Abstract
Improved therapies for the treatment of Trypanosoma brucei (T. brucei), the etiological agent of the neglected tropical disease human African trypanosomiasis, are urgently needed. We targeted T. brucei methionyl-tRNA synthetase (MetRS), an aminoacyl-tRNA synthase (aaRS), which is considered an important drug target due to its role in protein synthesis, cell survival and its significant differences in structure from its mammalian ortholog. Previous work using RNA interference of MetRS demonstrated growth inhibition of T. brucei, further validating it as an attractive target. We report the development and implementation of two orthogonal high throughput screening assays to identify inhibitors of T. brucei MetRS. First, a chemiluminescence assay was implemented in 1536-well plate format and used to monitor ATP depletion during the aminoacylation reaction. Hit confirmation then used a counterscreen in which AMP production was assessed using fluorescence polarization technology. In addition, a miniaturized cell viability assay was used to triage cytotoxic compounds. Finally, lower throughput assays involving whole parasite growth inhibition of both human and parasite MetRS were used to analyze compound selectivity and efficacy. The outcome of this HTS campaign has led to the discovery of nineteen potent and selective T. brucei MetRS inhibitors.
Keywords: aminoacyl-tRNA synthetases, high-throughput screening, human African trypanosomiasis, orthogonal screening, Trypanosoma brucei
Introduction
Aminoacyl-tRNA synthases (aaRS) are essential in protein synthesis, catalyzing the synthesis of aminoacyl-tRNAs. The catalyzed reaction of aaRS usually proceeds via two steps: first the recognition of a specific amino acid and ATP to form the aminoacyl-adenylate intermediate and secondly, the recognition of associated tRNA and the transfer of the aminoacyl group to the terminal adenosine of the tRNA. Inhibition of either step results in disruption of protein biosynthesis, attenuating cell growth and compromising its viability. aaRS are thus excellent drug targets for inhibition, delivering selective cytotoxicity vs. pathogens. This has been demonstrated by the development of the drug mupirocin, which selectively inhibits IleRS from archaea and eubacteria but, does not inhibit eukaryotic IleRS.1 Other aaRSs have been targeted in high throughput screening (HTS) efforts, such as PheRS and MetRS from S. aureus.2 Herein we describe a HTS program targeting MetRS from the protozoan parasite Trypanosoma brucei, including an orthogonal counterscreen.
T. brucei is the causative agent of the neglected tropical disease human African trypanosomiasis (HAT) which threatens ~60 million people in sub-Saharan Africa.3 When left untreated HAT is fatal and current treatment outcomes are unsatisfactory due to high toxicity, difficult administration regimes and poor efficacy of existing drugs.4 Hence, new drugs that are effective, non-toxic and affordable are needed. Early studies using RNAi demonstrated that MetRS from T. brucei is essential for parasite survival, making MetRS inhibition an attractive strategy for treating HAT.5,6,7 Potent aminoquinolone-based compounds, like 2-((3-((3,5-dichlorobenzyl)amino)propyl)amino)quinolin-4(1H)-one (CID 18353708, Figure 2), have been identified but present poor bioavailability.8 Moderately potent urea-based compounds and oxazolone–dipeptides have been reported.9,10,11 The goal of the HTS campaign presented here is to identify selective, potent and drug like small molecule T. brucei MetRS inhibitors, with the ultimate potential to become novel therapeutics for HAT.
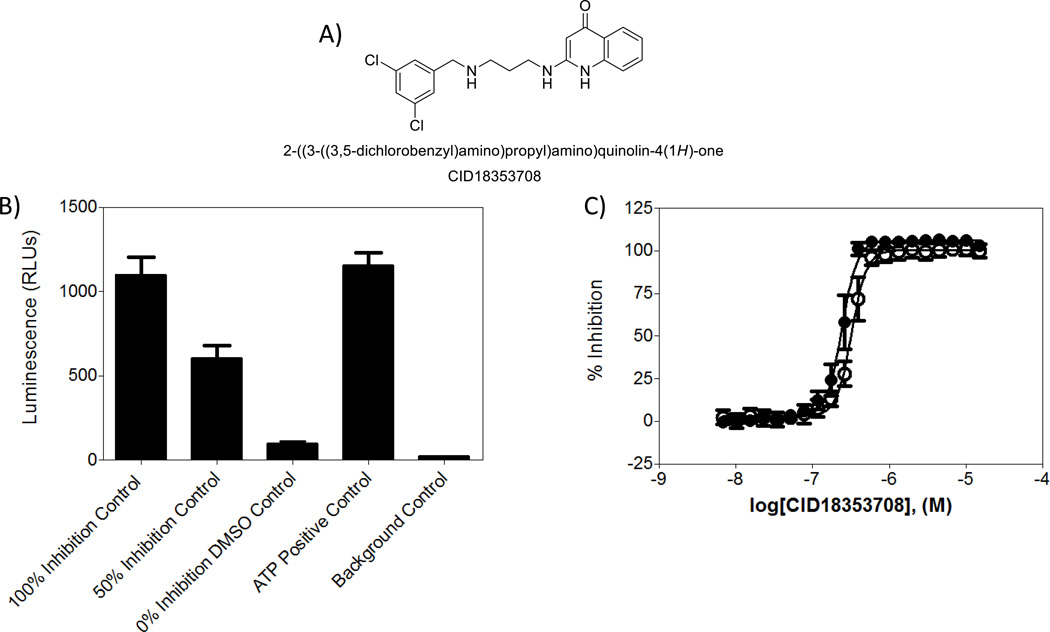
Figure 2. Assay controls.
