Abstract
In the present study we report on interactions of and competition between monovalent ions for two DNA sequences in MD simulations. Efforts included the development and validation of parameters for interactions among the first-group monovalent cations, Li+, Na+, K+ and Rb+, and DNA in the Drude polarizable and additive CHARMM36 force fields (FF). The optimization process targeted gas-phase QM interaction energies of various model compounds with ions and osmotic pressures of bulk electrolyte solutions of chemically relevant ions. The optimized ionic parameters are validated against counterion condensation theory and buffer exchange-atomic emission spectroscopy measurements providing quantitative data on the competitive association of different monovalent ions with DNA. Comparison between experimental and MD simulation results demonstrates that, compared to the additive CHARMM36 model, the Drude FF provides an improved description of the general features of the ionic atmosphere around DNA and leads to closer agreement with experiment on the ionic competition within the ion atmosphere. Results indicate the importance of extended simulation systems on the order of 25 Å beyond the DNA surface to obtain proper convergence of ion distributions.
INTRODUCTION
Mobile ions are known to regulate conformational behavior and functional dynamics of nucleic acids.1–2 For example, on the level of several nucleotides cations affect the local hydrogen bond network in DNA grooves, leading to significant local deviations of the DNA geometry from the canonical form, one of the proposed mechanisms regulating sequence-specific protein-DNA recognition.3 On a larger scale, counterions form a condensed layer around polyanionic DNA or RNA molecule (ionic “atmosphere”)4–5 which mitigates strong electrostatic repulsion between electronegative phosphate groups within the macromolecule, or between different macromolecules, to enable such vital biological processes as genomic packaging6 and RNA folding.7 From a physical viewpoint, ions regulate a number of critical polymeric large-scale properties of the nucleic acids, including persistence length and stiffness.2, 8–10
DNA under physiological conditions is exposed to a mixture of several (mono- and divalent) ionic species. Various experimental studies based on X-ray crystallography11–14 and solution NMR techniques15–17 have addressed sequence-specific details of the ion-DNA interactions and demonstrated that different ions vary in their propensity to reside in DNA grooves, or near certain DNA electronegative sites. At the same time, because of inherent limitations of these techniques associated with problems in distinguishing biologically relevant ions (such as Na+, K+, Mg+), there were disagreements in the interpretation of experimental results on the competition of the ions for the grooves of DNA.11, 18 Our recent molecular dynamics (MD) simulation study utilizing the first generation Drude polarizable force field for DNA19 has revealed differential modulation of the minor groove by different monovalent ionic species.20 In particular, the width of the minor groove strongly correlates with the size of the ion according to the following trend, Li+ < Na+ < K+ < Rb+.20 These results indicate that competition may occur among the first-group monovalent cations for the DNA minor groove.
Experiments focusing on macroscopic DNA properties in various ionic buffers, including measurements of DNA electrophoretic mobility21–22 and compaction of the long DNA chains monitored by fluorescent microscopy,23 have demonstrated that monovalent cations indeed have differential effects. However, these experiments do not provide a comprehensive picture of the ionic atmosphere and, more importantly, quantitative details on the competitiveness of different ions with respect to their propensity to neutralize DNA residual charge. Experimental techniques addressing competitive ion-DNA interactions have became available only recently. One such technique is anomalous small-angle X-ray scattering (ASAXS) enabling some general features of the ionic atmosphere around DNA, such as the number of ions in the atmosphere to be characterized.24–26 However, this technique possesses limitations in differentiating among similar cationic species (e.g., Li+, Na+ or K+) because of their low electron density.4 In contrast, a novel experimental approach, buffer equilibration–atomic emission spectroscopy (BE-AES), enables an accurate determination of the relative extent to which various ions occupy the atmosphere around DNA immersed in a mixture of two competing cations (e.g., Li+ and Na+) and one anion (Cl−).4 This information can be readily used for benchmarking or refinement of the computational models utilized in MD simulations. In the present study, we use BE-AES data to test optimized interaction parameters between DNA and the first-group monovalent cations, Li+, Na+, K+ and Rb+, in the all-atom polarizable force field based on the classical Drude oscillator formalism.19 Additionally, these data are used to test the ability of the CHARMM36 (C36) additive force field for DNA27 to capture competition of these monovalent cations for the ionic atmosphere around DNA.
The concept of ionic atmosphere, a region of enhanced cationic concentration around highly charged polyanions, originated from the counterion condensation (CC) theory by G. Manning.5 Despite inherent limitations of the theory associated with the continuum model for solvent and the mean-field description of the ionic distribution, its predictions for the size of the atmosphere (~9 Å), concentration of ions (~1M) and the fraction of the neutralized polyelectrolyte charge by the ions (~76%) within the atmosphere28 are widely used for assessment of the ionic distribution around DNA from MD simulations. For example, estimates from the commonly used AMBER and CHARMM additive (non-polarizable) models for nucleic acids are consistent with predictions from CC theory,29–30 with results from our recently developed Drude polarizable model for DNA being in even closer agreement with CC theory.31
Another notion of the ionic atmosphere based on the principles of electroneutrality emerged recently from BE-AES experiments.4 In particular, ionic atmosphere is considered to be a region around DNA where densities of both cations and anions deviate from their bulk values, with the number of excess cations plus the number of depleted anions (total charge of the atmosphere) completely neutralizing the charge of the DNA molecule. A recent MD simulation study based on the additive CHARMM27 force field for DNA32 and refined ionic parameters33 has demonstrated that the condition of electroneutrality required an ionic atmosphere ranging from 30 to 50 Å from the DNA helical axis.34 This means that for direct comparison with BE-AES experiment much larger DNA systems (~10 times bigger in terms of the number of atoms) need to be simulated compared to those typically used to study sequence-specific details of ionic binding, or related local phenomena.
Our recently developed polarizable model for DNA based on the classical Drude oscillator formalism was optimized to accurately reproduce a number of critically important conformational features in aqueous salt solution, including BI/BII transitions and the equilibrium between A and B forms of DNA.19 Achieving a proper balance of interactions among surrounding mobile ions, water and DNA molecule, one of the critical aspects of the entire parametrization process, was shown to stabilize the overall DNA structure and regulate the above mentioned subtler conformational phenomena.31 This was obtained utilizing gas-phase QM calculations on small model compound-metal ion interactions, the impact of the ions on DNA stability and hydration and osmotic properties of the relevant electrolyte solutions. In those studies19, 31 we used NaCl as the ionic buffer, one of the most commonly used buffers in experimental and computational studies. However, as mentioned above, physiological DNA solutions are composed of more than a single salt, with different ions competing for the environment around DNA, thereby modulating DNA structure and neutralizing of DNA’s residual charge. Therefore, to enable modelling of more realistic DNA systems, such as those involving mixed ionic solutions, our parametrization efforts need to be extended to cover more ionic species.
In the present study, we further optimize Drude polarizable parameters for the Li+, K+ and Rb+ ions35–36 for simulations of DNA in aqueous salt solutions. We test the optimized parameters against a range of experimental data and theoretical estimations, with one of the most critical comparisons made between MD simulations and BE-AES experiments on competition among Li+, Na+, K+ and Rb+ ions to occupy DNA’s ionic atmosphere.4 Analogous validations and, when necessary, adjustments of the interaction parameters between ions and DNA are carried out for the non-polarizable C36 force field. Overall, the Drude model is shown to perform better than the additive C36 model indicating the importance of inclusion of polarization effects to model such complex biological systems as polyanionic DNA in aqueous salt environments of various chemical contents. Our current efforts are based on the “default” ionic parameters developed by Roux and co-workers for the Drude polarizable model35–36 and the additive ionic parameters distributed with the C36 force field. As discussed below, the final ionic parameters presented here are identical to those utilized in our latest application study on differential impact of monovalent ions on DNA conformational properties.20
COMPUTATIONAL METHODS
Polarizable Force Field Based on Classical Drude Oscillators
The Drude oscillator model is used to represent electronic induction by introducing a massless charged particle attached to a polarizable atom by a harmonic spring.37 Charge redistribution in response to the change in the local electrostatic field is approximated by updating self-consistently the positions of these auxiliary particles to their local energy minima for any given configuration of the atoms in the system, thereby taking into account the permanent electric field due to the fixed charges and the contribution of the induced dipoles to the electric field.38 In the context of MD simulations, this self-consistent field (SCF) approach is substituted with an extended Lagrangian integration, which treats electronic degrees of freedom as additional classical dynamic variables.39–40 This allows for computational efficiency required for long-time MD simulations.
QM and MM Calculations
The energetics of ion-DNA interactions were studied using gas-phase QM calculations on model systems involving the ion of interest with individual nucleic acid bases or the dimethylphosphate (DMP) anion, a representative of the phosphate group in the DNA backbone. The ion (Li+, Na+, K+ or Rb+) was positioned at distances ranging from 1 Å to 10 Å from the chemically relevant atoms at various orientations. As previously performed for Na+ ions,31 interactions of Li+, K+ or Rb+ with hydrogen bond acceptors and some other atoms participating in Watson-Crick base-pairing, namely, N1 of adenine, N3 of cytosine, N1 of guanine and N3 of thymine were studied. Analogous calculations were carried out for the DMP−/Na+ dimers, probing interactions among the ions and phosphate group. The distance increments were 0.1 Å in the range of 1 to 4 Å, and 1 Å at larger separations. QM potential energy profiles of Li+, Na+ and K+ were based on structures optimized at MP2/6–31++G(2df, 2pd) model chemistry using Gaussian 0941 with counterpoise corrections42 applied to account for basis set superposition error (BSSE).43 For systems involving the heavier Rb+ ion, the def2-TZVPPD basis set including effective core potentials (ECP) for the core electrons was used.44–45 This basis set has been recently used to examine interactions of Rb+ with amino acids.46 The def2-TZVPPD is a balanced basis set on all atoms at the triple-ζ-valence level including polarization and diffusion functions and uses ECPs on rubidium developed by Bergner et al.47 As this basis set is not included in Gaussian distribution, it was obtained from the EMSL basis set exchange library.48–49
Equivalent single point calculations were performed on QM geometries using the CHARMM program50 and the Drude polarizable or additive C36 force fields. In case of the Drude systems, before calculation of the potential energy scans the models were minimized by self-consistently relaxing positions of the Drude particles in the local electric field using a combination of the steepest descent and ABNR minimization algorithms, while all other atoms were restrained (K=10000 kcal/mol·Å2). Interaction energies were then calculated based on the total dimer energies minus the sum of the self-consistently relaxed monomers.
