Abstract
The MC1R gene is a key regulator of skin pigmentation. We aimed to evaluate the association between MC1R variants and the risk of sporadic cutaneous melanoma (CM) within the M-SKIP project, an international pooled-analysis on MC1R, skin cancer and phenotypic characteristics. Data included 5,160 cases and 12,119 controls from 17 studies. We calculated a Summary Odds Ratio (SOR) for the association of each of the nine most studied MC1R variants and of variants combined with CM by using random-effects models. Stratified analysis by phenotypic characteristics were also performed.
Melanoma risk increased with presence of any of the main MC1R variants: the SOR for each variant ranged from 1.47 (95%CI: 1.17–1.84) for V60L to 2.74 (1.53–4.89) for D84E. Carriers of any MC1R variant had a 66% higher risk of developing melanoma compared to wild-type subjects (SOR; 95%CI: 1.66; 1.41–1.96), and the risk attributable to MC1R variants was 28%. When taking into account phenotypic characteristics, we found that MC1R–associated melanoma risk increased only for darker-pigmented Caucasians: SOR (95%CI) was 3.14 (2.06–4.80) for subjects with no freckles, no red hair and skin type III/IV.
Our study documents the important role of all the main MC1R variants in sporadic CM and suggests that they have a direct effect on melanoma risk, independently on the phenotypic characteristics of carriers. This is of particular importance for assessing preventive strategies, which may be directed to darker-pigmented Caucasians with MC1R variants as well as to lightly-pigmented, fair-skinned subjects.
Keywords: melanocortin-1 receptor, melanoma, meta-analysis, genetic epidemiology
Introduction
Starting from the 20th century, both incidence and mortality for cutaneous melanoma (CM) has increased among populations of European descent and the estimated annual melanoma incidence rate in the world is around 2.8 per 100.000.1 Of the three most prevalent types of skin cancer, melanoma is the most lethal, with a five-year survival rate for metastatic melanoma of 11%.2 Because of melanoma’s high incidence and poor treatment outcome for metastatic disease, development of early detection and preventive strategies are of the highest importance.
Among risk factors for CM, epidemiological studies highlighted the role of sun exposure, and of personal characteristics, such as fair skin, light hair and eyes color, high number of melanocytic naevi, and family history of melanoma.3–5 Twins studies estimated that genetic factors provide a contribution to CM risk of 18–55%, and familial melanoma was estimated to account for approximately 10% of new cases.6
The melanocortin-1-receptor gene (MC1R, MIM#155555) is the most common low risk susceptibility gene for melanoma and a key regulator of skin pigmentation. It is located on chromosome 16q24.3 and encodes for a seven pass transmembrane G-protein coupled-receptor of 317 amino acids, positively coupled to adenylate cyclase to increase cAMP levels upon binding its endogenous ligand, the α-melanocyte-stimulating hormone (α-MSH).7 Binding of α-MSH to the functional MC1R on melanocytes stimulates the synthesis of eumelanin pigments8 resulting in significant increases of the ratio of black/brown eumelanin to red/yellow phaeomelanin pigments. While eumelanin has been shown to be photoprotective, phaeomelanin is poorly photoprotective and may even contribute to cancer risk through the production of free radicals in response to UV exposure.9 The MC1R gene locus is highly polymorphic in populations of European origins, with more than 80 variants identified.10 MC1R variant alleles, resulting in amino acid substitutions that have been shown to reduce receptor function11–13, result in a quantitative shift of melanin synthesis from eumelanin to phaeomelanin,7 and determination of the so called “red hair color” (RHC) phenotype, characterized by the co-occurrence of fair skin, red hair, freckles and UV irradiation (UVR) sensitivity (poor tanning response and solar lentigines).
Several studies in different populations have reported that the risk of melanoma is higher among individuals who carry MC1R variant alleles. More recently, meta-analyses and genome-wide association studies (GWAS) confirmed this finding14–18. Although melanoma risk attributable to MC1R may arise through the determination of the tanning response of skin to UV light, some studies and a recent meta-analysis15 observed that MC1R–associated CM relative risk was stronger in darkly-pigmented subjects and for those with lower levels of recreational sun exposure. These results suggest that MC1R variants may partly mediate their effect through biological pathways that are independent of pigmentation and UV exposure. In keeping with this possibility, wild type (WT) MC1R has been shown to trigger DNA repair mechanisms and antioxidant defenses in UVR-exposed melanocytes, while inactivated MC1R, resulting in production of pheomelanin, increases damage from reactive oxygen species, even in the absence of UV-exposure.19,20 Those mechanisms may be of importance for at least some of the variant alleles.
Although the previous meta-analyses and GWAS gave reliable evidence of a role of MC1R in CM development, the lack of access to individual epidemiological information precluded in-depth investigations, including the assessment of the role of possible confounders, the estimation of melanoma risk according to different MC1R variants compared to WT subjects, and stratification for phenotypic characteristics. These investigations are in fact crucial for sporadic CM, which represents up to 95% of melanoma cases and is a complex and heterogeneous disease, probably the result of interactions between genetic, phenotypic and environmental factors.
The aim of this work is to evaluate the association between specific and combined MC1R variants and the risk of sporadic CM, and to evaluate whether risk estimates varied according to different phenotypic characteristics through a large multicenter pooled-analysis of individual data from the Melanocortin-1 receptor gene, SKin cancer and Phenotypic characteristics (M-SKIP) project.
Material and methods
Data for the present analyses were gathered through the M-SKIP project. A description of the project was previously published.21 Briefly, we searched for published and unpublished epidemiological studies on MC1R variants, sporadic CM, non-melanoma skin cancer (NMSC), and phenotypic characteristics associated with melanoma. Original individual data on participants in each identified study were requested from principal investigators. From May 2009 to December 2010, 43 investigators were contacted and 31 (72%) agreed to participate. Non-participant investigators where those who either did not reply to our invitation letter, were not able to retrieve the original dataset and/or were not interested in the project. More details are reported elsewhere21. Participant investigators sent their data along with a signed statement declaring that their original study was approved by an Ethics Committee and/or that study subjects provided a written consent to participate in the original study. Quality controls and data coding were performed, and the pooled database was created, including data on 7,806 CM cases, 3,151 NMSC cases and 14,875 controls.
For the purpose of the present study, we selected from the M-SKIP database all the melanoma case-control studies (N=17)15, 22–36, thus including data on 5,160 CM cases and 12,119 controls overall. Case-only or control-only studies, and studies on NMSC were excluded from the present analysis.
Statistical analysis
In order to evaluate the representativeness of the M-SKIP dataset, the main study population characteristics reported in publications of non-participating investigators were extracted and compared with those of studies included in the pooled-analysis, by the Chi-Square test for categorical variables and by the Wilcoxon two-sample test for continuous variables. We also assessed possible participation bias by drawing funnel plots and by Egger's test. We verified the departure of frequencies of each MC1R variant from expectation under Hardy-Weinberg (HW) equilibrium by the Chi Square test in controls for each included study.
We calculated study-specific Odds Ratio (OR) with 95% Confidence Interval (CI) by applying logistic regression to the data from each study. Beyond MC1R, each model included, if available, the following covariates: age, sex, intermittent and chronic sun exposure, lifetime and childhood sunburns, family history of melanoma, number of common total body naevi count and presence of atypical naevi. Because of different definitions of covariates between studies, particular attention was given to recoding and standardizing the variables in the M-SKIP database, as previously discussed.21 For each study, we imputed missing data with multiple imputation models for variables with less than 20% of missing data, by using the iterative Markov chain Monte Carlo method. For each of the nine most studied MC1R variants (V60L, D84E, V92M, R142H, R151C, I155T, R160W, R163Q, D294H), we tested different inheritance models and found that the dominant model was the one with the lowest Akaike's Information Criterion for almost all the studies and variants, therefore we assumed this model of inheritance in the pooled-analyses. We performed two different variant-specific analyses: the first one included the 12 studies in which MC1R was sequenced, for which we could identify the WT subjects as reference category; the second one included all the 17 studies and used as reference category for each variant the subjects without that variant. For studies which sequenced the MC1R gene, beyond the MC1R variant-specific OR, we estimated the OR associated with the most frequently observed haplotypes. Haplotype frequencies for the nine most common variants were calculated with the EM algorithm using UNPHASED, version 3.1.7. Rare haplotypes (frequency<0.005) were ignored. In addition, we took into account all the identified (common and rare) variants and calculated the OR for 1) carrying at least one MC1R variant compared to WT, 2) carrying just one and carrying ≥2 MC1R variants compared to WT. Finally, a MC1R score was calculated, based on classification of likely pathogenicity of variants using bioinformatics analysis as implemented by Davies et al:37 briefly, the score was calculated by summing across the MC1R alleles, giving a value of 1 to “r” and 2 to “R” variants.
Following the two-stage analysis approach, we pooled study-specific OR with random-effects model, using the DerSimonian-Laird method. When there were more than one OR calculated in a single study (i.e. analysis by MC1R score), the correlation between the ORs was taken into account by using a multivariate approach previously described.38 We evaluated homogeneity among study-specific estimates by the Q statistic and I-Square, which represents the percentage of total variation across studies that is attributable to heterogeneity rather than to chance. When a significant heterogeneity was detected, we performed meta-regression to assess the influence on Summary Odds Ratio (SOR) of different study features, such as publication year, study area, genotyping methodology, deviation from HW equilibrium, source of controls, and source of DNA. In order to evaluate the robustness of the results, we also examined changes in SOR after exclusion of specific studies, and we compared the pooled-OR obtained on the M-SKIP dataset with the meta-OR calculated by pooling risk estimates reported in studies from not-participating investigators. This latter meta-OR was also obtained with DerSimonian-Laird random-effects models.
We computed the attributable risk in the population for each of the nine MC1R variants listed above and for the presence of at least one MC1R variant by using the Miettinen’s formula: (OR-1/OR) × proportion of cases exposed, with the corresponding 95%CI.
Finally, in order to investigate whether the observed association between MC1R variants and melanoma varied according with different phenotypic characteristics, we performed stratified analysis. We defined subjects with RHC phenotype as those with either red hair color, freckles or high sun sensitivity (skin type I or II according to the Fitzpatrick classification), and as darker-pigmented individuals those subjects with none of the above phenotypic characteristics. The hypothesis of homogeneity of ORs among strata was tested by meta-regression models with random-effects and restricted maximum likelihood estimates, after the calculation of strata-specific OR in each study. The correlation between the ORs calculated in the same studies was taken into account by using the multivariate approach proposed by van Houwelingen et al.38
P-values <0.05 were considered statistically significant for all the tests but Q statistic, where p-values<0.10 were considered statistically significant. Both the false discovery rate approach and the more conservative Bonferroni correction were also used to take into account the problem of multiple comparisons. The analysis was carried out by using the software SAS (version 9.2) and STATA (version 11.2).
Results
Studies included in our pooled-analysis did not differ from studies from not-participating investigators according to publication period, study area, phenotype assessment, source of controls, genotyping methodology, mean age of cases and controls, sex distribution of cases and controls.
Among the 17 studies included in the pooled-analysis, no deviation from HW equilibrium was observed for the following MC1R variants: V60L, D84E, V92M, I155T, and R163Q. Deviation from HW equilibrium was observed in one study34 for the R142H variant, in four23,26,27,33 for R151C variant, in two studies29,34 for R160W variant, and in one study29 for D294H variant.
Complete sequencing analysis of the MC1R coding region was performed in 12 studies15,22,24,26–28,30,31,33,35,36, while in the remaining five studies MC1R was genotyped by SNaPshot23,29,34, Restriction Fragment Length Polymorphisms25 or allele discrimination assay.32
A description of the studies included in the pooled-analysis is presented in Table 1. Studies were published between 2001 and 2012; the majority were carried out in Europe (N=13, 76%), mainly in the southern countries (N=7, 41%), followed by USA (N=3, 18%) and Australia (N=1, 6%). In 13 studies (76%) healthy controls were recruited, while in the remaining four (24%) hospital controls were included. Overall, the average age of controls was higher than that of cases (62 versus 53 years), while the percentage of males was the same for both cases and controls (44%). Individual information on age, sex and family history of melanoma was available for each study, while further confounders differed between studies.
