Abstract
High-density single nucleotide polymorphism (SNP) mapping arrays have identified chromosomal features whose importance to cancer predisposition and progression is not yet clearly defined. Of interest is that the genomes of normal somatic cells (reflecting the combined parental germ-line contributions) often contain long homozygous stretches. These chromosomal segments may be explained by the common ancestry of the individual’s parents and thus may also be called autozygous. Several studies link consanguinity to higher rates of cancer, suggesting that autozygosity (a genomic consequence of consanguinity) may be a factor in cancer predisposition. SNP array analysis has also identified chromosomal regions of somatic uniparental disomy (UPD) in cancer genomes. These are chromosomal segments characterized by loss of heterozygosity (LOH) and a normal copy number (two) but which are not autozygous in the germ-line or normal somatic cell genome. In this review, we will also discuss a model [cancer gene activity model (CGAM)] that may explain how autozygosity influences cancer predisposition. CGAM can also explain how the occurrence of certain chromosomal aberrations (copy number gain, LOH, and somatic UPDs) during carcinogenesis may be dependent on the germ-line genotypes of important cancer-related genes (oncogenes and tumor suppressors) found in those chromosomal regions.
Introduction
High-density single nucleotide polymorphism (SNP) mapping technologies (or SNP arrays), although commercially introduced only several years ago (1, 2), have already contributed immensely to our understanding of the patterns of variation in human genome (3, 4). These techniques have paved the way for numerous genome-wide association studies, which aimed to further define the genetic basis of many common diseases, including cancer (5). The use of SNP arrays in studying chromosomal aberrations in cancer is also becoming routine (6). One advantage of SNP arrays over other techniques is the simultaneous measurement of DNA copy number and detection of genotype calls. This feature allows researchers to identify regions in the cancer genome characterized by both loss of heterozygosity (LOH) and neutral copy number. These are regions of somatic uniparental disomy (UPD; refs. 7, 8), although the term “gene conversion” may also be used when referring to smaller, gene locus–sized regions (9). It is believed that UPD/gene conversion regions arise through recombination of homologous chromosomes during carcinogenesis (10, 11; Fig. 1A).
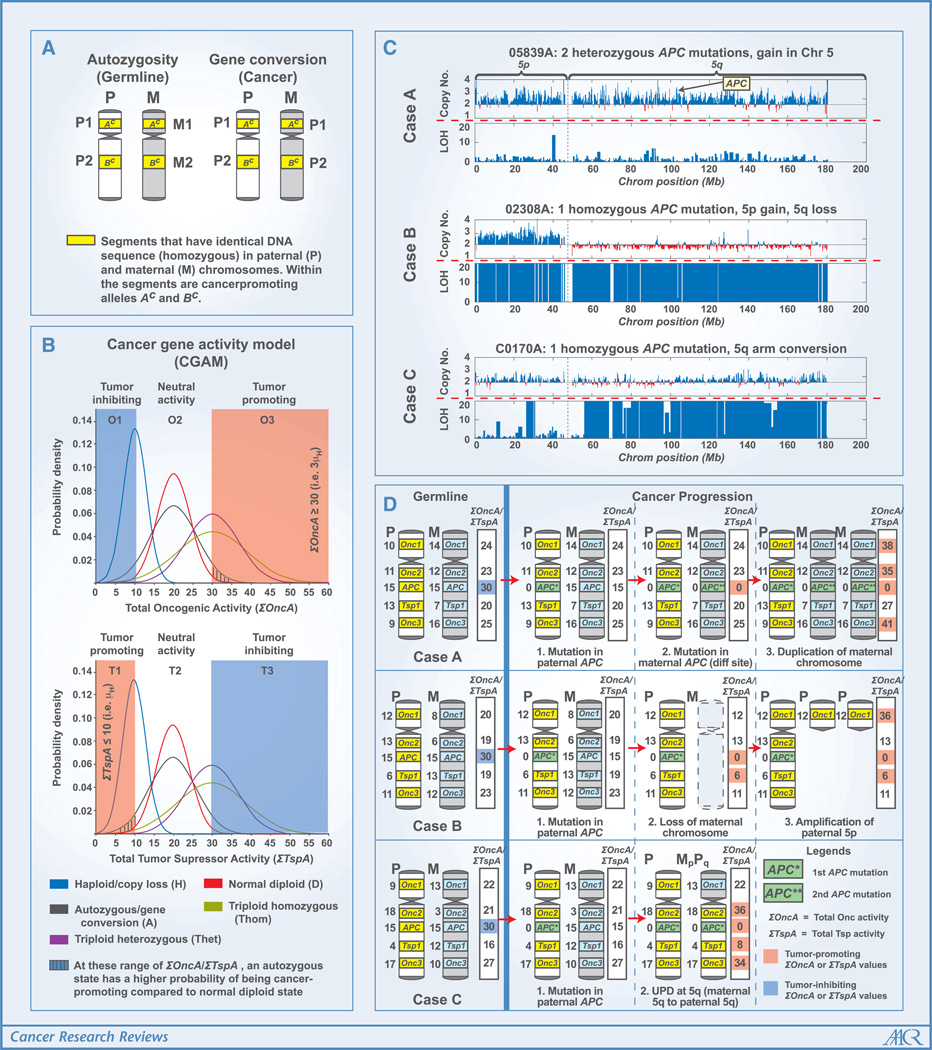
Figure 1.
