Abstract
High-throughput Affinity Purification Mass Spectrometry (AP-MS) experiments can identify a large number of protein interactions but only a fraction of these interactions are biologically relevant. Here, we describe a comprehensive computational strategy to process raw AP-MS data, perform quality controls and prioritize biologically relevant bait-prey pairs in a set of replicated AP-MS experiments with Mass spectrometry interaction STatistics (MiST). The MiST score is a linear combination of prey quantity (abundance), abundance invariability across repeated experiments (reproducibility), and prey uniqueness relative to other baits (specificity); We describe how to run the full MiST analysis pipeline in an R environment and discuss a number of configurable options that allow the lay user to convert any large-scale AP-MS data into an interpretable, biologically relevant protein-protein interaction network.
Keywords: Affinity Purification Mass Spectrometry, Protein Interactions, Scoring Algorithms, Interaction networks, Proteomics
Introduction
Affinity Purification Mass-Spectrometry (AP-MS) is one of the primary methods to discover the protein interactions in an unbiased manner. In recent years, due to advances in bottom-up mass-spectrometry and affinity tagging methods, this method has been applied in high throughput to chart the protein-protein interaction networks or ‘interactomes’ from entire pathways to complete eukaryotic, bacterial and even viral organisms (Arifuzzaman et al., 2006; Jäger et al., 2011; Sowa et al., 2009). A high-throughput dataset containing hundreds of replicated AP-MS samples poses a clear challenge for human processing, but also presents an opportunity to mine the data collectively with computational algorithms. To this end, a number of recent studies, which used AP-MS in a high-throughput fashion, developed computational algorithms that transform such a dataset into a list of bait-prey pairs ranked according their predicted biological significance (Jäger et al., 2011; Sowa et al., 2009; Choi et al., 2011). Knowing that a cellular protein is predicted to have on average 5-8 biologically relevant interactions (Grigoriev, 2003), prioritizing these from over hundreds of proteins and thousands of spectra identified by MS is far from trivial.
To understand the high number of ‘false positives’ identified by AP-MS it helps to categorize interactions into 4 broad classes: (I) biologically relevant interactions (II) specific, non-biologically relevant interactions between proteins from different cellular compartments in lysed cells (III) unspecific interactions with contaminants or highly abundant proteins and (IV) nonexisting interactions caused by residual peptides from previous runs or MS identification errors. Conversely, ‘false negatives’ occur because not every biologically relevant interaction is reproducibly detectable, especially if the protein is not very abundant, has peptides difficult to detect by MS or interacts only transiently (Yu et al., 2009; MacLean et al., 2010). To account for both false positive and false negative errors, high-throughput experimental setups need to be designed with proper controls and a sufficient amount of biological replicates (at least triplicates) and processed with a computation AP-MS scoring algorithm to separate signal from noise (Jäger et al., 2011).
The protocol we present here outlines our ‘best practices’ approach to convert a large data set of replicated AP-MS experiments into a list of bait-prey pairs ranked according to their predicted biological relevance. After installing MiST as described in the support protocol, the data is preprocessed in Basic Protocol 1 to generate a bait-prey matrix that can be subjected to the quality control protocol in Basic Protocol 2. Basic Protocol 3 is then used to calculate a MiST score.
Support Protocol 1: Installation of MiST
This MiST pipeline is implemented in R, an open-source programming language for statistical computing and graphics. Here, we describe how the most recent version of the MiST pipeline can be downloaded from GitHub and installed by any user with access to an online computer.
Necessary Resources
Hardware
Workstation running any current OS, Unix environment recommended
Software
R package (http://www.r-project.org)
R packages: getopt, optparse, reshape2, pheatmap, RcolorBrewer, ggplot2, MESS, yaml
MiST source code (https://github.com/everschueren/MiST)
Git (optional) (http://git-scm.com)
Setting up MiST
- Downloading the MiST source code to your workstation.
- Download the MiST package as a .zip archive from the public GitHub repository by clicking on the “Download ZIP” button on the bottom right, unzip the files and move the directory to a permanent location.
