Abstract
RNA interference (RNAi) has great potential to treat human disease1–3. However, in vivo delivery of short interfering RNAs (siRNAs), which are negatively charged double-stranded RNA macromolecules, remains a major hurdle4–9. Current siRNA delivery has begun to move away from large lipid and synthetic nanoparticles to more defined molecular conjugates9. Here we address this issue by synthesis of short interfering ribonucleic neutrals (siRNNs) whose phosphate backbone contains neutral phosphotriester groups, allowing for delivery into cells. Once inside cells, siRNNs are converted by cytoplasmic thioesterases into native, charged phosphodiester-backbone siRNAs, which induce robust RNAi responses. siRNNs have favorable drug-like properties, including high synthetic yields, serum stability and absence of innate immune responses. Unlike siRNAs, siRNNs avidly bind serum albumin to positively influence pharmacokinetic properties. Systemic delivery of siRNNs conjugated to a hepatocyte-specific targeting domain induced extended dose-dependent in vivo RNAi responses in mice. We believe that siRNNs represent a technology that will open new avenues for development of RNAi therapeutics.
Unlike small molecule inhibitors (<500 Da) that can passively diffuse across cellular membranes10, siRNAs are both too large (~14 kDa) and too charged (~40 charged phosphates) to enter cells on their own4–7. Consequently, siRNA delivery remains the major technological problem for development of RNAi therapeutics1–9. Improvements in siRNA delivery have begun to move away from large lipid and synthetic nanoparticles (~108 Da) that are >1,000× larger than the siRNA drug to more definable molecular conjugates9. However, because efficient RNAi responses require TRBP enzymatic loading of negatively charged double-stranded A-form siRNAs into Argonaute2 (Ago2), the catalytic core of the RNAi machinery11,12, the negatively charged phosphodiester siRNA backbone has remained recalcitrant to chemical manipulation and presents a significant unsolved problem for siRNA delivery. To address this problem, we synthesized siRNNs with neutralizing bioreversible phosphotriester groups that are intracellularly converted into a native charged phosphate backbone (Fig. 1a,b).
Figure 1.
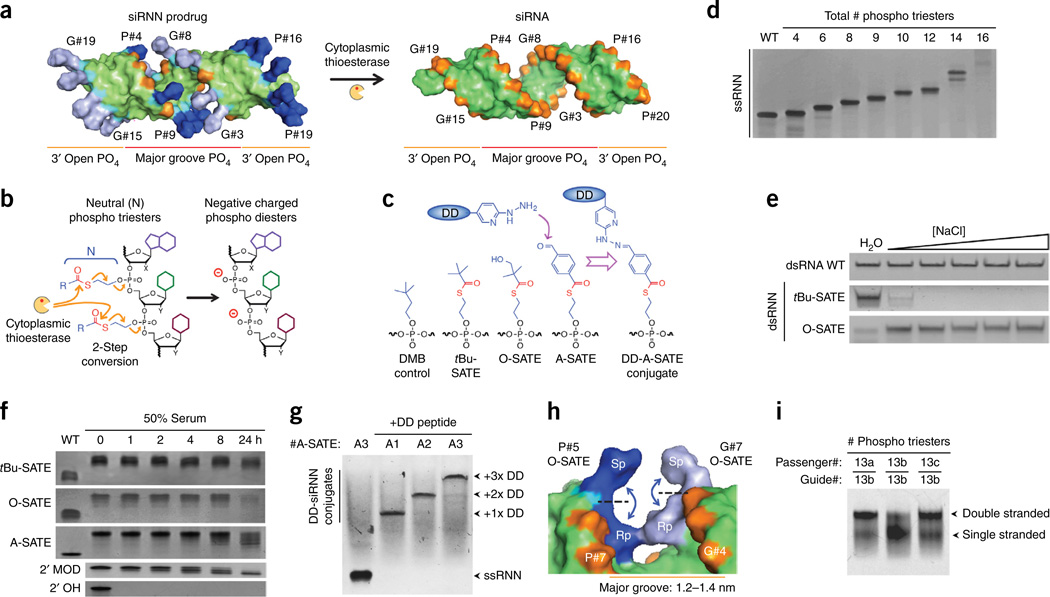
Synthesis and biophysical properties of bioreversible phosphotriester siRNN oligonucleotides. (a) The study concept. siRNN containing 26 phosphotriester groups (blue, gray) neutralize phosphate internucleotide linkages (cyan) and are converted by cytoplasmic thioesterases into negatively charged phosphodiester siRNAs (red, orange) inside cells. siRNAs contain two types of phosphates: major groove phosphates (red, #1–11) that oppose phosphates on the opposite strand, and 3′ open phosphates (orange, #12–20) on each strand. Phosphate labeling: G, guide strand; P, passenger strand. (b) Phosphotriester cleavage by cytoplasmic thioesterase initiates a two-step conversion that resolves as a charged phosphodiester linkage. (c) Structures of the main phosphotriester groups used in this study. Hydrazine containing delivery domain (DD) peptides are conjugated to siRNNs via chemically reactive aldehyde A-SATE phosphotriester group. Cleavage of DD-A-SATE by thioesterase removes both phosphotriester group and conjugated peptide. (d) Denaturing gel analysis of RNN passenger strands containing increasing numbers of tBu-SATE phosphotriesters showing decreasing electrophoretic mobility due to loss of negative charge. (e) Salt sensitivity analysis of double-stranded siRNNs containing 18× tBu-SATE phosphotriesters showing hydrophobic collapse with increasing salt. siRNNs containing 18× hydrophilic hydroxyl O-SATE phosphotriesters remained soluble. [NaCl] = 100, 200, 300, 400, 500 mM. (f) Serum stability analysis of single-stranded oligonucleotides containing 9× tBu-SATE, O-SATE or A-SATE phosphotriester groups vs. wild-type phosphodiester 2′-OH or 2′-F/O-Me (2′-Mod) RNA incubated in 50% human serum at 37 °C for indicated times. (g) Conjugation of delivery domain peptides to RNN oligonucleotides containing one (A1), two (A2) or three (A3) A-SATE phosphotriesters. (h) Model of Sp/Rp O-SATE phosphotriester diastereomers at passenger strand P#5 phosphate opposite guide strand G#7 phosphate. Note Rp/Rp phosphotriester steric congestion. (i) Placement of phosphotriester groups in major groove restricts extent of total phosphotriester numbers. Native gel analysis of dsRNNs containing phosphotriester groups as indicated (see Supplementary Fig. 2h). PO4, phosphate; WT, wild type.