A, the difference between autozygosity and gene conversion. Both are manifested as long stretches of homozygosity between the homologous chromosomes. Although autozygosity is acquired germinally, gene conversion occurs during carcinogenesis. B, a hypothetical Gaussian distribution of the activity of gene with oncogenic (Onc, upper chart) and tumor suppressor (Tsp) properties. There are 21 possible gene variants for the haploid (H) state (Onc0, Onc1, . . . Onc20 or Tsp0, Tsp1, . . . Tsp20) with oncogenic or tumor suppressor activity ranging from 0 to 20. There are also 212 possible genotypes for the autozygous (A; e.g., Onc1Onc1 and Onc2Onc2) and homozygous triploid (Thom; e.g., Onc1Onc1Onc1 and Onc2Onc2Onc2) states. On the other hand, there are 212 possible genotypes for the normal diploid (D; e.g., Onc1Onc1 and Onc1Onc2) and heterozygous triploid (Thet; e.g., Onc1Onc1Onc1 and Onc1Onc1Onc2) states. This model assumes that for the Onc to be tumor promoting, the total oncogenic activity (Σ OncA) needs to be ≥3 μH (or 30), and for Tsp to be tumor promoting, the total tumor suppressor activity (ΣTspA) needs to be ≤μH (or 10). Although the haploid (H), triploid homozygous (Thom), and triploid heterozygous (Thet) states may refer to copy number variations of Onc and Tsp in germ-line genomes, they may also refer to aberrations in cancer genomes (with haploid similar to a copy loss state, and triploid states similar to gene amplifications). Each probability distribution (H, D, A, Thom, and Thet) is a function of μH and σH. μD = 2 μH, ; μA = 2 μH, σA = 2 σH; μThom = 3 μH, σThom = 3 σH; μThet = 3 μH, . C, chromosome 5 (including APC) profiles of three colon cancer samples. These figures were generated from the SNP array (Affymetrix 50K Xba) data using the Affymetrix Copy Number Analysis Tool (42) and in-house programs. The copy number chart indicates deviations from the normal copy number of 2 (baseline of the chart). High LOH values (for the charts, the LOH value is capped at 20), indicated by tall blue bars, represent segments in the chromosome with contiguous homozygous SNPs. D, theoretical genetic pathways leading to chromosome 5 profiles of the actual colon cancer samples shown in C. Each case starts with a pair of chromosome 5 (P and M) with each Onc or Tsp allele having a defined gene activity (OncA and TspA) ranging from 0 to 20 (APC was assigned a value of 15 in the germ-line state and 0 in the mutated state).
