SUMMARY
X chromosome inactivation (XCI) depends on the long noncoding RNA Xist and its recruitment of Polycomb Repressive Complex 2 (PRC2). PRC2 is also targeted to other sites throughout the genome to effect transcriptional repression. Using XCI as a model, we apply an unbiased proteomics approach to isolate Xist and PRC2 regulators and identified ATRX. ATRX unexpectedly functions as a high-affinity RNA-binding protein that directly interacts with RepA/Xist RNA to promote loading of PRC2 in vivo. Without ATRX, PRC2 cannot load onto Xist RNA nor spread in cis along the X chromosome. Moreover, epigenomic profiling reveals that genome-wide targeting of PRC2 depends on ATRX, as loss of ATRX leads to spatial redistribution of PRC2 and derepression of Polycomb responsive genes. Thus, ATRX is a required specificity determinant for PRC2 targeting and function.
INTRODUCTION
In mammals, X chromosome inactivation (XCI) balances X chromosome gene dosages between the two sexes. During random XCI in the peri-implantation embryo, cells count X chromosomes and stochastically choose one X chromosome for inactivation (Dupont and Gribnau, 2013; Lee, 2012; Lee and Bartolomei, 2013; Starmer and Magnuson, 2009; Wutz, 2011). Once silenced, the inactive X chromosome (Xi) is maintained in a repressed state through subsequent cell divisions. XCI is controlled in cis by the “X inactivation center” (Xic), an X-linked region that harbors many noncoding genes, including the sense-antisense pair, Xist and Tsix (Brown et al., 1992; Lee et al., 1999), and the activator, Jpx (Sun et al., 2013; Tian et al., 2010). The 17 kb Xist RNA is transcribed exclusively from the Xi and initiates silencing as it spreads over the X chromosome in cis (Clemson et al., 1996). Xist is regulated negatively by the antisense Tsix RNA (Lee et al., 1999) and positively by Jpx RNA (Sun et al., 2013; Tian et al., 2010). Xist is also positively controlled by the 1.6 kb internal transcript, RepA, which shares the highly structured “Repeat A” motif with Xist RNA (Hoki et al., 2009; Maenner et al., 2010; Zhao et al., 2008). The outward spread of Xist RNA through the Xi leads to recruitment of silencing factors that, in turn, establish and maintain the repressed state (Dupont and Gribnau, 2013; Lee and Bartolomei, 2013).
A key recruited factor is Polycomb repressive complex 2 (PRC2) (Dupont and Gribnau, 2013; Lee, 2012; Lee and Bartolomei, 2013; Starmer and Magnuson, 2009; Wutz, 2011), the his-tone methyltransferase complex that trimethylates histone H3 at lysine 27 (H3K27me3) and establishes repressive chromatin (Müller and Verrijzer, 2009; Simon and Kingston, 2013).
Because PRC2 controls both normal development and the pathogenesis of disease, PRC2 has become a high-priority drug target (Helin and Dhanak, 2013). Apart from the Xi, PRC2 binds thousands of specific sites in the mammalian genome. Still not fully understood is how PRC2 is targeted when the core sub-units are not sequence-specific DNA-binding proteins. PRC2 preferentially occupies CpG-rich regions and is aided in recruitment by substoichiometric association with the Jumonji protein, JARID2, and the Zinc-finger protein, AEPB2 (Cifuentes-Rojas et al., 2014; da Rocha et al., 2014; Kaneko et al., 2014; Simon and Kingston, 2013). However, other specificity determinants must exist in vivo, given that JARID2 and AEPB2 are nonspecific DNA-binding proteins and cannot by themselves impart specificity to PRC2 localization.
The example of RepA/Xist RNA demonstrates that cis-regulatory RNAs can serve as locus-specific recruiting factors (Zhao et al., 2008). Because such transcripts are unique in the genome, their cis-action enables targeting of chromatin complexes to a singular location (Lee, 2012). During XCI, PRC2 is first targeted to the Xic by RepA via the Repeat A motif. Xist RNA then cotranscriptionally binds PRC2 via Repeat A and loads in cis onto a nucleation center before spreading outwardly to envelop the Xi (Jeon and Lee, 2011). PRC2 is now known to have a large RNA interactome, with membership exceeding 9,000 transcripts (Kaneko et al., 2013; Kanhere et al., 2010; Khalil et al., 2009; Zhao et al., 2010). In vitro, PRC2 can bind RNA with a range of affinities (Cifuentes-Rojas et al., 2014; Davidovich et al., 2013). The large RNA interactome raises the question of how PRC2 discriminates between RNA species in the physiological context. Here, we investigate this question by carrying out an unbiased screen for novel specificity determinants. We identify the chromatin remodeler, ATRX.
RESULTS
A Proteomics Screen Identifies ATRX as a Candidate XCI Regulator
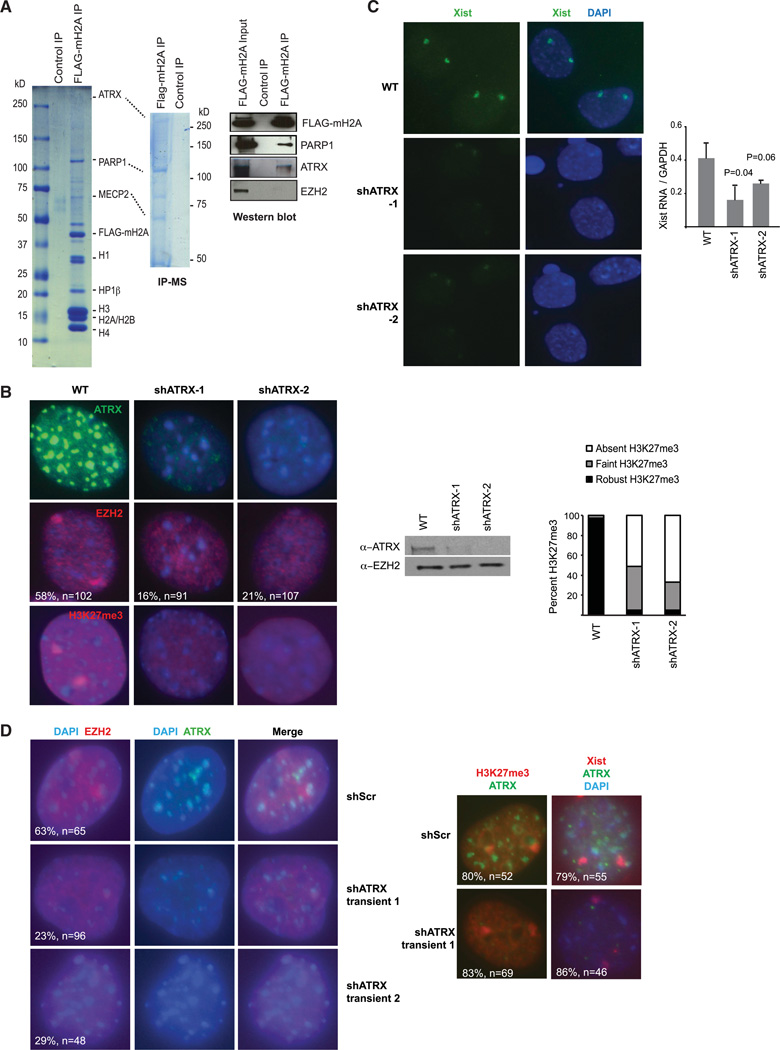
Because the macroH2A (mH2A) histone variant is enriched within gene-dense bands of the Xi together with Xist RNA and PRC2 (Chadwick and Willard, 2004; Costanzi and Pehrson, 1998), we performed an unbiased proteomics screen using mH2A as bait in an affinity purification. We expressed FLAG-tagged mH2A in 293, a human fibroblast cell line, and carried out FLAG immunoprecipitation followed by mass spectrometry (IP-MS). We observed several known interactors of mH2A, including PARP1 and linker histone H1 (Figure 1A, left and middle, and Figure S1A available online) (Nusinow et al., 2007), as well as MECP2 and HP1. In addition, IP-MS revealed the 280-kD ATRX protein (Figure 1A and S1B). We validated all interacting proteins by western blot after FLAG-mH2A IP (Figure 1A, right, and data not shown).
Figure 1. A Proteomics Screen Identifies ATRX as a Candidate XCI Regulator.
(A) IP-MS: Colloidal blue staining of FLAG IP from control (293F) and FLAG-mH2A-expressing 293 run on a 4%–20% (left) and a 6% (right) SDS gradient gel. FLAG IP was validated by western blot.
(B) Left: Immunostaining of ATRX, EZH2, and H3K27me3 in WT and two independent stable ATRX-KD MEF lines (shATRX-1, -2). Sample size (n) and %EZH2 association with Xi are shown.
Middle: western blot showing ATRX depletion but constant EZH2 levels in shATRX-1 and shATRTX-2 female MEFs.
Right: Patterns of H3K27me3 observed, n = 100–150 per experiment.
(C) Left: Xist RNA FISH in indicated fibroblast lines. Right: qRT-PCR analyses of Xist RNA levels. Standard error (SE) bars from three independent experiments are shown with Student’s t test P values.
(D) Left: Immunostaining of ATRX and EZH2 in MEFs transiently transfected with scrambled shRNA (shScr) and two shATRX constructs (shATRX-1, shATRX-2).
Right: H3K27me3 staining and Xist RNA FISH show no change in the intensity or foci number after transient ATRX KD.
