Abstract
A major goal of developmental biology is to explain the emergence of pattern in cell layers, tissues and organs. Developmental biologists now accept that reaction diffusion-based mechanisms are broadly employed in developing organisms to direct pattern formation. Here we briefly consider these mechanisms and then apply some of the concepts derived from them to several processes that occur in single cells: wound repair, yeast budding, and cytokinesis. Two conclusions emerge from this analysis: first, there is considerable overlap at the level of general mechanisms between developmental and single cell pattern formation; second, dynamic structures based on the actin cytoskeleton may be far more ordered than is generally recognized.
INTRODUCTION
The enduring focus of developmental biology is the reproducible emergence of organized form out of an apparently formless substrate. In contrast, cell biology tends to consider cells as organized containers populated by persistent machines that accomplish, more or less at steady state, their appointed tasks. In fact, however, nearly all single cells repeatedly undergo transient departures from steady state in a predictable way, and cell biologists increasingly recognize that changes in cell state are often spatially, as well as temporally, patterned. The assembly of the cytokinetic apparatus from a stripe of Rho activity is a pattern formation event just as is the development of an insect segment from a stripe of even-skipped expression (Fig. 1). Likewise, the formation of segregated cytoskeletal structures around single cell wounds or at the nascent bud of S. cerevisiae represent pattern formation.
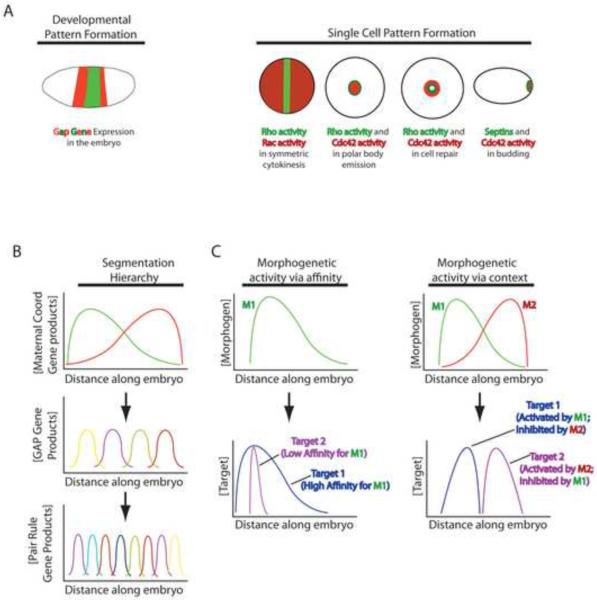
Figure 1.
Basic mechanisms underlying developmental and single cell pattern formation. A. Left: schematic showing stripe-like expression of different GAP genes in a Drosophila embryo. Each stripe will give rise to subsidiary stripes of gene expression and will ultimately direct the formation of a particular segment of the animal. Right: schematic showing examples of single cell pattern formation. For cytokinesis and wound healing, Rho activity zones shown in green and Rac or Cdc42 activity zones shown in red; each zone will specify a different part of the F-actin and myosin-2 arrays that form during cytokinesis and wound repair. For budding septins are shown in green and Cdc42 patch in red; each directs formation of a different region of the incipient bud. B. Stylized representation of the gene expression hierarchy in Drosophila segmentation. Each line represents the gradient of distribution of a particular gene product along the anterior (left)-posterior (right) axis of the embryo. Thus, the maternal coordinate gene product represented by the green line is in a gradient with its peak concentration near the anterior pole of the embryo. The maternal effect gene products form long range gradients that control the expression of the GAP genes, which are expressed in shorter range gradients and which control the expression patterns of the pair rule genes. Ultimately, the hierarchy results in the formation of distinct segments along the anterior-posterior axis of the embryo. C. Schematic representing two proposed mechanisms by which gradients of morphogens act in a qualitative manner. The red and green lines (M1 and M2) represent morphogen gradients; the blue and purple lines (target 1 and target 2) represent targets of the morphogen gradients. In morphogenetic activity via affinity, the low affinity target of M1 is only activated where the concentration of M1 is highest, while the high affinity target of M1 is activated proportionally along the entire M1 gradient. In morphogenetic activity via context, two overlapping morphogen gradients define the activity of targets. The activity of Target 1, which is activated by M1 and inhibited by M2 is confined to a narrow region within the M1 gradient while the activity of Target 2, which is activated by M2 and inhibited by M1 is confined to a narrow region within the M2 peak.
There are obvious differences between pattern formation in developing tissues and in single cells. Much developmental pattern formation is controlled by spatial differences in transcription compartmentalized within cell membranes, a strategy obviously impossible in a single cell. Intracellular signaling, meanwhile, often makes heavy use of cytoskeletally-mediated advection or convection, which do not feature prominently in developing tissues. Developmental pattern formation is largely predetermined (in animals, at any rate), such that frogs keep developing from frog eggs and flies result from fly eggs, while intracellular structures such as the wound array can – and must be able to – form anywhere within the cell in response to unanticipated stimuli. And, of course, developmental pattern formation is typically far slower, requiring hours, days or more to unfold, rather than seconds or minutes. But these are differences of implementation, not design, and it seems likely that good, robust patterning mechanisms on both scales might have a lot in common. We therefore think it might be useful to view these intracellular processes in roughly the same way that developmental biologists investigate morphogenetic fields in embryos.
CONTROL OF DEVELOPMENTAL PATTERN FORMATION BY MORPHOGEN GRADIENTS
Developing systems employ several general mechanisms for pattern formation, but conceptually the most important are “reaction diffusion” mechanisms, in which spatial patterns are created by reactions amongst agents that vary in diffusivity. The credit for this idea goes to Turing who showed that complex patterns could spontaneously arise from simple chemical reactions among factors he referred to as “morphogens” (1; Box 1). In Turing's formulation, a reaction produces an activator that stimulates its own production over a short length scale. The activator also stimulates the production of an inhibitor that counteracts the activator over a longer length scale. The difference in scales at which these two hypothetical factors work was assumed to result from differences in their diffusivity (Box 1). As anticipated by Turing, it is now clear that much of developmental pattern formation results from differential activation and inhibition of transcription and translation by diffusible factors that operate on various length scales. While the details don't necessarily conform to the mechanisms proposed by Turing, this is hardly surprising, as his work preceded the discovery of transcription, transcription factors, and translation by many years.
