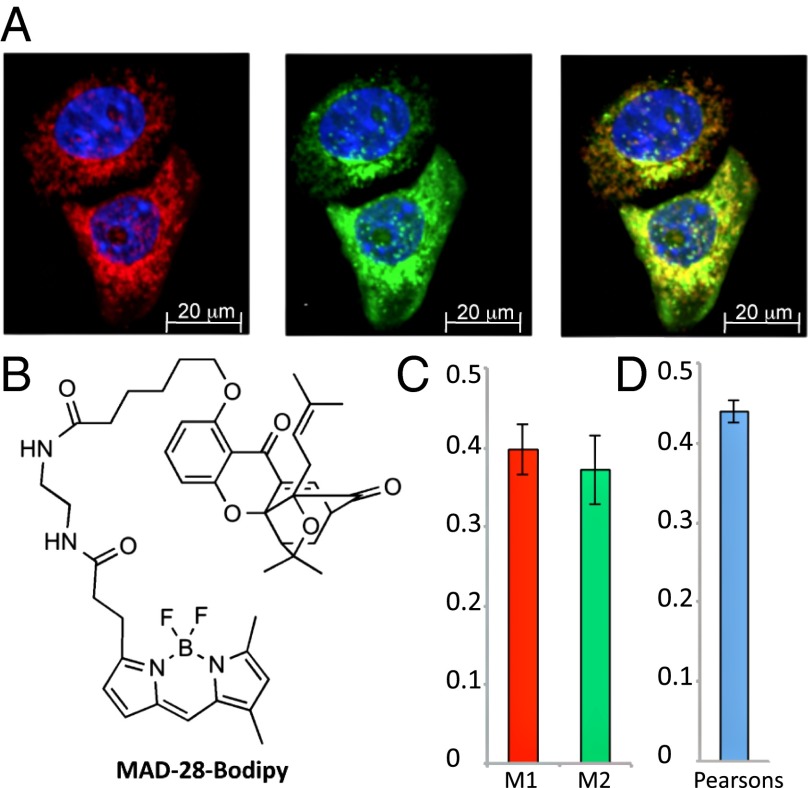
To gain information on the subcellular target of MAD-28, we used a Bodipy conjugate of this compound (i.e., MAD-28–Bodipy; chemical synthesis is described in SI Appendix, Fig. S1C). Mitochondria of MDA-MB-231 cells were visualized using the mitochondrial marker Tom20, and the conjugate MAD-28–Bodipy was added and co-imaged to reveal possible colocalization. As shown in Fig. 2, the green fluorescence of MAD-28–Bodipy clearly colocalized with the mitochondrial red fluorescent Tom20 marker. MAD-28 also displaced MAD-28–Bodipy localization in competition studies (SI Appendix, Fig. S3). Therefore, our data indicate that MAD-28 localizes to the mitochondria of cancer cells.
Fig. 2.
Subcellular localization of a MAD-28–Bodipy conjugate. (A) MDA-MB-231 cells labeled with mouse anti-Tom20 and visualized with Alexa Fluor 594 goat anti-mouse (Left) were then incubated with MAD-28–Bodipy (Middle). (Right) Merged image is shown. Nuclei are shown in blue. (B) Structure of the MAD-28–Bodipy. The synthesis of MAD-28–Bodipy is shown in SI Appendix, Fig. S1C. Colocalization efficiency was measured using ImageJ software and is shown as Manders’ coefficients M1 and M2 (C) and Pearson’s correlation coefficient (D). The average and SD were obtained by analysis of at least eight different images.