ATRX caught our attention, as it was shown to be enriched on the Xi by immunofluorescence (Baumann and De La Fuente, 2009) and has an ATPase and helicase domain (Clynes et al., 2013; Ratnakumar and Bernstein, 2013). ATRX is known as an unusual SNF2-like member of the SWI/SNF family of chromatin remodelers, as it appears to have only weak remodeling activity and does not affect nucleosome phasing in vitro. It does, however, have ATPase activity that is mildly stimulated by naked DNA and mononucleosomes (Tang et al., 2004; Xue et al., 2003), and a translocase activity that displaces a third strand of a DNA triplex (Mitson et al.,2011; Xue et al., 2003). The atypical N-terminal PHD finger domain of ATRX (a.k.a., ATRX-Dnmt3-Dnmt3L [ADD] domain) binds to unmodified histone H3-lysine 4 and to di- or tri-methylated H3-lysine 9 (Dhayalan et al., 2011; Eustermann et al., 2011; Iwase et al., 2011), in keeping with ATRX’s role in maintaining pericentric heterochromatin and telomeres (Goldberg et al., 2010; Lewis et al., 2010). Mutations in ATRX cause X-linked mental retardation and alpha-thalassemia in humans, with ~80% of mutations mapping to the PHD finger and helicase domains (Gibbons et al., 2008). Although ATRX appears enriched on the Xi and plays a role in peri-implantation development (Garrick et al., 2006), whether ATRX plays a role in XCI is unknown.
To investigate, we generated clones of mouse embryonic fibroblasts (MEF) with stable ATRX knockdown (KD) (shATRX-1, −2) and >90% depletion of ATRX protein (Figure 1B). We examined localization of EZH2, the PRC2 subunit that catalyzes the trimethylation of histone H3 at lysine-27 (H3K27me3). Interestingly, immunofluorescence showed that, while ~60% of WT cells showed prominent EZH2 foci on the Xi, only 16%-20% of ATRX KD cells retained EZH2 (Figure 1B). Moreover, H3K27me3 on the X chromosome was either absent or markedly reduced in shATRX cells. At the same time, RNA fluorescent in situ hybridization (FISH) showed that Xist RNA localized poorly, with quantitative RT-PCR showing that total Xist RNA levels were reduced 40%–60% (Figure 1C). Transient ATRX KDs in MEFs (48 hr) showed that EZH2 localization was similarly compromised (Figure 1D). In transient knockdowns, there was no obvious disturbance to Xist localization or H3K27me3, consistent with the short-term stability of Xist RNA and H3K27me3. Similar results were obtained using two independent ATRX shRNAs, whereas scrambled shRNA controls (Scr) resulted in no changes to Xist, EZH2, or H3K27me3 localization. Xist levels and localization were affected only in the long term, consistent with the notion that H3K27me3 at the Xist promoter targeted by RepA facilitates Xist upregulation (Sun et al., 2006; Zhao et al., 2008) and that the H3K27me3 mark is depleted only after several rounds of DNA replication. These findings hinted at a role of ATRX in recruiting and spreading PRC2 to form a “cloud” on the Xi.
Xist-Mediated Recruitment of PRC2 Depends on ATRX
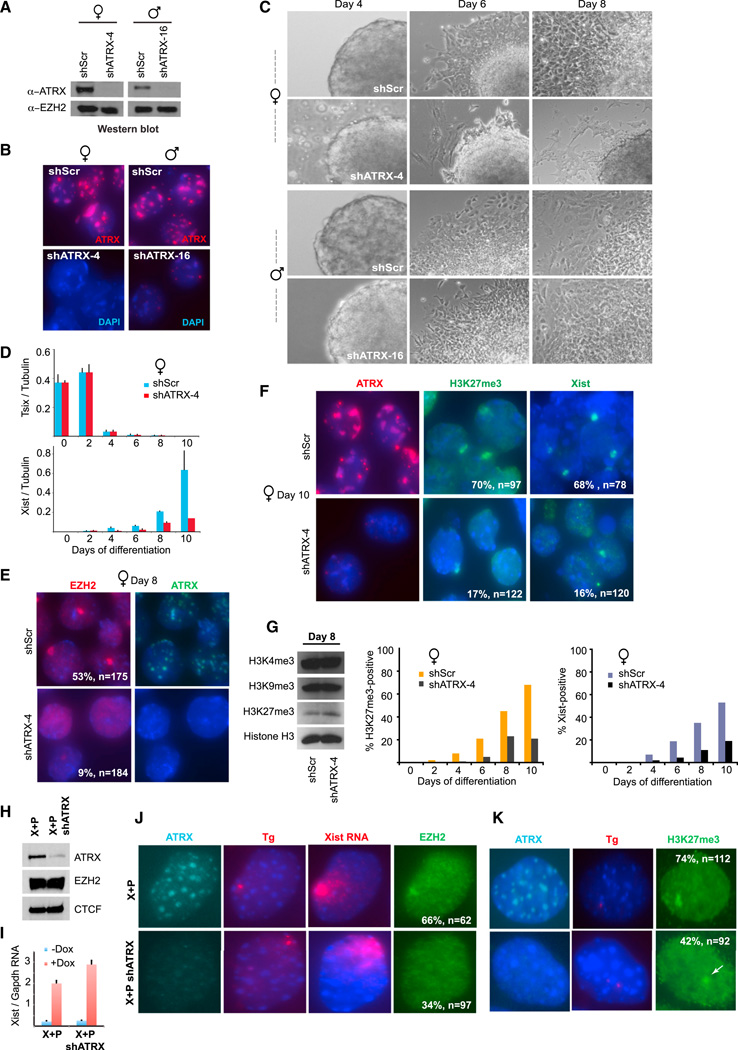
To examine whether there were sex-specific phenotypes of ATRX deficiency, we used a mouse embryonic stem cell (ESC) model to recapitulate random XCI during ex vivo cell differentiation. We generated male and female ESC clones with stable ATRX KD (Figures 2A and 2B; representative clones shown) and compared their behavior to shScr control clones in time course analyses from differentiation days 0–10. While no differences were evident from days 0–4, sex-specific differences emerged between days 4–8 (Figures 2C and 2D). shATRX female cells showed defective embryoid body (EB) outgrowth, whereas shScr control and male shATRX clones grew robustly (Figures 2C). Immunostaining showed appropriate downregulation of pluripotency markers, such as NANOG, between days 0–8 in all clones (Figure S2A and S2B), indicating that the defect was not caused by failed entry into the differentiation pathway per se. The Xist regulator, Tsix, was also appropriately downregulated in shATRX and shScr female ESC between days 0–10 (Figure 2D). These results indicated a developmental defect downstream of the differentiation signal. In spite of normal Tsix downregulation, however, Xist RNA was not appropriately upregulated in shATRX cells. Furthermore, EZH2 targeting was severely compromised, as only 9% of shATRX cells (n = 184) showed EZH2 enrichment (Figure 2E) and only 17% of cells showed H3K27me3 enrichment (Figures 2F and 2G) on the Xi, even though H3K27me3 levels were unperturbed globally (Figure 2G). Therefore, ATRX depletion resulted in female-specific outgrowth defects related to an inability to initiate XCI.
Figure 2. ATRX Is Required for Initiation of XCI during ES Cell Differentiation.
(A) Western blot of ATRX and EZH2 after stable ATRX KD.
(B) ATRX immunostaining after stable ATRX-KD.
(C) Representative phase contrast images of embryoid bodies during a differentiation time course.
(D) qRT-PCR analysis of Tsix and Xist RNA, normalized to tubulin RNA, during a differentiation time course. SE from three independent experiments shown.
(E) Immunostaining of EZH2 and ATRX at day 8 of differentiation, (n) and % with EZH2 Xi foci are indicated.
(F) Immunostaining of ATRX and H3K27me3 and Xist RNA FISH at day 10 of differentiation, (n) and % positive are shown.
(G) Left: western blot of indicated histone marks in day 8 ESCs. Graphs: Time course of Xist upregulation and acquisition of H3K27me3foci. n = 80–120 per time point.
(H) Western blot for ATRX, EZH2, and CTCF (control) in transgene (X+P) cells and in the same cell line expressing shATRX (X+P shATRX).
(I) qRT-PCR of Xist RNA before and after doxycycline induction in transgenic cells. SE from three independent experiments shown.
(J,K) Immunostaining of ATRX, EZH2 (J), and H3K27me3 (K), and Xist RNA/DNA FISH in indicated cells lines after 24 hr dox induction. % with shown pattern and sample size (n) are indicated.
To probe further, we circumvented a dependency of Xist expression on ATRX and used an inducible transgenic system in which Xist RNA is upregulated by addition of doxycycline (dox) (Jeon and Lee, 2011). In this “X+P” system, Xist is carried on an autosomal transgene in male cells and is upregulated >20-fold upon induction, accompanied by recruitment of PRC2 along the autosome in cis. In stable shATRX X+P clones (Figure 2H; representative clone #4 shown), ATRX KD did not affect total cellular EZH2 levels (Figure 2H) and dox-induction led to an appropriate increase of Xist expression (Figure 2I), indicating successful bypass of shATRX’s effect on Xist expression. Immuno-RNA FISH showed that EZH2 and H3K27me3 enrichment were observed on the Xist-coated chromosome in 66% and 74% of ATRX+ transgenic cells, respectively, confirming the sufficiency of ectopic Xist expression to recruit and spread EZH2 in cis (Figures 2J and 2K). However, on ATRX KD, EZH2 and H3K27me3 recruitment decreased to 34% and 42%, respectively. Thus, ATRX is required for Xist RNA-mediated recruitment of PRC2.
