Abstract
We compared handmade cloned (HMC) buffalo blastocysts produced from oocytes stained with Brilliant Cresyl Blue (BCB) and classified into those with blue (BCB+) or colorless cytoplasm (BCB−). The blastocyst rate was higher (p<0.001) for BCB+ than for BCB− oocytes (43.41±2.54 vs. 22.74±1.76%). BCB+ blastocysts had inner cell mass (ICM) cell number, ICM-to-trophectoderm ratio, global level of H3K18ac, apoptotic index, and expression level of BCL-XL, but not that of CASPASE-3, similar to that of blastocysts produced through in vitro fertilization (IVF), which was higher (p<0.05) than that of BCB− blastocysts. The global level of H3K9me2, which was similar in BCB+ and BCB− blastocysts, was higher (p<0.01) than that in IVF blastocysts. The expression level of OCT4 and SOX2 was higher (p<0.05) and that of GATA2 was lower (p<0.05) in BCB+ than that in BCB− blastocysts, whereas that of DNMT1, DNMT3a, NANOG, and CDX2 was not significantly different between the two groups. The expression level of DNMT1, OCT4, NANOG, and SOX2 was lower (p<0.05) and that of CDX2 was higher (p<0.05) in BCB+ than in IVF blastocysts. In conclusion, because BCB+ blastocysts have better developmental competence and are closer to IVF blastocysts in terms of quality, epigenetic status, and gene expression than BCB− blastocysts, BCB staining can be used effectively for selection of developmentally competent oocytes for HMC.
Introduction
Both the yield and the quality of embryos produced are important criteria for the evaluation of in vitro embryo production systems, whether these involve a combination of in vitro maturation (IVM), in vitro fertilization (IVF), and in vitro culture (IVC) or whether these are carried out through somatic cell nuclear transfer (SCNT) for producing cloned embryos. Because oocyte quality plays an important role in determining the developmental competence and quality of the embryos produced in vitro, criteria that enable distinguishing good-quality oocytes could be used for their selection from a pool of oocytes. A scheme for classification of oocytes that involves visual assessment of morphological features such as thickness and compactness of the cumulus investment and the homogeneity of the ooplasm (de Loos et al., 1989) is used routinely for producing IVF and SCNT embryos. Morphological criteria used for selection of competent oocytes are, however, subjective and often inaccurate. Additional selection criteria have, therefore, been developed which are based on the physiological status of the oocyte in addition to its morphological appearance.
One of such methods is based on the ability of glucose-6-phosphate dehydrogenase (G6PDH) to degrade Brilliant Cresyl Blue (BCB), a nontoxic dye, to a colorless product. G6PDH, which is synthesized within the oocytes (Mangia and Epstein, 1975), is active in the growing oocyte but has decreased activity in oocytes that have finished their growth phase and are likely to have achieved developmental competence (Wassarman, 1988). Thus, oocytes that have completed their growth phase show decreased G6PDH and exhibit cytoplasm with blue coloration (BCB+), whereas growing oocytes, which are expected to have a high level of active G6PDH, remain colorless (BCB−) after staining. BCB staining has been successfully used for the selection of oocytes for production of IVF embryos in several mammalian species such as pig (Roca et al., 1998), goat (Rodriguez-Gonzalez et al., 2002), sheep (Catalá et al., 2011), bovine (Pujol et al., 2004), and buffalo (Manjunatha et al., 2007).
Despite availability of a plethora of information on the use of BCB staining for selection of developmentally competent oocytes for IVF, there are only two reports available regarding the use of BCB staining of cytoplasm for selection of competent oocytes for SCNT (Bhojwani et al., 2007; Su et al., 2012). In both of these studies, conventional micromanipulation-based SCNT was used in which there is minimal loss of cytoplasm because the enucleation of zona-enclosed oocytes is carried out with the help of a micromanipulator. However, the conventional micromanipulation-based SCNT has now been replaced by handmade cloning (HMC) technique due to its overwhelming advantages, such as simplicity, higher efficiency, and lack of need for skilled manpower and expensive equipment (Vajta et al., 2003). We have found that almost 15–50% of cytoplasmic volume is lost in using the protrusion cone–guided enucleation process during HMC (Panda et al., 2011). When the cytoplasmic volume of the recipient oocyte is sufficiently reduced, it results in a decrease in blastocyst development rate (Zakhartchenko et al., 1997). The quality of cytoplasm is especially important in SCNT because the overall cloning efficiency is directly related to the ability of the cytoplast to reprogram a terminally differentiated somatic cell. Coupled with the higher amount cytoplasm loss associated with HMC compared to that in micromanipulation-based SCNT, it becomes even more important that the cytoplasm quality should be good. Therefore, noninvasive oocyte quality assessment for early selection of developmentally competent oocytes remains a major challenge for HMC.
In the present study, we compared the developmental competence, as indicated by the blastocyst rate, and embryo quality, as indicated by total cell number (TCN), inner cell mass (ICM), of ICM-to-trophectoderm (ICM:TE) cell numbers, and apoptotic index following terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay in buffalo HMC blastocysts produced from BCB+ and BCB− oocytes and those produced through IVF. We also compared the global acetylation level of histone H3 at lysine 18 (H3K18ac), global level of histone H3 dimethylated at lysine 9 (H3K9me2), and relative expression levels of pluripotency-related (OCT4, SOX2, NANOG), apoptosis-related (BCL-XL, CASPASE 3), trophectoderm lineage-specific (GATA2, CDX2), and epigenetics-related (DNMT1, DNMT3a) genes among blastocysts of the three groups.
Materials and Methods
All of the chemicals were obtained from Sigma Chemical Co. (St. Louis, MO, USA). The media were from GIBCO (Grand Island, NY, USA), and the disposable plasticware was purchased from Nunc (Roskilde, Denmark), unless otherwise stated. Fetal bovine serum (FBS) was obtained from Hyclone (Logan, UT, USA). The buffalo ovary samples were collected at an abattoir, and therefore ethical approval was not required.
Production of embryos by IVF
Cumulus–oocyte complexes (COCs) collected from abattoir buffalo ovaries were subjected to IVM and IVF, as described earlier (Sharma et al., 2011). For IVC, the presumed zygotes were washed several times with Research Vitro Cleave Medium (K-RVCL-50, Cook®, Queensland, Australia) supplemented with 1% fatty acid–free bovoine serum albumin (BSA) and were cultured in this medium for up to 8 days after insemination in a CO2 incubator at 38.5°C.
