Summary
Rod-shaped bacteria typically elongate at a uniform width. To investigate the genetic and physiological determinants involved in this process, we studied a mutation in the morphogenetic protein MreB in Caulobacter crescentus that gives rise to cells with a variable-width phenotype, where cells have regions that are both thinner and wider than wild-type. During growth, individual cells develop a balance of wide and thin regions, and mutant MreB dynamically localizes to poles and thin regions. Surprisingly, the surface area to volume ratio of these irregularly-shaped cells is, on average, very similar to wild-type. We propose that, while mutant MreB localizes to thin regions and promotes rod-like growth there, wide regions develop as a compensatory mechanism, allowing cells to maintain a wild-type-like surface area to volume ratio. To support this model, we have shown that cell widening is abrogated in growth conditions that promote higher surface area to volume ratios, and we have observed individual cells with high ratios return to wild-type levels over several hours by developing wide regions, suggesting that compensation can take place at the level of individual cells.
Keywords: cell wall biosynthesis, cell width, growth rate, morphogenesis, peptidoglycan
Introduction
Understanding how bacterial cells determine their size and shape is a fundamental challenge in biology (Young, 2006). The individual proteins that are the genetic determinants of cell shape are orders of magnitude smaller than the structures they help build, and the specific roles these proteins play in the process of shape determination remain largely unclear. It is therefore critical to define the impact each of these genetic players has on cell shape, as well as understand any physiological constraints that might limit the range of shapes that cells can adopt.
Cell shape in most bacteria is dictated by the cell wall, which is composed of a covalently-linked meshwork of peptidoglycan that encloses the cytoplasm and counteracts osmotic pressure (for reviews, see Cabeen and Jacobs-Wagner, 2005; Typas et al., 2011). In order for growth to occur, this elastic meshwork must be broken apart and new material inserted; peptidoglycan precursors synthesized in the cytoplasm are exported across the inner membrane and incorporated into the growing network, while previously existing connections are hydrolyzed, making room for newly inserted material. Due to the covalent nature of the peptidoglycan meshwork, the spatial and temporal patterning of these synthetic and hydrolytic activities is central to the determination and maintenance of cell shape.
Rod-shaped bacteria typically grow by elongation, adding new lateral cell wall material while maintaining a characteristic width, and then dividing at or near the middle to form two daughter cells (Höltje, 1998). One protein that has been implicated in coordinating elongation in rod-shaped bacteria is the bacterial actin homolog MreB, which is present in most rod-shaped bacteria but not in coccoids (Jones et al., 2001; Daniel and Errington, 2003). When MreB is depleted genetically or by the addition of chemical inhibitors, cells typically stop elongating and instead increase in width, becoming spherical in some species, e.g. Escherichia coli (Wachi et al., 1987; Doi et al., 1988; Iwai et al., 2002; Kruse et al., 2005), and lemon-shaped in others, e.g. the curved rod Caulobacter crescentus (Figge et al., 2004; Gitai et al., 2004; Gitai et al., 2005). The loss of width control upon MreB depletion, coupled with evidence that MreB can interact with many components of the peptidoglycan synthesis machinery (Carballido-López et al., 2006; Mohammadi et al., 2007; Kawai et al., 2009; White et al., 2010; Favini-Stabile et al., 2013), suggests that MreB might be directing the spatial pattern of cell wall insertion such that cells elongate as rods. Additionally, MreB has a membrane-binding motif that targets it to the inner face of the cytoplasmic membrane (Salje et al., 2011), and as an actin homolog it is thought to form short filaments that may potentially assemble into higher-order structures (van den Ent et al., 2001; Popp et al., 2010; van den Ent et al., 2014). Small assemblies of MreB have been shown to move circumferentially around rod-shaped cells, perpendicular to the cell’s long axis, and this motion is dependent on peptidoglycan synthesis (Kim et al., 2006; Domónguez-Escobar et al., 2011; Garner et al., 2011; van Teeffelen et al., 2011). Additional studies in E. coli have shown spatial and temporal correlation between the localization of MreB and the insertion of new cell wall material (Ursell et al., 2014).Taken together, this evidence suggests that MreB coordinates insertion of new cell wall material in circumferential arcs, and thus promotes rod-like elongation at a characteristic width.
What dictates this characteristic width is still unclear. There is some evidence that MreB itself could be determining this cellular dimension. Point mutations in MreB have been isolated that cause cells to become either thinner (Kruse et al., 2003; Gitai et al., 2005; Charbon et al., 2009; Dye et al., 2011) or wider (Jones et al., 2001; Kruse et al., 2003; Dye et al., 2011) than wild-type. This finding, coupled with the loss of width control when MreB is depleted, makes MreB an attractive candidate for setting cell width. However, the picture cannot be this simple. Classic observations revealed that cells growing quickly in rich media are bigger in both volume and diameter than those growing slowly in minimal media (Schaechter et al., 1958; Zaritsky and Pritchard, 1973; Woldringh et al., 1977), and intermediate sizes are observed for intermediate growth rates (Volkmer and Heinemann, 2011). Because genetically identical cells grown in different conditions can adopt a wide variety of diameters, width is likely not a precisely genetically encoded property but rather a result of a combination of many factors including growth conditions. Thus it is unclear what role MreB plays in determining a precise cell width under a given set of environmental conditions.
