Abstract
Despite increased comprehension of AML pathogenesis, current treatment strategies have done little to improve upon standard induction chemotherapy to induce long-term remissions. Since the identification of the leukemic stem cell, efforts have been placed on identifying therapeutically actionable pathways that distinguish this increasingly important cellular compartment. With the advent of increased genome sequencing efforts and phenotypic characterization, opportunities for personalized treatment strategies are rapidly emerging. In this review, we highlight recent advances in the understanding of leukemic stem cell biology and their potential for translation into clinically relevant therapeutics. NF-kappa B activation, Bcl-2 expression, oxidative and metabolic state, and epigenetic modifications all bear their own clinical implications. With advancements in genetic, epigenetic, and metabolic profiling, personalized strategies may be feasible in the near future to improve outcomes for AML patients.
Keywords: Leukemia stem cells, translational medicine, molecular targeted therapies, hematopoietic stem cells, acute myelogenous leukemia
Introduction
Acute myelogenous leukemia (AML) comprises a heterogeneous group of diseases defined by specific morphologic, genetic and clinical characteristics. Identification of recurrent cytogenetic abnormalities and somatic mutations within the AML genome has proven useful in delineating AML prognosis and have thereby been incorporated into the World Health Organization classification of myeloid leukemias [1, 2].
Standard induction therapy for AML is comprised of cytarabine arabinoside (Ara-C) in combination with an anthracycline such as daunorubicin, followed by high dose Ara-C consolidation [3]. Recently, the German AML Intergroup completed a large prospective study comparing 5 different induction strategies vs. the standard induction arm and found no significant difference in overall survival (OS), disease free survival (DFS), or event free survival (EFS) when compared to standard treatment [4]. This data revealed that variations and intensifications based on standard therapy have done little to improve upon survival and may signify the limited efficacy of chemotherapy-based induction strategies. Therefore, to improve outcomes, personalized strategies built upon a better understanding of the oncogenesis of AML are likely needed.
There is growing evidence that a subset of AML cells, comprised of early stem/progenitor cells termed leukemic stem cells (LSCs), give rise to the leukemic blast. Inadequate eradication of the LSC population is thought to contribute to the high incidence of relapse and poor overall survival observed in AML. Recent data has highlighted subtle molecular differences between normal hematopoietic stem cells (HSCs) and LSCs that serve as targets for therapeutic intervention, including genetic and epigenetic alterations, dependence on survival pathways, and levels of reactive oxygen species (ROS) [5].
Targeting leukemia stem cells (LSCs) through their aberrant immunophenotype
LSCs were initially characterized in 1994 when it was discovered that a subpopulation of CD34+CD38- cells were capable of serial engraftment in non-obese severe combined immune deficient (NOD/SCID) mice [6]. Subsequent studies have further expanded this characterization, detailing the repertoire of phenotypic markers and transcriptional signatures associated with the LSC. Among these markers, CD34+ CD38-, CD71-, HLA-DR-, CD123+, and CD25+ have been described to define the LSC population of most AML patients [6-8]. Specifically, CD123 and CD25 have demonstrated potential as both therapeutic and prognostic markers. Antibody-mediated targeting of CD123 (interleukin-3 receptor alpha) has been shown to eliminate LSC in preclinical animal models [9]. In addition, CD123-specific, single chain Fv antibody fragments coupled to a truncated Pseudomonas exotoxin A and FC receptor III have proven effective in targeting AML cell lines bearing CD123 [10]. Currently, there are two early phase 1 clinical trials targeting CD123 in patients with relapsed refractory AML, and in patients with de novo AML in first complete remission (CR) {NCT00397579, NCT01632852}.
Moreover, CD25 (interleukin-2 receptor alpha) expression on LSCs has also been recently shown to independently predict early treatment failure in AML [11]. Currently, immunotoxin-based therapies targeting CD25 have had early success as single agents in Phase II trials in patients with newly diagnosed T-Cell Lymphoma and refractory T-cell and B-cell lymphomas [12-14]. Given the recent evidence that CD25 can be found expressed on LSCs, anti-CD25-based therapies are of potential utility in the AML setting.
Other phenotypic markers are also currently under evaluation. Targets such as C-type lectin-like molecule-1 (CLL-1) positive LSCs have been detected in patients while in remission and high expression of CLL-1 has correlated with quicker relapse [15]. Antibodies targeting this marker have shown effective antibody dependent and complement dependent cytotoxicity in AML cell lines and fresh primary samples [16].
Likewise CD44 and CD47 antibody mediated targeting have also demonstrated effectiveness in xenotransplantation models [17, 18]. Use of an activating monoclonal antibody targeting CD44 has been reported to eliminate LSCs through distinct mechanisms, by disrupting their homing capacity to LSC supportive microenvironment and by inducing differentiation [17]. The CD47 blockade appears to eliminate LSCs by enabling macrophage-mediated ingestion via disruption of CD47 and macrophage SIRP-alpha protein.
