Abstract
Carrier proteins (CPs) play a central role in nonribosomal peptide synthetases (NRPSs) as they shuttle covalently attached substrates between active sites. Understanding how the covalent attachment of a substrate (loading) influences the molecular properties of CPs is key to determining the mechanism of NRPS synthesis. However, structural studies have been impaired by substrate hydrolysis. Here, we used nuclear magnetic resonance spectroscopy to monitor substrate loading of a CP and to overcome hydrolysis. Our results reveal the spectroscopic signature of substrate loading and provide evidence of molecular communication between an NRPS carrier protein and its covalently attached substrate.
Nonribosomal peptide synthetases (NRPSs) are bacterial and fungal enzymatic systems that produce complex secondary metabolites from simple starting materials such as amino or aryl acids, many of which have found use as antibiotics and anti-cancer agents1. NRPSs possess a remarkable assembly line architecture, in which substrates are covalently attached to contiguous modules and condensed to form the final product. Each module is comprised of a core set of conserved domains and a long standing goal of the field is swapping domains or modules with differing substrate specificities so as to generate novel pharmaceuticals2. Unfortunately the molecular mechanisms of NRPS synthesis, and particularly domain communication, remain largely unknown, impeding progress in reprogramming NRPS assembly lines. Amongst NRPS domains, carrier proteins (CPs) play a central role as they tether the substrates to the assembly line and, hence, they visit many catalytic domains during NRPS synthesis. CPs are first converted from an inactive apo to an active holo form via covalent attachment of a 4’-phosphopantetheine arm (4’-PP) onto a conserved serine. Next, adenylation (A) domains catalyze both substrate adenylation and thioester bond formation between the activated substrate and the 4’-PP of holo carrier proteins to generate a substrate loaded CP. Finally, condensation domains catalyze the peptide bond formation between two substrates loaded on neighboring CPs to extend the peptide. NMR and crystallographic studies indicate that NRPS modules are not rigid, but their domains are subject to inter- and intra-domain dynamics3–7. Moreover, attachment of the 4’-PP altered the structure and dynamics of an isolated CP5. Studies of structurally related fatty acid synthases (FAS) and polyketide synthases (PKS) have implicated substrate loading in influencing large scale domain rearrangements8, 9. However, the lability of NRPS substrate thioester bonds has precluded similar studies of loaded NRPS carrier proteins. Understanding how CPs efficiently orchestrate sequential, transient interactions with partner domains and elucidating the role of tethered substrates in modulating these interactions is of vital importance to understanding NRPS assembly line synthesis and, ultimately, rationally redesigning these systems. Here, we exploited the non-invasive nature of nuclear magnetic resonance (NMR) to overcome hydrolysis and study a loaded aryl carrier protein (ArCP) from yersiniabactin synthetase. Our results reveal that NRPS ArCPs interact either directly or indirectly with the substrates attached at the end of the 20 Å long 4’-PP.
In the yersiniabactin synthetase system, the free-standing A domain YbtE initiates synthesis by loading salicylate (Sal) onto the holo aryl carrier protein of the multidomain protein HMWP210. The excised ArCP was overexpressed in E. coli BL21 (DE3) ΔEntD cells (a gift of Drs. Chalut and Guilhot) and purified to yield pure, homogeneous apo ArCP. Apo ArCP was phosphopantetheinylated in vitro and purified to obtain holo ArCP. To study the loaded form of ArCP, two major obstacles had to be addressed: hydrolysis and transthiolation from thiol containing reducing agents,11 which were necessary to prevent disulfide bond formation in holo ArCP. Transthiolation was avoided by using tris(2-carboxyethyl)phosphine as a reducing agent. Although hydrolysis of thioesters is slower than transthiolation, it was rapid enough to preclude quantitative analysis of loaded ArCP.
Indeed, when ArCP was loaded with Sal (confirmed by MALDI mass spectrometry (MALDI MS)) and freshly purified, NMR spectra featured signals of both holo ArCP and a previously unobserved form. Unfortunately, the new signals decreased over time, raising the possibility that they were an artifact of sample preparation and not reporters of substrate loading. Purified, loaded ArCP was therefore unsuitable for NMR studies.