To investigate MreB’s role in promoting elongation at a characteristic width, we have chosen to study a point mutation in Caulobacter MreB that gives rise to cells of a variable width. Isolated from a screen for mutations that confer resistance to the MreB inhibitor A22, this mutation – alanine 325 to proline (A325P) – is one of several that cause Caulobacter cells to adopt a variable-width phenotype (Dye et al., 2011). Unlike other mutations in MreB that uniformly change the width of cells, this point mutation causes cells to have some regions that are much thinner and some that are much wider than wild-type, often within a single cell. By studying this mutant that is unable to promote elongation at a single characteristic width, we have gained insight into the shape-determining functions of MreB. Here, we present our findings that A325P MreB dynamically localizes to thin parts of cells and seems to direct rod-like elongation there, and that this mislocalization is the main difference we observe between the mutant and wild-type. We further show that wide regions develop through an MreB- and FtsZ-independent cell wall growth mechanism, and that expansion in wide regions surprisingly appears to compensate for elongation in thin regions, allowing cells to maintain a wild-type-like surface area to volume ratio despite their extremely aberrant morphology. These findings demonstrate the integral role that surface area to volume ratio maintenance must play in the determination of cell shape.
Results
A325P cells grow in wide and thin regions, but Venus-MreBA325P localizes only to thin regions
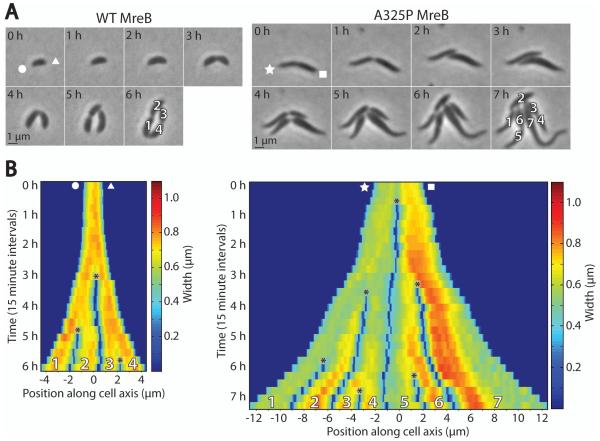
To probe MreB’s potential roles in setting cell width and directing rod-like growth, we chose to study a representative mutant, mreBA325P (referred to hereafter as A325P), from the class of MreB point mutants exhibiting a variable-width phenotype (Dye et al., 2011). To assess the growth pattern associated with this phenotype, we used time-lapse phase contrast microscopy to compare the growth of wild-type C. crescentus (CB15N) to A325P (Fig. 1 and Movies S1-2). Wild-type cells elongated while maintaining a constant width of about 0.7 μm, while A325P cells elongated in thin regions (0.5-0.6 μm) and also developed wide regions (> 0.7 μm). Often, A325P cells were wide on one side and thin on the other, and division in the center produced one wide and one thin daughter cell. This pattern is visible in Fig. 1B, where at the start of imaging the representative A325P cell was thin on the left and wide on the right; when it divided, it formed one thin and one wide daughter. During the remaining hours of the time-lapse sequence, the thin daughter on the left developed new wide regions, while the wide daughter on the right extended new thin regions from both poles. In this way, after several generations both lineages returned to an even mix of wide and thin parts. After observing this pattern of growth many times, we concluded that this tendency of individual cells to develop a mix of wide and thin regions was preventing the strain from diverging into separate wide and thin populations, and promoting the maintenance of a variable-width phenotype.
Fig. 1.
mreBA325P cells maintain a variable-width phenotype over time.
A. Time-lapse phase contrast images of representative wild-type C. crescentus (CB15N) and mreBA325P cells. White shapes (circle, triangle, star, square) denote which cell pole corresponds to which side of the kymographs in (B). See also Supplementary Movies S1 and S2.
B. Kymographs of width along the major cell axes from (A). Cells were laid out end to end from left to right. Asterisks denote cell divisions. Color represents cell width at each position along the major axis. Numbers at bottom correspond to cells in the final frame of (A).
We investigated the dynamic subcellular localization of MreB using merodiploid strains expressing either wild-type or A325P MreB from the native locus in addition to either wild-type or A325P MreB fused to the fluorescent protein Venus (Hsu et al., 2009) from the inducible xylose promoter (Meisenzahl et al., 1997). While overexpression of MreB with an N-terminal YFP tag in E. coli has been shown to cause artifactual immobile helical structures (Vats and Rothfield, 2007; Swulius and Jensen, 2012), single molecule imaging of fluorescent MreB expressed at a very low level has been used to observe the dynamic circumferential motion of MreB in Caulobacter (Kim et al., 2006). To demonstrate that our induction conditions produced fluorescent MreB that dynamically tracked with the motion of native MreB and thus cellular growth, we performed time-lapse epifluorescence imaging of Venus-MreBA325P (Fig. S1 and Movie S3). From this data it was clear that the majority of A325P MreB patches were moving in a manner consistent with circumferential motion, i.e. back and forth across the cell, perpendicular to the long axis. Furthermore, estimates of patch speed (~6-12 nm/sec, Fig. S1) were consistent with previous single-molecule measurements of MreB velocity in Caulobacter (Kim et al., 2006). We were therefore confident that in these conditions MreB fluorescence could be used as a proxy for native MreB localization and activity.
