Summary
Early development depends heavily on accurate control of maternally inherited mRNAs, and yet it remains unknown how maternal microRNAs are regulated during maternal to zygotic transition (MZT). We here find that maternal microRNAs are highly adenylated at their 3′ ends in mature oocytes and early embryos. Maternal microRNA adenylation is widely conserved in fly, sea urchin, and mouse. We identify Wispy, one of noncanonical poly(A) polymerases, as the enzyme responsible for microRNA adenylation in flies. Knockout of wispy abrogates adenylation and results in microRNA accumulation in eggs whereas overexpression of Wispy increases adenylation and reduces microRNA levels in S2 cells. Wispy interacts with Ago1 through protein-protein interaction, which may allow the effective and selective adenylation of microRNAs. Thus, adenylation may contribute to the clearance of maternally deposited microRNAs during MZT. Our work provides mechanistic insights into the regulation of maternal microRNAs and illustrates the importance of RNA tailing in development.
Keywords: microRNA, miRNA, adenylation, RNA tailing, Wispy, oocyte, embryo development, maternal to zygotic transition
Introduction
Embryos inherit from oocytes numerous maternal transcripts which play critical roles in early development. Transcription is silenced in late stage oocytes and remains largely suppressed throughout the initial stage of embryogenesis (Baroux et al., 2008; Tadros and Lipshitz, 2009). Thus, maternally deposited mRNAs direct the early events in embryogenesis and are under extensive posttranscriptional regulation for their stability, localization, and translation (Tadros and Lipshitz, 2005). Upon zygotic genome activation and transcription, maternal mRNAs are massively eliminated during maternal to zygotic transition (MZT) in which the developmental control shifts from maternal genome to zygotic genome (Bashirullah et al., 1999; Walser and Lipshitz, 2011). Thus, oocytes and early embryos represent a unique developmental window where posttranscriptional machineries dominate the regulatory network, and offer an excellent model system for the studies of RNA-mediated gene regulation.
Non-templated nucleotidyl addition of RNA (“RNA tailing”) plays a key role in early development. Maternally deposited mRNAs have shortened A tails, whose translation requires cytoplasmic polyadenylation (Barckmann and Simonelig, 2013; Richter, 1999). In vertebrates, GLD2 (also known as PAPD4, TUTase 2 or Hs1) mediates cytoplasmic polyadenylation and is required for the oocyte maturation (Mendez and Richter, 2001; Nakanishi et al., 2006). In Drosophila, Wispy is a putative functional homologue of GLD2 although the homology at the amino acid and domain organization level is not sufficient to make a reliable prediction. Wispy elongates the poly(A) tail of specific maternal mRNAs that are critical for early development (Benoit et al., 2008; Brent et al., 2000; Cui et al., 2008).
MicroRNAs (miRNAs) are ~22 nt small non-coding RNAs that regulate gene expression by base-pairing with target mRNAs in diverse developmental and pathological processes (Bartel, 2009; Bushati and Cohen, 2007; Kim et al., 2009; Sotiropoulou et al., 2009). MiRNA biogenesis typically involves two processing events (Ha and Kim, 2014). The primary transcript (pri-miRNA) is transcribed by RNA polymerase II and is initially processed by Drosha in the nucleus. Drosha processing releases a hairpin-shaped precursor (pre-miRNA). Pre-miRNA is then exported to the cytoplasm by Exportin-5 and further processed by Dicer to generate a short RNA duplex. The miRNA duplex is loaded onto the Argonaute (AGO) protein. After one strand is removed, the remaining single-stranded miRNA and AGO constitute the effector complex called RNA-induced silencing complex (RISC). Although one strand is usually more preferentially selected than the other strand from a given miRNA duplex, both strands end up in AGO albeit in different frequencies.
MiRNAs are subject to RNA tailing, which results in multiple isoforms (“isomiRs”) with varying 3′ ends. Although the frequency of miRNA tailing is usually kept low (below 5%) and such infrequent tailing is unlikely to affect the functionality of miRNA, growing evidences suggest that at least some miRNAs are highly modified with functional consequences under certain conditions (Kim et al., 2010; Krol et al., 2010; Neilsen et al., 2012). In plants, miRNAs are uridylated by nucleotidyl transferases such as HESO1 and MUT68 (in Arabidopsis and Chlamydomonas, respectively) and are subsequently degraded unless their 3′ ends are protected by 2′-O-methylation (Ibrahim et al., 2010; Ramachandran and Chen, 2008; Zhao et al., 2012). In animals, U/A tailing of miRNA is induced by highly complementary targets, which in turn causes trimming and decay of miRNAs (Ameres et al., 2010). The enzyme(s) responsible for the template-induced miRNA tailing remains unknown. In mammalian and fly cells infected by poxvirus, viral poly(A) polymerase (VP55) induces adenylation and decay of host miRNAs (Backes et al., 2012). Mammalian GLD2 (TUTase 2, PAPD4 and Hs1) also adenylates miR-122, but GLD2-mediated adenylation appears to have an opposite consequence and increases the stability of miR-122 (Burns et al., 2011; D’Ambrogio et al., 2012; Katoh et al., 2009). Unlike adenylation, uridylation occurs more frequently on pre-miRNAs than mature miRNAs in animal cells, with the most extensively studied example being pre-let-7. Mono-uridylation of pre-let-7 promotes Dicer processing whereas oligo-uridylation of pre-let-7 blocks Dicer processing and facilitates decay (Chang et al., 2013; Heo et al., 2012; Heo et al., 2008; Heo et al., 2009). LIN28 binds to the terminal loop of pre-let-7 and stably recruits TUTases 4 (ZCCHC11) and TUTase 7 (ZCCHC6) to allow oligo-uridylation in embryonic stem cells and cancer (Hagan et al., 2009; Heo et al., 2009; Newman et al., 2008; Rybak et al., 2008; Thornton et al., 2012; Viswanathan et al., 2008; Yeom et al., 2011). These observations collectively suggest that miRNA tailing may serve as an important mechanism for developmental and pathological regulations of the miRNA pathway.