Osmotic Pressure Calculations
Detailed description of the osmotic pressure calculations is provided in previous studies.31,33, 36, 51 Briefly, an aqueous electrolyte solution is partitioned into three compartments separated by two virtual semipermeable membranes aligned with the xy-plane, with the electrolyte compartment (ions plus water) being centered on the absolute origin of the coordinates and enveloped by compartments containing pure water. While water can freely pass through the membranes separating the electrolyte and water solutions, the ions are subject to the force of the half-harmonic planar potential applied to the membrane on each side of the ionic compartment (see Fig. S1, Supporting Information, SI). As a result, ions produce instantaneous osmotic pressure, which equals the instantaneous total force, Fmemb, they impose on the membranes divided by the total area of the membranes, A:
| (1) |
Terms in the above expression are the force constant of the membrane potential, k, set to be 10 kcal/(mol·Å) in our MD simulations, the linear size of the cubic ionic compartment, D=48 Å, and the z-coordinate of the i’s ion, zi.
In the present study we simulated aqueous electrolyte solutions comprised of the cation of interest (i.e. Li+, Na+, K+ or Rb+) and (1) Cl− or (2) DMP − anion. Chloride solutions were simulated at two concentrations, ~1M and ~3M, while solutions including DMP− anion were studied at low concentrations only (~1M) as experimental osmotic pressures are only available at that concentration. The MD simulation protocol is elaborated below.
Direct comparison of MD simulation results with experimental osmotic pressure measurements requires knowledge of the molal concentration of the electrolyte, m (in contrast with the traditionally used molar concentration, M), which needs to be computed from MD simulations as follows,
| (2) |
The above expressions operate with the molar concentration of the salt, Msalt, and the values of mean densities of the cations, anions and water in the ionic compartment,ρ̅cation, ρ̅anion and ρ̅water, respectively, computed from ensemble averages. The resulting molal concentration is then used to obtain the osmotic pressure of an ideal solution,
| (3) |
which, in turn, is used to determine experimental value of the osmotic pressure based on knowledge of the osmotic pressure coefficient at the same concentration m,
| (4) |
In Eq. (3), R is the gas constant, T is temperature in Kelvin, Vm is the molar volume of pure water at 1 bar and room temperature, and ν is the total number of cations and anions per formula unit (in our case of 1/1 electrolyte solutions ν=2).52
MD Simulation Protocols
Simulated DNA systems included the EcoRI dodecamer and the 10-base-pair 1DCV structure in various ionic aqueous buffers (see Table 1). One set of calculations was carried out with ionic buffers composed of a single salt, (1) LiCl, (2) NaCl, (3) KCl and (4) RbCl, while additional simulations were conducted with buffers representing a mixture of two competing cations and Cl− anion: (1) Na+, Li+ and Cl−; (2) Na+, K+ and Cl− and (3) Na+, Rb+ and Cl−. The mixed systems were, in turn, studied at two different DNA and ionic concentrations. In particular, the first set of (mixed) simulations included 1DCV and EcoRI oligomers solvated in cubic boxes of l~60 Å with addition of ~100 mM of the chloride salt containing equal numbers of the competing cations (1:1 mixture, e.g., ~50mM of Na+ and 50 mM of Li+). Another set of simulations involved 1DCV solvated cubic box of l~120 Å with ionic concentrations chosen to match those used in BE-AES experiments.4 In what follows, we refer to these mixed sets as small and large DNA systems, respectively.
Table 1.
DNA systems simulated in this studya
| System | Ionic Composition | Number of Ions | Bulk Ionic Concentration (mM) |
|||||
|---|---|---|---|---|---|---|---|---|
| BC | CI | anion | BC | CI | Cl− | BC | CI | |
|
Small systems (l~60 Å): |
||||||||
| 1DCV (single salt) | Li+ | Cl− | 28 | 10 | ~110 | |||
| Na+ | Cl− | 28 | 10 | ~110 | ||||
| K+ | Cl− | 28 | 10 | ~110 | ||||
| Rb+ | Cl− | 28 | 10 | ~110 | ||||
| EcoRI (single salt) | Li+ | Cl− | 36 | 14 | ~130 | |||
| Na+ | Cl− | 36 | 14 | ~130 | ||||
| K+ | Cl− | 36 | 14 | ~130 | ||||
| Rb+ | Cl− | 36 | 14 | ~130 | ||||
| 1DCV (mixed salt) | Na+ | Li+ | Cl− | 14 | 14 | 10 | ~55 | ~55 |
| Na+ | K+ | Cl− | 14 | 14 | 10 | ~55 | ~55 | |
| Na+ | Rb+ | Cl− | 14 | 14 | 10 | ~55 | ~55 | |
| EcoRI (mixed salt) | Na+ | Li+ | Cl− | 18 | 18 | 14 | ~65 | ~65 |
| Na+ | K+ | Cl− | 18 | 18 | 14 | ~65 | ~65 | |
| Na+ | Rb+ | Cl− | 18 | 18 | 14 | ~65 | ~65 | |
|
Large systems (l~120 Å): |
||||||||
| 1DCV (single salt) | Na+ | Cl− | 54 | 36 | ~50 | |||
| 1DCV (mixed salt) | Na+ | Li+ | Cl− | 54 | 38 | 74 | ~50 | ~36 |
| Na+ | K+ | Cl− | 54 | 52 | 88 | ~50 | ~51 | |
| Na+ | Rb+ | Cl− | 54 | 62 | 98 | ~50 | ~60 | |
BC and CI stand for backround cation and competing ion, respectively.
Initial configurations for all polarizable and non-polarizable DNA systems were generated using the additive C36 force field. Each duplex in the canonical B form conformation was solvated in a cubic box of size l~60 Å or l~120 Å with the additive TIP3P53 water model, neutralizing ions and the corresponding chloride salt (Table 1) and then simulated for several nanoseconds, as described elsewhere.27 Briefly, NPT ensembles at 298 K and 1 atm were simulated with periodic boundary conditions, with the particle mesh Ewald (PME) method54 used to treat electrostatic interactions, and with the SETTLE algorithm55 used to constrain covalent bonds involving hydrogen atoms enabling the use of the 2 fs time step. The last snapshots from these short additive runs were taken as inputs for subsequent Drude polarizable MD simulations, while production runs of all additive simulations were continued for another 150 ns. NAMD56 (v. 2.9) was used for MD simulations and the CHARMM-GUI57 was utilized for the additive system setup.
All polarizable systems were solvated with the SWM4-NDP58 water model; the number of water molecules, as well as ions, were identical to those in the additive systems. Polarizable systems were generated using the CHARMM utility 'GENERATE DRUDE' that automatically adds the Drude particles and lone pairs to the polarizable (non-hydrogen) atoms. A subsequent self-consistent relaxation of the Drude positions in the electric field was carried out using a combination of the steepest descent and ABNR minimization algorithms while all non-hydrogen atoms were restrained (K=10000 kcal/mol·Å2). For each system a short equilibration of the solvent and mobile ions was then performed by running a 500 ps MD simulation with all DNA atoms (but not the Drude particles) restrained (K=1000 kcal/mol·Å2), followed by an additional 1 ns equilibration of the entire system without any restraints, after which the production MD simulations were initiated. All equilibration runs were conducted with the CHARMM program, utilizing the Velocity-Verlet integrator (VV2)39 in conjunction with the TPCONTROL (Temperature-Pressure Control) command, allowing for efficient simulation of the motion of Drude oscillators. In particular, a Nose-Hoover thermostat was applied to all real atoms to control the global system temperature at 300K, and a separate low-temperature thermostat at 1K was applied to Drude particles to ensure that their time course approximates the self-consistent field (SCF) regimen.39 A constant pressure (1 atm) was maintained via a modified Andersen-Hoover barostat, and SHAKE59 was used to constrain covalent bonds involving hydrogens. Electrostatic interactions were treated using PME summation54 with a coupling parameter of 0.34 and a sixth-order spline for mesh interpolation. Non-bonded pair lists were maintained out to 16 Å, and a real space cutoff of 12 Å was used for the electrostatic and Lennard-Jones (LJ) terms, with a long-range correction applied to LJ interactions.60 The ‘HARDWALL’ feature of the CHARMM program enabled use of a 1 fs time step in MD simulations.61 This feature is associated with a “hard wall” reflective term in the potential energy function that has been added to resolve the possibility of polarization catastrophe in Drude MD simulations. This term was invoked only when Drude particles moved >0.2 Å away from their parent nuclei during MD simulations. An analogous simulation protocol was used in all production MD runs with NAMD where an alternate dual thermostating approach is applied based on Langevin dynamics.40 The SETLLE algorithm55 was used to constrain covalent bonds involving hydrogens. The longevity of all production runs was identical to those of the additive systems, 150 ns.