ATRX Is a High-Affinity RNA-Binding Protein that Loads PRC2 onto Xist RNA
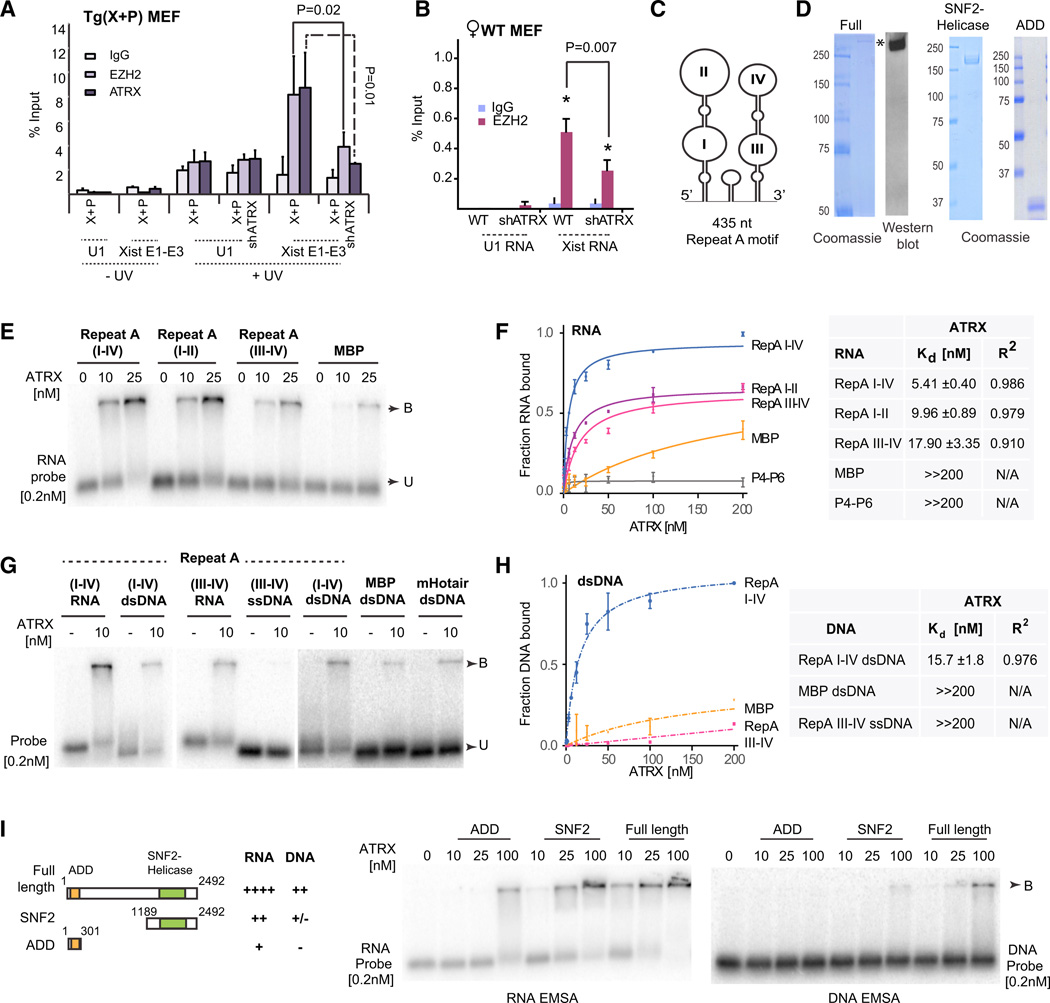
To ask if Xist and ATRX directly interact in vivo, we performed UV-crosslink RNA immunoprecipitation (UV-RIP; Figure 3A). As expected, in ATRX+ transgenic system (X+P), EZH2 pulled down Xist RNA under UV crosslinking conditions, consistent with a direct interaction between EZH2 and Xist RNA (Zhao et al., 2008). Intriguingly, ATRX also pulled down Xist RNA. This association was detectable only when cells were subjected to UV-crosslinking, suggesting that Xist is directly bound to ATRX. Neither EZH2 nor ATRX pulled down the nonspecific RNA control, U1 RNA. Significantly, when ATRX was knocked down (X+P shATRX), the ability of EZH2 to interact with Xist RNA was compromised (p = 0.02; Figure 3A). Similarly, EZH2-Xist RNA interactions were disrupted in wild-type (WT) female MEF cells when ATRX was depleted (p = 0.007; Figure 3B). These data demonstrate that ATRX-Xist interaction facilitates binding of PRC2 to Xist RNA in vivo.
Figure 3. ATRX Binds RNA and Stimulates RNA Binding to PRC2.
(A,B) RIP ± UV crosslinking in X+P transgenic MEFs after dox-induction for 24 hr (A) or WTMEF(B) Primers spanning Xist exons 1–3 and U1 snRNAs were used for qPCR. One percent input was processed in parallel. SE from three independent experiments. P values calculated using Student’s t test.
(C) One possible structure of Repeat A (Maenner et al., 2010).
(D) Coomassie stain of purified full-length FLAG-ATRX-HA (left), C-terminal SNF2/helicase (middle), and N-terminal ADD (right) domains.
(E) RNA EMSA with ATRX at indicated concentrations and 0.2 nM of each probe. B, bound; U, unbound probe.
(F) Left: Binding Isotherms of ATRX for indicated RNAs. SE from three independent experiments shown. Right: table summarizing Kd’s and R2 values. ≫200 nM indicates Kd’s out of the assay range. N/A, not applicable.
(G) EMSA of ATRX with different RNA, dsDNA, or ssDNA probes.
(H) Left: Binding Isotherms of ATRX for indicated DNAs. SE for three independent experiments shown for each point. Right: table summarizing Kd’s and R2 values. ≫200 nM indicates Kd’s out of the assay range. N/A, not applicable.
(I) RNA and DNA EMSAs using ATRX truncation mutants, with summary of results.
To understand mechanism, we characterized ATRX-RNA interactions in vitro. PRC2 binds RepA/Xist via Repeat A (Zhao et al., 2008), a 435 nt motif consisting of 8.5 repeats of a 28-nucleotide sequence, with the potential to form several structures, including two long stem-loop structures, I-II and III-IV (Figure 3C)(Duszczyk et al., 2011; Maenner et al., 2010; Wutz et al., 2002; Zhao et al., 2008). To ask how ATRX facilitates PRC2 interaction with Repeat A, we first performed RNA electromobility shift assays (EMSA) using purified full-length ATRX (Figure 3D, left, S3B). ATRX robustly shifted Repeat A in a concentration-dependent manner (Figure 3E), with ~100% probe binding at <100 nM ATRX protein (Figure S3C). To determine which region of Repeat A is sufficient for ATRX binding, we split Repeat A into I-II versus III-IV (Figure S3A) and found that binding still occurred, albeit at lower levels. We calculated dissociation constants (Kd) from binding isotherms plotted from densitometric analysis of double-filter binding assays (Figure S3D). Binding curves were fitted using a nonlinear regression model, with high R2 values showing excellent fit of data points to the curve (Figure 3F). ATRX strongly bound Repeat A I-IV, with a Kd of 5.41 ± 0.40 nM. Subfragments I-II and III-IV retained binding with Kd’s of 9.96 ± 0.89 nM and 17.90 ± 3.35 nM, respectively. To test the specificity of ATRX-RNA interactions, we used the first 300 nt of the Maltose Binding Protein (MBP) mRNA of Escherichia coli and the P4-P6 ribozyme of Tetrahymena, two noncognate RNAs. Neither bound ATRX to an appreciable extent (Kd ≫200 nM). We conclude that ATRX has high affinity and specificity for Repeat A RNA.
Interestingly, ATRX could also bind Repeat A double-stranded DNA (dsDNA; Kd = 15.7)(Figure 3G and 3H). However, it could not bind well to the single-stranded DNA (ssDNA), nor MBP and mouse Hotair dsDNAs (Figures 3G and 3H; Kd ≫200 nM). Therefore, ATRX has high affinity for both Repeat A RNA and dsDNA, and belongs to an emerging class of “bivalent” chromatin factors capable of binding both RNA and DNA [e.g., CTCF (Sun et al., 2013) and YY1 (Jeon and Lee, 2011)]. Competition analysis showed that 10–1,000× molar excess of cold Repeat A dsDNA could not titrate away ATRX from Repeat A I-IV RNA probe (Figure S3E) and, reciprocally, cold Repeat A I-IV RNA could not titrate ATRX away from the dsDNA probe (Figure S3F), suggesting that ATRX interacts with RNA and dsDNA via distinct domains. To delineate binding domains, we generated subfragments of ATRX (Figure 3D and 3I). The helicase domain could bind Repeat A RNA at 10–25 nM, but not so well as full-length ATRX. On the other hand, it bound dsDNA poorly, if at all (Figure 3I). Gel shifts with the ADD domain showed binding to RNA at 100 nM, but not to DNA at the same concentration.