- Alternatively, you can check out the MiST package through Git as follows: git clone https://github.com/everschueren/MiST.git MiST
The MiST pipeline is designed to run from a terminal using R. This requires the user to have executable permissions. To set these permissions in a Unix environment, navigate in the terminal to the MiST directory, hereafter referred to as the $INSTALL_DIR, then type: sudo chmod -R 775 *
Basic Protocol 1: Data pre-processing
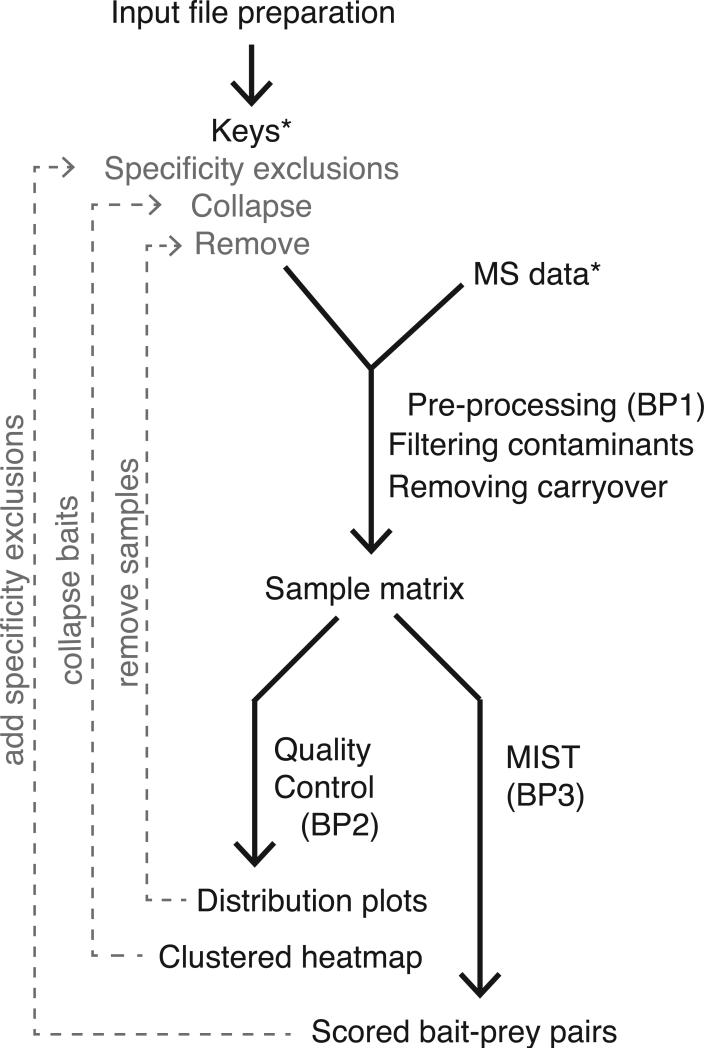
Prior to computing MiST scores, it is required to convert the search results into a format compatible with the MiST algorithm. The MiST pre-processing pipeline was initially designed to work with a Prospector (Clauser et al., 1999) (http://prospector.ucsf.edu) protein report file but virtually any report file that lists uniquely identified proteins, their observed peptide frequencies in tabular format is supported currently. Additionally, we built a number of filtering steps, such as contaminant removal and carryover removal, into the pre-processing and formatting steps. The result of the first basic protocol is a bait-prey matrix that can be subjected to the quality control protocol (Basic Protocol 2) and scored by the MiST protocol (Basic Protocol 3) (See Figure 1).
Figure 1.
Flow chart of the Mass Spectrometry Interaction Statistics (MiST) analysis pipeline. Required input files are marked with (*). Optional steps in the protocol and their corresponding input files are indicated in light gray.
Necessary Resources
Hardware
Workstation running any current OS, Unix environment recommended
Software
MiST pipeline (installed as described in Support Protocol)
Files
Data file and Keys file, see below
Remove file and Collapse file, optional, see below
Data preparation
Prior to running the MiST pre-processing protocol the user needs to have at least the two following files available (See Figure 2A and Table 1):
- Data file (required): the data file should be in tab-delimited format with a descriptive header for each column. The number of features in this file is in principal unconstrained but following are required:
- Sample identifier: a unique identifier per AP-MS run
- Protein identifier: a unique identifier per protein (i.e. Uniprot accession code)
- Observed peptide frequency: a quantitative value per protein, for example (from less to most quantitative):
- Number of unique peptides per protein or
- Summed spectral counts per protein or
- Summed MS1-intensities per protein, preferably log2-transformed
- Protein molecular weight: The molecular weight will be used as a scaling factor to normalize the quantitative value for each protein to its size
- Keys file (required): The goal of the keys file is two-fold: 1) describing which bait and experimental conditions are in the sample with a human readable description and 2) provide a unique name to group biological replicates. This file also needs to be tab-separated with two columns created by the user:
- Sample identifier: a unique identifier per AP-MS experiment that matches a sample identifier in the data file
- Bait name: A unique identifier per bait, grouping all replicates. This can be the bait's Uniprot accession number or a more easily readable name (i.e. gene name).
Remove file (optional): The remove option is a feature we put in place to dynamically exclude entire samples from the scoring process while keeping the original data files intact. Single samples might need to be removed for quality reasons, which we will address later, while an entire range of samples might be excluded to score subsets of the complete data set. This file is formatted as a single column, consisting of all sample identifiers that need to be excluded (one sample per line).
Collapse file (optional): The collapse option serves to merge samples belonging to different baits into a single group while keeping the original data files intact. This is useful when a particular experimental condition (i.e. compound addition, bait mutation, different affinity tags, differently tagged termini, organelle extraction) shows no perceivable difference to the wild type experiment. In this case it might be desirable to treat these samples as additional replicates of the wild type bait to improve reproducibility estimates. In the following section we will discuss how you can use a clustered heat map to reveal such patterns. The collapse is formatted as tab-separated entries: one for the original bait name, corresponding to an entry in the keys file, followed by the new name or composite group name.
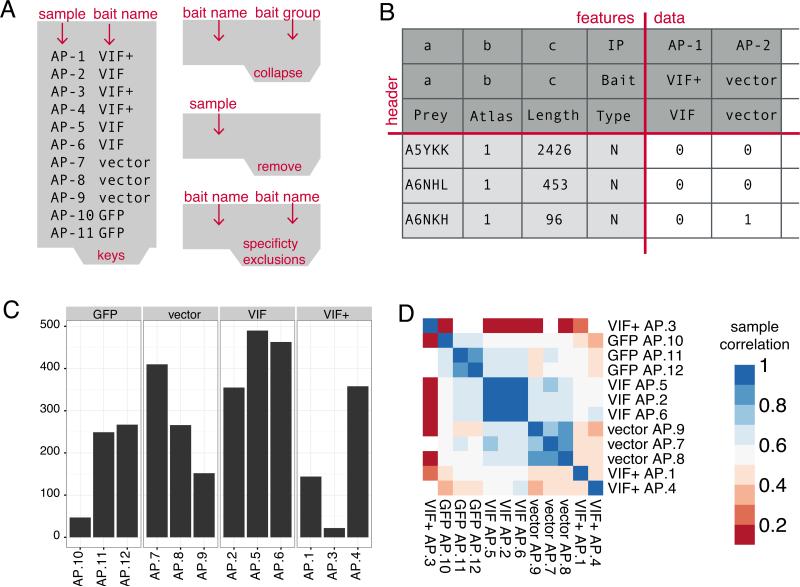
Figure 2.