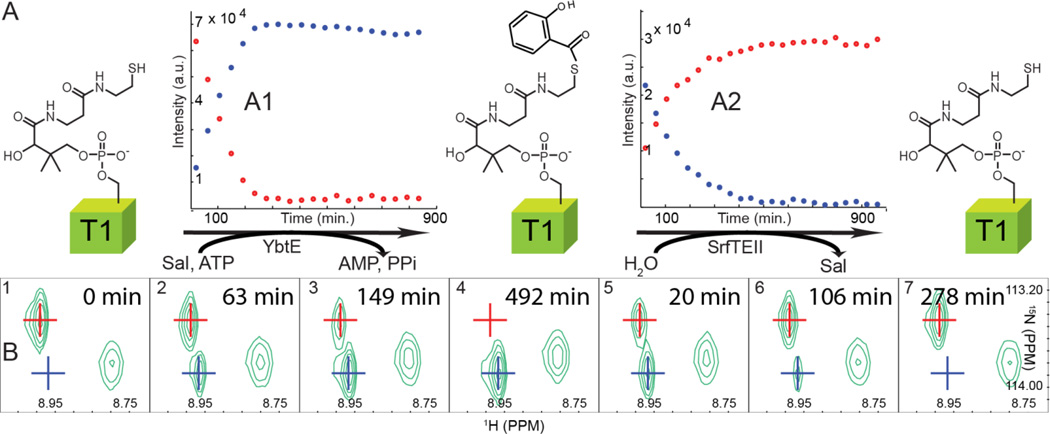
To allow for prolonged detection of loaded ArCP, we exploited the isotope editing ability of NMR, and we generated the loaded form in situ to bypass the need for purifying loaded ArCP. Sal and ATP (2 mM each) were added in large excess to 15N-labeled holo ArCP (300 µM). No interaction between holo ArCP and the reagents was detected in HN-HSQC spectra (SI, Figure S1). We then added catalytic quantities of YbtE (100 nM) and collected a series of HN-HSQCs. At such concentrations, the binding of YbtE to ArCP does not induce shifts in NMR signals. During the reaction, signals of loaded ArCP increase while signals of holo ArCP decay (as shown for the phosphopantetheinylation site, Ser52, in Figure 1 A1, B1-4). MALDI MS confirmed the conversion into ArCP loaded with Sal. To demonstrate that all signals observed by NMR report on substrate loading and not some undesired side effects, we induced substrate hydrolysis by enzymatic catalysis. We added the promiscuous thioesterase SrfTEII12 to purified loaded ArCP, and the NMR signals of loaded ArCP disappeared as those of holo ArCP reappeared (Figure 1 A2, B5-7). The HN-HSQC collected following the completion of SrfTEII-catalyzed hydrolysis (confirmed by MALDI MS) overlays perfectly with that of holo ArCP, demonstrating that the new peaks observed upon incubation of holo ArCP with ATP, Sal, and YbtE are indeed the spectroscopic signature of substrate loading (Figure S5). Comparison between the kinetics of uncatalyzed hydrolysis (Figure S4) and substrate loading suggested that hydrolysis may be compensated by continuous reloading of Sal on ArCP. Indeed, spectra of loaded ArCP recorded after 5 days displayed no significant regeneration of holo ArCP in presence of YbtE, Sal, and ATP, whereas in the absence of YbtE, Sal, and ATP, ~57% of purified loaded ArCP hydrolyses over that time (Figures S4 and S6). Thus, adding catalytic amounts of the cognate A domain overcomes hydrolysis of loaded monomers for a duration amenable for NMR studies. This method complements other strategies used to circumvent hydrolysis in loaded carrier proteins (SI, S6).
Figure 1.
(A) Reactions catalyzed by YbtE and SrfTEII and their time-course (A1, A2, respectively), here monitored by the signal of Ser52, the phosphopantetheinylation site. (B) Signal of Ser52 during the loading reaction (B1-B4) and following the addition of SrfTEII after purification to remove YbtE and substrates (B5-B7). Blue: loaded ArCP; red: holo ArCP.