Handmade cloning
COCs of usable quality were incubated in 26 μM BCB, prepared in the washing medium [tissue culture medium-199 (TCM-199), 10% FBS, 0.81 mM sodium pyruvate, 2 mM l-glutamine, and 50 μg/mL gentamicin sulfate], for 90 min in a CO2 incubator (5% CO2 in air) at 38.5°C. The COCs were washed once with the washing medium and examined under a stereomicroscope. These were then divided into two groups according to their cytoplasmic coloration: BCB+, oocytes with any degree of blue coloration to the cytoplasm, and BCB−, oocytes without blue cytoplasm. The oocytes of both the groups were washed twice with the IVM medium [TCM-199, 10% FBS, 5 μg/mL porcine follicle-stimulating hormone (pFSH),1 μg/mL estradiol-17β, 0.81 mM sodium pyruvate, 10% buffalo follicular fluid, 50 μg/mL gentamicin sulfate), following which groups of 18–20 COCs were placed in 100-μL droplets of the IVM medium, overlaid with sterile mineral oil in 35-mm Petri dishes, and cultured for 21 h in a CO2 incubator at 38.5°C for IVM. Primary cell culture of skin cells derived from the ear biopsy of an adult Murrah buffalo was established, and donor cell preparation was performed for HMC as reported earlier (Selokar et al., 2012a). HMC was performed as described previously with some modifications (Selokar et al., 2012b).
TUNEL assay
The level of apoptosis was determined by TUNEL staining using an In Situ Cell Death Detection Kit, Fluorescein (cat. no. 11684795910, Roche, Germany) to assess the quality of blastocysts as described earlier (Selokar et al., 2014). Briefly, the blastocysts were fixed with 4% paraformaldehyde for 1 h and permeabilized by incubating with 0.5% Triton X-100 for 1 h. The blastocysts were then incubated with fluorescein isothiocyanate (FITC)-conjugated deoxyuridine triphosphate (dUTP) and terminal deoxynucleotidyl transferase (TdT) enzyme for 1 h at 37°C in the dark. The treated blastocysts were incubated in RNase (50 μg/mL) for 20 min at 37°C and were stained with Hochest 33342 (5 μg/mL) for 5 min at 37°C in the dark. For the positive control, the cells and blastocysts were treated with DNase solution (100 U/mL) for 20 min at 37°C prior to incubation with FITC-conjugated dUTP and TdT. For the negative control, the assay was carried out without addition of TdT enzyme. The stained blastocysts were washed with Dulbecco's Phosphate-Buffered Saline (DPBS) (Ca2+ and Mg2+-free) and were mounted on glass slides in 3-μL droplets of antifade solution and flattened with a coverslip. Cell counting was performed from the digital images obtained on an inverted Nikon fluorescence microscope. Each experiment was repeated at least three times. The apoptotic index=(number of TUNEL positive nuclei/total number of nuclei counted in that blastocysts)×100.
Immunofluorescence staining
Immunofluorescence staining for examining the global levels of H3K18ac and H3K9me2 was performed as described earlier (Selokar et al., 2014) with some modifications. Briefly, blastocysts of the three groups were fixed in 4% paraformaldehyde, permeabilized in 0.5% Triton X-100, and blocked in 5% BSA. The blastocysts were then incubated overnight at 4°C with the respective rabbit primary antibody (anti-H3K9me, 1:200, Millipore MA, USA; anti-H3K18ac, 1:1500, Millipore) diluted in blocking reagent. For CDX2 staining, mouse primary anti-CDX2 antibody (ready-to-use, AM392-10M, Bio-Genex Inc., San Ramon, CA, USA) was used. After washing three times with DPBS containing 0.1% Triton X-100 (DPBST), the blastocysts were incubated with FITC-conjugated goat anti-rabbit (Sigma) for epigenetic markers and Alexa Fluor 594-conjugated donkey anti-mouse immunoglobulin G (IgG) (H+L) secondary antibody (A21203, Invitrogen) for CDX2, diluted 1:1000 in DPBS. After washing five times with DPBST, the nuclei were counterstained with Hochest 33342 (10 μg/mL) and rinsed in DPBST. The blastocysts were then mounted on slides in mounting medium (2.5% DABCO in glycerol). The slides were observed under a fluorescence microscope, and the images were captured keeping the same optical conditions. NIS-Elements Basic Research image processing software (Nikon, Tokyo, Japan) included with the microscope was used for image acquisition and quantitative measurements of the mean pixel intensity emitted by each individual nucleus. The mean pixel intensity for each blastocyst for each epigenetic marker was normalized by dividing with mean pixel intensity of Hoechst 33342 nuclei. The images were merged by NIS-Elements BR 3.0 software (Nikon, Tokyo, Japan). At least 15 blastocysts (60–80 nuclei/blastocyst) were analyzed for each epigenetic modification.
Quantitative real-time PCR
RNA was isolated from pools of 10 blastocysts each using an RNAqueous Micro Kit (Ambion, Austin, TX, USA) as per the manufacturer's protocol. Following DNase treatment, the reverse transcriptase (RT) reaction was performed for cDNA preparation using SuperScript Reverse Transcriptase III (Invitrogen). Quantification of mRNA was carried out by quantitative real-time qPCR using CFX 96 I Cycler (Bio-Rad). The reaction mixture (10 μL) contained 5 μL of SYBR Green PCR Master Mix (Maxima SYBR Green Mastermix, Thermo Scientific), 0.2 μL of 10 μM of each primer, and 2× diluted cDNA. Thermal cycling conditions consisted of initial denaturation at 95°C for 5 min, followed by 40 cycles of 15 sec at 95°C, 15 sec at the corresponding annealing temperature, and 15 sec at 72°C, followed by 95°C for 10 sec (Table S1) (Supplementary Data are available at www.liebertpub.com/cell/). All of the primer pairs used were confirmed for their PCR efficiency, and specific products were checked by melt curve analysis and for the appropriateness of size by 2% agarose gel electrophoresis. Primer sequences are provided in the supplementary data (Table S1). The expression data were normalized to the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and were analyzed with CFX Manager software (Bio-Rad). In all of the experiments, three trials were carried out, each in duplicate.
Experimental design and statistical analysis
Immediately after aspiration, the oocytes were stained with BCB and were divided into BCB+ and BCB− groups in each experiment. Blastocysts produced though IVF were used as controls in experiments 3, 4, and 5.