A) Tab-separated input files required to run a first pass of MiST B) Output bait-prey matrix after the pre-processing protocol (1) that serves as input for the MiST protocol (3) C) Output of the quality control protocol depicting the number of detected proteins across replicates for a single bait. Samples AP-10 and AP-3 are candidates for removal from the data set D) Output of the quality control protocol depicting a hierarchically clustered heatmap of the pairwise sample correlation matrix. The signal from sample AP-3 is clearly uncorrelated with the VIF+ replicates while hIP34-15 correlation with the GFP group is acceptable.
Table 1.
configurable parameters for file input/output
| name | type | description |
|---|---|---|
| data | string | path to the data file with the identified and quantified proteins (see data preparation) |
| keys | string | path to the keys file matching samples to bait names (see data preparation) |
| remove | string | path to the file listing samples that should be excluded from analysis |
| collapse | string | path to the file listing groups of baits that can be treated as biological replicates |
| specificity_exclusions | string | path to the file listing baits to mutually exclude when computing specificity (see basic protocol 2) |
| output_dir | string | directory to write output files |
To guide the reader through the various protocols we prepared an example project in $INSTALL_DIR/tests/small/, including example input files and a sample configuration file mist_small_test.yml, which can be run after installation to verify that all necessary resources are in place. The configuration file follows the Yaml Ain't Markup Language (YAML) (http://www.yaml.org) format that allows defining conceptual blocks of parameters. The first parameter block deals with file input/output (See Table 1) and the consecutive blocks correspond to configuration options for the three basic protocols outlined in this unit. The enabled [0/1] parameter in each parameter block turns each basic protocol off or on respectively. Finally, the MiST pipeline is run as follows:
$INSTALL_DIR/main.R --config [path to YML file]
Running the pre-processing protocol (See Table 2)
Table 2.
configurable parameters for pre-processing
| name | type | description |
|---|---|---|
| remove_carryover | boolean | enables the attempted removal of carryover proteins from a previous run |
| flter_contaminants | boolean | enables the removal of known contaminants from the prey list |
| contaminants_file | string | path to file listing all known contaminants in FASTA format |
| ld_colname | string | column identifier for sample identifier |
| prey_colname | string | column identifier for identified proteins in data file |
| pepcount_colname | string | column identifier for observed peptides per protein in data file |
| mw_colname | string | column identifier for protein molecular weight in data file |
- The first pre-processing step is to remove common contaminants such as all the keratins and known cross-reacting proteins with beads or affinity tags during purification. To enable this option turn on filter_contaminants in the configuration file and define the path to a text file listing all contaminants using the same identifiers as in the data file.Even though it's up to the user to define a custom set of contaminants we provided a minimal list, based on the Maxquant (Cox and Mann, 2008)contaminant file augmented with most Keratin proteins we regularly come across and a ‘decoy’ entry for Prospector false hits. Since contaminants might be condition and even machine specific we encourage users to search the recently published Crapome (Mellacheruvu et al., 2013) database for cell-, tag- or bead- specific contaminants matching their experimental setup.
- The second pre-processing step is to computationally remove peptide counts of proteins that are due to sample ‘carryover’ from a previous run on the MS. To enable this feature turn remove_carryover on.Accurately preventing and detecting sample carryover between consecutive MS runs is not a trivial problem and an active topic of discussion in the MS community. Carryover is caused by residual peptides from a previous sample and is generally serial in nature, often affecting several samples in a sequence (Hughes et al., 2007). We empirically observed that hydrophobic proteins in combination with over-expression of the bait protein can lead to a higher number of carryover peptides. We recommend addressing this issue by 1) testing various experimental conditions that minimize sample carryover and 2) shuffling the order in which biological replicates of samples are run. A third option is to remove any false hits that are detectable after the facts by a computational procedure similar to the one we implemented. The current procedure checks for carryover in up to four samples following the current sample. Carryover is determined if the following conditions are met in the samples tailing a sample: i) there is a positive number of unique peptides for a specific protein, ii) the number of unique peptides is less than half of the current sample, iii) the number of occurrences of this prey in the data set is less than one third of the number of total experiments.
- Map the values of the column names in the tab-delimited data file to the column description parameters (See Data preparation section for details):
- id_colname : Sample identifier
- prey_colname : Protein identifier
- pepcount_colname : Observed peptide frequency
- mw_colname : Protein molecular weight
Inspect the bait-prey matrix called preprocessed_MAT.txt in the output folder defined by output_dir, or the processed directory in the executable path if no output directory was defined. The bait-prey matrix is formatted according the data preparation guidelines in the next section (See Figure 2B).
Basic Protocol 2: Quality Control
We built a number of quality control and data summary plots such as MS yield statistics and a hierarchically clustered heat map of the experiment matrix into the MiST package. The output of these quality control scripts can help to decide critical parameters that influence the MiST scores, such as samples to remove or conditions to group together as replicates.