The best understood system of reaction diffusion-based pattern formation is segmentation of the Drosophila embryo – ironically, a largely intracellular process – wherein a hierarchy of gradients progressively regionalize the anterior-posterior axis (Fig 1; 2). At the top of the hierarchy are the products of the maternal coordinate genes bicoid and nanos, which are deposited at opposite poles as mRNA and whose protein products form shallow, opposite gradients, with Bicoid concentrated in the anterior and Nanos in the posterior. Bicoid activates transcription of several genes and represses translation of Caudal. Caudal is another maternal coordinate gene and as a result of its repression by Bicoid, its product ends up forming a posterior gradient. Similarly, Nanos negatively regulates translation of the maternal coordinate gene hunchback, whose product therefore accumulates in an anterior gradient. The overlapping gradients of maternal coordinate gene products regulate zygotic expression of the gap gene products, which are thereby expressed in broad bands. The gap gene products, which are all transcription factors, in turn regulate expression of the pair rule genes which become expressed in narrower stripes, and these stripes of expression define even narrower stripes of segment polarity gene expression (3).
While the details of these events are not important for this discussion, several general principles are. First, at each level of the hierarchy, spatial order increases as the characteristic length scale decreases: the gap gene products form more and sharper gradients than the maternal coordinate gene products, the pair rule gene products form more and sharper gradients than gap genes, and so forth (Fig. 1; 2,3). Second, a given participant can act at more than one tier of the hierarchy and may both positively and negatively affect other participants (4). Third, overlapping stimulatory (e.g. Bicoid) and inhibitory (e.g. Hunchback) gradients work together to sculpt target patterns (**5). Fourth, positive feedback is important for sharpening gradients – Hunchback, for example positively regulates its own transcription (6). Fifth, negative cross-talk sharpens and positions boundaries between gradients. For example, loss of Krüppel results in expansion of giant expression toward the region that would normally be occupied by Krüppel (7).
Bicoid and Nanos are morphogens as originally envisioned: they diffuse and react with specific targets – gene regulatory elements (mostly) for Bicoid, mRNA for Nanos – and provide positional information by modulating target activity. But they are also morphogens as the term has come to be accepted in the last few decades: they are deposited in gradients, and they have the curious property of exerting qualitatively different effects at different concentrations (3, 8, 9). Thus, rather than Bicoid simply promoting more expression of a particular gap gene at the top if its gradient and less in the middle of its gradient, it promotes the expression of different gap genes in these areas. This is a critical point: gradients of signaling molecules in single cells are typically assumed to recruit the same set of target proteins along their entire lengths. Thus, during cytokinesis for example, the top of the Rho gradient would be envisioned to have high concentrations of every available Rho target, while the middle would simply have lower concentrations of all those same targets. The analogy to classical morphogens suggests, however, that such intracellular gradients might have more complicated outputs.
Morphogens generate qualitative differences along their gradients through two idealized principles: differential affinity or local context. The affinity hypothesis proposes that distinct targets responses stem from differences in the affinity or sensitivity of those targets for the morphogens (Fig. 1). For example, it has been proposed that target genes whose upstream regulatory elements have high affinity for Bicoid will be activated at greater distances from the peak of the gradient, whereas those with the lowest affinity are activated only within the peak of the gradient (10). In the “context” model, qualitative differences are achieved by the action of two or more overlapping gradients that modulate the same target (Fig. 1; 3,**5). Real instances of morphogen gradient interpretation likely employ both idealizations to varying extents.
SINGLE CELL PATTERN FORMATION
To be meaningfully analogous to reaction diffusion-based patterning of the Drosophila embryo, single cell pattern formation would require gradients that form and act on the appropriate space and time scales and whose constituents have morphogen-like properties. The Rho GTPases – Rho, Rac, and Cdc42 – clearly satisfy the first condition in that they form gradients (also known as “zones”) associated with and required for wound repair, yeast polarization and cytokinesis (11). This is notable in its own right, because although these signaling molecules were originally described as cell state switches which, when activated, coordinate multiple signaling pathways to cause cells to adopt distinct, more or less global, states of cytoskeletal organization, it is increasingly clear that in many instances their essential function is to create or maintain spatial differentiation of the cytoskeleton within cells.
Whether the Rho GTPases satisfy the second condition is harder to assess but the following observations are consistent with them acting as intracellular morphogens: first, they have multiple targets that differ in affinity for the active GTPases. Rho, for example, has at least 10 targets (12) that vary by more than an order of magnitude in their affinity for active Rho (13). Affinity differences among effectors, possibly magnified by distinct effector abundances and varying diffusivities, provide the raw ingredients for elaborate morphogen gradient interpretation, but do not guarantee it (trivially: what if there were an overwhelming amount of the putative morphogen relative to effectors?). To the best of our knowledge, it is not known whether Rho GTPase targets are expressed at levels appropriate for them to respond in a morphogen-like manner, but there is evidence that multiple effectors for a given GTPase can be active in the cell at the same time (**14).
In addition, many GTPase targets are modulated by other agents that form intracellular gradients, such as signaling lipids or intracellular free calcium. For example, PKN is activated by both Rho and signaling lipids (15,16) while N-WASP requires both active Cdc42 and PIP2 for activation (17,18). Further, the Rho GTPases not only engage in cross talk with each other (19), they have the potential to act on other gradients and vice versa. Rho, for example, directly inhibits diacylglycerol kinase (20) and activates phospholipase C epsilon (21); Rac and Cdc42 activate PI 4,5 kinases (22); while myosin-2 interacts with and localizes Rho GEFs (23). Finally, it is becoming clear that the distal targets of the Rho GTPases have considerable potential for self-organization. For example, myosin-2 and cofilin, each a downstream target of Rho GTPases, compete for binding sites on F-actin (**24).
