Highlight
Persistence of intracellular rhizobia in legumes involves plant and bacterial genes. Here we show that DNF2, bacA, SymCRK/RSD and nifA/nifH successively prevent bacterial death during the symbiotic process.
Key words: bacA, CRK, DNF2, innate immunity, nifA, nifH, nitrogen fixation.
Abstract
Medicago truncatula belongs to the legume family and forms symbiotic associations with nitrogen fixing bacteria, the rhizobia. During these interactions, the plants develop root nodules in which bacteria invade the plant cells and fix nitrogen for the benefit of the plant. Despite massive infection, legume nodules do not develop visible defence reactions, suggesting a special immune status of these organs. Some factors influencing rhizobium maintenance within the plant cells have been previously identified, such as the M. truncatula NCR peptides whose toxic effects are reduced by the bacterial protein BacA. In addition, DNF2, SymCRK, and RSD are M. truncatula genes required to avoid rhizobial death within the symbiotic cells. DNF2 and SymCRK are essential to prevent defence-like reactions in nodules after bacteria internalization into the symbiotic cells. Herein, we used a combination of genetics, histology and molecular biology approaches to investigate the relationship between the factors preventing bacterial death in the nodule cells. We show that the RSD gene is also required to repress plant defences in nodules. Upon inoculation with the bacA mutant, defence responses are observed only in the dnf2 mutant and not in the symCRK and rsd mutants. In addition, our data suggest that lack of nitrogen fixation by the bacterial partner triggers bacterial death in nodule cells after bacteroid differentiation. Together our data indicate that, after internalization, at least four independent mechanisms prevent bacterial death in the plant cell. These mechanisms involve successively: DNF2, BacA, SymCRK/RSD and bacterial ability to fix nitrogen.
Introduction
Legume plants can be cultivated without application of nitrogen fertilizers on soil poor in nitrogen. This capacity is the result of the associations with soil bacteria, referred to as rhizobia, that fix nitrogen for the benefit of the legume (for recent review, see Udvardi and Poole, 2013). These symbioses are efficient thanks to the spectacular bacterial density within the symbiotic organ, the nodule. Despite the massive and chronic colonization by bacteria, legume nodules do not develop defences. As an illustration of this, expression of defence-related genes is drastically reduced in nodule infected cells as compared to non-infected cells (Limpens et al., 2013). A growing body of recent publications indicates that (the control of) plant innate immunity plays a major role during the early steps of rhizobium/legume interactions (Jones et al., 2008; Yang et al., 2010; Gough and Jacquet, 2013; Liang et al., 2013; Lopez-Gomez et al., 2012; Okazaki et al., 2013).
However, the control of legume immunity is not only critical during the first steps of the symbiotic process but is also required once the nodule is formed when bacteria are released into the plant cells. In Medicago truncatula Gaertn., one of the favourite model legumes, two genes required to repress defence-like reactions into the nodules have been identified recently: DNF2 and SymCRK (Bourcy et al., 2013; Berrabah et al., 2014a ). These genes encode for a phosphatidyl inositol specific phospholipase C-like protein and a cysteine-rich receptor-like kinase respectively. Mutants altered in these genes develop nodules in which transcription of defence-related genes is induced and phenolic compounds accumulate (Bourcy et al., 2013; Berrabah et al., 2014a ). Rhizobia viability is drastically reduced in these mutants maybe as a consequence of the observed defence-like reactions.
Interestingly, defences are not totally abolished in M. truncatula wild type (WT) nodules. Indeed, in indeterminate nodules (i.e. those harbouring a persistent meristem), legumes produce nodule-specific cysteine-rich (NCR) peptides that are defensin-like peptides (Mergaert et al., 2003; Alunni et al., 2007). NCR peptides are specifically produced in infected cells and have a toxic effect on bacteria and prevent their division. As a consequence, in M. truncatula, intracellular rhizobia (called bacteroids) are elongated bacteria, up to 10 µm long, that contain multiple genome copies, up to 24C (Mergaert et al., 2006). NCR toxicity is partially countered by the bacterial BacA protein (Haag et al., 2011), which is believed to act as a peptide importer. The bacterial bacA mutants are more susceptible to NCR peptides in vitro as well as in planta (Van de Velde et al., 2010; Haag et al., 2011). Indeed, once released into the plant cells the bacA mutant does not elongate as the WT bacteria and is rapidly killed (Haag et al., 2011). Interestingly, the ability of the bacA mutant to survive within the plant cells is restored in the M. truncatula dnf1 mutant (Haag et al., 2011). DNF1 is required for bacteroid differentiation and encodes a nodule-specific subunit of a signal peptidase necessary to address plant proteins (including NCR peptides) to the cellular compartments containing the bacteroids (Wang et al., 2010). In addition to DNF1, three other genes (RSD, DNF2 and SymCRK) are also required for bacteroid elongation in M. truncatula (Bourcy et al., 2013; Sinharoy et al., 2013; Berrabah et al., 2014a ). These three genes are specifically expressed in nodules and the symbiotic organs of the rsd, dnf2 and symCRK mutants accumulate brown material (Bourcy et al., 2013; Sinharoy et al., 2013; Berrabah et al., 2014a ), reminiscent of defence reactions.
Defence-like reactions are not developed in bacA-triggered nodules indicating that the lack of bacteroid elongation and the lack of nitrogen fixation do not elicit dnf2- and symCRK-like defences (Berrabah et al., 2014b ). However, the bacA mutant is unable to colonize massively the plant cells and the question remains as to whether the lack of nitrogen fixation associated with a massive bacterial intracellular colonization could elicit the plant defences.