MD simulation protocol for studying electrolyte solutions is analogous to the above protocol for simulating DNA duplexes. A few differences are represented by the use of ‘useFlexibleCel’ and ‘useConstantArea’ options of NAMD. The first option allows for the three dimensions of the periodic cell to fluctuate independently, while the second one keeps the ratio of the unit cell constant in the xy-plane while allowing fluctuations along z axes in the course of MD simulation. In addition, the ‘TclForce’ script of NAMD was utilized to impose semi-harmonic restraints on ionic species during MD simulations, as elaborated above. Simulations were carried out with the CHARMM Drude polarizable and the additive C36 force fields. Each simulated system was approximately composed of ~45,000 particles in case of the polarizable model and ~26,000 atoms when additive model was employed. MD simulation run was 70 ns for each simulated electrolyte system. The convergence of MD simulations was verified by calculating osmotic pressures from the two halves of the MD trajectory. Standard deviations around the averaged values were obtained based on seven 10 ns blocks of the total MD trajectories.
Analysis of Ionic Distributions around DNA
Ion-DNA radial distribution functions (RDFs) were computed following the approach developed by Savelyev et al.30 in which RDF is based first on defining ion–DNA separation as the closest distance between the DNA molecule and the particular ion, with subsequent normalization of the resulting ion-DNA distance histogram, N(r), by the numerically calculated volume Jacobian, J(r), leading to the following expression for the RDF:
| (5) |
Here, V and N are the volume of the simulation box and the number of ions, respectively. One of the principal differences between our definition of the ion-DNA RDF and those used in other studies is the method of computing the volume Jacobian. Particularly, the latter is defined as the volume of a shell, equidistant from the DNA surface (see Fig. S2, SI), providing a more robust measure of the ionic entropy than in commonly used standard procedures relying on cylindrical or spherical symmetries.29, 34 As discussed previously,30 the latter techniques lead to an overestimation of the counterion condensation at small distances from the DNA surface, which, in turn, may overestimate the absolute number of ions contributing to the various RDF peaks used to determine the fraction of the DNA charge neutralized by condensed counterions. Additionally, to minimize the end effects, only the inner fraction of the DNA segment (approximately eight internal base pairs in the case of the 12-bais-pair EcorI dodecamer) was used for computing the RDFs. This was achieved by “clipping” the periodic boundary cell with two parallel planes that pass through the centers of the upper four and the lower four DNA base pairs and perpendicular to the line connecting these centers.
Ion-DNA distance histograms and volume Jacobian were computed over the MD simulation course for every snapshot. Three-dimensional grids with lattice spacing of 0.5 Å were used to calculate both the ion-DNA distance histograms and the volume Jacobian. The biochemical algorithm library (BALL)62 was used to implement the computational analysis subroutines. Additionally, BALL was used for writing code for reimaging/recentering of MD trajectories in cases when DNA strands (treated as independent segments) appeared separated in the course of simulation because of the periodic boundary conditions. This post-processing was necessary for subsequent calculations of the volume Jacobian and RDFs, and also other overall structural DNA properties, such as RMS deviations and Watson-Crick base-pairing.
Given the ion-DNA RDFs computed from Eq. (5), we estimate the number of ions associated with DNA (i.e., those contributing to the ionic atmosphere), an experimentally accessible value from BE-AES measurements, as follows,
| (6) |
by performing integration of the excess ionic concentration (relative to bulk) over the entire simulation box. These calculations and comparison with the BE-AES experiment have been performed for large systems (l~120 Å) only, as discussed below.
RESULTS AND DISCUSSION
Optimization of the LJ Interactions Between Ions, Nucleic Acid Bases and DNA Phosphate Group based on QM data and Osmotic Pressure calculations
Our prior efforts in tuning the interactions between DNA and Na+ ions have demonstrated that application of the default Drude nonbond parameters for individual ions and nucleic acid bases led to instabilities of the duplex DNA in MD simulations.31 This was due to the Lennard-Jones (LJ) parameters for direct interactions of the ions with the bases being derived from the Lorenzt-Berthelot combining rules,63 whose application together with the electrostatic parameters for the individual moieties was shown to be problematic in the Drude force field.36, 64 In the case of DNA, imbalance between Na+ ions and certain sites of the nucleic acids led to significant overestimation of their interactions, resulting in destabilization of DNA. An extensive analysis revealed that DNA structural disruption was caused by Na+ interacting strongly with a number of DNA electronegative atoms – N1 of adenine, N3 of cytosine, N1 of guanine and N3 of thymine – which participate in the Watson-Crick base-pairing (N3–H⋯N1 and O4⋯H–N6 hydrogen bonds).31 Initial MD simulations of DNA oligomer in the ionic buffers composed of the monovalent cations Li+, K+ and Rb+ led to the same outcome indicating the need for additional optimization of the ion-DNA LJ parameters in the Drude polarizable force field. The optimization was performed targeting both QM interaction energy data and experimental osmotic pressure data. Interestingly, the default additive C36 parameters did not lead to any DNA instabilities, however, adjustments of selected ion-DNA LJ parameters were necessary to improve consistency with the QM data and experimental osmotic pressure measurements.
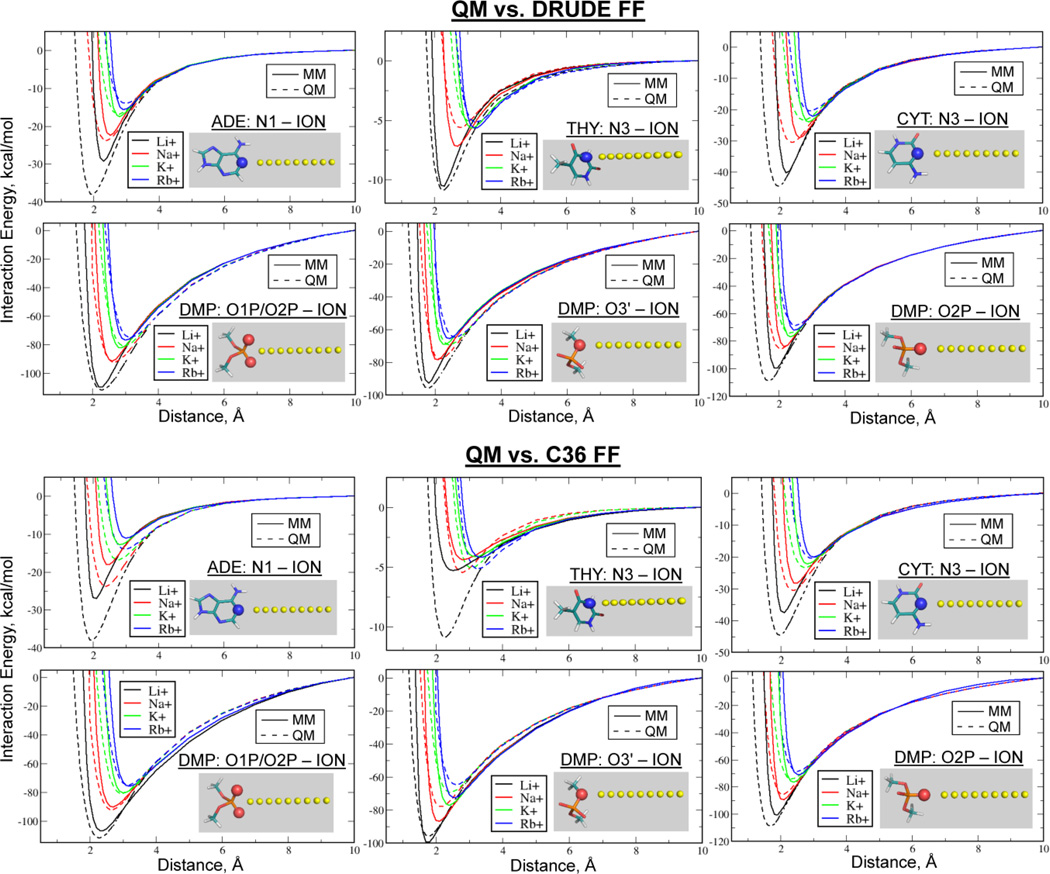
Gas phase QM data on interactions between individual bases and Li+, Na+, K+ and Rb+ as a function of distance were used to initially calibrate the LJ parameters. Similar data was generated for ion-DMP− complexes probing interactions of the different ions with the oxygens of the phosphate group. Shown in the Fig. 1 are the QM and MM potential energy scans, with the Drude and C36 empirical profiles obtained with the final sets of parameters. Both force fields provide a similar level of consistency with the QM data, preserving overall trends as a function of ion. At the same time, there are some noticeable differences between the two FFs, such as for the THY: N3–Li+ potential energy profiles. This is an important example as it deals with the out-of-plane relative orientation of the Li+-THY complex, which may be compared to the in-plane CYT:N3-Li+ complex. For the in-plane complex, both FFs are in reasonable agreement with the QM data, while only the Drude model is in satisfactory agreement for the out-of-plane interaction with Thy. Similar results are seen for in-plane and out-of-plane interactions with the GUA:O6-K+ complex (Fig. S3, SI). These examples demonstrate an importance of inclusion of polarization as well as lone pairs for satisfying simultaneously several mutual ion-base orientations.
Figure 1.
QM and empirical Drude and C36 potential energy surfaces as functions of the distance between Li+, Na+, K+ and Rb+ ions and selected atoms of the nucleic acid bases or of the dimethylphosphate (DMP) anion.
Following the approach used to optimize Na+–bases Drude interactions,31 we utilized the NBFIX option in CHARMM50 to introduce pair-specific LJ terms for interactions of Li+, K+ and Rb+ ions with the different nitrogen or oxygen atoms. Modifications were only made to the combining rule values of Rmin with the LJ depths (ε) not altered. Such modifications were made for LJ interactions between Li+, K+ and Rb+ ions and the following atoms of the nucleic acid bases: (1) N1, N3, N6 of adenine; (2) N1, N2, N3 of guanine; (3) N1, N3, N6 of thymine; and (4) N4, N3 of cytosine. In practice, because of the repeating atom types in all four bases, only 3 unique NBFIX modifications were introduced for each ion type. Additionally, NBFIXes were introduced for the Drude interactions of Li+ and Na+ ions with the (1) anionic O1P/O2P and (2) ester O3/O4 oxygen atoms of the DMP− anion. For the additive C36 parameters, a single modification was made for interactions between Li+ and O1P/O2P atoms of DMP−. It is important to note that these modifications have been made based on targeting both QM data and osmotic pressure calculations, as discussed below.
