Abstract
Aberrant epigenetic reprogramming, especially genomic hypermethylation, is implicated as the primary reason behind the failure of the cloning process during somatic cell nuclear transfer (SCNT). We transfected one-cell-stage zona-free buffalo embryos produced by handmade cloning with 50 nM DNMT1 small interfering RNA (siRNA), using lipofectamine, to knockdown the DNA methyltransferase 1 (DNMT1) gene. siRNA treatment decreased (p<0.001) the expression level of DNMT1 mRNA and DNMT1 protein in the one-cell-stage embryos and increased (p<0.05) the blastocyst rate (52.3±1.3% vs. 45.3±2.5%) compared to that in the controls, but did not reduce the DNA methylation level similar to the in vitro–fertilized (IVF) embryos. It also increased (p<0.05) the relative mRNA abundance of P53 and CASPASE 3, but not that of HDAC1, DNMT1, and DNMT3a, in the blastocysts of the siRNA group compared to the controls. The global level of H3K18ac was higher (p<0.05) in the blastocysts of the siRNA group than in the controls, whereas that of H3K9ac and H3K27me3 was not significantly different between the two groups. In conclusion, lipofection can be successfully used for transfection of DNMT1 siRNA into one-cell-stage zona-free cloned buffalo embryos. It results in a concomitant decrease in the DNMT1 mRNA and protein levels in the one-cell-stage embryos. siRNA-mediated knockdown increases the blastocyst rate but does not alter the DNA methylation level.
Introduction
Compared to the more than 40% birth rate obtained with embryos produced by in vitro fertilization (IVF), at <5%, the rate of live offspring obtained from cloned blastocysts of most of mammalian species is very low (Campbell et al., 2007). This is believed to be due to incomplete or incorrect nuclear reprogramming of the donor somatic cell by the oocyte, resulting in very high mortality during pre- and postimplantation development in vitro and in vivo and a high incidence of abnormalities in the offspring born (Yang et al., 2007; Young et al., 1998). Following normal fertilization, the methylation patterns of the paternal and maternal genome are erased during preimplantation embryonic development, following which the embryos are methylated de novo by DNA methyltransferases (DNMTs) again to establish a new pattern (Dean et al., 2001). In contrast, during somatic cell nuclear transfer (SCNT), highly methylated somatic donor cells are used to generate cloned embryos, which, as a consequence, have higher levels of DNA methylation than embryos from natural reproduction, possibly due to incomplete erasure of pre-existing methylation in the donor cells (Niemann et al., 2008; Yang et al., 2007). The level of methylation of cloned embryos has been found to be much higher than in those produced through normal fertilization (Bourc'his et al., 2001). Lowering the methylation level of cloned embryos could, therefore, be expected to decrease abnormalities and improve cloning efficiency. Therefore, we attempted silencing of the DNMT1 gene by transfection of DNMT1 siRNA into one-cell-stage zona-free buffalo embryos produced by handmade cloning and investigated its effects on the yield, quality, DNA methylation, gene expression, and epigenetic status of cloned blastocysts.
Materials and Methods
All chemicals were purchased from Sigma Chemical Company, St. Louis, MO (USA); media were obtained from GIBCO, NY (USA), and the disposable plasticware was purchased from Nunc (Roskilde, Denmark), unless otherwise stated. In vitro culture of cells, oocytes, and embryos was carried out in a CO2 incubator (5% CO2 in air) at 38.5°C.
Production of cloned buffalo embryos
Buffalo embryos were produced by handmade cloning, and their quality was evaluated by determining the total cell number (TCN) and the level of apoptosis by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay, as described earlier (Selokar et al., 2012). Reconstructed embryos at the one-cell stage were divided randomly into two groups and were either treated with DNMT1-specific small interfering RNA (siRNA) or not subjected to this treatment, following which the embryos were cultured in a CO2 incubator for 8 days.