In this study, we discover pervasive and conserved adenylation of maternally inherited miRNAs. We identify Wispy, a fly noncanonical poly(A) polymerase, as the enzyme responsible for miRNA adenylation during MZT. Adenylation is abolished in wispy mutant while Wispy overexpression induces miRNA adenylation and reduces miRNA levels. Our study shows that RNA tailing may play an integral role in miRNA regulation during MZT.
Results
MicroRNAs are highly adenylated in Drosophila eggs and early embryos
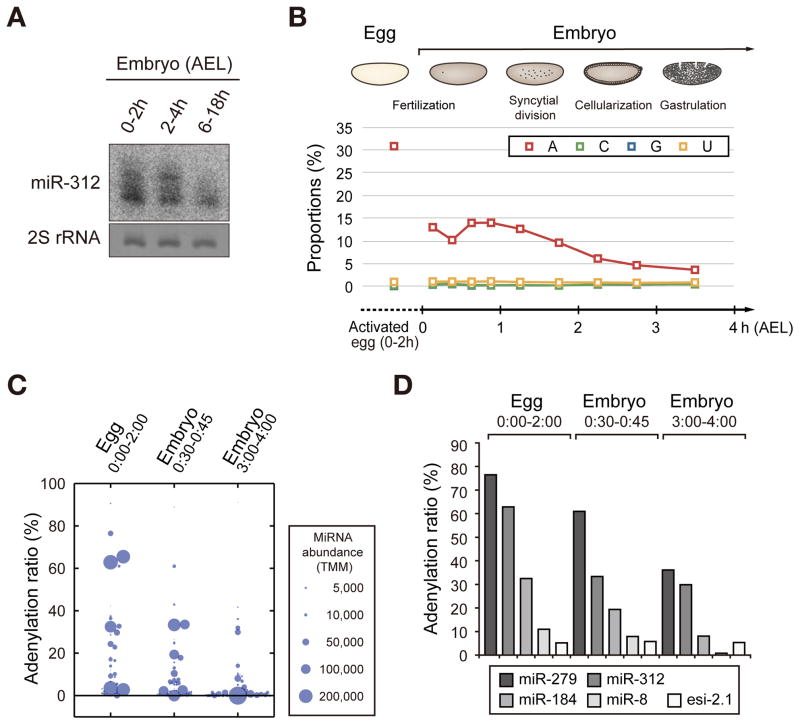
In a recent report, Mattick and colleagues collected and analyzed small RNA sequencing data and noticed a high level of nontemplated adenylyl groups on some miRNAs present in early embryos of Drosophila (Fernandez-Valverde et al., 2010). Intrigued by this report, we examined miRNA modification pattern by northern blotting using embryos collected at various time points. Consistent with the previous sequencing data, we indeed observed a heterogeneous miR-312 population with longer isoforms in early embryos (0–4 h after egg laying (AEL)) (Figure 1A). The long isoforms disappeared in late stage embryo (6–18 h AEL), indicating that modification of miR-312 is tightly regulated during embryonic development (Figure 1A).
Figure 1. Adenylation of miRNA in Drosophila eggs and embryos.
(A) Northern blot showing the miR-312 expression pattern in Drosophila embryos. Long isomiRs are detected strongly in early embryos (0–4 h AEL) but not in later stage embryos (6–18 h AEL). (B) Small RNA sequencing from activated eggs and embryos at different time points. The modification ratio is calculated by dividing the modified miRNA reads by total miRNA reads. Nontemplated C, G, U additions are constantly rare throughout development, while adenylation ratio is specifically high in eggs and early embryos. (C) A dot plot showing the adenylation rate of individual miRNA species in indicated time frames. The size of the circle reflects trimmed mean of M-values (TMM) which is the normalized abundance of miRNA by total miRNA read count. Ninety two miRNA species were analyzed (TMM > 100 in at least one of the three samples). (D) Adenylation of representative small RNAs in deep sequencing data at different developmental stages. Adenylation frequency of endo-siRNA 2.1 is relatively low and constant, which contrasts with those of miRNAs. See also Figure S1, Table S1 and Table S2.
To investigate miRNA modification patterns more closely, we next performed small RNA sequencing from activated eggs as well as early embryos (Figure S1A) because the published sequencing data are from only embryos collected at sparse intervals. It is noted that due to technical limitation, we prepared unfertilized activated oocytes for 0–2 h AEL instead of the oocytes under maturation in the ovary. Drosophila oocytes are activated by physical pressure during egg-laying and resume meiosis even without fertilization (Heifetz et al., 2001).
The sequencing data show that the relative read number of miRNA is low in eggs and increases in later stages after the major surge of zygotic transcription (2–4 h AEL) (Figures S1A and S1B). Surprisingly, over 30% of total miRNA reads carry nontemplated A addition in activated eggs (Figure 1B and Table S1). The proportion of adenylated miRNAs is ~15% in 0–1 h AEL embryos and declines to below 10% after 2 h AEL and eventually to 3.7% at ~4 h AEL. In contrast to adenylation, other modifications such as cytidylation, guanylation and uridylation are rare: the proportions of miRNAs with C, G or U are below 1.5% in all stages (Figure 1B). Compared to the reported adenylation ratios in mammalian cells (2–4%) and Drosophila adult tissues (~3%) (Berezikov et al., 2011; Burroughs et al., 2010; Wyman et al., 2011), the adenylation ratio observed in this study is remarkably high. About a half (45%) of the miRNA species detected in eggs (TMM > 50) shows adenylation rate of over 5% (Figure 1C and Table S1). Unlike miRNAs, endogenous siRNAs (endo-siRNAs) are not highly adenylated. We could hardly detect elongated isoforms of esi-2.1 by northern blotting (Figure S1C). We also analyzed our sequencing data for endo-siRNAs (Czech et al., 2008; Kawamura et al., 2008). The abundance of siRNA was generally too low for reliable analysis, but abundant siRNAs showed low adenylation levels, which does not change significantly throughout embryo development (Figures 1D, S1D and Table S2).