Unlike the 5′ or 3′ end of an oligonucleotide, native negatively charged phosphodiester linkages cannot be modified after oligonucleotide synthesis without inadvertently alkylating the nucleobases13,14. Therefore, we pursued a chemical approach that placed the bioreversible phosphotriester group on phosphoramidite building blocks used for RNN oligonucleotide synthesis. Several groups have previously developed single-stranded deoxyoligonucleotide charge neutralization approaches using irreversible and reversible phosphotriester groups15–19, but none of these approaches has been applied to double-stranded DNA or RNA molecules. After reviewing the literature, we focused our efforts on a thioester bond containing S-acyl-2-thioethyl (SATE) phosphotriester group that was originally developed in the early 1990s in Imbach’s laboratory as a monophosphate nucleotide prodrug inhibitor of HIV and hepatitis B virus20–22. Extracellular esterases are unable to cleave thioester bonds; however, once inside cells, cytoplasmic thioesterases cleave the SATE thioester bond to initiate a rapid two-step conversion that unmasks the charged phosphate group20 (Fig. 1b). Unfortunately, expansion of SATE phosphotriesters to single-stranded DNA oligonucleotides in the late 1990s resulted in poor synthetic yields that made phenotypic biological analyses or synthesis of double-stranded RNA (or DNA) molecules impossible23–25. In our hands, synthesizing RNN oligonucleotides required addressing multiple chemical complexities, including dipole moment destabilization of the thioester bond, 2′ hydroxyl nucleophilic attack on the phosphotriester linkage, nucleobase protecting group stability, deprotection and high-performance liquid chromatography (HPLC) purification conditions that avoided phosphotriester cleavage or strand scission, and hydrophobicity. We exhaustively addressed these problems in an orthogonally compatible fashion that maintained high yields and used standard solid-phase oligonucleotide synthesis conditions (Supplementary Fig. 1a).
To develop siRNNs, we synthesized a library of >40 phosphotriester groups on RNN phosphoramidite building blocks (Supplementary Fig. 1b). To prevent a cis 2′ hydroxyl nucleophilic attack on the phosphotriester linkage26, RNN phosphoramidites contain 2′ modifications (2′-F, pyrimidines; 2′-O-Me, purines) that are highly tolerated by the RNAi machinery and used on all therapeutic siRNAs in clinical trials2,3,6,8,9. To prevent base-mediated phosphotriester cleavage during nucleobase deprotection, RNN phosphoramidites contain phenoxyacetyl chloride protecting groups27 on adenine and cytosine, and isopropyl-phenoxyacetyl chloride on guanosine that allowed for deprotection using extremely mild basic diisopropylamine (DIA) in methanol (Supplementary Fig. 2a). Stabilization of the thioester bond to DIA/methanol required substituting electron-donating groups at the distal α-carbon position or lengthening the proximal ethyl linker to a butyl linker (Supplementary Fig. 1b). RNN oligonucleotide synthesis results in high yields (93.0% yield; 99.6% coupling efficiency) that are comparable to RNA synthesis (85.7% yield; 99.2% coupling efficiency; Supplementary Fig. 2b,c). After screening the RNN phosphoramidite library, we focused on three bioreversible phosphotriester groups, namely, tBu-SATE, hydroxyl O-SATE and a conjugatable aldehyde A-SATE, as well as a control irreversible phosphotriester dimethyl-butyl (DMB) group (Fig. 1c). Biophysical analyses by gel electrophoresis showed that increasing the number of phosphotriester groups resulted in a concomitant RNN charge neutralization, especially at 14 and 16 phosphotriester insertions (Fig. 1d and Supplementary Fig. 2d). Synthesis of double-stranded siRNNs with 18× tBu-SATE phosphotriesters resulted in a salt-mediated hydrophobic collapse, but substitution of 18× hydrophilic hydroxyl O-SATE phosphotriesters rescued solubility (Fig. 1c,e and Supplementary Fig. 2e). Phosphotriester groups on RNN oligonucleotides were markedly stable in serum (Fig. 1f and Supplementary Fig. 2f). We believe that the generation of siRNNs with chemically reactive aldehyde A-SATE phosphotriester groups will be very useful as this allows for efficient conjugation to hydrazine-containing delivery domain peptides28,29 and targeting domains at defined locations on siRNNs (Fig. 1c,g and Supplementary Fig. 2g).
siRNAs contain a central major groove core that is bordered by phosphates #1 to #11 from both strands (Fig. 1a). Passenger strand phosphate #1 (P#1) sterically opposes guide strand phosphate G#11 across the major groove. Likewise, P#2 opposes G#10, and so on. In contrast, phosphates #12 to #20 on the 3′ ends of both strands are not opposed to other phosphates and have a greater solution accessibility, thus we term them 3′ open phosphates (Fig. 1a). During oligonucleotide synthesis, oxidation of the chiral phosphorous randomly generates either an Sp (outward) or an Rp (inward) diastereomer of the phosphotriester group (Fig. 1h). While there are no apparent steric conflicts with Sp or Rp phosphotriester diastereomers on 3′ open phosphates, major groove phosphates have sufficient area for two Sp paired phosphotriesters or Sp/Rp paired phosphotriesters, but not two Rp paired phosphotriesters (25% frequency) that can potentially result in steric congestion. To test the phosphate rules, we synthesized a collection of passenger and guide RNN oligonucleotides with increasing O-SATE phosphotriesters. Placement of 26× O-SATE phosphotriester groups (~65% of total phosphates) at selected locations was the upper limit to maintain a double-stranded siRNN helix of 19 base pairs (Fig. 1i and Supplementary Fig. 2h). However, moving a single phosphotriester from a 3′ open phosphate position into the major groove destabilized duplex formation (Fig. 1i). Thus, phosphotriester placement has an impact on siRNN duplex formation.
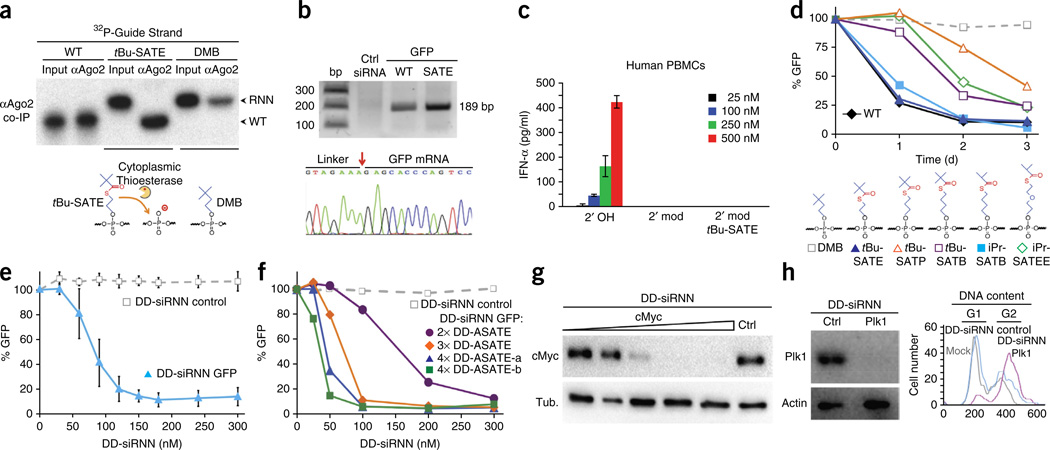
To assay for intracellular conversion of phosphotriester siRNNs into charged phosphodiester siRNAs, we used 32P to label the 5′ end of guide strand oligonucleotides containing wild-type phosphodiesters, six bioreversible tBu-SATE phosphotriesters or six control irreversible DMB phosphotriesters that were then duplexed to unlabeled wild-type passenger strands (Supplementary Fig. 3a). The 32P-labeled siRNNs were transfected into cells, then lysed, and Ago2-associated, 32P-labeled guide strand oligonucleotides were co-immunoprecipitated with anti-Ago2 antibodies (Fig. 2a). As expected, the positive control 32P-labeled wild-type phosphodiester siRNA guide strand co-immunoprecipitated with Ago2 as a full-length guide strand. However, Ago2-associated 32P-labeled tBu-SATE siRNN guide strand migrated as a quantitatively converted, native charged phosphodiester siRNA guide strand (Fig. 2a). In contrast, we detected only background levels of control 32P-labeled irreversible DMB guide strand associated with Ago2 that had not undergone conversion. To determine if in vivo conversion of siRNNs into siRNAs induced a characteristic Ago2 mRNA cleavage, we performed a 5′ rapid amplification of cDNA ends (RACE) analysis4,6 of GFP mRNA from cells transfected with bioreversible tBu-SATE phosphotriester GFP siRNN and positive control GFP siRNA (Supplementary Fig. 2e). Both GFP siRNN and siRNA induced correct Ago2-dependent cleavage, whereas nontargeting control siRNA failed to cleave GFP mRNA (Fig. 2b). Consistent with these observations, transfection of tBu-SATE and O-SATE phosphotriester GFP and luciferase siRNNs (Supplementary Fig. 2e) induced robust dose-dependent RNAi responses, whereas control irreversible DMB siRNNs did not (Supplementary Fig. 3b–g).