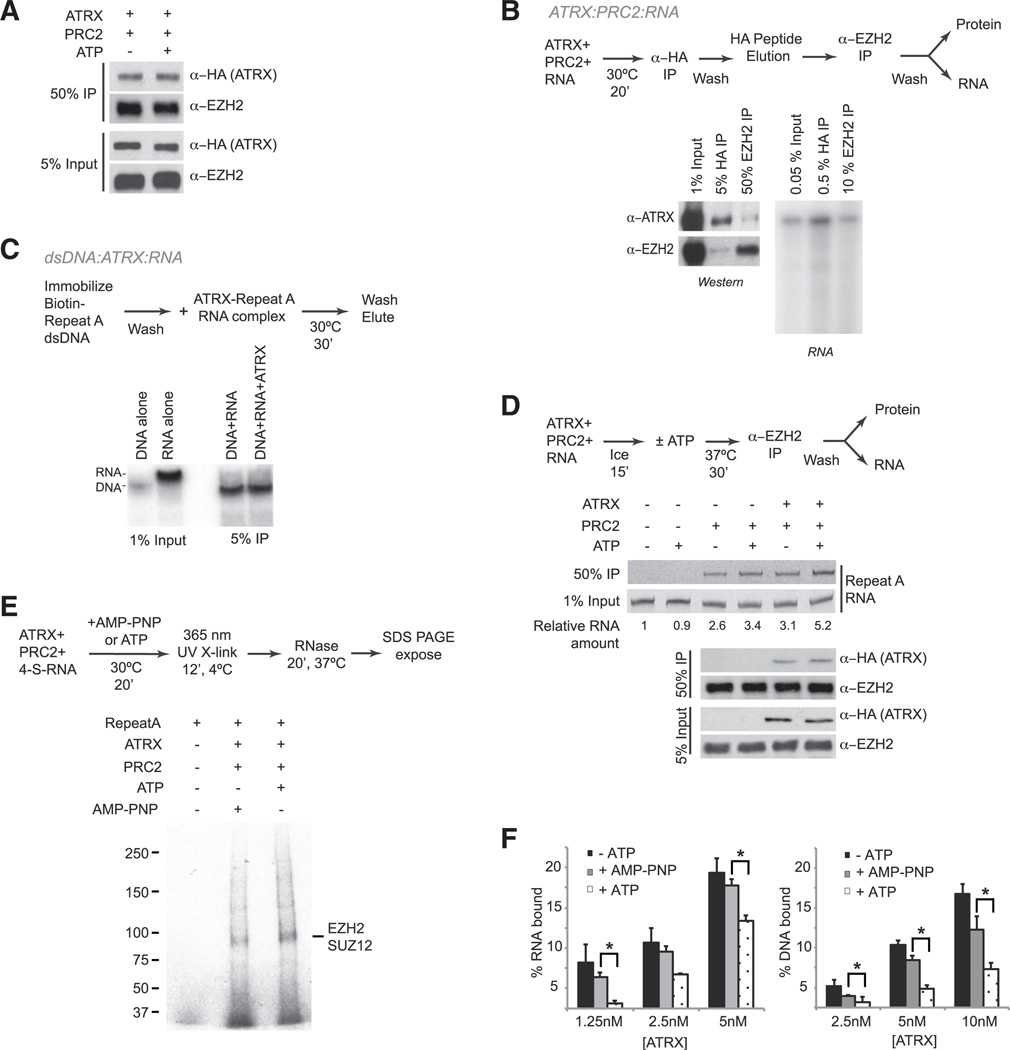
Because ATRX and PRC2 both bind Repeat A RNA, we asked whether ATRX and PRC2 could make direct contact to create an ATRX-RNA-PRC2 ternary complex. Using purified proteins, we observed that they do (Figure 4A). This finding is consistent with a yeast two-hybrid screen in which EZH2 was identified as a directly interacting partner for ATRX (Cardoso et al., 1998), but which has not been verified by in vivo analyses. Notably, PRC2 components were not identified by our IP-MS analysis, though ATRX was a clear interacting partner (Figure 1A). This apparent discrepancy may be due to substoichiometric ATRX-PRC2 associations in vivo and to mH2A being used as a bait in the proteomics screen. To confirm the ternary complex in vitro, we performed tandem IP, first pulling down ATRX via the HA tag, then immunoprecipitating PRC2 with an EZH2 antibody. Indeed, we observed end-labeled Repeat A RNA in the tandem IP (Figure 4B), indicating that ATRX, PRC2, and Repeat A RNA formed a ternary complex.
Figure 4. A Ternary Complex of ATRX, PRC2, and RNA, with Stimulation of PRC2 Binding in an ATP-Dependent Manner.
(A) FLAG-ATRX-HA binding to PRC2 ± ATP in the absence of RNA. HA-immunoprecipitated material was probed with α-EZH2 antibodies for western.
(B) Tandem IP to detect ternary complex formation between ATRX, Repeat A RNA, and PRC2.
(C) Binding reaction to test for simultaneous association of ATRX with RNA and DNA. RNA and DNA can be discerned by their different electrophoretic mobilities.
(D) Top: Inverted image of RNA gel showing Repeat A RNA recovered after EZH2 IP ± ATRX and ± ATP. Bottom: western blot showing protein levels in input and recovered after IP. Representative results from three independent experiments shown.
(E) Photocrosslinking of PRC2 and Repeat A RNA in the presence of ATRX with ATP or AMP-PNP. Representative results from 5 biological replicates shown.
(F) Filter binding of ATRX binding to Repeat A RNA (left) or DNA (right) ± ATP and +AMP-PNP. ATRX concentrations are indicated and SE from three independent experiments shown. *p < 0.05.
Given pairwise interactions between ATRX-RNA and ATRX-DNA (Figures 3E-3I), we also explored the possibility of an ATRX-RNA-DNA ternary complex. We immobilized biotinylated Repeat A dsDNA on streptavidin beads and tested its ability to bind the ATRX-Repeat A RNA complex. We reasoned that, if the ATRX-RNA complex could engage dsDNA, RNA would be visualized after elution from the column. Under these conditions, RNA was minimally present, if at all, in the eluate (Figure 4C). Thus, if an RNA:ATRX:dsDNA ternary complex could form, it may be too transient to detect in vitro. We conclude that ATRX, PRC2, and Repeat A RNA forms a stable ternary complex, but that ATRX likely does not form a stable ternary complex with RNA and dsDNA in vitro.
With ATRX possessing an ATPase domain, we examined how interaction dynamics might be changed by ATP. We assessed PRC2-RNA binding in the presence or absence of ATRX and/or ATP by immunoprecipitating EZH2 to quantitate the amount of RNA bound (Figure 4D). Interestingly, ATRX’s stimulatory effect was modestly increased (2-fold) in the presence of ATP, raising the possibility that ATRX may act by remodeling Repeat A to a configuration that promotes PRC2 binding. To test this idea, we carried out an RNase protection assay with photocrosslinking. We incubated PRC2 with thiolated radiolabeled Repeat A RNA in the presence of ATRX and ATP or the nonhydrolysable analog, AMP-PNP. The resulting complexes were subjected to UV crosslinking and treated with RNase A, T1, and V1. In principle, protein-bound radiolabeled RNA would be protected from digestion and detected in an SDS-PAGE autoradiograph. In five biological replicates, the presence of ATRX and ATP consistently resulted in greater protection of Repeat A RNA from RNases, as visualized by the increased RNA signal bound to EZH2/SUZ12 (Figure 4E), two known RNA-binding subunits of PRC2 (Cifuentes-Rojas et al., 2014). Together, these experiments show that ATRX stimulates binding of PRC2 to Repeat A RNA in a manner that is enhanced by ATP.
Intriguingly, ATRX bound RNA and DNA less well in the presence of ATP (Figure 4F). However, when AMP-PNP was used, binding was not affected. This observation suggests that the nucleic acids are released by ATRX upon ATP hydrolysis. That is, ATRX may release RNA (and DNA) once the nucleic acid is reconfigured for PRC2 binding. The transient nature of the interactions may explain why a dsDNA-ATRX-RNA ternary complex was difficult to detect (Figure 4C). The ATRX:RNA:PRC2 ternary complex may be easier to detect (Figure 4B) because it is held in place by both PRC2-RNA and PRC2-ATRX interactions. Combined, our data demonstrate that ATRX directly promotes binding of PRC2 to RepA/Xist RNA in vivo and in vitro.