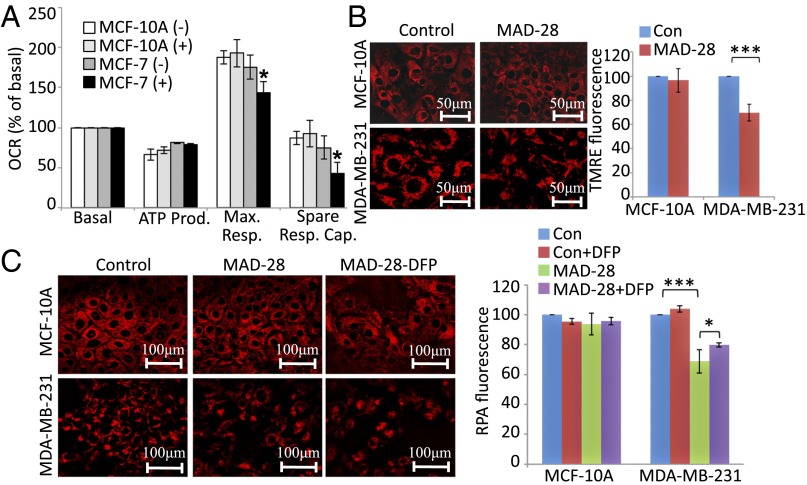
To test how the localization of MAD-28 to mitochondria affects cancer cell metabolism, we measured its effect on mitochondrial function and glycolysis using a Seahorse XF24 analyzer (Seahorse Bioscience). Application of MAD-28 to MCF-7 breast cancer cells resulted in diminished spare respiratory capacity of mitochondria (Fig. 3A) and enhanced glycolytic activity (SI Appendix, Fig. S4). In contrast, MAD-28 had no significant effects on the metabolism of control MCF-10A cells (Fig. 3A and SI Appendix, Fig. S4).
Fig. 3.
Measurements of mitochondrial function of control MCF-10A and malignant MCF-7 human epithelial breast cells treated or untreated with MAD-28. (A) Measurements of maximal respiration (Max. Resp.) and spare respiratory capacity (Spare Resp. Cap.) of MCF-10A and MCF-7 cells treated with MAD-28 (+) or DMSO (−), showing that MAD-28 causes a selective decrease in the mitochondrial activity of cancer cells. Measurements of mitochondrial respiration shown were performed as described in Materials and Methods using an XF-24 Seahorse apparatus. *P < 0.05. OCR, oxygen consumption rate. (B) Images (Left) and a quantitative BAR graph (Right) showing the effect of MAD-28 on MMP in control MCF-10A and breast cancer MDA-MB-231 cells [TMRE fluorescence is shown as percentage of control (Con)]. MAD-28 is shown to have a specific effect on cancer cells, decreasing their MMP. (C) Images (Left) and a quantitative BAR graph (Right) showing mitochondrial iron levels in MDA-MB-231 and MCF-10A cells treated or untreated with MAD-28 (RPA fluorescence is shown as percentage of control). MAD-28 is shown to have a specific effect on cancer cells, causing an increase in their mitochondrial iron levels. Error bars represent SD obtained from three individual experiments. (Scale bars: B, 50 μm and C, 100 μm.) *P < 0.05; **P < 0.01; ***P < 0.001.
MAD-28 Causes a Decrease in Mitochondrial Membrane Potential and an Increase in Mitochondrial Iron Accumulation in Breast Cancer Cells.
The targeting and physiological effects of MAD-28 suggested that this compound alters mitochondrial function in cancer cells. We therefore tested its effects on the mitochondrial membrane potential (MMP) of breast cells using tetramethylrhodamine ethyl ester (TMRE), a positively charged red-orange dye that readily accumulates in active mitochondria, as well as its effect on mitochondrial iron accumulation using rhodamine B-[(1, 10-phenanthrolin-5-yl aminocarbonyl] (RPA), a fluorescent iron sensor (21). As shown in Fig. 3 B and C, MAD-28 caused a significant decrease in MMP, as well as a significant increase in the accumulation of iron in mitochondria of breast cancer cells (MDA-MB-231), without affecting control MCF-10A cells. Iron accumulation in the mitochondria of cancer cells following MAD-28 treatment for 48 h was partially blocked by pretreatment of cells with the iron chelator deferiprone (DFP; 1,2-dimethyl-3-hydroxypyridin-4-one), suggesting that MAD-28 caused an alteration in the iron homeostasis of cancer cell mitochondria. These findings provided strong evidence for the function of MAD-28 as a mitocan that selectively targets the mitochondria of cancer cells.
MAD-28 Destabilizes the 2Fe-2S Clusters of NEET Proteins, and Its Biological Activity Depends on the Level of NEET Proteins in Cancer Cells.