We aim to better understand the repression of defence-like reactions in nodules by identifying the factors involved in this process and by determining precisely the sequence of events that conduct defence development in the mutants. Herein we show that early bacterial death is associated with bacterial inability to fix nitrogen and that RSD is also required to prevent defence-like reactions in nodules. In addition, by combining the use of the dnf2, symCRK and rsd mutants together with various bacterial mutants, we define two steps in the symbiotic control of immunity after rhizobia internalization.
Materials and methods
Bacteria strains, plant lines, cultivation and inoculation methods
Sinorhizobium meliloti strain 1021 (Galibert et al., 2001), bacA (Ferguson et al., 2002), nifH, nifA, Sm2011, Rm41 (Kondorosi et al., 1984), strain AK631 (eps -) (Putnoky et al., 1988), strain PP711(kps -) (Becquart-de Kozak et al., 1997) were cultivated in YEB medium (Krall et al., 2002) for 24h at 30°C. M. truncatula ecotype R108, also referred to as M. truncatula ssp. tricycla, and its derived lines NF0737, i.e. symCRK, MS240, dnf2-4, NF11265, rsd-1, NF8776 and NF17452 (Tadege et al., 2009; Pislariu et al., 2012; Bourcy et al., 2013; Sinharoy et al., 2013; Berrabah et al., 2014a ; Cheng et al., 2014) as well as M. truncatula cv. Jemalong, line A17 and the dnf1 (Mitra and Long, 2004; Starker et al., 2006; Wang et al., 2010; Young et al., 2011) derived line were cultivated as previously described (Berrabah et al., 2014a ). Briefly, seeds were scarified by incubation in sulphuric acid for 7 mins followed by 4 washing steps in sterile distilled water. After scarification, seeds were surface sterilized by incubation in 15ml of 0.3% chlorine followed by four washing steps in 50ml of sterile distilled water. Sterile seeds were then incubated at 4°C onto 1% agar (water) plates for 48h at 4°C. Seeds were then transferred to room temperature for 24 to 48h. After germination, seedlings were transferred onto buffered nodulation medium (BNM) (Ehrhardt et al., 1992) supplemented with AVG 1 µM and immediately inoculated. Bacterial cells were washed with distilled sterile water before seedling inoculation. Optical densities of the bacterial cell suspension were adjusted to OD600=0.1 and 1ml of this suspension was used per 12×12cm plate containing eight seedlings. After 30 mins, liquid excess was removed by pipetting and the plates were sealed on three sides. Plants were cultivated in a growth chamber at 26°C at 60% humidity with a photoperiod of 16h/8h light/dark respectively.
Molecular methods
Nodules were collected on plants with forceps and bistoury and immediately frozen in liquid nitrogen. Samples were reduced to powder by grinding material with steel beads in 2ml tubes using a Qiagen tissues lyser 2×30 s at 25 pulsations/second. RNAs were extracted using GeneJET Plant RNA Purification Mini kit (Thermo Scientific) as recommended by the manufacturer, RNA extracts were treated with Turbo DNAse ambion (Life Technology) to remove DNA traces. RNAs were reverse transcripted using SuperScriptII (Life Technology) as recommended by the manufacturer. qPCR was then carried out on Light Cycler 480 using the LightCycler 480 SYBER Green I (Roche) device as previously described (Gonzalez-Rizzo et al., 2006). Samples were normalized using the constitutive MtACTIN2 as a reference gene (Limpens et al., 2005). Progeny of mutant lines altered in DNF1 was genotyped using primers listed in Supplementary Table S1 following the method described in Ratet et al. (2010). Primers used are listed in Supplementary Table S1.
Imaging and histological analysis
Entire nodules were imaged using Nikon macroscope AZ100. To produce sections, nodules were embedded in 6% agarose (water) and 70 µm sections were produced using vibratome VT1200S from Leica. To observe necrosis, nodule sections were also imaged with a macroscope AZ100. Sections mounted between slide and slipcover were bright light illuminated and observed with CIS illumination. For live and dead staining, we used the protocol previously described in Haag et al. (2011). Briefly, nodule sections were incubated in a 50mM Tris-HCl buffer (pH 7.2) containing 30 µM propidium iodide and 5 µM Cyto9 (Life Technology) for 20 mins. Stained sections were then mounted between slide and slipcover with a few Tris-HCl buffer drops and observed using a Leica SP8 confocal microscope.
For phenolic staining, a protocol derived from (Vasse et al., 1993) was used. Briefly, nodule sections were fixed for 30 to 40 mins using potassium permanganate 0.02%. Sections were washed as necessary using PIPES 100mM (pH 7.2) buffer to remove precipitate. Phenolics were then stained with methylene blue 0.01% for 10 to 15 mins and nodule sections were washed in 6.1% chlorine solution prior observation.
Acetylene reduction assay
An acetylene reduction assay was conducted as previously described (Berrabah et al., 2014a, b ). The method is derived from the one described in Koch and Evans (1966). Briefly, entire plants were individually incubated in 10ml vials containing air supplemented with 250 µl acetylene. After at least 1h at room temperature, 200 µl of the gas phase were analysed by gas chromatography coupled with flame ionization detection.
In situ hybridization
In situ hybridizations followed the protocol described in Boualem et al. (2008). Twenty-one dpi nodules of M. truncatula cultivated in vitro on BNM-agar were analysed using RNA probes labelled with dioxygenin and revealed using antibodies coupled to alkaline phosphatase. The anti-sense probe was designed on extracellular cysteine-rich SymCRK domain to specifically target the SymCRK transcript. The sense probe was used as a negative control; probes were synthethized using T7/SP6 primers after target sequence cloning in the pGEMT easy vector. The primers used to amplify the target region are listed in Supplementary Table S1.