In experiment 1, the percentage of BCB+ and BCB− oocytes was determined among those obtained from slaughterhouse buffalo ovaries, following staining with BCB. Also, the percentage of oocytes in which a clear protrusion cone was formed was examined in this experiment. In experiment 2, the development competence of BCB+ and BCB− oocytes was compared following HMC. In experiment 3, the quality of blastocysts of BCB+ and BCB− groups and those produced through IVF (control) was determined by examining their TCN, ICM cell number, and ICM:TE ratio. The total number of nuclei in blastocysts was determined following staining with Hoechst 33342, whereas the number of TE nuclei was determined following CDX2 immunofluorescence. The ICM cell number was calculated by subtracting the number of TE nuclei from the number of total nuclei. In addition, the apoptotic index was determined in blastocysts of the three groups by TUNEL assay. In experiment 4, the global level of H3K9me2 and H3K18ac was compared among blastocysts of the three groups using immunofluorescence staining. In experiment 5, the relative mRNA abundance of some pluripotency-related (OCT4, SOX2, NANOG), apoptosis-related (BCL-XL, CASPASE 3), trophectoderm lineage-specific (GATA2, CDX2), and epigenetics-related genes (DNMT1, DNMT3a) was compared among blastocysts of the three groups.
Statistical analysis was carried out using Sigma Stat version 3.1 (Aspire Software International, VA, USA). The datasets were analyzed by one-way analysis of variance (ANOVA) followed by the Holm–Sidak test. Percentage values were subjected to arcsine transformation prior to analysis. The differences were considered to be statistically significant at p<0.05. Data were presented as mean±standard error of the mean (SEM).
Results
The oocytes selected as BCB+ and BCB− after staining with BCB are shown in Figure 1. In experiment 1, out of 3327 (range 210–345 among individual trials) COCs examined in the present study, the percentage of BCB+ oocytes (2093, 65.1±2.11%, range 50.2–77.9%) was significantly higher (p<0.001) than that of BCB− oocytes (1134, 34.9±2.12%, range 22.1–48.8%). The percentage of oocytes in which a clear protrusion cone was formed was significantly higher (p<0.05) for BCB+ oocytes (77.1±5.6%, range 58.2–91.2%) than that for BCB− oocytes (62.0±5.4%, range 41.1–72.5%).
FIG. 1.
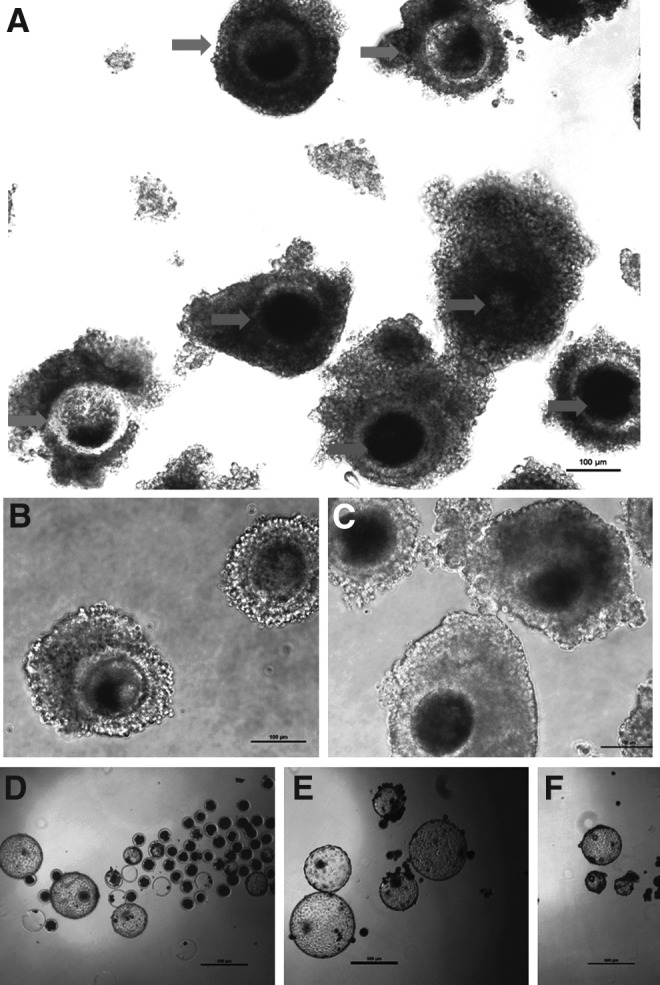
BCB staining. (A) Oocytes observed under phase-contrast microscope (magnification, 100×) after incubation with 26 μM BCB stain. Dark and light arrows indicate BCB− and BCB+ oocytes, respectively. BCB− (B) and BCB+ (C) oocytes at 200× magnification. Scale bar, 100 μm. Blastocysts from IVF (D), BCB+ (E), and BCB− (F) oocytes.
In experiment 2, which was aimed at comparing the developmental competence, the blastocyst rate was significantly higher (p<0.001) for BCB+ than that for BCB− oocytes following HMC although the cleavage rate was not statistically different (Table 1, Fig. 1).
Table 1.
Developmental Competence of Oocytes Selected for Handmade Cloning by Brilliant Cresyl Blue Staining
| Group | Oocytes taken for IVF/reconstructs taken for HMC (n) | Cleavage n (%) | Blastocysts n (%) |
|---|---|---|---|
| IVF | 506 | 339 (68.20±3.41)x | 69 (14.63±4.05)a |
| HMC (BCB+) | 418 | 393 (93.63±2.17)y | 178 (43.41±2.54)b |
| HMC (BCB−) | 247 | 227 (91.35±1.98)y | 55 (22.74±1.76)c |
Data from 15 trials.
Values are mean±standard error of the mean (SEM).
Values with different superscripts within the same column differ significantly (p<0.001).
Unstained oocytes were used as controls for IVF.
IVF, in vitro fertilization; HMC, handmade cloning; BCB, Brilliant Cresyl Blue.