Adenylation of microRNA is conserved in sea urchin and mouse
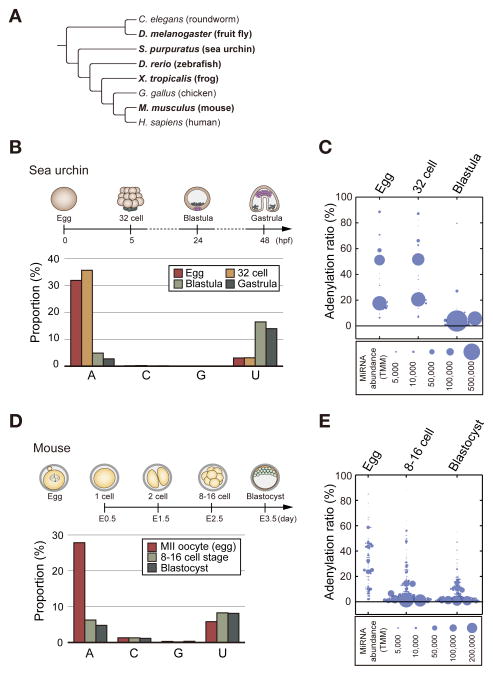
Next, to address whether miRNA adenylation is an evolutionarily conserved process, we collected published sequencing data from eggs and embryos of diverse model organisms including sea urchin (Strongylocentrotus purpuratus), frog (Xenopus tropicalis), and mouse (Mus musculus) (Harding et al., 2014; Ohnishi et al., 2010; Song et al., 2012) (Figure 2A). Additionally, we carried out deep sequencing of zebrafish (Danio rerio) unfertilized eggs (squeezed and not exposed to water), embryos collected at 0.5 h (first cell division) and 24 h post-fertilization because there is no public data available for eggs and early embryos before 2 h post-fertilization.
Figure 2. Conservation of maternal miRNA adenylation.
(A) A phylogenetic tree of model organisms. The organisms we tested are indicated in bold. (B) MiRNA modification ratios in sea urchin at different developmental stages. The adenylation rates are notably high in egg and 32 cell embryo. (C) A dot plot showing the adenylation rate of each miRNA species in egg, 32 cell embryo, and blastula of sea urchin. The size of the circle indicates the normalized abundance of miRNA (TMM). 34 mature miRNA species were analyzed (TMM > 100 in at least one of the three samples). (D) Modification ratios of miRNAs in mouse at different developmental stages. The adenylation rate is specifically high in unfertilized (metaphase II: MII) oocytes. (E) A dot plot showing the adenylation rate of each miRNA species in egg, 8–16 cell embryo, and blastocyst of mouse. The size of the circle indicates the normalized abundance of miRNA (TMM). 166 mature miRNA species were analyzed (TMM > 500 in at least one of the three samples). See also Figure S2.
All sequencing data showed that miRNA contents (relative to other RNA species) are substantially smaller in eggs and early embryos, compared to those in the later stages of development (Figure S2), which is similar to Drosophila (Figures S1A and S1B). Because miRNAs comprise less than 1% of total small RNA reads in eggs and early embryos of frog and zebrafish, it was not possible to reliably measure modification frequencies (Figure S2). Instead, we analyzed miRNA sequencing data from sea urchin and mouse, and found that their modification patterns are surprisingly similar to that of Drosophila. In eggs and 32-cell embryos of sea urchin, miRNAs show a high percentage of adenylation over 30% (Figures 2B and 2C). In mouse, the total adenylation ratio reaches almost 30% in metaphase II oocytes, and drops down to about 5% in later embryonic stages (Figures 2D and 2E). In contrast, the ratios of C or G addition remain low (under 2%) in all data we analyzed, and the ratio of uridylation tends to increase in later stages (Figures 2B and 2D). Like in flies, most miRNA species are adenylated in eggs albeit at varying frequencies (Figures 2C and 2E). Thus, maternal miRNA adenylation is conserved from fly to mouse.
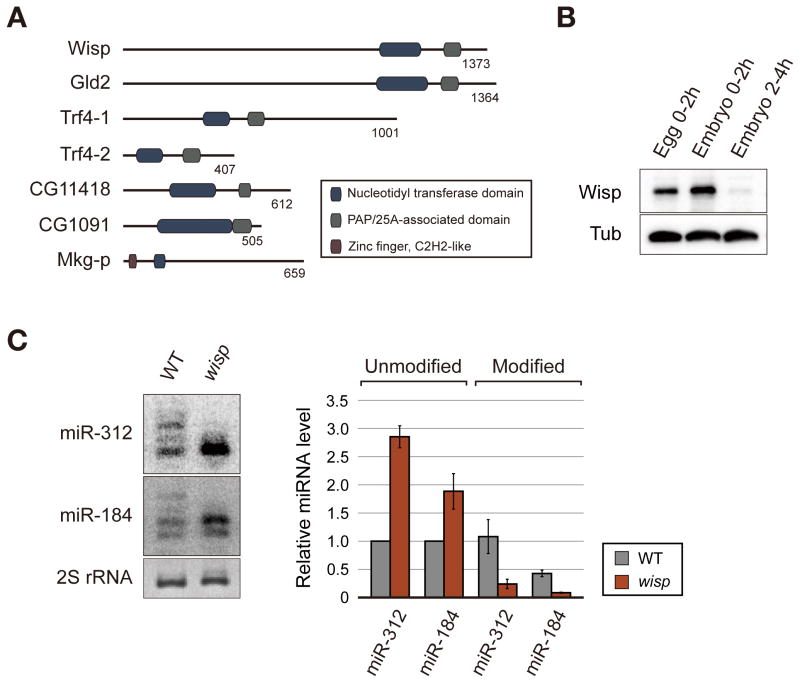
Wispy is required for the adenylation of maternal microRNAs
To identify the enzyme responsible for miRNA adenylation, we searched for proteins that contain a conserved nucleotidyl transferase domain (Schmidt and Norbury, 2010). Among the seven predicted noncanonical poly(A) polymerases in Drosophila (Figure 3A), Wispy (Wisp) shows the most exclusive expression in early embryos based on modENCODE RNA-Seq data (data not shown). Consistent with the RNA-Seq data and previous reports (Benoit et al., 2008; Cui et al., 2008), we observed by western blotting that Wisp protein is highly expressed in egg and early embryos (0–2 h AEL) and rapidly disappears in later stage (2–4 h AEL) (Figure 3B). Wisp is well known to have poly(A) polymerase activity on maternal mRNAs (Benoit et al., 2008; Cui et al., 2008; Cui et al., 2013). Therefore, we tested whether Wisp is involved in miRNA modification as well. Because wisp mutant embryos show an early developmental arrest (Brent et al., 2000), we could not analyze miRNAs in embryo samples at diverse time points. Thus, we collected unfertilized eggs of wild type and wisp mutant, both of which are activated by egg-laying, and compared the modification patterns by northern blot analysis. In wisp mutant eggs, we found a decrease of miRNA modification and an increase of miRNA abundance (Figure 3C).