A Hot Spot of ATRX Localization at the X/c
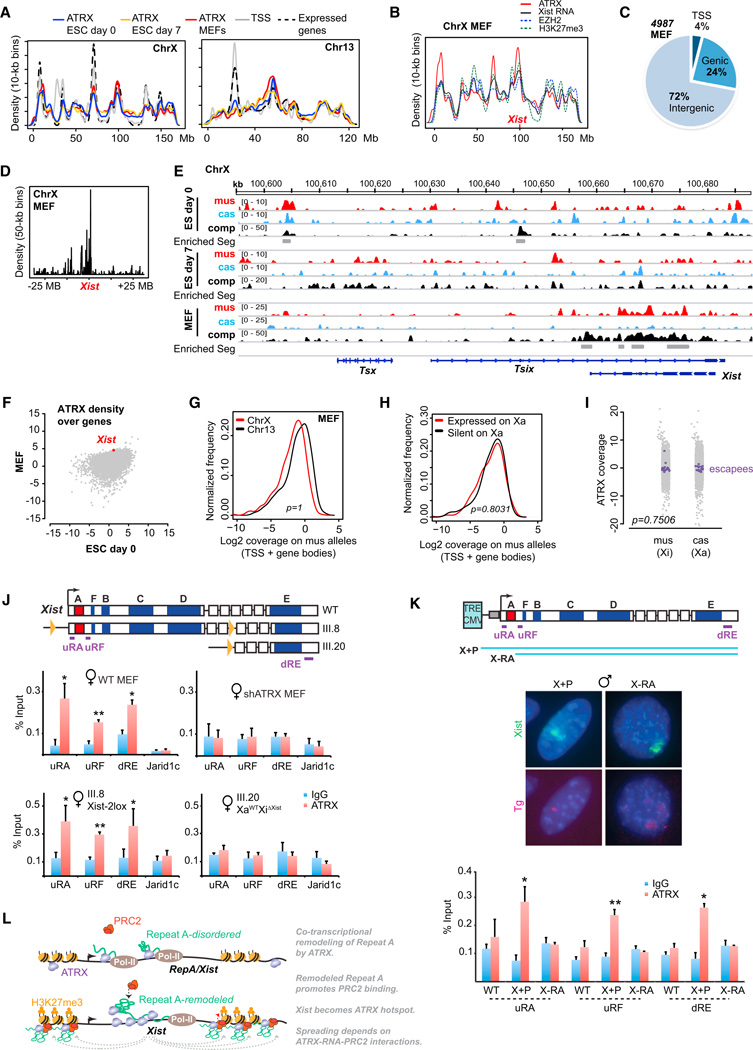
We investigated genomic binding patterns for ATRX by performing chromatin immunoprecipitation with deep sequencing (ChIP-seq). To distinguish between Xa and Xi, we used genetically marked hybrid cell lines in which the Xi is invariably of Mus musculus (mus) origin and the Xa of Mus castaneus (cas) origin (Ogawa et al., 2008; Pinter et al., 2012). Analysis of two biological replicates revealed that, in MEFs and ESC, ATRX was broadly distributed along all chromosomes (Chr)—including ChrX and a representative autosome, Chr13—with preferential enrichment in gene-dense regions (Figure 5A). Allele-specific analysis of ChIP-seq coverage density plots for ATRX, EZH2, and H3K27me3 (Pinter et al., 2012) and CHART-seq analysis of Xist RNA (Simon et al., 2013) revealed that the four epitopes were concentrated in the same regions of the Xi in female MEFs (Figure 5B), supporting the idea that ATRX, PRC2, and Xist RNA are functionally linked. Strong ATRX binding occurred at ~4,987 sites, with 4% in promoter regions (transcription start site [TSS] ± 3 kb), 24% in coding gene bodies, and the rest in intergenic space (including enhancers, unannotated Inc RNA transcription units) (Figure 5C). This distribution is consistent with previous ChIP-seq analysis in male ESCs (Law et al., 2010).
Figure 5. An ATRX Hot Spot at the Xist Locus Requires Function of Repeat A.
(A) Chromosome-wide profiles of ATRX coverage on Xi and Chr13 show input-normalized ATRX density over 10 Kb bins, gene (TSS) density, and the density of transcriptionally active genes based on previous data (Yang et al., 2010).
(B) Allele-specific analysis: Normalized densities (10-kb bins, smoothened) of Xist RNA, EZH2, H3K27me3 (Pinter et al., 2012; Simon et al., 2013), and ATRX in female MEFs along X.
(C) ATRX peak distribution in MEFs across promoter (TSS+/−3 kb), genie, and intergenic regions.
(D) ATRX densities at 50-kb resolution across 50 Mb of the Xic, centered on Xist.
(E) Allele-specific ATRX ChIP-seq tracks showing M. musculus (mus), M. castaneus (cas), and composite (comp) reads across the Xist locus in day 0 and 7 ESC and MEFs. Comp track includes all aligned reads, both allelic and nonallelic. Positions of peaks/enriched segments (gray bars) are shown under comp tracks. Representative autosomal tracks (Chr13) shown in Figure S4C.
(F) ATRX genie densities for all genes in MEFs (y axis) and ESC (x axis).
(G) ATRX coverages on gene bodies of mus alleles for ChrX and Chr13 in female MEFs. KS test, p = 1.
(H) Log2 ATRX coverage densities for Xi genes classified as “expressed on Xa” (FPKM ≥ 1) or “silent on Xa” (FPKM < 1). KS test, p = 0.80
(I) ATRX coverage on XCI escapees (purple) on mus or cas alleles. Grey, all other ChrX genes. P value was computed between mus and cas escapees using KS test.
(J) Map of Xist alleles in WT, XaWT Xi2lox (III.8), and the conditional Xist deletion XaWT XiΔXist (III.20) MEF lines, with ChIP-qPCR amplicon positions and ChIP-qPCR results (graphs) for ATRX. Averages of three independent ATRX ChIP expressed as % of input, with SE shown. *p < 0.05. **p < 0.005 (Student’s t test).
(K) RNA/DNA FISH for Xist RNA and transgenic DNA in the transgenic X+P and X-RA male MEF lines after 24h dox-induction. Map of transgenes shown on top. Bottom graph: ChIP-pPCR of corresponding WT and transgenic MEF lines 24h after dox-induction of Xist. Averages of three independent ChIP-qPCR experiments shown with SE. *p < 0.05. **p < 0.005 (Student’s t test).
(L) Model: RepA/Xist RNAs cotranscriptionally recruit ATRX. ATRX may be “poised” to bind RNA via Repeat A dsDNA. ATRX remodels Repeat A RNA motif and enhances binding of PRC2. The Xist locus becomes an ATRX hot spot. Spreading depends on the ATRX-RNA-PRC2 interactions.
Among thousands of ATRX sites on ChrX, one locus stood out (Figure 5D-5E, S4A, and S4B). Xist was among the top genic hits in female MEFs (Figure 5F) and was covered by ATRX along its entire length, contrasting with the restricted peaks characteristic of other loci (Figure S4C). Allele-specific ChIP-seq of shATRX MEFs showed loss of enriched signals at Xist (Figure S4D), confirming the specificity of the ATRX ChIP-seq analysis. ATRX enrichment at Xist was developmentally specific, being absent in pre-XCI ESC and present in post-XCI MEF. Furthermore, enrichment was observed only on the Xi (mus, Figure 5E). However, ATRX coverage across Xi alleles was not enriched relative to the genomic average (Figure 5G, representative Chr13 shown; KS test, p = 1). Nor was ATRX coverage different on average between two classes of Xi genes—those expressed on Xa versus those that are silent (Figure 5H; KS test, p = 0.8031), indicating that ATRX was no more enriched on X-genes that are expressed in MEFs. Furthermore, ATRX coverage in genes that escape XCI was not significantly different between Xa and Xi (Figure 5I, purple dots; KS test, p = 0.75).
ChIP-qPCR confirmed that, in WT female MEFs, ATRX was significantly enriched (relative to IgG pulldowns) within the Xist gene regardless of what positions were queried (Figure 5J). By contrast, Jarid1c—an X-linked gene that escapes XCI—did not show significant ATRX enrichment. Enrichment at all Xist positions, including the nucleation center (uRF) was lost upon ATRX KD (Figure 5J). ChIP-qPCR also verified the allele-specific nature of ATRX localization. The MEF line, XaWT Xi2LOX (clone III.8), carried two loxP sites flanking a 16 kb Xist sequence on the Xi, whereas XaWT Xi ΔXist (clone 111.20) carried a 16 kb conditional deletion of Xist on the Xi (Zhang et al., 2007). ATRX was significantly enriched at Xist only in the XaWT Xi2lox, the cell line bearing an intact Xist gene on the Xi (Figure 5J). Given that ATRX-RNA interactions are mediated by Repeat A RNA (Figures 1, 2, 3, 4), we asked whether the localization of ATRX to the chromatin hot spot depended on Repeat A DNA. In the Xist transgene model, ChIP-qPCR showed that, while deleting Repeat A (X-RA) did not affect spreading of Xist RNA along the chromosome as shown by RNA FISH (Figure 5K, top)(Jeon and Lee, 2011), it abolished ATRX localization to the Xist locus (Figure 5K, bottom). Repeat A is therefore essential for ATRX-Xist chromatin interactions in vivo.
We conclude that the Xi allele of Xist is a hot spot of ATRX binding. These data lead to a model in which ATRX binds the Repeat A motif and reconfigures the RepA/Xist transcript to promote PRC2 binding (Figure 5L), with poising of ATRX at the Repeat A DNA potentially aiding ATRX-RNA interactions. Following nucleation at the Xist hot spot, the ATRX-RNA-PRC2 ternary complex spreads outwardly along the Xi.