Clearly, then, Rho GTPases have the potential to act in a morphogen-like manner. Many of the same arguments hold for calcium and lipids, and other GTPases such as Ran (25) and the Arfs (26). But do they do so in cells? And are related features of developmental pattern formation such as gradient hierarchies evident in single cells?
Single cell wound repair
The response of frog oocytes to a membrane breach provides our first example of single cell pattern formation. Wounding these cells triggers rapid (15–20s) local activation of Rho and Cdc42 (27). Following activation, Rho and Cdc42 sort into complementary activity zones with active Cdc42 circumscribing active Rho (Fig. 1). The complementary zones direct the formation of a ring-like array of F-actin and myosin-2, with the bulk of the myosin-2 concentrated in the Rho zone and the bulk of the dynamic actin concentrated in the Cdc42 zone (27,28). After assembling, the actomyosin array closes inward, in concert with the GTPase zones. While the cytoskeletal response to cell wounding has been most extensively studied in frog oocytes, recent work indicates that its basic features are shared by budding yeast (29), Drosophila (30), and mammals (31).
The manner in which this process unfolds has striking parallels to the Drosophila segmentation cascade. Within seconds, wounding triggers formation of a relatively broad calcium gradient around the wound which is necessary for activation of Rho and Cdc42 (Fig. 2; 27,32). Initially, the gradients of active Rho and Cdc42 are shallow, and overlap each other almost completely. Subsequently, however, they become steeper, more intense, and more segregated until they form the characteristic concentric pattern at about 90 s post wounding (27). How the calcium gradient is coupled to the initial activation of Rho and Cdc42 is unknown, but at least part of the pattern resolution amongst the active GTPases results from participation of the dual GEF-GAP Abr (**33). In vitro Abr acts as a GEF for (i.e. activates) Rho, Rac and Cdc42 but as a GAP (i.e. inhibits) only for Rac and Cdc42 (34). Abr is recruited to the Rho zone by active Rho, where it establishes a positive feedback loop with active Rho via its GEF activity and suppresses Cdc42 via its GAP activity (**33). A mathematical model shows that Abr's involvement explains the basic features of the Rho GTPase response and makes accurate, nonintuitive predictions about pattern formation, as long as the model includes nonlinear positive feedback not only for the Rho-Abr interaction but also for Cdc42 activation (**35). The nature of the feedback to Cdc42 is unknown, but based on other systems, a loop running from Cdc42 to dynamic actin seems plausible (**36).
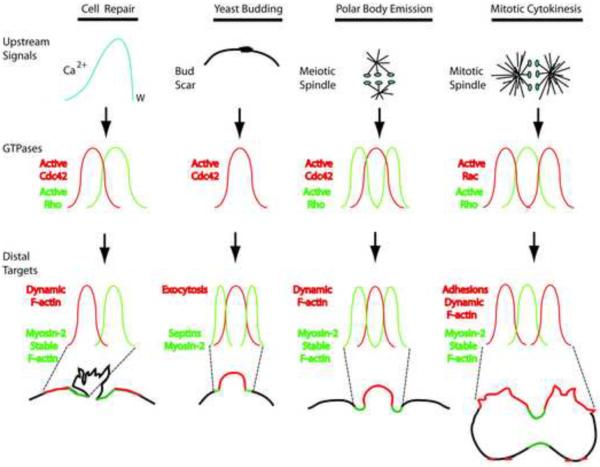
Figure 2.
Hierarchies of signaling gradients in single cell pattern formation. In single cell repair, wounding triggers formation of a long range gradient of elevated intracellular free calcium (blue line), with the peak concentration of calcium at the edge of the wound (w). This gradient is subsequently transduced into narrower (relative to the calcium gradient) gradients (zones) of active Rho (green line) and active Cdc42 (red line). These gradients ultimately direct the formation of gradients of distal cytoskeletal targets with different positions around wounds: stable F-actin and myosin-2 (green line) and dynamic F-actin (red line). The ring-like array of F-actin and myosin-2 closes inward, pinching off the damaged material and membrane of the wound surface (irregular black line).
In yeast budding, the bud scar direct formation of a gradient of Cdc42 activity (red line) which is transduced into gradients of exocytosis (red line) and septin accumulation (green line); the septins help direct formation of the cytokinetic apparatus which contains, among other things, myosin-2. The region of high exocytotic activity grows outward; the cytokinetic apparatus pinches inward.
In both meiotic (polar body emission) and mitotic cytokinesis, the spindle provides the initial cues (likely in the form of gradients of activity of Ect2 and other GEF and GAPs) which lead to activation of GTPases in downstream gradients. In the case of polar body emission, a ring-like gradient of active Rho (green line) surrounds a disc-like gradient of active Cdc42 (red line); in the case of mitotic cytokinesis, a stripe-like gradient of active Rho (green line) is flanked by stripe-like gradients of active Rac red line). In both cases the Rho zones direct formation gradients of myosin-2 and relatively stable F-actin (the cytokinetic apparatus; green line); these pinch inward. In polar body emission, the Cdc42 zone directs formation of a gradient of highly dynamic F-actin (red line); this evaginates to become the surface of the nascent polar body. In mitotic cytokinesis, the Rac zone directs formation of a gradient of adhesions and very dynamic actin (red line).
Regardless of the precise details, the features of this system parallel those of GAP gene expression control with initiation depending on a relatively long range upstream gradient that is ultimately converted into shorter-range downstream gradients as a result of autocatalysis, with downstream gradient segregation enhancement occurring through mutually suppressive cross-talk. Further similarities are apparent from the dynamics of GTPase turnover: the Rho zone is apparently subdivided, such that Rho is preferentially inactivated at the trailing edge of the zone, indicating that yes, different domains within the Rho zone may differ qualitatively in Rho target population (**37).