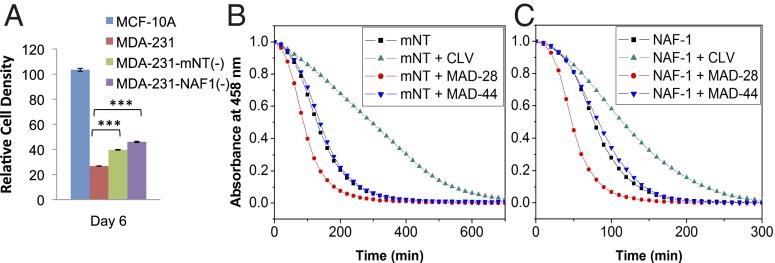
The biological activity of MAD-28 on MMP, iron accumulation, respiration, and glycolysis of human breast cancer cells (Fig. 3 and SI Appendix, Fig. S4) resembled the phenotype of cells observed when the expression of the NEET protein mNT or NAF-1 was suppressed using shRNA in epithelial breast cancer cells (21). This similarity could suggest that at least part of the effects of MAD-28 on cancer cells is mediated via these proteins. To test whether MAD-28 functions in cells by interacting with NEET proteins, we studied its effect on human breast cancer cells with suppressed levels of expression of NAF-1 or mNT, obtained as described by Sohn et al. (21). As shown in Fig. 4A, the cytotoxicity of MAD-28 toward breast cancer cells was lower in cells with suppressed expression of the NEET protein mNT or NAF-1.
Fig. 4.
Effect of MAD-28 on the survival of cells with suppressed levels of mNT or NAF-1 and on the cluster stability of mNT and NAF-1. (A) Survival assays of MCF-10A and MDA-MB-231 cells with suppressed levels of mNT [MDA-MB-231–mNT(−)] or NAF-1 [MDA-MB-231–NAF-1(−)], showing that the killing effect of MAD-28 on cancer cells is attenuated in cells with suppressed expression of the NEET protein mNT or NAF-1. ***P < 0.001. (B and C) 2Fe-2S cluster stability of mNT (B) or NAF-1 (C) in the absence or presence of stoichiometric concentrations of CLV, MAD-28, or MAD-44 monitored by UV-Vis spectroscopy at 458 nm, showing that MAD-28 has a destabilizing effect and CLV has a stabilizing effect on the clusters of mNT or NAF-1.
The function of NEET proteins in cells was demonstrated to involve their 2Fe-2S clusters (19, 21, 22); therefore, we studied the effects of MAD-28 on the stability of mNT or NAF-1 clusters and compared it with the effects of CLV or MAD-44 (Fig. 4 B and C). NEET 2Fe-2S cluster stability can be assessed by monitoring the decay in absorbance of its 458-nm peak (characteristic of the oxidized 2Fe-2S cluster of mNT or NAF-1) over time. We incubated each protein (mNT or NAF-1) in the absence and presence of stoichiometric concentrations of CLV, MAD-28, or MAD-44. The rate of cluster release was compared for each compound relative to each protein alone. Effects on cluster stability were observed for both mNT and NAF-1 with CLV or MAD-28. Although CLV increased 2Fe-2S cluster stability by 50–100% (green traces in Fig. 4 B and C), MAD-28 decreased cluster stability by 30–50% (red traces in Fig. 4 B and C). In contrast, MAD-44 had no effect above uncertainty (∼5%; blue traces in Fig. 4 B and C).
Identifying Potential NEET Protein Binding Pockets for MAD-28, MAD-44, and CLV.
The opposing effects of MAD-28 and CLV on the stability of NEET proteins’ 2Fe-2S clusters prompted us to identify possible binding sites of these drugs to the mNT and NAF-1 structures (23, 24). Eleven druggable “hot spots” were identified for the mNT structure [Protein Data Bank (PDB) ID code 2QH7] using the small-molecule mapping server FTMap (25, 26). We picked the hot spots with the largest number of small molecules bound and grouped them into two main consensus sites as described in Materials and Methods and SI Appendix. The areas of mNT within the 6-Å vicinity of the two main consensus sites were defined as potential binding pockets on mNT. Pocket 1 covers patches of highly conserved residues (I102, K55′, V57′, and D64) and five of the nine conserved core amino acids of the [2Fe-2S] binding motif (C83, H87, Y71, F82, and D84). Moreover, a large portion of residues in NAF-1 (S92, A96, A109, C110, D111, and H114), corresponding to the residues (G62, A69, F82, C83, D84, and H87) in pocket 1 of mNT, overlap with putative interfacial binding residues between proteins Bcl-2 and NAF-1, which were identified in an integrated experimental and direct coupling analysis strategy (27). These overlapping residues interact with several residues of the BH4 domain and the BH2 domain of Bcl-2 (SI Appendix, Fig. S5). Therefore, this overlap gives further support to the hypothesis that a compound that binds pocket 1 can interfere and affect the function of NEET protein–protein interactions. In contrast, pocket 2, which is composed of the residues of I49′, Q50′, I56′, H58′, S77′, K78′, K79′, F80′, and P81′, is relatively small and polar, and does not seem to be the potential binding pocket of the three compounds compared with our computational results.