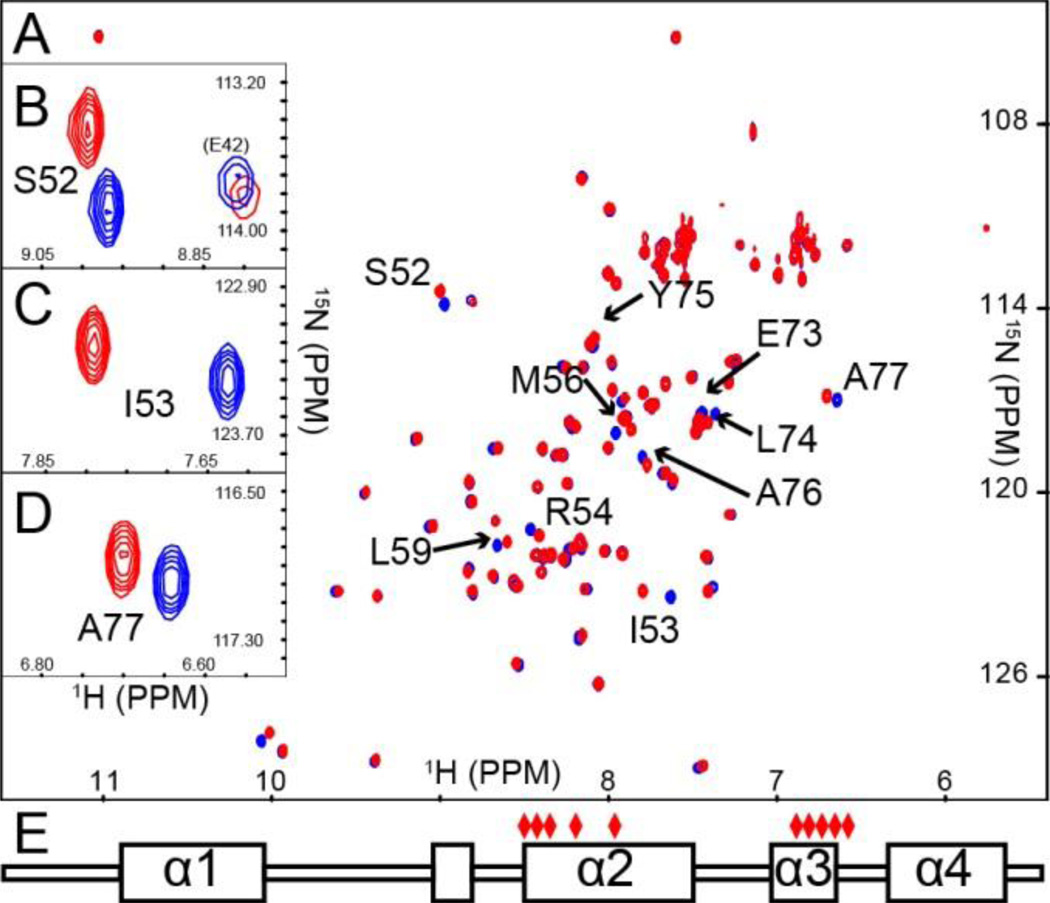
The changes we observe in NMR spectra of loaded ArCP may reflect many molecular events. NMR signals in HN-HSQC report on the local chemical environment of the amide group of each residue. Differences between spectra of holo and loaded ArCP, so-called chemical shift perturbations (CSPs), can indicate a variety of events, such as direct contact with the salicylate moiety, repositioning of the 4’-PP, structural alterations, and modulation of dynamics. Figure 2A shows an overlay of holo and loaded ArCP spectra. Simple inspection reveals that no massive structural rearrangement occurs upon loading, since most signals are the same in both spectra. However, a significant number of peaks shift markedly, indicating that loading Sal impacts the related amide groups. To gain insights into the origins of the CSPs of holo/loaded ArCP, we assigned the 1H and 15N resonances of apo 15N-13C-ArCP using conventional experiments13, 14 and transposed the assignment of apo ArCP to holo 15N-ArCP and to loaded 15N-ArCP with NOESY-HN-HSQC spectra. Figure 2B, C, and D highlight four peaks that show varying degrees of CSP. Mapping the largest CSPs (Figure S7) onto the secondary structure of ArCP as determined with chemical shift indexing,15, 16 reveals distinct clusters of residues affected by loading (Figure 2E). CSPs of residues around the phosphopantetheinylation site (Ser52, N-terminus of helix 2) likely reflect a change in conformation of the 4’-PP. CSPs of residues in helix 3 together with residues in the middle of helix 2 may indicate a direct substrate interaction reminiscent of that observed PKS and FAS acyl carrier proteins.17, 18 In both FAS and PKS, changing tethered substrates repositions helices 2 and 3,19, 20 and it was suggested that these structural variations may modulate binding events9. Various NMR experiments can test if salicylate alters the structure and/or dynamics of ArCP and modulates protein-protein interactions. Our newly designed conditions will permit such measurements while ArCP remains loaded.