Table 1.
Description of the 17 case-control studies included in the pooled-analysis
| First Author | Publication year |
Country |
MC1R sequencing |
Controls typea |
N Cases/ N Controls |
Mean Age (SD) |
Percent of males |
Available confoundersb |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |||||||
| Kennedy22 | 2001 | The Netherlands | Yes | Hospital | 115/378 | 49 (12) | 58 (11) | 37 | 42 | Sun exposure, sunburns, common and atypical naevi |
| Dwyer23 | 2004 | Australia | No | Healthy | 159/290 | 44 (10) | 44 (10) | 41 | 46 | Sun exposure, sunburns |
| Landi24 | 2005 | Italy | Yes | Healthy | 165/171 | 49 (15) | 46 (13) | 49 | 49 | Sun exposure, sunburns |
| Debniak25 | 2006 | Poland | No | Healthy | 349/313 | 53 (14) | 53 (13) | 32 | 24 | Sunburns |
| Fargnoli26 | 2006 | Italy | Yes | Hospital | 155/163 | 49 (14) | 49 (14) | 40 | 50 | Sun exposure, sunburns, common and atypical naevi |
| Stratigos27 | 2006 | Greece | Yes | Hospital | 123/155 | 52 (16) | 44 (15) | 51 | 54 | Sun exposure, sunburns, common and atypical naevi |
| Fernandez28 | 2007 | Spain | Yes | Healthy | 108/188 | 51 (15) | 53 (14) | 46 | 39 | Sunburns, common naevi |
| Brudnik29 | 2009 | Poland | No | Hospital | 116/489 | 62 (14) | 43 (19) | 35 | 40 | - |
| Casula30 | 2009 | Italy | Yes | Healthy | 259/75 | 49 (14) | 61 (15) | 48 | 27 | - |
| Council31 | 2009 | USA | Yes | Healthy | 83/166 | 51 (15) | 77 (7) | 45 | 50 | - |
