Abstract
The genetic improvement of disease resistance in poultry continues to be a challenge. To identify candidate genes and loci responsible for these traits, genome-wide association studies using the chicken 60k high density single nucleotide polymorphism (SNP) array for six immune traits, total serum immunoglobulin Y (IgY) level, numbers of, and the ratio of heterophils and lymphocytes, and antibody responses against Avian Influenza Virus (AIV) and Sheep Red Blood Cell (SRBC), were performed. RT-qPCR was used to quantify the relative expression of the identified candidate genes. Nine significantly associated SNPs (P < 2.81E-06) and 30 SNPs reaching the suggestively significant level (P < 5.62E-05) were identified. Five of the 10 SNPs that were suggestively associated with the antibody response to SRBC were located within or close to previously reported QTL regions. Fifteen SNPs reached a suggestive significance level for AIV antibody titer and seven were found on the sex chromosome Z. Seven suggestive markers involving five different SNPs were identified for the numbers of heterophils and lymphocytes, and the heterophil/lymphocyte ratio. Nine significant SNPs, all on chromosome 16, were significantly associated with serum total IgY concentration, and the five most significant were located within a narrow region spanning 6.4kb to 253.4kb (P = 1.20E-14 to 5.33E-08). After testing expression of five candidate genes (IL4I1, CD1b, GNB2L1, TRIM27 and ZNF692) located in this region, changes in IL4I1, CD1b transcripts were consistent with the concentrations of IgY, while abundances of TRIM27 and ZNF692 showed reciprocal changes to those of IgY concentrations. This study has revealed 39 SNPs associated with six immune traits (total serum IgY level, numbers of, and the ratio of heterophils and lymphocytes, and antibody responses against AIV and SRBC) in Beijing-You chickens. The narrow region spanning 247kb on chromosome 16 is an important QTL for serum total IgY concentration. Five candidate genes related to IgY level validated here are novel and may play critical roles in the modulation of immune responses. Potentially useful candidate SNPs for marker-assisted selection for disease resistance are identified. It is highly likely that these candidate genes play roles in various aspects of the immune response in chickens.
Introduction
Great effort has been expended globally to understand and genetically improve disease resistance in domestic animals [1–4]. Immune capacity associated with specific diseases may be useful indicators for indirect selection for general disease resistance, because such traits can be evaluated and quantified in live animals [5–9].
Immunological characteristics such as antibody titers have been shown to be heritable in poultry [10–11], indicating the possibility of discover loci or genes related to immune or disease resistance traits. Previous studies have found immune traits located in several chromosomal regions in chickens by microsatellite markers [10–12], and quantitative trait loci (QTLs) have been reported to be linked to the immune traits on chicken (Gallus gallus) chromosomes (GGA) 2, 3, 4, 5, 9, 13, 16, 18, 19, 22 and Z [13–21]. Only a few causative genes, however, have been identified because of low map resolution [22].
More recently, genome wide association studies (GWASs) have become one of the most commonly used strategies for identifying genes for complex traits in humans, as well as in animals [22]. In chickens, some major loci associated with resistance to Marek’s disease [23] and immune response to Newcastle disease virus and infectious bronchitis virus were identified by GWASs [9, 22]. Despite these studies, there is still limited information about the multiple immune traits that underly the full immune response at the genome-wide level in chickens.
This study aimed to identify major genomic regions (loci) and candidate genes associated with the immune response using GWAS for an array of important immune traits, including total serum concentrations of immunoglobulin Y (IgY), numbers of, and the ratio of heterophils and lymphocytes (H/L), and antibody responses against Avian Influenza Virus (AIV) and Sheep Red Blood Cells (SRBC) in chickens. This approach may offer valuable information for understanding the molecular mechanisms of the regulation of immune traits and facilitate the application of marker-assisted selection in breeding programs for disease resistance in chickens.
Results and Discussion
Genome-wide association analysis
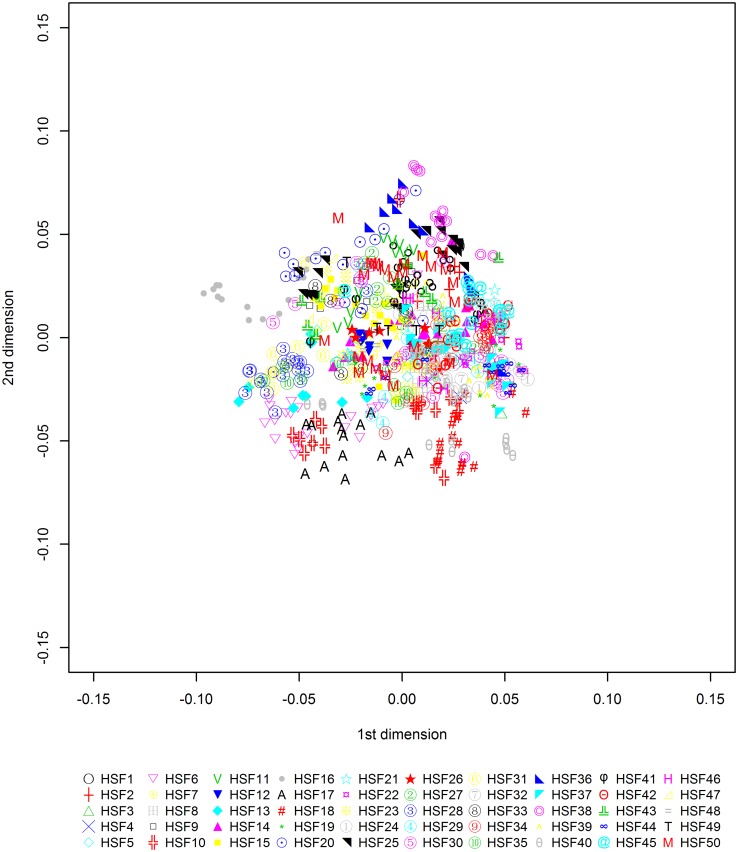
A total of 7,175 independent SNP markers were obtained with multidimensional scaling (MDS) analysis of these SNPs using the first two principal components (Fig. 1) indicating that chickens within each half-sibling family were clustered together. No obvious population substructure was detected. The analytical method was as described previously [24] and resulted in the elimination of approximately 13,506 SNPs from further analysis. The distribution of SNPs after this quality control is summarized in Table 1. All raw genotypes are available from the Dryad Repository (doi:10.5061/dryad.8m4r5).
Fig 1. Sample structure evaluated by the first two MDS dimensions.
HSF indicates half-sibling family.