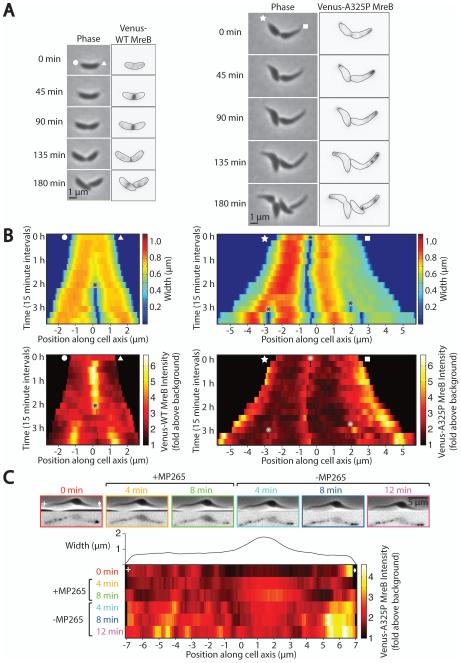
Using these strains and conditions we performed time-lapse imaging of live cells to observe the localization patterns of wild-type and A325P MreB during growth (Fig. 2A-B). The wild-type Venus-MreB localization pattern agreed with previously published results (Figge et al., 2004; Gitai et al., 2004): in Caulobacter, MreB is spread along the length of cells at the start of the cell cycle, accumulates at midcell after formation of the FtsZ ring, and disperses again late during cell constriction. This localization mirrors the pattern of new cell wall insertion in Caulobacter, where new material is initially inserted along the length of cells and then predominantly at midcell after FtsZ ring formation (Aaron et al., 2007). In contrast, Venus-MreBA325P did not accumulate at midcell, and instead appeared not only polarly localized, as previously described (Dye et al., 2011), but also enriched in thin regions of cells. MreB is thought to play a role in directing the insertion of new lateral cell wall material and is normally present at sites of insertion (Aaron et al., 2007). This mutant, however, appears to have an altered pattern (Fig. 1): A325P cells elongate in thin regions but also expand in wide regions, suggesting that these cells incorporate new wall material everywhere, despite the fact that MreB is only localized in thin regions.
Fig. 2.
Venus-MreBA325P is dynamically localized to thin regions of cells.
A. Representative time-lapse phase contrast and epifluorescence images of wild-type and mreBA325P cells expressing Venus-MreB (left) or Venus-MreBA325P (right). Expression of fluorescent protein was induced for 1 h prior to and during imaging. These and all subsequent fluorescence images have been inverted for ease of viewing. Indicated cell outlines were identified using MicrobeTracker (Sliusarenko et al., 2011). White shapes (circle, triangle, star, square) denote which cell pole corresponds to which side of the kymographs in (B).
B. Kymographs of cell width and Venus-MreB localization from (A). Color represents cell width (top) and Venus-MreB fluorescence intensity (bottom). Asterisks denote cell divisions.
C. Representative time-lapse phase contrast and epifluorescence images of an A325P cell prior to, during, and after treatment with 150 μg ml−1 of the MreB inhibitor MP265. Expression of fluorescent protein was induced for 1 h prior to but not during imaging. Images were taken every 4 minutes and MP265 was added immediately after the 0 min image (outlined in red) and removed immediately after the 8 min image (outlined in green). Venus-MreBA325P fluorescence intensity along the long cell axis at each timepoint is plotted in the kymograph below, where color represents fluorescence intensity, and cell width at each position is plotted directly above the kymograph. White shapes (plus, diamond) indicate the left and right cell poles respectively.
Wondering if A325P MreB was preferentially and dynamically associating with thin parts of cells, we delocalized Venus-MreBA325P using the MreB inhibitor MP265 and observed its relocalization after the inhibitor was removed (Fig. 2C). MP265 is a structural analog of the MreB inhibitor A22 that has comparable inhibitory effects on MreB and lower cytotoxicity (Takacs et al., 2010). Treatment with either inhibitor leads to the delocalization of MreB in Caulobacter and causes MreB to become diffuse (Gitai et al., 2005; Takacs et al., 2010). Although the A325P mutation in MreB confers resistance to low levels of A22 (2.5 μg ml−1) (Dye et al., 2011) and MP265 (data not shown), a higher concentration of MP265 (150 μg ml−1) was sufficient to delocalize Venus-MreBA325P puncta and make the fluorescent signal cytoplasmic (Fig. 2C). Because the wide region of the cell was much thicker and contained more cytoplasm, when the fluorescent signal was displaced to the cytoplasm by drug treatment, epifluorescence imaging showed enriched fluorescence in this wide part. Upon inhibitor removal, multiple puncta reformed in thin regions after several minutes and the cytoplasmic signal decreased. This result demonstrates that A325P MreB is able to dynamically localize to thin regions, suggesting that this point mutation in MreB causes it to bind preferentially to surfaces with high negative mean curvature, i.e. the inner face of the cell envelope in thin regions and at poles.