Osmotic pressure measurements provide valuable data that can be used to calibrate interactions among chemically relevant ions in aqueous solutions. In particular, to investigate interactions of ions with the DNA phosphodiester backbone and the Cl− in an ionic buffer, osmotic pressure coefficients were calculated from MD simulations of the bulk ion-DMP− and ion-Cl− electrolyte solutions for Li+, Na+, K+ or Rb+. It is known that the osmotic pressure coefficient is a measure of the solution non-ideality, describing how much the behavior of the real electrolyte solution deviates from that of an ideal solution at a particular concentration.65 With that deviation becoming increasingly larger at higher concentrations, it is important to estimate osmotic pressures at both low and high concentrations. However, whereas experimental data for chloride solutions are available for a wide range of salt concentrations, up to ~6M,66 for ion-DMP− electrolytes the data are known only at low concentrations, ≤ 1M.67 Moreover, data for Rb+-DMP− are not available. Therefore, chloride solutions were studied at ~1M and ~3M concentrations, with the DMP− solutions (except that containing Rb+) simulated at ~1M only. Based on osmotic pressure calculations, adjustments of selected pair-specific nonbond interactions among cations and DMP− anion and between cations and Cl− were carried out.
Experimental osmotic pressures and those computed from MD simulations using optimized Drude and additive C36 parameters are shown in the Table 2. Actual molal concentrations in the ionic slab computed from ensemble averages are provided in the Table 3. Those are needed for direct determination of the experimental values of osmotic pressures in solutions at exactly the same concentration as in MD simulations (see Method section). Overall, the level of agreement between both the Drude and C36 results with experiment is good, with the Drude generally being in better agreement. With the exception of LiCl at 3 M, the Drude values deviate by 7.2% or less while the C36 values were 10.6% or better. For LiCl at 3 M the percent differences are 9.3 and 24.3 for the Drude and C36 models, respectively. Thus, both models provide a plausible description of osmotic properties of chemically relevant ionic solutions. The final LJ parameters in the NBFIX format are shown in Table S1 of the supporting information.
Table 2.
Experimental and MD simulation results for the osmotic pressures (in bars) for different electrolyte solutions at ~1M and ~3M concentrations.
| EXPDRUDEa | DRUDE FF | EXPC36a | C36 FF | |||||
|---|---|---|---|---|---|---|---|---|
| ~1M | ~3M | ~1M | ~3M | ~1M | ~3M | ~1M | ~3M | |
| Li – Cl | 43.1 | 172.0 | 41.4 | 156.0 | 43.1 | 171.9 | 40.5 | 130.2 |
| Na – Cl | 41.1 | 154.5 | 42.1 | 155.7 | 40.5 | 149.0 | 44.0 | 149.5 |
| K – Cl | 40.0 | 138.9 | 39.5 | 137.3 | 40.0 | 136.5 | 41.3 | 135.8 |
| Rb – Cl | 40.1 | 138.5 | 37.2 | 140.3 | 40.0 | 138.1 | 40.2 | 123.4 |
| Li – DMP | 45.1 | N/a | 44.1 | N/a | 45.5 | N/a | 41.7 | N/a |
| Na – DMP | 48.0 | N/a | 49.3 | N/a | 48.0 | N/a | 45.4 | N/a |
| K – DMP | 48.8 | N/a | 50.5 | N/a | 48.8 | N/a | 47.3 | N/a |
Table 3.
Actual molal concentrations for all studied electrolyte solutions computed from MD simulations according to Eq. (2)
| DRUDE FF | C36 FF | |||
|---|---|---|---|---|
| ~1M | ~3M | ~1M | ~3M | |
| Li – Cl | 0.86 | 2.81 | 0.86 | 2.80 |
| Na – Cl | 0.88 | 2.96 | 0.80 | 2.81 |
| K – Cl | 0.87 | 2.95 | 0.87 | 2.87 |
| Rb – Cl | 0.90 | 3.01 | 0.87 | 2.94 |
| Li – DMP | 0.90 | N/a | 0.94 | N/a |
| Na – DMP | 0.90 | N/a | 0.91 | N/a |
| K – DMP | 0.92 | N/a | 0.92 | N/a |
General Features of Distributions of Various Ions around DNA
Having achieved good consistency with both gas phase QM data and experimental osmotic pressure measurements, we addressed macroscopic features of the ionic atmosphere around DNA. In this section we focus on details of ionic structuring around the whole DNA duplex based on MD simulations of small DNA systems (l~60 Å). Those systems include DNA molecules solvated in ~100mM ionic buffers composed of a single chloride salt, as well as 1:1 mixtures of NaCl and one of the competing chloride salt, LiCl, KCl or RbCl.
One of the commonly used macroscopic characteristics used for assessment of the ionic distribution around DNA from MD simulations is the fraction of the DNA residual charge neutralized by the ions within the ionic atmosphere. According to CC theory,5, 28 a region of “condensed” cations (the atmosphere) must occur in the vicinity of the linear rods that are uniformly charged above some critical charge density. Because DNA is a linear polyanion possessing high charge per unit length, it is well above this threshold. Applying arguments of CC theory to DNA, one can estimate that ~76% of its total charge is expected to be neutralized by condensed counterions within a ~9 Å Manning radius.
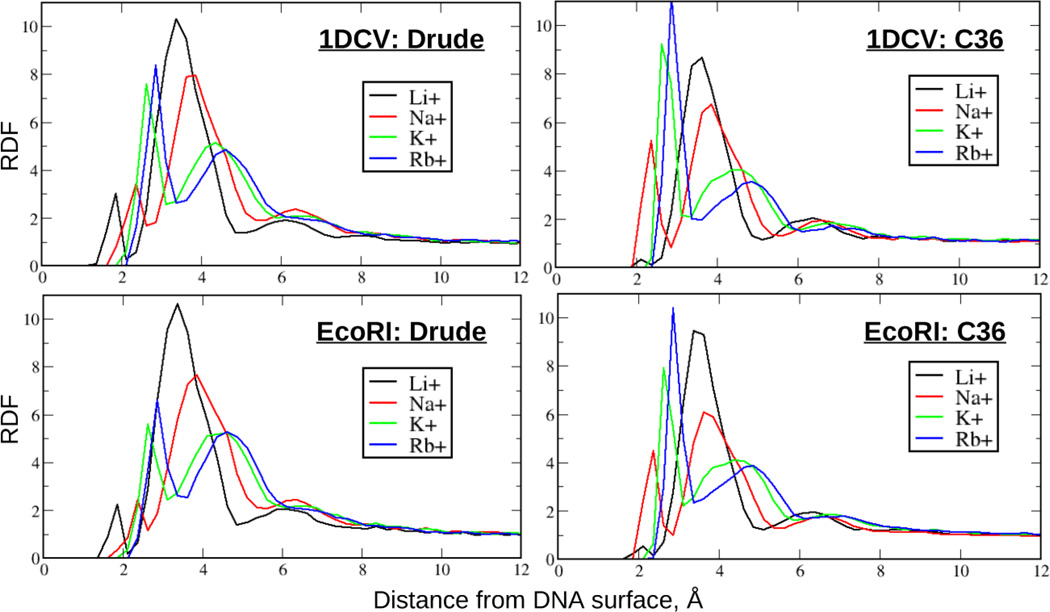
To compare MD simulation results with predictions from CC theory, we computed and analysed ion-DNA RDFs focusing on the overall ionic condensation around the central region of the molecule. Fig. 2 shows RDFs for Li+, Na+, K+ and Rb+ cations for EcoRI and 1DCV systems for both FFs. Analogous results inferred from 1:1 mixed salt DNA solutions are shown in the Fig. 3 that also includes and RDFs for the Cl− anions. It is seen that in all cases the cation RDFs are characterized by three prominent peaks indicating the formation of ionic ‘shells’ around polyanionic DNA. The onset of such an ionic structuring when approaching DNA from the bulk is commonly associated with the Manning’s radius in CC theory. As seen from the plots, counterions are structured within ~9 Å of the DNA surface, coinciding with the CC predictions. This is also consistent with the definition of Beveridge and co-workers29 for the condensed counterions as those lying within a 9 Å region from the DNA surface, where ions are structured with respect to DNA.
Figure 2.
Cation/DNA radial distribution functions (RDF) from the Drude and additive C36 MD simulations of the small size 1DCV and EcoRI systems in ~100 mM ionic buffers composed of a single salt, LiCl, NaCl, KCl or RbCl, computed based on the closest approach, as elaborated in the Methods section.