Table 1. Distribution of SNPs on each chromosome after quality control.
| Chromosome | Physical length (Mb) c | No. SNPs | Marker density (kb/SNP) |
|---|---|---|---|
| 1 | 200.9 | 6,876 | 29.2 |
| 2 | 154.8 | 5,172 | 29.9 |
| 3 | 113.6 | 3,925 | 28.9 |
| 4 | 94.2 | 3,155 | 29.9 |
| 5 | 62.2 | 2,096 | 29.7 |
| 6 | 37.4 | 1,613 | 23.2 |
| 7 | 38.3 | 1,761 | 21.7 |
| 8 | 30.6 | 1,329 | 23 |
| 9 | 25.5 | 1,162 | 21.9 |
| 10 | 22.5 | 1,259 | 17.9 |
| 11 | 21.9 | 1,163 | 18.8 |
| 12 | 20.5 | 1,314 | 15.6 |
| 13 | 18.9 | 1,068 | 17.7 |
| 14 | 15.8 | 970 | 16.3 |
| 15 | 12.9 | 947 | 13.6 |
| 16 | 0.43 | 20 | 21.5 |
| 17 | 11.1 | 831 | 13.4 |
| 18 | 10.9 | 803 | 13.6 |
| 19 | 9.8 | 792 | 12.4 |
| 20 | 13.9 | 1,428 | 9.7 |
| 21 | 6.9 | 771 | 8.9 |
| 22 | 3.9 | 269 | 14.5 |
| 23 | 6 | 587 | 10.2 |
| 24 | 6.4 | 653 | 9.8 |
| 25 | 2 | 167 | 12 |
| 26 | 5.1 | 632 | 8.1 |
| 27 | 4.8 | 430 | 11.2 |
| 28 | 4.5 | 542 | 8.3 |
| LGE22C19W28_E50C23 a | 0.89 | 98 | 9.1 |
| LGE64 a | 0.049 | 1 | 49 |
| Z | 74.6 | 1,737 | 42.9 |
| 0 b | — | 559 | — |
| Total | 1031.3 | 44,130 | 23.4 |
a Two linkage groups.
b These SNPs are not mapped to any chromosome.
c The physical length of the chromosome was based on the genome build Gallus_gallus-2.1(May, 2006).
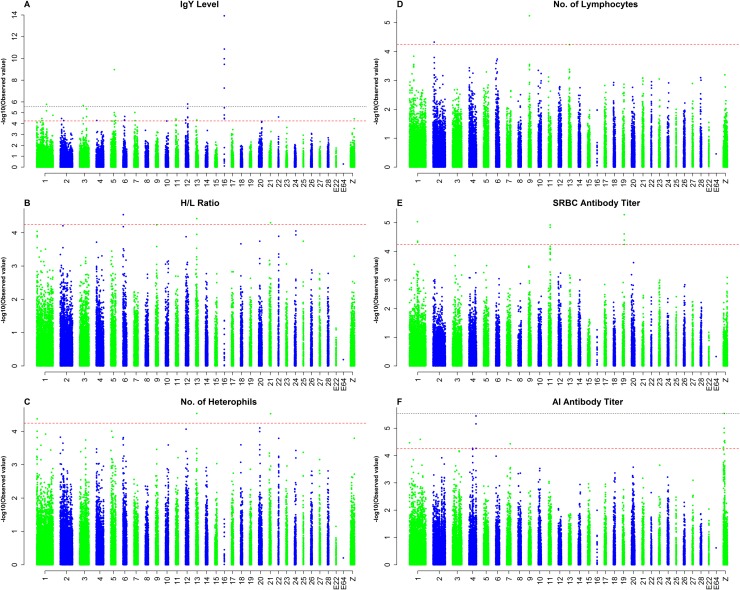
The descriptive statistics for six immune traits in the 720 Beijing-You chickens used for the present GWAS are shown in Table 2. The numbers of genome-wide significant SNPs detected for the six immune traits, and the details of these significant SNPs, including their positions in the genome, the nearest known genes and the P-values, are given in Table 3 and Table 4. The global view of P-values (in terms of—log10 (P-value)) for all SNP markers of each trait is represented by a Manhattan plot shown in Fig. 2. The genome-wide association analysis in this study identified 39 SNPs, consisting of nine significantly associated SNPs and 30 suggestively associated SNPs.
Table 2. Descriptive statistics of the six immune traits.
| Trait | Mean | Standard Deviation | Coefficient of Variation (%) |
|---|---|---|---|
| H/L Ratio | 0.39 | 0.21 | 53.84 |
| No. H | 25.11 | 10.94 | 43.56 |
| No. L | 70.33 | 10.51 | 14.94 |
| AIV Ab Titer(log2) | 9.22 | 1.7 | 18.44 |
| IgG Level (ng/ul) | 538.59 | 279.91 | 51.97 |
| SRBC Ab Titer(log2) | 4.24 | 1.21 | 28.54 |
Table 3. Genome-wise 5% significant SNPs associated with total serum IgY level.
| Trait | Chromosome | SNP ID | Position (bp) | Nearest Gene a | P-Value |
|---|---|---|---|---|---|
| IgY Level | 16 | Gga_rs16057130 | 6471 | CD1b & CD1c b | 1.20E-14 |
| 16 | Gga_rs14738106 | 253412 | 2.9kb D B-G c | 1.38E-11 | |
| 16 | Gga_rs15788101 | 110880 | GNB2L1 | 1.08E-10 | |
| 16 | GGaluGA111792 | 164922 | LAO | 3.50E-10 | |
| 16 | Gga_rs15788237 | 59984 | BMA1 | 5.33E-08 | |
| 5 | GGaluGA285957 | 46399742 | 39.7kb U TTC7B d | 1.08E-09 | |
| 12 | Gga_rs15661138 | 14395118 | no gene | 1.56E-06 | |
| 1 | Gga_rs15399236 | 121689702 | EIF2S3 | 1.66E-06 | |
| 3 | Gga_rs16261002 | 44438665 | 40.1kb D RNASET2 | 2.14E-06 |
a Genes located around 500kb upstream and downstream of the SNP were searched.
bGene name alone indicates the SNP is within the gene.
cD indicates that the SNP is downstream of the gene.
dU indicates that the SNP is upstream of the gene.
These definitions are also used in Table 4.
Table 4. Suggestively Associated SNPs for five immune traits.