Based on our docking analysis, CLV and MAD-28 selectively bind to the vicinity of the [2Fe-2S] cluster of pocket 1. However, unlike CLV or MAD-28, MAD-44 binds toward K68 of pocket 1. We further clustered the binding modes in pocket 1 for each compound based on their binding orientation, picked the lowest energy bind mode for each of them from the cluster with the dominant binding orientation, and performed energy minimization for the selected binding modes to refine their structures. The refined binding modes of these three compounds to mNT and NAF-1 are illustrated in Fig. 5A and SI Appendix, Fig. S6, and the evaluated binding energies corresponding to the binding modes are shown in SI Appendix, Table S1. In a parallel analysis performed for NAF-1, we found that CLV, MAD-28, and MAD-44 present very similar binding modes and molecular interactions to mNT, as shown in SI Appendix, Fig. S6. The estimated binding energies for the predicted binding modes of CLV, MAD-28, and MAD-44 to the NEET proteins mNT and NAF-1 are summarized in SI Appendix, Table S1.
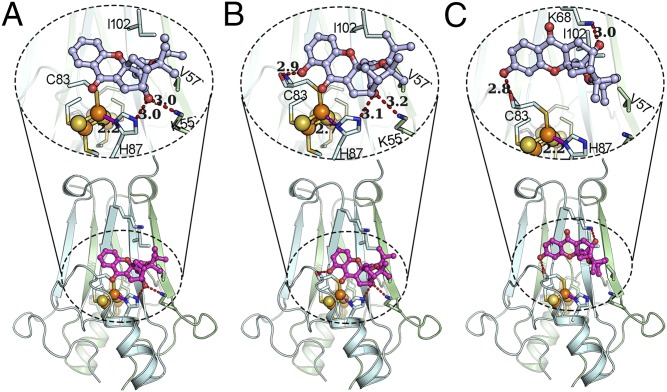
Fig. 5.
CLV, MAD-28, and MAD-44 are different in their binding to and potential effects on the [2Fe-2S] clusters of mNT. The predicted binding modes of CLV (A), MAD-28 (B), and MAD-44 (C) to mNT (PDB ID code 2QH7) are shown. The lower part highlights the docking of the compound on mNT, the upper part expands the region near the docking site, and the hydrogen bonds emphasized by dashed lines. mNT is shown in cartoon representation, where sticks show the critical residues of mNT, spheres represent the [2Fe-2S] cluster, and light gray stick ball models are the compounds. Hydrogen bonds are shown as red dashed lines, and coordination bonds are shown as solid magenta lines. When MAD-28 binds to mNT, a coordination bond between the iron ion and the H87 is broken (a dashed magenta line shows an elongated distance of 2.7 Å), potentially destabilizing the [2Fe-2S] cluster.
Discussion
Biological and Biophysical Action of CLV, MAD-44, and MAD-28 on Cancer Cells and NEET Proteins.
Our characterization of MAD-28, a derivative of CLV containing a phenolic group at the C6 position of its A ring, revealed that compared with CLV or MAD-44, this compound is highly potent in its selective cytotoxicity toward human breast cancer cells (Fig. 1 and SI Appendix, Fig. S2). CLV was previously found to be an effective and selective drug to target cancer cells (8–10). Nevertheless, our analysis of its derivative MAD-28 demonstrated that it had significantly higher cytotoxicity toward cancer cells, without affecting control human epithelial breast cells. The addition of a phenolic group at the C6 position of the CLV A ring therefore appears to enhance the potency of this compound significantly without affecting its targeting to the mitochondria of cancer cells (Figs. 1 and 2).
The physiological and cellular effects of MAD-28 on human breast cancer cells included some of the hallmarks observed in these cells when the expression of the NEET protein mNT or NAF-1 was suppressed via shRNA (21). These effects included suppression of cancer cell proliferation, a decrease in respiration and MMP, enhanced glycolysis, and an increase in the accumulation of iron in mitochondria. The expression of NEET proteins was found to be enhanced in many different types of cancer cells, promoting a hypothesis that their function is essential to support the higher metabolic rates of cancer cells by protecting mitochondria from overaccumulation of iron, which would result in enhanced reactive oxygen accumulation and cell death (16). The central role of mNT and NAF-1 in cancer metabolism and regulation (28), as well as the similarity observed between the phenotype of breast cancer cells treated with MAD-28 and breast cancer cells with suppressed expression of mNT or NAF-1 (21), suggests that part of the mitocan function of MAD-28 on cancer cells is executed by binding to, and affecting the function of, NEET proteins.