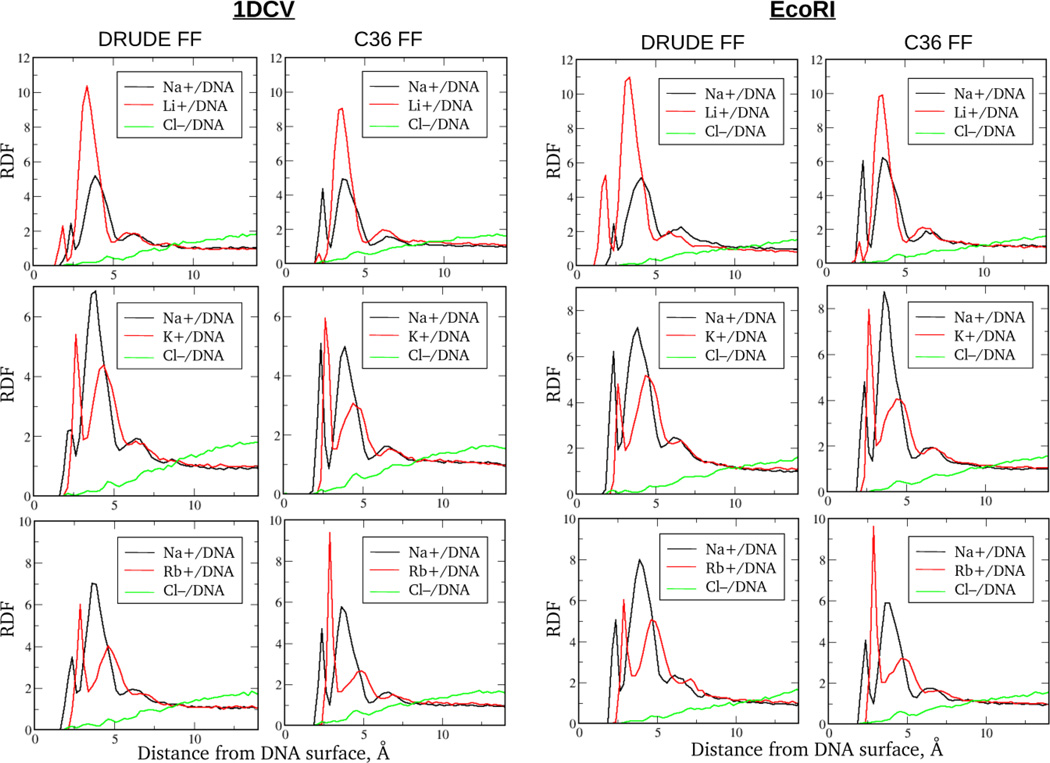
Figure 3.
Cation/DNA and Cl− /DNA radial distribution functions (RDF) from the Drude and additive C36 MD simulations of the small size 1DCV and EcoRI systems in ~100 mM ionic buffers of the mixed (1:1) salt composition.
Based on the RDFs computed for DNA systems in buffers composed of a single salt (Fig. 2), both additive and Drude models generate qualitatively similar ionic trends. This reflects the effect of ionic size and hydration pattern on the relative height of different peaks and their positions. As seen from Fig. 2, the second RDF peak is the biggest one for smaller cations, Li+ and Na+, indicating these ions to be predominantly hydrated within the atmosphere and separated from DNA by a single water molecule. In contrast, for the larger K+ and Rb+ cations the first RDF peak possesses the largest amplitude reflecting a higher probability of formation of direct ion-DNA contacts due to the ion's smaller dehydration penalty. It is worth noting that considerable deviations from this trend were observed for additive C36 systems containing Li+ when the standard LJ parameters for the cation-phosphate interactions were used. In particular the osmotic pressure calculations indicated the binding of Li+ to phosphate groups to be very favourable when no NBFIX correction was applied between Li+ and O1P/O2P oxygen atoms, leading to excessive formation of direct contacts between DNA backbone and Li+ (see Fig. S4, SI). Such behavior was also observed in prior computational studies,33, 68 including the direct comparison of MD simulations with BE-AES experiment on competitive ionic binding to DNA. We discuss this in the following section.34
To calculate the fraction of the DNA charge neutralized by the ions within the Manning radius RDFs for Li+, Na+, K+, Rb+ and Cl− have been integrated from 0 to 9 Å, with the resulting number of coions and counterions summed and divided by DNA residual charge. Such calculations were carried out on both pure and mixed systems since the continuous mean-field CC theory does not distinguish among chemically different monovalent ionic species. While this is a limitation of the theory, comparison between MD simulations and CC theory allows judging the adequacy of atomic-detailed computational models to describe ionic distribution around DNA. Results for the Drude and additive MD simulations of the EcoRI and 1DCV systems are presented in the Table 4. It is seen that the fraction of the neutralized DNA charge is systematically underestimated by ~5–10% in the additive model compared to the Drude model. Results on both DNA systems were very similar, indicating the overall DNA charge neutralization to be a sequence-independent phenomena. This suggestion is consistent with inconclusive experimental data on whether Na+ or K+ condenses stronger to DNA, as discussed previously.30 In particular, while NMR experiments16 suggest K+ to have slightly higher affinity to bind to the DNA minor groove, electrophoretic measurements of DNA mobility in different ionic buffers22 may be interpreted to suggest the binding propensity to follow an opposite trend, Na+ > K+. Because DNA mobility measurements are macroscopic in nature, with similar trends obtained for different sequences (with and without A-tract22), the above arguments suggest that preferential (i.e., sequence-specific) binding of counterions to DNA grooves may not affect overall charge DNA neutralization.
Table 4.
Comparison between MD simulation results and counterion condensation theory based on analysis of ion/DNA RDFs computed from small DNA systems.
| % DNA charge neutralized by all ions within 9 Å | |||||
|---|---|---|---|---|---|
| 1DCV | EcoRI | CC theory | |||
| DRUDE | C36 | DRUDE | C36 | ||
| LiCl | 78.0% | 66.5% | 78.1% | 73.4% | 76% |
| NaCl | 76.2% | 68.0% | 76.6% | 69.2% | 76% |
| KCl | 76.3% | 67.6% | 78.3% | 66.1% | 76% |
| RbCl | 74.8% | 67.2% | 77.1% | 65.6% | 76% |
| NaCl & LiCl (1:1) | 73.3% | 68.0% | 71.2% | 69.7% | 76% |
| NaCl & KCl (1:1) | 72.2% | 65.5% | 73.8% | 69.4% | 76% |
| NaCl & RbCl (1:1) | 71.6% | 67.1% | 69.8% | 64.5% | 76% |
Another test was performed on small DNA systems in mixed ionic solutions involving qualitative assessment of the competitive distribution of different cations around DNA. In particular, we compared contributions of the chemically different ionic species to the ionic atmosphere. Ratios of Na+ to other competing cations, Li+, K+ and Rb+, obtained by integrating the corresponding ionic RDFs over the distance extending up to 9 Å from DNA surface are provided in Table 5. Results show the propensity of ions to occupy ionic atmosphere follows the trend, Li+ > Na+ > K+ > Rb+, consistent with that obtained from BE-AES experiments4 and DNA mobility measurements.22 Analysis performed on EcoRI and 1DCV sequences yield very similar ratios for the Drude and additive models. This is another indication that effects from specific ion-binding motifs, while important for studying local phenomena, are negligible on the overall outcome of the ionic structuring and competition around DNA.
Table 5.
Qualitative assessment of the ionic competition from MD simulations of small DNA systems based on ion-counting within the Manning radius from the DNA surface.
| Ion/Ion ratio within 9 Å | ||||
|---|---|---|---|---|
| 1DCV | EcoRI | |||
| DRUDE | C36 | DRUDE | C36 | |
| Na+/Li+ | 0.72 | 0.95 | 0.70 | 0.96 |
| Na+/K+ | 1.12 | 1.06 | 1.14 | 1.12 |
| Na+/Rb+ | 1.18 | 1.13 | 1.16 | 1.05 |
Competition among Cations within the Ionic Atmosphere: Comparison with BE-AES Experiment
As evidenced above, general features and qualitative trends of ionic distributions can be inferred from MD simulations of small DNA systems. However, quantitative comparisons with BE-AES experiments on competitive association of cations require simulating much larger DNA systems.