Figure 3. Wisp mediates the adenylation of maternal miRNA.
(A) Domain structures of fly noncanonical poly (A) polymerases. Dark blue, nucleotidyl transferase domain; gray, PAP/25A-associated domain; purple, zinc finger C2H2-like domain. (B) Expression of Wisp in eggs and embryos. Wisp protein level was visualized by western blotting using anti-Wisp antibody. (C) Northern blot analysis of adenylated miRNAs. Modified isoforms of miR-312 and miR-184 are clearly observed in wild type eggs (0–2 h AEL) while they are strongly reduced in wisp mutant eggs (0–2 h AEL). The right panel shows the quantitation from three biological replicates. Data are presented as mean ± standard error. The p values for the differences between wild type and wisp mutant in unmodified miR-312, unmodified miR-184, modified miR-312, and modified miR-184 are 0.0007, 0.0480, 0.0548 and 0.0042, respectively.
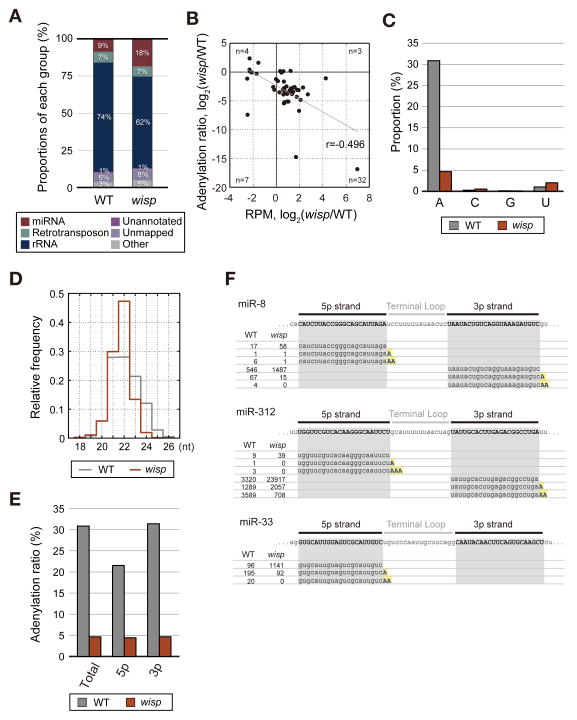
We further examined the global effect of wisp mutation by small RNA sequencing (Figure 4). The proportion of miRNA reads increased in wisp mutant eggs (Figure 4A). Consistent with northern blotting results (Figure 3C), the majority of miRNAs are reduced in adenylation (Figure 4B, y-axis) and upregulated in abundance (Figure 4B, x-axis) in wisp mutant. Total adenylation ratio is reduced to 4.6% in wisp mutant (Figure 4C). The effect was specific to adenylation as the other modifications were not decreased. Overall length of miRNA population was reduced and the length distribution became more homogenous in wisp mutant, as expected (Figure 4D).
Figure 4. Adenylation of miRNAs is globally decreased in wisp mutant.
(A) Proportion of each RNA class in the sequencing libraries from wild type and wisp mutant eggs (0–2 h AEL). (B) A scatter plot showing the differences of adenylation ratio versus those of miRNA read counts between wild type and wisp mutant eggs. The Spearman correlation coefficient is −0.496. The number of miRNA species (n) in each quadrant is presented. Forty six miRNA species were analyzed (reads per million (RPM) > 50 in at least one of the two samples). (C) MiRNA modification determined by sequencing. Adenylation ratio is significantly reduced in wisp mutant eggs, but other modification ratios were not altered. (D) Length distribution of miRNAs in wild type and wisp mutant eggs. The mean lengths are 22.1 nt in wild type, and 21.7 nt in wisp mutant. (E) Adenylation of 5p miRNAs and 3p strand miRNAs. (F) Examples of sequencing reads mapped to the representative miRNAs in eggs. Reference sequences according to miRBase are highlighted in gray boxes. Nontemplated A addition is indicated in blue letters highlighted in yellow. The RPM values of each sequence are presented on the left side.
Our analysis also revealed that both the 5p and 3p strands are adenylated by Wisp (Figure 4E). Because the 3′ end of 5p strand is exposed only after Dicer cleavage, this result indicates that Wisp acts at the downstream of Dicer processing step. Closer examination of individual sequencing reads shows that mono-, di-, and tri-adenylation indeed occurs on both the 5p and 3p strands in wild type eggs (Figure 4F). In wisp mutant, unmodified miRNA reads increase at the expense of adenylated reads. The residual adenylation in wisp mutant (~4%) may be produced by another enzyme that is active ubiquitously in all developmental stages as the adenylation frequency in wisp mutant is comparable to that in later stage embryos (3–4 h AEL) (Figure 1B).