Results
The nifA fix- mutant undergoes bacterial death that is not associated with symCRK/dnf2-like defence reactions
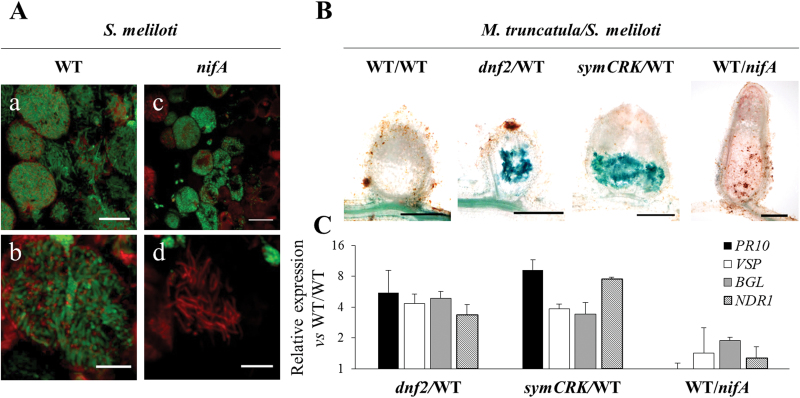
Using the bacterial bacA mutant that is unable to differentiate elongated bacteroids, we previously showed that lack of bacteroid differentiation does not elicit expression of defence genes or phenolics accumulation that are observed in the dnf2 and the symCRK nodules (Berrabah et al., 2014b ). Nevertheless, the bacA mutant is hypersensitive to NCR peptides and this sensitivity does not allow massive colonization of the plant cells (Haag et al., 2011). In order to determine if a massive colonization by ineffective bacteria triggers defence reactions, M. truncatula WT plants were inoculated with the S. meliloti nifA mutant. NifA is a key regulator of the nitrogenase synthesis and the corresponding mutant does not fix nitrogen (Zimmerman et al., 1983; Dixon and Kahn, 2004). Nodules induced by nifA are infected and bacteroids clearly undergo terminal differentiation (Fig. 1). However, in contrast to the WT, the mutant bacteria prematurely died as revealed by the live/dead assay that stains viable bacteria green and dead bacteria red (Fig. 1). The bacterial death occurs only after bacteroid elongation in the nifA mutant (Supplementary Fig. S1). Beyond genes responsible for nitrogenase synthesis, NifA regulates additional genes (Bobik et al., 2006) and the nifA mutant displays pleiotropic phenotypes (Gong et al., 2007). To determine whether nifA bacteroid death is caused by the lack of nitrogen fixation or to another nifA-regulated process, nifH bacteroid viability was also studied; nifH encodes a subunit of the nitrogenase. Like for the nifA mutant, bacteroid death was observed for elongated nifH bacteroids (Supplementary Fig. S1) suggesting that lack of nitrogen fixation is responsible for the death of nifA and nifH elongated bacteroids. In order to determine if defence reactions such as those observed in the nodules of the symCRK and the dnf2 mutants could be responsible for nifA bacterial death, the induction of these defences was evaluated by RT-qPCR and histological approaches. In contrast to the dnf2 and symCRK mutant nodules, the expression of the defence-related genes PR10, VSP, BGL and NDR1 was not induced in the nifA-triggered nodules, which also do not accumulate phenolics (Fig. 1). In agreement of a role of nifA after DNF2 and SymCRK, mutation in the nifA gene does not prevent the development of defence-like reactions in dnf2 and symCRK nodules (Supplementary Fig. S2). Together these results suggest that lack of nitrogen fixation induces early death of the bacteria. Furthermore this bacterial death is not associated with defence-like reactions observed in the dnf2 and symCRK mutants (Fig. 1).
Fig. 1.
Mutation in nifA elicits early bacterial death that is not mediated by symCRK/dnf2-dependant defence reactions. (A): a–d: On these sections of 26dpi nodules imaged using a confocal microscope after live/dead staining assay, the early death of the nifA mutant within the plant cell is observable. Green and red stain alive and dead bacteria respectively. a and b represent WT bacteria induced nodules, c and d: nifA induced nodules, note that like the WT bacteria, the nifA mutant undergoes terminal differentiation, scale bars correspond to, 20µm (a, c) and 10µm (b, d). (B): nifA mutant does not elicit the accumulation of phenolics in nodules. Sections of WT plant nodules induced by WT bacteria or by nifA mutant as well as dnf2 and symCRK nodules induced by WT bacteria were stained with methylene blue to reveal phenolics in blue. Scale bars correspond to 500 µm. (C): PR10, VSP, BGL and NDR1 defence-related genes are not induced in nifA-triggered nodules. In contrast these defence-related genes are induced in dnf2 and symCRK nodules as revealed by RT-qPCR analysis. Results are expressed as induction fold vs R108 nodules induced by WT bacteria after normalization using Mtactin2 constitutive gene as an internal standard.