Yeast Budding
Specification of the new bud during the yeast cell division cycle has proven an enormously powerful and informative system for understanding of how Rho GTPases are controlled and in turn control specialization of different regions of the plasma membrane (38,39). In contrast to the wound response, which can be elicited anywhere on the cell surface, the yeast bud pattern is normally initiated around an established spatial cue: next to the bud scar, the site of the last round of budding. This results from the deposition of developmental information in the form the protein Rsr1p which is necessary for concentrating Bem1p near the bud scar. Bem1p is a scaffold protein that binds active Cdc42, a Cdc42 target (Pak) and a Cdc42 GEF, Cdc24 (39). These interactions serve as the basis of a positive feedback loop that drives formation of a small (~1 um), disc-like gradient of active Cdc42, the key event of budding (39). Localization of Rga1, a Cdc42 GAP, to the bud scar ensures that the Cdc42 gradient is centered next to, not directly at,the previous division site (40). Two aspects of this process are of particular interest here: the formation of the gradient, and its subsequent encirclement by septins.
While the site of the gradient source is normally ensured by the localization of Rsr1p, the generation of a single gradient of Cdc42 is not. Rather, it is the outcome of a competitive process in which multiple local clusters of Cdc42 activity vie with each other for cortical supremacy. This was first anticipated in a modeling study that discovered that Cdc42 clustering and gradient formation is a Turing-like process with Cdc42 serving as both the substrate and the activator (**41). In the model, clusters of Cdc42 form spontaneously at the plasma membrane and compete with each other for soluble Bem1p, with one winner eventually emerging and forming a stable gradient at the plasma membrane. A modified form of this model was confirmed empirically (**42), and subsequently further modified to incorporate negative feedback from GAPs, which renders the process less sensitive to variations in the concentrations of the various participants (*43).
Once established, the Cdc42 gradient defines the site where budding occurs and thus where the daughter cell forms (Fig. 2). An essential step in this process is the formation of a ring of septin filaments around the Cdc42 gradient (Fig. 2). This ring acts as a diffusion barrier (44), which may help corral the active Cdc42, and ultimately defines the site where the cytokinetic apparatus will assemble.
How do the septins end up at the outer edges of the Cdc42 gradient? A fascinating and plausible model was developed in a recent study that combined modeling and experimental work (**45): the septins are recruited to the Cdc42 gradient as a result of their interaction with Gic1 and Gic2, Cdc42 targets. The binding of Cdc42 to the Gics apparently causes release of the septins which thus accumulate as a diffuse cap overlapping the Cdc42 gradient. Modeling and direct tests showed that polarized exocytosis hollows out the cap of septins, presumably by driving the relatively insoluble septin filaments away from the sight of membrane insertion. Polarized exocytosis is targeted to the center of the Cdc42 gradient by recruitment of the exocyst complex, which is a target of active Cdc42 (Fig. 2). In other words, the gradient ends up with qualitative differences in target protein distribution – the exocyst in the center and septins at the periphery – with some help from exocytosis.
Cytokinesis
Cytokinesis in animal cells is controlled by the spindle, which regulates the distribution of active Rho GTPases at the plasma membrane. It thus differs from both wound repair and budding, where the specification information (wound, bud scar) is already present at the plasma membrane. In mitotic cells the spindle directs formation of a stripe-like zone of Rho activity at the equator; in meiotic cells it directs formation of a ring-like zone of Rho activity at the animal pole (46). In mitotic cytokinesis in at least some cell types the Rho zone is flanked by gradients of high Rac activity (**14), while in meiotic cytokinesis the Rho zone encircles a patch of active Cdc42 (47). The Rho zones in both cases direct formation of the F-actin and myosin-2 rich cytokinetic apparatus. The Rac zones in mitotic cells direct local adhesion, whereas the Cdc42 zone in meiotic cells directs formation of a dynamic actin cap which will become the surface of the evaginating polar body (Fig. 2; 48).
The situation parallels wound repair, in that complementary gradients of GTPase activity direct recruitment of different targets to distinct regions of the cell surface. The distinctive regions thus created are not merely decorative: loss of Rho activity prevents furrow ingression while failure to exclude Rac from the Rho zone results in formation of ectopic adhesions which retard cytokinetic progress (**16). Similarly, in meiotic cytokinesis, the complementary Rho and Cdc42 zones make distinct functional contributions to polar body emission: the Rho zone directs furrow ingression, the Cdc42 zone promotes evagination (Fig. 2; 48).
How are the cytokinetic zones generated and segregated? Rho activation is controlled by the RhoGEF Ect2, which localizes to both the central spindle and the equatorial cortex (49, 50*). Rho activity is maintained within its zone by at least three processes: rapid flux through the GTPase cycle (11,51; 52**), suppression of Rho activity outside the zone by astral microtubules (52**, 53) and, potentially, negative cross talk with Rac (**14). How the Rac and Cdc42 zones are generated is unclear although Ect2 is also a GEF for Rac and Cdc42 (54) and it has been suggested that astral microtubules may stimulate Rac at the poles (55). Rac activity may be excluded from the Rho zone by the action of MgcRacGAP, a GAP for the Rho class GTPases (56). While this idea is controversial (see 57), it received direct support from the demonstration that expression of a GAP-inactive version of MgcRacGAP results in Rac activation at the cell equator (**14). MgcRacGAP is an Ect2 binding partner that localizes to the central spindle and to the ends of astral microtubules and which is essential for cytokinesis (50, 58,59). If this model is correct, it indicates that cytokinesis, like wound healing, employes a dual GEF-GAP to generate complementary Rho GTPase zones, but with the distinction that the GEF and GAP activities (MgcRacGAP and Ect2) are in separate polypeptides that associate transiently, in contrast to Abr, in which they are integrated into a single polypeptide.