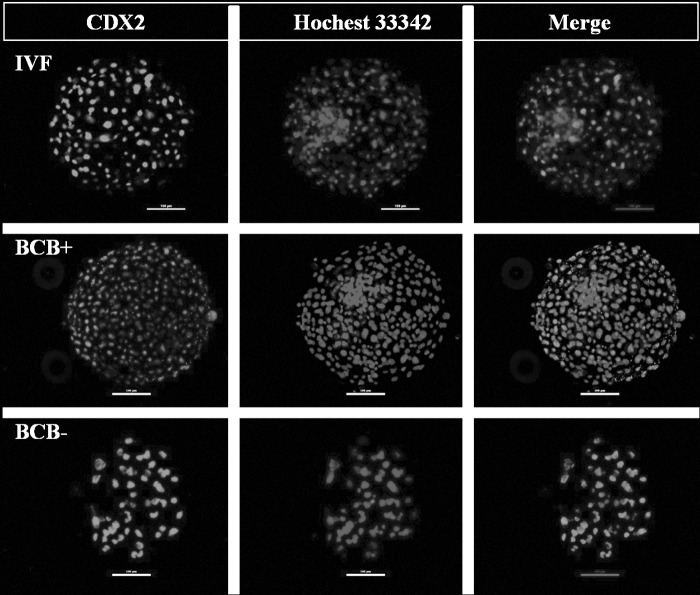
In experiment 3, which was aimed at comparing the quality of blastocysts of the three groups, the TCN of BCB+ blastocysts was found to be significantly higher (p<0.05) than that of IVF blastocysts which, in turn, was higher (p<0.05) than that of BCB− blastocysts (Table 2, Fig. 2). The ICM cell number and the ratio of ICM:TE cell number, which was similar for IVF and BCB+ blastocysts, was significantly higher (p<0.05) than that of BCB− blastocysts. Following TUNEL assay, the apoptotic index, which was similar for BCB+ and IVF blastocysts, was significantly higher (p<0.001) than that for BCB- blastocysts (Fig. 3).
Table 2.
Quality of In Vitro Fertilized Blastocysts and of Handmade Cloning Blastocysts Produced from Oocytes Selected by Brilliant Cresyl Blue Staining
| Source of blastocyst | Blastocysts examined (n) | TCN | Blastocysts examined (n) | Number of ICM cells | ICM:TE (%) |
|---|---|---|---|---|---|
| IVF | 47 | 198.91±08.77a | 15 | 38.83±4.67a | 20.60±1.78a |
| HMC (BCB+) | 46 | 242.78±16.08b | 14 | 31.78±3.41a | 17.20±1.46a |
| HMC (BCB-) | 34 | 160.94±09.24c | 12 | 19.75±2.61b | 11.90±0.99b |
Values are mean±standard error of the mean (SEM).
Values with different superscripts within the same column differ significantly (p<0.05).
TCN, total cell number; ICM, inner cell mass; TE, trophectoderm; IVF, in vitro fertilization; BCB, Brilliant Cresyl Blue.
FIG. 2.
Immunofluorescence staining of blastocysts for CDX2. Scale bar, 100 μm.
FIG. 3.
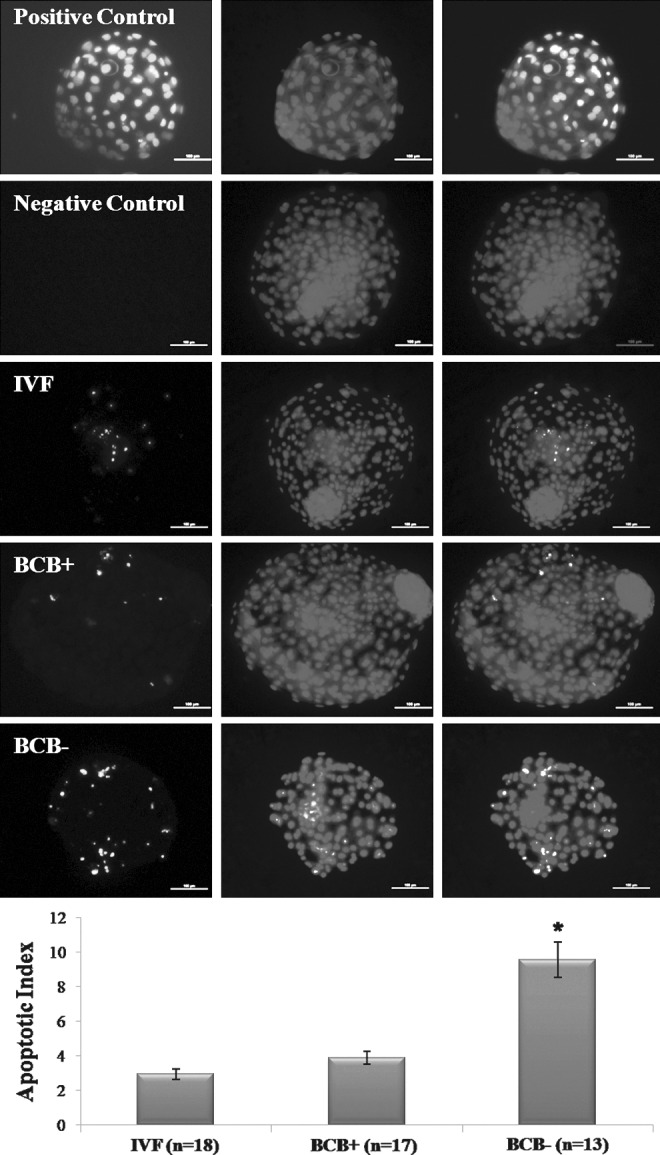
TUNEL staining for determination of apoptotic index of BCB+, BCB−, and IVF blastocysts. Scale bar, 100 μm. Bars marked with an asterisk differ significantly from other group (p<0.001).
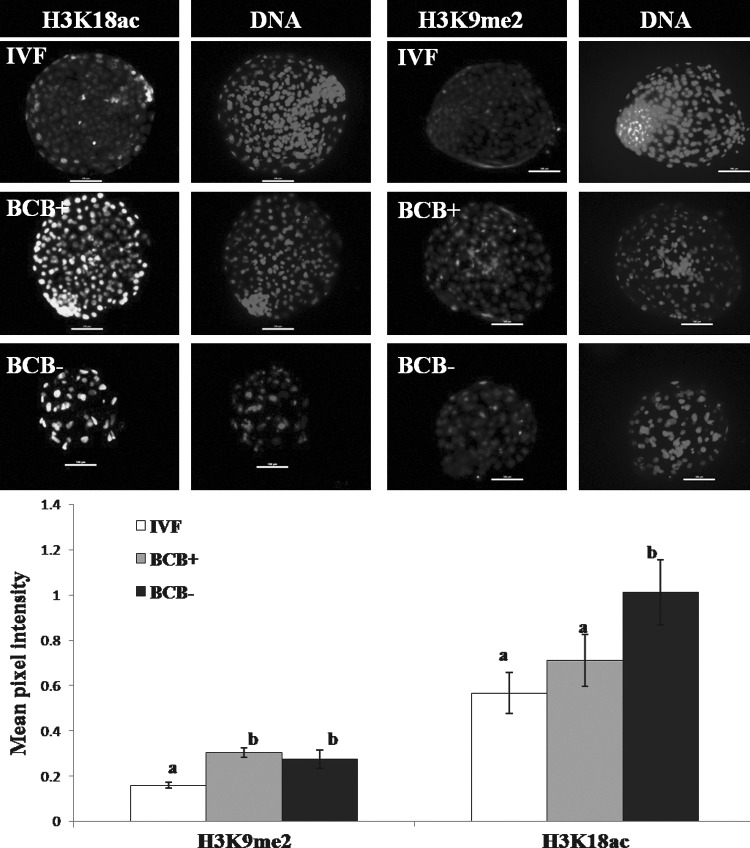
In experiment 4, the global level of H3K18ac was found to be similar for IVF and BCB+ blastocysts, and was significantly higher (p<0.05) than that of BCB− blastocysts (Fig. 4). However, the global level of H3K9me2, which was not significantly different between the BCB+ and BCB− blastocysts, was significantly higher (p<0.01) than that in IVF blastocysts.