ATRX-Dependent Recruitment of PRC2 on a Global Scale
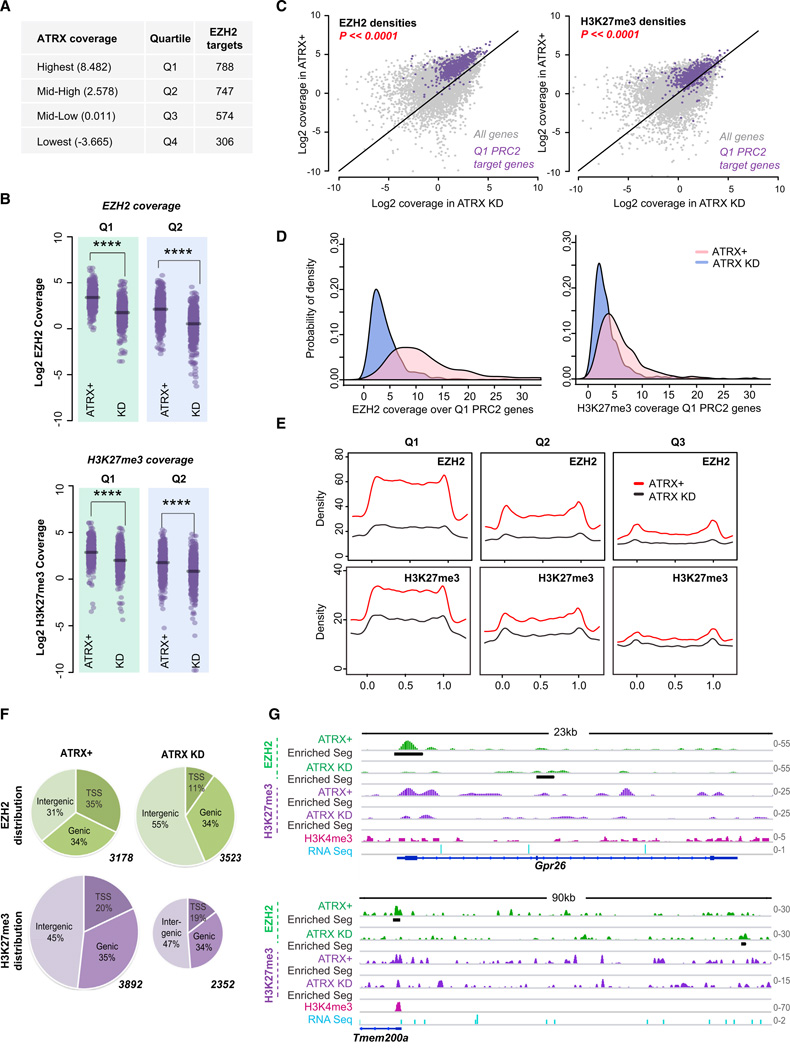
We next investigated whether ATRX’s effect on PRC2 localization extended beyond the Xi. We classified 21,677 annotated unique RefSeq genes into equal quartiles (Q1-Q4) based on ATRX coverage (Figure 6A; Tables S1 and S2), with Q1 having high positive ATRX coverage and Q2 moderate coverage. Interestingly, Q1 and Q2 harbored a large number of PRC2 target genes, together accounting for 63% of all autosomal PRC2 target genes. By contrast, Q3 and Q4 had near-zero or negative ATRX coverage and harbored fewer PRC2 target genes. To determine whether loss of ATRX affected PRC2 recruitment to genes marked by ATRX binding, we compared EZH2 and H3K27me3 coverages in ATRX+ versus ATRX-deficient cells (Figure 6B). Indeed, ATRX KD affected PRC2 localization and function, with the 2- to 4-fold drop in EZH2 and H3K27me3 coverages being highly significant (Figure 6B). Scatterplots showed an obvious shift in EZH2 and H3K27me3 coverage for Q1 PRC2 targets (Figure 6C, purple dots; Table S1). We also examined probability density functions for EZH2 and H3K27me3 to determine the likelihood that a given gene in ATRX-deficient cells will have decreased EZH2 and H3K27me3 coverage based on the population behavior of ATRX-PRC2 target genes (Figure 6D). The distribution for ATRX KD is left and upwardly shifted, indicating that the probability of a target gene in ATRX KD having a low EZH2 (or H3K27me3) coverage is greater than that of the same gene in ATRX+. To examine how EZH2 and H3K27me3 patterns change within gene bodies, we performed metagene analysis (Figure 6E). In ATRX+, EZH2 and H3K27me3 patterns showed characteristic gene-body enrichment. Upon ATRX KD, EZH2 and H3K27me3 densities dramatically decreased in Q1 and Q2 genes, with decrease occurring at TSS’s and within gene bodies. Minimal changes were observed for Q3 and Q4 genes (Figure 6E and data not shown). Analysis of biological replicates showed good reproducibility (Figure S5, data not shown). Collectively, these data demonstrate that ATRX is also required to target PRC2 to autosomal genes.
Figure 6. Global Roles of ATRX in Regulating PRC2 Localization and Function.
(A) RefSeq genes (21,677) divided into equal quartiles (Q1-Q4) based on ATRX coverage. Average ATRX coverages and number of EZH2 target genes (peaks) in each quartile are shown.
(B) Dot plots showing Log2 densities of EZH2 and H3K27me3 in ATRX+ versus ATRX KD MEFs for each quartile. ****p ≪ 0.0001, as calculated by the Student’s t test.
(C) Scatterplot of EZH2 and H3K27me3 densities in Log2 scale overall genes (gray dots) and Q1 PRC2 target genes (purple dots) in ATRX+ or ATRX KD MEFs. The difference between ATRX+ and ATRX KD cells of Q1 PRC2 target genes is highly significant (p < 2.2 × 10−16, Student’s t test) for both epitopes.
(D) Probability density function for EZH2 and H3K27me3 based on coverages over the Q1-PRC2 target genes in the indicated MEF samples.
(E) Metagene profiles of EZH2 (top) and H3K27me3 (bottom) coverage. The metagenes are scaled 0 to 1 from genie start (TSS, x = 0) to end (TTS, x = 1).
(F) Distribution of EZH2 and H3K27me3 peaks in indicated MEF samples. Peaks are categorized as TSS (+/— 3Kb), genie, or intergenic. Total number of peaks called for each data set is indicated next to each chart.
(G) ChIP-seq tracks showing EZH2 and H3K27me3 densities in ATRX+ and ATRX-KD MEFs. Black bars, significantly enriched segments. H3K4me3 (pink) and RNA Seq (blue) tracks are as published (Yang et al., 2010; Yildirim et al., 2012).
Given the loss of PRC2 from genic regions upon ATRX KD, we asked whether PRC2 became ectopically localized elsewhere. Examination of global EZH2 and H3K27me3 ChIP-seq patterns revealed an intriguing shift to intergenic space (Figure 6F). In ATRX+, EZH2 was bound to 3,178 sites, roughly distributed equally among gene bodies, promoters, and intergenic space. On the other hand, in ATRX KD, EZH2 shifted away from promoters and expanded into intergenic space. Thus, ATRX is required to specify PRC2 localization to genes in general.
Curiously, the distribution pattern for H3K27me3 peaks did not change in ATRX-depleted cells, as the percentages occurring in TSS, gene bodies, and intergenic space remained the same (Figure 6F, purple pie charts). At the same time, bulk H3K27me3 levels also did not change on ATRX KD in MEFs and ES cells (Figure 2G and data not shown). In spite of unchanged overall levels of H3K27me3 and genomic ChIP-seq density, there was an apparent loss of H3K27me3 peaks in many regions (Figure 6F). Combined, these observations suggest a significant change in H3K27me3 patterns concurrent with mistargeting of PRC2. At the Polycomb target, Gpr26, loss of ATRX resulted in loss of EZH2 binding to the promoter and a relative reduction in H3K27me3 across the gene body (Figure 6G). At Tmem200a, loss of EZH2 binding at the promoter was accompanied by a new EZH2-binding site ~70 kb upstream in intergenic space. However, in spite of the newly acquired EZH2 site, H3K27me3 densities were not increased in the intergenic region. Altogether, these data demonstrate that loss of ATRX function causes improper PRC2 targeting, shifting bulk PRC2 to ectopic intergenic space where chaotic or unstable PRC2 accumulation results in decreased selectivity of H3K27 methylation. We conclude that ATRX is a specificity determinant for PRC2 localization and function.