A) Compound CID 18353708 used as both assay and pharmacological control. B) Average raw luminescence units of controls used during primary screen to assess assay quality. 100% inhibition control wells contain compound CID 18353708 at 1.5µM. 50% inhibition wells contain CID 18353708 at a concentration corresponding to the IC50 0% inhibition control wells contain assay components and DMSO, ATP positive control correspond to wells not containing L-methionine, and background control refers to wells containing no ATP. C) CRC results of the reference control CID 18353708 in orthogonal assays. (●) Luminescence curve (○) fluorescence polarization assay curve. Reference compound was tested as a 20-point, 2:1 serial dilution starting at 15µM.
To minimize false leads, a key aspect of our effort was to use two orthogonal screens early in the program, using different assay technologies, to identify novel inhibitors of the MetRS catalyzed reaction in vitro. This reaction is monitored by measuring the amount of residual ATP or AMP from the MetRS reaction via luminescence and fluorescence polarization (FP), respectively. Excellent performance of both assays in 1536-well plate format was achieved throughout the screen of the MLSMR collection of 364,131 compounds. More than a dozen tractable compounds with sub micromolar potency were identified using these assays.
Materials and Methods
Protein expression and purification
The full-length gene for T. brucei MetRS (Tb927.10.1500) was amplified from T. brucei brucei strain 427, expressed in E. coli, and purified as previously described.8 The N-terminal 6-His tag was retained. The human mitochondrial MetRS enzyme was expressed, as previously reported, with a plasmid provided by Dr. L. Spremulli.12
Optimization and Miniaturization of MetRS inhibition assays
The assay for high-throughput screening required a robust, non-radiometric methodology. An ATP depletion assay based on a chemiluminescence was developed for the initial screen.13,14 Optimization with the Kinase-Glo® reagent (Promega) was initially done in 96-well format using buffer conditions previously utilized for the radiometric aminoacylation assay:8 10 mM MgCl2, 25 mM HEPES-KOH pH 7.9, 50 mM KCl, 2.5 mM dithiothreitol, 0.1 mg/mL bovine serum albumin, 0.2 mM spermine, and 10 U/mL pyrophosphatase. To achieve separation between the high controls (wells not containing cold L-methionine and inhibitors) and the low controls (wells not containing inhibitors) different variables (incubation time, substrate concentration, and enzyme concentration) were manipulated independently while all other conditions were held constant. Optimized substrate concentrations were as follows: L-methionine 32 µM, bulk E. coli tRNA (Sigma-Aldrich) 200 µg/mL, and ATP 100 nM. The L-methionine is in the range of the reported Km of 54±42 µM15 whereas the concentration of ATP is lower than the Km reported for related MetRS enzyme of E. coli (20 µM).12 It was necessary to use low concentration of ATP to achieve optimal separation between the high and low controls. Once the substrate concentrations were established, enzyme reaction progress curves (Supplement Figure 1A) were completed to determine the optimal enzyme concentration and incubation time, 50 nM and 120 minutes respectively. Under these conditions the amount of ATP consumed was around 85% and resulted in excellent assay reproducibility and statistics with an average Z’-factor of 0.84 (Supplement Figure 1B). DMSO tolerance was also tested and no significant decrease in ATP consumption was observed up to 2% DMSO. For the HTS, pyrophosphatase was decreased from 10 to 0.1 U/mL with no significant decrease in ATP consumption. The assay was miniaturized to 384-well format at the Scripps Research Institute (La Jolla, CA) and a pilot screen of 5,120 Maybridge compounds tested at 1 µM was completed. The pilot screen had excellent Z’-factors of 0.75 – 0.89. Finally, the assay was further miniaturized to 1536-well format as discussed in the results section.
A stepwise protocol is depicted in Supplement Table 1. During the primary luminescence-based screen, test compounds were analyzed in singlicate at a final nominal concentration of 12 µM using the automated Kalypsys/GNF platform (GNF Systems, San Diego, CA) at the Scripps Research Institute Molecular Screening Center (SRIMSC, Jupiter, FL). The cutoff used to qualify active compounds was calculated as the average percent inhibition of all the compounds tested plus three times their standard deviation.16 The confirmation assay and the orthogonal FP-based counterscreen tested each compound in triplicate at a single concentration (~12 µM). The primary assay hit-cutoff was applied to the confirmation assay. The counterscreen employed the same algorithm to determine its hit cut off. Actives were progressed forward to determine their concentration response curves (CRC) (Table 1). CRC experiments were also performed as above, testing the compounds in triplicate as 10-point serial dilution starting at 120 µM. ATP levels were measured via luminescence using Kinase-Glo® detection reagent (Promega) on a ViewLux microplate reader (PerkinElmer). In the orthogonal assay, AMP levels were measured by FP using Transcreener® AMP2/GMP2 reagent (BellBrook Labs) on an EnVision microplate reader (PerkinElmer) using a Cy5 FP filter set and a Cy5 dichroic mirror (excitation = 620 nm, emission = 688 nm). FP was read using 50 flashes per well. Further details for each of these assays can be found at the PubChem website using the unique assay identifiers listed in Table 1.
Table 1.