FIG. 4.
Immunofluorescence staining and mean pixel intensity of H3K18ac and H3K9me2 in cloned BCB+ and BCB− and in IVF blastocysts. Scale bar, 100 μm. Bars with different superscripts differ significantly (p<0.05).
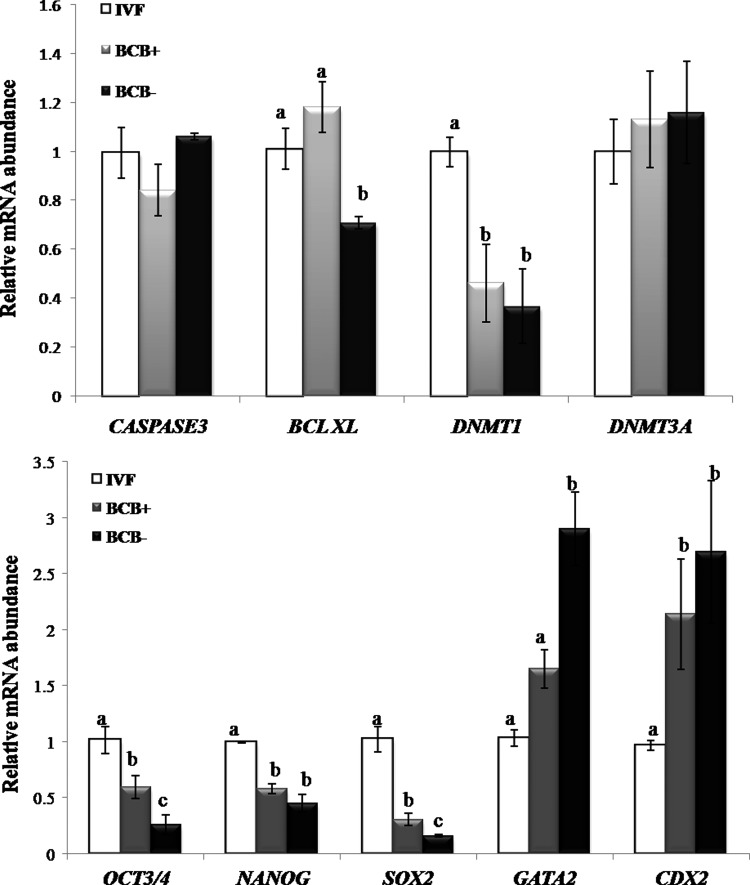
In experiment 5, among apoptosis-related genes, the relative mRNA abundance of BCL-XL was similar in IVF and BCB+ blastocysts, and was higher (p<0.05) than that in BCB− blastocysts, whereas that of CASPASE 3 was not significantly different among the three groups (Fig. 5). Among pluripotency-related genes, the expression level of OCT4 and SOX2 was higher (p<0.05) in IVF blastocysts than that in BCB+ blastocysts which, in turn, was higher (p<0.05) than that in BCB− blastocysts. The expression level of NANOG was higher (p<0.05) in IVF than that in BCB+ and BCB− blastocysts. Among epigenetics-related genes, the expression level of DNMT1, which was similar in BCB+ and BCB− blastocysts, was lower (p<0.05) than that in IVF blastocysts, whereas the transcript level of DNMT3a was not significantly different among the three groups. Among trophectoderm lineage-specific genes, the expression level of GATA2 was similar in BCB+ and IVF blastocysts, and was significantly lower (p<0.05) than that in BCB− blastocysts. However, the relative mRNA abundance of CDX2 was similar in BCB+ and BCB− blastocysts, and was higher (p<0.05) than that in IVF blastocysts.
FIG. 5.
Relative mRNA abundance of some important genes in cloned BCB+ and BCB− and in IVF blastocysts. Bars with different superscripts differ significantly (p<0.05).
Discussion
To our knowledge, this is the first report on the use of BCB staining for the selection of competent oocytes for HMC. We found that following HMC, the percentage of oocytes in which a clear protrusion cone was visible was significantly higher and the blastocyst rate was nearly two-fold higher for BCB+ than that for BCB− oocytes. Also, the blastocysts produced from BCB+ oocytes were closer to those produced by IVF as indicated by similar apoptotic index, ICM cell number, ICM:TE ratio, global level of H3K18ac, and expression levels of BCL-XL and GATA2, which were significantly different from those in blastocysts produced from BCB− oocytes.
Because enucleation in HMC is carried out manually with the help of a microblade, formation of a clear protrusion cone, which contains the oocyte's nuclear material, is essential for successful enucleation. The oocytes in which a protrusion cone is not visible are rejected; thus, their numbers contribute to the cloning efficiency. We found that the percentage of oocytes in which a clear protrusion cone was formed was higher (p<0.05) for BCB+ than that for BCB− oocytes (77.1% vs. 62.0%, respectively). A higher blastocyst rate for BCB+ than that for BCB− oocytes has been reported following IVF in many studies in cattle (Alm et al., 2005; Mirshamsi et al., 2013; Pujol et al., 2004; Silva et al., 2013) and buffalo (Manjunatha et al., 2007). It has been established through these studies that the level of G6PDH in the oocytes can be effectively used as a marker of their developmental competence following fertilization. We found that the blastocyst rate was nearly two-fold higher (p<0.001) for BCB+ than that for BCB− oocytes (43.4% vs. 22.7%) following SCNT using HMC, which agrees well with previous studies by Bhojwani et al. (2007) (30.7% vs. 2.0%) and Su et al. (2012) (39.0% vs. 12.1%,) in which SCNT was done by conventional micromanipulation-based technique. The process of SCNT involves reprogramming of a differentiated somatic cell by the oocyte cytoplasm into a totipotent state through a mechanism that differs drastically from that of fertilization. The results of our study and those of earlier studies mentioned above suggest that the G6PDH level of the oocyte is a valid marker of the ability of cytoplasm to reprogram a differentiated somatic cell.