Necessary Resources
Hardware
Workstation running any current OS, Unix environment recommended
Software
MiST pipeline (installed as described in Support Protocol)
Files
Data pre-processed as described in Basic Protocol 1
Data preparation
After the preprocessing protocol of the MiST pipeline has been run, the input data should be properly reformatted for the quality control algorithm. For convenience, the quality control and scoring input follows the same formatting guidelines as the SAINT algorithm bait-prey input matrix. If the pre-processing step was skipped, the user should make sure to format the input for all subsequent steps as follows (See Figure 2B):
- Columns:
- (Preys) all distinct protein identifiers found as preys in the complete set of samples. These correspond to the unique identifiers in the prey_colname of the data file
- (PepAtlas) If Peptide Atlas counts are not known set this to 1. PepAtlas counts are for SAINT compatibility.(Choi et al., 2012).
- (Length) scaling factor derived from the indicated molecular weight per protein in the mw_colname column
- (PreyType) prey type is set to “N” to maintain matrix structure compatible with SAINT.
- Header rows:
- (row 1) all unique sample identifiers. Sample identifiers correspond to the unique identifiers in the id_colname of the data file and the first column in the keys file.
- (row 2) bait names for samples on row 1. Bait names correspond to the mapping of baits to samples described in the keys file.
- (row 3) specificity exclusions for baits on row 2. Specificity exclusions are described in the specificity_exclusions file and will be discussed in the section on MiST scoring (Basic Protocol 3)
Row 4-[unique preys] x Column 5-[samples]: peptide counts per prey/sample from the pepcount_colname column in the data file
Running the quality control protocol (See Table 3)
Table 3.
configurable parameters for quality control
| name | type | description |
|---|---|---|
| matrix_file | string | Path to preprocessed matrix file. The matrix created in the preprocess step will be used if this is left blank |
| cluster | boolean | Whether to perform a hierarchical cluster analysis on the data |
| cluster_font_scale | integer | Row/column font adjustment factor for the clustered heat map |
| ip_distributions | string | Whether to perform a protein count and peptide distribution analysis per group of biological replicates |
If matrix_file is left blank then the output matrix produced by the pre-processing step (Basic Protocol 1) is used, otherwise this parameter should point to the path of a correctly formatted bait-prey matrix as described above.
- If ip_distributions is enabled then a number of plots summarizing sample features per group of biological replicates (See Figure X) are saved into the output_dir:
- _proteincounts.pdf shows the total number of identified proteins with unique peptides. (See Figure 2C)
- _NumUniqPep.pdf shows the distribution via boxplot of the peptide counts by the replicates grouped by bait. Replicate distributions that are very different may imply something went wrong with that sample, leading to it being removed in future scoring.
- If cluster is enabled then a hierarchically clustered heatmap showing the pair-wise signal correlation between all samples is saved into the output_dir (See Figure 2D).The correlation between the observed peptide counts for each pair of samples is measured by the Pearson correlation coefficient. Proteins that were not identified in a sample are given a zero peptide count. The resulting symmetric correlation matrix is then clustered with R's ‘hclust’ algorithm using the default Euclidian distance metric and visualized with R's ‘pheatmap’ library. Except the cluster_font_scale parameter, which can be decreased or increased for larger or smaller datasets respectively, the remaining described parameters are currently not configurable.
Basic Protocol 3: Calculating the MiST score
The MiST score is a weighted sum of three features: 1) normalized protein abundance measured by peak intensities, spectral counts or unique number of peptide per protein (abundance); 2) invariability of abundance over replicated experiments (reproducibility); and 3) a measure of how unique a bait-prey pair is compared to all other baits (specificity). The weights of the three features are configurable in three different ways: first, pre-configured fixed weights can be used; second, they can be trained de-novo on a custom list of trusted bait-prey pairs identified in the data set; lastly, a principal component analysis (PCA) can be run to assign the feature weights according their contribution to the variance in the data set.
Necessary Resources
Hardware
Workstation running any current OS, Unix environment recommended
Software
MiST pipeline (installed as described in Support Protocol)
Data preparation
After the preprocessing protocol of the MiST pipeline has been run, the input data should be properly reformatted for the main scoring algorithm. For convenience, the scoring input follows the same formatting guidelines as the SAINT algorithm bait-prey input matrix. If the preprocessing step was skipped, the user should make sure to format the input for all subsequent steps as described in the data preparation section of the Quality Control step (See also Figure 2B).
In addition to the bait-prey matrix input file, the user also has the option of setting bait exclusion rules when calculating the MiST scores. These rules only apply when computing MiST's specificity feature value. In brief, every exclusion rule defines which baits should be excluded from the specificity denominator. Even though the definition of bait exclusion rules is an optional parameter, it can highly influence the results. Therefore, we further discuss its proper in the critical parameters section of this protocol.
To apply specificity exclusion rules, create a tab-delimited file (See Figure 2A) where every row consists of:
○ (column 1): the name of the bait, whose specificity exclusion rules you would like to define. Make sure that the bait name corresponds to the name that was used in the keys or collapse file.
○ (column 2): the names of the baits that you would like to exclude when specificity is being computed for the bait listed in column 1. Multiple baits can be excluded by separating them with a pipe (‘|’) symbol. Again, make sure that all bait names correspond to names that were used in the keys or collapse file.