| Nan32 | 2009 | USA | No | Healthy | 219/241 | 64 (8) | 58 (7) | 0 | 0 | Sunburns |
| Hoiom33 | 2009 | Sweden | Partiallyc | Healthy | 675/477 | 53 (19) | 42 (12) | 47 | 64 | - |
| Rotterdam Study34 | 2009 | The Netherlands | No | Healthy | 68/6,559 | 70 (8) | 72 (9) | 47 | 41 | - |
| Scherer (G)35 | 2009 | Germany | Yes | Healthy | 512/1,064 | 58 (15) | 54 (12) | 56 | 56 | - |
| Scherer (S)35 | 2009 | Spain | Yes | Healthy | 1,031/558 | 52 (16) | 37 (12) | 46 | 62 | - |
| Kanetsky15 | 2010 | USA | Yes | Healthy | 769/325 | 49 (14) | 48 (13) | 49 | 43 | Sun exposure, sunburns, atypical naevi |
| Ghiorzo36 | 2012 | Italy | Yes | Healthy | 254/507 | 52 (16) | 51 (17) | 50 | 47 | Sunburns |
| Total | 5,160/12,119 | 53 (16) | 62 (16) | 44 | 44 | |||||
G= German study; S= Spanish study
Healthy controls are population controls, blood donors, friends or relatives of cases.
Beyond age, sex and family history of melanoma, which were available in all the 17 studies. Sun exposure includes separate information on chronic and intermittent sun exposure.
MC1R sequencing performed on a subsample of 65 cases and 30 controls.
Association between single MC1R variants and melanoma
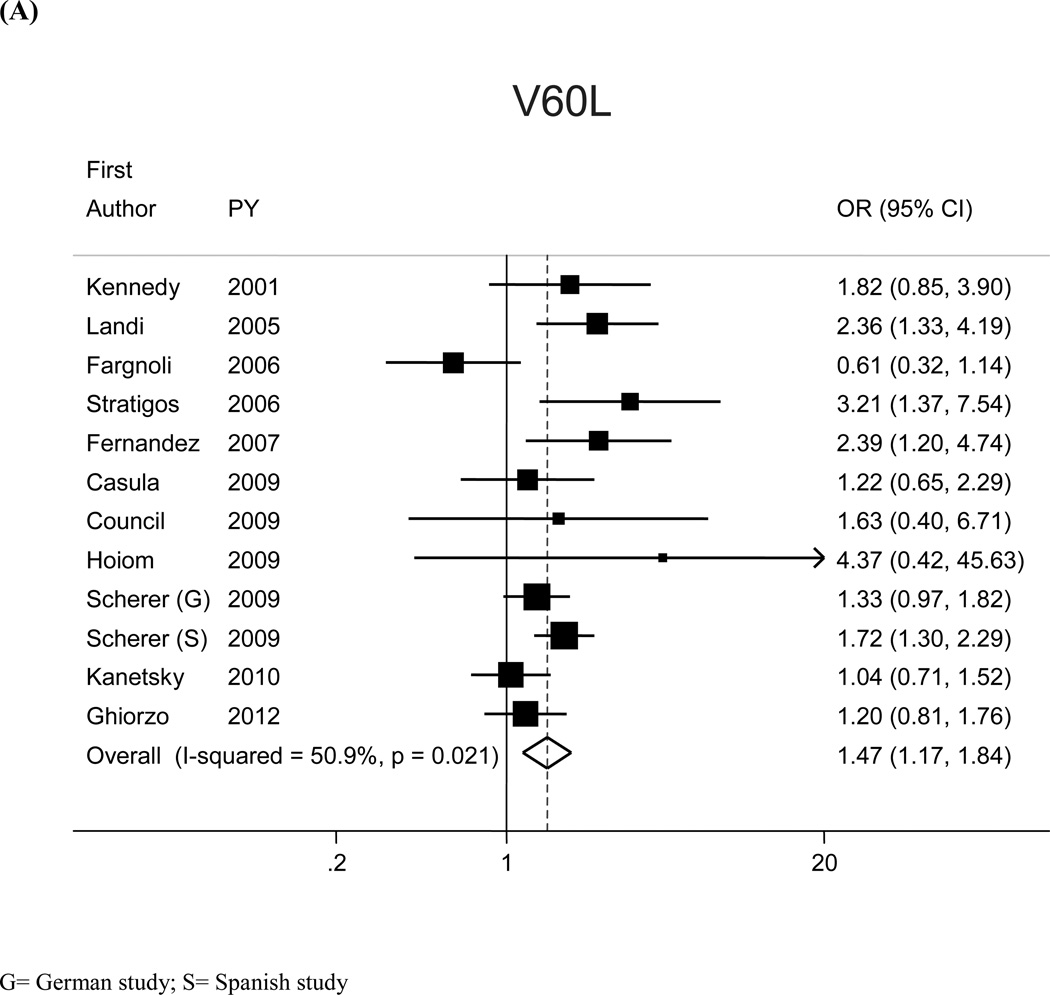
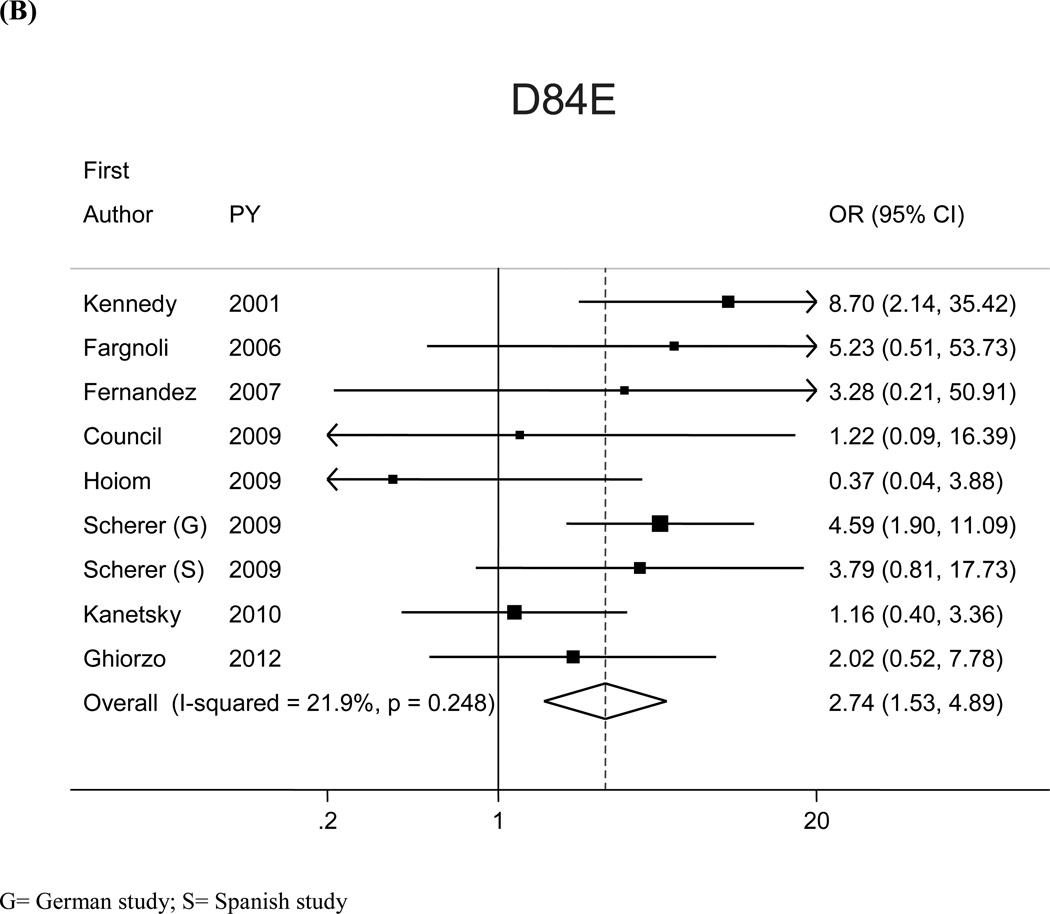
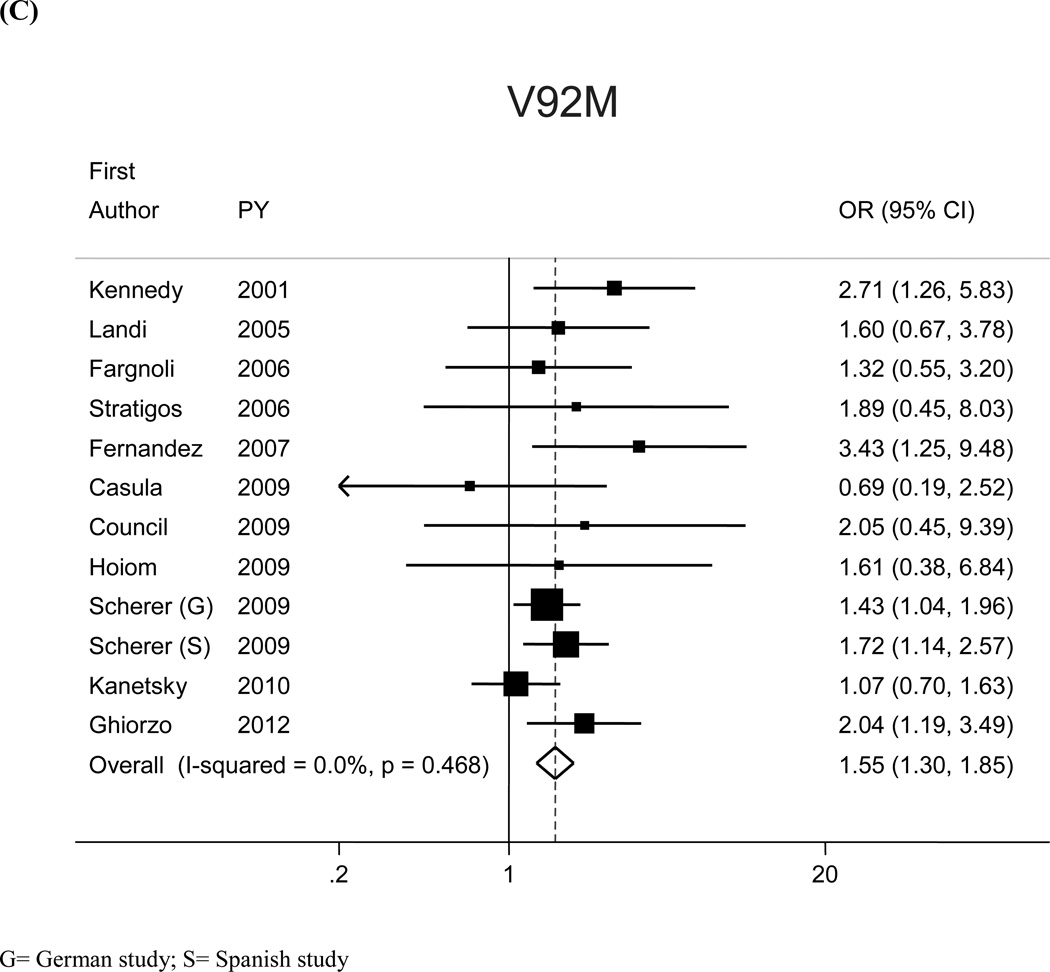
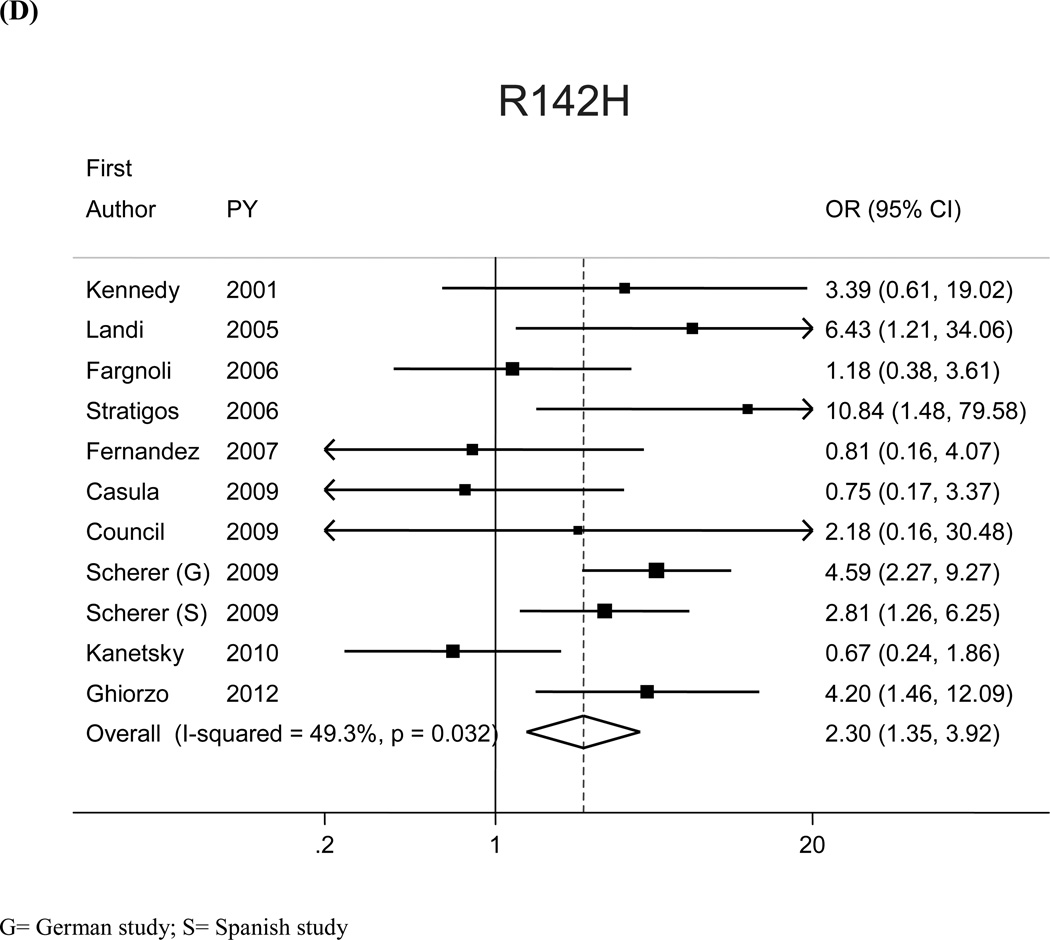
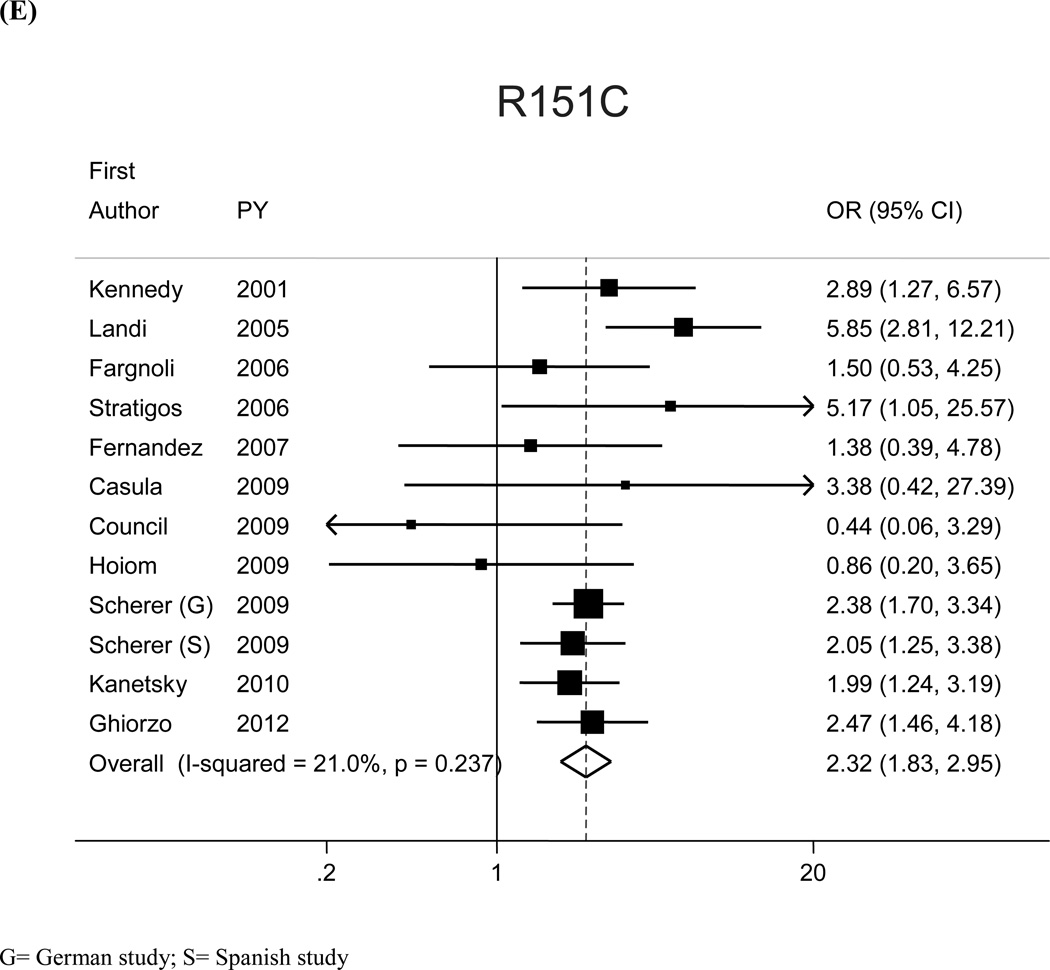
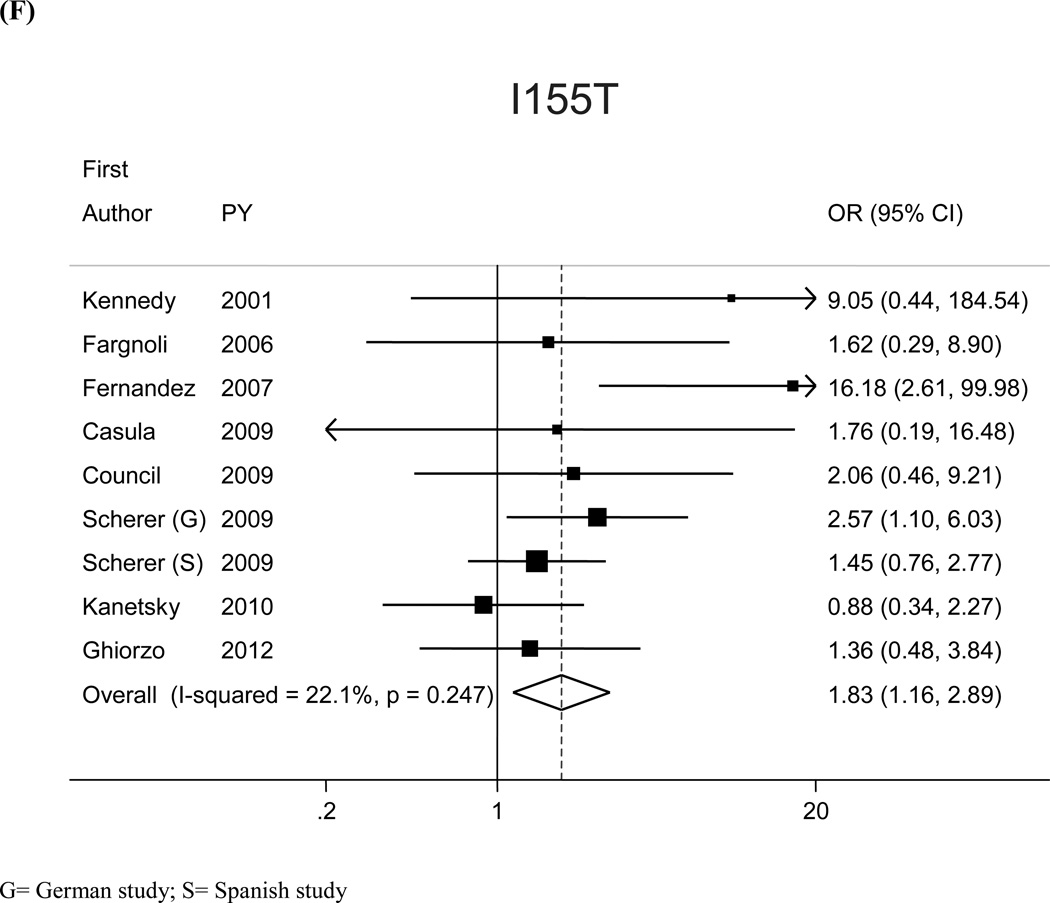
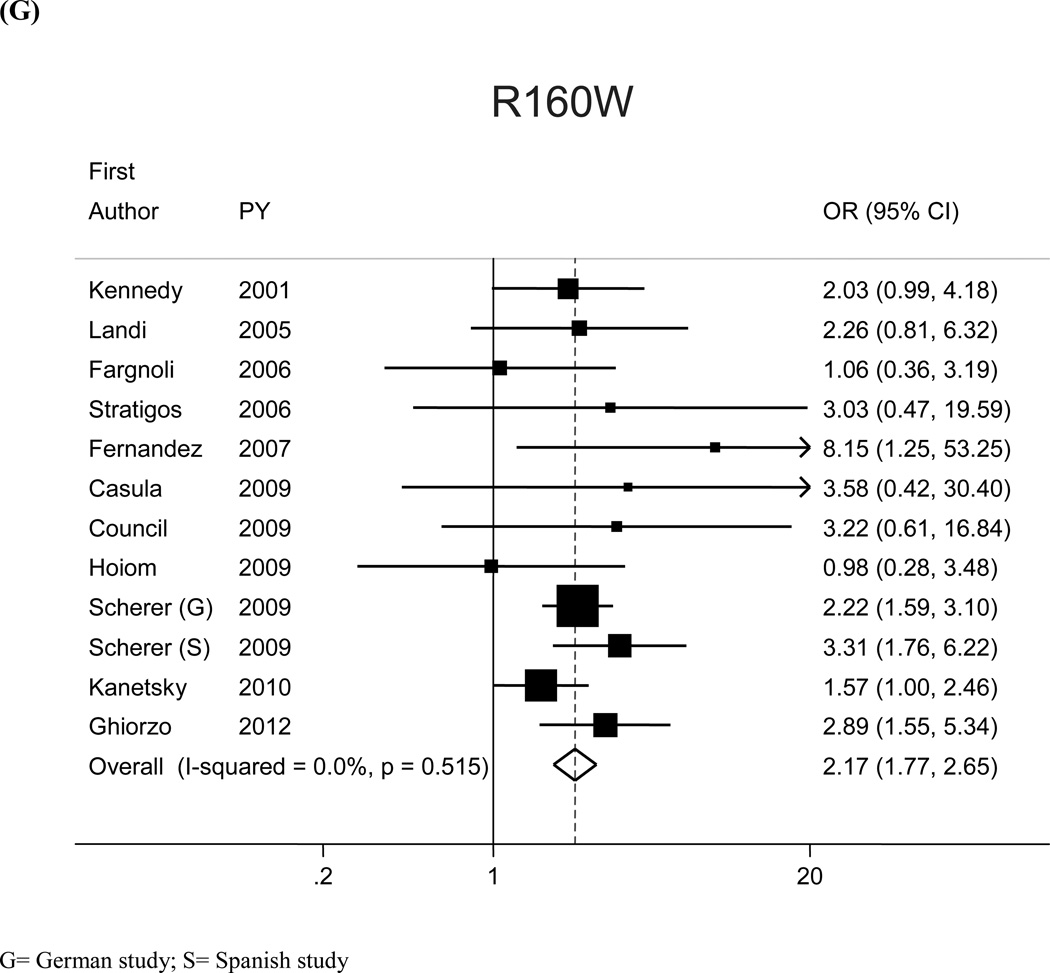
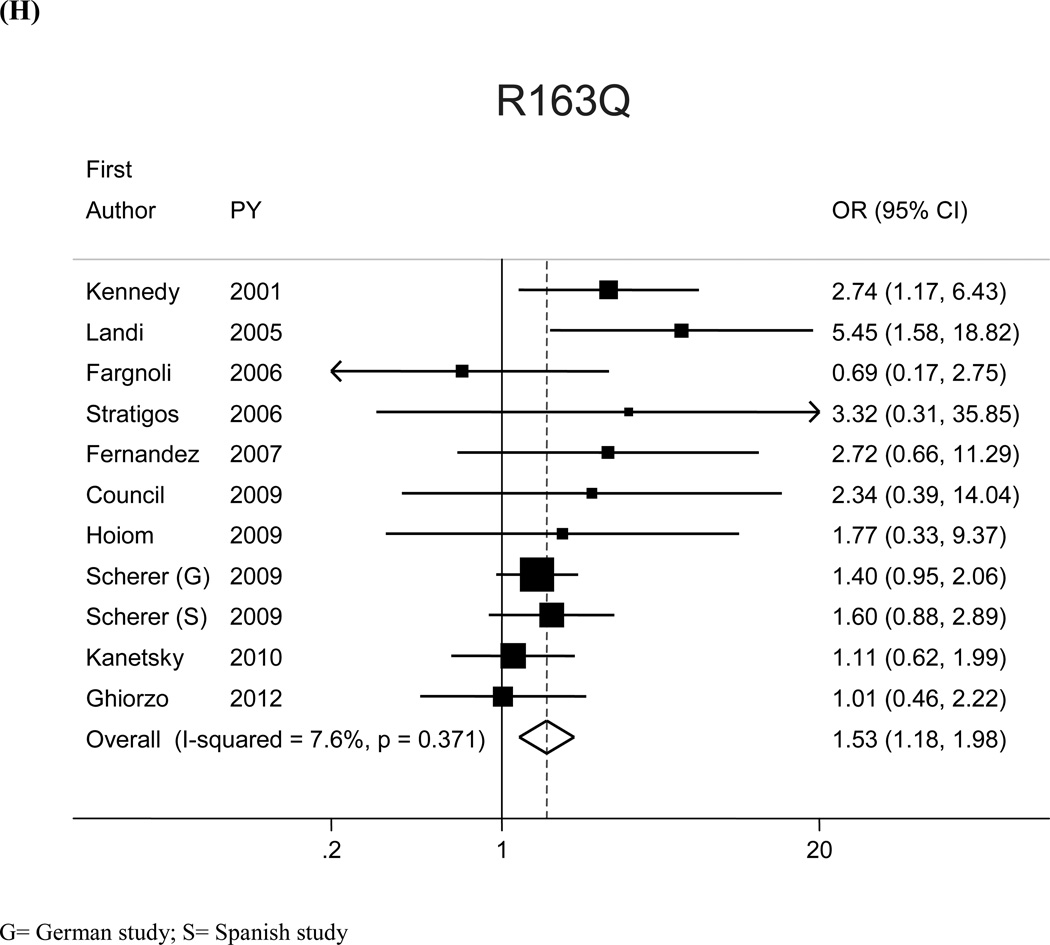