Wisp reduces microRNA abundance and affects target repression
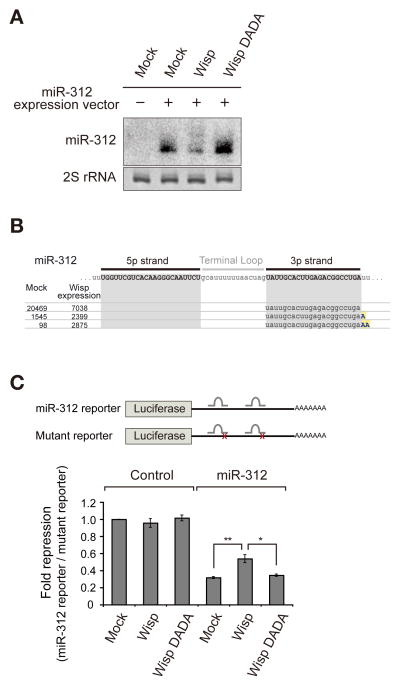
To confirm the miRNA adenylation activity of Wisp, we ectopically introduced Wisp into S2 cells by co-transfecting plasmids expressing Wisp and miR-312, neither of which is normally detected in S2 cells. The miRNA modification was monitored by northern blotting (Figure 5A). The effect of Wisp overexpression was the opposite of that of wisp knockout. Wisp overexpression resulted in the elongation and reduction of mature miRNA, without obvious modification or accumulation of pre-miRNA (Figures 5A, S3A, and S3B). A catalytically dead mutant (DADA) that has point mutations at 1029 and 1031 residues (aspartate to alanine) in the catalytic domain failed to induce modification, confirming that the nucleotidyl transferase domain of Wisp directs this modification (Benoit et al., 2008; Martin and Keller, 1996) (Figure S3C). To further confirm that the modified isoforms detected by northern blotting correspond to adenylation products, we performed small RNA deep sequencing (Figures 5B and S3D). Indeed, when Wisp was expressed, the adenylated miR-312 increased while the unmodified counterpart decreased. Endogenous miRNAs were not significantly affected by Wisp overexpression, presumably because only a small fraction of S2 cells were transfected due to low transfection efficiency.
Figure 5. Wisp overexpression leads to miRNA adenylation and impairs target repression activity.
(A) Northern blot analysis following co-transfection of the miR-312 and Wisp expression plasmids into S2 cells. miR-312 is highly modified and decreased in cells overexpressing Wisp, but not affected by the catalytic mutant (DADA). This is a representative result of three independent experiments. (B) Small RNA sequencing of the following samples: “Mock” (S2 cell transfected with only miR-312) and “Wisp expression” (S2 cell transfected with miR-312 and Wisp). Reads of miR-312 aligned to the reference sequence. The RPM values of each sequence are presented on the left side. (C) Dual luciferase reporter assay to measure miR-312 activity. A firefly luciferase reporter containing two miR-312 bulged sites was used. The relative activity was calculated by firefly luciferase activity from the miR-312 reporter (normalized to renilla luciferase activity) divided by that of firefly luciferase activity from the mutant reporter (normalized to renilla luciferase activity). Data are presented as mean ± standard error of four biological replicates. The p value was determined by two-tailed T-test (*: p < 0.05, **: p < 0.01). See also Figure S3.
Next, we tested whether Wisp has an effect on the activity of miRNA. For this, we generated a firefly luciferase reporter containing two intact miR-312 binding sites with a bulge at the center (Figure 5C). A control reporter contains two mutated miR-312 binding sites with 4 nucleotide mismatches. The miR-312 reporter was suppressed by miR-312. Wisp co-expression significantly de-repressed the reporter (Figure 5C) while the catalytic mutant (DADA) did not have an impact on the suppression. Taken together, our results indicate that Wisp downregulates miRNA abundance and consequently affects target repression.
Wisp interacts with Ago1
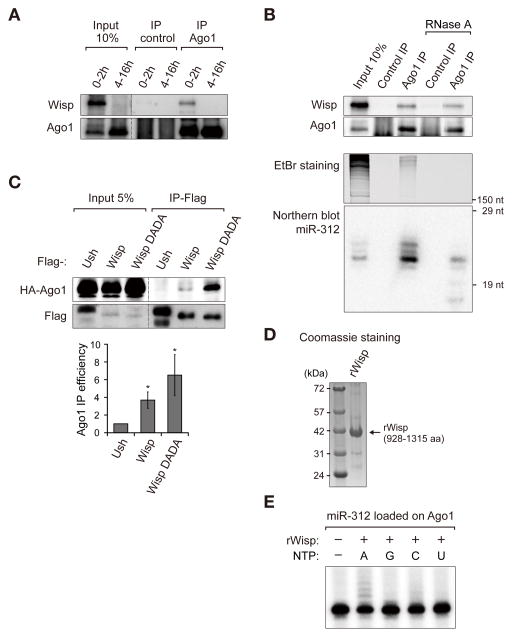
How does Wisp recognize miRNAs? It was shown that, during cytoplasmic mRNA polyadenylation, Wisp is recruited to mRNAs through the interaction with RNA binding proteins (Benoit et al., 2008). Because Wisp does not have any putative RNA binding domain, it may need a partner to recognize its substrate miRNAs. In our analysis of sequencing data, we learned that Wisp acts on miRNAs after Dicer cleavage. Because miRNA duplex is loaded onto Ago1 for their action after Dicer cleavage (Czech and Hannon, 2011; Kawamata and Tomari, 2010), we tested whether Wisp interacts with Ago1. Co-immunoprecipitation experiment demonstrates that Wisp physically associates with Ago1 (Figure 6A). Furthermore, this interaction was not abolished by RNase A treatment which thoroughly removed long RNAs (Figure 6B, EtBr staining) and degraded most miRNA bound to Ago1 (Figure 6B, Northern blotting). Thus, the Ago1-Wisp interaction is unlikely to be bridged by mRNAs or miRNAs, but rather mediated mainly by protein-protein interaction. We further confirmed their interaction in S2 cells after transfection of Flag-Wisp and HA-Ago1 expression plasmids. HA-Ago1 was co-immunoprecipitated with Flag-Wisp, but not with a control protein Flag-Ush. Interestingly, we found that Wisp DADA mutant binds more strongly to Ago1 compared to wild type Wisp (Figure 6C). The enhanced binding of the catalytic mutant implies that Wisp may bind to Ago1 transiently and that, following successful catalysis, Wisp may be released from Ago1.
Figure 6. Wisp physically associates with Ago1.