Extended Homozygous Segments Can Also Be Detected in Germline Human DNA
SNP array analyses have also detected the presence of extended homozygous segments in normal human DNA (Fig. 1A). However, the size of these segments was dependent on the density of the arrays as well as the variables set by the investigators (4, 12–14). For instance, Gibson and colleagues (12), in their analysis of phase I HapMap genotype data (209 unrelated individuals, 1 SNP per 5 kb), were able to identify 1,393 homozygous tracks with a minimum length of 1 Mb. The presence of these germ-line homozygous segments may be explained by consanguinity somewhere in the individual’s ancestry. After all, 1/16 and 1/64 of a child’s genome is expected to be identical by descent if his/her parents are first and second cousins, respectively (15). By analyzing short tandem repeat polymorphism genotyping data, Broman and Weber (16) initially observed the presence of these long stretches of homozygosity among members of CEPH reference families. Their calculations showed that these segments are most likely homozygous by descent (or autozygous). Li and colleagues (14) showed that these long continuous homozygous segments were more pronounced among offspring of consanguineous marriages compared with children of unrelated individuals. Very recently, Clarimon and colleagues (also using SNP arrays) observed the prevalence of homozygous segments among early-onset Alzheimer’s disease–inflicted children of parents who were first cousins (17).
Autozygous Segments: Are They Useful in Genetic Studies of Diseases Such As Cancer?
The idea that long homozygous by descent regions may harbor disease-causing genes (particularly recessive alleles) has been explored in several studies, including patients with autism (18), Alzheimer’s disease (17), Parkinson’s disease (19), schizophrenia (20), and bipolar affective disorder (21). When our group examined the germ-line SNP array data of 74 colorectal cancer patients, we found that the percentage of those having autozygous segments greater or equal to 4 Mb is at least twice as high as control groups (22). Our analysis, as well as that by another laboratory (23), also discovered that these long homozygous segments are more common among individuals of Ashkenazi Jewish ancestry, a population group with one of the world’s highest incidence of colorectal cancer (24). A similar study by Assie and colleagues (25) found that 16 markers of homozygosity were common in the germline DNA of three cancer patient groups (147 breast, 116 prostate, and 122 head and neck carcinomas). However, due to the limitation of the technique they used in the genotyping experiment (345 autosomal markers), they may have missed the detection of extended homozygous tracks.
The Link between Consanguinity and Cancer
In certain parts of the world, the rate of consanguineous marriages (those between second cousins or closer) may exceed 50% (26). Accepted in certain cultures, consanguinity may also help proliferate the expression of disease-causing recessive alleles in the population. Evidence has also shown that level of consanguinity positively correlates to incidence of adult-onset complex diseases. In one example, Rudan and colleagues (27) showed that the genetic isolation of coastal island populations living in middle Dalmatia, Croatia is likely to be a factor in the high incidence of diseases such as cancer, heart disease, and stroke. Other investigators have linked the elevated levels of certain cancers among the Hutterites (28), Syrian Jewish community in Brooklyn, New York (29), Pakistanis (30), and Louisiana Acadians (31) to high incidence of consanguinity within these groups. Recent case studies have shown that two copies of highly penetrant colorectal cancer–causing alleles [PMS2 deletion (32) and the MYH frameshift mutation (33)] may be passed down by consanguineous parents to their offspring. As illustrated in Fig. 1A, autozygous segments may harbor alleles (Ac and Bc), which when present in homozygous form can increase cancer risk for the individual. The simplest explanation is that these genes in the autozygous regions can either be (a) a pair of recessive mutant cancer genes (such as MYH and ATM; ref. 34), (b) highly penetrant dominant cancer genes (such as BRCA2, MSH2, and MSH6) whose biallelic mutations may lead to distinct cancer phenotypes (35), or (c) low-penetrance, dose-dependent cancer predisposition SNPs such as the 8q24 SNPs recently linked to colorectal cancer (36, 37). In addition, a large autozygous segment may contain multiple genes satisfying any of the above characteristics, with all of them effectively contributing to increased cancer predisposition.