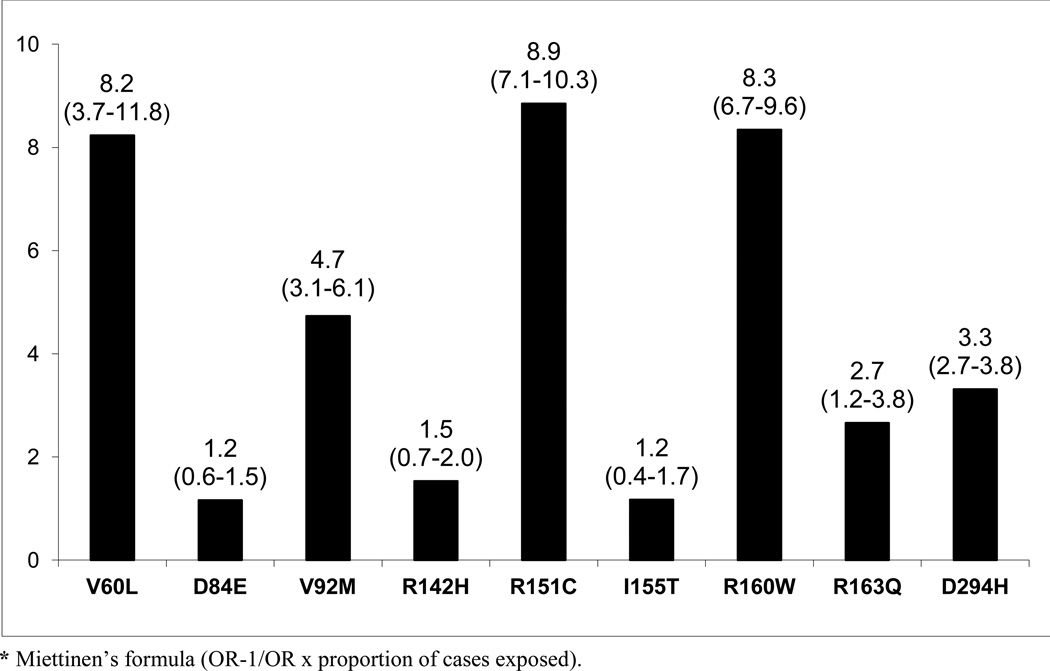
The most prevalent MC1R variants in the M-SKIP database were nine: V60L, D84E, V92M, R142H, R151C, I155T, R160W, R163Q, D294H. When we compared carriers of each variant with WT subjects using study-specific ORs and combined ORs, we found that CM risk significantly increased for carriers of any of the nine MC1R studied variants, with SOR (95%CI) ranging from 1.47 (1.17–1.84) for V60L to 2.74 (1.53–4.89) for D84E (Figure 1 and Table 2). After adjusting for multiple comparisons, all the p-values were still significant following the false discovery rate approach, while the association between I155T variant and melanoma was not confirmed after the more conservative Bonferroni correction (adjusted p-value: 0.08). The related attributable risk (AR) is presented in Figure 2. The highest AR was observed for R151 variant (8.9%), followed by R160W (8.3%) and V60L (8.2%) variants. In Table 2 we compared the results obtained in the 12 studies with WT as reference category with those obtained in the whole set of 17 studies and using, as reference group for each variant, the subjects without that variant. For this latter analysis, a significant association with CM was observed for all the MC1R variants but V60L, I155T and R163Q. It should be noted that the reference group of this latter analysis included, for each studied variant, both WT and any other MC1R variant.
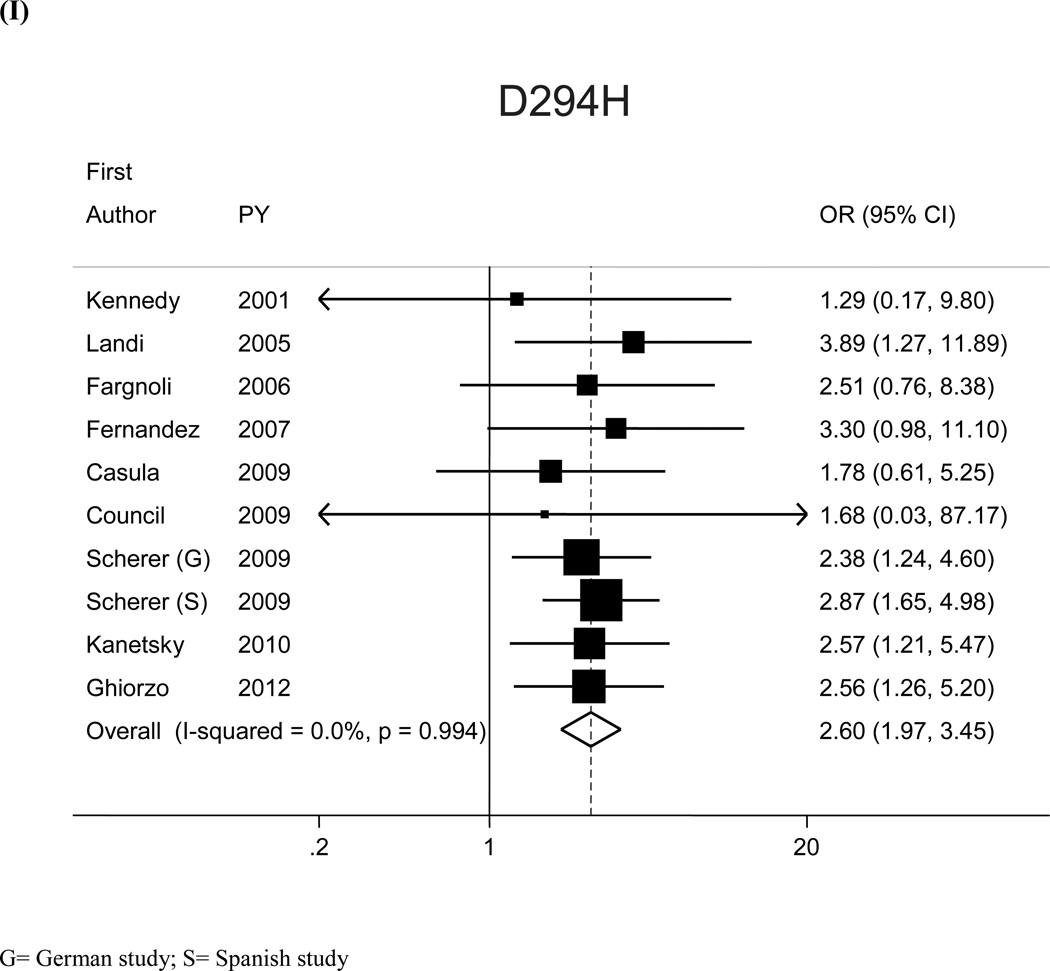
Figure 1.