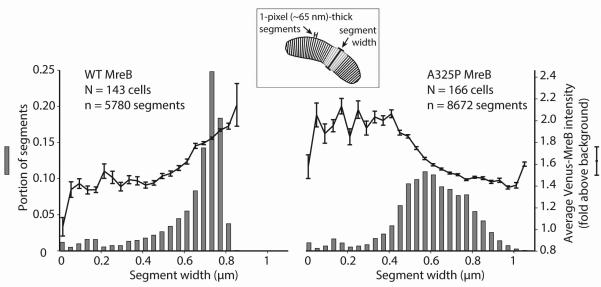
To determine if the enrichment of Venus-MreBA325P in thin regions held true for a large population of cells, we measured cell width and Venus-MreB fluorescence intensity for one-pixel (~65 nm)-thick segments along the long axes of many (N > 140) wild-type and A325P cells. Fig. 3 shows histograms of wild-type and A325P cell segments binned by width and the corresponding average fluorescence intensity for segments in each bin. Wild-type cells were primarily composed of segments that were the characteristic width for Caulobacter, 0.7 μm, and showed enriched fluorescence in wide segments, corresponding to cells where MreB was accumulating at midcell prior to constriction. A325P cells, however, had a much broader distribution of widths, ranging both above and below wild-type, and in contrast to wild-type cells where MreB fluorescence was excluded from the poles, A325P cells had enriched fluorescence in thin regions and at poles, confirming our previous observation that A325P MreB localizes to thin parts of cells and is largely absent from wide regions.
Fig. 3.
A population of mreBA325P cells contains a broad distribution of widths and Venus-MreBA325P localizes predominantly in thin regions.
Histograms of cell widths for large populations of wild-type and mreBA325P cells expressing Venus-MreB and Venus-MreBA325P respectively. Cells were divided into 1-pixel (~65 nm)-thick segments along their long axes, and segments were binned according to width. Note that the average number of segments per cell was greater for mreBA325P because these cells were often longer than wild-type. The mean Venus-MreB intensity of segments in each width bin was calculated and is overlaid on top of the histograms. Error bars represent the standard error of the mean.
We examined several other components of the peptidoglycan insertion machinery and also polar markers to determine whether A325P mislocalization led to other defects in protein localization and cell polarity. We found no measurable changes (Figs. S2-3).
Dynamic motion of Venus-MreBA325P is comparable to wild-type
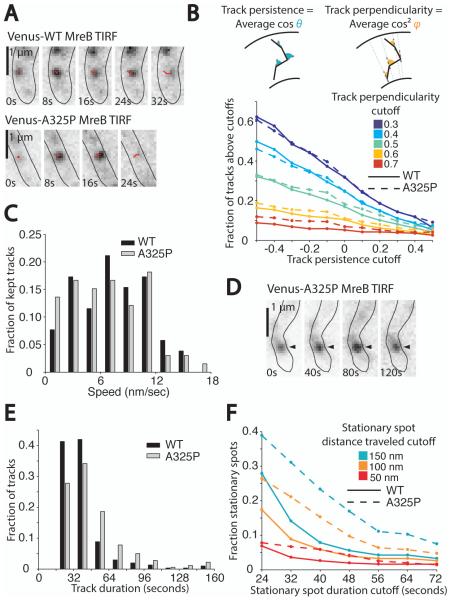
We wondered whether the dynamic motion of A325P MreB might be different from wild-type and if such a difference might contribute to the variable-width phenotype. To investigate the motion of MreB, we imaged wild-type and A325P cells expressing very low levels of Venus-MreB or Venus-MreBA325P respectively using a total internal reflection fluorescence (TIRF) microscope. This method allowed us to image only the bottom of cells and identify spots of MreB that were moving in a persistent, circumferential manner, perpendicular to the long axis of the cell. Single molecule induction levels (Kim et al., 2006) were used to produce well-separated fluorescent spots to facilitate tracking and to minimize any perturbation of the dynamic motion that might be caused by the presence of fluorescent fusion proteins. Consistent with previous single molecule imaging of Venus-MreB in Caulobacter (Kim et al., 2006), we observed many spots that were present for only a single frame (i.e. < 8 seconds) as well as a subset that appeared to move persistently across the cell diameters (Fig. 4A).
Fig. 4.
Circumferential motion of Venus-MreBA325P is similar to Venus-MreB.
A. Time-lapse TIRF images of wild-type and mreBA325P cells expressing very low levels of Venus-MreB and Venus-MreBA325P respectively. Images were taken every 8 s with 300 ms exposures. Cells were genetically depleted of FtsZ by growing a strain with FtsZ under an inducible promoter under non-inducing conditions so that wild-type MreB would not accumulate in a band at midcell, obscuring individual MreB puncta (see Experimental Procedures). Wild-type and mreBA325P cells were depleted of FtsZ for 3 h prior to imaging, and Venus-MreB and Venus-MreBA325P were induced for 1 h prior. Shown are examples of MreB puncta that moved persistently across cells, perpendicular to the cell axis. Red lines show the progress of tracks over time.