Conclusions
We highlighted the potential for Rho-family GTPases to function morphogenetically. Other agents that appear in intracellular gradients associated with the transient cytoskeletal arrays considered here– calcium (60, 61), signaling lipids (62*,63), and other GTPases such as Ran and Arfs – likely contribute to patterning the cytoskeleton. Mechanical gradients are also associated with cytokinesis (64–67), and are likely to be important during wound healing and budding, and potentially shape signaling to the cytoskeleton as well (68). As in the Drosophila segmentation cascade, one might imagine that collaboration between multiple gradients, each with different length scales, physical properties, and targets, could impart a high degree of order on transient F-actin based assemblies. But is such rich differentiation within the cell a common phenomenon, and if so, is it functional? With respect to the former, it is often the case that one must be looking for something in order to see it, and transient cytoskeletal arrays pose serious challenges for imaging because they are typically small, fast, and protean. Quantitative comparisons of effector distributions, quantities, and response to perturbation would be a valuable starting point to distinguish patterning from mere co-activity.
With respect to the second question – what for? – we suggest contrasting pattern formation with image formation. Opposite to pattern forming mechanisms, in which the information density progressively increases down the chain of events, image formation always degrades the original. Consider the way the specification of the cleavage plane is conventionally described: As part of centralspindlin, MgcRacGAP's distribution at the spindle midzone approximates the appropriate cleavage plane; because MgcRacGAP activates Ect2, so GEF activity in turn approximates the position of MgcRacGAP; Ect2, perhaps after diffusing around a bit, activates Rho at the cell surface; whereupon Rho promotes actin assembly and myosin recruitment through various effectors. At each step in this description – in which an image of the spindle mid-plane is imprinted on the cell cortex – positional information is inevitably lost. The metaphase plate is one of the most definite, finely-localized cues that a cell ever confronts, and yet simply transmitting its position necessarily blurs the picture. In optics, the clever contrivance of interference effects has often been used to recover information that is otherwise lost; likewise, pattern formation mechanisms are the biochemist's means to convert weak or transient cues into ordered and coupled differentiated states. The creation of such detail by pattern-forming mechanisms seems likely to endow transient contractile arrays with important systems-level traits like scalability, adaptation, and robustness to perturbation.
Box 1 Turing/reaction diffusion mechanisms and developmental pattern formation.
In “The chemical basis of morphogenesis” (1952. Phil. Trans. Royal Soc. 237:37–72), Turing suggested that interactions of diffusible molecules (“morphogens”) that control specific chemical reactions could form the basis of developmental morphogenesis. To illustrate this point, Turing discussed a simple, analytically-tractable set of chemical reactions in which one of the products – the activator (A) – promoted its own production but also stimulated the production of an inhibitor (I), which in turn suppressed production of the activator. Thus, the reaction has intrinsic positive and negative feedback. Turing showed that to produce patterns, as represented by different local concentrations of the activator, he needed only to assume that A and I differed significantly with respect to diffusivity. In particular he showed that if A dispersed more slowly than I, strikingly varied patterns could be produced even though the starting conditions of the reaction are homogeneous. The patterns produced included both dynamic and standing waves, and showed dramatic variation in response to changes in the rates of A or I production, degradation and diffusion.
Turing's legacy to developmental biology has been complicated by the fact that considerable effort was expended by others to show that developmental pattern formation arises in embryos in the absence of pre-existing cues and as a result of a simple activator-inhibitor system. In fact, it is now apparent that embryos come with a liberal sprinkling of pre-existing developmental information, and that the real system for making stripes in a fly embryo is vastly more elaborate than a single activator and inhibitor. This has led many to dismiss Turing's work as largely irrelevant to developmental biology except in one or two specialized instances. However, it is important to keep in mind the historical context of Turing's paper which, a decade before the lac operon, demonstrated that even a simple set of chemical reactions could be profoundly creative. Further, with some modifications, his basic vision was remarkably prescient: development is in fact controlled by diffusible factors, many of which operate autocatalytically, and whose mode of action is critically dependent on differences in diffusibility. Moreover, he also recognized that developmental information might be laid down in latent form, and subsequently interpreted or elaborated by reaction diffusion systems.
The immense diversity of developmental pattern formation obscures the fact that there are only a few distinct categories of pattern-forming mechanism. Broadly, there are three alternatives to reaction-diffusion. First, cells in embryos can sort determinants such that descendants possess distinct sets of information. Well-known examples range from simple (P granules) to elaborate (mRNA in snails (Lambert JD, Nagy LM. Nature. 2002 420(6916):682–6.) and chromatin diminution in Ascaris (Boveri, T. Anat. Anz. 1887 2:688–693.)). From a conceptual point of view, sorting of cytoplasmic determinants is almost a null hypothesis – the cells become different by starting out different – but it is by now clear that even the most regulative of embryos use asymmetric inheritance of determinants to differentiate cells. Second, cells within fields could conduct signal relays. Textbook examples include specification of photoreceptor fates within Drosophila ommatidia, or selection of neural from epidermal cells in the Drosophila neuroectoderm. Because in such cases the signaling is juxtacrine, with cells signaling directly to immediate neighbors, diffusion and concentration gradients are largely irrelevant. This makes signal relays the natural antithesis to reaction-diffusion mechanisms. Finally, during induction, one population of cells physically brings information to another, and thereby elicits changes in fate. The original example is induction of the lens within the epidermis of a tadpole by the optic cup, which grows out from the infolded neural plate. This is conceptually distinct from the rest because of the key role of physical shape change in bringing distinct informational states together, and its intracellular analog is not clear.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.The chemical basis of morphogenesis. Turing, AM. Phil Trans Royal Soc. 1952;237:37–72. [Google Scholar]
- 2.Generating patterns from fields of cells. Examples from Drosophila segmentation. Sanson B. EMBO Rep. 2001 Dec;2(12):1083–8. doi: 10.1093/embo-reports/kve255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The gap gene network. Jaeger J. Cell Mol Life Sci. 2011 Jan;68(2):243–74. doi: 10.1007/s00018-010-0536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996 Feb 22;379(6567):694–9. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Xu Z, Mei C, Yu D, Small S. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell. 2012 Apr 27;149(3):618–29. doi: 10.1016/j.cell.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; **A comprehensive analysis of bicoid regulated genes shows that in Drosophila, boundaries of expression in the anterior are controlled by the collaboration of bicoid with gradients of different transcription repressors. The results support the “context” model of morphogen function and provide a serious challenge to the idea that bicoid alone defines expression boundaries in a threshold dependent manner.