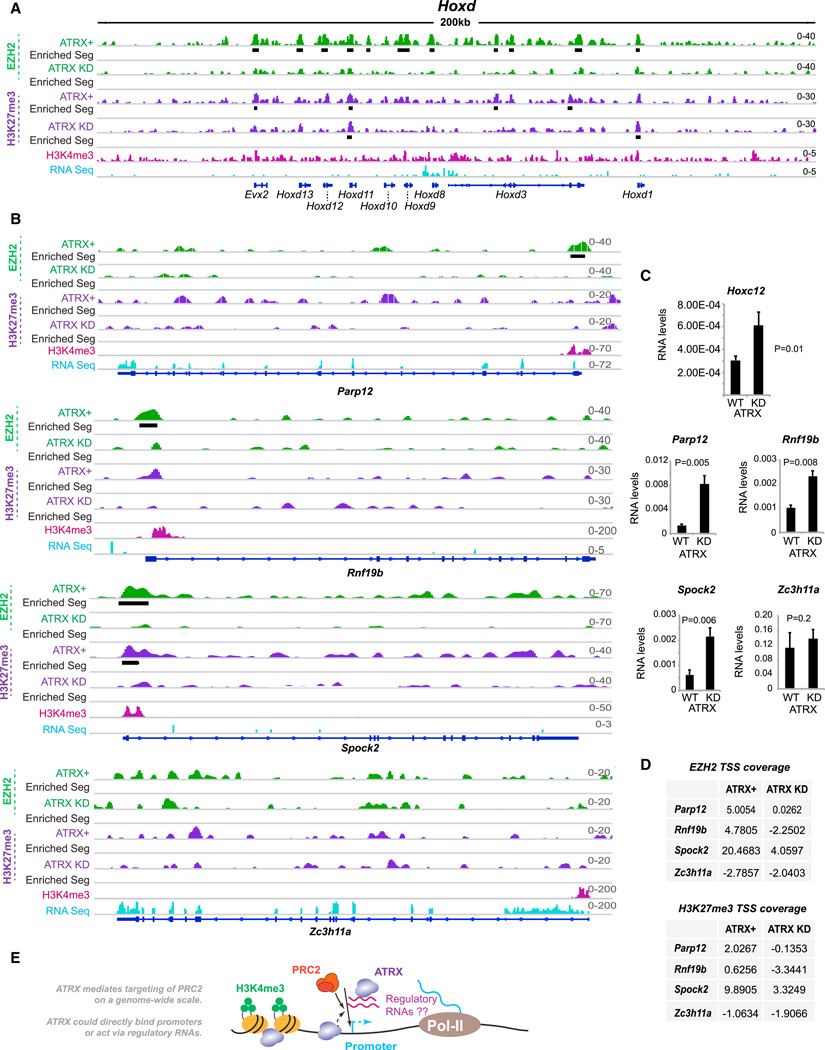
To determine whether failure of PRC2 targeting results in gene expression changes, we tested specific Polycomb targets (Figures 7A, 7B, S6A, and S6B). Within the Hoxc and Hoxd clusters, EZH2 and H3K27me3 were highly enriched in ATRX+ MEFs. Upon ATRX KD, EZH2 and H3K27me3 levels were significantly reduced, as evidenced by both decreased peak numbers as well as overall coverages across the locus. Quantitative RT-PCR analysis revealed concomitant gene upregulation (Figure 7C and Figure S6C). At Parp12, Rnf19b, and Spock2, EZH2 bound the TSS region in ATRX+ cells (Figure 7B). On ATRX KD, EZH2 and H3K27me3 were consistently lost at the promoters (Figure 7B and 7D; biological replicate shown in Figure S6D) and gene expression increased 2- to 5-fold at all loci (Figure 7C). In contrast, expression of Zc3h11a—a control that is not a Polycomb target—did not change significantly after ATRX KD. We conclude that ATRX plays a global role in gene regulation by controlling PRC2 localization.
Figure 7. Loss of EZH2 from Target Genes as a Result of ATRX Knockdown Results in Activation of These Genes.
(A, B) Left panel: ChIP-seq tracks of the Hoxd cluster (A) or individual genes (B) showing EZH2 and H3K27me3 patterns in ATRX+ and ATRX-KD MEFs. Black bars, statistically significant enriched segments. H3K4me3 (pink) and RNA Seq (blue) tracks were previously published (Yang et al., 2010; Yildirim et al., 2012).
(C) qRT-PCR analysis of expression levels before and after ATRX KD. Averages and SE of three independent experiments shown with Student’s t test P values.
(D) EZH2 and H3K27me3 coverages over TSS before and after ATRX KD.
(E) Model: ATRX-dependent targeting of PRC2 on a genome-wide scale. ATRX mediates targeting either by directly binding DNA, or via regulatory RNAs such as those in the PRC2 interactome.
DISCUSSION
Although in vitro analysis shows that PRC2 is intrinsically capable of distinguishing between cognate and nonspecific transcripts (Cifuentes-Rojas et al., 2014), the thousands of possible RNA partners in vivo (Zhao et al., 2010) argue that additional specificity determinants must exist in the physiological context in order for PRC2 to be targeted in a locus-specific manner. Our proteomics screen has identified ATRX as an essential discrimination factor for PRC2 localization and function on a global scale. During XCI, ATRX interacts with RepA/Xist RNA and promotes loading of PRC2 (Figure 5L). ATRX is an avid but also specific RNA-binding protein, with an impressive Kd of ~5 nM for Repeat A RNA. ATRX is also an avid DNA-binding protein (Kd ~15 nM). ATRX’s interaction with Repeat A DNA leaves it poised, enabling ATRX to bind RepA/Xist RNAs in a cotranscriptional manner. Augmentation of ATRX’s effect on Repeat A-PRC2 interactions by ATP suggests a role, potentially as an RNA helicase, to remodel Repeat A RNA to a permissive structure. This activity may parallel that of the MLE helicase for the MSL-roX dosage compensation complex of the fruitfly (Ilik et al., 2013; Maenner et al., 2013). In the case of XCI, absence of ATRX action at both Repeat A RNA and DNA precludes PRC2 targeting and spread along the Xi in cis.
ATRX’s specificity function extends beyond XCI. Without ATRX, PRC2 cannot discriminate true targets from nonspecific sequences. Loss of ATRX causes global shifts in PRC2 localization and function, with PRC2 redistributing en masse to ectopic sites in intergenic space and to noncanonical sites within coding genes. At ectopic loci, PRC2 seems to lack proper context and cofactors to carry out H3K27 methylation in a specific manner. While fewer enriched H3K27me3 peaks are observed, overall H3K27me3 levels are unchanged. Loss of ATRX-dependent PRC2 function results in upregulation of Polycomb target genes (Figure 7 and Figure S6). We do not know whether ATRX action at autosomal genes depends on noncoding transcripts, such as RepA/Xist in the case of XCI. Because ATRX can bind DNA directly (Figure 3H), ATRX-dependent targeting of PRC2 at autosomal loci may be mediated by either RNA or DNA (Figures 7E). The large RNA interactome of PRC2 may also play a role. Finally the presence of EZH2 peaks in Q3 and Q4 genes (Figure 6A) suggests the presence of an ATRX-independent mechanism for PRC2 recruitment as well: for example, at the Q4 Polycomb targets, Onecut1 and KIhI31, loss of ATRX did not affect EZH2 targeting (Figure S7).
ATRX’s role in PRC2 targeting and function may help elucidate the pathogenesis of the X-linked alpha-thalassemia mental retardation (ATR-X) syndrome (Gibbons et al., 2008), a disease associated with aberrant chromatin at telomeres and pericentric heterochromatin (Clynes et al., 2013). Mutations in ATRX and its interacting partner, DAXX, are also frequently mutated in tumors that rely on homologous recombination instead of telomerase activation to circumvent telomere shortening. Our present study suggests that, in addition to abnormalities in constitutive heterochromatin caused by abnormal H3K9 methylation, ATRX-associated diseases may also be ascribed to dysregulated PRC2 function in facultative heterochromatin. ATRX-mediated targeting of PRC2 thus provides a new framework for understanding Polycomb biology and human disease.
EXPERIMENTAL PROCEDURES
Cell Lines
Clonal Xist knockout MEFs, TsixTST/+ ESC, and X+P and X-RA male MEF lines were described previously (Jeon and Lee, 2011; Ogawa et al., 2008; Zhang et al., 2007). Mouse embryonic fibroblasts (MEFs) were immortalized (SV40T) and selected for clonal cell lines with Xi of mus origin. The TsixTST allele ensures exclusive inactivation of the mus X chromosome in ESC.
RNA FISH, DNA FISH and Immunostaining
RNA FISH, DNA FISH, or sequential RNA-DNA FISH and immunostaining were described previously (Zhang et al., 2007).
Immunoprecipitation and Mass Spectrometry
Nuclear extract from 293F FLAG macroH2A cell lines was prepared and dialyzed into BC100 buffer (20 mM Tris CI pH 7.6, 2 mM EDTA, 100 mM KCI, 10% glycerol, and 0.2 mM PMSF), incubated with M2 agarose beads and washed 3X with BC300. Flag peptide eluted complexes were run SDS-PAGE gels and colloidal blue stained. MacroH2A IP specific bands were sent for mass spectrometry. Proteins were digested and extracted peptides (Wilker et al., 2007) loaded on a precolumn and separated by reverse phase HPLC (Agilent) over a 75 min gradient before nanoelectrospray using Orbitrap XL mass spectrometer (Thermo). Raw mass spectral data files were processed as described (Johnson et al., 2012). Mascot peptide identifications were verified manually with the assistance of CAMV (Curran et al., 2013).
qRT-PCR, ChIP, and UV-RNA IP
RNA isolation and RT-PCR were performed according to manufacturer’s protocols (Invitrogen). ChIP and UV crosslinked RNA immunoprecipitation were performed as described previously (Jeon and Lee, 2011). Primers and antibodies are listed in Table S3.
Protein Expression and Purification
Full-length ATRX containing an N term FLAG tag and C term HA tag was cloned into a pfastbac vector (Invitrogen). Sf9 cells were infected with virus for 60 hr and protein was purified using Flag M2 agarose. The helicase domain of ATRX with an N term FLAG was cloned into the pdest10 vector (invitrogen). The ADD domain of ATRX was cloned into pet101 vector and purified via a C term His tag using Ni-NTA resin (QIAGEN) according to manufacturer’s instructions.
RNA and DNA EMSA and Double Filter Binding Assays
EMSAs and filter binding assays were performed as described previously (Cifuentes-Rojas et al., 2014).
In Vitro Protein-RNA Interaction Assays
Two micrograms ATRX, 2 µg PRC2, and 1 µg Repeat A RNA were combined in buffer containing 50 mM Tris-HCI pH 8.0, 100 mM NaCl, 1.5 mM MgCl2, 10 µg/ml BSA, 0.05% NP40, 1 mM DTT, 20U RNaseOUT (Invitrogen), 50 ng/µl yeast tRNA (Ambion Cat # AM7119), and 1 mM ATP in a 20 µl reaction and incubated at 37°C for 30 min. Reactions were added to Protein G coupled α-EZH2 at 4°C for 2h and washed with 1X binding buffer. Beads were split in 2: for RNA and protein analyses. RNA was eluted with for mamide- urea buffer, loaded on a 6% Urea-PAGE and SYBR Gold stained. Proteins were eluted by boiling beads in SDS loading buffer.