Running the MiST scoring protocol (See Table 4)
Table 4.
configurable parameters for the MiST scoring algorithm
| name | type | description |
|---|---|---|
| matrix_file | string | Path to preprocessed matrix file. The matrix created in the preprocess step will be used if this is left blank |
| weights | string | One of three possible values: fixed/training/PCA (See MiST section) |
| training_file | string | path the the file containing bait-prey pairs for training (only if weights : training) |
| reproducibility | double | MiST weight [0-1] for the reproducibility feature |
| abundance | double | MiST weight [0-1] for the abundance feature |
| specificity | double | MiST weight [0-1] for the specificity feature |
(Optional) Create a specificity exclusion file (See data preparation) and define the path to this file through the specificity_exclusions entry in the files block of the configuration file.
If matrix_file is left blank then the output matrix produced by the pre-processing step (Basic Protocol 1) is used, otherwise this parameter should point to the path of a correctly formatted bait-prey matrix.
- Decide on a strategy to combine the abundance, reproducibility and specificity features into a single MiST score. To do so, set the weights parameter to either:
- fixed: Choose a decimal value between 0 and 1 for reproducibility, abundance and specificity.When choosing fixed values, make sure the sum of these values sums up to 1. If no weight values are chosen the weights default to 0.309 for reproducibility, 0.685 for specificity and 0.006 for abundance. These weights were established in the first MiST publication (Jäger et al., 2011)and are a good choice to select reproducible, specific bait-prey pairs.
- training: MiST will use a training_file to exhaustively test the performance of different parameter combinations and select the optimal configuration.We recommend using this option only when a sufficiently large benchmark set is available. See the advanced parameter section below for details.
- PCA: MiST will perform a Principal Component Analysis (PCA) on the three-dimensional feature matrix and select weights that project the feature values on the first principal component.We recommend using this option only when the suggested fixed weights do not perform well and insufficient training data is available.
Result file
The result file is tab-delimited file with a unique entry for observed bait-prey pair organized in the following columns:
○ Bait: Bait name as described in the keys or collapse file
○ Prey: Protein identifier as listed in the prey_colname column of the data file
○ Abundance: MiST abundance feature value of the bait-prey pair
○ Reproducibility: MiST reproducibility feature value of the bait-prey pair
○ Specificity: MiST specificity feature value of the bait-prey pair
○ MiST: The total MiST score value of the bait-prey pair
○ Ip: All samples where the bait-prey was observed in.
Guidelines for Understanding Results
Quality Control Plots
Different protein baits can have drastically different amounts of interacting proteins.
Nevertheless, these levels should be consistent across biological replicates. A simple bar plot grouping all replicates for a single bait can help spotting samples of lower quality, which can consequently be removed from the dataset (See Figure 2C and 3A).
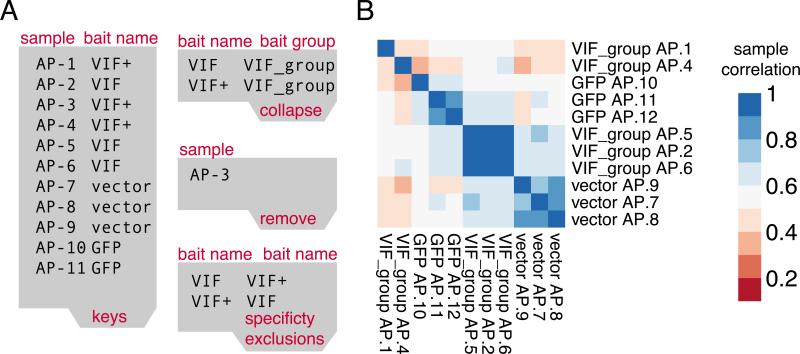
Figure 3.
A) Tab-separated input files adjusted based on the Quality Control observations: (1) sample AP-3 is ‘removed’ from the data. (2) VIF and VIF+ are ‘collapsed’ into a VIF_group and. Optionally VIF and VIF+ could have been mutually excluded for specificity calculations B) The clustered heatmap after adjustments shows no low quality data and clearly distinct replicate clusters aligned with the 3 different bait groups.
The primary use of the hierarchically clustered heatmap is to validate that samples are indeed more correlated within their group of replicates compared to negative controls and different baits. If this is not the case and a sufficient number of replicates are available, the sample can be removed from the data by adding its identifier to the remove file. In addition, carefully inspecting clusters can reveal accidentally mislabeled samples. For example, if one sample clusters more tightly with an unrelated group of baits compared to the other samples in its group this could be an indication that the sample was accidentally mislabeled. If this is the case, it is helpful to inspect the number of bait peptides that should be detected at high levels in the sample. Lastly, if a bait is purified under multiple experimental conditions (mutations, beads, tags, drugs, etc.), the correlation between these conditions can be an indication of the effect of the condition. Instead of discarding these samples the user can make a conscious choice to treat the conditional purifications as replicates of the wild type (See Figure 2D and 3B).
MiST Score
MiST aims to predict whether a protein-protein interaction detected in a set of repeated AP-MS experiments is biologically relevant using three features: reproducibility, abundance and specificity. To get as close to the optimal MiST score of 1 for an interaction, it is important that this interaction scores well across all features (See Figure 4A). Since values of 1 or close to 1 are rare, a minimum threshold can be applied to separate the predicted true interactions from the rest. However, choosing the appropriate threshold that makes a good trade-off between prediction sensitivity (detecting all true interactions) and specificity (minimizing the number of false positives) can be challenging and depends on the choice of feature weights.