- 6.Wimmer EA, Carleton A, Harjes P, Turner T, Desplan C. Bicoid-independent formation of thoracic segments in Drosophila. Science. 2000 Mar 31;287(5462):2476–9. doi: 10.1126/science.287.5462.2476. [DOI] [PubMed] [Google Scholar]
- 7.Kraut R, Levine M. Mutually repressive interactions between the gap genes giant and Krüppel define middle body regions of the Drosophila embryo. Development. 1991 Feb;111(2):611–21. doi: 10.1242/dev.111.2.611. [DOI] [PubMed] [Google Scholar]
- 8.Driever W, Nüsslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988 Jul 1;54(1):95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- 9.Wharton RP, Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell. 1991 Nov 29;67(5):955–67. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- 10.Driever W, Nüsslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988 Jul 1;54(1):95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- 11.Bement WM, Miller AL, von Dassow G. Rho GTPase activity zones and transient contractile arrays. Bioessays. 2006 Oct;28(10):983–93. doi: 10.1002/bies.20477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J. 2000 Jun 1;348(Pt 2):241–55. [PMC free article] [PubMed] [Google Scholar]
- 13.Blumenstein L, Ahmadian MR. Models of the cooperative mechanism for Rho effector recognition: implications for RhoA-mediated effector activation. J Biol Chem. 2004 Dec 17;279(51):53419–26. doi: 10.1074/jbc.M409551200. [DOI] [PubMed] [Google Scholar]
- 14.Bastos RN, Penate X, Bates M, Hammond D, Barr FA. CYK4 inhibits Rac1-dependent PAK1 and ARHGEF7 effector pathways during cytokinesis. J Cell Biol. 2012 Sep 3;198(5):865–80. doi: 10.1083/jcb.201204107. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This study provides clear evidence for zones of Rac activity that complement the cytokinetic Rho zone, show that exclusion of active Rac from the Rho zone is due to the GAP activity of MgcRacGAP, and also indicates that Rho, Rac and Cdc42 each engage multiple effectors during cytokinesis.
- 15.Amano M, Mukai H, Ono Y, Chihara K, Matsui T, Hamajima Y, Okawa K, Iwamatsu A, Kaibuchi K. Identification of a putative target for Rho as the serine-threonine kinase protein kinase N. Science. 1996 Feb 2;271(5249):648–50. doi: 10.1126/science.271.5249.648. [DOI] [PubMed] [Google Scholar]
- 16.Yoshinaga C, Mukai H, Toshimori M, Miyamoto M, Ono Y. Mutational analysis of the regulatory mechanism of PKN: the regulatory region of PKN contains an arachidonic acid-sensitive autoinhibitory domain. J Biochem. 1999 Sep;126(3):475–84. doi: 10.1093/oxfordjournals.jbchem.a022476. [DOI] [PubMed] [Google Scholar]
- 17.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999 Apr 16;97(2):221–31. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 18.Higgs HN, Pollard TD. Activation by Cdc42 and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J Cell Biol. 2000 Sep 18;150(6):1311–20. doi: 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guilluy C, Garcia-Mata R, Burridge K. Rho protein crosstalk: another social network? Trends Cell Biol. 2011 Dec;21(12):718–26. doi: 10.1016/j.tcb.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houssa B, de Widt J, Kranenburg O, Moolenaar WH, van Blitterswijk WJ. Diacylglycerol kinase theta binds to and is negatively regulated by active RhoA. J Biol Chem. 1999 Mar 12;274(11):6820–2. doi: 10.1074/jbc.274.11.6820. [DOI] [PubMed] [Google Scholar]
- 21.Wing MR, Snyder JT, Sondek J, Harden TK. Direct activation of phospholipase C-epsilon by Rho. J Biol Chem. 2003 Oct 17;278(42):41253–8. doi: 10.1074/jbc.M306904200. [DOI] [PubMed] [Google Scholar]
- 22.Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, Diefenbacher M, Stamp G, Downward J. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell. 2013 May 23;153(5):1050–63. doi: 10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee CS, Choi CK, Shin EY, Schwartz MA, Kim EG. Myosin II directly binds and inhibits Dbl family guanine nucleotide exchange factors: a possible link to Rho family GTPases. J Cell Biol. 2010 Aug 23;190(4):663–74. doi: 10.1083/jcb.201003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **24.Wiggan O, Shaw AE, DeLuca JG, Bamburg JR. ADF/cofilin regulates actomyosin assembly through competitive inhibition of myosin II binding to F-actin. Dev Cell. 2012 Mar 13;22(3):530–43. doi: 10.1016/j.devcel.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows nicely that cofilin competes with myosin-2 for binding to F-actin, raising the possibility that this may be one reason that myosin-2 is often excluded from regions of highly dynamic actin filaments.
- 25.Athale CA, Dinarina A, Mora-Coral M, Pugieux C, Nedelec F, Karsenti E. Regulation of microtubule dynamics by reaction cascades around chromosomes. Science. 2008 Nov 21;322(5905):1243–7. doi: 10.1126/science.1161820. [DOI] [PubMed] [Google Scholar]
- 26.Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–23. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 27.Benink HA, Bement WM. Concentric zones of active RhoA and Cdc42 around single cell wounds. J Cell Biol. 2005 Jan 31;168(3):429–39. doi: 10.1083/jcb.200411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandato CA, Bement WM. Contraction and polymerization cooperate to assemble and close actomyosin rings around Xenopus oocyte wounds. J Cell Biol. 2001 Aug 20;154(4):785–97. doi: 10.1083/jcb.200103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kono K, Saeki Y, Yoshida S, Tanaka K, Pellman D. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell. 2012 Jul 6;150(1):151–64. doi: 10.1016/j.cell.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 30.Abreu-Blanco MT, Verboon JM, Parkhurst SM. Cell wound repair in Drosophila occurs through three distinct phases of membrane and cytoskeletal remodeling. J Cell Biol. 2011 May 2;193(3):455–64. doi: 10.1083/jcb.201011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin P, Zhu H, Cai C, Wang X, Cao C, Xiao R, Pan Z, Weisleder N, Takeshima H, Ma J. Nonmuscle myosin IIA facilitates vesicle trafficking for MG53-mediated cell membrane repair. FASEB J. 2012 May;26(5):1875–83. doi: 10.1096/fj.11-188599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark AG, Miller AL, Vaughan E, Yu HY, Penkert R, Bement WM. Integration of single and multicellular wound responses. Curr Biol. 2009 Aug 25;19(16):1389–95. doi: 10.1016/j.cub.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **33.Vaughan EM, Miller AL, Yu HY, Bement WM. Control of local Rho GTPase crosstalk by Abr. Curr Biol. 2011 Feb 22;21(4):270–7. doi: 10.1016/j.cub.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; The dual GEF-GAP Abr is identified as an agent of both autoamplification (for Rho) and negative cross talk (from Rho to Cdc42) during oocyte wound healing. It is shown that Manipulation of Abr levels, GAP function, or GEF function expands, eliminates or blends the Rho and Cdc42 zones.