Given the importance of immunophenotypic markers in identifying patients who may do poorly, targeting these molecules and designing rational clinical trials incorporating this data will be important next steps going forward. The current clinical trials targeting CD123 are designed in both the relapsed refractory setting and in patients in first remission. The potential of these therapies to seek and destroy low volume minimal residual disease (MRD) makes them attractive candidates to use in combination, consolidation, and, or as single agent maintenance regimens.
Targeting NF-kappa B
The role of NF-kappa B and disruption of its regulatory pathways which stand to promote cell survival and proliferation have been described in a variety of malignancies [19-21]. Guzman et al first reported in 2001 that NF-kappa B is constitutively active in the LSC population but not in the normal HSC population and that this difference could be used to selectively eradicate LSCs via proteasome-inhibitor based inhibition of NF-kappa B [22, 23]. Later studies revealed that parthenolide (PTL), a naturally occurring small molecule and well-established single agent NF-kappa B inhibitor, could preferentially induce apoptosis in the LSC population via a robust increase in ROS, NF-kappa B inhibition, and pro-apoptotic activation of p53 [24]. These data suggested the utility of NF-kappa B disruption in the clinic.
Indeed, a recent clinical trial has corroborated this utility. In a recent Phase II trial, 95 patients aged 60-75 with previously untreated de novo or secondary AML were treated with bortezomib in combination with standard induction therapy. Sixty-five percent of patients achieved a CR, and DFS and OS rates of 7.5 and 17.5 months, respectively [25, 26]. This is in comparison to a similar control group of elderly patients in a separate Phase II study who received standard induction, revealing a CR rate of 55% and DFS and OS rates of 8 and 9 months, respectively [27]. While cross study comparisons should be evaluated with caution, the results with bortezomib are encouraging. Whether these improvements arose from a reduction of LSC burden remains to be determined in future studies.
Therapeutic implications of dysregulation of ROS metabolism
ROS metabolism has been shown to be critical to the HSC population. Studies have demonstrated that ATM null mice experience rising levels of ROS with advancing age, leading to activation of p38/Mitogen Activated Protein Kinase (MAPK) pathway and thus diminished lifespan of HSCs in vivo [28]. In addition, deficiency of the Forkhead transcription factor (FOXO) family of genes was later shown to result in a severely defective HSC compartment associated with emergence from quiescence, terminal differentiation, and with resultant increases in ROS [29]. The FOXO genes encode for a family of transcription factors that are thought to be tumor suppressors in a wide range of malignancies. The disruption of their function via phosphorylation and nuclear exclusion secondary to phosphoinositide 3-kinase (PI3K)/Akt kinase activity can promote tumorigenesis, and/or, resistance to therapies [30-32]. However, in AML the role of FOXO regulation is contrary to that described in other solid tumors and lymphoid malignancies referenced above.
Sykes et al found that FOXO activity was increased in 40% of evaluated primary human AML samples regardless of subtype. The group noted FOXO activity was required for LSC function and described a resistance mechanism to constitutive Akt activity, via upregulation of c-Jun N-terminal Kinase (JNK)/c-Jun signaling pathways. FOXO inhibition with silencing RNA and gene deletion decreased disease burden, diminished leukemia stem cell function and increased survival in human allele MLL-AF9 AML mouse models. Pharmacologic JNK inhibition further augmented myeloid maturation and apoptosis [33]. The increased activity of FOXO transcription factors in AML appears to be involved in a wide range of AML subtypes, and inhibition is associated with improved outcomes in preclinical models. While the exact role and regulation of FOXO transcription factors in AML remains unclear, this pathway undoubtedly remains important for LSC function and warrants further development and characterization with the hope that therapies can be tailored to AML subtypes reliant on this pathway.
LSCs generally reside in a low ROS niche within the bone marrow [34]. This population of cells relies heavily on oxidative phosphorylation and lack of an ability to utilize glycolysis for energy production. This is in stark contrast to normal HSC that can switch to glycolysis when oxidative phosphorylation is inhibited [35]. Lagadinou et al found that B-cell Lymphoma-2 (Bcl-2) is upregulated in the LSC population and regulates oxidative phosphorylation. Importantly, targeted inhibition of Bcl-2, lead to decreased oxidative phosphorylation, decreased ATP levels and increased apoptosis in LSCs. These agents effectively targeted the LSC population, decreased engraftment of pretreated LSCs and also decreased secondary engraftment of xenotransplanted mice exposed to the inhibitors.