Our findings that CLV and MAD-28 alter the cluster stability of NEET proteins in vitro (Fig. 4 B and C) show that CLV and MAD-28 bind to these proteins. According to the above computational results, the predicted binding pocket of CLV and MAD-28 is also involved in the protein–protein interface of NAF-1 and Bcl-2. Therefore, altering the stability of NEET proteins’ clusters could modify their cellular function and/or protein–protein interactions. For example, a derivative of NAF-1 with an altered cluster-binding ability was found to have an altered interaction with Bcl-2 that affected its ability to regulate autophagy (19). Interestingly, MAD-28 was found to have an opposite effect on NEET cluster stability (destabilize) compared with CLV (stabilize). It is possible that this difference could be responsible for the higher potency of this particular derivative. Destabilizing the clusters of NAF-1 could, for example, cause it to dissociate from Bcl-2 and decrease the viability of cancer cells. To test further whether MAD-28 functions via affecting NEET proteins, we tested its effect on cancer cells in which the elevated levels of mNT or NAF-1 were suppressed via shRNA. MAD-28 is less effective in killing these cells (Fig. 4A). At least two different models could explain this result. The lower level of a NEET protein in these cells could present a smaller target for MAD-28 binding and function, or the pathways activated in cells with lower levels of mNT or NAF-1 could be mitigating the function of MAD-28. Both explanations are consistent with our hypothesis that MAD-28 works, in part, via NEET proteins or NEET-related pathways.
Docking Analysis of CLV, MAD-28, and MAD-44 Provides Insight into the Differences in Their Biological Function.
To decipher the mechanism by which CLV, MAD-28, and MAD-44 affect the stability of NEET protein clusters, we performed docking analysis for these compounds to mNT and NAF-1 (PDB ID codes 2QH7 and 4OO7). Our analysis revealed that in the predicted binding mode of CLV to mNT (Fig. 5A), a hydrogen bond network is formed between the CLV and residues K55 and H87 with equal hydrogen bond lengths of 3.0 Å and that the coordination bond between the H87 and the outer Fe, which plays an important role in stabilizing the 2Fe-2S cluster (29), remains with a bond length of 2.2 Å. The formation of the hydrogen bond between the CLV and H87 can lead to an increase in the negativity of the Nδ1 atom (coordination bond acceptor) in the imidazole ring of H87, and hence contributes to increasing the coordinate bond strength and stabilizes the clusters. In addition, a series of hydrophobic contacts are observed between CLV and mNT, including the interactions between the hydrophobic tail group of CLV and residues V57 and I102 as well as the interactions between the tricyclogroup and beta-strands of mNT, which can further help CLV to stabilize the cluster via increasing the overall protein stability through binding energy.
In contrast to the effect of CLV, the observed conformation in the lowest energy binding mode of MAD-28 to mNT (Fig. 5B) contains an additional hydrogen bond between the hydroxyl group in the 6-position of MAD-28 and the backbone carbonyl of C83 and, at the same time, maintains a hydrogen network similar to CLV against mNT, resulting in a strain energy penalty of conformation of MAD-28 (∆Estrain = 5.73 kcal/mol; SI Appendix, Table S1) relative to the binding conformation of CLV (∆Estrain = 1.85 kcal/mol; SI Appendix, Table S1). Even more importantly, the conformation of the side chain of residue H87 was induced to flip up nearly 28° relative to the conformation of this side chain observed for CLV, which breaks the coordination bond between the H87 and outer Fe of the 2Fe-2S cluster (the bond length is elongated to 2.7 Å). This observation supports our biophysical results (Fig. 4 B and C), showing that the 2Fe-2S clusters of mNT and NAF-1 are significantly destabilized by binding of MAD-28. Furthermore, structural NMR studies (SI Appendix, Fig. S7) support the proposed model for MAD-28 binding. Contrary to CLV and MAD-28, the binding mode of MAD-44 to mNT depicted in Fig. 5C indicates that when the hydroxyl group is in the C18 position, the rigidity of the tricyclogroup cannot maintain the hydrogen bond network with H87 but forms a hydrogen bond with K68 to keep a relatively extended binding conformation (∆Estrain = 1.33 kcal/mol; SI Appendix, Table S1). In this case, H87 is not involved in the binding interaction of MAD-44 to mNT. Therefore, the stabilization of [2Fe-2S] clusters is not affected by MAD-44. In summary, with these docking models, we now provide a molecular explanation for the observed stabilization of NEET cluster-containing proteins with CLV and the destabilization with the more selective anticancer agent MAD-28.