Ultra-High-Throughput Screening Campaign Summary and Results
| Assay Statistics | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Step | Screen type | Target | Number of compounds tested |
Number of replicates/ concentrations |
Selection Criteria |
Number of selected compounds (Hits) |
*PubChem AID |
Z’ | S/B | |
| 1 | Primary | MetRS | 364,131 | 1/1 | %inh> 5.61% | 1,456 | 624268 | 0.90 ± 0.03 | 16.33 ± 13.13 | |
| 2a | Confirmation | MetRS | 1,370 | 3/1 | %inh> 5.61% | 1,270 (522, 266) | 624412 | 0.92 ± 0.02 | 7.10 ± 0.20 | |
| 2b | Cytotoxicity | Cell growth | 1,370 | 3/1 | %inh> 14.97% 161 | 624413 | 0.83 ± 0.02 | 19.79 ± 1.44 | ||
| 2c | Orthogonal | MetRS | 1,370 | 3/1 | %inh> 16.67% | 599 (522) | 651607 | 0.70 ± 0.02 | 1.86 ± 0.02 | |
| 3a | Titration | MetRS | 249 | 3/10 | IC50<10µM | 137 (54) | 651971 | 0.90 ± 0.01 | 5.82 ± 0.18 | |
| 3b | Cytotoxicity | Cell growth | 249 | 3/10 | IC50<10µM | 2 (54) | 651972 | 0.71 ± 0.05 | 23.73 ± 0.92 | |
| 3c | Orthogonal | MetRS | 249 | 3/10 | IC50<10µM | 101 (54) | 651989 | 0.60 ± 0.04 | 1.61 ± 0.05 | |
PubChem AID = the unique assay identifier for use at http://pubchem.ncbi.nlm.nih.gov/
Cytotoxicity counterscreen
Jurkat cells (clone E6.1, ATCC- TIB-152) were cultured in suspension with growth media containing RPMI-1640, 10% dialyzed fetal bovine serum, 0.1 mM NEAA, 1 mM sodium pyruvate, 25 mM HEPES, 5 mM L-glutamine and 1× Anti-Anti (Life Technologies, Carlsbad, CA.) at 37°C in 95% relative humidity and 5% CO2. Five hundred cells per well were added into 1536-well plates and incubated with compounds or DMSO alone for 18hrs at 37°C in 95% relative humidity and 5% CO2, at which time 5µl of CellTiter-Glo (Promega, Madison, WI) were added to all wells. Plates were centrifuged and incubated for 10 minutes at room temperature. Luminescence was measured on a ViewLux microplate reader (PerkinElmer, Waltham, MA). Doxorubicin was used a positive control. For more information, see Table 1 and PubChem AID 364.
Aminoacylation assay (T. brucei and human mitochondrial MetRS)
To confirm the specificity of the hits from the HTS, a follow-up screen was performed using the aminoacylation assay that quantifies incorporation of [3H]L-methionine into tRNA.8 Reactions were performed in 96-well filter plates with Durapore membranes (MSHVN4B10; Millipore, Billerica, MA) in volumes of 75 µl. The reaction buffer was 25 mM HEPES-KOH pH 7.9, 10 mM MgCl2, 50 mM KCl, 0.2 mM spermine, 0.1 mg/ml bovine serum albumin, 2.5 mM dithiothreitol, 0.1 mM ATP, 240 nM [3H]L-methionine (83 Ci/mmol), 2% DMSO, and 0.1 U/ml pyrophosphatase (I1643; Sigma-Aldrich, St. Louis, MO). Recombinant enzyme (10 nM) and test compounds (10 µM or 1 µM) were mixed with the buffer and preincubated for 15 min. To start the reaction, 400 µg/ml bulk Escherichia coli tRNA (R4251; Sigma-Aldrich, St. Louis, MO) was added and the plate was incubated without shaking at room temperature for 120 min. The reactions were stopped by the addition of 100 µl/well cold 10% trichloroacetic acid. The reaction components were separated from tRNA by filtration through a vacuum manifold and washed three times with 300 µl/well cold 10% trichloroacetic acid. The filter plates were dried, 25 µl/well scintillation fluid was added, and the counts on the plates were determined using a scintillation plate counter. Samples were run in triplicate, and the average activity of inhibitors was compared to that in control wells without inhibitors. Percent inhibition was calculated for the two concentrations (10 µM and 1 µM).
As a selectivity counterscreen, hit compounds were assayed against the human mitochondrial MetRS enzyme using the aminoacylation methodology as described above with a few differences. The assay was performed in a reaction buffer containing 50 mM Tris-HCl pH 8.0, 6 mM MgCl2, 2.5 mM KCl, 0.2 mM spermine, 0.2 mg/ml bovine serum albumin, 2.5 mM dithiothreitol, 2.5 mM ATP, 240 nM [3H]L-methionine (83 Ci/mmol), 2% DMSO, and 0.1 U/ml pyrophosphatase. Recombinant enzyme (13 nM) and inhibitors (10 µM or 1 µM) were mixed with the buffer and pre-incubated for 15 min. To start the reaction, 200 µg/ml bulk Escherichia coli tRNA was added. The plate was incubated without shaking at room temperature for 60 min and processed as described above.