Cancer Gene Activity Model Explains How Autozygosity Increases Cancer Predisposition
The presence of autozygosity may not automatically lead to cancer. Our own analysis has shown that autozygous segments are also found in genomes of control (noncancer) individuals, although at lower frequency compared with those of colorectal cancer patients (22). An autozygous segment may not influence cancer predisposition if it does not include a gene (Onc or Tsp) that affects the probability of cancer. Even if a cancer-related gene locus is within an autozygous segment, the allele may not be the one that contributes to tumor promotion. The low frequency of cancer-promoting alleles in certain populations may also limit the influence of autozygosity on cancer predisposition. To explain this further, imagine that the cancer-related gene (Onc or Tsp) has a sizeable number of variants (polymorphisms at different positions in the gene, including regulatory regions) whose cancer-causing activities (OncA and TspA) are quantifiable. The OncA or TspA values may then depend on the structure of the protein or its expression level. In colon tumors, the expression level of many genes is related to DNA copy number, as shown in genome-wide analyses of cancer and matched normal samples (38).4 However, expression may also be influenced by transcription factors from other chromosomal locations, as well as epigenetics, such as promoter methylation. Another assumption we can make is that in terms of OncA (or TspA), the distribution of Onc (or Tsp) among the population is Gaussian.5 Shown as the blue curves in Fig. 1B, OncA and TspA were arbitrarily assigned values ranging from 0 to 20 with mean (μH) of 10 and SD (σH) of 3. In cases when Onc or Tsp must be present as two (in the case of autozygosity or gene conversion) or three (triploid homozygous) identical copies, both the mean and SD of the activity distribution would be doubled and tripled, respectively. In the normal diploid state (i.e., a pair of alleles is not necessarily identical), μD would still be 20, but σD = 4.24 (which is ).6 In a triploid heterozygous state (i.e., a pair of genes is always identical alleles, but the third copy can be any of the possible variants), μThet = 30 and σThet = 6.71 (which is ).6 Assume that an increase in activity of an oncogene by at least 50% (ΣOncA ≥ 30, region O3) would be tumor promoting. Assume also that a loss in activity of a tumor suppressor (ΣTspA ≤ 10, region T1) would similarly favor cancer formation. On the other hand, a decrease in activity of an oncogene (ΣOncA ≤ 10, region O1), or increase in activity of tumor suppressor (ΣTspA ≥ 30, region T3), would be tumor inhibiting. Given these assumptions, how does autozygosity provide an advantage over the diploid state when it comes to influencing cancer predisposition? If we carefully examine the regions O3 and T1 in the graphs (Fig. 1B), we can see that an Onc or Tsp always has a higher probability of being tumor promoting in the autozygous than in the normal diploid state. However, we can also see that only a small fraction of possible allele combinations (ΣTspA ≤ 10 or ΣOncA ≥ 30) would be tumor promoting for both states. This is consistent with our prior assumption that for every cancer-related gene, only a fraction of the possible variants in the autozygous state would add to cancer predisposition. At the other tail of the normal distribution (O1 and T3), an Onc or Tsp is more likely to be tumor inhibiting in the autozygous than in the normal diploid state. For the central areas in the graphs (O2 and T2), the difference in probabilities between autozygous and normal diploid states is unimportant because the activity values are neither tumor promoting nor tumor inhibiting. These last two scenarios may explain why certain studies did not see clear correlations between consanguinity and cancer or even concluded that consanguinity may decrease cancer incidence (39).
The Germline Status of Cancer Genes and How They May Affect Ensuing Chromosomal Aberrations
Although generation of chromosomal aberrations is most likely a stochastic process, those aberrations providing either a survival or growth advantage are selected during the progression of a cancer cell from normal to neoplastic state. Based on our understanding of cancer gene activity (above), it would be possible to hypothesize the sequence of events (or “genetic pathways”) leading to certain types of chromosomal aberrations. Consider the chromosome 5 (which includes APC) profiles of three actual colon cancer samples (cases A, B, and C) shown in Fig. 1C.7 First, assume that the genetic pathways that occur in chromosome 5 are entirely dependent on the status of APC mutation, and the OncA and TspA values (see Fig. 1B) of each allele of the oncogene Onc1 in the p arm, as well as the oncogenes Onc2 and Onc3, and tumor suppressor Tsp1 in the q arm. As illustrated in Fig. 1D, germ-line activity numbers are assigned to each copy (paternal and maternal) of Onc1, Onc2, Onc3, and Tsp1 such that the total activity (ΣOncA, ΣTspA) for each gene is not high enough to be tumor promoting. Each copy of APC gene, because of its known cancer-inhibitory function (if not mutated), is given a TspA value of 15, so that a ΣTspA value of 30 in the germ-line state of each sample would be good enough to be tumor inhibiting. A mutated APC is then assigned a TspA of 0, indicating the high-level penetrance of such mutation. As we follow the sequence of events for each case, we can see that the location of resulting aberrations largely depends on the assigned gene activity for each allele. For each case, the first event in cancer progression is the mutation of APC in the paternal chromosome (reducing its paternal TspA to 0). In case A, it is followed by mutation of the maternal APC copy leading to ΣTspA value of 0 for APC, rendering it a tumor-promoting state. The next step would have to be the duplication of either chromosome. Based on the germ-line activities of the non-APC genes, we can see that duplicating the maternal rather than the paternal chromosome would lead to higher ΣOncA values for all the oncogenes. For case B, the second event is the loss of maternal chromosome (whose APC is not mutated), which would also make both APC (ΣTspA = 0) and Tsp1 (ΣTspA = 6) both cancer promoting. The loss of the maternal chromosome 5 is then followed by amplification (two additional copies) of the paternal 5p arm, resulting in an additional cancer-promoting gene (Onc1). Case C is characterized by UPD in the 5q arm. Because the initial mutation occurred in the paternal arm, the next event would have to be the loss of the maternal 5q arm and the duplication of the paternal 5q arm (to totally inactivate APC). We can see that the paternal Onc2, Tsp1, and Onc3 were all assigned haploid activity values at the tail of normal distribution, closer to cancer-promoting activity values. Thus, after the conversion of maternal q arm to paternal q arm, all the four genes in the q arm would have cancer-promoting activity values. For case C, other options to totally inactivate APC is a mutation in the maternal copy, or a loss of the maternal 5q. However, both of these possibilities would result in fewer genes with ΣTspA or ΣOncA values appropriate for tumor promotion. Case C is different from the first two cases: the somatic UPD event results in all oncogenes and tumor suppressors in the q arm having tumor-promoting activities. Thus, whereas copy number gain and LOH may increase the tumor-promoting activities of oncogenes and tumor suppressors, respectively, a somatic UPD enhances the tumor-promoting activities of both. Further, a somatic UPD event (as in case C) may contribute to onset of cancer by disrupting the imprint patterns of certain genes. Imagine that the germ-line maternal copy of Onc2 is normally imprinted (i.e., hypermethylated at its promoter region), resulting in a lower expression level and OncA value (equal to 3). In contrast, the paternal Onc2 is not imprinted and highly expressed (OncA equal to 18). The UPD that followed would have caused a loss of imprinting in maternal Onc2, resulting in two copies of unmethylated alleles and ΣOncA value that is tumor promoting. This is analogous to germinally acquired UPDs that result in genetic disorders such as Prader-Willi syndrome (40). Lastly, we must consider the possibility that a Tsp can be tumor promoting while haploinsufficient (e.g., the p27Kip1 gene; see ref. 41 for review).
Future Directions and Conclusion
We present a model to help understand how autozygosity and UPD may play a role in the general context of chromosomal instability and cancer progression. We acknowledge that chromosomal instability is complex, with many other factors influencing the type of chromosomal aberrations that occur during cancer progression. Our model explains how ensuing aberration (gain, LOH, and UPD) may depend on the cancer-promoting activities of the alleles within these chromosomal regions. Further, this model considers how autozygous regions may initiate cancer progression. Advances in acquiring SNP array data on an exponentially increasing number of tumors and matched normal samples not only distinguishes between autozygosity and somatic UPDs but will help further elucidate the role of autozygosity in cancers.
Acknowledgments
Grant support: National Cancer Institute grant P01-CA65930, Ludwig Institute for Cancer Research/Conrad N. Hilton Foundation joint Hilton-Ludwig Cancer Metastasis Initiative, and Gilbert Family Foundation.
Footnotes
A separate manuscript (Bacolod et al., in preparation) will describe in detail the genome-wide analysis that relates DNA copy number and gene expression level in colon cancer samples. The 20q (gain), 13q (gain), and 18p (loss) arms have the highest percentages of dysregulated genes (or genes exhibiting up-regulated/down-regulated expression and copy number gain/loss at the same time).