Mitochondrial biogenesis and mitochondrial proteins as LSC targets
Relative mitochondrial readiness for apoptosis (“priming”) has been utilized to evaluate the sensitivity of AML cells to chemotherapeutic agents. Vo and colleagues evaluated mitochondrial priming by characterizing BCL-2 family member proteins and their accompanying BCL-2 Homology 3(BH3) only family members in AML patient samples [36]. BH3 only family members represent proapoptotic activators that can interact with and negate antiapoptotic BCL-2 activity [36]. They reason that cellular stress induced by chemotherapy induces proapoptotic activators, thereby overwhelming cells that may already have high levels of BH3-only family members (primed for death; “mitochondrial priming”). In contrast, cells that have higher levels of anti-apoptotic proteins will not be able to respond to therapy as efficiently. Thus, the authors demonstrate that mitochondrial priming was a determinant of topoisomerase II inhibitor efficacy in vitro. Pretreatment AML samples were isolated and mitochondrial priming was correlated with chemotherapy induction success, defined as achieving a 5-year CR. Within each risk category established by European Leukemia Network criteria, it was found that patients with low priming required an allogeneic transplant for long-term survival. Furthermore, using BH3 profiling, they identified a dependency of AML cells to Bcl-2. Thus, with inhibition of Bcl-2, chemotherapeutic efficacy was significantly increased. Importantly, these studies noted that normal HSCs rely more on Mcl-1 thus providing a therapeutic window for Bcl-2 inhibitor ABT-737 [36]. This study suggests that the use of mitochondrial priming may aid to identify patients more likely to respond to chemotherapy, patients that may benefit from Bcl-2 inhibitor combination therapy, or patients that may benefit from bone marrow transplant.
Additional studies by Goff et al further highlighted the importance of the Bcl-2 anti-apoptotic proteins in LSCs [37]. In these studies, blast crisis chronic myeloid leukemia (bcCML) LSCs were found to over-express several anti-apoptotic Bcl-2 family member splice isoforms (i.e. Bcl-2L, Mcl1L, Bcl-xL, Bfl-1L) when compared to chronic phase progenitors. In addition, LSCs further demonstrated higher levels of Bcl-2L, Bcl-xL, and Bfl-1L compared to normal HSCs. Among these proteins, only Bcl-xL elevation was correlated to Bcr-Abl expression, suggesting a secondary mechanism for overexpression of the other anti-apoptotic family members. These secondary mechanisms may be targetable and are potential reasons for the observed resistance of LSCs to TKIs [38]. Quiescent LSCs isolated from these patients demonstrated engraftment in mouse bone marrow and resistance to TKI-induced killing despite adequate inhibition of Bcr-Abl, consistent with previously reported LSC resistance to TKIs [38]. Given the dependence of CML LSCs on pro-survival Bcl-2 family members, the ability of the pan-active Bcl-2 protein family antagonist, sabutoclax, to sensitize CML LSCs was determined. These experiments revealed that sabutoclax-treated LSCs exhibited decreased survival, colony forming capability, and capacity for serial xenotransplantation. Moreover, tumor burden was reduced in established CML xenografts upon dual treatment with sabutoclax and dasatanib relative to dasatinib alone. Thus, the dependence of CML LSCs on pro-survival Bcl-2 family member signaling represents a potentially important consideration for patients who require change in TKI therapy due to a lack of response.
In other studies by Konopleva and colleagues, it was noted that treatment of AML cell lines with ABT-737, a specific inhibitor of Bcl-2 and Bcl-xL, resulted in the upregulation of Mcl-1 with an increased binding to pro-apoptotic Bim. In lymphoid malignancies, acquired resistance to ABT-737 seems in part due to this upregulation of Mcl-1 [39]. Mcl-1 is known to accumulate via MAPK/ERK signaling, which phosphorylates and stabilizes Mcl-1 [40]. Notably, dual treatment with ABT-737 and a MEK inhibitor prevented Mcl-1 upregulation and increased apoptosis of LSCs derived from patient samples [41]. Further efforts by Rahmani et al support the importance of Mcl-1 suppression in overcoming acquired resistance to Bcl-2 inhibition. These studies demonstrated that the efficacy of the BH3 mimetic, obutoclax, was enhanced by sorafenib-mediated suppression of Mcl-1 [42]. The combination resulted in enhanced killing of AML cell lines and prolonged survival of mice bearing established AML cell line xenotransplants.
Recent evidence has shown that mitochondrial translation represents a target to ablate AML cells and LSCs [43]. A chemical screen identified the FDA-approved antimicrobial compound tigecycline as a potential therapeutic agent. The authors reveal that pretreatment with tigecycline effectively reduced the repopulating capacity of AML cells in NOD/SCID mice while leaving normal HSCs repopulating capacity relatively unaffected. The mechanism of action appears to be potent inhibition of mitochondrial ribosomes, resulting in the inhibition of mitochondrial translation. They found that LSCs have larger mitochondrial mass, higher mitochondrial DNA copy number, and increased rate of oxygen consumption compared to their normal counterparts. AML cells exhibited a direct correlation between higher mitochondrial mass and sensitivity to tigecycline suggesting that evaluation of mitochondrial mass may be useful to identify patients that could benefit from therapies targeting mitochondrial function. These effects were evident in the stem, progenitor and AML bulk populations. Combination therapy with tigecycline and other agents that induce ROS may be an effective therapeutic strategy to further consolidate the LSC compartment when in remission. It will be important to correlate outcomes with mitochondrial mass and if directly correlated to poor outcome, providing an assay that can be potentially used to direct mitrochondrial translation inhibition in an individual's treatment regimen.