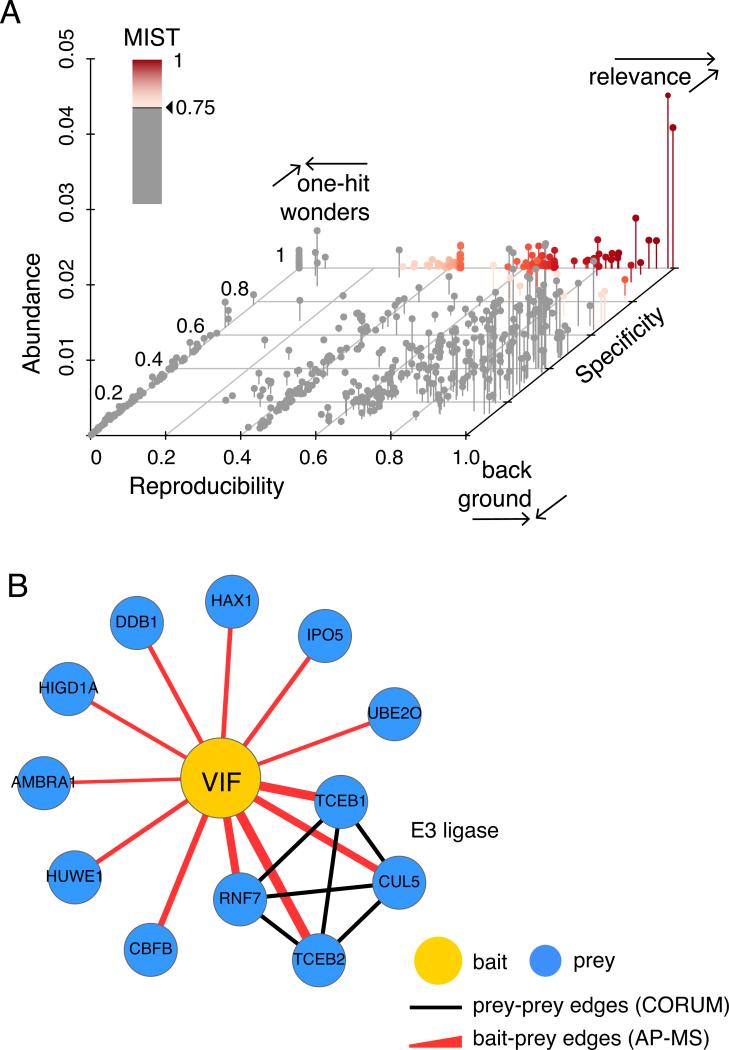
Figure 4.
A) A 3d scatterplot illustrating three different areas in the three-dimensional MiST feature space: (1) biologically relevant interactions (red gradient, MiST scores > 0.75) are specific and reproducible with variable abundance (2) non-specific, reproducible interactions are often background proteins and (3) specific, irreproducible interactions are often one-hit-wonders including contaminants and MS artifacts B) The scored bait-prey list at a high MiST score threshold visualized as a protein interaction network with Cytoscape. Red edges depict interactions identified by AP-MS while black edges are mined from the CORUM database. Edge width corresponds to the MiST score. High MiST scores for multiple subunits of a described complex add confidence to observed AP-MS interactions.
The easiest way to pick a threshold is therefore to stick to the recommended value for specific weights. For example, for the previously published HIV–host interaction network, a MiST lower bound threshold of 0.75 was recommended. When your dataset is comparable to a reference set this is an easy and sound solution.
A slightly harder but more preferred way of picking a threshold is by making prediction plots such as a Receiver Operating Curve (ROC) or a precision-recall curve and computing prediction accuracy statistics like the f1 score. The main drawback of this approach is that these metrics depend on the presence of ‘true’ positive and negative bait-prey interactions in the MS data set. While the so-called ‘negative’ interactions are often picked randomly from the data, the absence of known ‘positive’ interactions for a protein is often the very reason to perform an AP-MS experiment. When compiling a benchmark set we recommend using a positive set of at least 20 interactions and picking a negative set roughly 100 times larger than the positive set.
Finally, if neither of the aforementioned strategies for choosing a threshold is feasible, we advise to respect two rules of thumb. First, never pick a threshold lower than the highest feature weight value. For example, if specificity has the highest weight of 0.68, the MiST threshold should be strictly greater than 0.68. As explained before, biologically relevant interactions are expected to be reproducible and specific (See Figure 4A). Interactions with a perfect reproducibility score but zero specificity score are most likely ‘background’ interactions with highly abundant proteins. Conversely, interactions with a perfect specificity score and zero reproducibility score are likely to be ‘one-hit-wonders’. Second, keep in mind that the goal of scoring an AP-MS data set should be to approach the true biologically relevant interaction network as close as possible. Even though it is known that some proteins act as hubs and others have just one single interaction partner, current studies estimate the average number of interactions to be around 5-8 per protein. This expected bait-prey ratio could therefore be used to determine a reasonable cutoff for the MiST scores. Since the primary purpose of an AP-MS experiment is to discover new interactions and these studies are often followed up with a more targeted experiment, it is still acceptable to consciously allow a higher number of potential false positives.
Commentary
Background Information
The MiST score was originally developed to rank bait-prey pairs in an ex vivo HIV-human data set (Jäger et al., 2011). When this data set was being produced, SAINT (Choi et al., 2011, 2012; UNIT 8.15) and the CompPASS-D (Sowa et al., 2009) score were the two main computational algorithms that were suited to analyze large-scale AP-MS data sets. However, their prediction performance was only reported on a data set of human-human protein interactions. The uniqueness of the HIV-human data sets became the primary reason to develop MiST, a custom AP-MS scoring algorithm. Although MiST and CompPASS both use exclusively abundance, reproducibility and specificity as predictive features, making them therefore somewhat comparable, MiST scores are easier to interpret because their feature value and total score varies between 0 and 1. Even though a ranked bait-prey list based on CompPASS scores is quite accurate, the actual scores are by definition harder to interpret because they vary between 0 and extremely high numbers.