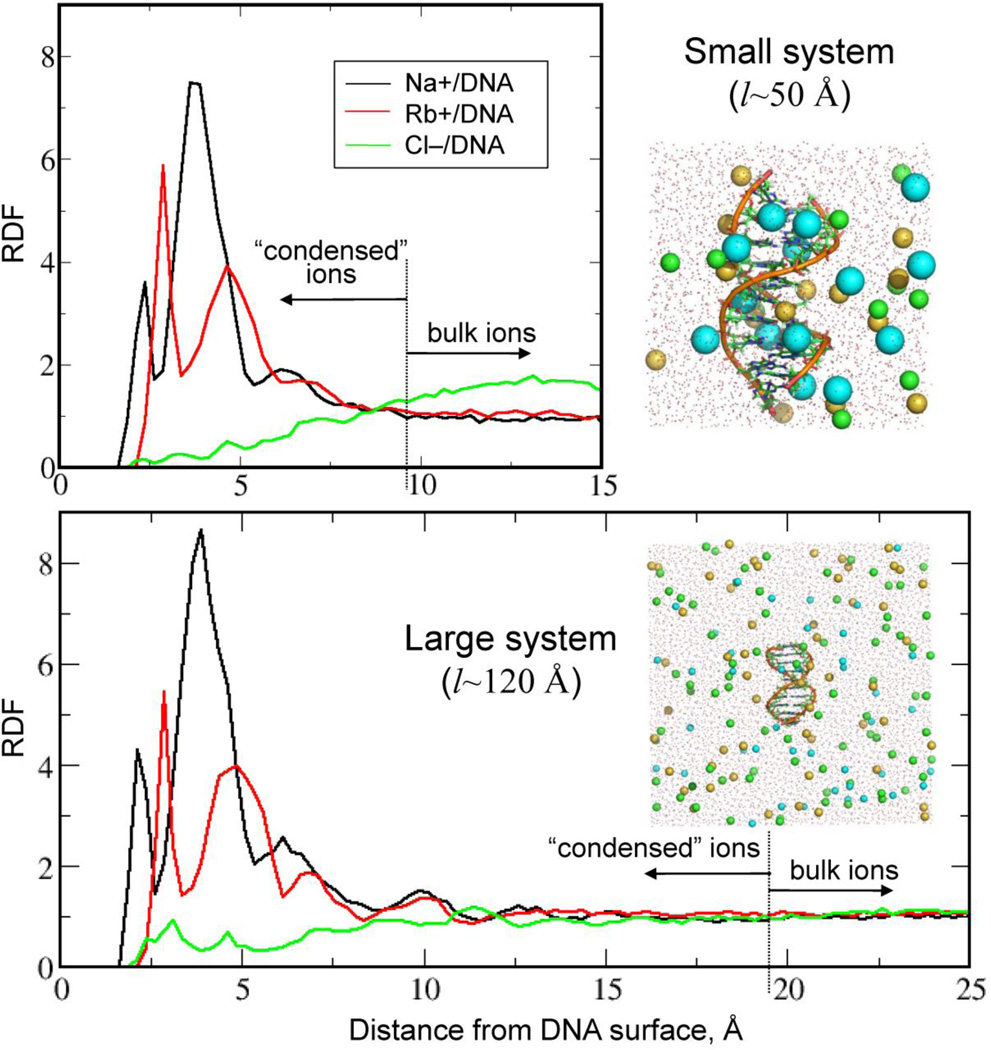
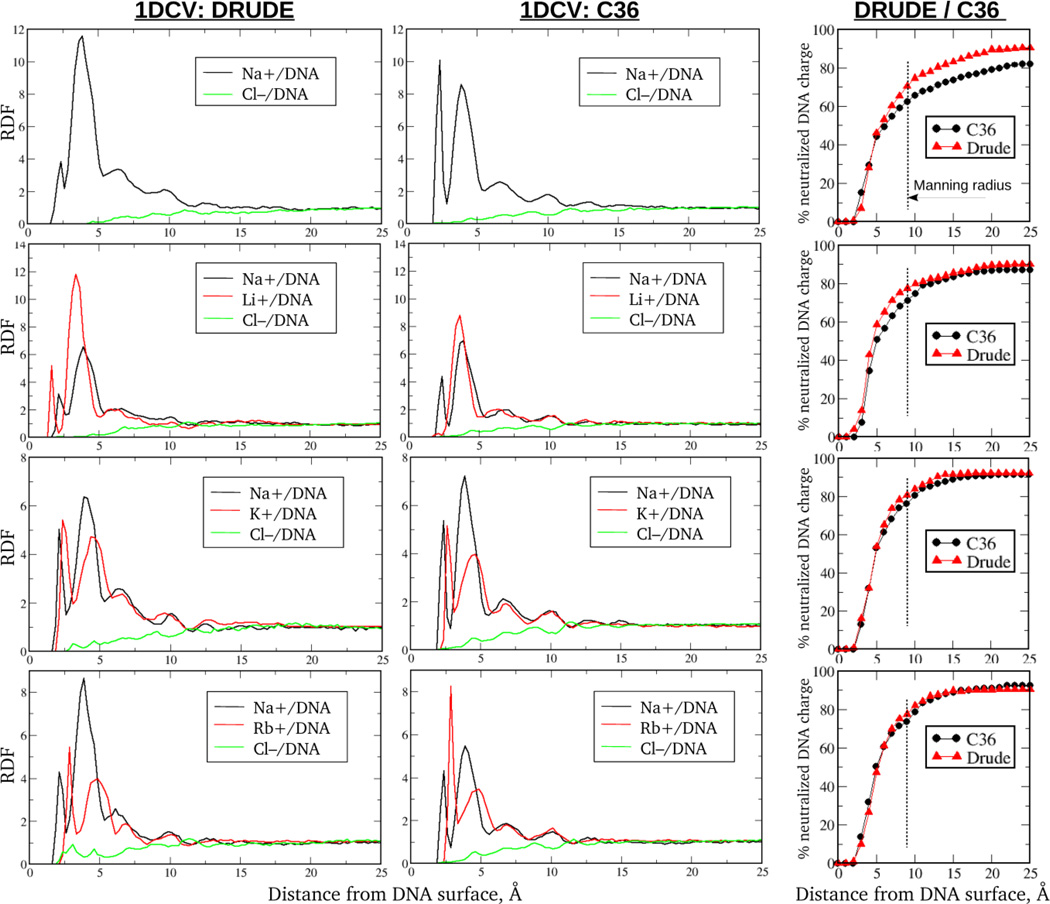
As mentioned in the Introduction, the method of ion counting in BE-AES technique is based on the principle of electroneutrality requiring the ionic atmosphere to completely neutralize the DNA molecule. The BE-AES technique detects the cations associated with and anions depleted from DNA which cover a region where the concentration of ions deviates from that of the bulk.4 In terms of MD simulations, the requirement of electroneutrality requires the size of DNA simulation box to have an edge length of ~120 Å (versus ~60 Å for small systems) to allow for DNA neutralization by the ions located as far as ~50 Å from the DNA helical axis.34 As seen in Fig. 4 for the Drude FF ions differ in their structuring with respect to DNA in the small versus the large systems. In particular, cationic RDFs saturate at distances ~9Å and ~20Å from DNA surface, respectively, indicating the ionic atmosphere to extend twice as far from the DNA in the large systems. As for the Cl− RDFs, the size effects render impossible a proper normalization (to unity) of the anionic RDFs in small systems as also present in Figs. 2 and 3.
Figure 4.
Comparison of small and large DNA systems simulated in the present study including ion radial distributions and images of the small and large systems. Demarcation of the distances defining the condensed versus bulk ions for the two types of systems are shown.
Another important aspect of establishing the consistency between MD simulations and BE-AES experiment is the way the concentration of the ions is computed. As discussed below, the notion of the ion's competition constant, a molar concentration by definition, is devised from BE-AES experiments to characterize ionic competitiveness for association with DNA.4 However, defining a concentration of the cations in MD simulations is rather ambiguous, especially for smaller systems. For example, the lion's share of all cations in small systems comes from the ions which directly neutralize DNA, with the extra cations from ~100mM of chloride salt constituting only ~35% of the total number of cations. While adding both neutralizing counterions and extra salt is a standard practice in MD simulations, it is not clear whether total number of cations, or just their salt portion needs to be used in defining ionic concentration for direct comparison with BE-AES experiment. Dealing with large DNA systems possessing larger number of ions, with the fraction of the neutralizing counterions not exceeding ~20% (Table 1), makes definition of ionic concentration much less ambiguous.
Because of computationally demanding calculations (i.e. ~150000 and 240000 particles, including Drudes and lone pairs, in the additive and polarizable systems, respectively), we conducted Drude and C36 MD simulations on the large 1DCV system only. Although this molecule is of different length and sequential content than that used in BE-AES experiment, the character of the overall distribution of ions and their competition turned out to be independent on DNA sequence, as evidenced in the previous section. The same tendency was observed from BE-AES experiments, when control measurements on several different DNA duplexes (with and without A-tract and CG-rich sequences) have led to identical results for ionic competition.4 These observations suggest that comparison of the results from our large 1DCV simulations with BE-AES experiments is appropriate.
The simulations focus on four sets of experimental data: (1) 50mM of Na+ only; (2) 50mM of Na+ and 36mM of Li+; (3) 50mM of Na+ and 51mM of K+; and (4) 50mM of Na+ and 60mM of Rb+. In all systems sodium is the background cation (BC) of constant concentration. Concentrations of the extra competing ions (CI), Li+, K+ and Rb+, were chosen to be their competition constants determined from experiment and defined as competing cation concentrations at which half the amount of the BCs associated with DNA in the absence of CIs is excluded. The numbers of associated cations are computed from Eq. (6) by integrating the deviations of the corresponding RDFs from unity over the entire simulation box. These numbers should not be confused with the absolute numbers of ions, which were used in the previous section for qualitative analysis of ionic competition.
Fig. 5 shows RDFs for cations and anions computed from the Drude and additive C36 MD simulations. In all cases, distributions converge to the bulk value (unity) after ~20 Å from the DNA, an indication that simulations reached equilibrium. Compared to the small systems, the number of prominent peaks reflecting ionic structuring increased from three to five in all simulated systems. Importantly, RDF profiles do not exhibit substantial modulations after ~60 ns of simulation time. Also shown are the fractions of the DNA charge neutralized by all ions (co- and counterions) as functions of the distance from DNA surface. Consistent with the RDFs, the fraction plots saturate after ~20 Å from the DNA, defining the size of ionic atmosphere. It is important to comment on the following. The plateau values of the neutralized charge never reach 100%, meaning no complete neutralization of the DNA by the ionic atmosphere occurs. This can be explained by two factors. First, the above discussed issue of defining ionic concentration may still contribute to the overall outcome of DNA neutralization. Second, RDFs were computed by averaging over the central region of the DNA duplex to avoid end effects (see Method section and Fig. S2). This contrasts with the BE-AES experiment where ions from the entire DNA environment contributed to neutralization. Indeed, when the ions from all space around DNA are considered in our computational systems, they completely neutralize DNA charge, as exemplified in the Fig. S5, SI. It is important to realize, however, that such an approach is not appropriate for rigorous determination of the excess number of ions (i.e., associated with DNA) since the bulk ionic concentration becomes an anisotropic parameter with respect to DNA which cannot be conveniently reduced to the dependence on a single (closest) distance from DNA surface.
Figure 5.
Radial distribution functions (RDF) for co- and counterions around DNA computed from the Drude and C36 MD simulations of the large size 1DCV systems in ionic buffers containing only background cations (Na+), or mixtures of background and competing cations (Li+, K+, or Rb+). Ionic concentrations are chosen to match BE-AES experimental conditions. The right column shows the percent fraction of the DNA charge neutralized by all ions within different distances from DNA surface. The Manning radius is schematically shown by dashed lines.
It is also instructive to look at the neutralized charge fraction by the ions within the Manning radius and compare with results from the previous section. As seen from the Fig. 5, ions within ~9Å from DNA surface neutralize ~72–80% and ~65–75% of its residual charge in the Drude and additive models, respectively. These numbers are consistent with the data in Table 4. One should keep in mind, however, that similarity of the results obtained from small and large simulations is a consequence of very different size or edge effects imposed on the systems. While those effects are negligible in large systems, they ‘deform’ the anionic concentration profiles in smaller systems, leading to the overestimated depletion of the coions from DNA and the associated enhanced DNA charge neutralization. Despite these artefacts, the obtained consistency still suggests that small-to-medium size computational systems are suitable for qualitative assessment of ionic distribution around DNA though larger systems are required for quantitative comparisons.