B. Plot of the fraction of automatically detected wild-type and A325P MreB tracks that met various cutoffs for track persistence and perpendicularity (see Experimental Procedures). For wild-type, N = 147 cells and n = 305 total tracks, and for A325P, N = 39 cells and n = 360 total tracks. Tracks were filtered by persistence, or the average of the cosines of the angles between successive steps in a track, and by perpendicularity, or the average of the cosines squared of the angles made between each track segment and a line perpendicular to the long axis of the cell. Inset diagrams depict the angles used to calculate persistence and perpendicularity.
C. Histogram of track speeds. The track persistence cutoff used was 0 and the track perpendicularity cutoff was 0.5. The average speed for wild-type was 7.3±3.7 nm/sec, and the average speed for A325P was 6.5±4.0 nm/sec (± standard deviation). These distributions are not significantly different (p = 0.33 as determined by two-sample Kolmogorov-Smirnov test).
D. Time-lapse TIRF images of Venus-MreBA325P acquired as described in (A). The black arrowhead marks a long-lived, stationary spot.
E. Histogram of track duration for all automatically detected tracks. Note that to be detected tracks were required to last for at least 24 seconds. The average duration for wild-type was 36±20 seconds, and the average duration for A325P was 46±27 seconds (± standard deviation). These distributions are significantly different (p < 0.001 as determined by two-sample Kolmogorov-Smirnov test).
F. Plot of the fraction of wild-type and A325P tracks counted as stationary spots using various cutoffs for stationary spot duration and total distance traveled.
To quantify differences between wild-type and A325P spot motion we focused on spots moving circumferentially around the cells. We automatically detected tracks and then filtered them to identify tracks that were both persistent and perpendicular to the long axis of cells. We used MATLAB-based analysis software to automatically detect MreB spots (Sliusarenko et al., 2011) and link spots between frames into tracks (Jaqaman et al., 2008). We then defined two metrics: persistence, or the mean of the cosines of the angles formed between successive steps in the track; and perpendicularity, or the mean of the cosines squared of the angles formed between each step of the track and a line perpendicular to the cell axis at that point. Figure 4B shows the fraction of wild-type and A325P tracks meeting various cutoffs for persistence and perpendicularity. No matter how stringent we made these cutoffs, we could not detect a significant difference between the portion of automatically detected tracks meeting these cutoffs in wild-type and A325P, suggesting that wild-type and A325P MreB spots have comparable persistence and perpendicularity. Furthermore, using a persistence cutoff of 0 and perpendicularity cutoff of 0.5 and plotting the distribution of speeds for tracks meeting these criteria revealed no significant difference in speed between wild-type and A325P (Fig. 4C).
The main difference we observed between wild-type and A325P was that A325P had more long-lived, stationary spots often occurring near cell poles. An example is shown in Fig. 4D. We quantified this observation by binning automatically detected tracks by duration and confirming that A325P tracks were generally more long-lived (Fig. 4E). To identify what fraction of tracks were long-lived, stationary spots, we filtered all automatically detected tracks by duration and final displacement (for a plot of duration and displacement for all tracks, see Fig. S4). Fig. 4F shows that for a variety of duration and distance cutoffs, A325P had a much larger portion of tracks classified as stationary spots. These observations together support the conclusion that a larger subset of A325P spots are stationary compared to wild-type, but that dynamic spots move circumferentially with the same speed, persistence, and perpendicularity as wild-type.
FtsZ is not required for growth in wide regions of A325P cells
Because A325P cells grow in both wide and thin regions despite the fact that MreB is only localized in thin parts of cells, we wondered if something else could be directing new wall insertion in wide regions. Specifically, there is evidence that upon formation of the FtsZ ring, synthesis of new lateral wall material is concentrated around midcell prior to invagination and division, and that this FtsZ-directed growth is independent of MreB (Aaron et al., 2007; Varma and Young, 2009). Based on this evidence, we wondered whether FtsZ could be responsible for directing insertion of new material in wide regions, while A325P MreB directs insertion in thin regions.
To test this idea, we genetically depleted FtsZ by transducing into the A325P background previously published constructs that express FtsZ solely from an inducible promoter (Wang et al., 2001; Alyahya et al., 2009). The resulting strains must be grown in the presence of inducer; when inducer is removed, FtsZ is > 90% depleted within two hours, and the cells grow as long filaments (Wang et al., 2001). When FtsZ was depleted in the A325P background, the cells became filamentous and each cell developed extremely long, thin extensions at both poles with a wide region in the middle (Fig. 5A). If FtsZ were responsible for growth in wide regions, A325P cells lacking FtsZ would continue growing in thin regions but not in wide regions. We found, however, that wide regions continued expanding even without FtsZ, and uniformly thin cells that were depleted of FtsZ were able to develop wide bulges over time (Fig. 5B-C, Movie S4). We also confirmed that Venus-MreBA325P was still dynamically localized in the thin regions of FtsZ-depleted cells by imaging cells during induction of fluorescent MreB (Fig. 5D-E). Upon induction, Venus-MreBA325P appeared and remained solely in the thin regions of cells. Indeed, the width kymograph of cell 1 revealed that this cell expanded its wide region over the course of imaging, and that MreB was largely absent from this region during that process. Together, these data suggest that growth in wide regions is independent of both MreB and FtsZ.
Fig. 5.
FtsZ is not required for the development or expansion of wide regions in mreBA325P cells.