(A) Co-immunoprecipitation of endogenous Ago1 and Wisp in embryos. Polyclonal Ago1 antibody or normal rabbit serum was used for immunoprecipitation. Wisp is specifically detected in Ago1 immunoprecipitate. The dashed line indicates discontinuous lanes from the same blot. (B) The interaction between Ago1 and Wisp is RNA-independent. Wisp is co-immunoprecipitated with Ago1 from 0–2 h embryo lysate in the presence of RNase A. Total RNA and miRNA from input and IP samples were visualized with EtBr gel staining and northern blotting. RNase A treatment effectively degrades cellular RNAs including miRNA loaded on Ago1. This is a representative result of three independent experiments. (C) Co-immunoprecipitation of HA-Ago1 and Flag-Wisp in S2 cells transfected with the HA-Ago1 and Flag-Wisp expression plasmids. HA-Ago1 is more efficiently co-precipitated with Flag-Wisp DADA (catalytic mutant) than Flag-Wisp. Flag-Ush was used as a negative control. The efficiency of Ago1 immunoprecipitation is shown in the lower panel. The quantification is from three biological replicates and presented as mean ± standard error. The p value was calculated by one-tailed T-test (*: p < 0.05). (D) Quality of recombinant protein visualized by SDS-PAGE and coomassie staining. (E) In vitro adenylation assay. HA-Ago1 was expressed in S2 cells, immunopurified and incubated with miR-312 duplex of which only 3p strand was 5′-end-radiolabeled. MiRNA loaded on Ago1 was incubated with purified recombinant Wisp protein in the presence of ATP, GTP, CTP or UTP. RNA was extracted and analyzed by UREA-PAGE and autoradiography. See also Figure S4.
Next, to examine whether Wisp directly adenylates miRNA, in vitro adenylation assay was performed. For this, we expressed and purified recombinant Wisp protein from E.coli (Figure 6D). The recombinant protein encompasses the C-terminal part of Wisp (928-1315 aa) including the catalytic domain. The C-terminal part induces miRNA adenylation when overexpressed in S2 cell (Figure S4). When miRNA-loaded Ago1 was incubated with Wisp protein in the presence of ATP, GTP, CTP, or UTP, Wisp selectively used ATP to elongate miR-312 by 1–4 nt (Figure 6E), demonstrating that Wisp directly adenylates miRNA.
Discussion
Our study provides the first mechanistic insights into the regulation of maternally deposited miRNAs. The majority of maternal miRNAs are subject to adenylation during early development in Drosophila (Figures 1B, 1C). The sequencing and northern blotting data indicate that the levels of adenylated miRNAs begin to drop at ~1.5 h AEL with an average half-life of ~2 h (Figures S5A and S5D). This is substantially shorter than those estimated in mammalian cell lines (> 100 h considering dilution caused by cell division) (Gantier et al., 2011). Our results collectively suggest that Wisp induces adenylation, and facilitates miRNA downregulation. Thus, miRNA adenylation may provide a molecular basis for the clearance of maternally inherited miRNAs during MZT. MiRNA tailing is often associated with miRNA destabilization (Ameres et al., 2010; Backes et al., 2012; Ibrahim et al., 2010; Zhao et al., 2012). An interesting example is poxvirus whose adenylyl transferase VP55 downregulates host miRNAs (Backes et al., 2012). It is conceivable that poxviruses have adopted the cellular adenylation/decay machineries to their own benefits. It is noted that adenylation may have an opposing effect in different contexts as mammalian poly(A) polymerase GLD2 is known to stabilize certain miRNAs (Burns et al., 2011; D’Ambrogio et al., 2012; Katoh et al., 2009). It is yet unclear why the seemingly related modifications result in such different consequences. To dissect the mechanism behind the link between tailing and decay, it will be important to identify the enzyme that executes decay. We have tested exoribonuclease candidates including exosome components and a known fly miRNA trimming factor, Nibbler, by individual knockdown in S2 cells (Liu et al., 2011), but have not been successful in finding the enzyme(s), implying that multiple factors may act redundantly, as in Arabidopsis (Ramachandran and Chen, 2008).
It is intriguing that three distant species (fly, sea urchin and mouse) show a similar pattern of miRNA expression and adenylation. In all species examined, relative contents of miRNA are low in mature oocytes (Harding et al., 2014; Ohnishi et al., 2010; Song et al., 2012). In frogs and zebrafish, the miRNA proportion was particularly small in oocytes and early embryos. Thus, maternal miRNAs may be cleared out at an early stage(s) of oocyte maturation in frogs and fish so that maternal miRNAs may not be transmitted to the next generation. In flies, maternal miRNA clearance is relatively delayed and overlaps with zygotic transcription. Therefore, while the clearance of maternal miRNA appears to be a universal phenomenon, the precise timing of the event varies among species. Consistently, previous studies have suggested that miRNAs may be inactive in oocytes and early development (Svoboda, 2010). Maternal and zygotic Dgcr8-null mouse embryos develop normally to the blastocyst stage, indicating that miRNA function is suppressed in oocytes and early embryos (Suh et al., 2010). In parallel, reporter assays for endogenous miRNA activity in mouse oocytes revealed a dramatic decrease in miRNA activity along oocyte maturation (Ma et al., 2010). Our current data suggest that active clearance of miRNA via adenylation may be necessary for normal gene regulation in early embryo development.
Wisp physically associates with Ago1, which may explain why miRNAs are effectively adenylated. However, adenylation frequency varies among miRNA species, and does not show a strong correlation with abundance (Figures 1C, S5A, and S5B). We searched for common features among the highly adenylated miRNAs, but failed to find a sequence motif that is significantly enriched in adenylated miRNAs (data not shown). When we ectopically expressed miR-312 (that is highly adenylated in embryos) and miR-286 (that is not adenylated in embryos) in S2 cells, both miRNAs were similarly adenylated by Wisp (Figure S6), suggesting that adenylation is not determined by intrinsic sequences of miRNA. We next examined temporal differential expression in embryos (Figure S5A). Note that, due to an inevitable normalization issue, the heat map does not strictly reflect the changes in absolute abundance (Figure S5A, red color indicating the unit of standard deviation from the mean value of the given miRNA species across developmental stages). Nevertheless, this analysis was useful to classify miRNAs according to their expression patterns during development. MiRNAs are clustered into three groups. The “early” miRNAs show higher expression at early stage and decline after major zygotic genome activation (~1.5 h AEL), indicating that these miRNAs are reduced during MZT. The “late” miRNAs are mainly induced after zygotic activation in the later stage. The “biphasic” miRNAs also show the highest levels in late embryos, but they are detected in early stage to some degrees. By comparing adenylation ratios of three groups, we found that the “early” group tends to be more frequently adenylated than the other groups (Figure S5C, p < 3.8 x 10−3, Mann-Whitney U test). This result was validated by northern blotting of two representative miRNAs (Figure S5D). miR-312 (an “early” miRNA) is highly adenylated and gradually disappears while miR-286 (a “late” miRNA) is not modified and is induced zygotically. The data collectively suggest an intriguing possibility that “early” maternal miRNAs are downregulated during maternal to zygotic transition although the mechanism underlying the preference of adenylation is unclear at this point.