| Trait | Chromosome | SNP ID | Position (bp) | Nearest Gene | P- Value |
|---|---|---|---|---|---|
| AIV Ab Titer | 1 | Gga_rs13937877 | 133493971 | 234kb D SLC25A6 | 2.54E-05 |
| 1 | Gga_rs15995401 | 111556 | 28kb D HCLS1 | 3.37E-05 | |
| 1 | Gga_rs15995401 | 111556 | 28kb D HCLS1 | 3.37E-05 | |
| 4 | GGaluGA269205 | 88150596 | 48kb U FGFRL1 | 3.53E-06 | |
| 4 | GGaluGA269230 | 88264974 | FGFRL1 | 6.88E-06 | |
| 4 | GGaluGA255778 | 44000064 | 357kb U AGA | 5.41E-05 | |
| 4 | GGaluGA269388 | 88690383 | 24kb U IMMT | 5.46E-05 | |
| 7 | Gga_rs14626540 | 32964405 | no gene | 3.69E-05 | |
| Z | GGaluGA348626 | 17004336 | no gene | 2.86E-06 | |
| Z | Gga_rs16098994 | 17002454 | no gene | 9.90E-06 | |
| Z | Gga_rs16099280 | 17569998 | no gene | 1.45E-05 | |
| Z | Gga_rs16099440 | 17736692 | no gene | 3.00E-05 | |
| Z | Gga_rs14778781 | 30762177 | 200kb D TYRP1 | 3.36E-05 | |
| Z | Gga_rs14066816 | 3078791 | no gene | 4.92E-05 | |
| Z | Gga_rs15648827 | 3147673 | no gene | 4.92E-05 | |
| SRBC Ab | 1 | Gga_rs13905265 | 95938436 | 68kb D GPR89B | 9.15E-06 |
| Titer | 1 | Gga_rs13907209 | 98680358 | no gene | 4.41E-05 |
| 1 | Gga_rs13907411 | 98949817 | no gene | 4.78E-05 | |
| 11 | Gga_rs14025779 | 13991523 | no gene | 1.19E-05 | |
| 11 | Gga_rs15619912 | 14576402 | no gene | 1.45E-05 | |
| 19 | GGaluGA128939 | 9229902 | 48kb D iNOS | 5.21E-06 | |
| 19 | GGaluGA128957 | 9272295 | 1kb U TNFAIP1 | 5.21E-06 | |
| 19 | Gga_rs15050742 | 9306653 | 21kb D TNFAIP1 | 5.21E-06 | |
| 19 | Gga_rs10725789 | 9162804 | iNOS | 2.46E-05 | |
| 19 | Gga_rs13576070 | 9265157 | 8kb U TNFAIP1 | 4.02E-05 | |
| H/L Ratio | 6 | Gga_rs14562554 | 4 673 503 | 559kb D GHITM | 2.87E-05 |
| 13 | Gga_rs14051163 | 7 248 634 | 192kb D GABRA6 | 3.78E-05 | |
| 21 | Gga_rs16743572 | 5 008 458 | 21kb D DNAJC16 | 5.03E-05 | |
| No. H | 13 | Gga_rs14051163 | 7 248 634 | 192kb D GABRA6 | 2.89E-05 |
| 21 | Gga_rs16743572 | 5 008 458 | 21kb D DNAJC16 | 2.94E-05 | |
| No. L | 2 | GGaluGA134321 | 14 283 920 | 11kb U ZEB1 | 4.73E-05 |
| 9 | Gga_rs15950814 | 10 984 983 | PID1 | 5.82E-06 |
Fig 2. Manhattan plots showing association of all SNPs with six immune traits.
SNPs are plotted on the x-axis according to their position on each chromosome against association with these traits on the y-axis (shown as-log10 p-value). The red dashed line indicates suggestive genome-wise significance (p-value = 5.62E-05), and the black dashed line shows genome-wise 5% significance with a p-value threshold of 2.81E-06. Fig. 2-A, 2-B, 2-C, 2-D, 2-E and 2-F refer to plots for total serum IgY level, H/L ratio, number of Heterophils, number of Lymphocytes, SRBC antibody titer and AI antibody titer, respectively.
Serum Concentrations of IgY at d 80
Five SNPs, associated with the serum total IgY level, were clustered within a 0.26 Mb region on chromosome 16 (S1 Table). One SNP, rs14738106, was located 2.9 kb downstream of the B-G gene (within the MHC region). Four of the SNPs (rs16057130, rs14738106, rs15788101 and rs15788237) were located within CD1b and CD1c genes, an intron of GNB2L1 gene, an intron of IL4I1 gene and an intron of BMA1 gene, respectively. A QTL for titres of Natural antibodies (NA) was detected at the SNP rs15788216 on chromosome 16 in chickens [14–15], near the SNP rs15788237 found here.
The chicken MHC, which is composed of the B and Y complexes [25] and plays a central role in the immune response, has also been mapped to this NOR (Nucleolus Organizing Region, cluster of genes for ribosomal RNA 18S, 5.8S and 28S)-bearing microchromosome 16 [26].
CD1b and CD1c encode two members of the CD1 family, which are structurally similar to the MHC class I antigen and have been shown to present lipid antigens for recognition by T lymphocytes [27]. The expression of the IL4I1 gene can be induced by interleukin 4 in B cells. The IL4I1 protein has L-phenylalanine oxidase activity, and oxidative deamination of phenylalanine by IL4I1 produces H2O2, and functions in the innate immune defenses of vertebrates. Previous studies have revealed that the IL4I1 protein inhibits T-cell proliferation with a preference for memory T cells, and H2O2 mediates this inhibition [28–29].
In the current study, significant SNPs on GGA1, 3, 5 and 12 were also identified that related to serum total IgY level. Those were not located within the previously known QTLs for immune traits, for example, five QTLs associated with humoral innate immune response in chickens (NA titers binding LPS, located on GGA5, GGA9, GGA14, GGA18, and GGAZ) were recently validated in a cross between Green-Legged Partridge-like and White Leghorn [19, 21]. The reason for the discrepancy might result from the different chicken populations being studied.
Heterophils and Lymphocytes
The heterophil to lymphocyte (H/L) ratio in the peripheral blood of chickens has been widely accepted as a reliable and accurate physiological indicator of the stress response in chickens [30–31]. The number of lymphocytes decrease and the number of heterophils increase in response to stressors such as high temperature, hypothermy, excessive NH3, bacterial infection, inter alia [32–33]. The ratio, and individual numbers of heterophils and lymphocytes are highly heritable traits, with heritability estimates exceeding 0.5 [34], suggesting that good responses to selection should be possible.
There were three SNPs detected as being suggestively associated with the H/L ratio (S2 Table), two of which were simultaneously associated with the number of heterophils (S3 Table). One SNP (rs14562554) was mapped to 559kb downstream of the growth hormone inducible transmembrane protein (GHITM) gene on chromosome 6. The GHITM protein is part of the interferon signaling system that includes Janus kinases and their downstream target STAT proteins [35]. One SNP at 7.2Mb on chromosome 13 is located 192kb downstream of gamma-aminobutyric acid receptor subunit alpha-6 (GABRA6). This gene is associated with increased production of cortisol and increased blood pressure in response to psychological stress [36]. The rs16743572 SNP is located downstream of DnaJ homolog, subfamily C, member 16 (DNAJC16) in chromosome 21; the gene is a member of the DnaJ/Hsp40 (heat shock protein 40) family. These proteins are molecular chaperones induced in response to cellular stress [37] and have been preserved throughout evolution, functioning in protein translation, folding, unfolding, translocation, and degradation, primarily by stimulating the ATPase activity of Hsp70s chaperone proteins [38]. Human Hsp40s have an immune-modulatory effect on T cells and cytokine secretion in rheumatoid arthritis patients [39].
There were two SNPs, rs15950814 and GGaluGA134321, that were suggestively associated with the number of lymphocytes (S4 Table). The former is located within phosphotyrosine interaction domain containing 1 (PID1), also called NYGGF4, on chromosome 9, which encodes a protein possessing a phosphotyrosine interaction domain (PID). Mouse and human NYGGF4 are involved in the regulation of proliferation of adipocyte precursors [40]. The gene is more widely expressed in chicken tissues than in those of mice and humans [41]. Proteins with PID are widely involved in physiological processes, such as neural development, immune function, homeostasis and cell growth [42], which indicates that the chicken NYGGF4 homolog possibly possesses multiple functions. The GGaluGA134321 SNP, is located 11kb downstream of the zinc finger E-box binding homeobox 1 (ZEB1) gene on chromosome 2. In human and mouse studies, ZEB1 is a transcriptional repressor and plays an important role in regulating lymphoid differentiation [43]. It has been implicated in the regulation of other important genes in the immune system such as the Ig heavy chain enhancer, CD4, GATA-3, α7-integrin and IL-2 [44–45]. It has been shown that ZEB1-deficient mice have poorly developed thymuses and significant decreases in their T cell population [46].