T. brucei growth inhibition assay
T. brucei (bloodstream form strain 427 from K. Stuart, Seattle Biomedical Research Institute, Seattle, WA) was cultured in HMI-9 medium17 containing 10% fetal bovine serum, penicillin, and streptomycin at 37°C with 5% CO2. Drug sensitivity of the T. brucei strain was determined in 96-well microtiter plates in triplicate with an initial inoculum of 1×104 trypomastigotes per well. Compound stock solutions were prepared in DMSO at 20 mM and/or 10 mM and added in serial dilutions for a final volume of 200 µl/well. Parasite growth was quantified at 48 h by the addition of AlamarBlue (Alamar Biosciences, Sacramento, CA).18 Pentamidine isethionate (Aventis, Dagenham, United Kingdom) was included in each assay as a positive control. Compounds were tested in triplicate using a 4-point serial three fold dilution system starting at a test concentration of 10 µM. Inhibition at various concentrations were determined and the EC50 value were calculated by the online service CDD (www.collaborativedrug.com) using an equation to describe the sigmoidal dose-response curve.
Screening data acquisition, normalization, representation and analysis
Raw luminescence data or FP values were uploaded into the SRIMSC institutional HTS database (Symyx Technologies Inc., Santa Clara, CA). Raw luminescence data was imported directly while FP values were calculated using the following equation prior to upload:
Where Raw1 is defined as polarized fluorescence in the P channel and Raw2 is defined as polarized fluorescence in the S channel.
In either case, response data was normalized using the following equation:
where High Control represents wells from the same plate containing 1.5 µM control compound CID 18353708, or in the case of the cytotoxicity assay, 8 µM doxorubicin, and Low Control represents wells treated with DMSO only.
Data was normalized on a per plate basis and each assay plate underwent a quality control check; a Z′ value greater than 0.5 was required for acceptance of data.19 CRC data were analyzed by plotting the average percent inhibition of the triplicate results against the compound concentration. A four-parameter equation describing a sigmoidal concentration-response curve was then fitted with adjustable baseline using Assay Explorer software (Symyx). Similarly, for the graphs shown in this manuscript, CRCs and pIC50 values were generated by Prism (GraphPad Software, San Diego, CA). All further results from the screens can be found at NIH’s PubChem website (http://pubchem.ncbi.nlm.nih.gov/) utilizing the AID listed in Table 1.
Cheminformatics
Shared scaffolds of active compound families from the confirmation screen and CRC experiments were identified using a Maximum Common Substructure hierarchical clustering (ChemAxon LibraryMCS 5.10.2). The physical properties (i.e. molecular mass, topological polar surface area, chiral atoms, H-bond acceptors/donors, ring count and rotatable bonds) of the compounds tested in dose response format were calculated (ChemAxon Instant JChem 6.2.2).
Chemicals
The MLSMR library was provided by BioFocus DPI (South San Francisco, CA) through the NIH’s Roadmap Molecular Libraries Initiative. Details regarding compound selection for this library can be found online at http://mli.nih.gov/mli/compound-repository/mlsmr-compounds/. Briefly, this library is a highly diversified collection of small molecules (more than 50% in the molecular weight range 350–410 g/mol) and is comprised of both synthetic and natural products, from either commercial or academic sources, that can be grouped into the 4 following categories: (1) specialty sets of known bioactive compounds such as drugs and toxins (0.65%), (2) focused libraries aimed at specific target classes (2.85%), (3) non-commercial sources (7.4%) and (4) diversity sets covering a large area of the chemical space (89.1%).
The positive control compound CID18353708 was synthesized as described elsewhere.8
Results
Implementation of the orthogonal HTS assays
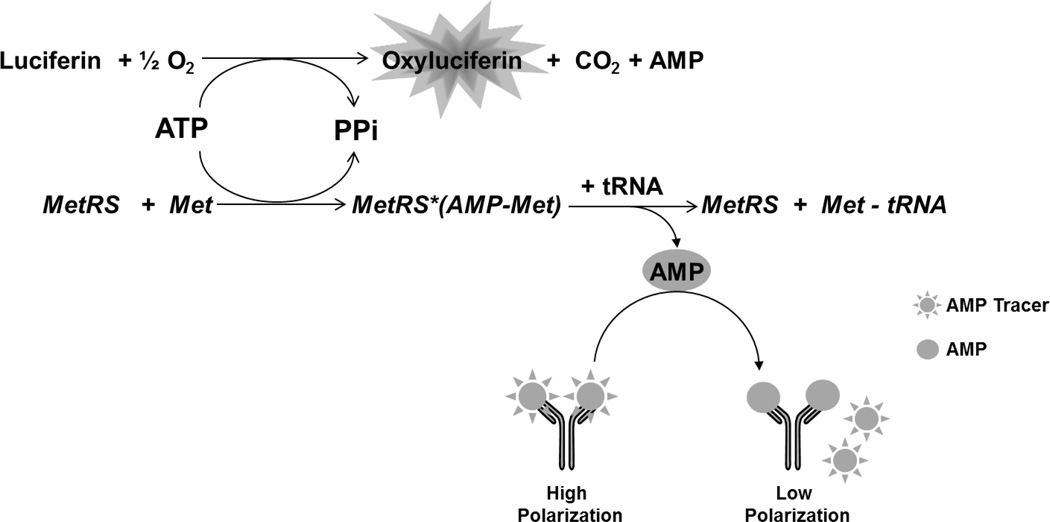
A luminescence based assay was designed and run as a primary screen to identify inhibitors of T. brucei MetRS by measuring the ATP depletion during the aminoacylation reaction. Residual ATP was measured through the luciferase catalyzed reaction of the mono-oxygenation of luciferin substrate (Kinase-Glo®) in which the luminescence signal is inversely proportional to MetRS activity (Figure 1).
Figure 1.