Figure 2.
Biological activity of siRNNs. (a) Anti-Ago2 co-immunoprecipitation from cells transfected with 32P-labeled guide strand containing control wild-type charged phosphodiester (WT), six tBu-SATE or six control irreversible DMB phosphotriester oligonucleotides, duplexed to wild-type passenger strands and analyzed by denaturing gel electrophoresis. Note conversion of 32P-labeled tBu-SATE phosphotriester guide strand into wild-type phosphodiester guide strand. Input, 32P-labeled single guide strand. (b) 5′ RACE analysis of GFP mRNA from cells transfected with GFP tBu-SATE (18×) siRNN, control GFP siRNA and nontargeting control siRNA. The correct calculated cDNA RACE fragment size is 189 bp. Red arrow indicates correct Ago2 cleavage site. (c) Analysis of IFN-α induction in human PBMCs at 24 h post-treatment with highly stimulatory β-gal 2′-OH siRNA (2′ OH), 2′-modified siRNA (2′ mod) and O-SATE (14×) phosphotriester siRNN (2′ mod tBU-SATE). (d) Chemical determination of structures surrounding the thioester bond required for intracellular conversion by transfection of GFP siRNNs containing six phosphotriester groups on guide strand duplexed to wild-type passenger strands into GFP-expressing cells, followed by analysis for GFP RNAi responses by flow cytometry at 1–3 days. (e) Dose-dependent GFP RNAi responses by self-delivering GFP DD-siRNNA4 conjugates vs. nontargeting control DD-siRNNA4 conjugates analyzed for GFP RNAi responses by flow cytometry at 48 h. Error bar indicates s.d. (f) Dose-dependent GFP RNAi response comparison of self-delivering GFP siRNNs conjugated to two delivery domains (DD-siRNNA2), three delivery domains (DD-siRNNA3) or two location variations of four delivery domains (DD-siRNNA4) versus nontargeting control DD-siRNNA4 by flow cytometry at 48 h. (g) Dose-dependent cMyc RNAi response (1, 5, 25, 50, 100 nM) in MDA-MB-231-cMyc-HA breast cancer cells by self-delivering cMyc DD-siRNNA4 vs. nontargeting control DD-siRNNA4 (ctrl) conjugate (100 nM) at 48 h. (h) Treatment of U2OS osteosarcoma cells with self-delivering Plk1 DD-siRNNA4 conjugates induced Plk1 RNAi response and induced G2 phase cell cycle arrest, whereas nontargeting control DD-siRNNA4 (ctrl) conjugate did not.
Recognition of exogenous RNAs by the innate immune system’s Toll-like receptors remains problematic for development of RNAi therapeutics30,31. Exposure of primary human peripheral blood mononuclear cells (PBMCs) to a highly stimulatory β-gal siRNA (2′ hydroxyl) sequence32 resulted in a strong dose-dependent stimulation of both interferon-α and tumor necrosis factor-α (Fig. 2c and Supplementary Fig. 4a,b). However, both a fully 2′-modified β-gal siRNA and a β-gal siRNN containing 14 phosphotriesters failed to stimulate an innate immune response, suggesting that siRNNs containing phosphotriesters are not recognized by the innate immune system (Fig. 2c).
To determine the requirements for efficient intracellular phosphotriester conversion, we synthesized GFP siRNNs containing phosphotriester groups that were altered surrounding the thioester bond. Transfection of GFP siRNNs containing six tBu-SATE phosphotriesters on the guide strand with an ethyl linker between the phosphate oxygen and thioester bond, which uses a favored three-membered ring intermediate, efficiently converted to active siRNAs to induce GFP RNAi responses, whereas placement of six irreversible control DMB phosphotriesters did not (Fig. 2d and Supplementary Fig. 4c). GFP siRNNs containing phosphotriesters with a propyl linker (tBu-S-acyl-thiopropyl [tBu-SATP]), which uses a geometrically unfavored four-membered ring during conversion, showed poor induction of GFP RNAi. Addition of a butyl phosphotriester linker (tBu-S-acyl-thiobutyl [tBu-SATB]), which uses a favored five-membered ring intermediate, resulted in an intermediate RNAi response. However, reducing the distal electron donor from a tbutyl group to an isopropyl group (iPr-S-acyl-thiobutyl [iPr-SATB]) restored full GFP RNAi activity (Fig. 2d). By contrast, the combination of a distal isopropyl group and a pentyl linker33 (iPr-S-acyl-thioethoxyethyl [iPr-SATEE]), which uses a six-member ring intermediate, performed poorly, suggesting that the distance between the reactive sulfur and the phosphorus is critical for efficient conversion.