- 34.Chuang TH, Xu X, Kaartinen V, Heisterkamp N, Groffen J, Bokoch GM. Abr and Bcr are multifunctional regulators of the Rho GTP-binding protein family. Proc Natl Acad Sci U S A. 1995 Oct 24;92(22):10282–6. doi: 10.1073/pnas.92.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Simon CM, Vaughan EM, Bement WM, Edelstein-Keshet L. Pattern formation of Rho GTPases in single cell wound healing. Mol Biol Cell. 2013 Feb;24(3):421–32. doi: 10.1091/mbc.E12-08-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]; A combination of modeling and experiment shows that a mathematical model based on the known in vitro and in vivo features of Abr, Rho and Cdc42 can explain the basic events of the Rho and Cdc42 patterning during the wound response if nonlinearity is incorporated into the activation of Rho and Cdc42. The model makes accurate, nonintuitive predictions about the consequences of multiple wounds spaced different distances apart.
- 36**.Orchard RC, Kittisopikul M, Altschuler SJ, Wu LF, Süel GM, Alto NM. Identification of F-actin as the dynamic hub in a microbial-induced GTPase polarity circuit. Cell. 2012 Feb 17;148(4):803–15. doi: 10.1016/j.cell.2011.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]; An interaction between a Cdc42 GEF from a bacterial pathogen and host cell F-actin is discovered and exploited to show that a relatively simple positive feedback loop between F-actin and the GEF is sufficient to drive stable polarization of active Cdc42 and its targets.
- 37**.Burkel BM, Benink HA, Vaughan EM, von Dassow G, Bement WM. A Rho GTPase signal treadmill backs a contractile array. Dev Cell. 2012 Aug 14;23(2):384–96. doi: 10.1016/j.devcel.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]; The Rho and Cdc42 zones were found to close inward even as a result of differential turnover of Rho and Cdc42 within distinct regions of the zones; thus the zones are in fact inward moving waves that direct inward moving waves of actin and myosin assembly.
- 38.Slaughter BD, Smith SE, Li R. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb Perspect Biol. 2009 Sep;1(3):a003384. doi: 10.1101/cshperspect.a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bi E, Park HO. Cell polarization and cytokinesis in budding yeast. Genetics. 2012 Jun;191(2):347–87. doi: 10.1534/genetics.111.132886. doi: 10.1534/genetics.111.132886. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong Z, Gao XD, Howell AS, Bose I, Lew DJ, Bi E. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J Cell Biol. 2007 Dec 31;179(7):1375–84. doi: 10.1083/jcb.200705160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Goryachev AB, Pokhilko AV. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008 Apr 30;582(10):1437–43. doi: 10.1016/j.febslet.2008.03.029. [DOI] [PubMed] [Google Scholar]; A modeling study making the remarkable demonstration that stable GTPase polarity can be the consequence of a Turing-like mechanism in silico. The patch of active Cdc42 arises spontaneously following competition between smaller patches.
- 42**.Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, Lew DJ. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 2009 Nov 13;139(4):731–43. doi: 10.1016/j.cell.2009.10.024. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; A modified version of the model from 41 accounts for basic features of Cdc42 polarization during budding. The role of competition is confirmed by experiments in which the yeast are genetically modified to produce more than one patch of active Cdc42.
- 43.Howell AS, Jin M, Wu CF, Zyla TR, Elston TC, Lew DJ. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell. 2012 Apr 13;149(2):322–33. doi: 10.1016/j.cell.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; *A follow up to 39 in which the authors uncover and then demonstrate the importance of negative feedback in the yeast budding mechanism--it ensures that polarization is stable even in the face of major changes in the concentrations of the key polarization participants.
- 44.Barral Y, Mermall V, Mooseker MS, Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000 May;5(5):841–51. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- 45**.Okada S, Leda M, Hanna J, Savage NS, Bi E, Goryachev AB. Daughter cell identity emerges from the interplay of cdc42, septins, and exocytosis. Dev Cell. 2013 Jul 29;26(2):148–61. doi: 10.1016/j.devcel.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; A demonstration that a GTPase-dependent cell patterning event--formation of the septin ring around the perimeter of the active patch of Cdc42--can be explained as the outcome of collaboration between a gradient of Cdc42 activity, diffusion, and local exocytosis.
- 46.Bement WM, Benink HA, von Dassow G. A microtubule-dependent zone of active RhoA during cleavage plane specification. J Cell Biol. 2005 Jul 4;170(1):91–101. doi: 10.1083/jcb.200501131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma C, Benink HA, Cheng D, Montplaisir V, Wang L, Xi Y, Zheng PP, Bement WM, Liu XJ. Cdc42 activation couples spindle positioning to first polar body formation in oocyte maturation. Curr Biol. 2006 Jan 24;16(2):214–20. doi: 10.1016/j.cub.2005.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Ma C, Miller AL, Katbi HA, Bement WM, Liu XJ. Polar body emission requires a RhoA contractile ring and Cdc42-mediated membrane protrusion. Dev Cell. 2008 Sep;15(3):386–400. doi: 10.1016/j.devcel.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somers WG, Saint R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev Cell. 2003 Jan;4(1):29–39. doi: 10.1016/s1534-5807(02)00402-1. [DOI] [PubMed] [Google Scholar]
- 50.Su KC, Takaki T, Petronczki M. Targeting of the RhoGEF Ect2 to the equatorial membrane controls cleavage furrow formation during cytokinesis. Dev Cell. 2011 Dec 13;21(6):1104–15. doi: 10.1016/j.devcel.2011.11.003. [DOI] [PubMed] [Google Scholar]; *The first demonstration of localization of Ect2 to the equatorial plasma membrane before the onset of furrowing.