T. brucei MetRS catalyzed reaction. The primary screen assay focuses on the luminescence signal generated from the luciferase catalyzed reaction using residual ATP. The orthogonal counterscreen focuses on the fluorescence polarization signal generated from the displacement of a labeled AMP molecule from a selective antibody when AMP is generated from the T. brucei MetRS catalyzed reaction.
To reduce the cost and increase the throughput of the HTS campaign, the assay was miniaturized and implemented in 1536-well plate format. Optimization of the assay was performed by assessing the effect of critical variables that affect the aminoacylation reaction; (i.e.; MetRS concentration, ATP concentration, and DMSO tolerance; see Supplement). Three sets of controls, N=24 per set, were placed on every assay plate: the high control (1.5 µM CID18353708 final), the low control containing DMSO (0.9% final) and a 50% inhibition control containing CID18353708 at its IC50 concentration (40 nM). These controls were used to (1) ensure T. brucei MetRS activity, (2) normalize the data, and (3) monitor the data quality by measuring Z’ and S:B. Two additional controls were placed on every plate: a positive control containing ATP and tRNA but no L-methionine, and also a background control without ATP to ensure proper compound and reagent dispensing. Good separation of the controls was observed as shown in the plot of the raw luminescence values measured during the primary screen (Figure 2). All primary HTS data was normalized to the inhibition of compound CID18353708 vs. the DMSO only wells, and was used to produce a scatterplot to aid in visualization of the activity across the HTS campaign (Figure 3). Notably the positive control compound has been formerly shown to produce sub-micromolar inhibition of T. brucei MetRS.18 Under the final conditions listed in Supplement Table 1, excellent assay performance was observed with an average Z’ factor of 0.93 ± 0.02 and a pIC50=7.62±0.05 (IC50=24.20 ± 2.74 nM) with a Hill slope of 4.41 ± 0.56 for CID18353708 (N=11).
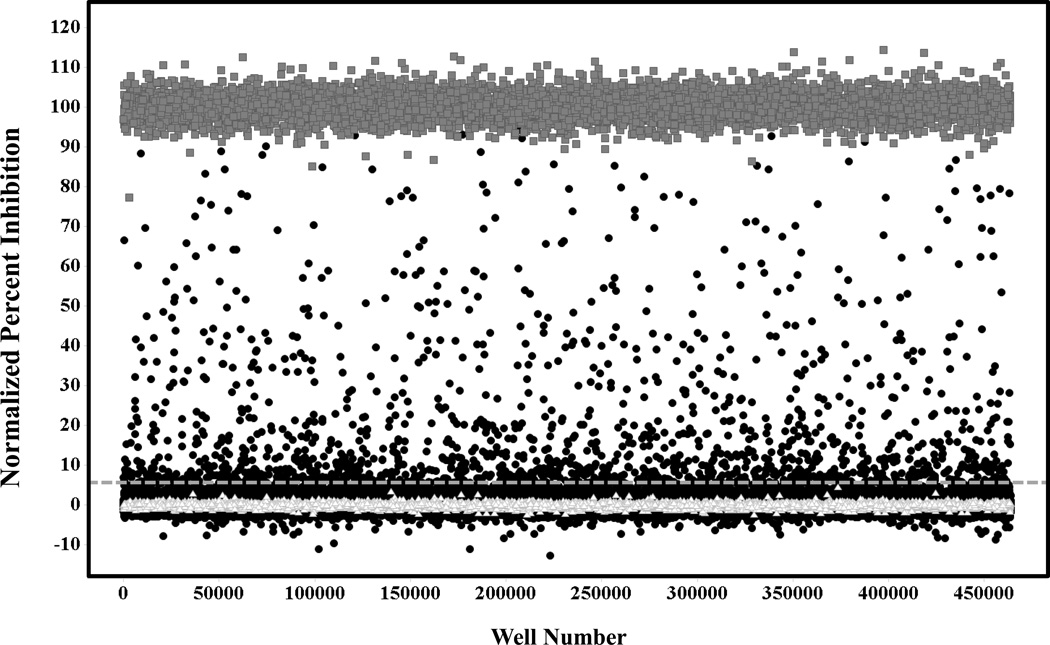
Figure 3. Primary screen scatter plot.
(∆) Represent low control wells containing DMSO only, (●) represent data wells containing compounds and (■) represent high control wells containing compound CID18353708. The dashed line represents the hit cut-off. Compounds with inhibition above this line were considered primary actives or hits.
The orthogonal FP counterscreen was implemented to help prioritize hit selection by removing potential artifacts from the luminescent detection system (quenchers, luciferase inhibitors, etc.). This assay measures AMP production in the T. brucei MetRS catalyzed aminoacylation reaction as a function that is proportional to MetRS activity. Optimal assay conditions were in accord with the manufacturer’s recommendations (BellBrook Labs, Madison, WI) and all subsequent experiments were performed using the conditions listed in Supplement Table 1. In miniaturized format, the Z’ factor was 0.71 ± 0.02 and a pIC50=7.52±0.07 (IC50 = 30.34 ± 4.81) nM with a Hill slope of 4.34 ± 0.17 for CID18353708 (N=3). Inhibition of MetRS by CID18353708 was nearly identical in both assays, an indication of good correlation between the sensitivity of the two assay formats (Figure 2).