SAINT scores, on the other hand, vary between 0 and 1 but are conceptually very different because they describe the probability that a bait-prey pair is true based on a distribution model of its abundance values. For optimal SAINT performance it is therefore recommended to have a well-defined set of negative control affinity purifications to compare against. Neither MIST nor CompPASS requires explicit definition of negative controls; in fact negative controls are treated just the same as any another bait purification in the dataset.
To assess the accuracy of MiST, we compared it to the SAINT, and CompPASS scores. The accuracy of each score was evaluated by its recall rate for the set of 39 well-characterized biologically relevant HIV-human bait-prey pairs. The MiST score was the most accurate among all the tested scores (Jäger et al., 2011). For example, at the threshold of 0.75, the recall number of known bait-prey pairs for the CompPASS, and MiST scores was 19, 29, and 32, respectively. Furthermore, 97 out of 127 (76% recall) top-ranked interactions predicted by MiST were validated using co-immunoprecipitation followed by Western-blot as an orthogonal assay. For an additional test, we counted bait-prey pairs involving ribosomal proteins, which are a good indicator of biologically irrelevant bait-prey pair predictions (Ewing et al., 2007). Again, MiST was the most accurate score, resulting in only 3 HIV-ribosomal protein bait-prey pairs, compared to 32 and 75 for SAINT and CompPASS, respectively.
Critical Parameters
Specificity exclusions
We repeatedly observed in gold-standard data sets that good prediction performance goes hand in hand with a high weight for specificity. Preys exclusively identified with a single bait protein will receive a high specificity value, therefore these preys will overall get high MiST scores. Conversely, prey proteins identified with several baits are not bait specific and will receive overall lower scores. However, often large-scale interrogations of biological systems are designed in a way that inherently introduces a bias into the number of times preys are identified across multiple baits. For ‘specificity’ to optimally work the way it is intended to, it is therefore important to remove design bias from the calculations. Here, we illustrate how you can achieve this by describing the three most common examples:
○ Conditional interactions: When an experiment is designed to determine whether a drug or mutation influences one specific interaction between two proteins, most of the unaffected interactions will still be detected in the samples with the drug or mutation. To ensure the scores of both the wild type and conditional sample are not incorrectly penalized by their shared interactions, we add exclusion rules between the (bait, bait + condition) pair and the (bait + condition, bait) pair (See Figure 3B).
○ Highly homologous baits: When two baits in a data set share a more than expected degree of sequence identity, they likely share a significant amount of interactions too. The most common examples are intra- or inter-species homologs, different isoforms of the same protein and cleaved protein products from poly-proteins.
○ Complex subunits: When subunits of a stable multi-subunit complex are all used as baits, the remaining subunits of the full complex will be identified recurrently in all samples.
Beyond these obvious cases, we recommend to not get carried away with specificity exclusions. Only data sets that have a high percentage of baits that match the conditions above should be scored with a specificity exclusion list. If there “might” be set of common interactions between baits that could throw of the specificity score, then score the data without excluding the baits from each other and look whether the MiST scores are affected by a low specificity component.
Advanced parameters
MiST training with gold-standard interactions
The training file contains known true interactions that were identified in the data. This file should be tab-delimited, listing baits with the bait name described in the keys or collapse file and preys with their name in the prey_colname column of the data file. MiST will label these bait-prey pairs as positive interactions set and compile a 100 times larger negative interaction set randomly selected from the data.
MiST will then run a simulation that cycles through all possible assignments of the three weights with 0.01 increments that together sum to 1, and 0.01 increments of threshold between 0 and 1. In every simulation cycle, MiST scores using these weights are computed and the subset greater than the threshold is compared to the positive and negative set. We compare the performance of the simulated weights at a given threshold by computing the precision and recall rates along with the ‘f1 score’ to measure overall accuracy.
Finally, the combination of weights with the best f1 score will be selected to compute the MiST scores and an additional file with the summary of all simulations is written to the output_dir.
Suggestions for Further Analysis
Here we describe a number of suggestions for further analysis, starting from the MiST scores output file.
Protein interaction networks
After MiST outputs the scored list of bait-prey pairs and the appropriate threshold to select the high-confidence pairs is determined, the next step is usually to convert this filtered list into a more visual representation as a protein interaction network. Cytoscape (Shannon, 2003; Su et al., 2014; UNIT 8.13) is by far the most popular piece of software used to create such networks. An in-depth tutorial on how to create interaction networks using AP-MS data is recently reviewed by Morris et al. (Manuscript in press)
Connecting protein complexes
Unlike yeast-two-hybrid, which is binary in nature, interactions identified by AP-MS can be either in direct contact with the bait or indirectly mediated through members of the same protein complex. By overlaying published interaction data as edges between two ‘prey’ proteins, it is easier to see which protein complexes a particular bait protein interacts with. There are many online resources that collect and curate large-scale interaction data from different experimental sources such as the Biogrid (Stark et al., 2006), STRING (Mering, 2003), Corum (Ruepp et al., 2009) and Compleat (Vinayagam et al., 2013) that allow queries with a list of proteins to return all observed interactions between them (See Figure 4B). If the bait-prey list is imported in Cytoscape, this process can be done automatically by using the BisoGenet plugin (Martin et al., 2010).