DNF2, bacA and SymCRK act successively during the symbiotic process
In order to get an insight into the development of the symbiotic process and the timing of immunity suppression during symbiosis, nodulation tests were conducted using dnf2 and symCRK mutants in combination with bacterial mutants altered in surface components or in the symbiotic process. Defence-like reactions were evaluated 14 days after inoculation in the nodules induced by S. meliloti strain Rm41 eps- and kps- mutants (altered in exopolysaccharide and capsular polysaccharide respectively) as well as S. meliloti Sm1021 bacA, and nifH mutants. Defence reactions were monitored by following the expression of the NDR1 and PR10 genes using RT-qPCR. In addition, nodule sections were observed under bright field illumination to reveal the presence of necrotic zones accumulating phenolics. The eps and kps mutants elicit a moderate induction of the expression of NDR1 and PR10 genes in nodules of the WT plants (Fig. 2A). These inductions were not associated with the development of necrotic zones in the WT plant nodules (Fig. 2B). In contrast, the expression of defence-related genes was observed when these bacterial mutants were nodulating dnf2 and symCRK plant mutants and this was associated to the presence of necrotic zones (Fig. 2B). In nodules developed on WT plants inoculated by Sm1021 or its nifH or bacA mutant derivatives, the NDR1 and PR10 genes are not induced (Fig. 2A) and these nodules do not accumulate phenolics. In contrast to the WT plants, dnf2 and symCRK nodules display necrosis and induction of the defence markers with these strains, except for symCRK inoculated with the bacA mutant; in this plant/bacteria mutant combination the expression of defence-related genes and nodule necrosis were not observed (Fig. 2A, B). This absence of defence reactions in symCRK/bacA nodules was further confirmed in 21 and 27 day-old nodules (Fig. 2C, Supplementary Fig. S3). However, acetylene reduction assays indicate that nitrogen fixation was not restored in the symCRK/bacA nodules (Supplementary Fig. S4). Furthermore, despite the reduction in the expression of late NCR genes (Berrabah et al., 2014a ) (Supplementary Fig. S5) and the absence of defence-like reactions in the symCRK mutant upon bacA infection, the bacA mutant viability was not restored in these symCRK/bacA nodules (Supplementary Fig. S6) contrasting with what was observed in dnf1/bacA nodules (Haag et al., 2011).
Fig. 2.
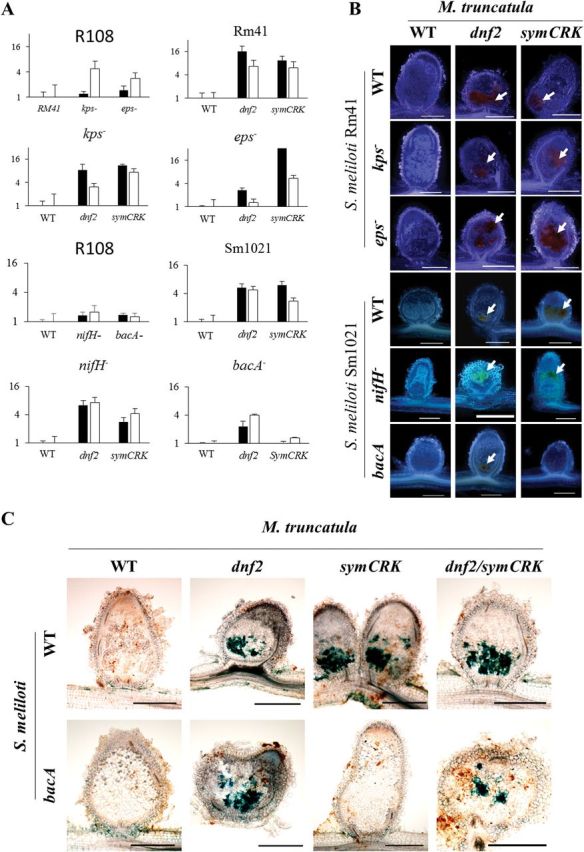
DNF2, bacA and SymCRK act successively during the symbiotic process. (A) Development of defence reactions was investigated in nodules by evaluating the expression of defence-related genes PR10 and NDR1. The four top sub-panels correspond to plants inoculated with Rm41 WT or the corresponding mutant strains. The four bottom sub-panels correspond to plants inoculated with Sm1021 WT or the corresponding mutant strains. The top left sub-panel of each four sub-panels corresponds to gene expression in the R108 WT plants inoculated by the WT bacteria or its mutant derivatives. The three other sub-panels correspond to gene expression in the R108 WT or dnf2 or symCRK mutants inoculated by different bacteria. (B) The development of defence reactions was investigated in nodules by examination of necrotic zone in nodule section. Columns represent the plant genotypes and rows the bacterial genotypes. Arrows in (B) point to the necrotic zones. (C) Accumulation of phenolic compounds is revealed by the blue color on nodule sections stained with methylene blue. B, C: scale bars correspond to 500 µm.
The double mutant dnf2symCRK was analysed with respect to defence development in nodules elicited with WT bacteria or bacA mutant strain. We did not observe any additive effect of the mutation in the double mutant nodules upon inoculation with the WT bacteria (Fig. 2C, Supplementary Figs S3 and S6). When inoculated with the bacA mutant, defence reactions were observed in the dnf2symCRK nodules but not on the symCRK mutant (Fig. 2C, Supplementary Fig. S3). These results thus indicate that the double mutant behaves as the dnf2 mutant in terms of defence reactions and that DNF2 is epistatic to SymCRK in the M. truncatula R108 background. Together, these results suggest that DNF2, bacA and SymCRK act successively during the symbiotic process.