T. brucei MetRS inhibition screening campaign results
The T. brucei MetRS inhibition screening campaign is summarized in Table 1. A total of 364,131 distinct samples from the MLSMR were tested during the primary screen at a nominal concentration of 12 µM. While the Z’ data indicate robustness at each step throughout the HTS campaign, we did observe a high degree of variation within the signal to basal ratio (S/B) for the MetRS luminescence assay. This variability between batches was due to basal signal levels approaching zero in terms of raw data relative light units. Hence, even a very small shift in the raw data, resulted in a fairly significant shift in the normalized S/B. From the primary screen 1,456 compounds (i.e. 0.4% of the total library) were identified as actives, exhibiting a percent inhibition greater than the nominal hit cutoff of 5.61% (average background + 3SD). Of these, 1,370 were available from the MLSMR for testing at the confirmation and counterscreen stages.
In order to confirm inhibitory activity, to eliminate luminescent artifacts, and to eliminate broadly cytotoxic compounds, the 1,370 samples were retested in triplicate in three different assays: (1) the luminescence MetRS ATP depletion assay, (2) the fluorescence polarization AMP production assay, and (3) the luminescence-based Jurkat cell cytotoxicity assay. The 161 compounds found to be active in the cytotoxicity assay were eliminated from further follow-up. Non-cytotoxic compounds with activity in both the ATP depletion and AMP formation assays were prioritized for CRC studies (522 compounds). To facilitate compound selection for titration studies, the 522 compounds were classified into 86 clusters based on their most common substructure. The compounds for CRC studies were selected based on two main criteria: (1) the most potent compounds from each cluster were selected to ensure chemical diversity and (2) all compounds that exhibited a 1:1 ± 0.6 correlation between the IC50s of the two target-based assays were selected to proceed. Under these criteria, 266 compounds were selected for IC50 titration, of which 249 compounds were available from the MLSMR.
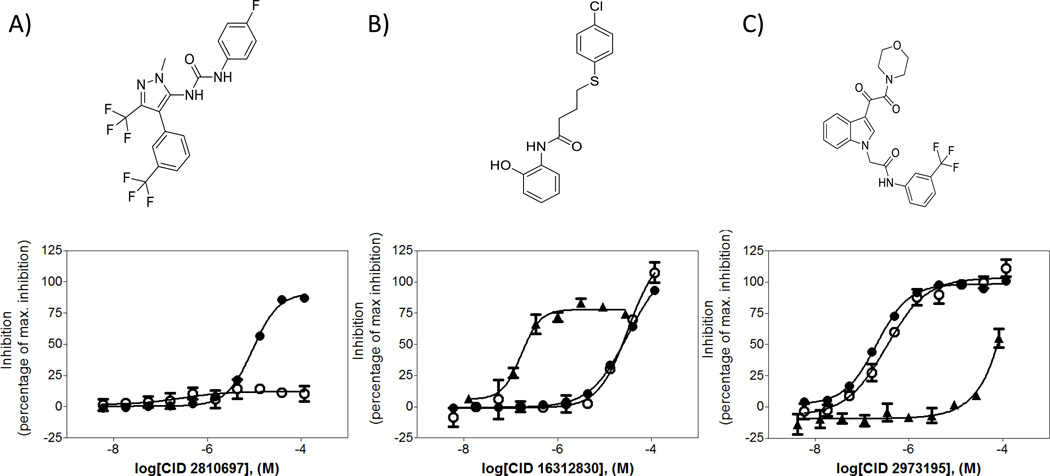
The CRCs for the 249 compounds were determined via testing in triplicate 10 point three fold serial dilution format against the same three assays. Individual CRCs offered better insight into the selectivity of the compounds and their inhibition profiles between the different assays, facilitating the triage of cytotoxic compounds such as CID16312830 and luminescent artifacts such as CID2810697 (Figure 4 and Supplement Table 2). From these results, compounds, were selected for further follow-up based on three criteria: (1) IC50 below 10 µM in both luminescence and FP assays, (2) overlapping dose response curves in luminescence and FP assays (maximum percent response within 10 percent of each other) and (3) a relatively high cytotoxicity index (i.e.; Jurkat cytotoxicity assay resulting cytotoxic concentration (CC50) greater than 10 times the IC50 in luminescence assay). A total of 54 compounds met these criteria and were subjected to further characterization, including secondary assays and structure activity relationship (SAR) studies (Supplement Table 2).
Figure 4. Compound dose response curves.
(●) Luminescence curve (○) fluorescence polarization assay curve (▲) cytotoxicity assay curve. A) Compound CID 2810697 flagged as a luminescent artifact due to inactivity in fluorescence polarization assay. B) Compound CID 16312830 flagged as cytotoxic compound due to activity in the Jurkat cytotoxicity assay. C) Compound CID 2973195 identified as a potent hit due to high potency against T. brucei MetRS in both assay modes and low cytotoxic effect against Jurkat cells.
Low Throughput Secondary assays
The 54 confirmed active compounds were tested for activity in an aminoacylation assay vs. T. brucei cells grown in culture. The data are shown in Supplement Table 2 with compounds ranked by pIC50 in the luminescence dose-response assay done in 1536-well format. Nineteen of these compounds had sub-micromolar IC50 values in this ATP depletion assay with the most potent compound (CID 24793614) having an IC50 of 44 nM. Results from the orthogonal FP assay showed strong correlation between the luminescent assay with an R2=0.895. The activity of these compounds was determined in a low-throughput aminoacylation assay that measures incorporation of [3H]L-methionine into tRNA. All compounds demonstrated enzyme inhibition in this secondary assay with only a single compound (CID 4656759) having a somewhat discordant result (28% inhibition at 10 M compared to the IC50 value of 2.2 M from the ATP depletion assay).