Transfection with siRNA
siRNA against the coding sequence of bovine DNMT1 mRNA (acc. no. NM_1826582.2) was designed and prepared commercially from Sigma Aldrich (Table S1; Supplementary Data are available at www.liebertpub.com/cell/). A BLAST search was performed at the National Center for Biotechnology Information (NCBI) against the DNMT1 gene to confirm the target specificity of designed siRNA. It was labeled with carboxyfluorescein (FAM) for determining the transfection efficiency. siRNA was diluted in 100 μL of nuclease-free water to make 100 μM stocks. For optimization of transfection conditions, zona-free one-cell-stage cloned embryos were incubated in 200 μL of Opti-MEM medium containing 50 nM siRNA with different concentrations of lipofectamine (1, 2, 4, or 8 μL) for different time intervals (1, 3, or 6 h) in four-well dishes. The transfection efficiency was examined by observing a fluorescence signal under an epifluorescence microscope and examination of the lysis rate. The effects of siRNA were examined both at the mRNA and protein levels. For the former, total RNA was isolated from one-cell-stage embryos (n=10–15) using RNAqueous-Micro Kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions. Real-time RT-PCR was performed using the optimized primer DNMT1 shown in Table S1 on a CFX96 real-time system (Bio-Rad, Hercules, CA, USA) with Maxima SYBR Green Master Mix (Fermentas, St. Leon-Rot, Germany) at the following thermal cycling conditions: 95°C for 5 min, followed by 40 PCR cycles of 95°C for 15 sec, 58°C for 30 sec, and 72°C for 30 sec, as described earlier (Selokar et al., 2014), using β-actin as the housekeeping gene. Melting peaks were determined using melting curve analysis to ensure the specific amplification. For comparison, the δδCT method was used. The average expression level of each gene from control embryo was set as 1, and three separate experiments with three replicates were performed.
For protein level, a commercially available enzyme-linked immunosorbent assay (ELISA)-based kit (EpiQuik™, catalog no. P-3011-96, Epigentek) was used according to the manufacturer's protocol to determine the DNMT1 enzyme level. Briefly, total nuclear extract isolated from one-cell-stage embryos (n=200), according to the method provided in the Nuclear Extraction Kit (catalog no. OP-0002-1, Epigentek), was diluted in 100 μL of dilution buffer, loaded into each well of an eight-well strip, and incubated for 120 min at 37°C. The samples were blocked with 150 μL of blocking solution and incubated with the capture (1:1200) and detection (1:1200) antibody for 60 and 30 min, respectively. Addition of 100 μL of the developing solution to each well led to the development of blue color, which turned to yellow after addition of the stop solution. The absorbance was read immediately at 450 nm. The experiment had three replicates.
Analysis of epigenetic markers and gene expression in cloned blastocysts
The global levels of H3K9/14ac, H3K18ac, and H3K27me3 were determined by immunofluorescence staining in cloned blastocysts produced from siRNA-treated and nontreated control one-cell-stage embryos, as described earlier (Selokar et al., 2014). Briefly, the blastocysts were fixed in 4% paraformaldehyde for 1 h at 37°C, washed three times with Dulbecco's phosphate-buffered saline (DPBS), and then permeabilized in 0.5% Triton X-100 for 20 min at 37°C. The cells were blocked for 1 h in 3% bovine serum albumin (BSA) and then incubated overnight at 4°C with the respective rabbit primary antibody. Primary antibodies, which included anti-H3K9/14ac (1:1000, Santa Cruz Biotechnology, CA, USA), anti-H3K18ac (1:1500, Millipore, MA, USA), and anti-H3K27me3 (1:1500, Millipore), were diluted in 3% BSA. After washing five times with DPBS containing 0.1% Triton X-100 and 0.3% polyvinyl alcohol (PVA) (DPBSTP), the cells were incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit secondary antibody diluted 1:700 in DPBS. After five washings with DPBSTP, the nuclei were counterstained with H33342 (10 μg/mL) and rinsed in DPBSTP. The blastocysts were then mounted on slides in mounting medium [2.5% 1,4-diazabicyclo[2.2.2]octane (DABCO) in glycerol]. The slides were observed under a fluorescence microscope, and the images were captured keeping the same optical conditions. For DNA methylation analysis, all steps were similar as described above except that 4 M hydrochloric acid treatment was given before incubation with mouse anti-5-methyl cytosine primary antibody (NA81, Calbiochem).
NIS-element basic research image processing software (Nikon, Tokyo, Japan) supplied with the microscope was used for image acquisition and quantitative measurements of the mean pixel intensity emitted by each individual nucleus. At least 15 blastocyst images (50 nuclei from each image) were analyzed for each epigenetic marker. In case of gene expression analysis, the relative mRNA abundance of HDAC1, DNMT1, DNMT3a, P53, and CASPASE3 was determined in blastocysts by qPCR as described above using β-actin as the housekeeping gene (Table S1). Three separate experiments were performed with three replicates for each gene.