Like siRNAs (~14 kDa), siRNNs (~17 kDa) are too large to passively diffuse across the cell membrane10. Therefore, we selectively conjugated a TAT peptide delivery domain28,29 to siRNNs via conjugatable A-SATE phosphotriesters (Fig. 1c,g). In contrast to the confined small chemical motifs placed distal to SATE groups on monophosphate nucleoside inhibitors20–22, we discovered that regardless of length, the entire delivery domain-conjugated A-SATE phosphotriester group is removed by intracellular thioesterase cleavage at the thioester bond. Based on the steric restrictions of the phosphate rules from above, we synthesized a chimeric passenger strand containing four A-SATE phosphotriesters that was duplexed to an RNN guide strand and conjugated to delivery domain peptides (DD-siRNNA4; Supplementary Fig. 5a). GFP DD-siRNNA4 conjugates self-delivered into cells to induce a dose-dependent GFP knockdown in the entire population in a noncytotoxic fashion, whereas nontargeting control DD-siRNNA4 conjugates failed to knock down GFP (Fig. 2e and Supplementary Fig. 5b,c). To ascertain the requirements for the number and location of A-SATE phosphotriesters, we synthesized a series of RNN oligonucleotides containing passenger and guide strand A-SATE (Supplementary Fig. 5a). Regardless of position, delivery domain conjugation to two A-SATE phosphotriester siRNNs (DD-siRNNA2) induced an intermediate GFP RNAi response compared with DD-siRNNs containing three (DD-siRNNA3) or four (DD-siRNNA4) DD-conjugated A-SATE phosphotriester groups (Fig. 2f). Self-delivered c-Myc DD-siRNNs efficiently induced a dose-dependent c-Myc RNAi response in a breast carcinoma cell line, whereas nontargeting control DD-siRNN conjugates did not (Fig. 2g and Supplementary Fig. 5a). Likewise, self-delivered Plk1 DD-siRNN conjugates containing hydroxyl O-SATE phosphotriesters induced a Plk1 RNAi response and appropriate G2 phase cell cycle arrest in a human osteosarcoma tumor cell line, whereas nontargeting control DD-siRNN conjugates did not (Fig. 2h and Supplementary Fig. 5a).
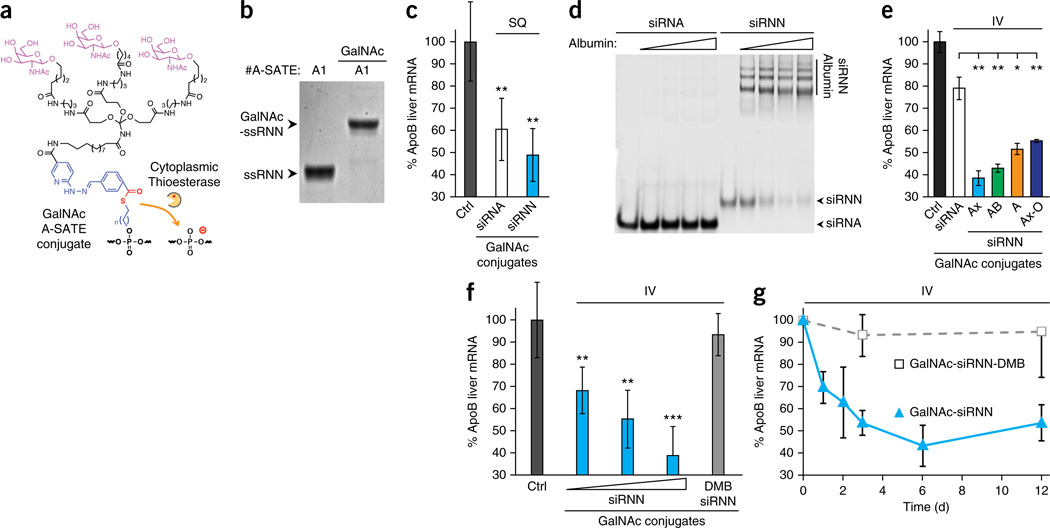
We next tested the in vivo ability of siRNNs to induce RNAi responses versus wild-type siRNAs. To do so, we synthesized a hepatocyte-specific tris-N-acetylgalactosamine (GalNAc) targeting domain (Fig. 3a) that is currently being tested in RNAi clinical trials34. siRNNs targeting liver-specific ApoB contained a 5′ A-SATE (P#1) for conjugation to GalNAc plus nuclease stabilizing 5′ and 3′ terminal phosphorothioates (Fig. 3b and Supplementary Fig. 6). Control charged ApoB siRNAs contained a passenger strand with a single 5′ A-SATE phosphotriester (P#1) for conjugation to tris-GalNAc plus 2′ modifications matched to siRNNs and stabilizing 5′ and 3′ terminal phosphorothioates. In vivo, due to biophysical attributes of the target ApoB mRNA—including Ago2 accessibility and mRNA synthesis rates combined with the siRNA sequence used—a single subcutaneous 25 mg/kg dose into mice of both charged ApoB GalNAc-siRNAs and neutral ApoB GalNAc-siRNNs was required to induce significant ApoB RNAi responses at 72 h, although GalNAc-siRNNs induced a stronger RNAi response (Fig. 3c). Charged siRNAs do not bind serum albumin (Fig. 3d) and are rapidly cleared by the kidneys (half-life <5 min)35, preventing effective intravenous administration. In contrast, we found that charge neutralized siRNNs avidly bound serum albumin (Fig. 3d). A single intravenous 25 mg/kg dose of four variants of ApoB GalNAc-siRNNs into mice induced strong ApoB RNAi responses at 72 h, whereas a 25 mg/kg intravenous dose of GalNAc-siRNAs induced a poor RNAi response (Fig. 3e and Supplementary Fig. 6). GalNAc-siRNNs induced significant ApoB RNAi responses compared with GalNAc-siRNA treated animals (P < 0.05 to <0.01). GalNAc-siRNNs also induced a significant dose-dependent ApoB RNAi response (median effective dose, ED50 ~10 mg/kg) compared with control GalNAc-siRNN-DMBs (P < 0.01 to <0.001) that contained six irreversible DMB phosphotriester groups on the guide strand (Fig. 3f). Lastly, kinetic analyses of single intravenous dose of GalNAc-siRNN into ApoB-treated mice showed partial ApoB RNAi responses at 24 and 48 h, which reached a near maximum at 72 h and were maintained for >12 days (Fig. 3g). These observations demonstrate for the first time the ability to synthesize and self-deliver phosphotriester siRNN conjugates that are intracellularly converted into charged phosphodiester siRNAs that induce robust RNAi responses in vivo.
Figure 3.