Quantitative results on competitive ionic association from simulations and BE-AES experiment are provided in Table 6. Shown are the ratios of the number of associated BCs (Na+) from the reference simulation with no competing cations added (system 1) versus the same from the mixed salt solutions (systems 2–4). According to the definition of the ion competition constant, BE-AES experiment predicts the ratios to be 2. Computational results from the Drude and additive simulations both deviate from the experimental value. However, the level of consistency is clearly better for the Drude model. For example, despite decent agreement with QM data and osmotic pressure measurements, Li+ and K+ ions are not competitive enough in the additive force field, indicating that the current parametrization has room for further improvement. At the same time, Rb+ in the additive model possesses enhanced tendency compared with Na+ for association with DNA. This contrasts with a near quantitative agreement of the Drude results with experiment.
Table 6.
Comparison of MD simulation results with BE-AES experimental data on the competitive ionic association with DNA. Shown are the ratios of excess numbers of background cations (Na+) in the absence and presence of the competing cationa
| Competing Ion | ||||
|---|---|---|---|---|
| DRUDE | C36 | EXP | ||
| Li+ | 2.11 ± 0.09 | 1.87 ± 0.08 | ||
| K+ | 2.06 ± 0.11 | 1.49 ± 0.01 | 2.0 | |
| Rb+ | 1.95 ± 0.05 | 2.29 ± 0.10 | ||
BC and CI stand for background cation and competing ion, respectively. Standard errors are estimated as standard deviations of the mean values computed based on trajectory decomposition into 4 blocks of 30 ns each. Last 120 ns of the production run were analysed.
CONCLUDING REMARKS
Aqueous salt environments are crucially important in determining the structure and functional dynamics of all biological molecules including proteins and nucleic acids. In the present study we have optimized and tested nonbond parameters for the first-group monovalent cations, Li+, Na+, K+ and Rb+, in the Drude polarizable and CHARMM36 additive force fields for use in studies of DNA in aqueous solution. The complex nature of interactions between polyanionic DNA, water molecules and mobile ions required careful optimization of parameters to satisfy a broad range of experimental data and theoretical estimations, such as gas-phase QM calculations on energetics of various model compounds with ions, osmotic pressure measurements for aqueous solutions of biologically relevant ions, and theoretically and experimentally observed trends for the overall ionic condensation around DNA, as well as for competition of various monovalent ions for association with DNA.
Comparison between Drude and C36 models reveals that, whereas both force fields reproduced QM data and osmotic pressure measurements at similar level of agreement, the Drude model demonstrated markedly improved description of the ion atmosphere and ionic competition in the vicinity of DNA, based on predictions from CC theory and BE-AES experiments. Our recent computational study20 demonstrated that the Drude force field reproduced the solution X-ray diffraction patterns for the EcoRI and 1DCV sequences in NaCl ionic buffer at a level of accuracy exceeding that of the additive C36 model – an indication that not only are the overall features of the ionic distribution improved but also the interplay between ionic atmosphere and DNA conformational dynamics is better captured by the Drude model. In addition, it was predicted that interactions of Li+, Na+, K+ or Rb+ ions with DNA leads to different DNA structural behavior associated largely with minor grove width modulation when using the Drude model.20 Notably, no effect was observed in the additive C36 model.20 Although not tested experimentally, this prediction is strengthened by the present near quantitative agreement of the results from the Drude simulations with BE-AES experiments. Overall, results from the present and earlier computational studies19–20, 31, 69 indicate that Drude polarizable MD simulations provide a more realistic model of the physical forces involved in the interactions of DNA with its ionic environment, offering the potential to yield new insights into salt-mediated biological processes involving DNA.
Supplementary Material
Acknowledgments
The NIH (GM051501) is thanked for financial support and we would like to thank Dr. Ignacio Soteras Gutierres for helpful discussions. The University of Maryland Computer-Aided Drug Design Center and the XSEDE resources are gratefully acknowledged for their generous allocations of computer time.
Footnotes
Supporting Information Available: Supplementary tables and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Draper DE. A Guide to Ions and RNA Structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters JP, Maher LJ., III DNA Curvature and Flexibility In Vitro and In Vivo. Q. Rev. Biophys. 2010;43:23–63. doi: 10.1017/S0033583510000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris L-A, Williams LD, Koudelka GB. Specific Minor Groove Solvation is a Crucial Determinant of DNA Binding Site Recognition. Nucleic Acids Res. 2014;42:14053–14059. doi: 10.1093/nar/gku1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai Y, Greenfeld M, Travers KJ, Chu VB, Lipfert J, Doniach S, Herschlag D. Quantitative and Comprehensive Decomposition of the Ion Atmosphere Around Nucleic Acids. J. Am. Chem. Soc. 2007;129:14981–14988. doi: 10.1021/ja075020g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manning GS. Limiting Laws and Counterion Condensation in Polyelectrolyte Solutions I. Colligative Properties. J. Chem. Phys. 1969;51:924–933. [Google Scholar]
- 6.Knobler CM, Gelbart WM. Annu. Rev. Phys. Chem. 2009;60:367–383. doi: 10.1146/annurev.physchem.59.032607.093728. [DOI] [PubMed] [Google Scholar]
- 7.Koculi E, Hyeon C, Thirumalai D, Woodson SA. Charge Density of Divalent Metal Cations Determines RNA Stability. J. Am. Chem. Soc. 2007;129:2676–2682. doi: 10.1021/ja068027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savelyev A, Materese CK, Papoian GA. Is DNA's Rigidity Dominated by Electrostatic or Nonelectrostatic Interactions? J. Am. Chem. Soc. 2011;133:19290–19293. doi: 10.1021/ja207984z. [DOI] [PubMed] [Google Scholar]
- 9.Savelyev A. Do Monovalent Mobile Ions Affect DNA's Flexibility at High Salt Content? Phys. Chem. Chem. Phys. 2012;14:2250–2254. doi: 10.1039/c2cp23499h. [DOI] [PubMed] [Google Scholar]
- 10.Savelyev A, Papoian GA. Chemically Accurate Coarse Graining of Double-Stranded DNA. Proc. Natl. Acad. Sci. U S A. 2010;107:20340–20345. doi: 10.1073/pnas.1001163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shui XQ, McFail-Isom L, Hu GG, Williams LD. The B-DNA Dodecamer at High Resolution Reveals a Spine of Water on Sodium. Biochemistry. 1998;37:8341–8355. doi: 10.1021/bi973073c. [DOI] [PubMed] [Google Scholar]
- 12.Tereshko V, Minasov G, Egli M. A "Hydrat-Ion" Spine in a B-DNA Minor Groove. J. Am. Chem. Soc. 1999;121:3590–3595. [Google Scholar]
- 13.Tereshko V, Minasov G, Egli M. The Dickerson-Drew B-DNA Dodecamer Revisited at Atomic Resolution. J. Am. Chem. Soc. 1999;121:470–471. [Google Scholar]
- 14.Tereshko V, Wilds CJ, Minasov G, Prakash TP, Maier MA, Howard A, Wawrzak Z, Manoharan M, Egli M. Detection of Alkali Metal Ions in DNA Crystals using state-of-the-art X-ray Diffraction Experiments. Nucleic Acids Res. 2001;29:1208–1215. doi: 10.1093/nar/29.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halle B, Denisov VP. Water and Monovalent Ions in the Minor Groove of B-DNA Oligonucleotides as Seen by NMR. Biopolymers. 1998;48:210–233. doi: 10.1002/(sici)1097-0282(1998)48:4<210::aid-bip3>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 16.Denisov VP, Halle B. Sequence-Specific Binding of Counterions to B-DNA. Proc. Nat. Acad. Sci. USA. 2000;97:629–633. doi: 10.1073/pnas.97.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marincola FC, Denisov VP, Halle B. Competitive Na+ and Rb+ Binding in the Minor Groove of DNA. J. Am. Chem. Soc. 2004;126:6739–6750. doi: 10.1021/ja049930z. [DOI] [PubMed] [Google Scholar]
- 18.Chiu TK, Kaczor-Grzeskowiak M, Dickerson RE. Absence of Minor Groove Monovalent Cations in the Crosslinked Dodecamer C-G-C-G-A-A-T-T-C-G-C-G. J. Mol. Biol. 1999;292:589–608. doi: 10.1006/jmbi.1999.3075. [DOI] [PubMed] [Google Scholar]
- 19.Savelyev A, MacKerell AD., Jr All- Atom Polarizable Force Field for DNA Based on the Classical Drude Oscillator Model. J. Comput. Chem. 2014;35:1219–1239. doi: 10.1002/jcc.23611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savelyev A, MacKerell AD., Jr Differential Impact of the Monovalent Ions Li+, Na+, K+, and Rb+ on DNA Conformational Properties. J Phys Chem Lett. 2015;6:212–216. doi: 10.1021/jz5024543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stellwagen E, Stellwagen NC. Probing the Electrostatic Shielding of DNA with Capillary Electrophoresis. Biophys. J. 2003;84:1855–1866. doi: 10.1016/S0006-3495(03)74993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stellwagen E, Dong Q, Stellwagen NC. Monovalent Cations Affect the Free Solution Mobility of DNA by Perturbing the Hydrogen-Bonded Structure of Water. Biopolymers. 2005;78:62–68. doi: 10.1002/bip.20260. [DOI] [PubMed] [Google Scholar]