For ternary complex IPs, 3µg ATRX, PRC2 and Repeat A RNA were incubated at 30°C in an RNA-EMSA-binding reaction (see above) and precipitated using α-HA beads. Beads were washed with 1X binding buffer and complexes eluted with 1 µg/µl HA peptide. HA eluate was incubated with α-EZH2-protein G dynabeads. After washing, beads were split into two parts: for RNA and protein western blot. RNA was eluted with 0.2 M glycine pH 2.0, neurtralized with Tris.CI pH 8.0 and end labeled in a PNK reaction for visualization.
Two hundred nanograms of 5′ Biotin tagged Repeat A dsDNA was immobilized to Streptavidin Myone dynabeads; 25 nM ATRX and 35 nM RNA were combined in an EMSA reaction at 30°C for 20 min, added to DNA and incubated for 20 min at 30°C. Beads were washed (as above) and nucleic acids were end labeled and resolved on an 18% Urea-PAGE gel.
Photocrosslinking
In vitro transcription reactions were done as previously described (Hernandez et al., 2008) for 16 hr at 37°C with addition of 1 nM 4-Thio-UTP and 32Pα-GTP and 32Pα-CTP at 0.6 µM. Binding reactions were performed as described previously (Cifuentes-Rojas et al., 2014), irradiated for 12 min with 365 nm UV and digested with 0.1 ng RNase A, 1U RNase T1 and 1U RNase V1 for 20 min. SDS loading buffer was added and samples run on a 3%–8% Tris-acetate SDS gradient gel.
ChIP-Seq Read Mapping
ChIP-seq samples were sequenced using Illumina HiSeq2000 resulting 25–50 million 50 bp paired-end reads per sample. Using ASAP tool (http://www.bioinformatics.babraham.ac.uk/projects/ASAP/), reads were aligned allele-specifically to CAST/EiJ and 129S1/SvJm genomes constructed using high quality polymorphisms to the C57/BI6 reference genome (mm9). Twenty-five percent of reads mapped uniquely and allele specifically and 45% equally well to both genomes. Composite coverage tracks were generated using alignment against mm9 with ≤2 alternative mappings allowed. Eighty percent of reads were aligned, with 10%–15% of them discarded due to multiple alternate mappings. Input-normalized read densities were computed using SPP (Kharchenko et al., 2008) after PCR duplicate removal.
Peak Calling
Regions of ATRX enrichment were determined based on tag counts in a 1 kb window sliding with a 200 bp step. Statistical significance of enrichment of ChIP versus input was estimated using negative binomial distribution, with mean based on input tag count in input and size parameter (s) selected based on manual inspection of resulting peak calls. Regions of enrichment were defined by merging adjacent significantly enriched windows separated by ≤ 1 Kb.
Coverage over Genomic Elements
Coverage over TSS-proximal regions, gene bodies, or nonoverlapping genome-wide 1Kb bins was calculated from input-normalized SPP read densities. Actively expressed genes were identified using the cutoff of FPKM ≥ 1.0 based on RNA-Seq data from (Yang et al., 2010). Input-normalized coverage over 1 Kb windows for Xist CHART and EZH2 ChIP-seq was computed based on data from (Pinter et al., 2012; Simon et al., 2013). Significance of change in EZH2 and H3K27me3 densities produced by ATRX KD was assessed by t test in separate quartiles of unique RefSeq genes ranked by WT ATRX density over gene body.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ulandt Kim for next generation sequencing, all lab members for helpful advice, and Drs. David Picketts (Ottawa Hospital Research Institute) and Xiaolu Yang (University of Pennsylvania) for the kind gift of ATRX plasmids. This work was supported by National Institutes of Health Grants F32-GM090765 (to K.S.), F32-GM101828 (to CCR), and RO1-GM090278 (to J.T.L.). J.T.L is an Investigator of the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBERS
The Gene Expression Omnibus accession numbers for the sequencing data reported in this paper is GSE56981.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.10.019.
REFERENCES
- Baumann C, De La Fuente R. ATRX marks the inactive X chromosome (Xi) in somatic cells and during imprinted X chromosome inactivation in trophoblast stem cells. Chromosoma. 2009;118:209–222. doi: 10.1007/s00412-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Cardoso C, Timsit S, Villard L, Khrestchatisky M, Fontés M, Colleaux L. Specific interaction between the XNP/ATR-X gene product and the SET domain of the human EZH2 protein. Hum. Mol. Genet. 1998;7:679–684. doi: 10.1093/hmg/7.4.679. [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl. Acad. Sci. USA. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell. 2014;55:171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes D, Higgs DR, Gibbons RJ. The chromatin remodeller ATRX: a repeat offender in human disease. Trends Biochem. Sci. 2013;38:461–466. doi: 10.1016/j.tibs.2013.06.011. [DOI] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Curran TG, Bryson BD, Reigelhaupt M, Johnson H, White FM. Computer aided manual validation of mass spectrometry-based proteomic data. Methods. 2013;61:219–226. doi: 10.1016/j.ymeth.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, et al. Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Mol. Cell. 2014;53:301–316. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A, Tamas R, Bock I, Tattermusch A, Dimitrova E, Kudithipudi S, Ragozin S, Jeltsch A. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum. Mol. Genet. 2011;20:2195–2203. doi: 10.1093/hmg/ddr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C, Gribnau J. Different flavors of X-chromosome inactivation in mammals. Curr. Opin. Cell Biol. 2013;25:314–321. doi: 10.1016/j.ceb.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Duszczyk MM, Wutz A, Rybin V, Sattler M. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA. 2011;17:1973–1982. doi: 10.1261/rna.2747411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, Jelinska C, Garrick D, Clynes D, Gibbons RJ, Rhodes D, et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 2011;18:777–782. doi: 10.1038/nsmb.2070. [DOI] [PubMed] [Google Scholar]
- Garrick D, Sharpe JA, Arkell R, Dobbie L, Smith AJ, Wood WG, Higgs DR, Gibbons RJ. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2006;2:e58. doi: 10.1371/journal.pgen.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, Wada T, Fisher CA, Malik N, Mitson MJ, Steensma DP, Fryer A, Goudie DR, Krantz ID, Traeger-Synodinos J. Mutations in the chromatin-associated protein ATRX. Hum. Mutat. 2008;29:796–802. doi: 10.1002/humu.20734. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K, Dhanak D. Chromatin proteins and modifications as drug targets. Nature. 2013;502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- Hernandez A, Panigrahi A, Cifuentes-Rojas C, Sacharidou A, Stuart K, Cruz-Reyes J. Determinants for association and guide RNA-directed endonuclease cleavage by purified RNA editing complexes from Trypanosoma brucei. J. Mol. Biol. 2008;381:35–48. doi: 10.1016/j.jmb.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoki Y, Kimura N, Kanbayashi M, Amakawa Y, Ohhata T, Sasaki H, Sado T. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development. 2009;136:139–146. doi: 10.1242/dev.026427. [DOI] [PubMed] [Google Scholar]
- Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, Toscano S, Wan Y, Spitale RC, Luscombe N, Backofen R, et al. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol. Cell. 2013;51:156–173. doi: 10.1016/j.molcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, Shi Y. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 2011;18:769–776. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H, Del Rosario AM, Bryson BD, Schroeder MA, Sarkaria JN, White FM. Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts. Mol. Cell Proteomics. 2012;11:1724–1740. doi: 10.1074/mcp.M112.019984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and non-coding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner S, Müller M, Fröhlich J, Langer D, Becker PB. ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol. Cell. 2013;51:174–184. doi: 10.1016/j.molcel.2013.06.011. [DOI] [PubMed] [Google Scholar]
- Mitson M, Kelley LA, Sternberg MJ, Higgs DR, Gibbons RJ. Functional significance of mutations in the Snf2 domain of ATRX. Hum. Mol. Genet. 2011;20:2603–2610. doi: 10.1093/hmg/ddr163. [DOI] [PubMed] [Google Scholar]
- Müller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 2009;19:150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Hernández-Muñoz I, Fazzio TG, Shah GM, Kraus WL, Panning B. Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J. Biol. Chem. 2007;282:12851–12859. doi: 10.1074/jbc.M610502200. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter SF, Sadreyev RI, Yildirim E, Jeon Y, Ohsumi TK, Borowsky M, Lee JT. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 2012;22:1864–1876. doi: 10.1101/gr.133751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakumar K, Bernstein E. ATRX: the case of a peculiar chromatin remodeler. Epigenetics. 2013;8:3–9. doi: 10.4161/epi.23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starmer J, Magnuson T. A new model for random X chromosome inactivation. Development. 2009;136:1–10. doi: 10.1242/dev.025908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, Picketts DJ, Yang X. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J. Biol. Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker EW, van Vugt MA, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, Yaffe MB. 14-3-3sigma controls mitotic translation to facilitate cytokinesis. Nature. 2007;446:329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. USA. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E, Sadreyev RI, Pinter SF, Lee JT. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat. Struct. Mol. Biol. 2012;19:56–61. doi: 10.1038/nsmb.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.