A. Phase contrast image of filamentous mreBA325P cells genetically depleted of FtsZ. To deplete FtsZ in this strain, FtsZ was expressed under an inducible promoter and cells were grown in non-inducing conditions (see Experimental Procedures).
B. Time-lapse phase contrast images of an mreBA325P cell depleted of FtsZ as in (A). FtsZ was completely depleted prior to imaging and depletion continued throughout. White shapes (plus and diamond) mark the left and right poles of the cell and correspond to kymograph in (C).
C. Kymograph of cell width during growth of the FtsZ-depleted mreBA325P cell shown in (B). See also Supplementary Movie S4.
D. Time-lapse phase contrast and epifluorescence images of mreBA325P cells genetically depleted of FtsZ as in (A) during induction of Venus-MreBA325P. Fluorescent MreB was induced at the start of imaging, and time stamps reflect time since imaging and induction began. Red and blue cell number labels refer to the left and right cells respectively and are referred to in parts (E) and (F). White shapes (circle, triangle, square, star) refer to the left and right poles of cells 1 and 2 and correspond to the kymographs in E.
E. Kymographs of cell width and Venus-MreBA325P fluorescence intensity in cells 1 and 2 from (D) during growth and induction of fluorescent protein. Color represents cell width in the top kymograph and Venus-MreBA325P fluorescence intensity in the bottom kymograph.
F. Scatter plot of the surface area to volume ratios of cells 1 and 2 from (D) and (E) over time. Surface area to volume ratios were calculated using MicrobeTracker (Sliusarenko et al., 2011) and treating each pixel-thick segment as a 3D disk with cylindrical symmetry.
Maintenance of a wild-type-like surface area to volume ratio underlies the variable-width phenotype of A325P cells
The ratio of cell surface area to cell volume is an important aspect of bacterial physiology, and is often cited as a key parameter in determining the rate of nutrient uptake (Koch, 1996; Young, 2006). However, the interplay between the surface area to volume ratio and cell shape is still largely unexplored, though these properties are inherently linked. Noting that elongation of thin regions and expansion of wide regions in A325P cells would have competing effects on cell surface area to volume ratio, we wondered how the surface area to volume ratio was changing during the growth of these cells. Using MicrobeTracker (Sliusarenko et al., 2011) and assuming cylindrical symmetry at each pixel-thick segment along the backbone of cells, we calculated the surface area to volume ratios of cells 1 and 2 from Fig. 5D-E and plotted the evolution of these ratios over the course of growth (Fig. 5F). Remarkably, both cells moved toward the same surface area to volume ratio over time (6.0 μm−1), and this appeared to be due to the cell with a higher initial ratio (cell 1) expanding its wide region, and the cell with a lower initial ratio (cell 2) elongating almost exclusively in thin regions (Fig. 5E-F).
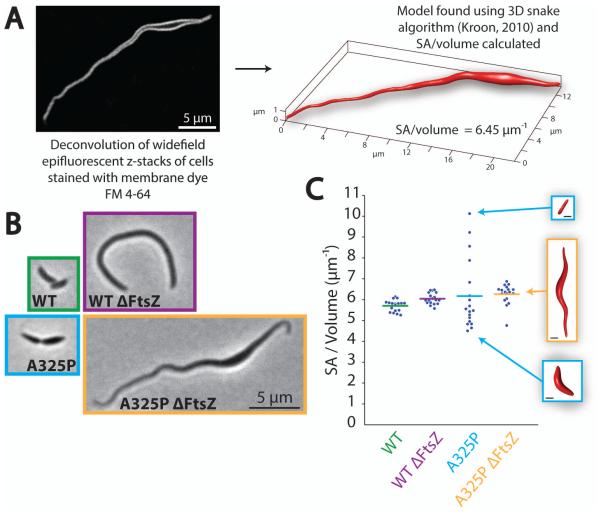
Wondering how this surface area to volume ratio compared to wild-type, we developed a method to accurately determine the three-dimensional shape of cells and used this shape to calculate cell surface area to volume ratio. Briefly, deconvolution of widefield epifluorescent z-stacks of cells stained with membrane dye FM 4-64 was used to visualize the cell surface. This data was fit using a three-dimensional snake algorithm (Kroon, 2010) and the surface area to volume ratio of the resulting mesh was calculated (Legland, 2009) (Fig. 6A). We performed surface area to volume ratio measurements on wild-type and A325P cells with and without FtsZ (representative cells shown at the same scale in Fig. 6B), and the results are plotted in Fig. 6C. Amazingly, wild-type cells with and without FtsZ both had the same surface area to volume ratio that we had observed A325P FtsZ depletion cells moving toward in Fig. 5F (6.0 μm−1), and the averages for A325P cells with and without FtsZ were also very comparable, despite dramatic variations in overall cell size and shape. Notably, the spread of surface area to volume ratio values in dividing A325P cells was much larger than any other strain. As shown in the insets in Fig. 6C, this was because this strain often produced cells that were entirely thin or entirely wide. The average ratio, however, remained similar to wild-type. In the A325P FtsZ depletion cells, however, division was no longer a factor, and all cells developed a mixture of wide and thin parts, resulting in a collapse of the surface area to volume ratios into a tighter distribution close to the wild-type value. Together, these findings suggest that expansion in wide regions of A325P cells exactly balances growth of thin regions to produce a surface area to volume ratio comparable to wild-type. This observation led us to hypothesize the following explanation of the variable-width phenotype: while A325P MreB directs insertion exclusively in thin regions, individual cells develop wide regions of an appropriate size that expand by an MreB- and FtsZ-independent mechanism so as to allow cells to maintain a wild-type-like surface area to volume ratio.