In summary, dependence of LSCs on mitochondrial function or mitochondrial proteins such as Bcl-2 for survival has important clinical implications. Analysis of mitochondrial mass or mitochondrial priming in primary patient samples may provide the ability to select patients more likely to respond to standard chemotherapy, sensitive to Bcl-2 inhibitors, or sensitive to mitochondrial translation inhibitors. BH3 profiling can now be rapidly incorporated into clinical decision making and help predict an individual's response to therapy [44].
Epigenetic regulators
Epigenetic modifications have a clear and established role in stratification of AML where specific epigenetic profiles are associated with clinical features of the disease [45]. However, recent studies have begun to further delineate the AML epigenome through comparisons of stem, progenitor and mature AML cells. In a recent small study, DNA methylation differences between stem (CD34+CD38-), progenitor (CD34+CD38+), and mature (CD34-) AML cells were evaluated focusing on genes known to be differentially methylated in AML. The group found no consistent differences in DNA methylation patterns between stem, progenitor or mature cells in the evaluated primary AML samples [46]. In contrast, chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) identified differences in chromatin associated with H3K4me3(K4) and H3K27me3(K27) marks when evaluated in purified stem, progenitor, and mature AML subpopulations derived from 2 individual AML patients as well as 3 pooled patient samples. Consistent with the known role of these methylated histones [47], K4 marks were mostly found at the promoter regions while K27 marks were found distributed from transcription start sites (TSS) and transcription end sites (TES) [46]. Importantly, these patterns were similar between stem and progenitor cells. Interestingly, when testing for differences between stem and progenitor populations, enrichment for ERK/MAPK, hypoxia and NRF2 mediated oxidative stress response pathways were revealed in one of the AML stem cell K4 analysis patient samples [46]. These pathways are very consistent with those reported in the ROS-low LSC population found in the hypoxic areas of the bone marrow and represent a chemoresistant LSC population [35, 37]. Further analysis with a larger cohort of samples is warranted to help define potential epigenetic profiles that may implicate candidate genes and pathways responsible for LSC biology that could potentially be exploited in the future.
Global epigenetic modifier agents such as DNA methyltransferase (DNMT) inhibitors and histone deacetylase (HDAC) inhibitors are currently being tested for efficacy in AML, alone or in combination with other therapies [48-50]. Thus, an overall interest in testing the effects of these agents in LSCs has increased. Recently, Craddock and colleagues serially measured the changes in the LSC population in patients undergoing treatment with 5’-azacitidine and sodium valproate (VAL-AZA) [51]. This study found that the treatment had no effect on LSC populations in non-responding patients. Interestingly, patients who achieved CR or complete remission with incomplete blood count (CRi) showed a decrease in LSCs. However, complete eradication of LSCs was not achieved [51]. The serial tracking of LSCs employed in this clinical trial indicated an expansion of LSCs just prior to overt relapse.
The lack of LSC eradication with DNMT inhibitors and HDAC inhibitors as well as the intra/inter-patient heterogeneity observed in epigenetic marks suggests the need for inhibitors that are better tailored to the specific pattern of genetic and epigenetic alterations evident in an individual's AML. Examples of new emerging therapeutic targets are DOT1L and LSD1.
DOT1L is a highly conserved histone H3K79 methyltransferase known to play a role in cell cycle regulation and transcriptional elongation [52]. In leukemia, DOT1L has been found to regulate genes critical to LSC self-renewal and survival. Importantly, DOT1L was also found to be required for initiation and maintenance of MLL-AF9-induced leukemia. DOT1L downregulation has been shown to decrease the expression of HOXA genes [53-55]. Therefore, specific inhibitors for DOT1L have been generated and are currently under evaluation, such as EPZ004777 [56]. DOT1L represents a novel target with the potential to ablate LSCs in MLL driven leukemias.
Another epigenetic regulator of interest is LSD1 (KDM1/AOF2), a lysine-specific demethylase. LSD1 demethylates H3K4 and H3K9 and has been found to be highly expressed in AML patients. In a recent study, it was demonstrated that a loss of RARα2 expression in AML was associated with a reduction in H3K4me2 on the RARA2 promoter. Thus, targeting LSD1 was determined to restore the expression of RARα2 and confer sensitivity to ATRA, causing differentiation and cell death of AML cells [57].