Conclusions
In this work, we present a designed mitocan drug, MAD-28, that shows selective targeting of cancer cells and evidence that it targets NEET proteins, which are, to our knowledge, the first Fe-S proteins shown to control human breast cancer cell proliferation (21). Our results suggest a unique mode of action for MAD-28 binding to the vicinity of the Fe-S clusters of NEET proteins and destabilizing them by breaking the coordinative bond between the mNT-H87 or NAF-1–H114 and the cluster Fe (Fig. 5 and SI Appendix, Fig. S5). The ability to explain the experimental observations theoretically opens up the possibility for rational drug design. The high selectivity of MAD-28 toward cancer cells, with no apparent effects on normal cells (Figs. 1 and 3), could be explained by the apparent dependency of cancer cells on NEET protein function (19, 21) and by the high levels of NEET proteins in cancer cells (21). Consistent with this model are the findings that MAD-28 is less effective on cells with suppressed mNT or NAF-1 expression (Fig. 4A) and that the effects of MAD-28 are partially alleviated by the addition of an iron-chelating agent (Fig. 3C). Because elevated levels of mNT and NAF-1 are nearly universal in many epithelial cancers (16, 21), targeting their clusters provides an unexplored and highly promising area for pharmaceutical development. MAD-28 could therefore be viewed as a first lead to a new class of anticancer drugs that target the metal clusters (2Fe-2S) of proteins.
Materials and Methods
Materials and Cell Cultures.
MCF-10A, MCF-7, and MDA-MB-231 epithelial breast cancer cells were obtained from the American Type Culture Collection and from Tamar Peretz-Yablonski (Sharett Institute of Oncology, Hadassah-Hebrew University Medical Center, Jerusalem, Israel). Synthesis of CLV, MAD-28, MAD-44, and the MAD-28–Bodipy conjugate is described in SI Appendix, Fig. S1. MDA-MB-231, MCF-10A, MCF7, MDA231-NAF-1(−), and MDA231-mNT(−) cell lines were grown or generated using shRNA transfections as described previously (21).
Cell Viability.
Cells were seeded in 96-well plates in triplicate at a density of 15,000 cells per plate and incubated with MAD-28, CLV, or MAD-44 at a concentration of 1.25 μM. This concentration was used based on previous IC50 studies (10, 13): We have previously reported that CLV exhibits potent growth inhibitory activity against the NCI60 cell panel, with an IC50 range of 0.1–2.7 μM (10). We have also reported that the IC50 values of CLV, MAD-28, and MAD-44 in the T-cell acute lymphoblastic leukemia (CEM) cell line are 0.2, 0.3, and 1.0 μM, respectively (13). We designed our experiments based on the above studies and selected an average concentration of 1.25 μM for our viability studies on epithelial breast cancer cells. Alamar-blue (Prestoblue cell viability reagent; Invitrogen) was used to determine cell viability 2, 3, and 6 d following addition of the drugs as described previously (21, 30). Fluorescence was measured using a plate reader after 1–4 h of incubation at 37 °C (excitation, 530–560 nm; emission, 590 nm).
Immunofluorescence Analysis.
Cells plated on 12-mm glass coverslips were fixed with 4% (vol/vol) formaldehyde, permeabilized with 0.1% Triton X-100 in PBS for 5 min, and incubated in blocking buffer (PBS containing 5% fetal goat serum) for 30 min at room temperature. Cells were then incubated for 1 h at room temperature with primary antibody diluted in blocking buffer. Cells were then washed three times with PBS and incubated with secondary antibody, diluted in blocking buffer, for 1 h at room temperature. Alexa Fluor 594 goat anti-mouse (1:500; Molecular Probes) was used. Cells were washed three times with PBS containing Hoechst (1:100,000, H33342; Molecular Probes) to stain DNA and visualized using a Zeiss Observer Z1 156 inverted microscope. The degree of colocalization between mitochondria (red) and gambogic acid (GA)-Bodipy (green) was quantified using the colocalization finder and the JaCoP plug-in of ImageJ software (rsb.info.nih.gov/ij/). Both Pearson’s correlation and Manders’ overlap coefficients were measured for at least five cells per sample.
MMP Measurement.