Study-specific and pooled-Odds Ratio (OR) with 95% Confidence Intervals (CI) for the association between cutaneous melanoma and MC1R variants (A) V60L, (B) D84E, (C) V92M, (D) R142H, (E) R151C, (F) I155T, (G) R160W, (H) R163Q, (I) D294H. Reference category for each variant comprises WT subjects
Table 2.
Allele frequency and Summary Odds Ratios (SOR) with 95% Confidence Intervals (CI) for the association between MC1R variants and cutaneous melanoma using two different reference categories.
|
MC1R variant |
Allele frequency in controls (%) |
1. Variant present vs WT |
2. Variant present vs absenta |
||||
|---|---|---|---|---|---|---|---|
| N studies | N cases/N controls |
SOR (95%CI) | N studies | N cases/N controls |
SOR (95%CI) | ||
| V60L | 10.6 | 12 | 3,636/3,772 | 1.47 (1.17, 1.84) | 17 | 5,111/11,854 | 1.07 (0.91–1.26) |
| D84E | 0.4 | 9 | 3,089/3,371 | 2.74 (1.53, 4.89) | 11 | 3,956/4,588 | 2.13 (1.44–3.17) |
| V92M | 7.0 | 12 | 3,636/3,772 | 1.55 (1.30, 1.85) | 14 | 4,577/4,948 | 1.15 (1.00–1.31) |
| R142H | 0.6 | 11 | 3,571/3,742 | 2.30 (1.35, 3.92) | 14 | 4,430/10,888 | 1.77 (1.14–2.75) |
| R151C | 5.7 | 12 | 3,636/3,772 | 2.32 (1.83, 2.95) | 17 | 5,146/11,943 | 1.62 (1.34–1.96) |
| I155T | 0.8 | 9 | 3,283/3,416 | 1.83 (1.16, 2.89) | 12 | 4,287/4,621 | 1.36 (0.97–1.90 |
| R160W | 7.2 | 12 | 3,636/3,772 | 2.17 (1.77, 2.65) | 17 | 5,144/11,968 | 1.74 (1.43–2.13) |
| R163Q | 4.7 | 11 | 3,377/3,697 | 1.53 (1.18, 1.98) | 15 | 4,734/11,739 | 1.10 (0.94–1.30) |
| D294H | 1.3 | 10 | 3,448/3,587 | 2.60 (1.97, 3.45) | 14 | 4,607/5,079 | 1.78 (1.40–2.28) |
Note: In addition to MC1R, each study-specific logistic regression model included, if available, the following covariates: age, sex, intermittent and chronic sun exposure, lifetime and childhood sunburns, family history of melanoma, number of common total body naevi count and presence of atypical naevi.
For each MC1R variant, reference category included subjects either WT or carriers of any other MC1R variant.
Figure 2.
Attributable risks* in the population for cutaneous melanoma according to each MC1R variant (percentages with 95% confidence intervals). Reference category for each variant comprises WT subjects
Funnel plots for each MC1R variant are presented in Supplemental Figure 1 for the whole set of studies. We found some evidence of participation bias for the R163Q variant, with a borderline p-value (0.05).
Significant heterogeneity among the 12 risk estimates with WT as reference category was found for V60L (I2:50.9%, Q statistic p-value:0.02) and R142H (I2:49.3%, Q statistic p-value:0.03) (Figure 1). Variability in publication year, study area, genotyping methodology, deviation from HW equilibrium, source of controls, and source of DNA did not seem to explain the observed heterogeneity. Sensitivity analyses showed that the heterogeneity may be attributable to single studies: when we excluded the studies that lied out of the corresponding funnel plots, we obtained similar pooled-ORs than the original analyses, but with no more evidence of heterogeneity among study-specific estimates. For the analysis with WT as reference category SORs (95%CI) increased to 1.54 (1.27–1.88) and 2.76 (1.71–4.45) for V60L (excluding26) and R142H (excluding15), respectively; I2 (Q statistic p-value) were, respectively, 33.6% (p=0.13) and 29.3%, (p=0.18).
Meta-ORs calculated for studies not included in the M-SKIP project were similar to those obtained from our pooled-analysis on the whole dataset for all but R151C variant, for which it was significantly higher than our pooled-OR (meta regression p-value=0.05).
Association between combined MC1R variants and melanoma
SOR for the most common haplotypes were reported in Supplemental Table 1 for the 12 studies in which MC1R was sequenced. Results were similar to those observed for the single-variant analysis, with the higher SOR (3.05; 95%CI: 1.56–5.98) observed for the haplotype corresponding to a single mutation at rs1805006 (D84E).
We found that subjects carrying at least one MC1R variant had a significantly increased risk of CM, with SOR (95%CI) = 1.66 (1.41, 1.96) (Table 3). The risk attributable to any MC1R variant was 28.3%.
Table 3.
Summary Odds Ratios (SOR) with 95% Confidence Intervals (CI) for the association between combined MC1R variants and cutaneous melanoma, and heterogeneity estimates
| Variant | N cases |
N controls |
SOR (95%CI) | Q test p-value |
I2 for heterogeneity (%) |
|---|---|---|---|---|---|
| Wild-type | 1,047 | 1,461 | Reference | - | - |
| Any variant | 2,589 | 2,311 | 1.66 (1.41–1.96) | 0.10 | 36.8 |
| 1 variant | 1,615 | 1,719 | 1.41 (1.07–1.87) | 0.15 | 30.8 |
| 2+ variants | 974 | 592 | 2.51 (1.83–3.44) | 0.02 | 52.9 |
| Scorea 1 | 904 | 1,116 | 1.24 (0.90–1.72) | 0.05 | 44.5 |
| Score 2 | 955 | 834 | 1.69 (1.21–2.35) | 0.44 | 0.6 |
| Score 3 | 478 | 245 | 3.28 (2.22–4.87) | 0.57 | 0 |
| Score ≥4 | 252 | 116 | 3.12 (1.99–4.91) | 0.02 | 53.1 |
Note: In addition to MC1R, each study-specific logistic regression model included, if available, the following covariates: age, sex, intermittent and chronic sun exposure, lifetime and childhood sunburns, family history of melanoma, number of common total body naevi count and presence of atypical naevi.
Score calculated as detailed in 37.
Individuals carrying just one MC1R variant had almost 40% increased risk of CM compared to WT homozygous subjects (SOR 1.41; 95%CI 1.07–1.87, Table 3), whereas carriers of two or more MC1R variants had more than a double risk of CM than WT subjects (SOR 2.51; 95%CI 1.83–3.44, Table 3).
A significant linear trend was observed for one point increase in MC1R score (per-point SOR 1.39; 95%CI 1.31–1.48, p-value <0.0001). SOR (95%CI) increased from 1.24 (0.90–1.72) to 1.69 (1.21–2.35) to 3.28 (2.22–4.87) and then slightly decrease to 3.12 (1.99–4.91) with increasing score from 1 to 4+ (Table 3).
Analyses stratified by phenotypic characteristics
Results from analyses stratified according by freckles, hair color, skin type and their combination for any MC1R variant and for the nine most prevalent MC1R variants are reported in Table 4 for seven studies with WT as reference category and information on phenotypic characteristics. Stratified analysis for the most common haplotypes are reported in Supplemental Table 2.
Table 4.
Stratified analysis for MC1R variants and cutaneous melanoma association, according with freckles, hair color, skin type, and their combination. Reference category for each variant are WT subjects
| Variant | Phenotypic characteristic |
Strata | N studies (N cases/ N controls) |
OR (95%CI) | P-valuea |
|---|---|---|---|---|---|
| Any MC1R variant |
Freckles | Any | 4 (951/498) | 1.16 (0.79–1.72) | 0.003 |
| None | 4 (326/655) | 2.39 (1.60–3.57) | |||
| Hair color | Red | 3 (114/40) | 0.90 (0.23–3.58) | 0.36 | |
| Other | 6 (1,424/1,621) | 1.70 (1.20–2.42) | |||
| Skin type | I/II | 6 (689/567) | 1.16 (0.73–1.85) | 0.03 | |
| III/IV | 6 (832/1,111) | 1.89 (1.29–2.78) | |||
| Freckles-red hair-skin type I/II |
Any | 4 (1,102/934) | 1.16 (0.88–1.52) | <0.0001 | |
| None | 4 (197/444) | 3.14 (2.06–4.80) | |||
| V60L | Freckles | Any | 4 (951/498) | 1.00 (0.63–1.56) | 0.01 |
| None | 4 (326/655) | 2.00 (1.25–3.19) | |||
| Hair color | Red | 2 (111/35) | 0.59 (0.09–3.63) | 0.33 | |
| Other | 6 (1,424/1,621) | 1.44 (0.94–2.19) | |||
| Skin type | I/II | 6 (689/567) | 0.72 (0.37–1.38) | 0.002 | |
| III/IV | 6 (832/1,111) | 1.79 (1.03–3.12) | |||
| Freckles-red hair-skin type I/II |
Any | 4 (1,102/934) | 0.95 (0.70–1.30) | 0.0008 | |
| None | 4 (197/444) | 2.51 (1.54–4.07) | |||
| D84E | Freckles | Any | 2 (806/341) | 0.87 (0.33–2.28) | 0.04 |
| None | 2 (207/485) | 5.23 (1.29–21.24) | |||
| Hair color | Red | 2 (111/35) | 1.25 (0.09–17.23) | 0.45 | |
| Other | 4 (1,160/1,308) | 3.39 (1.02–11.30) | |||
| Skin type | I/II | 4 (577/481) | 1.23 (0.32–4.67) | 0.41 | |
| III/IV | 4 (666/876) | 3.12 (0.33–29.13) | |||
| Freckles-red hair-skin type I/II |
Any | 3 (988/849) | 1.56 (0.49–4.93) | 0.51 | |
| None | 1 (26/167) | 4.13 (0.25–67.39) | |||
| V92M | Freckles | Any | 4 (951/498) | 0.87 (0.59–1.29) | 0.0002 |
| None | 4 (326/655) | 2.36 (1.41–3.95) | |||
| Hair color | Red | 1 (94/19) | 0.94 (0.09–9.78) | 0.66 | |
| Other | 6 (1,424/1,621) | 1.61 (1.12–2.29) | |||
| Skin type | I/II | 6 (689/567) | 1.21 (0.69–2.10) | 0.27 | |
| III/IV | 6 (832/1,111) | 1.71 (1.10–2.66) | |||
| Freckles-red hair-skin type I/II |
Any | 4 (1,102/934) | 1.04 (0.74–1.47) | 0.005 | |
| None | 4 (197/444) | 3.13 (1.59–6.14) | |||
| R142H | Freckles | Any | 3 (855/425) | 1.93 (0.35–10.55) | 0.85 |
| None | 2 (208/289) | 2.40 (0.31–18.36) | |||
| Hair color | Red | 2 (111/35) | 0.35 (0.03–4.55) | 0.08 | |
| Other | 5 (1,318/1,263) | 3.03 (0.86–10.69) | |||
| Skin type | I/II | 6 (689/567) | 1.76 (0.59–5.24) | 0.70 | |
| III/IV | 5 (796/908) | 2.24 (0.77–6.49) | |||
| Freckles-red hair-skin type I/II |
Any | 4 (1,102/934) | 1.84 (0.50–6.83) | 0.50 | |
| None | 2 (135/217) | 3.75 (0.44–32.02) | |||
| R151C | Freckles | Any | 4 (951/498) | 1.67 (1.00–2.80) | 0.01 |
| None | 4 (326/655) | 4.27 (2.25–8.10) | |||
| Hair color | Red | 3 (114/40) | 0.85 (0.20–3.67) | 0.12 | |
| Other | 6 (1,424/1,621) | 2.75 (1.90–3.97) | |||
| Skin type | I/II | 6 (689/567) | 1.60 (0.95–2.70) | 0.05 | |
| III/IV | 6 (832/1,111) | 3.05 (1.89–4.93) | |||
| Freckles-red hair-skin type I/II |
Any | 4 (1,102/934) | 1.77 (1.26–2.49) | 0.01 | |
| None | 4 (197/444) | 5.55 (2.47–12.48) | |||
| I155T | Freckles | Any | 2 (806/341) | 0.58 (0.25–1.35) | 0.16 |
| None | 2 (207/485) | 2.11 (0.43–10.46) | |||
| Hair color | Red | 1 (94/19) | 0.14 (0.01–2.52) | 0.15 | |
| Other | 4 (1,160/1,308) | 1.25 (0.63–2.48) | |||
| Skin type | I/II | 3 (498/306) | 0.52 (0.18–1.46) | 0.23 | |
| III/IV | 2 (563/565) | 1.29 (0.45–3.70) | |||
| Freckles-red hair-skin type I/II |
Any | 2 (899/639) | 0.76 (0.34–1.67) | 0.51 | |
| None | 2 (120/191) | 1.83 (0.15–22.80) | |||
| R160W | Freckles | Any | 4 (951/498) | 1.21 (0.79–1.84) | 0.0002 |
| None | 4 (326/655) | 3.72 (2.09–6.63) | |||
| Hair color | Red | 2 (111/35) | 0.85 (0.15–4.68) | 0.39 | |
| Other | 6 (1,424/1,621) | 1.82 (1.33–2.48) | |||
| Skin type | I/II | 6 (689/567) | 1.24 (0.70–2.19) | 0.10 | |
| III/IV | 5 (776/1,023) | 2.15 (1.31–3.53) | |||
| Freckles-red hair-skin type I/II |
Any | 4 (1,102/934) | 1.27 (0.88–1.84) | 0.0003 | |
| None | 4 (197/444) | 6.69 (2.92–15.35) | |||
| R163Q | Freckles | Any | 4 (951/498) | 1.45 (0.55–3.77) | 0.37 |
| None | 4 (326/655) | 2.31 (0.93–5.74) | |||
| Hair color | Red | 0 (0/0) | nc | nc | |
| Other | 6 (1,424/1,621) | 1.70 (0.93–3.14) | |||
| Skin type | I/II | 4 (577/481) | 1.66 (0.87–3.16) | 0.51 | |
| III/IV | 6 (832/1,111) | 1.24 (0.56–2.78) | |||
| Freckles-red hair-skin type I/II |
Any | 4 (1,102/934) | 1.56 (0.64–3.80) | 0.33 | |
| None | 4 (197/444) | 2.60 (0.86–7.89) | |||
| D294H | Freckles | Any | 4 (951/498) | 1.64 (0.92–2.91) | 0.04 |
| None | 2 (208/289) | 6.36 (2.00–20.21) | |||
| Hair color | Red | 2 (111/35) | 1.78 (0.22–14.52) | 0.73 | |