- 51.Miller AL, Bement WM. Regulation of cytokinesis by Rho GTPase flux. Nat Cell Biol. 2009 Jan;11(1):71–7. doi: 10.1038/ncb1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zanin E, Desai A, Poser I, Toyoda Y, Andree C, Moebius C, Bickle M, Conradt B, Piekny A, Oegema K. A Conserved RhoGAP Limits M Phase Contractility and Coordinates with Microtubule Asters to Confine RhoA during Cytokinesis. Dev Cell. 2013 Sep 4; doi: 10.1016/j.devcel.2013.08.005. doi:pii: S1534-5807(13)00474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; **In this study the authors identify a Rho GAP responsible for maintaining Rho GTPase flux (ie cycling) during cytokinesis. They find that depletion of this GAP results in formation of ectopic sites of presumptive Rho activation in regions outside the cell equator, while depletion of this GAP in combination with disruption of astral microtubules results in broadening of the region of presumptive Rho activation at the equator.
- 53.von Dassow G, Verbrugghe KJ, Miller AL, Sider JR, Bement WM. Action at a distance during cytokinesis. J Cell Biol. 2009 Dec 14;187(6):831–45. doi: 10.1083/jcb.200907090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999 Nov 29;147(5):921–8. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandato CA, Benink HA, Bement WM. Microtubule-actomyosin interactions in cortical flow and cytokinesis. Cell Motil Cytoskeleton. 2000 Feb;45(2):87–92. doi: 10.1002/(SICI)1097-0169(200002)45:2<87::AID-CM1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 56.Canman JC, Lewellyn L, Laband K, Smerdon SJ, Desai A, Bowerman B, Oegema K. Inhibition of Rac by the GAP activity of centralspindlin is essential for cytokinesis. Science. 2008 Dec 5;322(5907):1543–6. doi: 10.1126/science.1163086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loria A, Longhini KM, Glotzer M. The RhoGAP domain of CYK-4 has an essential role in RhoA activation. Curr Biol. 2012 Feb 7;22(3):213–9. doi: 10.1016/j.cub.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jantsch-Plunger V, Gönczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M. CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol. 2000 Jun 26;149(7):1391–404. doi: 10.1083/jcb.149.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishimura Y, Yonemura S. Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J Cell Sci. 2006 Jan 1;119(Pt 1):104–14. doi: 10.1242/jcs.02737. Epub 2005 Dec 13. [DOI] [PubMed] [Google Scholar]
- 60.Wong R, Hadjiyanni I, Wei HC, Polevoy G, McBride R, Sem KP, Brill JA. PIP2 hydrolysis and calcium release are required for cytokinesis in Drosophila spermatocytes. Curr Biol. 2005 Aug 9;15(15):1401–6. doi: 10.1016/j.cub.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 61.Webb SE, Lee KW, Karplus E, Miller AL. Localized calcium transients accompany furrow positioning, propagation, and deepening during the early cleavage period of zebrafish embryos. Dev Biol. 1997 Dec 1;192(1):78–92. doi: 10.1006/dbio.1997.8724. [DOI] [PubMed] [Google Scholar]
- 62.Das A, Slaughter BD, Unruh JR, Bradford WD, Alexander R, Rubinstein B, Li R. Flippase-mediated phospholipid asymmetry promotes fast Cdc42 recycling in dynamic maintenance of cell polarity. Nat Cell Biol. 2012 Feb 19;14(3):304–10. doi: 10.1038/ncb2444. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study links a lipid gradient to Cdc42 dynamics in budding yeast.
- 63.Field SJ, Madson N, Kerr ML, Galbraith KA, Kennedy CE, Tahiliani M, Wilkins A, Cantley LC. PtdIns(4,5)P2 functions at the cleavage furrow during cytokinesis. Curr Biol. 2005 Aug 9;15(15):1407–12. doi: 10.1016/j.cub.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 64.Kee YS, Ren Y, Dorfman D, Iijima M, Firtel R, Iglesias PA, Robinson DN. A mechanosensory system governs myosin II accumulation in dividing cells. Mol Biol Cell. 2012 Apr;23(8):1510–23. doi: 10.1091/mbc.E11-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lafaurie-Janvore J, Maiuri P, Wang I, Pinot M, Manneville JB, Betz T, Balland M, Piel M. ESCRT-III assembly and cytokinetic abscission are induced by tension release in the intercellular bridge. Science. 2013 Mar 29;339(6127):1625–9. doi: 10.1126/science.1233866. [DOI] [PubMed] [Google Scholar]
- 66.Burton K, Taylor DL. Traction forces of cytokinesis measured with optically modified elastic substrata. Nature. 1997 Jan 30;385(6615):450–4. doi: 10.1038/385450a0. [DOI] [PubMed] [Google Scholar]
- 67.Sedzinski J, Biro M, Oswald A, Tinevez JY, Salbreux G, Paluch E. Polar actomyosin contractility destabilizes the position of the cytokinetic furrow. Nature. 2011 Aug 7;476(7361):462–6. doi: 10.1038/nature10286. [DOI] [PubMed] [Google Scholar]
- 68.Goehring NW, Grill SW. Cell polarity: mechanochemical patterning. Trends Cell Biol. 2013 Feb;23(2):72–80. doi: 10.1016/j.tcb.2012.10.009. [DOI] [PubMed] [Google Scholar]