Further studies have shown that the histone methyltransferase inhibitor, 3-deazaneplanocin A (DZNep), disrupts the polycomb-repressive complex 2 (PRC2) and thereby inducing apoptosis in AML cells [58, 59]. It is known that PRC2 mediates gene silencing through H3K27 trimethylation [60]. The methyltransferase activity of PRC2 is conferred by the well-known component, EZH2 [61]. Target genes of PRC2 include transcription factors and signaling molecules important in cell differentiation [62, 63]. DZNep treatment of AML cells was shown to induce apoptosis of LSC subpopulations via reactivation of thioredoxin-binding protein 2 (TXNIP), resulting in an increase of ROS [59]. Reactivation of TXNIP by DZNep resulted from PRC2 depletion and the subsequent decrease of H3K27me3. Furthermore, DZNep had a negligible effect on normal HSC colony formation [59]. Recent studies have suggested that EZH2 may function as both an oncogene and a tumor suppressor in myeloid cells [64]. While further work is needed to improve our understanding of PRC2 function in leukemogenesis, inhibitors targeting this complex can affect LSC survival and need further investigation and development as they show promise in preclinical models.
Isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) are frequently mutated in AML and cause global changes in DNA methylation [65]. The commonly observed mutations in AML are in conserved arginine residues (IDH1-R132, IDH2-R140 and IDH2-R172) resulting in a neomorphic enzymatic activity that converts α-ketoglutarate (α-KG) to 2-hydroxyglutarate (2HG) [66]. The aberrant production of 2HG affects the DNA demethylase, Tet methylcytosine deoxygenase 2 (TET2), resulting in DNA hypermethylation and impaired hematopoietic differentiation [67]. Production of 2HG leads to increased ROS, while the decreased αKG results in stabilization of Hif-1α. These changes affect quiescence and self-renewal capabilities of HSCs and are thought to contribute to leukemic transformation [68].
IDH mutations and Tet2 depletion have been shown to disrupt hematopoietic differentiation and are associated with increased stem cell marker expression [67]. Importantly, new small molecule inhibitors are now available and can target specific IDH mutations representing a therapeutic opportunity to target AML harboring these mutations [69, 70].
Discussion
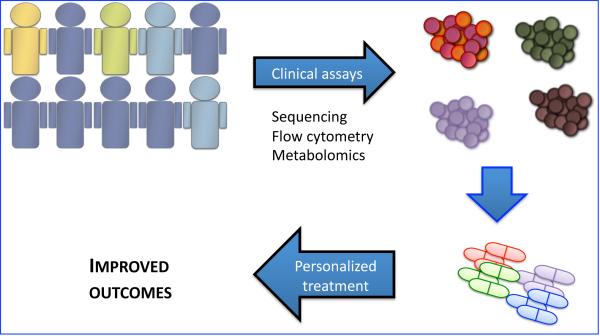
Despite advances in our understanding of the pathogenesis of AML we have failed to improve outcomes in patients over the past 20 years. While the limit of effectiveness for chemotherapy alone may have been met, it is encouraging to know that we continue to find opportunities to exploit within the AML genome. Targeting the LSC via its specific immunophenotype, oxidative stress pathways, NF-kappa B regulation, Bcl2 reliance and epigenetic mechanisms provide opportunities for combining targeted therapies with standard treatment. The targets outlined in this review are highlighted in Table 1. More important, the incorporation of LSC directed therapies will result from specific assays that will allow for tailored therapies to improve the disease outcome for AML patients, as highlighted in the schematic in Figure 1.
Table 1.
Targets of interest to target leukemia stem cells in AML patients.
| Target | Reference | Therapeutic approach | Stage |
|---|---|---|---|
| CD123 | [9-11] | DT388-IL3 (SL-401); anti-CD123 monoclonal antibody (CSL360) | Phase I/II |
| CD25 | [8] | Humanized anti-CD25 antibody (daclizumab), immunotoxin denileukin diftitox | Phase I/II |
| NF-kappaB | [23,26,27] | Bortezomib | Phase I/II |
| BCL-2 | [33] | Obatoclax, Sabutoclax | Phase I/II, Pre-clinical |
| DOT1L | [53] | EPZ004777 | Pre-clinical |
| LSD1 | [54] | Tranylcypromine (TCP) | Pre-Clinical |
| PRC2 complex | [56] | DZNep | Preclinical |
| IDH1/2 | [66,67] | AGI-6780, AGI-5198 | Pre-clinical |
| DNMT inhibitors | [46-47] | Decitabine, Azacitidine | Phase I/II/III |
| Mitochrondrial translation | [43] | Tigecycline | Pre-clinical |
Figure 1. Implementation of personalized strategies to target leukemia stem cells in AML.
Current technological advances will allow for tailored therapeutic approaches based on a full characterization of AML tumors. Genomic squencing, transcriptional profiling, flow cytometry and metabolomic assays (e.g. evaluation for IDH1/IDH2 mutations, stem cell immunophenotype, mitochondrial mass, mitochondrial priming) will allow for a classification of patients and better identification of actionable therapeutic targets.
Currently personalization of AML treatment targeting LSCs is limited by the emergent need for induction therapy and the large scope of sophisticated testing that is required to evaluate an individual's genetic, epigenetic, metabolic profile. While this is in no doubt a daunting task we feel it is by no means insurmountable.