| Other | 5 (1,315/1,474) | 2.59 (1.64–4.11) | |||
| Skin type | I/II | 5 (632/503) | 0.97 (0.49–1.92) | 0.001 | |
| III/IV | 4 (740/820) | 4.57 (2.45–8.53) | |||
| Freckles-red hair-skin type I/II |
Any | 4 (1,102/934) | 1.61 (0.96–2.68) | 0.02 | |
| None | 2 (135/217) | 14.36 (2.55–80.89) |
nc=not calculable; OR= Odds Ratio; CI=Confidence Intervals
Note: In addition to MC1R, each study-specific logistic regression model included, if available, the following covariates: age, sex, intermittent and chronic sun exposure, lifetime and childhood sunburns, family history of melanoma, number of common total body naevi count and presence of atypical naevi.
Overall p-value for any significant difference among strata-specific ORs.
For subjects with the RHC phenotype, the risk of CM was not independently predicted by having MC1R variants: SORs (95%CI) for carriers of any MC1R variant were 1.16 (0.79–1.72), 0.90 (0.23–3.58), 1.16 (0.73–1.85), and 1.16 (0.88–1.52) for subjects with freckles, red hair, skin type I/II, and any of the above phenotypic characteristics, respectively. On the contrary, darker-pigmented subjects presented a significantly higher risk of CM associated with MC1R variants: SORs (95%CI) for carriers of any MC1R variant were 2.39 (1.60–3.57), 1.70 (1.20–2.42), 1.89 (1.29–2.78) and 3.14 (2.06–4.80) for subjects with no freckles, no red hair, skin type III/IV, and all the above phenotypic characteristics, respectively. This trend was observed also when we analyzed each single variant and when we looked separately at the 5 southern European studies,24,26–28,36 the USA study15 and the northern European study.22 The difference between the strata-specific ORs was statistically significant for the analysis on freckles, skin type and the combination of the three RHC phenotypic characteristics for any MC1R variant.
Further stratified analyses were performed for skin color and eye color (Supplemental Table 3). Also for these variables, a constant trend was observed, with higher SORs obtained for darker-pigmented subjects compared to lighter-pigmented individuals.
Discussion
Our pooled-analysis identified a significant association with sporadic CM for all the most common MC1R variants compared to WT. An unbiased and precise estimate of the risk of melanoma in carriers of MC1R variants for different populations of European origins is provided for the first time by our group, since in previous large meta-analysis14–16,18 SORs were systematically biased toward the null, due to the inclusion of carriers of other MC1R variants in the reference category for each variant analysis. The three RHC variants R151C, R160W, and D294H were previously associated with CM risk.14,16,18,22,24–26 The association of D84E, R142H and I155T with CM was not always identified in single studies, probably because of the low frequency of these alleles, but it was confirmed by more powerful meta-analyses.14,16,18
An important question raised by these findings refers to the functional characteristics of mutant alleles accounting for their association with CM and for their different penetrance. The functional properties of WT MC1R, the most common RHC variants, and a few rarer variants have been addressed in several independent studies. WT MC1R triggers at least three major signal transduction pathways. The Gs protein-dependent activation of the cAMP pathway leads to induction of the master melanocyte transcription factor MITF and its downstream targets including the melanogenic enzymes.39 Efficient activation of the cAMP pathway appears essential for the synthesis of eumelanins and for a normal tanning response.40 In addition, MC1R activation triggers two mitogen-activated protein kinase (MAPK) modules, one leading to activation of p38 kinase,41 and the other to the extracellular signal-regulated kinases ERK1 and ERK2.42 The functional outputs in melanocytes of these two related pathways are poorly understood. Interestingly, whereas p38 activation downstream of the MC1R depends on cAMP,41 ERK activation in human melanocytic cells appears cAMP-independent.42 Accordingly natural mutations in the MC1R gene may have different effects on functional coupling to the cAMP and ERK pathways, as confirmed by several studies.43,44
Several independent studies have firmly established that the major RHC alleles are loss-of-function forms with decreased cAMP signaling,11–13 although they may retain a high efficacy for ERK activation.13,42,44 For the D84E, R151C, I155T and R160W receptors impairment in cAMP coupling is largely accounted for by reduced cell surface expression. Conversely, the R142H and D294H variants show normal or even slightly increased cell surface expression,11 and their loss-of-function phenotype is most likely related to an inability to properly undergo the agonist-induced transition to the active state and/or to impaired coupling to the Gs protein. In any case, inefficient or even absent activation of the cAMP pathway downstream of these forms is consistent with impaired eumelanogenesis, which may account for their frequent occurrence in individuals with red hair and fair skin. This suggests a pigmentary component for their contribution to CM development,14 whereby tanning ability would be compromised in these individuals and production of photoprotective eumelanin would switch towards biosynthesis of photosensitizing pheomelanin.
On the other hand, previous meta-analyses14,16,18 reported controversial results for the association of V60L, V92M and R163Q variants with CM. It is worthwhile to note that the SOR calculated in the present study for all the nine studied variants compared to WT were markedly higher than those reported in previous meta-analyses. These differences are probably attributable to a bias towards the null in meta-analyses, due to the fact that reference category for OR calculation often includes, for each studied variant, both WT and any other MC1R variant. Indeed, when we compared the results obtained in our pooled-analysis, we noted that SORs calculated with WT as the reference category were always higher than the corresponding ones obtained by a classical analysis based on presence/absence of each variant.
A marginal effect of the V92M substitution on the cell surface expression or the ability to activate the cAMP and ERK cascades has been reported.11,44 Concerning R163Q, a selective decrease in ERK activation has been recently described13 but its functional coupling to the cAMP pathway is normal or only slightly modified,45 and this variant is highly frequent in Asian populations. V60L allele has been shown to display reduced cell surface expression with a corresponding impairment in cAMP activation.11,44 In fact, its functional properties are comparable to those of the RHC forms R151C or R160W mentioned above.
When we analyzed MC1R variants combined, we observed a significantly higher risk of CM both for carriers of one and carriers of two or more variants compared to WT subjects. It has been previously suggested that RHC alleles may act in a recessive manner and that one fully functional copy of MC1R may be sufficient to provide normal function;12 however our results suggested that CM risk increased even for carriers of just one MC1R variant. When we assigned a score to each subject according with the number of R and r alleles, a dose-response relationship was noted, with SORs significantly higher with increasing score, as expected.
A very interesting result of our pooled-analysis was that we observed a significantly higher CM risk associated with MC1R variants only for darker-pigmented subjects of European origins, while carriers of MC1R variants with RHC phenotype had no increased risk of melanoma compared to non carriers. A similar result was observed in previous studies and meta-analysis,15,24 which highlighted that the association between some MC1R variants and CM was stronger in subjects with dark hair, dark eyes, skin type III/IV, and in subjects who reported low recreational sun exposure. A recent meta-analysis on darker-pigmented Southern European populations46 also showed that MC1R RHC variants were strong CM risk predictors. These results suggest that MC1R variants may mediate their effects, at least partially, through biological pathways that are independent on pigmentation, thus providing additional information about melanoma risk in people who would not be identified as high risk based on their phenotypic characteristics alone. Recent studies implicate MC1R signaling in a number of key biological pathways involved in cell cycle control,47 apoptosis,48 and activation of DNA repair mechanisms and antioxidant defenses.19 Production of pheomelanin pigments seems associated with increased oxidative DNA damage compared with synthesis of eumelanins.49 Further evidence for pheomelanin-associated increased cellular oxidative stress was obtained in studies of mice carrying a loss-of-function mutation of the Mc1r gene. These mice have a yellowish pheomelanic hair mimicking the human red hair and show higher levels of lipid peroxides, a product of reactive oxygen species (ROS)-mediated lipid damage.20 This study provided evidence in support of a melanogenetic effect independent of UV exposure. In addition, as discussed above, stimulation of MC1R also activates MAPK pathway and regulates target genes involved in inflammation through the NF-Kb pathway.50 Thus, interpretation of the effect of MC1R alleles in melanoma beyond its role in pigmentation is complex.
A strength of the present pooled-analysis is the large sample size, which gives power to detect associations with relatively rare MC1R variants, and to perform stratified analysis by phenotypic characteristics at an individual, rather than at a population, level. Moreover, the international collaborative nature of the M-SKIP project makes it possible to assess the MC1R–related CM risk in various populations and ancestries, thus providing evidence that the estimated risk is robust and consistent in different geographical areas. As it was previously pointed out,18 a source of heterogeneity between studies in meta-analysis was the choice of study comparator group, which is often characterized, for each variant, by people without that variant. This practice leads to underestimate the true risk of disease in meta-analysis, because MC1R variants are very common: 66% of our study population had at least one variant. Within our pooled-analysis we were able to compare each variant with WT in the 12 studies with MC1R sequenced, and we indeed obtained markedly higher SORs. Moreover we were able to study the combined effect of one or more MC1R variants compared to WT subjects, and this would not be usually possible in a meta-analytic context, where the analysis of variants combined usually differed from study to study. Finally, we could take into account all the available confounders in our centralized statistical analysis, with a homogeneous plan of analysis and homogeneous definition of confounders.