Antibody responses to SRBC
The direct response to selection on antibody titer to SRBC challenge is accompanied by correlated responses in production and disease-related traits [13, 16]. The heritability of anti-SRBC titer in chickens is moderate (0.20 to 0.25), and the trait responds to divergent selection [47].
In this study, of special interest, five SNPs in a narrow region (9.16Mb—9.31Mb) on chromosome 19 were suggestively associated with antibody titer against SRBC (S5 Table). The rs10725789 and GGaluGA128939 are located within and 48 kb downstream of the inducible nitric oxide synthase (iNOS) gene. The expression of iNOS is regulated by cytokines such as IFN-γ, IL-1β, IL-6, TNF-α, and iNOS-derived nitric oxide (NO) is well known to play important roles in infection and immunity [48]. The rs15050742, rs13576070 and GGaluGA128957 SNPs are located 21kb downstream, 1kb and 8kb upstream of the tumor necrosis factor, alpha-induced protein 1 (TNFAIP1). The TNFAIP1 protein is a TNF-α and IL-6 inducible protein. Research in humans and rats indicate that the protein may have roles in DNA synthesis and repair and in apoptosis [49]. Such results refine the detail of this genomic region and lead to more conclusive identification of the contributing loci involved and the putative causal mutations.
In addition, three SNPs on chromosome 1 were suggestively associated with anti-SRBC titer (S5 Table), and rs13905265 is 68 kb downstream of G protein-coupled receptor 89B (GPR89B), but no known genes were found within 500 kb of the other two SNPs. A QTL has been detected in the vicinity of this region in a F2 cross of two lines divergently selected for SRBC antibody responses [16]. Two SNPs of suggestive significance at 13.9 Mb and 14.6 Mb are consistent with a genomic region identified in a previous study in chickens [50]. There are several other genomic regions previously identified as QTLs for SRBC antibody response in chickens, including those on GGA2, GGA3, GGA5, GGA11, GGA16, GGA23 and GGA24 [13, 16–19, 21, 50–51].
Antibody responses to AIV
Two SNPs on chromosome 1 were suggestively associated with AIV antibody titer (S6 Table). One SNP, rs13937877 is located 234 kb downstream of solute carrier family 25, member 6 (SLC25A6). The present study found that the region at about 100 Mb from the proximal end of chicken chromosome 1, including the ROBO1 (Roundabout, axon guidance receptor, homolog 1) and ROBO2 (homolog 2) genes, has a strong effect on the antibody response to the NDV in chickens [22]. One SNP rs14527240 was located in the SPARC-related modular calcium-binding 1 (SMOC1) gene on GGA5 playing an important role in MD tumorigenesis [23]. GGA1 also had the most significantly associated SNPs on the antibody level against infectious bronchitis virus (IBV), but a genomic region containing a cluster of 13 beta-defensin (GAL1–13) and interleukin-17F genes on GGA3 probably plays an important role in the immune response against IBV [9]. The SLC25A6 gene encodes adenine nucleotide translocase 3 (ANT3), an enzyme which exchanges ATP and ADP through the mitochondrial membrane. Mitochondrial ANT3 is regulated by IL-4 and IFN-γ via STAT-dependent pathways [52]. Considering the critical role of mitochondrial ANTs in energy metabolism and apoptosis, ANT3 regulation by IL-4 and IFN-γ may have a functional implication in cytokine-mediated T cell survival [53].
One SNP (rs15995401) is located 28 kb downstream of hematopoietic cell-specific Lyn substrate 1 (HCLS1). HCLS1 is an intracellular protein expressed mainly in hematopoietic cells. It was reported to be an important actin regulator at the T-cell immunologic synapse and also to influence numerous functions of natural killer cells [54].
Eight SNPs on chromosome Z were suggestively novel and specific associated with AIV antibody titer in this study (S6 Table). One SNP (rs14778781) is located within multiple PDZ domain protein (MPDZ) which is involved in a diverse range of pathways that maintain cell integrity and function [55]. No known genes were found within 500kb of the remaining seven SNPs. These studies concerning immune responses against NDV, IBV and AIV add to basic knowledge of host attributes potentially relevant to improved methods for controlling outbreaks of viral diseases.
Relative expression of candidate genes in spleen
The present GWAS verified that the important immune-related locus was located on chromosome 16 [9, 22]. In this study, five of the significant SNPs for total serum IgY level were located within a narrow 247 kb region on chromosome 16. On this basis, the genes within this region were selected for further expression analysis, including CD1b, IL4I1 (Interleukin 4-induced 1), TRIM27 (tripartite motif-containing 27), GNB2L1 (guanine nucleotide binding protein, beta polypeptide 2-like 1), and ZNF692 (zinc finger protein 692).
Compared with SRBC, Salmonella enteritidis is more potent in inducing production of serum antibodies (IgY) in the infected chickens. For just that reason, it was used here to validate the association between total serum IgY level and the corresponding candidate genes identified in this study.
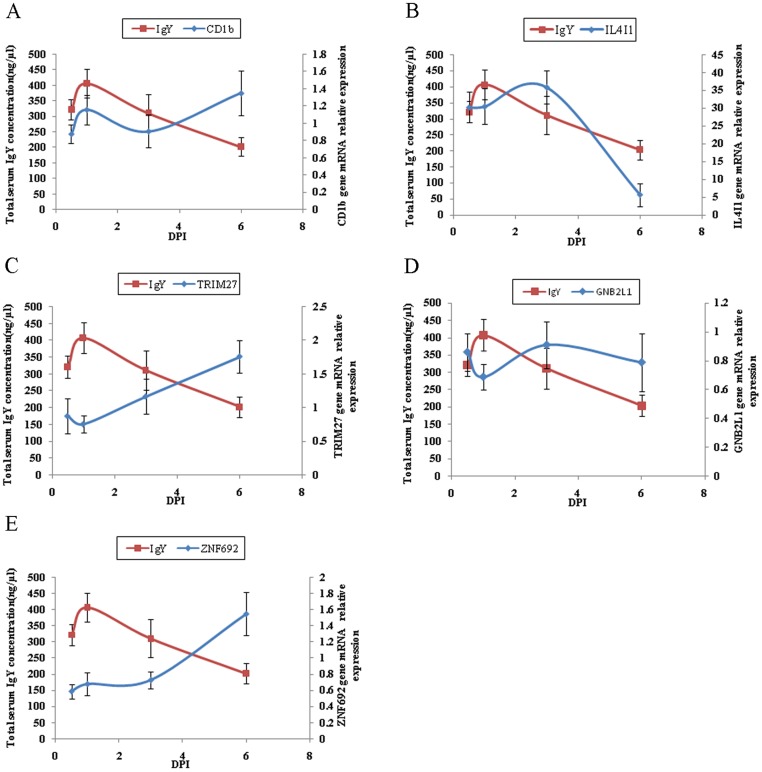
The relative expression of those five candidate genes near associated signals from the GWAS analysis was determined by RT-qPCR in chickens with and without challenge with Salmonella enteritidis. In this study, the changes across days in expression of IL4I1 and CD1b followed a similar pattern to the concentrations of IgY, while changes in TRIM27 (alternatively named RET finger protein, RFP) and ZNF692 expression were the reverse (mirror-image) of concentrations of IgY (Fig. 3).