Statistical analysis
Statistical analysis was performed using SYSTAT 12 (Systat Software, Inc. Chicago, IL, USA) software. Percentage values were analyzed after arcsine transformation. Student's t-test or Chi Square (×2) test were used to compare the means of different groups. The differences were considered to be significant at p<0.05.
Results and Discussion
In the present study, the DNMT1 gene was silenced by transfection of DNMT1 siRNA into one-cell-stage zona-free buffalo embryos produced by handmade cloning. This was found to result in a concomitant decrease in the DNMT1 mRNA and protein level in the one-cell-stage embryos. siRNA-mediated knockdown increased the blastocyst rate, but did not reduce DNA methylation level.
Because hypermethylation of the genome is a major epigenetic aberration associated with SCNT, artificial rebuilding of the abnormal methylation status could help minimize abnormalities and improve cloning efficiency. Many different approaches have been attempted for reducing DNA methylation in cloned embryos. These include treatment with inhibitors such as 5-aza-2′-deoxycytidine, a DNMT inhibitor that reduces DNA methylation in cloned embryos (Enright et al., 2005). However, such inhibitors may also target pathways other than those at which it is aimed; therefore, it is sometimes not clear whether the outcome observed was the result of an off-target effect. Complete inhibition of DNMT1 by gene knockout has been shown to be lethal during embryonic development (Li et al., 1992). Silencing of the DNMT1 gene by siRNA, which offers specificity comparable to that of gene knockout and is much less time-consuming and expensive, has recently been applied to reduce DNMT1 expression in cloned bovine embryos (Giraldo et al., 2009; Yamanaka et al., 2011). However, Giraldo et al. (2009) carried out gene silencing by transfection of donor fibroblast cells, which left 20–30% cells nontransfected. In such an approach, the isolation of transfected cells, which is very difficult, becomes necessary to ensure that cloned embryos are not produced from nontransfected donor cells. In an improvement over this method, Yamanaka et al. (2011) transferred siRNA to one-cell-stage bovine embryos. However, these authors used microinjection, which has several disadvantages, such as need for a high level of skill and expenses associated with the use of a micromanipulator. Recently, lipid-based transfection was reported to be more efficient than microinjection for transfer of siRNA into bovine zygotes (O'Meara et al., 2011). To date, there are no reports regarding the transfection of siRNA into zona-free cloned embryos. Hence, we used lipofection to transfer DNMT1 siRNA to one-cell-stage buffalo embryos.
Visual examination of the transfected zona-free cloned embryos revealed that the fluorescence level was similar following use of 2, 4, or 8 μL of lipofectamine for transfection. Following transfection of cloned embryos with siRNA, using 2 μL of lipofectamine for 1, 3, or 6 h, the lysis rate was significantly lower (p<0.001) at 1 h than that at 2 h, which, in turn, was lower (p<0.001) than that at 6h (Fig. 1). Thus, 2 μL of lipofectamine in 200 μL of Opti-MEM medium containing 50 nM siRNA with 1 h incubation time could transfer siRNA effectively with minimal lysis of embryos. Following qPCR, the relative mRNA abundance of DNMT1 was found to be significantly lower (p<0.001) in siRNA-treated one-cell-stage cloned embryos compared to that in controls (Fig. 1). siRNA treatment also significantly decreased (p<0.001) the DNMT1 protein level. This concomitant highly significant decrease in the mRNA and protein levels of the DNMT1 gene confirmed the efficacy of siRNA in silencing it in the treatment group. The effect of siRNA knockdown was transient and reversible since, unlike in one-cell-stage embryos, the relative mRNA abundance of DNMT1 was similar in the blastocysts of the treatment and control groups (see Fig. 3, below). This agrees with the report of Yamanaka et al. (2011), who observed that following microinjection of DNMT1 siRNA into one-cell-stage bovine embryos, the knockdown of DNMT1 was effective for only 72 h as indicated by DNMT1 mRNA and protein.
FIG. 1.
(A) Visual appearance of zona-free one-cell-stage handmade cloned buffalo embryos transfected with 50 nM DNMT1 siRNA using lipofectamine. (B) Lysis rate following transfection using 2 μL of lipofectamine. Relative abundance of DNMT1 mRNA (C) and DNMT1 protein (D) in siRNA-treated one-cell-stage cloned embryos. Bars with different superscripts differ significantly (p<0.001). UV, ultraviolet.