SymCRK correct expression requires initiation of bacteroids differentiation
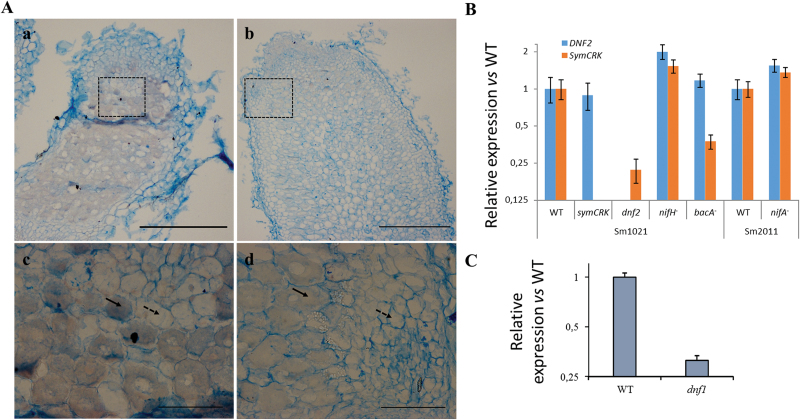
In order to improve our understanding of the SymCRK gene function, its expression pattern inside nodules was studied using in situ hybridization in WT nodules. This experiment (Fig. 3A) suggests that SymCRK is specifically expressed in infected cells in agreement with its timing of expression (Berrabah et al., 2014a ; see also below). In order to define more precisely the role of SymCRK and DNF2 during the symbiotic process, we investigated the expression of the two genes in M. truncatula nodules induced by the S. meliloti nifA and nifH mutants altered in the production of nitrogenase (for review, see Dixon and Kahn, 2004) as well as by the bacA mutant. In nodules induced by the nifA and nifH bacterial mutants (Fig. 3B), expression of the two symbiotic genes SymCRK and DNF2 was not altered, indicating that nitrogen fixation is not necessary for their expression. DNF2 transcript accumulation is not modified in nodules induced by the bacA mutant or in symCRK mutant nodules induced by WT bacteria placing DNF2 upstream or in a parallel pathway to BacA and SymCRK. In contrast, the level of SymCRK transcripts was reduced in WT nodules induced by the bacA mutant or in dnf2 mutant nodules induced by WT bacteria, in agreement with the data shown above, indicating that SymCRK act downstream of DNF2 and bacA during the symbiotic process. As mentioned previously, bacteroids do not differentiate and are not maintained viable in both the dnf2 and the bacA nodules. In contrast, in the dnf1M. truncatula mutant, bacteria do not undergo terminal differentiation but bacteroids remain viable during colonization (Haag et al., 2011). Thus, this mutant constitutes a good tool to discriminate whether the colonization defect and/or the differentiation defect is responsible for incorrect symCRK expression. We have used two Tnt1 insertion mutant lines for DNF1 (NF8776 and NF17452) in the M. truncatula R108 background (in which the dnf2 and symCRK mutants used in this study were generated). In these lines the Tnt1 retrotransposon is inserted in the first intron 131bp and 324bp after the predicted start codon. However, we could not observe any symbiotic phenotypes in the progeny, possibly as a result of the insertion in the intron or of the absence of homozygous mutant (Supplementary Fig. S7). The expression of SymCRK was thus studied in the only dnf1 available mutant (generated in the A17 background). This experiment shows that SymCRK expression was reduced in the dnf1 mutant, suggesting that not only bacterial colonization but also bacteroid differentiation is required for SymCRK expression (Fig. 3C).
Fig. 3.
SymCRK is expressed in infected cells. (A) Expression of SymCRK was investigated using in situ hybridization and (B, C) RT-qPCR. Using antisense probe (a, c), signal was detected in infected cells (indicated with plain arrows) of the nitrogen fixation zone and not in uninfected cells (indicated with dashed arrows); sense probe was used as a control of specificity (b, d). c, d represent magnification of the zones delimited by a dashed rectangle in a and b. Scale bars represent 300 and 30 µm in whole nodule sections and in enlargement respectively. (B) RT-qPCR analyses revealed that SymCRK expression is strongly reduced in nodules of the dnf2 mutant. In contrast, the expression of DNF2 is not altered in the symCRK nodules. Also, expression of SymCRK was strongly reduced in the bacA induced nodules (B) as well as in the 14dpi nodules of the dnf1 mutant (C). B, C: error bars represent standard errors of three biological experiments with two technical replicates.
RSD represses defence-like reactions and act downstream of bacA and DNF2
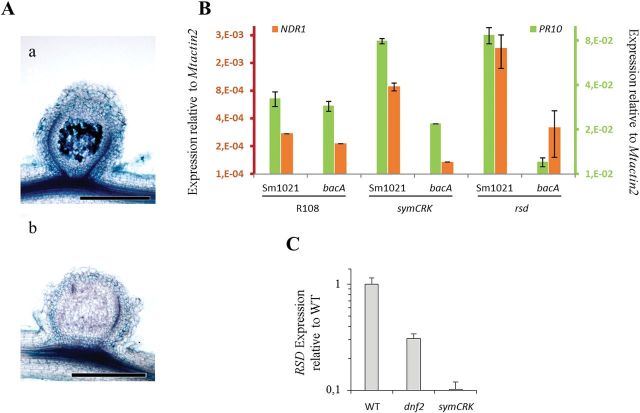
Like dnf2 and symCRK, rsd displays necrotic nodules in which bacteroids do not undergo terminal differentiation (Sinharoy et al., 2013). In order to determine if rsd also shares the defence-like reactions observed in the dnf2 and symCRK mutants, induction of defence-related genes was evaluated in rsd-1 nodules. Expression of the PR10 and NDR1 genes was evaluated by RT-qPCR and was found to be induced in rsd-1 as compared to WT nodules (Fig. 4). In addition, presence of phenolic compounds was detected in rsd-1 nodules (Fig. 4) indicating that rsd develops defence-like reactions similar to those of symCRK and dnf2. In order to position RSD with respect to SymCRK and DNF2 during the development of the symbiosis, we analysed the expression of RSD in the dnf2 and in the symCRK backgrounds. RSD expression is strongly reduced in both mutants (Fig. 4C) suggesting a role for RSD downstream of DNF2 and SymCRK.