Fig 3. Changes in relative transcript abundance of five genes in spleen at 12h, 24h, 3 or 6 days post-infection (DPI) and total serum IgY concentration.
The relative expression of CD1b (A), IL4I1 (B), TRIM27 (C), GNB2L1 (D), and ZNF692 (E) was measured in spleen at different times post-infection with Salmonella enteritidis (108 CFU) (n = 6). Serum concentrations of total IgY of SE-infected and uninfected birds were determined by chicken ELISA kits. Results were expressed as the mean fold-change in gene expression at each time point, from triplicate analyses, using the non-infected control sample at 0.5 dpi as the calibrator (assigned an expression level of 1). Each point-in-time represents the mean ± S.D. of 6 birds. There were barely detectable or no changes in relative expression of the five candidate genes and total IgY concentration in un-challenged controls (data not shown).
Chickens possess two CD1 genes designated chCD1–1 and chCD1–2 that are closely linked to the MHC locus on chromosome 16. Both genes are expressed in lymphoid tissues as evidenced by Northern analysis and PCR [56]. The spleen was examined here because it is the major lymphoid organ in birds and has previously been shown to have high level expression of CD1 [57].
IL4I1 modulates the number of antitumor cytotoxic T cells in vivo and affects their capacity to secrete IFN-γ. The expression of IL4I1 is restricted to lymphoid tissues, with the highest levels found in lymph nodes and spleen [58]. In the present study, the expression of CD1b and IL4I1 was up-regulated in spleens of infected birds.
TRIM27 is a member of the TRIM superfamily and exhibits the classical PRY-SPRY domain C-terminal of the RBCC motif. TRIM27 is highly expressed in mouse spleen and thymus [59] and in hematopoietic cells [60]. Roles for TRIM27 in transcriptional repression [60], the negative regulation of NF-κB and in IFN-signalling pathways, apoptosis and in cell cycle regulation have been reported [61], indicating its involvement the control of multiple cellular processes. These findings account for the present demonstration that the abundance of TRIM27 showed reciprocal changes to those of IgY concentrations.
The genes ZNF692 and GNB2L1 relate to the MHC genes, but little is known of their precise function in chickens. The changes in relative expression and relationship to changes in IgY after infection, support their having some role, probably negative regulatory, in the magnitude of the immune response.
Conclusions
This study has revealed 39 SNPs associated with six immune traits (total serum IgY level, numbers of, and the ratio of heterophils and lymphocytes, and antibody responses against AIV and SRBC) in Beijing-You chickens. Five of the significant SNPs for total serum IgY level were located within a narrow 247 kb region on chromosome 16. Some SNPs were located within or close to the previously reported QTL regions. Five candidate genes related to IgY level by GWAS, and validated here by experimental challenge, are novel and may play critical roles in the modulation of immune responses. It is highly likely that these candidate genes play an important role in the immune response. Findings herein lay an important foundation for further identifying the immunity-related genes and provide useful information for the molecular mechanisms underlying disease-resistance traits in chickens.
Materials and Methods
Ethics Statement
All of the animal experiments were conducted in accordance with the Guidelines for Experimental Animals established by the Ministry of Science and Technology (Beijing, China). Animal experiments were approved by the Science Research Department (in charge of animal welfare issue) of the Institute of Animal Sciences, CAAS (Beijing, China).
Animal resource
Beijing-You chickens (a Chinese indigenous breed) from the national conservation farm, Institute of Animal Sciences, CAAS, were used. A total of 728 male offspring from 50 sire families were produced in one hatch. The birds were reared in stair-step cages under standard conditions of humidity, temperature, and ventilation, and had access to water and feed ad libitum. All chickens received the conventional inoculation regimen (S7 Table), including vaccination against H5N1 avian influenza virus (AIV) on d 17.
Phenotypic measurements
All blood samples were collected from the brachial vein by venipuncture. At d 49, chickens were injected intramuscularly in the leg with 1 ml of a 0.25% suspension of SRBC. Sera were isolated from blood collected on d 55 (6d post-injection) for immediate determination of anti-SRBC titers by hemagglutination [62] using 96-well plates and the same preparation of SRBC as used for the immunization. Antibody titers were expressed as the log2 of the reciprocal of the highest dilution giving complete agglutination. On d 62, blood smears were prepared from each chicken, stained with May-Grunwald and Giemsa stains and differentially counted [63]. One hundred leukocytes, including granular (heterophils, eosinophils, and basophils) and nongranular (lymphocytes and monocytes), were counted on one slide from each bird, and the heterophil to lymphocyte ratio (H/L) was calculated. Concentrations of IgY and anti-AIV titers were measured in serum collected on d 80. A double antibody sandwich enzyme-linked immunosorbent assay (ELISA) was used for IgY (R&D Systerm, Minneapolis, USA) and a hemagglutination-inhibition diagnostic test was used for titering anti-AIV. The descriptive statistics of phenotypic values of all traits were analyzed using the MEANS procedure in SAS 9.2 software (SAS Institute Inc., Cary, NC).
Genotyping and quality control
Genomic DNA (gDNA) was extracted from blood samples using phenol-chloroform. The DNA samples were genotyped using the Illumina 60K chicken SNP Beadchip (DNA LandMarks Inc., Saint-Jean-sur-Richelieu, Quebec, Canada). The intensity files generated were processed with the GenomeStudio GT module with default settings [64].
Quality control was performed by PLINK 1.07 [65] and eight chickens were excluded because of high genotyping missing rate (> 95%). SNPs failing to meet any one of the following filtering criteria were excluded: call rate > 90%; minimum minor allele frequency (MAF) > 0.03; P-value for Hardy-Weinberg equilibrium test > 1×10–6.
Statistical analysis
To evaluate if population substructure existed among the individuals, classical MDS, as implemented by PLINK 1.07 [65], was performed. The procedures were as follows: first, to ensure uncorrected LD did not distort the analysis, all autosomal SNPs passing the quality control were subjected to LD-based pruning, thus the remaining SNPs within a given window size of 25 had pairwise r2 < 0.2; second, the pairwise IBS distance among the 720 individuals was calculated using the remaining 7,175 SNPs; finally, the first two MDS dimensions were extracted via the “MDS-plot” command and visualized in R v3.0.1 (www.r-project.org).
Association analyses were conducted to test the additive effects of allele dosage by PLINK, using the linear regression model. The fitted equation was:
Where: Y ij is the phenotypic value of a given immune trait for individual i in the j th sire family, b 0 is the intercept, b k-1, b 50 is the regression coefficient, f is the dummy variable for the sire family, k is an integer ranging from 2 to 50, X ij denote the genotype at the given SNP, and ε ij is independent, normally distributed random residual, with mean 0 and constant variance. For the family variable with 50 categories, 49 binary dummy variables f (coded 0/1), created by the dummy-coding option within PLINK, were fitted in the model.