FIG. 3.
Relative mRNA abundance of some important genes in the blastocysts produced from one-cell-stage cloned buffalo embryos that had been transfected with 50 nM DNMT1 siRNA. Bars with different superscripts differ significantly (p<0.05).
siRNA treatment of one-cell-stage cloned buffalo embryos significantly increased (p<0.05) the blastocyst rate compared to that in the controls, but the cleavage rate and TCN were not significantly different between the two groups (Table 1). This agrees with the results of Yamanaka et al. (2011), who reported improved cleavage and blastocyst rates but not a significant effect on TCN and trophectoderm-to-inner cell mass ratio in blastocysts produced from DNMT1 siRNA-treated one-cell-stage bovine embryos. In contrast, Giraldo et al. (2009) did not observe any effect on developmental competence of SCNT bovine embryos following use of fibroblasts transfected with DNMT1 siRNA. This discrepancy could be due to the use of a substantial population of nontransfected donor cells for cloning because the transfection efficiency in their study was only 70%. We found that despite a significant increase (p<0.05) in the relative mRNA abundance of CASPASE 3 and P53 in the blastocysts of the siRNA group compared to that of the control group, the apoptotic index was not significantly different between the blastocysts of the two groups. Caspase-3 is a member of group II caspases, which are responsible for destruction of structural and regulatory proteins and lead to DNA damage and cell demise (Chang and Yang, 2000). However, because we have not studied the expression levels of many other important pro-apoptotic and anti-apoptotic genes and microRNAs, which form a complex network leading to apoptosis, it is difficult to interpret these results.
Table 1.
Effect of Treatment of One-Cell-Stage Cloned Buffalo Embryos with DNMT1 siRNA on Their Developmental Competence and Quality
| Treatment | Reconstructed embryos | Cleaved n (%) | Blastocysts n (%) | Blastocysts examined | Total cell number | Apoptotic index |
|---|---|---|---|---|---|---|
| Control | 303 | 292 (96.3±1.0) | 132 (44.2±1.8)a | 35 | 197.6±13.5 | 3.3±0.8 |
| siRNA (50 nM) | 287 | 278 (96.6±1.0) | 150 (51.7±1.6)b | 35 | 210.9±13.2 | 3.0±0.0 |
| IVF | 470 | 235 (55.7±8.5) | 60 (14.5±3.16)c | 12 | 215.4±17.3 | 3.3±0.6 |
Data from 15 trials. Values are mean±standard error of the mean (SEM).
Values with different superscripts within the same column differ significantly (p<0.05).
siRNA, small interfering RNA; IVF, in vitro fertilization.
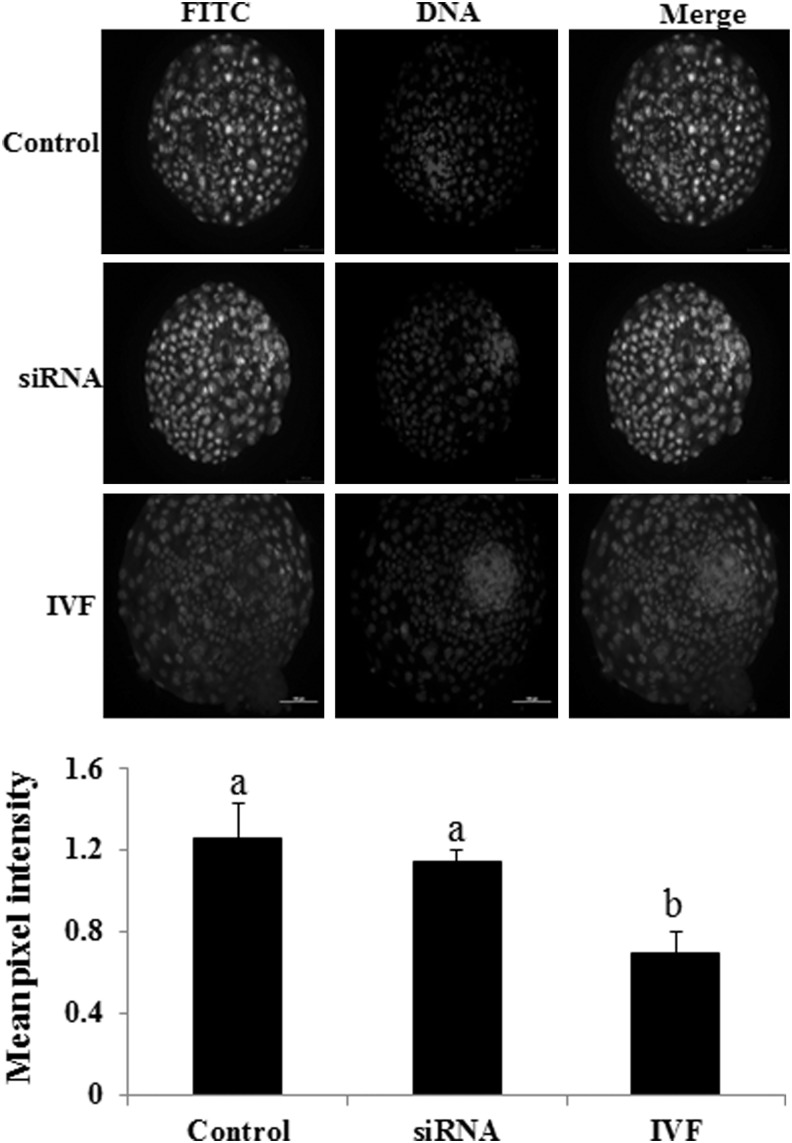