In vivo delivery of GalNAc-siRNN conjugates. (a) Structure of GalNAc targeting domain conjugated to A-SATE phosphotriester group. (b) Conjugation of GalNAc targeting domain to RNN oligonucleotide containing one A-SATE phosphotriester. (c) Single-dose subcutaneous (SQ) administration into C57BL/6 mice of charged GalNAc-siRNA and neutral GalNAc-siRNNs targeting ApoB at 72 h by qRT-PCR (25 mg/kg; n = 3, each group). Values normalized to β2-microglobulin from water-treated control (ctrl) group (n = 5). Error bar indicates s.d. **P < 0.01; one-tailed Student’s t-test. (d) Charged siRNA and neutral siRNNs were assayed for albumin binding by incubation with 0, 0.5, 1.0, 1.5 or 2.0 mg/ml serum albumin then separated by gel electrophoresis mobility shift and ethidium bromide staining. Note that due to charge neutralization, siRNNs do not stain as efficiently as charged siRNAs. (e) Single-dose intravenous (IV) administration into C57BL/6 mice of charged GalNAc-siRNA conjugates targeting ApoB compared with GalNAc conjugated neutral siRNNs via A-SATE (A), A-SATB (AB) or Ax-BOE (Ax) phosphotriester groups and containing tBu-SATE and O-SATE (O) phosphotriester groups, as indicated, at 72 h by qRT-PCR (25 mg/kg; n = 3, each group). Values normalized to β2-microglobulin from water-treated control (ctrl) group (n = 5). Error bar indicates s.d. *P < 0.05, **P < 0.01 one-tailed Student’s t-test. (f) Single intravenous dose curve of GalNAc-Ax-siRNN ApoB (10, 17.5, 25 mg/kg; n = 3 each group) compared with irreversible control GalNAc-Ax-siRNN-DMB ApoB (6 irreversible DMBs on guide strand; 25 mg/kg; n = 3) at 72 h by qRT-PCR. Values normalized to β2-microglobulin from water-treated control (ctrl) group (n = 5). Error bar indicates s.d. **P < 0.01; ***P < 0.001 one-tailed Student’s t-test. (g) Single intravenous dose kinetic comparison of GalNAc-siRNN ApoB vs. irreversible control GalNAc-siRNN-DMB ApoB by qRT-PCR (25 mg/kg; n = 3, each time point). Values normalized to β2-microglobulin from water treated control group (n = 5). Error bar indicates s.d. See Supplementary Figure 6 for oligonucleotide structures.
RNAi responses induced by siRNA and microRNA (miRNA) have great potential for treating human disease, especially cancer and viral infections that hold a myriad of unique genetic targets1–3. However, due to their size and highly charged phosphodiester backbone, cellular delivery remains the main technological problem for development of systemic RNAi therapeutics. Because TRBP and Ago2 have strict chemical requirements for binding to charged, A-form double-stranded siRNAs11,12, the siRNA phosphate backbone has—to the best of our knowledge—not previously been amendable to chemical manipulation. We addressed this problem by synthesizing RNAi prodrugs that contain neutral bioreversible phosphotriester groups that are intracellularly converted by thioesterases into charged phosphodiester siRNAs. Because siRNN oligonucleotide synthesis is modular, we synthesized RNN phosphoramidite building blocks that allow for the placement of phosphotriester groups at any location and chimeric combination on siRNNs. In the course of these studies, we synthesized >3,000 RNN oligonucleotides, demonstrating the robustness and versatility of our approach. We note that the phosphotriester chemistry can also be readily applied to miRNA mimetics as miRNN therapeutics. To further facilitate intracellular delivery, we also developed conjugatable A-SATE phosphotriester groups that allowed for siRNN conjugation to delivery and targeting domains. siRNNs have many drug-like properties such as high solubility and serum stability. They can be delivered systemically and produced with high synthetic yields using standard solid-phase oligonucleotide synthesis conditions. The collective attributes of siRNNs combined with distinct phosphotriester terminal groups and conjugation handles opens up entirely new avenues for molecularly sculpting the siRNN surface to optimize pharmacokinetics, cellular delivery and endosomal escape into a given tissue. We performed an in vivo siRNA to siRNN head-to-head comparison using the same ApoB sequence and modifications delivered by conjugation to the GalNAc targeting domain, and found that siRNNs were far superior to siRNAs (P < 0.001). Compared with other liver delivery approaches, such as lipid and synthetic nanoparticles that can achieve lower in vivo doses or cholesterol conjugations (which form large-micrometer particles in blood), GalNAc-siRNN conjugates have multiple positive attributes, including ease of synthesis, serum stability and a 5,000-fold smaller size that dramatically increases its diffusion coefficient compared with nanoparticle approaches. In summary, we believe siRNNs represent a technology that will open new avenues for RNAi therapeutics.
Methods
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Oligonucleotide synthesis and analysis
Unless otherwise noted, all positive control wild-type charged phosphodiester siRNAs used in this study contained the same pattern of 2′ modifications as corresponding phosphotriester siRNNs. The sequences of all oligonucleotides used in this study can be found in Supplementary Table 1. Oligonucleotides were synthesized on a BioAutomation Mermade-6 oligonucleotide synthesizer (Bioautomation). Commercially available phosphoramidites were coupled per manufacturer’s recommendation (R.I. Chemicals; Glen Research). All SATE and DMB phosphoramidites were coupled at 100 mM for 6 min. Standard Q-T columns36 (Glen Research) were used for CPGs. SATE-containing oligonucleotides were deprotected for 4 h in 10% DIA/methanol37 (room temperature) before evaporation. All oligonucleotides were purified by RP-HPLC with a 1200 Series Analytical HPLC (Agilent) on a Zorbax SB-C18 column (Agilent) with linear acetonitrile gradients. Oligonucleotides were analyzed by MALDI-TOF mass spec using a DE-Pro MALDI-TOF mass spec (Applied Biosystems). ssRNA oligonucleotides were analyzed by denaturing gel electrophoresis on 15% acrylamide/7 M Urea denaturing gels and stained with methylene blue. dsRNA analysis was performed by nondenaturing gel electrophoresis using 15% acrylamide nondenaturing gels and stained with ethidium bromide. Double-stranded 2′ fully modified siRNA was modeled and modified in PyMOL.
Secondary deprotection of O-SATE phosphotriesters was performed by addition of 0.75 M TEA.3HF in anhydrous DMSO. Following deprotection, O-SATE RNN oligonucletotides were butanol precipitated with the addition of sodium acetate and n-butanol. Resulting oligonucleotide pellets were redissolved in 20% acetonitrile and gel filtered through illustra NAP-10 columns (GE Healthcare), then purified by RP-HPLC. A-SATE RNN oligonucleotides were deprotected in 80% formic acid following acetal-on RP-HPLC purification. Deprotected oligonucleotide samples were frozen, lyophilized and gel filtered through illustra NAP-10 columns (GE Healthcare).
Peptide synthesis
All protected amino acids and coupling reagents were purchased from Nova Biochem, except Nε-benzyloxycarbonyl-l-lysine tert-butyl ester hydrochloride and Fmoc Aspertic acid (Bachem), BOC 6-hydrazino-nicotinic acid (Solulink) and Fmoc-N-amido-dPEG6-acid (Quanta Biodesign). Delivery domain peptides: HyNic-GG-RKKRRQRRR (TAT); HyNic-GG-(TAT)-PEG18-(TAT)-PEG18-(TAT). Peptide synthesis was performed at 25 µM scale using Fmoc solid phase peptide synthesis on Symphony Quartet peptide synthesizer (Ranin) and rink-amide MBHA resin as solid support. All Hynic peptides were cleaved and deprotected using standard conditions (92.5% TFA, 2.5% acetone, 2.5% water, 2.5% TIS) for 2 h. Crude peptides were precipitated using cold diethylether and purified using prep-scale RP-HPLC with an Agilent Prep C18 30 × 250 mm column. Peptide purity was confirmed by mass spectrometry using α-CHCA matrix (Voyager, Applied Biosystems DE-Pro MALDI-TOF).