Concerning the possible limitations of this study, one drawback is the exclusion of GWAS in the first phase of the project. This was done to avoid increasing the heterogeneity of the pooled estimate with a different approach. However GWAS may be included in the M-SKIP in a second step of the project. Unavailability of information on other genes in most studies prevents the analysis of possible gene-gene interactions. The interactions of other low-risk loci need to be taken into account for a full assessment of genetic susceptibility to CM. Other genes such as SLC45A2, TYR, TYRP1, ASIP, OCA2, XRCC and GSTP1 may also contribute to skin cancer susceptibility. Because we carried out a retrospective pooled-analysis, we did not perform centralized sequencing. However, previous studies37 have reported excellent concordance in sequencing data from different centres. Finally, differences in the assessment of sun exposure did not allow for the use of this variable in stratified analysis, although it remained possible to take it into account for the adjustment for confounders.
In conclusion our study remarks the important role of all the main MC1R variants in sporadic CM and suggests that they have a direct effect on melanoma risk, independently on the phenotypic characteristics of carriers. This is of particular importance for assessing preventive strategies, which may be directed to darker-pigmented individuals of European origins with MC1R variants as well as to lightly-pigmented, fair-skinned subjects.
Supplementary Material
Acknowledgements
This work was supported by the Italian Association for Cancer Research (grant number: MFAG 11831).
The M-SKIP study group consists of the following members: Principal Investigator: Sara Raimondi (European Institute of Oncology, Milan, Italy); Advisory Committee members: Philippe Autier (International Prevention Research Institute, Lyon, France), Maria Concetta Fargnoli (University of L'Aquila, Italy), José C. García-Borrón (University of Murcia, Spain), Jiali Han (Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA), Peter A. Kanetsky (Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL, USA), Maria Teresa Landi (National Cancer Institute, NIH, Bethesda, MD, USA), Julian Little (University of Ottawa, Canada), Julia Newton-Bishop (University of Leeds, UK), Francesco Sera (UCL Institute of Child Health, London, UK); Consultants: Saverio Caini (ISPO, Florence, Italy), Sara Gandini and Patrick Maisonneuve (European Institute of Oncology, Milan, Italy); Participant Investigators: Albert Hofman, Manfred Kayser, Fan Liu, Tamar Nijsten and Andre G. Uitterlinden (Erasmus MC University Medical Center, Rotterdam, The Netherlands), Rajiv Kumar and Dominique Scherer (German Cancer Research Center, Heidelberg, Germany), Eduardo Nagore (Instituto Valenciano de Oncologia, Valencia, Spain), Johan Hansson and Veronica Hoiom (Karolinska Institutet, Stockholm, Sweden), Paola Ghiorzo and Lorenza Pastorino (University of Genoa, Italy), Nelleke A. Gruis (Leiden University Medical Center, The Netherlands), Terry Dwyer (Murdoch Childrens Research Institute, Victoria, Australia), Leigh Blizzard and Jennifer Cochrane (Menzies Research Institute Tasmania, Hobart, Australia), Ricardo Fernandez-de-Misa (Hospital Universitario Nuestra Señora de Candelaria, Santa Cruz de Tenerife, Spain), Wojciech Branicki (Institute of Forensic Research, Krakow, Poland), Tadeusz Debniak (Pomeranian Medical University, Polabska, Poland), Niels Morling and Peter Johansen (University of Copenhagen, Denmark), Ruth Pfeiffer (National Cancer Institute, NIH, Bethesda, MD, USA), Giuseppe Palmieri (Istituto di Chimica Biomolecolare, CNR, Sassari, Italy), Gloria Ribas (Fundación Investigación Clínico de Valencia Instituto de Investigación Sanitaria- INCLIVA, Spain), Alexander Stratigos and Katerina Kypreou (University of Athens, Andreas Sygros Hospital, Athens, Greece), Anne Bowcock, Lynn Cornelius and M. Laurin Council (Washington University School of Medicine, St. Louis, MO, USA), Tomonori Motokawa (POLA Chemical Industries, Yokohama, Japan), Sumiko Anno (Shibaura Institute of Technology, Tokyo, Japan), Per Helsing and Per Arne Andresen (Oslo University Hospital, Norway), Terence H. Wong (University of Edinburgh, UK), and the GEM Study Group.
Participants in the GEM Study Group are as follows: Coordinating Center, Memorial Sloan-Kettering Cancer Center, New York, NY, USA: Marianne Berwick (PI, currently at the University of New Mexico), Colin Begg (Co-PI), Irene Orlow (Co-Investigator), Urvi Mujumdar (Project Coordinator), Amanda Hummer (Biostatistician), Klaus Busam (Dermatopathologist), Pampa Roy (Laboratory Technician), Rebecca Canchola (Laboratory Technician), Brian Clas (Laboratory Technician), Javiar Cotignola (Laboratory Technician), Yvette Monroe (Interviewer). Study Centers: The University of Sydney and The Cancer Council New South Wales, Sydney (Australia): Bruce Armstrong (PI), Anne Kricker (co-PI), Melisa Litchfield (Study Coordinator). Menzies Centre for Population Health Research, University of Tasmania, Hobart (Australia): Terence Dwyer (PI), Paul Tucker (Dermatopathologist), Nicola Stephens (Study Coordinator). British Columbia Cancer Agency, Vancouver (Canada): Richard Gallagher (PI), Teresa Switzer (Coordinator). Cancer Care Ontario, Toronto (Canada): Loraine Marrett (PI), Beth Theis (Co-Investigator), Lynn From (Dermatopathologist), Noori Chowdhury (Coordinator), Louise Vanasse (Coordinator), Mark Purdue (Research Officer). David Northrup (Manager for CATI). Centro per la Prevenzione Oncologia Torino, Piemonte (Italy): Roberto Zanetti (PI), Stefano Rosso (Data Manager), Carlotta Sacerdote (Coordinator). University of California, Irvine (USA): Hoda Anton-Culver (PI), Nancy Leighton (Coordinator), Maureen Gildea (Data Manager). University of Michigan, Ann Arbor (USA): Stephen Gruber (PI), Joe Bonner (Data Manager), Joanne Jeter (Coordinator). New Jersey Department of Health and Senior Services, Trenton (USA): Judith Klotz (PI), Homer Wilcox (Co-PI), Helen Weiss (Coordinator). University of North Carolina, Chapel Hill (USA): Robert Millikan (PI), Nancy Thomas (Co-Investigator), Dianne Mattingly (Coordinator), Jon Player (Laboratory Technician), Chiu-Kit Tse (Data Analyst). University of Pennsylvania, Philadelphia, PA (USA): Timothy Rebbeck (PI), Peter Kanetsky (Co-Investigator), Amy Walker (Laboratory Technician), Saarene Panossian (Laboratory Technician). Consultants: Harvey Mohrenweiser, University of California, Irvine, Irvine, CA (USA); Richard Setlow, Brookhaven National Laboratory, Upton, NY (USA).
Abbreviations
- α-MSH
α-melanocyte-stimulating hormone
- CI
confidence interval
- CM
cutaneous melanoma
- ERK
extracellular signal-regulated kinases
- GWAS
genome-wide association studies
- HW
Hardy-Weinberg
- MAPK
mitogen-activated protein kinase
- MC1R
melanocortin-1-receptor
- NMSC
non-melanoma skin cancer
- OR
odds ratio
- ROS
reactive oxygen species
- RHC
red hair color
- SOR
summary odds ratio
- WT
wild-type
Footnotes
Author’s disclosures of potential conflicts of interest: none for all authors.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. GLOBOCAN 2008 v2.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10. Lyon, France: International Agency for Research on Cancer; 2010. http://globocan.iarc.fr. [Google Scholar]
- 2.Thompson JF, Scolyer RA, Kefford RF. Cutaneous melanoma. Lancet. 2005;365:687–701. doi: 10.1016/S0140-6736(05)17951-3. [DOI] [PubMed] [Google Scholar]
- 3.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Zanetti R, Masini C, Boyle P, Melchi CF. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur J Cancer. 2005;41:2040–2059. doi: 10.1016/j.ejca.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 4.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Picconi O, Boyle P, Melchi CF. Meta-analysis of risk factors for cutaneous melanoma: II. Sun exposure. Eur J Cancer. 2005;41:45–60. doi: 10.1016/j.ejca.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Abeni D, Boyle P, Melchi CF. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur J Cancer. 2005;41:28–44. doi: 10.1016/j.ejca.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Hayward NK. Genetics of melanoma predisposition. Oncogene. 2003;22:3053–3062. doi: 10.1038/sj.onc.1206445. [DOI] [PubMed] [Google Scholar]
- 7.Duffy DL, Box NF, Chen W, Palmer JS, Montgomery GW, James MR, Hayward NK, Martin NG, Sturm RA. Interactive effects of MC1R and OCA2 on melanoma risk phenotypes. Hum Mol Genet. 2004;13:447–461. doi: 10.1093/hmg/ddh043. [DOI] [PubMed] [Google Scholar]
- 8.Burchill SA, Ito S, Thody AJ. Effects of melanocyte-stimulating hormone on tyrosinase expression and melanin synthesis in hair follicular melanocytes of the mouse. J Endocrinol. 1993;137:189–195. doi: 10.1677/joe.0.1370189. [DOI] [PubMed] [Google Scholar]
- 9.Rouzaud F, Kadekaro AL, Abdel-Malek ZA, Hearing VJ. MC1R and the response of melanocytes to ultraviolet radiation. Mutat Res. 2005;571:133–152. doi: 10.1016/j.mrfmmm.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Gerstenblith MR, Goldstein AM, Fargnoli MC, Peris K, Landi MT. Comprehensive evaluation of allele frequency differences of MC1R variants across populations. Hum Mutat. 2007;28:495–505. doi: 10.1002/humu.20476. [DOI] [PubMed] [Google Scholar]
- 11.Beaumont KA, Shekar SN, Newton RA, James MR, Stow JL, Duffy DL, Sturm RA. Receptor function, dominant negative activity and phenotype correlations for MC1R variant alleles. Hum Mol Genet. 2007;16:2249–2260. doi: 10.1093/hmg/ddm177. [DOI] [PubMed] [Google Scholar]
- 12.Beaumont KA, Shekar SN, Cook AL, Duffy DL, Sturm RA. Red hair is the null phenotype of MC1R. Hum Mutat. 2008;29:E88–E94. doi: 10.1002/humu.20788. [DOI] [PubMed] [Google Scholar]
- 13.Doyle JR, Fortin JP, Beinborn M, Kopin AS. Selected melanocortin 1 receptor single-nucleotide polymorphisms differentially alter multiple signaling pathways. J Pharmacol Exp Ther. 2012;342:318–326. doi: 10.1124/jpet.112.194548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raimondi S, Sera F, Gandini S, Iodice S, Caini S, Maisonneuve P, Fargnoli MC. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer. 2008;122:2753–2760. doi: 10.1002/ijc.23396. [DOI] [PubMed] [Google Scholar]