It is conceivable that miRNA targets may affect adenylation rate in vivo. MiRNAs bound to complementary targets may be more susceptible to Wisp-mediated adenylation because the 3′ end of guide RNA is thought to be released from the PAZ domain of AGO protein when the miRNA binds to a highly complementary target (Wang et al., 2009). Not mutually exclusively, some miRNAs may be more accessible to Wisp than others in vivo. Certain miRNAs may be localized separately from Wisp and the physical segregation may protect the miRNAs from adenylation and decay. It will be interesting in the future to dissect the mechanism underlying the selectivity of adenylation.
Knockout mutation of wisp results in a lethal phenotype with defects in late meiosis and early development (Benoit et al., 2008; Brent et al., 2000; Cui et al., 2008). MiRNA adenylation activity of Wisp may at least partly contribute to the phenotype although currently we cannot separate the contribution of miRNA adenylation from that of mRNA polyadenylation. Cytoplasmic polyadenylation of mRNA induces translation while miRNA adenylation facilitates miRNA decay. Because both events are expected to positively regulate protein synthesis, it will be interesting to ask if the two seemingly independent activities corroborate to enhance translation.
Thus far, the extensive miRNA adenylation in oocytes (over 30%) is unprecedented among uninfected cell types. MiRNAs typically show less than 2–4% of adenylation. But it is conceivable that adenylation may be used for gene regulation in diverse cellular contexts. Some specific cell types and/or subcellular locations may contain highly adenylated miRNA species. It will be interesting in the future to investigate miRNA tailing in various cell types such as in neurons as cytoplasmic polyadenylation is known to be active and important for local translation in neural synapses (Kwak et al., 2008; Udagawa et al., 2012).
Experimental Procedures
Drosophila stocks and egg/embryo collection
w1118 was used as wild type control. wispKG5287 obtained from Bloomington stock center was originally deposited by Hugo Bellen, and has been previously described as a null allele of wisp (Benoit et al., 2008). Unfertilized activated eggs were produced from w1118 virgin females mated to T(Y;2) sterile males (Semotok et al., 2008). Fly eggs and embryos were collected on grape juice plates for the designated time frame and kept for the additional time to set the developmental stage at 25°C.
RNA extraction and northern blot
Total RNA was prepared from eggs, embryos, S2 cells, or Ago1 immunoprecipitates with TRIzol reagent (Life Technologies). For RNA extraction, fly eggs and embryos were dechorionated with bleach before homogenization in TRIzol reagent. RNA (10–30 ug total RNA) was separated on 15% urea-PAGE gel and transferred to Zeta-probe GT membrane (Biorad) for UV crosslinking or Hybond-NX membrane (Amersham) for 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)- mediated chemical crosslinking (Pall and Hamilton, 2008). 5′ end radiolabeled oligonucleotides were used as probes for hybridization. The membrane was exposed to phosphor imaging plate (Fujifilm) and read by BAS-2500 (Fujifilm) or Typhoon FLA 7000 (GE Healthcare). The sequences of northern probes used in this study are presented in Table S3.
Immunoprecipitation
For Immunoprecipitation in fly embryo lysate, dechorionated embryos were homogenized in lysis buffer (200 mM KCl, 10 mM Tris-HCl [pH8.0], 0.2 mM EDTA) and the supernatant of lysate was obtained by centrifugation twice for 15 min at 4°C. After pre-clearing with Protein A Sepharose beads (GE Healthcare), the lysate was incubated with anti-Ago1 antibody immobilized on Protein A beads for 2 h at 4°C. To test RNA-independent interaction, RNase A was added to the pre-cleared extract before incubation with antibody-bound beads at 0.05 mg/ml concentration. The beads were washed six times with lysis buffer and boiled in SDS sample buffer for western blotting. To prepare lysates of S2 cells expressing Flag-Wisp and HA-Ago1, transfected S2 cells were incubated with lysis buffer (200 mM KCl, 10 mM Tris-HCl [pH8.0], 0.2 mM EDTA, 0.1% NP40) for 20 min on ice, followed by sonication and centrifugation. Then, the lysate was incubated with anti-Flag M2 affinity gel (Sigma) for 2 h at 4°C for immunoprecipitation. IP efficiency was calculated by dividing the band intensity of “IP” by that of “Input”.
Small RNA library preparation
Small RNA cDNA libraries were generated using TruSeq small RNA sample preparation kit (Illumina) according to manufacturer’s guide. For each library, 10 ug of total RNA was separated on 15% urea-PAGE. RNA of 15–29 nt was gel-purified and then ligated to the 3′ adaptor with T4 RNA ligase2 truncated (NEB). After gel purification of 3′ adaptor-ligated RNA, the 5′ adaptor ligation was performed using T4 RNA ligase1. The ligation product was reverse-transcribed by SuperScript III (Life Technologies) and amplified by PCR with Phusion DNA polymerase. The cDNA libraries were sequenced by HiSeq 2000 or HiSeq 2500.
Analysis of miRNA modification
We collected reads that perfectly match to the mature miRNA sequences annotated in miRBase as well as those that have 3′ additions to the perfectly matching sequences. When the annotated miRNA sequence is 22 nt or shorter, the additional sequences in the 3′ end of the read were considered as post-cleavage nucleotidyl addition. When the annotated miRNA sequence is 23 nt or longer, the additional sequences were considered as post-cleavage nucleotidyl addition when it does not match the genome. The total mature miRNA count was calculated by adding all unmodified and modified read counts. The nontemplated modification ratio was defined as the percent proportion of the modified read count over total read count, with pseudocount 0.001.
Supplementary Material
Acknowledgments
We are grateful to the members of our laboratory for discussion and technical help. We thank Dr. Elisa Izaurralde for the pAc5.1 luciferase vector, and Dr. Mikiko Siomi for Ago1 antibody. This work was supported by IBS-R008-D1 of Institute for Basic Science from the Ministry of Science, ICT and Future Planning of Korea (M.L., Y.C., K.K., H.J., J.L., T.A.N., J.Y., and V.N.K.), and by the BK21 Research Fellowships from the Ministry of Education of Korea (Y.C., K.K., and J.L.).