siRNN conjugation
For DD peptide conjugations, duplexed siRNN oligonucleotides were conjugated with DD-HyNic peptides (1:5) in the presence of 10 mM HEPES (pH 5.5), 20 mM aniline and 50 mM NaCl for 1 h at RT. Conjugate reaction mixtures were solubilized with 5.1 M acetic acid. After conjugation, samples were frozen and lyophilized. Resulting conjugated siRNN pellets were resuspended in water and spun with Amicon Ultra-0.5 ml 30 kDa Spin Filters (Millipore) to remove unconjugated DD peptide. siRNN conjugates were analyzed by silver stained SDS-PAGE. For tris-GalNAc conjugation, single-stranded RNN oligonucleotides and GalNAc (1:5 per A-SATE phosphotriester group) were conjugated with 50% acetonitrile plus 100 mM aniline for 1 h at RT. After conjugation, samples were frozen, lyophilized and pellets were washed with ethanol to remove unconjugated tris-GalNAc. GalNAc-siRNN conjugates were resuspended in water, frozen, lyophilized, resuspended in 50% acetonitrile and duplexed with complementary ssRNN oligonucleotides. GalNAc-siRNN conjugates were analyzed by PAGE and methylene blue staining.
Cell culture and siRNA transfection
All cell lines were obtained from the ATCC and routinely tested negative for mycoplasma. H1299-dGFP, U2OS and MDA-MB-231-cMyc-HA cells were grown in 10% FBS and high glucose DMEM (Life Sciences). H1299-dGFP cells38 express a destabilized dGFP with a short half-life (<2 h). For transfection, cells were plated in 24-well plates at 40,000 cells/well. Before transfection cells were washed twice with serum-free DMEM. 100 µl samples of corresponding siRNA concentrations in serum-free DMEM were mixed with 100 µl transfection mixture (98 µl serum-free DMEM/2 µl of Lipofectamine 2000) and left at room temperature for 30 min before treatments. Transfection solutions were left on cells for 8 h before removal and the addition of DMEM with 5% FBS. Cells were trypsinized and analyzed by flow cytometry on a BD Biosciences LSRII (BD Sciences, San Jose, CA). Error bar indicates s.d. from three biological repeats.
IFN-α and TNF-α analyses
With approval from the University of California Institutional Review Board, blood from healthy human volunteers was collected into EDTA-containing tubes, and human PBMCs were isolated by standard density gradient centrifugation with Ficoll-Paque PLUS (Amersham Biosciences) at 2,000 rpm for 20 min at 20 °C. To remove platelets, PBMCs were washed 4× in 50 ml PBS, centrifuged at 1,500 rpm for 8 min at 4 °C. 8 × 105 freshly isolated PBMCs were seeded into 96 well-plate (4 × 105 cells/well) and treated with 25, 100, 250 or 500 nM β-gal29 siRNA or siRNN. Culture supernatants were collected at 24 h postaddition, and assayed for IFN-α and TNF-α by ELISA (R&D Systems).
Preparation and transfection of 32P-labeled siRNA
400 pmol of guide strand was 5′ labeled with 2.5 units T4 polynucleotide kinase (NEB) and 200 µCi γ−32P-ATP (6000 Ci/mmol) in a 50 µl volume for 20 min. Reaction was inactivated for 20 min at 65 °C and free γ-32P-ATP was removed from the reaction mixture using Amicon Ultra 3K centrifugal filter (Millipore). 400 pmol of wild-type passenger strand was added and annealed by heating reaction mixture to 65 °C and cooling to room temperature. 400 pMol of labeled siRNA was diluted to a volume of 250 µl in Optimem-I and mixed with 250 µl Optimem-I containing 10 µl Lipofectamine 2000. After a 20 min incubation the mixture was added to 2.4 × 106 H1299-dGFP cells in 8 ml DMEM containing 5% FBS and seeded in a 10 cm tissue culture dish. Cells were transfected for 12 h and washed with PBS before medium replacement.
Ago2 co-immunoprecipitation
36 h post-transfection, cells were washed with ice cold PBS and lysed in 1 ml ice cold lysis buffer (20 mM Tris-Hcl [pH 7.9], 250 mM NaCl, 0.5% Triton X-100, 0.1% DEPC, 0.5% protease inhibitor cocktail and 1% phosphatase inhibitor cocktail II [Sigma]) for 10 min on ice. Lysates were cleared at 16,000 RCF for 10 min at 4 °C. 900 µl of cleared lysate was incubated with 100 µl of washed protein G-Sepharose 4B slurry (Invitrogen) on rotator at 4 °C. After 2 h, beads were spun down and 850 µl of lysate was transferred to a tube containing 75 µl of washed protein G-Sepharose 4B slurry preincubated for 2 h with 10 µl anti-human Argonaute 2 antibody (4G8, 0.9 mg/ml, Wako Chemical USA) and incubated on a rotary shaker for 4 h. Beads were washed 4 times with ice cold lysis buffer, eluted with 50 µl 7 M urea/ 0.1% bromophenol at 95 °C for 1 min. Eluate and input siRNA were counted on a scintillation counter and input counts were normalized to IP sample, except for DMB RNN sample, of which an equal volume of eluate to tBu-SATE RNN sample was loaded on 15%/7 M Urea PAGE. Gels were exposed to film overnight or 30 min to a phosphoimager screen at −20 °C.
Immunoblot analysis and cell cycle analysis
All immunoblots were performed as described28,38 and used the following primary antibodies, anti-Plk1 (3F8, Santa Cruz Biotechnology), anti-cMyc-HA (3F10, Roche), and secondary antibodies, goat anti-mouse IgG HRP (SC-2005, Santa Cruz Biotechnology). Immunoblots were developed on ChemiDoc (Bio-Rad) and quantified using ChemiDoc XRS (Bio-Rad) sub-saturating linear signals. Cell cycle position for Plk1 knockdown was determined by flow cytometry of 104 cells stained with propidium iodide using a BD LSR-II FACS (Becton Dickinson).
5′ RACE
For 5′ RACE, 200 × 103 cells were reverse transfected with siRNA and siRNN at 100 nM using Lipofectamine 2000. Cells were harvested in Trizol (Invitrogen) 48 h post-transfection. The aqueous phase was mixed with equal volume of 70% ethanol, loaded on an RNAeasy column (Qiagen) and RNA was prepared according to the manufacturer’s instruction. 3 µg RNA was ligated to 0.25 µg GeneRacer RNA oligonucleotide (Invitrogen). Ligated RNA was reverse transcribed with superscript III and dT(20). cDNA was amplified with the GeneRacer 5′ primer and a gene-specific primer (5′ TGGGCAGCGTGCCATCATCC for GFP, ATCATCCCCCTCGGGTGTAATCAGAA for GL3) using a step down PCR protocol. Second round amplification was performed using the GeneRacer 5′ nested primer and a gene-specific nested primer (5′-ATCATCCTGCTCCTCCACCTCCGG for GFP, TCAGTGAGCCCATATCCTTGCCTGAT for GL3) at 60 °C. PCR products were analyzed on a 1.2% Agarose/TBE gel and bands of predicted size were excised, purified on S.N.A.P. columns, cloned into TOPO-PCR4 vector (Invitrogen) and sequenced.