Fig. 6.
The average surface area to volume ratio of mreBA325P cells is comparable to wild-type.
A. Summary of scheme for determining accurate surface area to volume ratios. Shown is one non-inverted slice of a deconvolved epifluorescent z-stack of an FtsZ-depleted mreBA325P cell stained with membrane dye FM 4-64, as well as the 3D mesh of this cell calculated using a 3D snake algorithm (Kroon, 2010).
B. Representative phase contrast images of dividing and FtsZ-depleted wild-type and mreBA325P cells. All images are at the same scale.
C. Plot of surface area to volume ratios for N = 18 cells from each condition in (B) determined using the scheme in (A). Horizontal bars mark the mean of each distribution and inset images show the 3D meshes corresponding to the indicated data points. All inset images are at the same scale and the scale bars represent 1 μm.
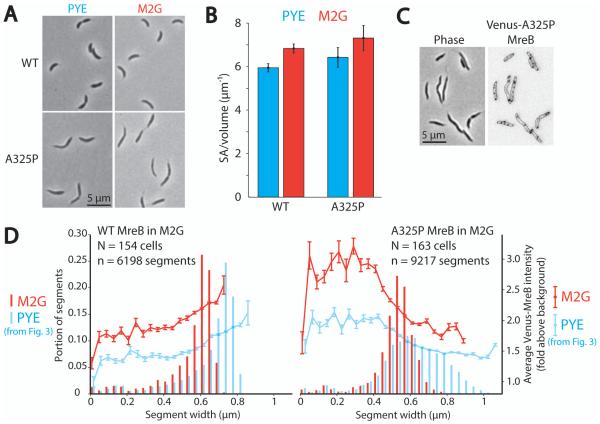
To test this model, we increased the surface area to volume ratio by growing cells in minimal media and assessed the effect on cell shape. It has long been known that bacteria grown in minimal media have higher surface area to volume ratios than those grown in rich media, and that rod-shaped cells achieve this change by decreasing their radius (Schaechter et al., 1958; Zaritsky and Pritchard, 1973; Woldringh et al., 1977). Our model predicts that if A325P cells are grown in conditions that increase the surface area to volume ratio, thin regions of cells will remain unchanged, and the prevalence of wide regions will decrease because they are no longer required to depress the surface area to volume ratio. To test these predictions, we grew wild-type and A325P cells in rich media (PYE) and minimal media (M2G), and compared the resulting cell shapes, average surface area to volume ratios, MreB localization patterns, and width distributions (Fig. 6A-D). Note that these and all subsequent surface area to volume ratio measurements were performed using phase contrast microscopy and assuming cylindrical symmetry along the cell axis because this method was more time-efficient and gave the same results as the three-dimensional analysis of deconvolved epifluorescent z-stacks (Fig. S5).
As expected, wild-type cells increased their surface area to volume ratio in minimal media by decreasing their width from about 0.7 to 0.6 μm (Fig. 7A-B and D). A325P cells likewise increased their surface area to volume ratio by an equivalent amount in minimal media (Fig. 7B), and inspection of these cells revealed that they were almost entirely thin and lacked the wide bulges observed in rich media (Fig. 7A). The width profile of A325P cells grown in minimal media supported this observation, peaking between 0.5 and 0.6 μm (Fig. 7D), the same range defined as thin in Fig. 1. Additionally, images of A325P cells grown in M2G and expressing Venus-MreBA325P showed that A325P MreB could localize throughout most of these entirely thin cells (Fig. 7C). Plotting the average Venus-MreB fluorescence intensity at different widths for many wild-type and A325P cells grown in rich and minimal media (Fig. 7D) revealed that in wild-type cells, the fluorescence intensity always peaked in the widest regions of cells, shifting from about 0.8 μm in rich media to 0.7 μm in minimal media (note that fluorescence intensity quantified as fold above background was overall lower in rich media because of increased background due to media autofluorescence). Conversely, the width preference of A325P MreB was unperturbed by the change in media; in both rich and minimal media, Venus-MreBA325P showed an increasing localization preference for successively thinner regions, peaking in segments < 0.4 μm that corresponded to cell poles.
Fig. 7.
Growth in minimal media increases cell surface area to volume ratio and in mreBA325P results in the loss of wide regions.
A. Representative phase contrast images of wild-type and mreBA325P cells grown in PYE (rich media) or M2G (minimal media).
B. Bar graph of average surface area to volume ratio of n > 84 cells from each condition in (A). Error bars represent the standard deviation.
C. Phase contrast and epifluorescence image of mreBA325P cells grown in M2G and expressing Venus-MreBA325P. Fluorescent MreB was induced 1 h prior to imaging.