Given the correlation between SNPs in linkage disequilibrium, the strict Bonferroni adjustment appears to be overly conservative [66]. The sum of independent LD blocks plus singleton markers was used to define the number of independent statistical comparisons [67], as described in [68]. Finally, 17,794 independent tests were used to determine the P-value thresholds, including 5% genome-wide significance (2.81E-6, 0.05/17,794) and suggestive association (5.62E-5, 1/17,794). A global view of the p-value results for the six traits from the association analysis was created by the “gap” package [66] in R v3.0.1 (www.r-project.org).
Relative expression of candidate genes identified by the GWAS
Birds and Treatment. Twelve-day-old Beijing-You chickens were from the same conservation stock and were reared as described earlier, in separate cages. Birds were confirmed to be free of Salmonella by culturing fecal samples in buffered peptone water (BPW) overnight with shaking at 150 rpm and spreading on brilliant green agar containing 100 mg/ml nalidixic acid (37°C, 18–24 h). Chickens were randomly divided into 2 groups of 30 each (Salmonella enteritidis challenge and un-challenged controls). Birds in the challenged group were injected intramuscularly into the leg with 0.5 ml of SE bacterial suspension containing 108 CFU and the control group was given 0.5 ml saline. Six chickens from each group were randomly chosen, and sacrificed at 12h, 24h, 3 or 6 days post-infection. Blood was collected, allowed to clot, and serum was stored. The spleen was rapidly removed, snap-frozen in liquid nitrogen, and stored at-80°C for mRNA extraction.
Serological test. Concentrations of total IgY in all sampled birds were determined in triplicate by ELISA, as described earlier.
mRNA abundance of candidate genes by RT-qPCR. Total RNA was isolated from each spleen sample with the RNAsimple Total RNA kit (TIANGEN BIOTECH, Beijing, China) and first-strand cDNA was synthesised from 2 μg total RNA using the Promega Reverse Transcription Kit (Promega, Beijing, China), according to the manufacturer’s instructions. Power SYBR Green PCR Master Mix (Applied Biosystems, USA) was used to quantify mRNA abundance of the genes by Reverse Transcription-Quantitative PCR (RT-qPCR) with an ABI 7500 Real-time Detection System (Applied Biosystems, USA). The primers were designed with Primer Premier 5.0 from the GenBank sequences (Table 5). The PCR amplification was performed, as previously described [24]. To determine fold-changes in gene expression, the comparative CT method was used [69], calculated as 2-ΔΔCT. Results were expressed as the mean fold-change in gene expression at each time, from triplicate analyses, using the non-infected control sample at 0.5dpi as the calibrator (assigned an expression level of 1). All analyses were performed using Student’s t-test and data are presented as means ± S.D.
Table 5. Primers used for the RT-qPCR.
| Gene | Primer | (5’-3’)Primers sequence | Product/bp | GenBank No. |
|---|---|---|---|---|
| β-actin | F | GAGAAATTGTGCGTGACATCA | 152 | NM_205518.1 |
| R | CCTGAACCTCTCATTGCCA | |||
| CD1b | F | CATTGCTCTTGGGCAACGTC | 113 | NM_001024582 |
| R | TGGCCGGCAGATGTTTAAGT | |||
| IL4I1 | F | GAAGCACGTTGGCAGGAAAG | 138 | NC_006103.3 |
| R | AACCTGCACGAAGTCGGAAT | |||
| GNB2L1 | F | GCAGCAACCCCATCATTGTC | 188 | NM_001004378 |
| R | ATTCAGGTCCCACAGCATGG | |||
| TRIM27 | F | GCCGAGCTGGAGAAGAAACT | 247 | NM-0010099354.1 |
| R | CCAGCACGCAGAAGGAGTAA | |||
| ZNF692 | F | GGAAAGCAGCGAAACGTGAA | 116 | NM_001099356.1 |
| R | GGTCTTCTGGTGGACGTGTT |
Supporting Information
The raw results from the linear regression analysis by PLINK. Its basic format is: CHR, Chromosome; SNP, SNP identifier; BP, Physical position (base-pair); A1, Tested allele (minor allele by default); TEST, Code for the test (the additive effects of SNPs); NMISS, Number of non-missing individuals included in analysis; BETA, Regression coefficient (the direction of BETA represents the effect of each extra minor allele); STAT, Coefficient t-statistic; P, Asymptotic p-value for t-statistic. The same abbreviations are used in the following tables.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
The authors would like to acknowledge Dr. W. Bruce Currie (Emeritus Professor, Cornell University, USA) for his assistance in preparing the manuscript.
Data Availability
All raw genotypes are available in the Dryad Repository (doi:10.5061/dryad.8m4r5).
Funding Statement
The research was supported by grants: the National Key Technology R&D Program (2011BAD28B03); the Agricultural Science and Technology Innovation Program (ASTIP-IAS04); the earmarked fund for modern agro-industry technology research system (CARS-42). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Shook GE (1989) Selection for disease resistance. J Dairy Sci 72(5):1349–1362. [DOI] [PubMed] [Google Scholar]
- 2. Muller M, Brem G (1991) Disease resistance in farm animals. Experientia 47(9):923–934. [DOI] [PubMed] [Google Scholar]
- 3. Stear MJ, Bishop SC, Mallard BA, Raadsma H (2001) The sustainability, feasibility and desirability of breeding livestock for disease resistance. Res Vet Sci 71(1):1–7. [DOI] [PubMed] [Google Scholar]
- 4. Goddard ME, Hayes BJ (2009) Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat Rev Genet 10(6):381–391. 10.1038/nrg2575 [DOI] [PubMed] [Google Scholar]
- 5. Warner CM, Meeker DL, Rothschild MF (1987) Genetic control of immune responsiveness: a review of its use as a tool for selection for disease resistance. J Anim Sci 64(2):394–406. [DOI] [PubMed] [Google Scholar]
- 6. Pinard MH, van Arendonk JA, Nieuwland MG, van der Zijpp AJ (1992) Divergent selection for immune responsiveness in chickens: estimation of realized heritability with an animal model. J Anim Sci 70(10):2986–2993. [DOI] [PubMed] [Google Scholar]
- 7. Swaggerty CL, Pevzner IY, He H, Genovese KJ, Nisbet DJ, et al. (2009) Selection of broilers with improved innate immune responsiveness to reduce on-farm infection by foodborne pathogens. Foodborne Pathog Dis 6(7):777–783. 10.1089/fpd.2009.0307 [DOI] [PubMed] [Google Scholar]
- 8. Lu X, Liu J, Fu W, Zhou J, Luo Y, et al. (2013) Genome-wide association study for cytokines and immunoglobulin G in swine. PLoS One 8(10):e74846 10.1371/journal.pone.0074846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luo C, Qu H, Ma J, Wang J, Hu X, et al. (2014) A genome-wide association study identifies major loci affecting the immune response against infectious bronchitis virus in chicken. Infect Genet Evol 21:351–358. 10.1016/j.meegid.2013.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yonash N, Cheng HH, Hillel J, Heller DE, Cahaner A (2001) DNA microsatellites linked to quantitative trait loci affecting antibody response and survival rate in meat-type chickens. Poult Sci 80(1):22–28. [DOI] [PubMed] [Google Scholar]