We found that the DNA methylation level was similar among cloned blastocysts produced from control and DNMT1 siRNA-treated one-cell-stage embryos, which was significantly higher than that of IVF blastocysts (Fig. 2). In contrast, Giraldo et al. (2009) reported reduction of methylation in cloned blastocysts produced from fibroblasts transfected with DNMT1 siRNA than that of nontransfected cells. This might be due to the use of a pre-demethylated donor genome before SCNT by Giraldo et al. (2009). In addition, the incompletely reprogrammed cloned embryos may re-establish the somatic cell level of DNA methylation in the absence of effective concentrations of siRNA, as we have used single dose of siRNA.
FIG. 2.
DNA methylation in IVF and cloned blastocysts produced from one-cell-stage cloned buffalo embryos that had been transfected with 50 nM DNMT1 siRNA. Bars with different superscripts differ significantly (p<0.05).
We found that the relative mRNA abundance of DNMT1, DNMT3a, and HDAC1 was not significantly different between the blastocysts of DNMT1 siRNA-treated and control groups (Fig. 3). However, further studies are required to establish the absence of any effect of DNMT1 siRNA treatment on these genes because in our study and in other studies (Yamanaka et al., 2011) siRNA treatment, which was given to one-cell-stage embryos, was of a transient nature, and the possibility of its effect having worn off at the blastocyst stage cannot be ruled out.
Recent studies suggest that the primary mechanism through which DNMT1 siRNA elicits its effects is through demethylation of the satellite I region (Yamanaka et al., 2011). We found that siRNA treatment increased (p<0.05) the global level of H3K18ac but did not affect that of H3K9ac and H3K27me3 in blastocysts (Fig. 4). This suggests that DNMT1 siRNA treatment may also alter histone acetylation, pointing to a link between DNA methylation and histone acetylation. Previous studies in which trichostatin A, a histone deacetylase inhibitor, was found to decrease levels of DNA methylation in the satellite 1 region (Wang et al., 2011; Wee et al., 2007) also suggest such a link. However, further studies are needed to unravel its mechanism.
FIG. 4.
Global level of H3K9ac, H3K18ac, and H3K27me3 in the blastocysts produced from one-cell-stage cloned buffalo embryos that had been transfected with 50 nM DNMT1 siRNA. Bars with different superscripts differ significantly (p<0.05).
In conclusion, these results suggest that lipofection can be used successfully for transfection of DNMT1 siRNA into one-cell-stage zona-free buffalo embryos produced by handmade cloning. This results in a concomitant decrease in the DNMT1 mRNA and protein levels in the one-cell-stage embryos. The siRNA-mediated knockdown increased the blastocyst rate, but did not alter methylation level in the blastocysts.