Although the blastocyst rate and the TCN of blastocysts obtained with SCNT are higher than those achieved with IVF, as shown in this and in earlier studies (Selokar et al., 2013), the in vivo developmental competence of SCNT embryos is severely compromised. In comparison to the more than 40% birth rate obtained with IVF embryos, the live offspring rate obtained from cloned blastocysts of most of mammalian species is <5% (Campbell et al., 2007). This is primarily due to very high mortality during pre- and postimplantation development in vitro and in vivo. Moreover, the offspring born suffers from a high incidence of abnormalities such as large offspring syndrome (LOS), short life span, severe placental deficiency, respiratory problems, prolonged gestation, and dystocia (Yang et al., 2007; Young et al., 1998). These factors are believed to be due to incomplete or incorrect nuclear reprogramming of the donor somatic cell by the oocyte that affects the quality of SCNT embryos adversely. The quality of the SCNT embryos produced is, therefore, perhaps the most important factor influencing the cloning efficiency.
Cell number, especially the ICM:TE ratio, is an important criterion for evaluating blastocyst quality, as it might directly influence development after implantation in mammals (Yu et al., 2007). We found that the TCN, ICM cell number, and the ratio of the ICM:TE cell number were higher in BCB+ than in BCB− blastocysts, which agrees well with the results of Su et al. (2012) in bovine SCNT embryos. Our study extends these results by showing that the quality of BCB+ blastocysts was comparable to that of IVF blastocysts in terms of the TCN, ICM cell number, and the ratio of the ICM:TE cell number. This is important because placental abnormalities and early embryonic mortality, which is common in SCNT embryos, has been attributed to aberrant allocation of ICM and TE cells in preimplantation stages (Im et al., 2006). Higher ICM and lower TE cell numbers in cloned embryos may be responsible for insufficient placental development (Koo et al., 2002), and reduced TCN and ICM cell numbers in cloned embryos compared with IVF embryos have been reported to be correlated with abnormal Oct4 expression (Boiani et al., 2003).
Furthermore, our results show that in terms of apoptotic index BCB+ blastocysts were comparable to those produced by IVF and were superior to BCB− blastocysts. A lower level of apoptosis was further indicated by a higher expression level of BCL-XL in BCB+ and IVF blastocysts than that in BCB− blastocysts. BCL-XL is an antiapoptotic gene of the Bcl-2 family, which protects the cells from apoptosis. However, no difference was observed in the expression level of CASPASE-3, a proapoptotic gene among the three groups. The lowering of apoptotic index is particularly important for SCNT embryos because the higher apoptotic morphology and/or TUNEL labeling reported in SCNT bovine embryos compared to that in IVF embryos (Cui et al., 2011; Selokar et al., 2013) may be one of the important causes of the lower conception rate obtained with the former. The in vivo developmental competence of bovine SCNT-derived BCB+ blastocysts has been shown to be higher than that of BCB− blastocysts in terms of live offspring rate in a recent study (Su et al., 2012).
Aberrant epigenetic nuclear reprogramming of the donor somatic cell by the oocyte is believed to be the primary reason for low cloning efficiency. Some of the major epigenetic modifications of the genome, which affect the outcome of reprogramming and, therefore, the development epigenetic status of SCNT embryos profoundly, include the acetylation and methylation of histone tails (Yamanaka et al., 2009). We found that the global level of H3K18ac was higher in BCB+ than in BCB− blastocysts, which agrees with the results of Su et al. (2012), who reported a similar pattern in bovine SCNT blastocysts. Our results extend these by demonstrating that the BCB+ blastocysts are closer to IVF blastocysts in terms of similar global levels of H3K18ac. Increasing the global acetylation of histones is believed to alleviate the repression of transcription by facilitating chromatin remodeling and relieving methylated CpG sites, whereas hyperacetylation of histones is believed to facilitate the access of various factors to nucleosomes (Lee et al., 1993; Jones et al., 1998). These results indicate that an acetylation status similar to that of IVF blastocysts could be one of the reasons behind higher developmental competence of BCB+ blastocysts compared to BCB− blastocysts. However, further studies are required on other important epigenetic modifications to find out why BCB+ SCNT embryos are superior to BCB− embryos.
The methylation of the K9 residue of H3 histone is associated with inactive (constitutive heterochromatin) (Rice and Allis, 2001). We found that the level of dimethylation of H3K9 was not significantly different between BCB+ and BCB− blastocysts and was higher (p<0.05) than that in IVF blastocysts, which agrees with a previous report in sheep (Fu et al., 2012). The reasons behind hypermethylation of H3K9 in SCNT embryos relative to IVF embryos are not clear. However, this may have a significant influence on the outcome of reprogramming because a majority of cloned embryos exhibit H3-K9 hypermethylation, which is reported to be associated with DNA hypermethylation, suggesting a genome-wide failure of reprogramming, whereas H3-K9 methylation is reprogrammed in parallel with DNA methylation in IVF embryos (Santos et al., 2003). Moreover, H3K9 dimethylation plays an important role in a novel multistep program of inactivation of Oct4 following implantation (Feldman et al., 2006), suggesting that the effects hypermethylation of H3K9 may extend beyond early embryogenesis.
Successful embryonic development is dependent upon proper expression of specific genes, including OCT4, NANOG, and SOX2, which are closely associated with pluripotency and early embryonic development. We found that the expression level of these three pluripotency-related genes, which are expressed mainly in the ICM of blastocysts, was lower and that of two trophectoderm lineage-specific genes GATA2 and CDX2 was higher in the cloned than in IVF blastocysts. This agrees with the report of Zhang et al. (2014), who found OCT4 and SOX2 transcript levels to be higher in IVF than in SCNT bovine embryos. Aberrant expression of genes during early embryonic developmental stages is believed to be an important cause of not only embryonic losses but also for fetal abnormalities that continue during gestation and parturition resulting in abnormal offspring syndrome (Arnold et al., 2008; Walker et al., 1996). We found that the expression level of OCT4 and SOX2 was higher and that of GATA2 was lower in the BCB+ than in BCB− blastocysts. Whether this change in the expression of these important genes is sufficient to stimulate pluripotent embryonic cells to differentiate into those of trophoblast cell lineage needs to be studied further. Nevertheless, it indicates that besides the other reasons discussed above, BCB+ blastocysts may also be superior to BCB− blastocysts because of a lower magnitude of aberrance in the expression of these three genes.