We currently have the ability to immunophenotype and design clinical trials around targeting a specific immunophenotype defining poor prognostic AML and LSCs. Several monoclonal antibodies have been successful in preclinical models and are just now entering into the clinic as early Phase I trials. Going forward it will be imperative to trial these specific therapeutics in enriched populations both in the relapsed refractory setting and in combination with consolidation regimens or as maintenance therapies while in remission.
While targeting NF-kappa B and oxidative stress responses may be difficult to personalize, they still remain fundamental to LSC biology. Agents affecting these pathways and responses, like bortezomib, have recently been tested in the clinic and show promising results [26]. Likewise further development of FOXO pathway inhibitors could exploit LSC reliance on this pathway. Personalization, in this sense, encompasses a fundamental change in how we will treat AML going forward. Monitoring and eliminating LSCs should become an increasingly important endpoint and goal in prospective studies.
Several studies have shown the utility of gene expression profiling to help risk-stratify patients or predict response to induction chemotherapy [71-74]. We are just now beginning to understand the significance of multiple mutations and how they interact in AML. Designing clinical studies incorporating multi-gene mutational and expression analysis will help elucidate these answers and may eventually help strategize an individual's treatment course.
Epigenetic profiling with DNA methylation signatures, likewise, has demonstrated the ability to further delineate AML subtypes with specific recurrent mutations [45]. Histone methylation patterns are becoming increasingly important in the pathophysiology of AML. As such, specific histone methylation profiles have been correlated with outcomes [75]. The proteins involved in these processes have served as targets in several of the studies outlined in this review. It holds potential that individual epigenetic profiles may be associated with specific epigenetic regulation pathways, leading to a precise use of specific inhibitors and treatments. The dynamic nature of the epigenome holds promise as a possible biomarker that may also dictate treatment strategies.
There is a large body of data suggesting that targeting mitochondrial regulation of apoptosis is important and effective in AML. It is currently possible to perform same day BH3 profiling on patient samples, and this technology could be incorporated into prospective clinical trials [44]. BCL-2 inhibitors may be best utilized in selected populations in combination with induction chemotherapy, as they have been shown to be chemosensitizing agents [36].
While true personalization of AML is still years away, we have developed the individual pieces that, if coordinated and placed strategically within a developed open infrastructure could set the stage for improved long-term outcomes in patients who have been treated the same way for nearly 30 years.
Acknowledgments
M.L.G. is funded by the US National Institutes of Health (NIH) through the NIH Director's New Innovator Award Program, 1 DP2 OD007399-01, National Cancer Institute (R21 CA158728-01A1), Leukemia and Lymphoma Foundation (LLS 6330-11 and LLS 6427-13) and M.L.G is a V Foundation Scholar. M.L.G. is a 2010 V-Foundation Scholar.
Footnotes
Author contributions:
Monica L. Guzman: Conception and design, financial support, wrote the manuscript. John N. Allan: Conception and design, wrote the manuscript.
References
- 1.Grimwade D, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92(7):2322–33. [PubMed] [Google Scholar]
- 2.Vardiman JW, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–51. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 3.Rai KR, et al. Treatment of acute myelocytic leukemia: a study by cancer and leukemia group B. Blood. 1981;58(6):1203–12. [PubMed] [Google Scholar]
- 4.Buchner T, et al. Acute Myeloid Leukemia (AML): different treatment strategies versus a common standard arm--combined prospective analysis by the German AML Intergroup. J Clin Oncol. 2012;30(29):3604–10. doi: 10.1200/JCO.2012.42.2907. [DOI] [PubMed] [Google Scholar]
- 5.Felipe Rico J, Hassane DC, Guzman ML. Acute myelogenous leukemia stem cells: From Bench to Bedside. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 7.Blair A, Hogge DE, Sutherland HJ. Most acute myeloid leukemia progenitor cells with long-term proliferative ability in vitro and in vivo have the phenotype CD34(+)/CD71(−)/HLA-DR. Blood. 1998;92(11):4325–35. [PubMed] [Google Scholar]
- 8.Saito Y, et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci Transl Med. 2010;2(17):17ra9. doi: 10.1126/scitranslmed.3000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin L, et al. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 2009;5(1):31–42. doi: 10.1016/j.stem.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 10.Stein C, et al. Novel conjugates of single-chain Fv antibody fragments specific for stem cell antigen CD123 mediate potent death of acute myeloid leukaemia cells. Br J Haematol. 2010;148(6):879–89. doi: 10.1111/j.1365-2141.2009.08033.x. [DOI] [PubMed] [Google Scholar]
- 11.Cerny J, et al. Expression of CD25 independently predicts early treatment failure of acute myeloid leukaemia (AML). Br J Haematol. 2013;160(2):262–6. doi: 10.1111/bjh.12109. [DOI] [PubMed] [Google Scholar]
- 12.Dang NH, et al. Phase II trial of denileukin diftitox for relapsed/refractory T-cell non-Hodgkin lymphoma. Br J Haematol. 2007;136(3):439–47. doi: 10.1111/j.1365-2141.2006.06457.x. [DOI] [PubMed] [Google Scholar]