Supplementary Material
Acknowledgment
The present work was funded by National Agriculture Innovative Project (NAIP) grant to S.K.S. (C 2-1-(5)/2007) and M.S.C. (C-2067 and 075). Naresh Selokar and Monika Saini are recipients of CSIR-SRF fellowship.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Bourc'his D., Le Bourhis D., Patin D., Niveleau A., Comizzoli P., Renard J.P., and Viegas-Pequignot E. (2001). Delayed and incomplete reprogramming of chromosome methylation patterns in bovine cloned embryos. Curr. Biol. 11, 1542–1546 [DOI] [PubMed] [Google Scholar]
- Campbell K.H.S., Fisher P., Chen W.C., Choi I., Kelly R.D., Lee J.H., and Xhu J. (2007). Somatic cell nuclear transfer: Past, present and future perspectives. Theriogenology 68, S214–S231 [DOI] [PubMed] [Google Scholar]
- Chang H.Y., and Yang X. (2000). Proteases for cell suicide: Functions and regulation of caspases. Microbiol. Mol. Biol. Rev. 64, 821–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W., Santos F., Stojkovic M., Zakhartchenko V., Walter J., Wolf E., and Reik W. (2001). Conservation of methylation reprogramming in mammalian development: Aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. USA 98, 13734–13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright B.P., Sung L.Y., Chang C.C., Yang X., and Tian X.C. (2005). Methylation and acetylation characteristics of cloned bovine embryos from donor cells treated with 5-aza-20-deoxycytidine. Biol. Reprod. 72, 944–948 [DOI] [PubMed] [Google Scholar]
- Giraldo A.M., Lynn J.W., Purpera M.N., Vaught T.D., Ayares D.L., Godke R.A., and Bondioli K.R. (2009). Inhibition of DNA methyltransferase 1 expression in bovine fibroblast cells used for nuclear transfer. Reprod. Fertil. Dev. 21, 785–795 [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T.H., and Jaenisch R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 [DOI] [PubMed] [Google Scholar]
- Niemann H., Tian X.C., King W.A., and Lee R.S. (2008). Epigenetic reprogramming in embryonic and foetal development upon somatic cell nuclear transfer cloning. Reproduction 135, 151–163 [DOI] [PubMed] [Google Scholar]
- O'Meara C.M., Murray J.D., Mamo S., Gallagher E., Roche J., and Lonergan P. (2011). Gene silencing in bovine zygotes: siRNA transfection versus microinjection. Reprod. Fertil. Dev. 23, 534–543 [DOI] [PubMed] [Google Scholar]
- Selokar N.L., Saini M., Muzaffer M., Krishnakanth G., Saha A.P., Chauhan M.S., Manik R., Palta P., Madan P., and Singla S.K. (2012). Roscovitine treatment improves synchronization of donor cell cycle in G0/G1 stage and in vitro development of Handmade cloned buffalo (Bubalus bubalis) embryos. Cell. Reprogram. 14, 146–154 [DOI] [PubMed] [Google Scholar]
- Selokar N.L., Saini M., Palta P., Chauhan M.S., Manik R., and Singla S.K. (2014). Hope for restoration of dead valuable bulls through cloning using donor somatic cells isolated from cryopreserved semen. PLoS One 9, e90755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Su J., Wang L., Xu W., Quan F., Liu J., and Zhang Y. (2011). The effects of 5-aza-2′-deoxycytidine and trichostatin A on gene expression and DNA methylation status in cloned bovine blastocysts. Cell. Reprogram. 13, 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee G., Shim J.J., Koo D.B., Chae J.I., Lee K.K., and Han Y.M. (2007). Epigenetic alteration of the donor cells does not recapitulate the reprogramming of DNA methylation in cloned embryos. Reproduction 134, 781–787 [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Sakatani M., Kubota K., Balboula A.Z., Sawai K., and Takahashi M. (2011). Effects of down regulating DNA methyltransferase 1 transcript by RNA interference on DNA methylation status of the satellite I region and in vitro development of bovine somatic cell nuclear transfer embryos. J. Reprod. Dev. 57, 393–402 [DOI] [PubMed] [Google Scholar]
- Yang X.Z., Smith S.L., Tian XC, Lewin HA, Renard JP, and Wakayama T. (2007). Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat. Genet. 39, 295–302 [DOI] [PubMed] [Google Scholar]
- Young L.E., Sinclair K.D., and Wilmut I. (1998). Large offspring syndrome in cattle and sheep. Rev. Reprod. 3, 155–163 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.