- 11. Kaiser MG, Deeb N, Lamont SJ (2002) Microsatellite markers linked to Salmonella enterica serovar enteritidis vaccine response in young F1 broiler-cross chicks. Poult Sci 81:193–201. [DOI] [PubMed] [Google Scholar]
- 12. Yunis R, Heller ED, Hillel J, Cahaner A (2002) Microsatellite markers associated with quantitative trait loci controlling antibody response to Escherichia coliand Salmonella enteritidis in young broilers. Animal Genetics 33:407–414. [DOI] [PubMed] [Google Scholar]
- 13. Zhou H, Li H, Lamont SJ (2003) Genetic markers associated with antibody response kinetics in adult chickens. Poult Sci 82:699–708. [DOI] [PubMed] [Google Scholar]
- 14. Biscarini F, Bovenhuis H, van Arendonk JA, Parmentier HK, Jungerius AP, et al. (2010) Across-line SNP association study of innate and adaptive immune response in laying hens. Anim Genet 41(1):26–38. 10.1111/j.1365-2052.2009.01960.x [DOI] [PubMed] [Google Scholar]
- 15. van der Poel JJ, Biscarini F, Rodenburg BT, van Arendonk JA, Parmentier HK, et al. (2011) Across-line SNP association study for (innate) immune and behavioral traits in laying hens. BMC Proc 5 Suppl 4:S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siwek M, Cornelissen S, Nieuwland M, Buitenhuis A, Bovenhuis H, et al. (2003) Detection of QTL for immune response to sheep red blood cells in laying hens. Anim Genet 34:422–428. [DOI] [PubMed] [Google Scholar]
- 17. Siwek M, Buitenhuis AJ, Cornelissen SJB, Nieuwland MGB, Bovenhuis H, et al. (2003b) Detection of different QTL for antibody response to Keyhole Lymphet Haemocynain and Mycobacterium butiricum in two unrelated populations of laying hens. Poult Sci 82:1845–1842. [DOI] [PubMed] [Google Scholar]
- 18. Siwek M, Buitenhuis B, Cornelissen S, Nieuwland M, Knol EF, et al. (2006) Detection of QTL for innate: non-specific antibody levels binding LPS and LTA in two independent populations of laying hens. Dev Comp Immunol 30:659–666. [DOI] [PubMed] [Google Scholar]
- 19. Slawinska A, Witkowski A, Nieuwland M, Minozzi G, Bednarczyk M, et al. (2011) Quantitative trait loci associated with the humoral innate immune response in chickens were confirmed in a cross between Green-Legged Partridgelike and White Leghorn. Poult Sci 90:1909–1915. 10.3382/ps.2011-01465 [DOI] [PubMed] [Google Scholar]
- 20. Siwek M, Szyda J, Sławińska A, Bednarczyk M (2012) Detection of two QTL on chicken chromosome 14 for keyhole lymphet haemocyanin. J Appl Genet 53(1):115–119. 10.1007/s13353-011-0074-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Slawinska A, Siwek M (2013) Meta—and combined—QTL analysis of different experiments on immune traits in chickens. J Appl Genetics 54:483–487. 10.1007/s13353-013-0177-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luo C, Qu H, Ma J, Wang J, Li C, et al. (2013) Genome-wide association study of antibody response to Newcastle disease virus in chicken. BMC Genet 14:42 10.1186/1471-2156-14-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li DF, Lian L, Qu LJ, Chen YM, Liu WB, et al. (2012) A genome-wide SNP scan reveals two loci associated with the chicken resistance to Marek’s disease. Anim. Genet 44:217–222. 10.1111/j.1365-2052.2012.02395.x [DOI] [PubMed] [Google Scholar]
- 24. Sun Y, Zhao G, Liu R, Zheng M, Hu Y, et al. (2013) The identification of 14 new genes for meat quality traits in chicken using a genome-wide association study. BMC Genomics 8(14):458 10.1186/1471-2164-14-458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller MM, Bacon LD, Hala K, Hunt HD, Ewald SJ, et al. (2004) Nomenclature for the chicken major histocompatibility (B and Y) complex. Immunogenetics 56:261–279. [DOI] [PubMed] [Google Scholar]
- 26. Delany ME, Robinson CM, Goto RM, Miller MM (2009) Architecture and organization of chicken microchromosome 16: order of the NOR, MHC-Y, and MHC-B subregions. J Hered 100:507–514. 10.1093/jhered/esp044 [DOI] [PubMed] [Google Scholar]
- 27. Ly N, Danzl NM, Wang J, Zajonc DM, Dascher CC (2010) Conservation of CD1 protein expression patterns in the chicken. Dev & Comp Immunol 34:123–132. 10.1016/j.asd.2015.01.001 [DOI] [PubMed] [Google Scholar]
- 28. Hughes AL (2010) Origin and diversification of the L-amino oxidase family in innate immune defenses of animals. Immunogenetics 62:753–759. 10.1007/s00251-010-0482-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lasoudris F, Cousin C, Prevost-Blondel A, Martin-Garcia N, Abd-Alsamad I, et al. (2011) IL4I1: an inhibitor of the CD8+ antitumor T-cell response in vivo. Eur J Immunol 41:1629–1638. 10.1002/eji.201041119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Al-Murrani WK, Kassab A, Al-Sam HZ, Al-Athari AM (1997) Heterophil/lymphocyte ratio as a selection criterion for heat resistance in domestic fowls. Brit Poul Sci 38:159–163. [DOI] [PubMed] [Google Scholar]
- 31. Gross W, Siegel H (1983) Evaluation of the heterophil/lymphocyte ratio as a measure of stress in chickens. Avian Dis 27(4):972–979. [PubMed] [Google Scholar]
- 32. McFarlane JM, Curtis SE (1989) Multiple concurrent stressors in chicks. Effects on plasma corticosterone and the heterophil: lymphocyte ratio. Poul Sci 68(4):522–527. [DOI] [PubMed] [Google Scholar]
- 33. Gross W (1989) Factors affecting chicken thrombocyte morphology and the relationship with heterophil: lymphocyte ratios. Brit Poul Sci 30:919–925. [DOI] [PubMed] [Google Scholar]
- 34. Campo J, Davila S (2002) Estimation of heritability for heterophil: lymphocyte ratio in chickens by restricted maximum likelihood. Effects of age, sex, and crossing. Poul Sci 81:1448–1453. [DOI] [PubMed] [Google Scholar]
- 35. Nagel J, Smith R, Shaw L, Bertak D, Dixit V, et al. (2004) Identification of genes differentially expressed in T cells following stimulation with the chemokines CXCL12 and CXCL10. BMC Immunol 5:17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uhart M, McCaul M, Oswald L, Choi L, Wand G (2004) GABRA6 gene polymorphism and an attenuated stress response. Mol Psychiatry 9: 998–1006. [DOI] [PubMed] [Google Scholar]
- 37. Akerfelt M, Morimoto RI, Sistonen L (2010) Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Bio 11:545–555. 10.1038/nrm2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qiu XB, Shao YM, Miao S, Wang L (2006) The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci 63:2560–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tukaj S, Kotlarz A, Jozwik A, Smolenska Z, Bryl E, et al. (2010) Hsp40 proteins modulate humoral and cellular immune response in rheumatoid arthritis patients. Cell Stress Chaperones 15(5):555–566. 10.1007/s12192-010-0168-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qiu J, Ni YH, Gong HX, Fei L, Pan XQ, et al. (2007) Identification of differentially expressed genes in omental adipose tissues of obese patients by suppression subtractive hybridization. Biochem Biophys Res Commun 352(2):469–478. [DOI] [PubMed] [Google Scholar]