Fig. 4.
RSD is required to suppress immunity in nodules and act downstream of bacA. (Aa) Upon inoculation with the WT Sinorhizobium meliloti, the rsd-1 mutant nodules (32dpi) accumulate phenolics as revealed by methylene blue staining of nodules sections. (Ab) In contrast, bacA triggered nodules do not accumulate phenolics; scale bars represent 500 µm. (B) In agreement, RT-qPCR analyses indicate that defences are activated in rsd nodules induced by the WT bacteria but not in those triggered by the bacA mutant. (C) RSD expression is strongly reduced in dnf2 and symCRK mutant nodules. B, C: error bars represent standard errors.
In order to determine whether the development of defence-like reactions in the rsd-1 mutant is dependent on bacterial invasion and differentiation, we evaluated defences in this mutant upon nodulation with the bacA mutant. In contrast to dnf2 and, similarly to symCRK, the rsd mutant does not accumulate phenolics upon infection with bacA (Fig. 4A). In agreement with these results, defence-related gene expression was strongly reduced in rsd/bacA nodules as compared to rsd nodules triggered by WT bacteria (Fig. 4B). Thus, similar to what is observed in the symCRK mutant, development of defence-like reactions in rsd relies on the presence of a functional bacA.
Discussion
Herein we have investigated the sequence leading to the symbiotic suppression of immunity in legume nodules after rhizobia internalization within the plant cells (chronic infection). We showed that the bacterial mutants nifA and nifH died prematurely within the plant cells despite that defence reactions similar to those observed in symCRK and dnf2 nodules were not detected in these nodules. In addition, we showed that DNF2 and SymCRK act successively during the symbiotic process and that bacA is required after DNF2 and before SymCRK. Furthermore, our data indicate that rsd-1 behave like the symCRK mutant and acts after bacA. These results are in agreement with the observation that SymCRK and RSD expression is reduced in WT plants nodulated with bacA, a condition in which expression of DNF2 is not modified (Fig. 3B; Sinharoy et al., 2013). In addition, results obtained with the dnf1 mutant suggest that the initiation of bacteroid differentiation rather than the massive intracellular colonization is important for SymCRK activity. Interestingly, in the distant phylogenetic system, soybean/Bradyrhizobium japonicum, the nifA mutant triggers nodules displaying dark brown zones of necrotic appearance (Fischer et al., 1986) reminiscent of symCRK/dnf2-like defences. Altogether our results indicate that multiple steps are required for the symbiotic suppression of immunity after bacterial internalization. Furthermore the present study indicates that at least two types of defence can be the cause of bacterial death in the plant cells, one associated with nodule necrosis and induction of PR10 and NDR1 defence genes and another observed upon nifA- or nifH-triggered nodulation.
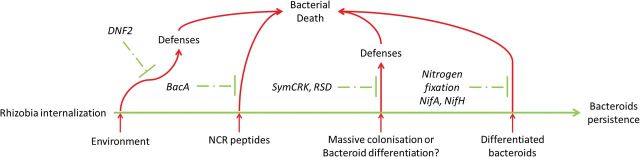
Based on our results, we propose a model focused on the intracellular stage of symbiosis, which describes the actors preventing bacteroid death (Fig. 5). In this model, DNF2 is the earliest actor identified as required for symbiotic suppression of immunity. Its requirement is determined by environmental conditions that influence the development of defence-like reactions in nodules (Berrabah et al., 2014b ). After DNF2, the bacA bacterial gene prevents the killing activity of NCR peptides before SymCRK and RSD prevent defence-like reactions possibly triggered by massive intracellular invasion or initiation of bacteroid differentiation. Finally, the production of nitrogenase prevents bacteroid death. In order to add more actors to this model, necrotic mutants (such as those described in Pislariu et al., 2012; Domonkos et al., 2013) will probably represent good tools. Such mutants could in future studies be classified according to their behaviour upon inoculation in combination with bacterial symbiotic mutants (amongst which is bacA) but also based on their symbiotic properties using dnf2 permissive media (Berrabah et al., 2014b ).
Fig. 5.
Bacteroid death is prevented by multiple actors acting successively. DNF2 is the earliest actor identified as required for symbiotic suppression of immunity at the intracellular stage of the symbiosis. Its requirement is determined by environmental conditions that influence the development of defence-like reactions in nodules (Berrabah et al., 2014a ). After DNF2, the bacA bacterial gene prevents the NCR triggered bacteroid death (Haag et al., 2011). Later, SymCRK and RSD prevent defence-like reactions possibly triggered by massive intracellular invasion or initiation of bacteroid differentiation. Finally, nitrogen fixation is required to prevent the death of elongated bacteroids.