Real-time qRT-PCR
200 × 103 cells were reverse transfected in a 12-well plate. RNA was isolated with Trizol according to the manufacturer. 10 ng of total RNA was used for real-time PCR using the Taqman RNA-to-Ct 1 step kit and Taqman pre-designed probe and primers (Applied Biosystems). GFP primers were as described38. For liver RNA isolation, 50 mg of liver from mice was homogenized with a 21G needle in Trizol. 1 µg RNA was used to synthesize cDNA with iScript cDNA Synthesis Kit (Bio-Rad) and qRT-PCR was performed on 10 ng cDNA with SYBR Green PCR Master Mix (Applied Biosystems) on Applied Biosystems 7300 Real-Time PCR System. ApoB primers: forward, 5′-ACAGGAGCTTACTCCAACGC; reverse, 5′-AGCTCATACCTTGTGTCCCC. B2M primers: forward, 5′-ACCGTCTACTGGGATCGAGA; reverse, 5′-TGCTATTTCTTTCTGCGTGCAT.
Preclinical animal models
Purified GalNAc-siRNN and GalNAc-siRNA conjugates were administered subcutaneously or intravenously into randomly chosen 6- to 8-week-old female C57B/6 mice (Jackson Laboratory) in doses of 10, 17.5 or 25 mg/kg diluted in water for a total volume injection of 10 µl/g mouse. All animals in a given treatment protocol were analyzed at the same time point; therefore, no blinding was performed. Mice were maintained and euthanized according to the UCSD committee on animal welfare. Livers were isolated for mRNA extraction. For statistical analyses, data are expressed as mean ± s.d., as indicated, and compared by Student’s t-test. We assign statistical significance at P < 0.05. All animals were treated in accordance with the UCSD Institutional Animal Care and Use Committee.
Serum stability
50 pmol of siRNA and siRNN GFP guide strands were incubated in 50% human complement active serum (Innovative Research) at 37 °C. At indicated time points an aliquot was taken and diluted 10-fold in 7M urea loading buffer. Samples were heated to 95 °C for 1 min, snap cooled on dry ice and stored at −80 °C until analysis on a 15%/7 M urea PAGE. Gels were analyzed on an Odyssey imager (LI-COR).
Luciferase assay
H1299 cells stably expressing firefly luciferase (pGL3; Promega) were maintained and transfected as described38. 24 h post-transfection, cells were split into 96-well plates in triplicate to reach 90% confluency at the day of luciferase measurement. Luciferase assay was performed using Promega’s dual-glow kit (E2920) according to manufacturer’s instructions. Briefly, cells were washed in PBS twice, 100 µl of passive lysis buffer was added and incubated for 20 min. 20 µl of lysate was transferred to a white wall 96-well assay plate (duplicate readings). Luminescence was counted in with the injector set to 80 µl of solution 1 (d-luciferin) and signals were integrated over a 5 s read on an IVIS Spectrum from PerkinElmer. Luciferase signals were corrected over protein concentration as determined by triplicate Bradford reading.
Phosphoramidite synthesis and reagents
Detailed phosphoramidite synthesis protocols are presented below (Supplementary Note). All phosphoramidites synthesized were fully characterized by ESI mass spectrometry, 1H and 31P NMR (Supplementary Figs. 7–24). All commercial chemicals were reagent grade and were used without further purification except where otherwise specified. All nucleoside precursors were purchased from R.I. Chemical Inc. and Chemgene Corporation. Bis(N,N-diisopropyl-amino)chlorophosphine and N,N-diisopropylethylamine were from Sigma-Aldrich. 0.25 M Ethyl thio tetrazole solution in acetonitrile was from Chemgene. All oligonucleotide synthesis reagents were purchased from Glen Research and R.I. Chemical Inc.
Analytical and preparative methods
All experiments involving air- and/or moisture sensitive compounds were carried out under an argon atmosphere. Thin-layer chromatography was performed on precoated silica gel plates (Kieselgel 60F254, Merck) and preparative silica gel flash chromatography was performed on Combi Flash Rf (Teledyne Isco) using Redi Sep Rf Columns. 1H NMR spectra were recorded on a Varian 300 and Varian 400 Mercury spectrometer at 300 and 400 MHz in CDCl3. Chemical shifts are expressed in parts per million relative to the central line of CDCl3 set at 7.26 p.p.m. and the spectra were processed using Mnova NMR software, Mestrelab Research. 31P-NMR spectra were recorded on Varian 300 Mercury spectrometer relative to 85% phosphoric acid as an external standard. Mass spectra were recorded at UCSD Chemistry and Biochemistry Mass Spectroscopy Facility using either an LCQDECA (Finnigan) ESI with a quadrupole ion trap or an MAT900XL (ThermoFinnigan) FAB double focusing mass spectrometer or a Quattro Ultima Triple Quadrupole mass spectrometer.
Acknowledgments
We thank Y. Tor (UCSD) for critical input, Y. Su (UCSD) for help with mass spectrometry, and Z. Rudolph and L.D.F. Vasconcelos for technical assistance. B.R.M., A.S.H. and A.D.S. were supported by a T32 Cancer Biology Training grant (NCI); A.S.H. was also supported by a Blasker Award from The San Diego Foundation; A.P. was supported by a C.I.R.M. Fellowship; C.P.-A. was supported by fellowships from Knut and Alice Wallenberg Foundation and Sweden-America Foundation; J.C.H. was supported by the CT2 training grant (N.C.I.); P.L. was supported by a Swedish Research Council grant; A.D.K. was supported by a IRACDA Training grant (N.I.H.). This work was supported by the W.M. Keck Foundation (S.F.D.), the Department of Defense (S.F.D.), SCOR grant from the Leukemia & Lymphoma Society (S.F.D.), the Pardee Foundation (S.F.D.), a grant from an anonymous donor (S.F.D.) and the Howard Hughes Medical Institute (S.F.D.).