- 13.Dang NH, et al. Phase II study of denileukin diftitox for relapsed/refractory B-Cell non-Hodgkin's lymphoma. J Clin Oncol. 2004;22(20):4095–102. doi: 10.1200/JCO.2004.03.071. [DOI] [PubMed] [Google Scholar]
- 14.Foss FM, et al. A multicenter phase II trial to determine the safety and efficacy of combination therapy with denileukin diftitox and cyclophosphamide, doxorubicin, vincristine and prednisone in untreated peripheral T-cell lymphoma: the CONCEPT study. Leuk Lymphoma. 2013 doi: 10.3109/10428194.2012.742521. [DOI] [PubMed] [Google Scholar]
- 15.van Rhenen A, et al. The novel AML stem cell associated antigen CLL-1 aids in discrimination between normal and leukemic stem cells. Blood. 2007;110(7):2659–66. doi: 10.1182/blood-2007-03-083048. [DOI] [PubMed] [Google Scholar]
- 16.Zhao X, et al. Targeting C-type lectin-like molecule-1 for antibody-mediated immunotherapy in acute myeloid leukemia. Haematologica. 2010;95(1):71–8. doi: 10.3324/haematol.2009.009811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin L, et al. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12(10):1167–74. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 18.Majeti R, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138(2):286–99. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin B, Cheng K. Silencing of the IKKepsilon gene by siRNA inhibits invasiveness and growth of breast cancer cells. Breast Cancer Res. 2010;12(5):R74. doi: 10.1186/bcr2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang MR, et al. NF-kappaB signalling proteins p50/p105, p52/p100, RelA, and IKKepsilon are over-expressed in oesophageal squamous cell carcinomas. Pathology. 2009;41(7):622–5. doi: 10.3109/00313020903257756. [DOI] [PubMed] [Google Scholar]
- 21.Hideshima T, et al. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem. 2002;277(19):16639–47. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- 22.Guzman ML, et al. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood. 2001;98(8):2301–7. doi: 10.1182/blood.v98.8.2301. [DOI] [PubMed] [Google Scholar]
- 23.Guzman ML, et al. Preferential induction of apoptosis for primary human leukemic stem cells. Proc Natl Acad Sci U S A. 2002;99(25):16220–5. doi: 10.1073/pnas.252462599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guzman ML, et al. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood. 2005;105(11):4163–9. doi: 10.1182/blood-2004-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Attar EC, et al. Phase I and pharmacokinetic study of bortezomib in combination with idarubicin and cytarabine in patients with acute myelogenous leukemia. Clin Cancer Res. 2008;14(5):1446–54. doi: 10.1158/1078-0432.CCR-07-4626. [DOI] [PubMed] [Google Scholar]
- 26.Attar EC, et al. Bortezomib added to daunorubicin and cytarabine during induction therapy and to intermediate-dose cytarabine for consolidation in patients with previously untreated acute myeloid leukemia age 60 to 75 years: CALGB (Alliance) study 10502. J Clin Oncol. 2013;31(7):923–9. doi: 10.1200/JCO.2012.45.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunnberg U, et al. Induction therapy of AML with ara-C plus daunorubicin versus ara-C plus gemtuzumab ozogamicin: a randomized phase II trial in elderly patients. Ann Oncol. 2012;23(4):990–6. doi: 10.1093/annonc/mdr346. [DOI] [PubMed] [Google Scholar]
- 28.Ito K, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12(4):446–51. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 29.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–39. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Chakrabarty A, et al. Trastuzumab-resistant cells rely on a HER2-PI3K-FoxO-survivin axis and are sensitive to PI3K inhibitors. Cancer Res. 2013;73(3):1190–200. doi: 10.1158/0008-5472.CAN-12-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaballos MA, Santisteban P. FOXO1 controls thyroid cell proliferation in response to TSH and IGF-I and is involved in thyroid tumorigenesis. Mol Endocrinol. 2013;27(1):50–62. doi: 10.1210/me.2012-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obrador-Hevia A, et al. The tumour suppressor FOXO3 is a key regulator of mantle cell lymphoma proliferation and survival. Br J Haematol. 2012;156(3):334–45. doi: 10.1111/j.1365-2141.2011.08951.x. [DOI] [PubMed] [Google Scholar]
- 33.Sykes SM, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146(5):697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nwajei F, Konopleva M. The bone marrow microenvironment as niche retreats for hematopoietic and leukemic stem cells. Adv Hematol. 2013:953982. doi: 10.1155/2013/953982. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagadinou ED, et al. BCL-2 Inhibition Targets Oxidative Phosphorylation and Selectively Eradicates Quiescent Human Leukemia Stem Cells. Cell Stem Cell. 2013;12(3):329–41. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vo TT, et al. Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell. 2012;151(2):344–55. doi: 10.1016/j.cell.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goff DJ, et al. A Pan-BCL2 Inhibitor Renders Bone-Marrow-Resident Human Leukemia Stem Cells Sensitive to Tyrosine Kinase Inhibition. Cell Stem Cell. 2013;12(3):316–28. doi: 10.1016/j.stem.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corbin AS, et al. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121(1):396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yecies D, et al. Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood. 2010;115(16):3304–13. doi: 10.1182/blood-2009-07-233304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domina AM, et al. MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene. 2004;23(31):5301–15. doi: 10.1038/sj.onc.1207692. [DOI] [PubMed] [Google Scholar]