The epigenetic abnormality of cloned blastocysts relative to those produced by IVF was further indicated by a higher transcript level of DNMT1, which maintains established patterns of methylation during DNA replication, although that of DNMT3a, which participates in establishing the methylation pattern de novo during preimplantation, was not different among the three groups. Drastically reduced transcript levels of Dnmt1 have been reported in cloned than in IVF or parthenote bovine embryos by Golding et al. (2011), who suggested that the hypermethylated status of the genome in cloned embryos could be because of attenuation of transcription of Dnmt1 in clones as a result of high levels of CpG methylation within the donor genome.
In conclusion, the results of the present study demonstrate that following SCNT by HMC, oocytes classified as BCB+ had higher in vitro developmental competence and resulted in formation of blastocysts with quality comparable to that of IVF blastocysts as indicated by apoptotic index, ICM cell number, ICM:TE ratio, and global level of H3K18ac, which were significantly different from those in blastocysts produced from BCB− oocytes. These results suggest that BCB staining can be effectively used for selection of developmentally competent oocytes for HMC.
Supplementary Material
Acknowledgments
The present work was funded by National Agriculture Innovative Project (NAIP) grant to S.K.S. (C 2-1-(5)/2007) and M.S.C. (C-2067 and 075). S.K.M., A.S., and N.L.S. are recipients of a Council of Scientific & Industrial Research Senior Research Fellowship (CSIR-SRF).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Alm H., Torner H., Lohrke B., Viergutz T., Ghoneim I.M., and Kanitz W. (2005). Bovine blastocyst development rate in vitro is influenced by selection of oocytes by brillant cresyl blue staining before IVM as indicator for glucose-6-phosphate dehydrogenase activity. Theriogenology 63, 2194–2205 [DOI] [PubMed] [Google Scholar]
- Arnold D.R., Fortier A.L., Lefebvre R., Miglino M.A., Pfarrer C., and Smith L.C. (2008). Placental insufficiencies in cloned animals—a workshop report. Placenta 29 (Suppl A), S108–S110 [DOI] [PubMed] [Google Scholar]
- Boiani M., Eckardt S., Leu N.A., Scholer H.R., and McLaughlin K.J. (2003). Pluripotency deficit in clones overcome by clone–clone aggregation: Epigenetic complementation? EMBO J 22, 5304–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhojwani S., Alm H., Torner H., Kanitz W., and Poehland R. (2007). Selection of developmentally competent oocytes through brilliant cresyl blue stain enhances blastocyst development rate after bovine nuclear transfer. Theriogenology 67, 341–345 [DOI] [PubMed] [Google Scholar]
- Campbell K.H.S., Fisher P., Chen W.C., Choi I., Kelly R.D., Lee J.H., and Xhu J. (2007). Somatic cell nuclear transfer: Past, present and future perspectives. Theriogenology 68, S214–S231 [DOI] [PubMed] [Google Scholar]
- Catalá M.G., Izquierdo D., Uzbekova S., Morató R., Roura M., Romaguera R., Papillier P., and Paramio M.T. (2011). Brilliant Cresyl Blue stain selects largest oocytes with highest mitochondrial activity, maturation-promoting factor activity and embryo developmental competence in prepubertal sheep. Reproduction 142, 517–527 [DOI] [PubMed] [Google Scholar]
- Cui X.S., Xu Y.N., Shen X.H., Zhang L.Q., Zhang J.B., and Kim N.H. (2011). Trichostatin A modulates apoptotic-related gene expression and improves embryo viability in cloned bovine embryos. Cell. Reprogram. 13, 179–189 [DOI] [PubMed] [Google Scholar]
- de Loos F., van Vliet C., van Maurik P., and Kruip T.A. (1989). Morphology of immature bovine oocytes. Gamete Res. 24, 197–204 [DOI] [PubMed] [Google Scholar]
- Feldman N., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., and Bergman Y. (2006). G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 8, 188–194 [DOI] [PubMed] [Google Scholar]
- Fu L., Zhang J., Yan F.X., Guan H., An X.R., and Hou J. (2012). Abnormal histone H3K9 dimethylation but normal dimethyltransferase EHMT2 expression in cloned sheep embryos. Theriogenology 78, 1929–1938 [DOI] [PubMed] [Google Scholar]
- Golding M.C., Williamson G.L., Stroud T.K., Westhusin M.E., and Long C.R. (2011). Examination of DNA methyltransferase expression in cloned embryos reveals an essential role for Dnmt1 in bovine development. Mol. Reprod. Dev. 78, 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im G.S., Seo J.S., Hwang I.S., Kim D.H., Kim S.W., Yang B.C., Yang B.S., Lai L., and Prather R.S. (2006). Development and apoptosis of pre-implantation porcine nuclear transfer embryos activated with different combination of chemicals. Mol. Reprod. Dev. 73, 1094–1101 [DOI] [PubMed] [Google Scholar]
- Jones P.L., Veenstra G.J.C., Wade P.A., Vermaak D., Kass S.U., Landsberger N., Strouboulis J., and Wolffe A.P. (1998). Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19, 187–191 [DOI] [PubMed] [Google Scholar]
- Koo D.B., Kang Y.K., Choi Y.H., Park J.S., Kim H.N., Oh K.B., Son D.S., Park H., Lee K.K., and Han Y.M. (2002). Aberrant allocations of inner cell mass and trophectoderm cells in bovine nuclear transfer blastocysts. Biol. Reprod. 67, 487–492 [DOI] [PubMed] [Google Scholar]
- Lee D.Y., Hayes J.J., Pruss D., and Wolffe A.P. (1993). A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72, 73–84 [DOI] [PubMed] [Google Scholar]
- Mangia F., and Epstein CJ. (1975). Biochemical studies of growing mouse oocytes: Preparation of oocytes and analysis of glucose-6-phosphate dehydrogenase and lactate dehydrogenase activities. Dev. Biol. 45, 211–220 [DOI] [PubMed] [Google Scholar]
- Manjunatha B.M., Gupta P.S.P., Devaraj M., Ravindra J.P., and Nandi S. (2007). Selection of developmentally competent buffalo oocytes by brilliant cresyl blue staining before IVM. Theriogenology 68, 1299–1304 [DOI] [PubMed] [Google Scholar]