Footnotes
Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
B.R.M. designed and performed experiments, analyzed and interpreted data, synthesized RNN oligonucleotides; K.G. designed and synthesized phosphotriesters, phosphoramidites, conjugating approaches, and analyzed and interpreted data; A.S.H. performed in vivo experiments, analyzed and interpreted data, and synthesized RNN oligonucleotides; C.P.-A. and A.D.S. performed experiments, analyzed and interpreted data, and synthesized RNN oligonucleotides; A.v.d.B., J.C.H., A.E., A.D.K. A.P., P.L. and M.K. performed experiments and analyzed data; C.F.D. synthesized RNN oligonucleotides; N.Y. and E.G. helped with data analysis; X.-S.C. synthesized peptides; S.F.D. conceived of siRNN concept, designed experiments, and analyzed and interpreted data. B.R.M., K.G. and S.F.D. wrote the manuscript, and all authors refined the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat. Rev. Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson BL, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat. Rev. Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–320. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 5.Rettig GR, Behlke MA. Progress toward in vivo use of siRNAs-II. Mol. Ther. 2012;20:483–512. doi: 10.1038/mt.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bramsen JB, Kjems J. Chemical modification of small interfering RNA. Methods Mol. Biol. 2011;721:77–103. doi: 10.1007/978-1-61779-037-9_5. [DOI] [PubMed] [Google Scholar]
- 7.Meade BR, Dowdy SF. The road to therapeutic RNA interference (RNAi): tackling the 800 pound siRNA delivery gorilla. Discov. Med. 2009;8:253–256. [PubMed] [Google Scholar]
- 8.Zhou J, Shum K-T, Burnett JC, Rossi JJ. Nanoparticle-based delivery of RNAi therapeutics: progress and challenges. Pharmaceuticals. 2013;6:85–107. doi: 10.3390/ph6010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 10.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 11.Joshua-Tor L, Hannon GJ. Ancestral roles of small RNAs: an Ago-centric perspective. Cold Spring Harb. Perspect. Biol. 2011;3:a003772. doi: 10.1101/cshperspect.a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reese CB. Oligo- and poly-nucleotides: 50 years of chemical synthesis. Org. Biomol. Chem. 2005;3:3851–3868. doi: 10.1039/b510458k. [DOI] [PubMed] [Google Scholar]
- 14.Gates KS. An overview of chemical processes that damage the cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem. Res. Toxicol. 2009;22:1747–1760. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grajkowski A, Wilk A, Chmielewski MK, Phillips LR, Beaucage SL. The 2-(N-formyl-N-methyl) aminoethyl group as a potential phosphate/thiophosphate protecting group in solid-phase oligodeoxyribonucleotide synthesis. Org. Lett. 2001;3:1287–1290. doi: 10.1021/ol0156852. [DOI] [PubMed] [Google Scholar]
- 16.Dellinger DJ, Sheehan DM, Christensen N, Lindberg JG, Caruthers MH. Solid phase chemical synthesis of phosphonoacetate and thiophosphonoacetate oligodeoxynucleotides. J. Am. Chem. Soc. 2003;125:940–950. doi: 10.1021/ja027983f. [DOI] [PubMed] [Google Scholar]
- 17.Tanabe K, Ando Y, Nishimoto S. Reversible modification of oligodeoxynucleotides: click reaction at phosphate group and alkali treatment. Tetrahedron Lett. 2011;52:7135–7137. [Google Scholar]
- 18.Krishna H, Caruthers MH. Alkynyl phosphonate DNA: a versatile “click”able backbone for DNA-based biological applications. J. Am. Chem. Soc. 2012;134:11618–11631. doi: 10.1021/ja3026714. [DOI] [PubMed] [Google Scholar]
- 19.Tosquellas G, et al. The pro-oligonucleotide approach: solid phase synthesis and preliminary evaluation of model pro-dodecathymidylates. Nucleic Acids Res. 1998;26:2069–2074. doi: 10.1093/nar/26.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Périgaud C, et al. Rational design for cytosolic delivery of nucleoside monophosphates: “SATE” and “DTE” as enzyme-labile transient phosphate protecting groups. Bioorg. Med. Chem. Lett. 1993;3:2521–2526. [Google Scholar]
- 21.Lefebvre I, et al. Mononucleoside phosphotriester derivatives with S-acyl-2-thioethyl bioreversible phosphate-protecting groups: intracellular delivery of 3′-azido-2’,3′-dideoxythymidine 5′-monophosphate. J. Med. Chem. 1995;38:3941–3950. doi: 10.1021/jm00020a007. [DOI] [PubMed] [Google Scholar]
- 22.Faraj A, et al. Intracellular metabolism of beta-L-ddAMP-bis(tbutylSATE), a potent inhibitor of hepatitis B virus replication. Nucleosides Nucleotides. 1999;18:987–988. doi: 10.1080/15257779908041623. [DOI] [PubMed] [Google Scholar]
- 23.Bologna JC, Morvan F, Imbach JL. The prooligonucleotide approach: synthesis of mixed phosphodiester and SATE phosphotriester prooligonucleotides using H-Phosphonate and phosphoramidite chemistries. Eur. J. Org. Chem. 1999;9:2353–2358. [Google Scholar]
- 24.Guzaev AP, Balow G, Manoharan M. Synthesis of chimerical oligonucleotides containing internucleosidic phosphodiester and S-pivaloyl mercaptoethyl phosphotriester linkages. Nucleosides Nucleotides. 1999;18:1391–1392. doi: 10.1081/NCN-100002481. [DOI] [PubMed] [Google Scholar]
- 25.Peyrottes S, et al. SATE pronucleotide approaches: an overview. Mini Rev. Med. Chem. 2004;4:395–408. doi: 10.2174/1389557043404007. [DOI] [PubMed] [Google Scholar]
- 26.Breslow R, Xu R. Recognition and catalysis in nucleic acid chemistry. Proc. Natl. Acad. Sci. USA. 1993;90:1201–1207. doi: 10.1073/pnas.90.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beaucage SL. Solid-phase synthesis of siRNA oligonucleotides. Curr. Opin. Drug Discov. Devel. 2008;11:203–216. [PubMed] [Google Scholar]
- 28.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 29.van den Berg A, Dowdy SF. Protein transduction domain delivery of therapeutic macromolecules. Curr. Opin. Biotechnol. 2011;22:888–893. doi: 10.1016/j.copbio.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 31.Whitehead KA, Dahlman JE, Langer RS, Anderson DG. Silencing or stimulation? siRNA delivery and the immune system. Annu. Rev. Chem. Biomol. Eng. 2011;2:77–96. doi: 10.1146/annurev-chembioeng-061010-114133. [DOI] [PubMed] [Google Scholar]
- 32.Judge AD, et al. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Petersen S. Self-delivering bio-labile phosphate protected pro-oligos for oligonucleotide based therapeutics and mediating RNA interference. USPTO. 2014;8:691–971. [Google Scholar]
- 34.Sliedregt LJAM, et al. Design and synthesis of novel amphiphilic dendritic galactosides for selective targeting of liposomes to the hepatic asialoglycoprotein receptor. J. Med. Chem. 1999;42:609–618. doi: 10.1021/jm981078h. [DOI] [PubMed] [Google Scholar]
- 35.Gao S, et al. The effect of chemical modification and nanoparticle formulation on stability and biodistribution of siRNA in mice. Mol. Ther. 2009;17:1225–1233. doi: 10.1038/mt.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pon RT, Yu S. Hydroquinone-O,O’-diacetic acid (‘Q-linker’) as a replacement for succinyl and oxalyl linker arms in solid phase oligonucleotide synthesis. Nucleic Acids Res. 1997;25:3629–3635. doi: 10.1093/nar/25.18.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuijpers WH, Huskens J, Koole LH, van Boeckel CA. Synthesis of well-defined phosphate-methylated DNA fragments: the application of potassium carbonate in methanol as deprotecting reagent. Nucleic Acids Res. 1990;18:5197–5205. doi: 10.1093/nar/18.17.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eguchi A, et al. Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein. Nat. Biotechnol. 2009;27:567–571. doi: 10.1038/nbt.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]