- 41. Man C, Li X, Zhao D (2011) Cloning, sequence identification, and tissue expression analysis of novel chicken NYGGF4 gene. Mol Cell Biochem 346:117–124. 10.1007/s11010-010-0598-z [DOI] [PubMed] [Google Scholar]
- 42. Wang J, Lee S, Teh CEY, Bunting K, Ma L, et al. (2009) The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Int. Immunol 21(3):227–235. 10.1093/intimm/dxn143 [DOI] [PubMed] [Google Scholar]
- 43. Brabletz T, Jung A, Hlubek F, Lhberg C, Meiler J, et al. (1999) Negative regulation of CD4 expression in T cells by the transcriptional repressor ZEB. Int. Immunol 11:1701–1708. [DOI] [PubMed] [Google Scholar]
- 44. Postigo AA, Dean DC (1999) Independent repressor domains in ZEB regulate muscle and T-cell differentiation. Mol Cell Biol 19:7961–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jethanandani P, Kramer RH (2005) α7 integrin expression is negatively regulated by δEF1 during skeletal myogenesis. J Biol Chem 280: 36037–36046. [DOI] [PubMed] [Google Scholar]
- 46. Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, et al. (1997) Impairment of T cell development in δEF1 mutant mice. J Exp Med 185:1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bovenhuis H, Bralten H, Nieuwland M, Parmentier H (2002) Genetic parameters for antibody response of chickens to sheep red blood cells based on a selection experiment. Poul Sci 81:309–315. [DOI] [PubMed] [Google Scholar]
- 48. Bogdan C (2001) Nitric oxide and the immune response. Nat Immunol 2:907–916. [DOI] [PubMed] [Google Scholar]
- 49. Yang L, Liu N, Hu X, Zhang W, Wang T, et al. (2010) CK2 phosphorylates TNFAIP1 to affect its subcellular localization and interaction with PCNA. Mol Biol Rep 37:2967–2973. 10.1007/s11033-009-9863-1 [DOI] [PubMed] [Google Scholar]
- 50. Dorshorst B, Siegel P, Ashwell C (2011) Genomic regions associated with antibody response to sheep red blood cells in the chicken. Anim Genet 42:300–308. 10.1111/j.1365-2052.2010.02146.x [DOI] [PubMed] [Google Scholar]
- 51. Zhao XL, Honaker CF, Siegel PB (2012) Phenotypic responses of chickens to long-term selection for high or low antibody titers to sheep red blood cells. Poult Sci 91:1047–1056 10.3382/ps.2011-01892 [DOI] [PubMed] [Google Scholar]
- 52. Jang JY, Lee CE (2003) Mitochondrial adenine nucleotide translocator 3 is regulated by IL-4 and IFN-γ via STAT-dependent pathways. Cell Immunol 226:11–19. [DOI] [PubMed] [Google Scholar]
- 53. Jang JY, Lee CE (2006) IL-4-induced upregulation of adenine nucleotide translocase 3 and its role in Th cell survival from apoptosis. Cell Immunol 241:14–25. [DOI] [PubMed] [Google Scholar]
- 54. Butler B, Kastendieck DH, Cooper JA (2008) Differently phosphorylated forms of the cortactin homolog HS1 mediate distinct functions in natural killer cells. Nat Immunl 9:887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ali M, Hocking PM, McKibbin M, Finnegan S, Shires M, et al. (2011) Mpdz null allele in an avian model of retinal degeneration and mutations in human leber congenital amaurosis and retinitis pigmentosa. Invest Ophthalmol Vis Sci 52:7432–7440. 10.1167/iovs.11-7872 [DOI] [PubMed] [Google Scholar]
- 56. Salomonsen J, Sorensen MR, Marston DA, Rogers SL, Collen T, et al. (2005) Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc Natl Acad Sci USA 102(24):8668–8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ly N, Danzl NM, Wang J, Zajonc DM, Dascher CC (2010) Conservation of CD1 protein expression patterns in the chicken. Developmental and Comparative Immunology 34:123–132. 10.1016/j.dci.2009.08.015 [DOI] [PubMed] [Google Scholar]
- 58. Boulland ML, Marquet J, Molinier-Frenkel V (2007) Human IL4I1 is a secreted L-phenylalanine oxidase expressed by mature dendritic cells that inhibits T-lymphocyte proliferation. Blood 110(1):220–227. [DOI] [PubMed] [Google Scholar]
- 59. Rajsbaum R, Stoye JP, O’Garra A (2008) Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur J Immunol 38: 619–630. 10.1002/eji.200737916 [DOI] [PubMed] [Google Scholar]
- 60. Shimono Y, Murakami H, Hasegawa Y, Takahashi M (2000) RET finger protein is a transcriptional repressor and interacts with enhancer of polycomb that has dual transcriptional functions. J Biol Chem 275: 39411–39419. [DOI] [PubMed] [Google Scholar]
- 61. Zurek B, Schoultz I, Neerincx A, Napolitano LM (2012) TRIM27 negatively regulates NOD2 by ubiquitination and proteasomal degradation. PLoS One 7(7):e41255 10.1371/journal.pone.0041255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wegmann TG, Smithies O (1968) Improvement of the microtiter hemagglutination method. Transfusion 8:47 [DOI] [PubMed] [Google Scholar]
- 63. Lucas AM, Jamroz C (1961) Atlas of avian hematology. Washington, D.C., U.S. Dept. of Agriculture; pp 271. [Google Scholar]
- 64.Illumina website. Available: http://www.illumina.com/Documents/products/technotes/technote_infinium_genotyping_data_analysis.pdf. Accessed 2011 Jun 26.
- 65. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao JH (2007) gap: Genetic analysis package. J Stat Softw 14:1–18. [Google Scholar]
- 67. Johnson R, Nelson G, Troyer J, Lautenberger J, Kessing B, et al. (2010) Accounting for multiple comparisons in a genome-wide association study (GWAS). BMC genomics 11:724 10.1186/1471-2164-11-724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nicodemus KK, Liu W, Chase GA, Tsai YY, Fallin MD (2005) Comparison of type I error for multiple test corrections in large single-nucleotide polymorphism studies using principal components versus haplotype blocking algorithms. BMC Genet 6: Suppl 1S78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The raw results from the linear regression analysis by PLINK. Its basic format is: CHR, Chromosome; SNP, SNP identifier; BP, Physical position (base-pair); A1, Tested allele (minor allele by default); TEST, Code for the test (the additive effects of SNPs); NMISS, Number of non-missing individuals included in analysis; BETA, Regression coefficient (the direction of BETA represents the effect of each extra minor allele); STAT, Coefficient t-statistic; P, Asymptotic p-value for t-statistic. The same abbreviations are used in the following tables.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
All raw genotypes are available in the Dryad Repository (doi:10.5061/dryad.8m4r5).