For the haploid (H) case, the distribution of the gene activity (n) is defined as . The values of n range from 0 to 20 (μH = 10 and σH = 3) and are normally distributed. Although n can be a continuous variable within this range, the discussion is simplified by assigning only discrete values for n.
and , where E(X1 + X2) and E(2X1 + X2) are the expected values for the normal diploid (D) and heterozygous triploid (Thet) tumor activity models, respectively. These expressions make use of the general property Var(X) = E(X2) − [E(X)]2, where Var(X) is the variance of the random variable X. More specifically, . The random variables X1 and X2 have the same probability density function f(x) but are independent of each other. Simplification results into and .
Some of the data will be included in H. Pincas, et al., Genetic alterations and genomic instability in colorectal cancers: APC and p53 mutations both correlate with chromosomal instability, in preparation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Dong S, Wang E, Hsie L, Cao Y, Chen X, Gingeras TR. Flexible use of high-density oligonucleotide arrays for single-nucleotide polymorphism discovery and validation. Genome Res. 2001;11:1418–1424. doi: 10.1101/gr.171101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen R, Fan JB, Campbell D, et al. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 3.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutt A, Beroukhim R. Single nucleotide polymorphism array analysis of cancer. Curr Opin Oncol. 2007;19:43–49. doi: 10.1097/CCO.0b013e328011a8c1. [DOI] [PubMed] [Google Scholar]
- 7.Teh MT, Blaydon D, Chaplin T, et al. Genomewide single nucleotide polymorphism microarray mapping in basal cell carcinomas unveils uniparental disomy as a key somatic event. Cancer Res. 2005;65:8597–8603. doi: 10.1158/0008-5472.CAN-05-0842. [DOI] [PubMed] [Google Scholar]
- 8.Andersen CL, Wiuf C, Kruhoffer M, Korsgaard M, Laurberg S, Orntoft TF. Frequent occurrence of uniparental disomy in colorectal cancer. Carcinogenesis. 2007;28:38–48. doi: 10.1093/carcin/bgl086. [DOI] [PubMed] [Google Scholar]
- 9.Tischfield JA. Loss of heterozygosity or: how I learned to stop worrying and love mitotic recombination. Am J Hum Genet. 1997;61:995–999. doi: 10.1086/301617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop AJ, Schiestl RH. Role of homologous recombination in carcinogenesis. Exp Mol Pathol. 2003;74:94–105. doi: 10.1016/s0014-4800(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 11.Robinson WP. Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays. 2000;22:452–459. doi: 10.1002/(SICI)1521-1878(200005)22:5<452::AID-BIES7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 12.Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15:789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- 13.Simon-Sanchez J, Scholz S, Fung HC, et al. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- 14.Li LH, Ho SF, Chen CH, et al. Long contiguous stretches of homozygosity in the human genome. Hum Mutat. 2006;27:1115–1121. doi: 10.1002/humu.20399. [DOI] [PubMed] [Google Scholar]
- 15.Lander ES, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- 16.Broman KW, Weber JL. Long homozygous chromosomal segments in reference families from the Centre d’Etude du Polymorphisme Humain. Am J Hum Genet. 1999;65:1493–1500. doi: 10.1086/302661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarimon J, Djaldetti R, Lleo A, et al. Whole genome analysis in a consanguineous family with early onset Alzheimer’s disease. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrow EM, Yoo SY, Flavell SW, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Haynes C, Barany F, Ott J. Genome-wide autozygosity mapping in human populations. Genet Epidemiol. 2008 doi: 10.1002/gepi.20344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knight HM, Maclean A, Irfan M, et al. Homozygosity mapping in a family presenting with schizophrenia, epilepsy and hearing impairment. Eur J Hum Genet. 2008;16:750–758. doi: 10.1038/ejhg.2008.11. [DOI] [PubMed] [Google Scholar]