- 15.Kanetsky PA, Panossian S, Elder DE, Guerry D, Ming ME, Schuchter L, Rebbeck TR. Does MC1R genotype convey information about melanoma risk beyond risk phenotypes? Cancer. 2010;116:2416–2428. doi: 10.1002/cncr.24994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatzinasiou F, Lill CM, Kypreou K, Stefanaki I, Nicolaou V, Spyrou G, Evangelou E, Roehr JT, Kodela E, Katsambas A, Tsao H, Ioannidis JP, et al. Comprehensive field synopsis and systematic meta-analyses of genetic association studies in cutaneous melanoma. J Natl Cancer Inst. 2011;103:1227–1235. doi: 10.1093/jnci/djr219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amos CI, Wang LE, Lee JE, Gershenwald JE, Chen WV, Fang S, Kosoy R, Zhang M, Qureshi AA, Vattathil S, Schacherer CW, Gardner JM, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum Mol Genet. 2011;20:5012–5023. doi: 10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams PF, Olsen CM, Hayward NK, Whiteman DC. Melanocortin 1 receptor and risk of cutaneous melanoma: a meta-analysis and estimates of population burden. Int J Cancer. 2011;129:1730–1740. doi: 10.1002/ijc.25804. [DOI] [PubMed] [Google Scholar]
- 19.Kadekaro AL, Chen J, Yang J, Chen S, Jameson J, Swope VB, Cheng T, Kadakia M, Abdel-Malek Z. Alpha-melanocyte-stimulating hormone suppresses oxidative stress through a p53-mediated signaling pathway in human melanocytes. Mol Cancer Res. 2012;10:778–7786. doi: 10.1158/1541-7786.MCR-11-0436. [DOI] [PubMed] [Google Scholar]
- 20.Mitra D, Luo X, Morgan A, Wang J, Hoang MP, Lo J, Guerrero CR, Lennerz JK, Mihm MC, Wargo JA, Robinson KC, Devi SP, et al. An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. Nature. 2012;491:449–453. doi: 10.1038/nature11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raimondi S, Gandini S, Fargnoli MC, Bagnardi V, Maisonneuve P, Specchia C, Kumar R, Nagore E, Han J, Hansson J, Kanetsky PA, Ghiorzo P, et al. Melanocortin-1 receptor, skin cancer and phenotypic characteristics (M-SKIP) project: study design and methods for pooling results of genetic epidemiological studies. BMC Med Res Methodol. 2012;12:116. doi: 10.1186/1471-2288-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kennedy C, ter Huurne J, Berkhout M, Gruis N, Bastiaens M, Bergman W, Willemze R, Bavinck JN. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol. 2001;117:294–300. doi: 10.1046/j.0022-202x.2001.01421.x. [DOI] [PubMed] [Google Scholar]
- 23.Dwyer T, Stankovich JM, Blizzard L, FitzGerald LM, Dickinson JL, Reilly A, Williamson J, Ashbolt R, Berwick M, Sale MM. Does the addition of information on genotype improve prediction of the risk of melanoma and nonmelanoma skin cancer beyond that obtained from skin phenotype? Am J Epidemiol. 2004;159:826–833. doi: 10.1093/aje/kwh120. [DOI] [PubMed] [Google Scholar]
- 24.Landi MT, Kanetsky PA, Tsang S, Gold B, Munroe D, Rebbeck T, Swoyer J, Ter-Minassian M, Hedayati M, Grossman L, Goldstein AM, Calista D, et al. MC1R, ASIP, and DNA repair in sporadic and familial melanoma in a Mediterranean population. J Natl Cancer Inst. 2005;97:998–1007. doi: 10.1093/jnci/dji176. [DOI] [PubMed] [Google Scholar]
- 25.Debniak T, Scott R, Masojc B, Serrano-Fernandez P, Huzarski T, Byrski T, Debniak B, Gorski B, Cybulski C, Medrek K, Kurzawski G, van de Wetering T, et al. MC1R common variants, CDKN2A and their association with melanoma and breast cancer risk. Int J Cancer. 2006;119:2597–2602. doi: 10.1002/ijc.22210. [DOI] [PubMed] [Google Scholar]
- 26.Fargnoli MC, Altobelli E, Keller G, Chimenti S, Hofler H, Peris K. Contribution of melanocortin-1 receptor gene variants to sporadic cutaneous melanoma risk in a population in central Italy: a case-control study. Melanoma Res. 2006;16:175–182. doi: 10.1097/01.cmr.0000198454.11580.b5. [DOI] [PubMed] [Google Scholar]
- 27.Stratigos AJ, Dimisianos G, Nikolaou V, Poulou M, Sypsa V, Stefanaki I, Papadopoulos O, Polydorou D, Plaka M, Christofidou E, Gogas H, Tsoutsos D, et al. Melanocortin receptor-1 gene polymorphisms and the risk of cutaneous melanoma in a low-risk southern European population. J Invest Dermatol. 2006;126:1842–1849. doi: 10.1038/sj.jid.5700292. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez L, Milne R, Bravo J, Lopez J, Aviles J, Longo M, Benitez J, Lazaro P, Ribas G. MC1R: three novel variants identified in a malignant melanoma association study in the Spanish population. Carcinogenesis. 2007;28:1659–1664. doi: 10.1093/carcin/bgm084. [DOI] [PubMed] [Google Scholar]
- 29.Brudnik U, Branicki W, Wojas-Pelc A, Kanas P. The contribution of melanocortin 1 receptor gene polymorphisms and the agouti signalling protein gene 8818A>G polymorphism to cutaneous melanoma and basal cell carcinoma in a Polish population. Exp Dermatol. 2009;18:167–174. doi: 10.1111/j.1600-0625.2008.00760.x. [DOI] [PubMed] [Google Scholar]
- 30.Casula M, Muggiano A, Cossu A, Budroni M, Caraco C, Ascierto PA, Pagani E, Stanganelli I, Canzanella S, Sini M, Palomba G, et al. Italian Melanoma Intergroup (IMI) Role of key-regulator genes in melanoma susceptibility and pathogenesis among patients from South Italy. BMC Cancer. 2009;9:352. doi: 10.1186/1471-2407-9-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Council ML, Gardner JM, Helms C, Liu Y, Cornelius LA, Bowcock AM. Contribution of genetic factors for melanoma susceptibility in sporadic US melanoma patients. Exp Dermatol. 2006;18:485–487. doi: 10.1111/j.1600-0625.2008.00807.x. [DOI] [PubMed] [Google Scholar]
- 32.Nan H, Kraft P, Hunter DJ, Han J. Genetic variants in pigmentation genes, pigmentary phenotypes, and risk of skin cancer in Caucasians. Int J Cancer. 2009;125:909–917. doi: 10.1002/ijc.24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoiom V, Tuominen R, Kaller M, Linden D, Ahmadian A, Mansson-Brahme E, Egyhazi S, Sjoberg K, Lundeberg J, Hansson J. MC1R variation and melanoma risk in the Swedish population in relation to clinical and pathological parameters. Pigment Cell Melanoma Res. 2009;22:196–204. doi: 10.1111/j.1755-148X.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu F, van Duijn K, Vingerling JR, Hofman A, Uitterlinden AG, Janssens AC, Kayser M. Eye color and the prediction of complex phenotypes from genotypes. Curr Biol. 2009;19:R192–R193. doi: 10.1016/j.cub.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 35.Scherer D, Nagore E, Bermejo JL, Figl A, Botella-Estrada R, Thirumaran RK, Angelini S, Hemminki K, Schadendorf D, Kumar R. Melanocortin receptor 1 variants and melanoma risk: a study of 2 European populations. Int J Cancer. 2009;125:1868–1875. doi: 10.1002/ijc.24548. [DOI] [PubMed] [Google Scholar]
- 36.Ghiorzo P, Bonelli L, Pastorino L, Bruno W, Barile M, Andreotti V, Nasti S, Battistuzzi L, Grosso M, Bianchi-Scarra G, Queirolo P. MC1R variation and melanoma risk in relation to host/clinical and environmental factors in CDKN2A positive and negative melanoma patients. Exp Dermatol. 2012;21:718–720. doi: 10.1111/j.1600-0625.2012.01549.x. [DOI] [PubMed] [Google Scholar]
- 37.Davies JR, Randerson-Moor J, Kukalizch K, Harland M, Kumar R, Madhusudan S, Nagore E, Hansson J, Hoiom V, Ghiorzo P, Gruis NA, Kanetsky PA, et al. Inherited variants in the MC1R gene and survival from cutaneous melanoma: a BioGenoMEL study. Pigment Cell Melanoma Res. 2012;25:384–394. doi: 10.1111/j.1755-148X.2012.00982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Houwelingen HC, Arends LR, Stijnen T. Advanced methods in meta-analysis: multivariate approach and meta-regression. Stat Med. 2002;21:589–624. doi: 10.1002/sim.1040. [DOI] [PubMed] [Google Scholar]
- 39.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 40.D'Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, Ito S, Fisher DE. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 41.Smalley K, Eisen T. The involvement of p38 mitogen-activated protein kinase in the alpha-melanocyte stimulating hormone (alpha-MSH)-induced melanogenic and anti-proliferative effects in B16 murine melanoma cells. FEBS Lett. 2000;476:198–202. doi: 10.1016/s0014-5793(00)01726-9. [DOI] [PubMed] [Google Scholar]
- 42.Herraiz C, Journe F, Abdel-Malek Z, Ghanem G, Jimenez-Cervantes C, Garcia-Borron JC. Signaling from the human melanocortin 1 receptor to ERK1 and ERK2 mitogen-activated protein kinases involves transactivation of cKIT. Mol Endocrinol. 2006;25:138–156. doi: 10.1210/me.2010-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herraiz C, Sanchez-Laorden BL, Jimenez-Cervantes C, Garcia-Borron JC. N-glycosylation of the human melanocortin 1 receptor: occupancy of glycosylation sequons and functional role. Pigment Cell Melanoma Res. 2011;24:479–489. doi: 10.1111/j.1755-148X.2011.00848.x. [DOI] [PubMed] [Google Scholar]
- 44.Herraiz C, Journe F, Ghanem G, Jimenez-Cervantes C, Garcia-Borron JC. Functional status and relationships of melanocortin 1 receptor signaling to the cAMP and extracellular signal-regulated protein kinases 1 and 2 pathways in human melanoma cells. Int J Biochem Cell Biol. 2012;44:2244–2252. doi: 10.1016/j.biocel.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 45.Nakayama K, Soemantri A, Jin F, Dashnyam B, Ohtsuka R, Duanchang P, Isa MN, Settheetham-Ishida W, Harihara S, Ishida T. Identification of novel functional variants of the melanocortin 1 receptor gene originated from Asians. Hum Genet. 2006;119:322–330. doi: 10.1007/s00439-006-0141-1. [DOI] [PubMed] [Google Scholar]
- 46.Ibarrola-Villava M, Hu HH, Guedj M, Fernandez LP, Descamps V, Basset-Seguin N, Bagot M, Benssussan A, Saiag P, Fargnoli MC, Peris K, Aviles JA, et al. MC1R, SLC45A2 and TYR genetic variants involved in melanoma susceptibility in southern European populations: results from a meta-analysis. Eur J Cancer. 2012;48:2183–2191. doi: 10.1016/j.ejca.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 47.April CS, Barsh GS. Distinct pigmentary and melanocortin 1 receptor-dependent components of cutaneous defense against ultraviolet radiation. PLoS Genet. 2007;3:e9. doi: 10.1371/journal.pgen.0030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hauser JE, Kadekaro AL, Kavanagh RJ, Wakamatsu K, Terzieva S, Schwemberger S, Babcock G, Rao MB, Ito S, Abdel-Malek ZA. Melanin content and MC1R function independently affect UVR-induced DNA damage in cultured human melanocytes. Pigment Cell Res. 2006;19:303–314. doi: 10.1111/j.1600-0749.2006.00315.x. [DOI] [PubMed] [Google Scholar]
- 49.Wong SS, Ainger SA, Leonard JH, Sturm RA. MC1R variant allele effects on UVR-induced phosphorylation of p38, p53, and DDB2 repair protein responses in melanocytic cells in culture. J Invest Dermatol. 2012;132:1452–1461. doi: 10.1038/jid.2011.473. [DOI] [PubMed] [Google Scholar]
- 50.Wikberg JE, Muceniece R, Mandrika I, Prusis P, Lindblom J, Post C, Skottner A. New aspects on the melanocortins and their receptors. Pharmacol Res. 2000;42:393–420. doi: 10.1006/phrs.2000.0725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.