Cells were plated at a density of 150,000 per well on glass-bottomed, 24-well plates incubated with MAD-28 at a concentration of 1.25 μM for 24 h and washed with DMEM-Hepes for measurement of MMP. TMRE (excitation, 543 nm; emission, 633 nm) was used for measuring MMP as described by Sohn et al. (21) utilizing a Nikon TE 2000 microscope equipped with opti-grid device and a Hamamatsu Orca-Era CCD camera. Images were acquired and analyzed with the Volocity or ImageJ software program.
Mitochondrial Labile Iron Measurement.
Cells were plated and incubated with MAD-28 as described above. The iron status of the mitochondrion of cells was then measured using RPA as described by Sohn et al. (21). Images were captured by epifluorescence microscopy (equipped with an opti-grid device) and analyzed with ImageJ. Pretreatment of cells with the chelator DFP (50–100 μM) was performed to determine indirectly the mitochondrial labile iron level (21).
Physiological Measurements.
Rates of oxygen consumption and extracellular acidification were measured with a Seahorse XF24 analyzer using the XF Cell Mito Stress Test Kit and XF Glycolysis Stress Test Kit (all from Seahorse Bioscience) according to the method of Sohn et al. (21). All measurements were recorded at set interval time points. All compounds and materials above were obtained from Seahorse Bioscience.
Cluster Stability Assay.
mNT and NAF-1 were expressed and purified as previously described (22). A total of 50 μM (25 μM dimeric) mNT or NAF-1 was incubated alone or with 50 μM CLV, MAD-28, or MAD-44 at 37 °C in 50 mM Bis-Tris buffer at pH 6.5, with 100 mM NaCl, 0.1% (wt/vol) lauryldimethylamine N-oxide, and 0.5% (vol/vol) DMSO. Absorbance spectra were measured from 300 nm to 800 nm every 10 min on a Cary 50 UV-visible spectrophotometer with temperature control (Agilent). Absorbance at 458 nm was normalized and plotted vs. time.
Computational Calculations of the Molecular Mode of Binding of the Mitocans to the NEET Proteins.
To investigate the molecular mechanism of CLV, MAD-28, and MAD-44 binding to mNT or NAF-1, a four-step computational scheme was designed based on the FTMap fragment-based druggable hot spots identification approach (25, 26) and using the molecular modeling environment Maestro 9.9 (Schrödinger) (details are provided in SI Appendix).
Supplementary Material
Acknowledgments
J.N.O. thanks Schrödinger for providing a free evaluation version of the molecular modeling environment Maestro 9.9. This work was supported by Israel Science Foundation Grant (ISF) 865/13 (to R.N.), funds from the University of North Texas College of Arts and Sciences (to R.M.), the Coordination for the Improvement of Higher Education Personnel (CAPES) Foundation-Brazil for a Ph.D. Fellowship to COR (12487-12-0), and NIH Grant GM101467 (to P.A.J.). Work at the Center for Theoretical Biological Physics was sponsored by the National Science Foundation (Grants PHY-1427654 and MCB-1214457) and by the Cancer Prevention and Research Institute of Texas. F.B. was also supported by Welch Foundation Grant C-1792.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502960112/-/DCSupplemental.