- 21.Ewald H, Kruse TA, Mors O. Genome wide scan using homozygosity mapping and linkage analyses of a single pedigree with affective disorder suggests oligogenic inheritance. Am J Med Genet B Neuropsychiatr Genet. 2003;120B:63–71. doi: 10.1002/ajmg.b.20039. [DOI] [PubMed] [Google Scholar]
- 22.Bacolod MD, Schemmann GS, Wang S, et al. The signatures of autozygosity among patients with colorectal cancer. Cancer Res. 2008;68:2610–2621. doi: 10.1158/0008-5472.CAN-07-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olshen AB, Gold B, Lohmueller KE, et al. Analysis of genetic variation in Ashkenazi Jews by high density SNP genotyping. BMC Genet. 2008;9:14. doi: 10.1186/1471-2156-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldman GE. Do Ashkenazi Jews have a higher than expected cancer burden? Implications for cancer control prioritization efforts. Isr Med Assoc J. 2001;3:341–346. [PubMed] [Google Scholar]
- 25.Assie G, LaFramboise T, Platzer P, Eng C. Frequency of germline genomic homozygosity associated with cancer cases. JAMA. 2008;299:1437–1445. doi: 10.1001/jama.299.12.1437. [DOI] [PubMed] [Google Scholar]
- 26.Bittles A. Consanguinity and its relevance to clinical genetics. Clin Genet. 2001;60:89–98. doi: 10.1034/j.1399-0004.2001.600201.x. [DOI] [PubMed] [Google Scholar]
- 27.Rudan I, Rudan D, Campbell H, et al. Inbreeding and risk of late onset complex disease. J Med Genet. 2003;40:925–932. doi: 10.1136/jmg.40.12.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson JL, Martin AO, Elias S, Sarto GE, Dunn JK. Cancers of the breast and female genital system: search for recessive genetic factors through analysis of human isolate. Am J Obstet Gynecol. 1981;141:629–636. doi: 10.1016/s0002-9378(15)33302-0. [DOI] [PubMed] [Google Scholar]
- 29.Feldman JG, Lee SL, Seligman B. Occurrence of acute leukemia in females in a genetically isolated population. Cancer. 1976;38:2548–2550. doi: 10.1002/1097-0142(197612)38:6<2548::aid-cncr2820380644>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 30.Shami SA, Qaisar R, Bittles AH. Consanguinity and adult morbidity in Pakistan. Lancet. 1991;338:954. doi: 10.1016/0140-6736(91)91828-i. [DOI] [PubMed] [Google Scholar]
- 31.Thurmon TF, Robertson KP. Genetic considerations in human cancer incidence. Public Health Rep. 1979;94:471–476. [PMC free article] [PubMed] [Google Scholar]
- 32.Will O, Carvajal-Carmona LG, Gorman P, et al. Homozygous PMS2 deletion causes a severe colorectal cancer and multiple adenoma phenotype without extraintestinal cancer. Gastroenterology. 2007;132:527–530. doi: 10.1053/j.gastro.2006.11.043. [DOI] [PubMed] [Google Scholar]
- 33.Baglioni S, Melean G, Gensini F, et al. A kindred with MYH-associated polyposis and pilomatricomas. Am J Med Genet A. 2005;134A:212–214. doi: 10.1002/ajmg.a.30585. [DOI] [PubMed] [Google Scholar]
- 34.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 35.Rahman N, Scott RH. Cancer genes associated with phenotypes in monoallelic and biallelic mutation carriers: new lessons from old players. Hum Mol Genet. 2007;16(Spec No 1):R60–R66. doi: 10.1093/hmg/ddm026. [DOI] [PubMed] [Google Scholar]
- 36.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 37.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 38.Tsafrir D, Bacolod M, Selvanayagam Z, et al. Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res. 2006;66:2129–2137. doi: 10.1158/0008-5472.CAN-05-2569. [DOI] [PubMed] [Google Scholar]
- 39.Denic S, Bener A. Consanguinity decreases risk of breast cancer—cervical cancer unaffected. Br J Cancer. 2001;85:1675–1679. doi: 10.1054/bjoc.2001.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horsthemke B, Wagstaff J. Mechanisms of imprinting of the Prader-Willi/Angelman region. Am J Med Genet A. 2008;146A:2041–2052. doi: 10.1002/ajmg.a.32364. [DOI] [PubMed] [Google Scholar]
- 41.Santarosa M, Ashworth A. Haploinsufficiency for tumour suppressor genes: when you don’t need to go all the way. Biochim Biophys Acta. 2004;1654:105–122. doi: 10.1016/j.bbcan.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Huang J, Wei W, Zhang J, et al. Whole genome DNA copy number changes identified by high density oligonucleotide arrays. Hum Genomics. 2004;1:287–299. doi: 10.1186/1479-7364-1-4-287. [DOI] [PMC free article] [PubMed] [Google Scholar]