- Mirshamsi S.M., Karamishabankareh H., Ahmadi-Hamedani M., Soltani L., Hajarian H., and Abdolmohammadi A.R. (2013). Combination of oocyte and zygote selection by brilliant cresyl blue (BCB) test enhanced prediction of developmental potential to the blastocyst in cattle. Anim. Reprod. Sci. 136, 245–251 [DOI] [PubMed] [Google Scholar]
- Panda S.K., George A., Saha A.P., Sharma R., Manik R.S., Chauhan M.S., Palta P., and Singla S.K. (2011). Effect of cytoplasmic volume on developmental competence of buffalo (Bubalus bubalis) embryos produced through hand-made cloning. Cell. Reprogram. 13, 257–262 [DOI] [PubMed] [Google Scholar]
- Pujol M., Lopez-Bejar M., and Paramio M.T. (2004). Developmental competence of heifer oocytes selected using the brilliant cresyl blue (BCB) test. Theriogenology 61, 735–744 [DOI] [PubMed] [Google Scholar]
- Rice J.C., and Allis C.D. (2001). Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr. Opin. Cell. Biol. 13, 263–273 [DOI] [PubMed] [Google Scholar]
- Roca J., Martinez E., Vazquez J.M., and Lucas X. (1998). Selection of immature pig oocytes for homologous in vitro penetration assays with the brilliant cresyl blue test. Reprod. Fertil. Dev. 10, 479–485 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gonzalez E., Lopez-Bejar M., Velilla E., and Paramio M.T. (2002). Selection of prepubertal goat oocytes using the brilliant cresyl blue test. Theriogenology 57, 1397–1409 [DOI] [PubMed] [Google Scholar]
- Santos F., Zakhartchenko V., Stojkovic M., Peters A., Jenuwein T., Wolf E., Reik W., and Dean W. (2003). Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr. Biol. 13, 1116–1121 [DOI] [PubMed] [Google Scholar]
- Selokar N. L., Saini M., Muzaffer M., Krishnakanth G., Saha A.P., Chauhan M.S., Manik R., Palta P., Madan P., and Singla S.K. (2012a). Roscovitine treatment improves synchronization of donor cell cycle in G0/G1 stage and in vitro development of handmade cloned buffalo (Bubalus bubalis) embryos. Cell. Reprogram. 14, 146–154 [DOI] [PubMed] [Google Scholar]
- Selokar N. L., Shah R. A., Saha A. P., Muzaffar M., Saini M., Chauhan M.S., Manik R.S., Palta P., and Singla S.K. (2012b). Effect of post-fusion holding time, orientation and position of somatic cell-cytoplasts during electrofusion on the development of handmade cloned embryos in buffalo (Bubalus bubalis). Theriogenology 78, 930–936 [DOI] [PubMed] [Google Scholar]
- Selokar NL, St John L, Revay T, King W.A., Singla S.K., and Madan P. (2013). Effect of histone deacetylase inhibitor valproic acid treatment on donor cell growth characteristics, cell cycle arrest, apoptosis and handmade cloned bovine embryo production efficiency. Cell. Reprogram. 15, 531–542 [DOI] [PubMed] [Google Scholar]
- Selokar N.L., Saini M., Palta P., Chauhan M.S., Manik R., and Singla S.K. (2014). Hope for restoration of dead valuable bulls through cloning using donor somatic cells isolated from cryopreserved semen. PloS One 9, e90755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., George A., Kamble N.M., Singh K.P., Chauhan M.S., Singla S.K., Manik R.S., and Palta P. (2011). Optimization of culture conditions to support long-term self-renewal of buffalo (Bubalus bubalis) embryonic stem cell-like cells. Cell. Reprogram. 13, 539–549 [DOI] [PubMed] [Google Scholar]
- Silva D.S., Rodriguez P., Galuppo A., Arruda N.S., and Rodrigues J.L. (2013). Selection of bovine oocytes by brilliant cresyl blue staining: Effect on meiosis progression, organelle distribution and embryo development. Zygote 21, 250–255 [DOI] [PubMed] [Google Scholar]
- Su J., Wang Y., Li R., Peng H., Hua S., Li Q., Quan F., Guo Z., and Zhang Y. (2012). Oocytes selected using BCB staining enhance nuclear reprogramming and the in vivo development of SCNT embryos in cattle. PLoS One 7, e36181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajta G., Lewis I.M., Trounson A.O., Purup S., Maddox-Hyttel P., Schmidt M., Pedersen H.G., Greve T., and Callesen H. (2003). Handmade somatic cell cloning in cattle: Analysis of factors contributing to high efficiency in vitro. Biol. Reprod. 68, 571–578 [DOI] [PubMed] [Google Scholar]
- Walker S.K., Hartwich K.M., and Seamark R.F. (1996). The production of unusually large offspring following embryo manipulation: Concepts and changes. Theriogenology 45, 111–120 [Google Scholar]
- Wassarman M. (1988). The mammalian ovum. In: Knobil E, Neil D, eds. The Physiology of Reproduction, vol. 1 Raven Press, New York, pp. 69–102 [Google Scholar]
- Yamanaka K., Sugimura S., Wakai T., Kawahara M., and Sato E. (2009). Acetylation level of histone H3 in early embryonic stages affects subsequent development of miniature pig somatic cell nuclear transfer embryos. J. Reprod. Dev. 55, 638–644 [DOI] [PubMed] [Google Scholar]
- Yang X.Z., Smith S.L., Tian X.C., Lewin H.A., Renard J.P., and Wakayama T. (2007). Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat. Genet. 39, 295–302 [DOI] [PubMed] [Google Scholar]
- Young L.E., Sinclair K.D., and Wilmut I. (1998). Large offspring syndrome in cattle and sheep. Rev. Reprod. 3, 155–163 [DOI] [PubMed] [Google Scholar]
- Yu Y., Ding C., Wang E., Chen X., Li X., Zhao C., Fan Y., Wang L., Beaujean N., Zhou Q., Jouneau A., and Ji W. (2007). Piezo-assisted nuclear transfer affects cloning efficiency and may cause apoptosis. Reproduction 133, 947–954 [DOI] [PubMed] [Google Scholar]
- Zakhartchenko V., Stojkovic M., Palma G., Wolf E., and Brem G. (1997). Enucleation of bovine oocytes with minimal cytoplasmic volume: Effect on development of nuclear transfer embryos. Theriogenology 47, 238 [Google Scholar]
- Zhang H., Wang Y., Sang Y., Zhang Y., and Hua S. (2014). Combination of S-adenosylhomocysteine and Scriptaid, a non-toxic epigenetic modifying reagent, modulates the reprogramming of bovine somatic-cell nuclear transfer embryos. Mol. Reprod. Dev. 81, 87–97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.