To assess parasite vs. human selectivity, the 54 confirmed active compounds were assayed against the human mitochondrial MetRS using the aminoacylation methodology. Forty four of the compounds (81%) showed weak or no inhibition of the human mitochondrial MetRS (i.e.; <20% at 10 M). Five compounds had >30% inhibition against the human enzyme, indicating that the screen was successful in identifying numerous active compounds that are specific for the trypanosome over the mammalian MetRS. Only one compound (CID 46902334) was moderately potent towards the human mitochondrial MetRS (80.3% inhibition at 10 M, 14.3% inhibition at 1 M). As noted earlier, one of the selection criteria for compound progression was low cytotoxicity; a CC50 against Jurkat cells of >10 M. Supplement Table 2, shows Jurkat CC50 data; the majority of compounds (66.7%) had no inhibitory activity even at the highest dilution (83.3 M) and only a single compound (CID 3718852) had a CC50 <20 M.
Finally, the 54 compounds were tested for growth inhibition activity (EC50) of T. brucei cultures. Twelve compounds (22.2%) showed some degree of inhibition at 10 M. The most potent compounds had apparent EC50s in the 2 M range. These compounds are screening hits that can penetrate cells well, an important criterion in drug discovery efforts.
Discussion
Orthogonal screens were successfully developed and implemented to identify potent small molecule inhibitors of T. brucei MetRS, using ATP and AMP levels as reporters of MetRS activity via luminescence and fluorescence polarization detection methods, respectively. Both assays technologies demonstrated robust performance on the Kalypsys GNF µHTS platform. As noted, batch to batch variability was observed for the overall signal to basal ratio (S/B) of the primary screen. This finding was of little concern given that (i) the data was normalized on a plate per plate basis, (ii) the relative shift in the raw data was minor yet resulted in substantial shifts in S/B and (iii) the pharmacological control behaved well, giving the expected IC50. We maintained Z’ values > 0.8 and an average throughput of approximately 15,000 compounds per hour throughout the HTS campaign (Supplement Figure 4). The use of a substrate (ATP) depletion assay in the primary screen necessitated high fractional conversion of ~85% of the substrate for a robust signal (Supplement Figure 1A). A high fractional conversion of substrate has been associated with lower sensitivity to identifying inhibitors.20 Notably, with the enzyme and the ATP concentration being nearly the same, 50 nM and 100nM respectively, the assay is being run under conditions enabling only limited turnover which, may minimize the sensitivity to slow binding inhibitors. Assay operating conditions for HTS also predicated working at substrate concentrations significantly lower than Km; a condition that prevents the identification of ATP uncompetitive inhibitors. However, given the large number of confirmed hits (1,270) identified in our initial high-throughput screen, we felt that the assay methodology was more than sufficient to provide ample numbers of hits to move forward with the downstream analysis.
Use of an orthogonal FP-based counterscreen to eliminate detection format-specific hits (such as CID2810697) eliminated nuisance compounds early in the process and allowed a high confirmation rate in subsequent secondary assays. In addition, cytotoxic compounds (i.e.; CID16312830) were eliminated from advancement based upon results in the Jurkat assay. This battery of complementary assays proved to be an effective way to quickly identify inhibitors of aaRS. Initial SAR analysis for CID2973195, the indole oxalamide shown in Figure 4C, demonstrated excellent “out of the box” potency in the primary and FP assays and relatively no cytotoxicity; pIC50 values in respective order are: 6.77, 6.53, <4.08 (IC50 values of 171 nM, 292 nM, and >83 µM respectively).
Virtually all compounds selected for low-throughput screening (52/54) were confirmed to be active in the T. brucei aminoacylation assay. This methodology measures the whole MetRS enzyme reaction by quantifying the incorporation of [3H]L-methionine into tRNA. These confirmatory results were reassuring and showed that the HTS assay cascade used was effective.
Since human mitochondrial MetRS is a potential off target for these hits, we tested the same set of compounds against this human mitochondrial enzyme. It should be noted that human cells also contain a cytoplasmic MetRS, but the homology to the T. brucei MetRS in the active site is relatively low (46% identical residues), whereas the homology of the human mitochondrial MetRS to the active site of T. brucei MetRS is higher (79% identical residues). The human selectivity screen showed that 81% percent of compounds had ≤20% inhibition of the human mitochondrial MetRS, meaning that if developed as drugs, relatively few compounds are likely to cause host toxicity, inhibiting this human enzyme in HAT patients. This selectivity in the hit set was likely enhanced by earlier implementation of the Jurkat cytotoxicity screen. It can be concluded that the differences between the human mitochondrial and trypanosomal MetRS (21% amino acid sequence difference in the ATP-methionine binding sites) are sufficient to make selective inhibition by small molecules possible. Structural studies of the compounds bound to the T. brucei MetRS may aid understanding and guide further enhancements in potency and selectivity of the hit compounds.
Finally, 12 of the 54 HTS hit compounds (22%) inhibited growth of T. brucei grown in culture. It is assumed that in each case the growth inhibition observed is due to inhibition of the MetRS target, although further work will be necessary to confirm this hypothesis. We have shown in previous work that aminoquinolone MetRS inhibitors in fact block protein synthesis in cultured T. brucei cells.10 It is likely that many of 42 compounds inactive vs. cultured T. brucei have poor membrane permeability and thus do not reach the target. Also, the weaker MetRS inhibitors may not sufficiently bind the target to affect growth at the concentrations tested, even if they enter the cells.