Immunity suppression is not only required for the intracellular stage of the symbiosis; various actors have been identified or proposed to be important to prevent plant defences during the infection process. For example the nodulation (Nod) factors might participate in the suppression of plant immunity triggered by microbial motifs (Liang et al., 2013). In agreement with this novel Nod factor’s role, the type three secretion system (often used by phytopathogenic bacteria to suppress plant immunity) of Bradyrhizobium elkanii can bypass the requirement of Nod factors for the nodulation of soybean (Okazaki et al., 2013). Also, two soybean resistance genes modify the rhizobial host range of the plant (Yang et al., 2010) and exopolysaccharides were shown to be important to reduce the induction of defence-related genes at the early stages of the symbiosis (Jones et al., 2008). Finally, defence-related phytohormones also interfere with the nodulation process (Penmetsa and Cook, 1997; Stacey et al., 2006; Sun et al., 2006). These data indicate that suppression of immunity during the early steps of the symbiotic process is required for nodulation. In agreement with this, the expression of a PR10 gene is drastically reduced in bumps (young nodules) and mature nodules as compared to roots. We showed that DNF2 prevents defence-like reactions in nodules only after bacteria internalization (Bourcy et al., 2013) and that in bumps, the expression of the PR10 gene is similar in the dnf2 and WT nodules (Bourcy et al., 2013). These results indicate that a mechanism distinct from DNF2 and SymCRK suppresses immunity in early developing nodules and reinforce the hypothesis of the existence of multiple steps in the suppression of plant immunity during the symbiotic process. It is now a challenge to gather the identified actors of the symbiotic suppression of immunity within an integrated model.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Lack of nitrogen fixation triggers bacterial death after bacteroid elongation.
Supplementary Fig. S2. dnf2 and symCRK act before nifA.
Supplementary Fig. S3. Defence reactions are abolished in the symCRK bacA nodules.
Supplementary Fig. S4. Nitrogen fixation is not restored in symCRK/bacA nodules.
Supplementary Fig. S5. NCR99 expression is reduced in bacA-triggered nodules of the dnf2 and symCRK mutants.
Supplementary Fig. S6. bacA viability is not restored in the symCRK mutant.
Supplementary Fig. S7. DNF1 gene structure (based on MTR_3g027890 sequence) and position of the Tnt1 insertions present in mutant lines NF8776 and NF17452.
Supplementary Table S1. List of primers used during this study.
Acknowledgements
This work was supported by the Centre National de la Recherche Scientifique (CNRS) and the grant Agence Nationale de la Recherche (ANR) Blanc International SVSE 6.2010.1 (LEGUMICS) to PR. This work has benefited from a French State grant (reference ANR-10-LABX-0040-SPS) managed by the French National Research Agency under an Investments for the Future programme (reference ANR-11-IDEX-0003-02). This work has benefited from the facilities and expertise of the IMAGIF Cell Biology Unit of the Gif campus (www.imagif.cnrs.fr) which is supported by the Conseil Général de l’Essonne. We are grateful to Michael Udvardi, Jiangqi Wen and Kiran Mysore (Noble Foundation, Ardmore, USA) for seeds of the rsd mutant and for the NF8776 and NF17452 lines. We address our gratitude to our colleagues of the Institut des Sciences du Végétal, Gif sur Yvette, France: Viviane Jean, Marie Bourcy and Alexis Eschstruth for preliminary studies and Ibtissem Guefrachi for her assistance during the in situ hybridization experiment.
References
- Alunni B, Kevei Z, Redondo-Nieto M, Kondorosi A, Mergaert P, Kondorosi E. 2007. Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula . Molecular Plant Microbe Interactions 20, 1138–1148. [DOI] [PubMed] [Google Scholar]
- Becquart-de Kozak I, Reuhs BL, Buffard D, Breda C, Kim JS, Esnault R, Kondorosi A. 1997. Role of the K-Antigen Subgroup of Capsular Polysaccharides in the Early Recognition Process Between Rhizobium meliloti and Alfalfa Leaves. Molecular Plant-Microbe Interactions 10, 114–123. [Google Scholar]
- Berrabah F, Bourcy M, Eschstruth A, et al. 2014. a A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytologist 203, 1305–1314. [DOI] [PubMed] [Google Scholar]
- Berrabah F, Bourcy M, Cayrel A, Eschstruth A, Mondy S, Ratet P, Gourion B. 2014. b Growth conditions determine the DNF2 requirement for symbiosis. PLoS One 9, e91866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik C, Meilhoc E, Batut J. 2006. FixJ: a major regulator of the oxygen limitation response and late symbiotic functions of Sinorhizobium meliloti . Journal of Bacteriology 188, 4890–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F. 2008. MicroRNA166 controls root and nodule development in Medicago truncatula . Plant Journal 54, 876–887. [DOI] [PubMed] [Google Scholar]
- Bourcy M, Brocard L, Pislariu CI, Cosson V, Mergaert P, Tadege M, Mysore KS, Udvardi MK, Gourion B, Ratet P. 2013. Medicago truncatula DNF2 is a PI-PLC-XD-containing protein required for bacteroid persistence and prevention of nodule early senescence and defense-like reactions. New Phytologist 197, 1250–1261. [DOI] [PubMed] [Google Scholar]
- Cheng X, Wang M, Lee HK, Tadege M, Ratet P, Udvardi M, Mysore KS, Wen J. 2014. An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytologist 201, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nature Reviews Microbiology 2, 621–631. [DOI] [PubMed] [Google Scholar]
- Domonkos A, Horvath B, Marsh JF, Halasz G, Ayaydin F, Oldroyd GE, Kalo P. 2013. The identification of novel loci required for appropriate nodule development in Medicago truncatula . BMC Plant Biology 13, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt DW, Atkinson EM, Long SR. 1992. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256, 998–1000. [DOI] [PubMed] [Google Scholar]
- Ferguson GP, Roop RM, 2nd, Walker GC. 2002. Deficiency of a Sinorhizobium meliloti bacA mutant in alfalfa symbiosis correlates with alteration of the cell envelope. Journal of Bacteriology 184, 5625–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer HM, Alvarez-Morales A, Hennecke H. 1986. The pleiotropic nature of symbiotic regulatory mutants: Bradyrhizobium japonicum nifA gene is involved in control of nif gene expression and formation of determinate symbiosis. EMBO Journal 5, 1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F, Finan TM, Long SR, et al. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti . Science 293, 668–672. [DOI] [PubMed] [Google Scholar]