Footnotes
Accession numbers
Sequenced reads have been deposited in the NCBI Gene Expression Omnibus (GEO) database (accession numbers GSE61931)
References
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes S, Shapiro JS, Sabin LR, Pham AM, Reyes I, Moss B, Cherry S, tenOever BR. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell host & microbe. 2012;12:200–210. doi: 10.1016/j.chom.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barckmann B, Simonelig M. Control of maternal mRNA stability in germ cells and early embryos. Biochimica et biophysica acta. 2013;1829:714–724. doi: 10.1016/j.bbagrm.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Baroux C, Autran D, Gillmor CS, Grimanelli D, Grossniklaus U. The maternal to zygotic transition in animals and plants. Cold Spring Harbor symposia on quantitative biology. 2008;73:89–100. doi: 10.1101/sqb.2008.73.053. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. The EMBO journal. 1999;18:2610–2620. doi: 10.1093/emboj/18.9.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development. 2008;135:1969–1979. doi: 10.1242/dev.021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome research. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent AE, MacQueen A, Hazelrigg T. The Drosophila wispy gene is required for RNA localization and other microtubule-based events of meiosis and early embryogenesis. Genetics. 2000;154:1649–1662. doi: 10.1093/genetics/154.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns DM, D’Ambrogio A, Nottrott S, Richter JD. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011;473:105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome research. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annual review of cell and developmental biology. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Sackton KL, Horner VL, Kumar KE, Wolfner MF. Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation. Genetics. 2008;178:2017–2029. doi: 10.1534/genetics.107.084558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Sartain CV, Pleiss JA, Wolfner MF. Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Developmental biology. 2013;383:121–131. doi: 10.1016/j.ydbio.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nature reviews Genetics. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrogio A, Gu W, Udagawa T, Mello CC, Richter JD. Specific miRNA stabilization by Gld2-catalyzed monoadenylation. Cell reports. 2012;2:1537–1545. doi: 10.1016/j.celrep.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Valverde SL, Taft RJ, Mattick JS. Dynamic isomiR regulation in Drosophila development. Rna. 2010;16:1881–1888. doi: 10.1261/rna.2379610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic acids research. 2011;39:5692–5703. doi: 10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nature reviews Molecular cell biology. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature structural & molecular biology. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding JL, Horswell S, Heliot C, Armisen J, Zimmerman LB, Luscombe NM, Miska EA, Hill CS. Small RNA profiling of Xenopus embryos reveals novel miRNAs and a new class of small RNAs derived from intronic transposable elements. Genome research. 2014;24:96–106. doi: 10.1101/gr.144469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz Y, Yu J, Wolfner MF. Ovulation triggers activation of Drosophila oocytes. Developmental biology. 2001;234:416–424. doi: 10.1006/dbio.2001.0246. [DOI] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Molecular cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes & development. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y. Making RISC. Trends in biochemical sciences. 2010;35:368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell. 2010;143:703–709. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature reviews Genetics. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kwak JE, Drier E, Barbee SA, Ramaswami M, Yin JC, Wickens M. GLD2 poly(A) polymerase is required for long-term memory. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14644–14649. doi: 10.1073/pnas.0803185105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Abe M, Sabin LR, Hendriks GJ, Naqvi AS, Yu Z, Cherry S, Bonini NM. The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Current biology : CB. 2011;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM. MicroRNA activity is suppressed in mouse oocytes. Current biology : CB. 2010;20:265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. The EMBO journal. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nature reviews Molecular cell biology. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Nakanishi T, Kubota H, Ishibashi N, Kumagai S, Watanabe H, Yamashita M, Kashiwabara S, Miyado K, Baba T. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Developmental biology. 2006;289:115–126. doi: 10.1016/j.ydbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Neilsen CT, Goodall GJ, Bracken CP. IsomiRs--the overlooked repertoire in the dynamic microRNAome. Trends in genetics : TIG. 2012;28:544–549. doi: 10.1016/j.tig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. Rna. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, Sakaki Y, Sasaki H, Hohjoh H. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic acids research. 2010;38:5141–5151. doi: 10.1093/nar/gkq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. Cytoplasmic polyadenylation in development and beyond. Microbiology and molecular biology reviews : MMBR. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature cell biology. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Schmidt MJ, Norbury CJ. Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. Wiley interdisciplinary reviews. RNA. 2010;1:142–151. doi: 10.1002/wrna.16. [DOI] [PubMed] [Google Scholar]
- Semotok JL, Luo H, Cooperstock RL, Karaiskakis A, Vari HK, Smibert CA, Lipshitz HD. Drosophila maternal Hsp83 mRNA destabilization is directed by multiple SMAUG recognition elements in the open reading frame. Molecular and cellular biology. 2008;28:6757–6772. doi: 10.1128/MCB.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JL, Stoeckius M, Maaskola J, Friedlander M, Stepicheva N, Juliano C, Lebedeva S, Thompson W, Rajewsky N, Wessel GM. Select microRNAs are essential for early development in the sea urchin. Developmental biology. 2012;362:104–113. doi: 10.1016/j.ydbio.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. Rna. 2009;15:1443–1461. doi: 10.1261/rna.1534709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Current biology : CB. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P. Why mouse oocytes and early embryos ignore miRNAs? RNA biology. 2010;7:559–563. doi: 10.4161/rna.7.5.12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Developmental dynamics : an official publication of the American Association of Anatomists. 2005;232:593–608. doi: 10.1002/dvdy.20297. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) Rna. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Molecular cell. 2012;47:253–266. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser CB, Lipshitz HD. Transcript clearance during the maternal-to-zygotic transition. Current opinion in genetics & development. 2011;21:431–443. doi: 10.1016/j.gde.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome research. 2011;21:1450–1461. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom KH, Heo I, Lee J, Hohng S, Kim VN, Joo C. Single-molecule approach to immunoprecipitated protein complexes: insights into miRNA uridylation. EMBO reports. 2011;12:690–696. doi: 10.1038/embor.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Current biology : CB. 2012;22:689–694. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.