- 41.Konopleva M, et al. MEK inhibition enhances ABT-737-induced leukemia cell apoptosis via prevention of ERK-activated MCL-1 induction and modulation of MCL-1/BIM complex. Leukemia. 2012;26(4):778–87. doi: 10.1038/leu.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Rahmani M, et al. Inhibition of Bcl-2 antiapoptotic members by obatoclax potently enhances sorafenib-induced apoptosis in human myeloid leukemia cells through a Bim-dependent process. Blood. 2012;119(25):6089–98. doi: 10.1182/blood-2011-09-378141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skrtic M, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011;20(5):674–88. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryan J, Letai A. BH3 profiling in whole cells by fluorimeter or FACS. Methods. 2013 doi: 10.1016/j.ymeth.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Figueroa ME, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamazaki J, et al. The epigenome of AML stem and progenitor cells. Epigenetics. 2013;8(1):92–104. doi: 10.4161/epi.23243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6(11):838–49. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 48.Ritchie EK, et al. Decitabine in patients with newly diagnosed and relapsed acute myeloid leukemia. Leuk Lymphoma. 2013 doi: 10.3109/10428194.2012.762093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quintas-Cardama A, et al. Epigenetic therapy is associated with similar survival compared with intensive chemotherapy in older patients with newly diagnosed acute myeloid leukemia. Blood. 2012;120(24):4840–5. doi: 10.1182/blood-2012-06-436055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scandura JM, et al. Phase 1 study of epigenetic priming with decitabine prior to standard induction chemotherapy for patients with AML. Blood. 2011;118(6):1472–80. doi: 10.1182/blood-2010-11-320093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Craddock C, et al. Azacitidine fails to eradicate leukemic stem/progenitor cell populations in patients with acute myeloid leukemia and myelodysplasia. Leukemia. 2012 doi: 10.1038/leu.2012.312. [DOI] [PubMed] [Google Scholar]
- 52.Barry ER, et al. ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cells. 2009;27(7):1538–47. doi: 10.1002/stem.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang PY, et al. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J Mol Biol. 2010;400(2):137–44. doi: 10.1016/j.jmb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jo SY, et al. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117(18):4759–68. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernt KM, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schenk T, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med. 2012;18(4):605–11. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiskus W, et al. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009;114(13):2733–43. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou J, et al. The histone methyltransferase inhibitor, DZNep, up-regulates TXNIP, increases ROS production, and targets leukemia cells in AML. Blood. 2011;118(10):2830–9. doi: 10.1182/blood-2010-07-294827. [DOI] [PubMed] [Google Scholar]
- 60.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 61.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 62.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9(11):773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 63.Ringrose L. Polycomb comes of age: genome-wide profiling of target sites. Curr Opin Cell Biol. 2007;19(3):290–7. doi: 10.1016/j.ceb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 64.Herrera-Merchan A, et al. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat Commun. 2012;3:623. doi: 10.1038/ncomms1623. [DOI] [PubMed] [Google Scholar]
- 65.Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gross S, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207(2):339–44. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abdel-Wahab O, Levine RL. Metabolism and the leukemic stem cell. J Exp Med. 2010;207(4):677–80. doi: 10.1084/jem.20100523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang F, et al. Targeted Inhibition of Mutant IDH2 in Leukemia Cells Induces Cellular Differentiation. Science. 2013 doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 70.Rohle D, et al. An Inhibitor of Mutant IDH1 Delays Growth and Promotes Differentiation of Glioma Cells. Science. 2013 doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eppert K, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17(9):1086–93. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 72.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–89. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lacayo NJ, et al. Development and validation of a single-cell network profiling assay-based classifier to predict response to induction therapy in paediatric patients with de novo acute myeloid leukaemia: a report from the Children's Oncology Group. Br J Haematol. 2013;162(2):250–62. doi: 10.1111/bjh.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Z, et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J Clin Oncol. 2013;31(9):1172–81. doi: 10.1200/JCO.2012.44.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muller-Tidow C, et al. Profiling of histone H3 lysine 9 trimethylation levels predicts transcription factor activity and survival in acute myeloid leukemia. Blood. 2010;116(18):3564–71. doi: 10.1182/blood-2009-09-240978. [DOI] [PMC free article] [PubMed] [Google Scholar]