Biological annotation enrichment
To make sense out of larger protein interaction data sets it is useful to annotate all identified proteins with meaningful terms describing their biological function, domain composition, pathway involvement, disease involvement and cellular localization. Again, there are many well-established online resources that collect and curate protein annotation terms such as GO (Gene and Consortium, 2000; Blake and Harris, 2008; UNIT 7.2) KEGG (Ogata et al., 1999; Tanabe and Kanehisa, 2012; UNIT 1.12) or PFAM (Finn et al., 2014; Coggill et al., 2008; UNIT 2.5). Next, these annotated protein lists can then be analyzed to test whether ontology terms are overrepresented in the full set or specifically with respect to a single bait protein. The Cytoscape software can be a valuable tool for this purpose too because plugins such as BINGO (Maere et al., 2005) take care of network annotation and enrichment tests in a few easy steps. Alternatively, complete lists of scored protein interactions or shorter lists of interactions above a certain threshold can be uploaded to online tools such as DAVID (Dennis et al., 2003) or GORILLA (Eden et al., 2009) respectively.
Acknowledgement
EV, JVD and NK are supported by NIH grants P50 GM082250, P50 GM081879, P01 AI090935, P01 AI091575, P01 AI106754, R01 GM084279 and the DARPA grant HR0011-11-C-0094. AS is supported by NIH R01 GM083960. PC is supported by a Howard Hughes Medical Institute Predoctoral Fellowship.
Footnotes
Internet Resources
The Github repository (https://github.com/everschueren/MiST/) is the main online resource for this protocol. We have opened the MiST repository to the public but currently it is only editable by approved collaborators. In the near future we will also update the webpage at http://modbase.compbio.ucsf.edu/MiST/ to reflect the protocol described in this manuscript.
Literature Cited
- Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Huang H-C, Hirai A, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome research. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Harris MA. The Gene Ontology (GO) Project: Structured vocabularies for molecular biology and their application to genome and expression analysis. Current Protocols in Bioinformatics. 2008;23:7.2:7.2.1–7.2.9. doi: 10.1002/0471250953.bi0702s23. [DOI] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin Z-Y, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras A-C, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nature methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Liu G, Mellacheruvu D, Tyers M, Gingras AC, Nesvizhskii AI. Analyzing protein-protein interactions from affinity purification-mass spectrometry data with SAINT. Current Protocols in Bioinformatics. 2012;39:8.15.1–8.15.23. doi: 10.1002/0471250953.bi0815s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauser KR, Baker P, Burlingame AL. Role of Accurate Mass Measurement (±10 ppm) in Protein Identification Strategies Employing MS or MS/MS and Database Searching. Analytical Chemistry. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- Coggill P, Finn RD, Bateman A. Identifying protein domains with the Pfam database. Current Protocols in Bioinformatics. 2008;23:2.5:2.5.1–2.5.17. doi: 10.1002/0471250953.bi0205s23. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, et al. Large-scale mapping of human protein- protein interactions by mass spectrometry. Molecular systems biology. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: The protein families database. Nucleic Acids Research. 2014;42 doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene T, Consortium O. Gene Ontology: tool for the. nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev A. On the number of protein-protein interactions in the yeast proteome. [August 11, 2014];Nucleic Acids Research. 2003 31:4157–4161. doi: 10.1093/nar/gkg466. Available at: http://nar.oxfordjournals.org/content/31/14/4157.full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes NC, Wong EYK, Fan J, Bajaj N. Determination of carryover and contamination for mass spectrometry-based chromatographic assays. The AAPS journal. 2007;9:E353–E360. doi: 10.1208/aapsj0903042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV–human protein complexes. Nature. 2011 doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Martin A, Ochagavia ME, Rabasa LC, Miranda J, Fernandez-de-Cossio J, Bringas R. BisoGenet: a new tool for gene network building, visualization and analysis. BMC bioinformatics. 2010;11:91. doi: 10.1186/1471-2105-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert J-P, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nature Methods. 2013:1–11. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mering CV. STRING: a database of predicted functional associations between proteins. [March 10, 2011];Nucleic Acids Research. 2003 31:258–261. doi: 10.1093/nar/gkg034. Available at: http://www.nar.oupjournals.org/cgi/doi/10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Research. 2009;38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. Available at: http://www.genome.org/cgi/doi/10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the Human Deubiquitinating Enzyme Interaction Landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic acids research. 2006;34:D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G, Morris JH, Demchak B, Bader GD. Biological Network Exploration with Cytoscape 3. Current Protocols in Bioinformatics. 2014;47:8.13:8.13.1–8.13.24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M, Kanehisa M. Using the KEGG database resource. Current Protocols in Bioinformatics. 2012;38:1.12:1.12.1–1.12.43. doi: 10.1002/0471250953.bi0112s38. [DOI] [PubMed] [Google Scholar]
- Vinayagam A, Hu Y, Kulkarni M, Roesel C, Sopko R, Mohr SE, Perrimon N. Protein Complex-Based Analysis Framework for High-Throughput Data Sets. Science Signaling. 2013;6:rs5–rs5. doi: 10.1126/scisignal.2003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Ivanic J, Wallqvist A, Reifman J. A Novel Scoring Approach for Protein Co- Purification Data Reveals High Interaction Specificity. PLoS Computational Biology. 2009;5:e1000515. doi: 10.1371/journal.pcbi.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]