The screening approach used during this campaign exhibited great robustness and the ability to rapidly identify and triage potent, non-cytotoxic compounds. In combination with secondary assays, several promising selective compounds (with pIC50 values as high as 7.36 or IC50 as low as 44 nM) were identified and will be pursued in future studies for further development of as novel potent molecular probes, perhaps ultimately leading to a drug to treat HAT. This orthogonal screening approach, using in an automated platform, has proven to be an excellent tactic to screen large compound libraries that can certainly be extended to other aaRSs.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health’s Roadmap Initiative through grants U54MH084512 (LPR, CE, FM, SB, JL, TB, LS, TS, PH), PO1AI067921 [Medical Structural Genomics of Pathogenic Protozoa (MSGPP)] and RO1AI084004 to WGJH, and RO1 AI097177 to FB and EF). We thank Pierre Baillargeon and Lina DeLuca (Lead Identification, Scripps Florida) for compound management and our MSGPP co-workers for contributions to the early stages of this project.
References
- 1.Nakama T, Nureki O, Yokoyama S. Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. The Journal of biological chemistry. 2001;276(50):47387–47393. doi: 10.1074/jbc.M109089200. [DOI] [PubMed] [Google Scholar]
- 2.Payne DJ, Gwynn MN, Holmes DJ, et al. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6(1):29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 3.Hotez PJ, Molyneux DH, Fenwick A, et al. Control of neglected tropical diseases. N Engl J Med. 2007;357(10):1018–1027. doi: 10.1056/NEJMra064142. [DOI] [PubMed] [Google Scholar]
- 4.Brun R, Blum J, Chappuis F, et al. Human African trypanosomiasis. Lancet. 2010;375(9709):148–159. doi: 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- 5.Sheppard K, Yuan J, Hohn MJ, et al. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36(6):1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurdle JG, O'Neill AJ, Chopra I. Prospects for aminoacyl-tRNA synthetase inhibitors as new antimicrobial agents. Antimicrobial agents and chemotherapy. 2005;49(12):4821–4833. doi: 10.1128/AAC.49.12.4821-4833.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raczniak G, Ibba M, Soll D. Genomics-based identification of targets in pathogenic bacteria for potential therapeutic and diagnostic use. Toxicology. 2001;160(1–3):181–189. doi: 10.1016/s0300-483x(00)00454-6. [DOI] [PubMed] [Google Scholar]
- 8.Shibata S, Gillespie JR, Kelley AM, et al. Selective inhibitors of methionyl-tRNA synthetase have potent activity against Trypanosoma brucei Infection in Mice. Antimicrobial agents and chemotherapy. 2011;55(5):1982–1989. doi: 10.1128/AAC.01796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koh CY, Kim JE, Wetzel AB, et al. Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness. PLoS neglected tropical diseases. 2014;8(4):e2775. doi: 10.1371/journal.pntd.0002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibata S, Gillespie JR, Ranade RM, et al. Urea-based inhibitors of Trypanosoma brucei methionyl-tRNA synthetase: selectivity and in vivo characterization. J Med Chem. 2012;55(14):6342–6351. doi: 10.1021/jm300303e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tandon M, Coffen DL, Gallant P, et al. Potent and selective inhibitors of bacterial methionyl tRNA synthetase derived from an oxazolone-dipeptide scaffold. Bioorganic & medicinal chemistry letters. 2004;14(8):1909–1911. doi: 10.1016/j.bmcl.2004.01.094. [DOI] [PubMed] [Google Scholar]
- 12.Spencer AC, Heck A, Takeuchi N, et al. Characterization of the human mitochondrial methionyl-tRNA synthetase. Biochemistry. 2004;43(30):9743–9754. doi: 10.1021/bi049639w. [DOI] [PubMed] [Google Scholar]
- 13.Godeau JM, Charlier J. Adenosine triphosphate consumption by bacterial arginyl-transfer ribonucleic acid synthetases. The Biochemical journal. 1979;179(2):407–412. doi: 10.1042/bj1790407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y, Yu K, Xu B, et al. Potent and selective inhibitors of Staphylococcus epidermidis tryptophanyl-tRNA synthetase. The Journal of antimicrobial chemotherapy. 2007;60(3):502–509. doi: 10.1093/jac/dkm229. [DOI] [PubMed] [Google Scholar]
- 15.Cestari I, Kalidas S, Monnerat S, et al. multiple aminoacyl-tRNA synthetase complex that enhances tRNA-aminoacylation in African trypanosomes. Molecular and cellular biology. 2013;33(24):4872–4888. doi: 10.1128/MCB.00711-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madoux F, Simanski S, Chase P, et al. An ultra-high throughput cell-based screen for wee1 degradation inhibitors. Journal of biomolecular screening. 2010;15(8):907–917. doi: 10.1177/1087057110375848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. The Journal of parasitology. 1989;75(6):985–989. [PubMed] [Google Scholar]
- 18.Koh CY, Kim JE, Shibata S, et al. Distinct states of methionyl-tRNA synthetase indicate inhibitor binding by conformational selection. Structure. 2012;20(10):1681–1691. doi: 10.1016/j.str.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of biomolecular screening. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 20.Wu G, Yuan Y, Hodge CN. Determining appropriate substrate conversion for enzymatic assays in high-throughput screening. Journal of biomolecular screening. 2003;8(6):694–700. doi: 10.1177/1087057103260050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.