References
- 1.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281(5381):1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 2.Biasutto L, Dong LF, Zoratti M, Neuzil J. Mitochondrially targeted anti-cancer agents. Mitochondrion. 2010;10(6):670–681. doi: 10.1016/j.mito.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9(6):447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 4.Neuzil J, et al. Vitamin E analogues as a novel group of mitocans: Anti-cancer agents that act by targeting mitochondria. Mol Aspects Med. 2007;28(5-6):607–645. doi: 10.1016/j.mam.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Chantarasriwong O, Batova A, Chavasiri W, Theodorakis EA. Chemistry and biology of the caged Garcinia xanthones. Chemistry. 2010;16(33):9944–9962. doi: 10.1002/chem.201000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo QL, et al. Effect of gambogic acid on the hemopoietic and immune function in experimental animals. Chin J Nat Med. 2003;1:229–231. [Google Scholar]
- 7.Li X, et al. Gambogic acid is a tissue-specific proteasome inhibitor in vitro and in vivo. Cell Reports. 2013;3(1):211–222. doi: 10.1016/j.celrep.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 8.Batova A, et al. Synthesis and evaluation of caged Garcinia xanthones. Org Biomol Chem. 2007;5(3):494–500. doi: 10.1039/b612903j. [DOI] [PubMed] [Google Scholar]
- 9.Chantarasriwong O, et al. Evaluation of the pharmacophoric motif of the caged Garcinia xanthones. Org Biomol Chem. 2009;7(23):4886–4894. doi: 10.1039/b913496d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batova A, et al. The synthetic caged garcinia xanthone cluvenone induces cell stress and apoptosis and has immune modulatory activity. Mol Cancer Ther. 2010;9(11):2869–2878. doi: 10.1158/1535-7163.MCT-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guizzunti G, Batova A, Chantarasriwong O, Dakanali M, Theodorakis EA. Subcellular localization and activity of gambogic acid. ChemBioChem. 2012;13(8):1191–1198. doi: 10.1002/cbic.201200065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guizzunti G, Theodorakis EA, Yu AL, Zurzolo C, Batova A. Cluvenone induces apoptosis via a direct target in mitochondria: A possible mechanism to circumvent chemo-resistance? Invest New Drugs. 2012;30(5):1841–1848. doi: 10.1007/s10637-011-9745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbel KM, et al. A-ring oxygenation modulates the chemistry and bioactivity of caged Garcinia xanthones. Org Biomol Chem. 2013;11(20):3341–3348. doi: 10.1039/c3ob40395e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiley SE, et al. Wolfram Syndrome protein, Miner1, regulates sulphydryl redox status, the unfolded protein response, and Ca2+ homeostasis. EMBO Mol Med. 2013;5(6):904–918. doi: 10.1002/emmm.201201429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YF, et al. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 2009;23(10):1183–1194. doi: 10.1101/gad.1779509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamir S, et al. 2014. Structure-function analysis of NEET proteins uncovers their role as key regulators of iron and ROS homeostasis in health and disease. Biochim Biophys Acta, 10.1016/j.bbamcr.2014.10.014.
- 17.Chang NC, et al. Bcl-2-associated autophagy regulator Naf-1 required for maintenance of skeletal muscle. Hum Mol Genet. 2012;21(10):2277–2287. doi: 10.1093/hmg/dds048. [DOI] [PubMed] [Google Scholar]
- 18.Kusminski CM, et al. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat Med. 2012;18(10):1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang NC, Nguyen M, Germain M, Shore GC. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 2010;29(3):606–618. doi: 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nechushtai R, et al. Characterization of Arabidopsis NEET reveals an ancient role for NEET proteins in iron metabolism. Plant Cell. 2012;24(5):2139–2154. doi: 10.1105/tpc.112.097634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sohn YS, et al. NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc Natl Acad Sci USA. 2013;110(36):14676–14681. doi: 10.1073/pnas.1313198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuris JA, et al. Facile transfer of [2Fe-2S] clusters from the diabetes drug target mitoNEET to an apo-acceptor protein. Proc Natl Acad Sci USA. 2011;108(32):13047–13052. doi: 10.1073/pnas.1109986108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conlan AR, et al. Crystal structure of Miner1: The redox-active 2Fe-2S protein causative in Wolfram Syndrome 2. J Mol Biol. 2009;392(1):143–153. doi: 10.1016/j.jmb.2009.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paddock ML, et al. MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc Natl Acad Sci USA. 2007;104(36):14342–14347. doi: 10.1073/pnas.0707189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenke R, et al. Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25(5):621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozakov D, et al. Structural conservation of druggable hot spots in protein-protein interfaces. Proc Natl Acad Sci USA. 2011;108(33):13528–13533. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamir S, et al. Integrated strategy reveals the protein interface between cancer targets Bcl-2 and NAF-1. Proc Natl Acad Sci USA. 2014;111(14):5177–5182. doi: 10.1073/pnas.1403770111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salem AF, Whitaker-Menezes D, Howell A, Sotgia F, Lisanti MP. Mitochondrial biogenesis in epithelial cancer cells promotes breast cancer tumor growth and confers autophagy resistance. Cell Cycle. 2012;11(22):4174–4180. doi: 10.4161/cc.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamir S, et al. A point mutation in the [2Fe-2S] cluster binding region of the NAF-1 protein (H114C) dramatically hinders the cluster donor properties. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 6):1572–1578. doi: 10.1107/S1399004714005458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sohn YS, Mitterstiller AM, Breuer W, Weiss G, Cabantchik ZI. Rescuing iron-overloaded macrophages by conservative relocation of the accumulated metal. Br J Pharmacol. 2011;164(2b):406–418. doi: 10.1111/j.1476-5381.2010.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.