- 23.Zinchenko AA, Yoshikawa K. Na+ Shows a Markedly Higher Potential than K+ in DNA Compaction in a Crowded Environment. Biophys. J. 2005;88:4118–4123. doi: 10.1529/biophysj.104.057323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das R, Mills TT, Kwok LW, Maskel GS, Millett IS, Doniach S, Finkelstein KD, Herschlag D, Pollack L. Counterion Distribution around DNA Probed by Solution X-ray Scattering. Phys. Rev. Lett. 2003;90:188103. doi: 10.1103/PhysRevLett.90.188103. [DOI] [PubMed] [Google Scholar]
- 25.Pabit SA, Meisburger SP, Li L, Blose JM, Jones CD, Pollack L. Counting Ions around DNA with Anomalous Small-Angle X-ray Scattering. J. Am. Chem. Soc. 2010;132:16334–16336. doi: 10.1021/ja107259y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong GCL, Pollack L. In: Annu. Rev. Phys. Chem. Leone SR, Cremer PS, Groves JT, Johnson MA, Richmond G, editors. Vol. 61. 2010. pp. 171–189. [DOI] [PubMed] [Google Scholar]
- 27.Hart K, Foloppe N, Baker CM, Denning EJ, Nilsson L, MacKerell AD., Jr Optimization of the CHARMM Additive Force Field for DNA: Improved Treatment of the BI/BII Conformational Equilibrium. J. Chem. Theory Comput. 2012;8:348–362. doi: 10.1021/ct200723y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manning GS. The Molecular Theory of Polyelectrolyte Solutions with Applications to the Electrostatic Properties of Polynucleotides. Q. Rev. Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 29.Ponomarev SY, Thayer KM, Beveridge DL. Ion Motions in Molecular Dynamics Simulations on DNA. Proc. Natl. Acad. Sci. U S A. 2004;101:14771–14775. doi: 10.1073/pnas.0406435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savelyev A, Papoian GA. Electrostatic, Steric, and Hydration Interactions Favor Na+ Condensation around DNA Compared with K+ J. Am. Chem. Soc. 2006;128:14506–14518. doi: 10.1021/ja0629460. [DOI] [PubMed] [Google Scholar]
- 31.Savelyev A, MacKerell AD., Jr Balancing the Interactions of Ions, Water, and DNA in the Drude Polarizable Force Field. J. Phys. Chem. B. 2014;118:6742–6757. doi: 10.1021/jp503469s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacKerell AD, Jr, Banavali NK. All-atom Empirical Force Field for Nucleic Acids: 2) Application to Solution MD Simulations of DNA. J. Comput. Chem. 2000;21:105–120. [Google Scholar]
- 33.Yoo J, Aksimentiev A. Improved Parametrization of Li+, Na+, K+, and Mg2+ Ions for All-Atom Molecular Dynamics Simulations of Nucleic Acid Systems. J Phys Chem Lett. 2012;3:45–50. [Google Scholar]
- 34.Yoo J, Aksimentiev A. Competitive Binding of Cations to Duplex DNA Revealed through Molecular Dynamics Simulations. J Phys Chem B. 2012;116:12946–12954. doi: 10.1021/jp306598y. [DOI] [PubMed] [Google Scholar]
- 35.Yu HA, Whitfield TW, Harder E, Lamoureux G, Vorobyov I, Anisimov VM, MacKerell AD, Roux B. Simulating Monovalent and Divalent Ions in Aqueous Solution Using a Drude Polarizable Force Field. J. Chem. Theory Comput. 2010;6:774–786. doi: 10.1021/ct900576a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo Y, Jiang W, Yu HA, MacKerell AD, Roux B. Simulation Study of Ion Pairing in Concentrated Aqueous Salt Solutions with a Polarizable Force Field. Faraday Discuss. 2013;160:135–149. doi: 10.1039/c2fd20068f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drude P, Millikan RA, Mann RC. The Theory of Optics. New York: Longmans, Green, and Co.; 1902. p. 588. [Google Scholar]
- 38.Lopes PEM, Harder E, Roux B, MacKerell AD. In: Multi-scale Quantum Models for Biocatalysis. York DM, Lee T-S, editors. Vol. 7. Netherlands: Springer; 2009. pp. 219–257. [Google Scholar]
- 39.Lamoureux G, Roux B. Modelling Induced Polarizability with Drude Oscillators: Theory and Molecular Dynamics Simulation Algorithm. J. Chem. Phys. 2003;119:5185–5197. [Google Scholar]
- 40.Jiang W, Hardy DJ, Phillips JC, Mackerell AD, Jr, Schulten K, Roux B. High-Performance Scalable Molecular Dynamics Simulations of a Polarizable Force Field Based on Classical Drude Oscillators in NAMD. J Phys Chem Lett. 2011;2:87–92. doi: 10.1021/jz101461d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, et al. Gaussian 09. Wallingford CT: Gaussian, Inc.; 2009. [Google Scholar]
- 42.Boys SF, Bernardi F. The Calculation of Small Molecular Interactions by the Differences of Separate Total Energies. Some Procedures with Reduced Errors(Reprinted from Mol. Phys., vol 19, pg 553–566, 1970) Molecular Physics. 2002;100:65–73. [Google Scholar]
- 43.Ransil BJ. Studies in Molecular Structure. IV. Potential Curve for the Interaction of Two Helium Atoms in Single-Configuration LCAO MO SCF Approximation. J. Chem. Phys. 1961;34:2109–2118. [Google Scholar]
- 44.Weigend F, Ahlrichs R. Balanced Basis Sets of Split Valence, Triple Zeta Valence and Quadruple Zeta Valence Quality for H to Rn: Design and Assessment of Accuracy. Phys. Chem. Chem. Phys. 2005;7:3297–3305. doi: 10.1039/b508541a. [DOI] [PubMed] [Google Scholar]
- 45.Rappoport D, Furche F. Property-Optimized Gaussian Basis Sets for Molecular Response Calculations. J. Chem. Phys. 2010;133:134105. doi: 10.1063/1.3484283. [DOI] [PubMed] [Google Scholar]
- 46.Armentrout PB, Yang B, Rodgers MT. Metal Cation Dependence of Interactions with Amino Acids: Bond Energies of Rb+ and Cs+ to Met, Phe, Tyr, Trp. J Phys Chem B. 2013;117:3771–3781. doi: 10.1021/jp401366g. [DOI] [PubMed] [Google Scholar]
- 47.Leininger T, Nicklass A, Kuchle W, Stoll H, Dolg M, Bergner A. The Accuracy of the Pseudopotential Approximation: Non-Frozen-Core Effects for Spectroscopic Constants of Alkali Fluorides XF (X=K, Rb, Ca) Chem. Phys. Lett. 1996;255:274–280. [Google Scholar]
- 48.Feller D. The Role of Databases in Support of Computational Chemistry Calculations. J. Comput. Chem. 1996;17:1571–1586. [Google Scholar]
- 49.Schuchardt KL, Didier BT, Elsethagen T, Sun LS, Gurumoorthi V, Chase J, Li J, Windus TL. Basis Set Exchange: A Community Database for Computational Sciences. J Chem Inf Model. 2007;47:1045–1052. doi: 10.1021/ci600510j. [DOI] [PubMed] [Google Scholar]
- 50.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: The Biomolecular Simulation Program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo Y, Roux B. Simulation of Osmotic Pressure in Concentrated Aqueous Salt Solutions. J Phys Chem Lett. 2010;1:183–189. [Google Scholar]
- 52.McQuarrie DA, Simon JD. Physical Chemistry: A Molecular Approach. Sausalito, California: University Science Books; 1997. [Google Scholar]
- 53.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 54.Darden TA, York D, Pedersen LG. Particle Mesh Ewald: An Nlog(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 55.Miyamoto S, Kollman P. SETTLE: An Analytical Version of the SHAKE and RATTLE Algorithm for Rigid Water Models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 56.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 58.Lamoureux G, Harder E, Vorobyov IV, Roux B, MacKerell AD., Jr A Polarizable Model of Water for Molecular Dynamics Simulations of Biomolecules. Chem. Phys. Lett. 2006;418:245–249. [Google Scholar]
- 59.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of N-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 60.Allen MP, Tildesley DJ. Computer simulation of liquids. Oxford: Oxford University Press; 1987. [Google Scholar]
- 61.Chowdhary J, Harder E, Lopes PE, Huang L, MacKerell AD, Jr, Roux B. A Polarizable Force Field of Dipalmitoylphosphatidylcholine Based on the Classical Drude Model for Molecular Dynamics Simulations of Lipids. J. Phys. Chem. B. 2013;117:9142–9160. doi: 10.1021/jp402860e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kohlbacher O, Lenhof HP. BALL - Rapid Software Prototyping in Computational Molecular Biology. Bioinformatics. 2000;16:815–824. doi: 10.1093/bioinformatics/16.9.815. [DOI] [PubMed] [Google Scholar]
- 63.Stone AJ. The Theory of Intermolecular Forces. Oxford University Press; 1997. [Google Scholar]
- 64.Baker CM, Lopes PEM, Zhu X, Roux B, MacKerell AD., Jr Accurate Calculation of Hydration Free Energies using Pair-Specific Lennard-Jones Parameters in the CHARMM Drude Polarizable Force Field. J. Chem. Theory Comput. 2010;6:1181–1198. doi: 10.1021/ct9005773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McMillan WG, Mayer JE. The Statistical Thermodynamics of Multicomponent Systems. J. Chem. Phys. 1945;13:276–305. [Google Scholar]
- 66.Robinson RA, Stokes RH. Electrolyte Solutions. Dover Publications, Inc; 2002. [Google Scholar]
- 67.Tamaki K, Suga K, Tanihara E. Solution Properties of Dialkyl-Phosphate Salts - Apparent Molar Volumes, Viscosity B-Coefficients, Heats of Solution, And Osmotic Coefficients. Bull. Chem. Soc. Jpn. 1987;60:1225–1229. [Google Scholar]
- 68.Kowalczyk SW, Wells DB, Aksimentiev A, Dekker C. Slowing down DNA Translocation through a Nanopore in Lithium Chloride. Nano Lett. 2012;12:1038–1044. doi: 10.1021/nl204273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lemkul JA, Savelyev A, MacKerell AD., Jr Induced Polarization Influences the Fundamental Forces in DNA Base Flipping. J Phys Chem Lett. 2014;5:2077–2083. doi: 10.1021/jz5009517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.