- Gong Z, Zhu J, Yu G, Zou H. 2007. Disruption of nifA gene influences multiple cellular processes in Sinorhizobium meliloti . Journal of Genetics and Genomics 34, 783–789. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. 2006. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti . Plant Cell 18, 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough C, Jacquet C. 2013. Nod factor perception protein carries weight in biotic interactions. Trends in Plant Science 18, 566–574. [DOI] [PubMed] [Google Scholar]
- Haag AF, Baloban M, Sani M, et al. 2011. Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biology 9, e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Sharopova N, Lohar DP, Zhang JQ, VandenBosch KA, Walker GC. 2008. Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proceedings of the National Academy of Sciences of the United States of America 105, 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch B, Evans HJ. 1966. Reduction of acetylene to ethylene by soybean root nodules. Plant Physiology 41, 1748–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi E, Banfalvi Z, Kondorosi A. 1984. Physical and genetic analysis of a symbiotic region of Rhizobium meliloti: Identification of nodulation genes. Molecular and General Genetics 193, 445–452. [Google Scholar]
- Krall L, Wiedemann U, Unsin G, Weiss S, Domke N, Baron C. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens . Proceedings of the National Academy of Sciences of the United States of America 99, 11405–11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Cao Y, Tanaka K, Thibivilliers S, Wan J, Choi J, Kang C, Qiu J, Stacey G. 2013. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 341, 1384–1387. [DOI] [PubMed] [Google Scholar]
- Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, Geurts R. 2005. Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proceedings of the National Academy of Sciences of the United States of America 102, 10375–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Moling S, Hooiveld G, Pereira PA, Bisseling T, Becker JD, Kuster H. 2013. Cell- and tissue-specific transcriptome analyses of Medicago truncatula root nodules. PLoS One 8, e64377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gomez M, Sandal N, Stougaard J, Boller T. 2012. Interplay of flg22-induced defence responses and nodulation in Lotus japonicus . Journal of Experimental Botany 63, 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E. 2003. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiology 132, 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Uchiumi T, Alunni B, et al. 2006. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proceedings of the National Academy of Sciences of the United States of America 103, 5230–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Long SR. 2004. Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiology 134, 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki S, Kaneko T, Sato S, Saeki K. 2013. Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proceedings of the National Academy of Sciences of the United States of America 110, 17131–17136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR. 1997. A Legume Ethylene-Insensitive Mutant Hyperinfected by Its Rhizobial Symbiont. Science 275, 527–530. [DOI] [PubMed] [Google Scholar]
- Pislariu CI, Murray JD, Wen J, et al. 2012. A Medicago truncatula tobacco retrotransposon insertion mutant collection with defects in nodule development and symbiotic nitrogen fixation. Plant Physiology 159, 1686–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnoky P, Grosskopf E, Ha DT, Kiss GB, Kondorosi A. 1988. Rhizobium fix genes mediate at least two communication steps in symbiotic nodule development. The Journal of Cell Biology 106, 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratet P, Wen J, Cosson V, Tadege M, Mysore KS. 2010. Tnt1 Induced Mutations in Medicago: Characterization and Applications. The Handbook of Plant Mutation Screening: Wiley-VCH Verlag GmbH & Co. KGaA, 83–99. [Google Scholar]
- Sinharoy S, Torres-Jerez I, Bandyopadhyay K, Kereszt A, Pislariu CI, Nakashima J, Benedito VA, Kondorosi E, Udvardi MK. 2013. The C2H2 transcription factor regulator of symbiosome differentiation represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula . Plant Cell 25, 3584–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey G, McAlvin CB, Kim SY, Olivares J, Soto MJ. 2006. Effects of endogenous salicylic acid on nodulation in the model legumes Lotus japonicus and Medicago truncatula . Plant Physiology 141, 1473–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starker CG, Parra-Colmenares AL, Smith L, Mitra RM, Long SR. 2006. Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiology 140, 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Cardoza V, Mitchell DM, Bright L, Oldroyd G, Harris JM. 2006. Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integration of diverse inputs for regulation of nodulation. The Plant Journal 46, 961–970. [DOI] [PubMed] [Google Scholar]
- Tadege M, Wang TL, Wen J, Ratet P, Mysore KS. 2009. Mutagenesis and beyond! Tools for understanding legume biology. Plant Physiology 151, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M, Poole PS. 2013. Transport and metabolism in legume-rhizobia symbioses. Annual Review of Plant Biology 64, 781–805. [DOI] [PubMed] [Google Scholar]
- Van de Velde W, Zehirov G, Szatmari A, et al. 2010. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126. [DOI] [PubMed] [Google Scholar]
- Vasse J, de Billy F, Truchet G. 1993. Abortion of infection during the Rhizobium meliloti—alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. The Plant Journal 4, 555–566. [Google Scholar]
- Wang D, Griffitts J, Starker C, Fedorova E, Limpens E, Ivanov S, Bisseling T, Long S. 2010. A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science 327, 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Tang F, Gao M, Krishnan HB, Zhu H. 2010. R gene-controlled host specificity in the legume-rhizobia symbiosis. Proceedings of the National Academy of Sciences of the United States of America 107, 18735–18740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debelle F, Oldroyd GE, et al. 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JL, Szeto WW, Ausubel FM. 1983. Molecular characterization of Tn5-induced symbiotic (Fix-) mutants of Rhizobium meliloti . Journal of Bacteriology 156, 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.