Figure 2.
(A) Overlay of holo (red) and salicylate loaded (blue) ArCP HN-HSQCs. Signals with CSP greater than 1 standard deviation above the median are indicated. Insets show zooms on the signals of (B) Ser52, (C) Ile53, and (D) Ala77. (E) Mapping the residues with significant CSPs on the secondary structure of ArCP shows two distinct clusters of residues spanning both helices 2 and 3.
In summary, we used NMR to directly monitor NRPS substrate loading, thereby providing the first atomic level description of this process. We found that NRPS substrates directly or indirectly interact with their cognate carrier proteins. Whether substrates bind to CPs or induce conformational fluctuations, substrate loading is expected to modulate the binding affinity of CPs toward partner catalytic domains. Decades of biochemical studies have demonstrated interplay between carrier proteins and their substrates during catalytic steps involving various domains21, 22 and our method provides a framework for investigating the molecular determinants of this interplay.
Supplementary Material
ACKNOWLEDGMENT
We thank Drs. Christian Chalut and Christophe Guilhot for providing the E. coli BL21(DE3)ΔEntD strain. We thank Dr. Craig Townsend for discussions on the manuscript and NRPS substrate hydrolysis, and Scott Nichols for preparing SrfTEII. We thank the Mass Spectrometry and Proteomics Facility at the Johns Hopkins School of Medicine for use of their instrument.
Funding Sources
Work supported by National Institutes of Health (NIH) Grant RO1 GM104257. A.C.G. supported by American Heart Association Mid-Atlantic Affiliate Predoctoral Fellowship 14PRE20460253.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Supplementary figures, tables, and experimental procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Walsh CT. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 2.Stachelhaus T, Schneider A, Marahiel MA. Science. 1995;269:69–72. doi: 10.1126/science.7604280. [DOI] [PubMed] [Google Scholar]
- 3.Frueh D, Arthanari H, Koglin A, Vosburg D. Nature. 2008;454:903–906. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanovic A, Samel SA, Essen L-O, Marahiel MA. Science. 2008;321:659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 5.Koglin A, et al. Science. 2006;312:273–276. doi: 10.1126/science.1122928. [DOI] [PubMed] [Google Scholar]
- 6.Gulick AM. ACS Chem. Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloudoff K, Rodionov D, Schmeing TM. J. Mol. Biol. 2013;425:3137–3150. doi: 10.1016/j.jmb.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whicher JR, et al. Nature. 2014;510:560–564. doi: 10.1038/nature13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen C, et al. Nature. 2014;505:427–431. doi: 10.1038/nature12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gehring AM, Mori I, Perry RD, Walsh CT. Biochemistry. 1998;37:11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- 11.Suo Z, Walsh CT, Miller DA. Biochemistry. 1999;38:14023–14035. doi: 10.1021/bi991574n. [DOI] [PubMed] [Google Scholar]
- 12.Schwarzer D, Mootz HD, Linne U, Marahiel MA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14083–14088. doi: 10.1073/pnas.212382199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sattler M, Schleucher J. Prog. Nucl. Magn. Reson. Spectrosc. 1999;34:93–158. [Google Scholar]
- 14.Ferentz AE, Wagner G. Q. Rev. Biophys. 2002;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 15.Shen Y, Delaglio F, Cornilescu G, Bax A. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wishart D, Sykes B. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 17.Mayo KH, Prestegard JH. Biochemistry. 1985;24:7834–7838. doi: 10.1021/bi00347a049. [DOI] [PubMed] [Google Scholar]
- 18.Roujeinikova A, et al. Structure. 2002;10:825–835. doi: 10.1016/s0969-2126(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 19.Płoskoń E, et al. Chem. Biol. 2010;17:776–785. doi: 10.1016/j.chembiol.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 20.Evans SE, et al. J. Mol. Biol. 2009;389:511–528. doi: 10.1016/j.jmb.2009.03.072. [DOI] [PubMed] [Google Scholar]
- 21.Miller DA, Walsh CT. Biochemistry. 2001;40:5313–5321. doi: 10.1021/bi002905v. [DOI] [PubMed] [Google Scholar]
- 22.Marshall CG, Burkart MD, Keating TA, Walsh CT. Biochemistry. 2001;40:10655–10663. doi: 10.1021/bi010937s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.