D. Histograms of cell width and plots of width-specific mean Venus-MreB intensity for many wild-type and mreBA325P cells expressing Venus-MreB and Venus-MreBA325P respectively and grown in PYE and M2G. PYE data is the same as Fig. 3. Cells were divided into pixel-thick segments along their long axes and segments were binned according to width. The mean Venus-MreB intensity of segments in each width bin is plotted on top of the histograms. Error bars represent the standard error of the mean. Note that fluorescence signal quantified as fold above background is generally higher in M2G because the rich medium, PYE, has more autofluorescence and thus higher background.
These results further support the model that A325P MreB is dynamically localized to regions of high negative mean curvature such as thin regions and poles in both rich and minimal media and directs elongation at this thin radius in both conditions. This radius, however, is more compatible with the higher surface area to volume ratio associated with growth in minimal media, and thus A325P cells grown in minimal media need not develop the extensive compensatory wide regions observed in rich media. Interestingly, the width distribution of A325P cells in minimal media was still not as narrow as for wild-type cells, extending both above and below the characteristic width of wild-type cells grown in minimal media (Fig. 7D). This supports the model that in minimal media A325P MreB continues directing growth in thin regions, but because these regions are still slightly thinner than wild-type cells in minimal media, some compensatory widening occurs – though to a much lesser extent than in rich media – allowing A325P cells to achieve the same surface area to volume ratio as wild-type cells in identical growth conditions.
Wondering whether changes in cell shape in response to surface area to volume ratio requirements could occur at a single cell level, we dynamically tracked the surface area to volume ratios of individual cells during growth. We returned to the colonies of wild-type and A325P cells shown growing in Fig. 1. Surface area to volume ratios for all cells in each frame were calculated and plotted over time (Fig. 8A), with colored lines representing individual cells and new colors assigned to each daughter after division events. In wild-type cells, the surface area to volume ratio remained very close to 6 μm−1. For A325P, however, cells upon division often produced one daughter with a very high surface area to volume ratio (> 7 μm−1). These events correspond to cells that were thin on one side and divided to produce a daughter that was entirely thin. Over time, these entirely thin cells developed wide regions (Fig. 1B), thereby reducing their surface area to volume ratio to wild-type levels (Fig. 8A and Movie S5). Conversely, cells that already had a wide region and wild-type-like surface area to volume ratio elongated primarily in thin regions and maintained their surface area to volume ratio (see A325P cell 7 in Figs. 1B and 8A). Together, this evidence supports our model that A325P MreB directs elongation in thin regions but individual cells develop compensatory wide regions in order to maintain a wild-type-like surface area to volume ratio.
Fig. 8.
Entirely thin mreBA325P cells with elevated surface area to volume ratios return to wild-type-like levels by developing wide regions.
A. Plots of individual cell surface area to volume ratio trajectories over time. The surface area to volume ratios for the wild-type and mreBA325P cells in Fig. 1 were plotted over time. Colored lines represent individual cells, and after division daughters are assigned new colors. Numbers correspond to numbering in Fig. 1. See also Supplementary Movie S5.
B. Model for maintenance of the variable-width phenotype in mreBA325P cells. We propose that MreBA325P promotes rod-like growth in thin regions of cells, and that compensatory growth in wide regions allows cells to return to wild-type-like surface area to volume ratios.
Discussion
Molecular effects of A325P mutation
We have demonstrated that the A325P point mutation does not have an observable effect on the circumferential motion of MreB puncta, and indeed, the only major difference we noted between the behavior of wild-type and A325P MreB is the dynamic localization of A325P MreB to thin regions and poles, or regions of high negative mean curvature. This strongly suggests that MreB itself is able to detect local cell curvature, as has been reported in E. coli (Renner et al., 2013; Ursell et al., 2014). While it is possible that MreB’s curvature-sensing abilities arise from a binding partner, it seems unlikely that an internal point mutation like A325P would alter a binding partner interaction such that the mutant protein would become specifically localized to regions of high negative mean curvature. We believe it is much more probable that this point mutation changes the curvature preference of MreB itself, potentially by producing more highly curved MreB filaments. Consistent with this, recent molecular dynamics simulations suggest that deficiencies in ATP hydrolysis could give rise to more highly curved MreB filaments (Colavin et al., 2014). We therefore propose that A325P MreB is functioning to promote rod-like growth of the cell wall in the same way as wild-type, and that the aberrant overall shape phenotype arises simply because A325P MreB is mislocalized to thin regions and poles due to a preference for highly curved surfaces.
MreB localization and implications for cell shape
By mislocalizing to thin sections and poles and directing rod-like elongation there, A325P MreB appears to facilitate elongation only at a thin radius. When this radius is incompatible with the preferred surface area to volume ratio for a particular growth condition, i.e. in rich media, cells develop wide bulges that grow outward in a geometrically unconstrained manner that is both MreB- and FtsZ-independent. These observations are summarized in Fig. 8B. In dividing A325P cells, divisions of variable-width cells often produce cells that are entirely thin or entirely wide, and the surface area to volume ratios of these cells are higher or lower than wild-type. Over time, thin cells with high ratios return to normal by increasing their width through an MreB- and FtsZ-independent mechanism, while wide cells with low ratios increase their ratio by elongating in thin regions in an A325P MreB-directed fashion.
Notably, A325P cells usually only develop one bulge, even when filamented by FtsZ depletion. This suggests that once one bulge forms, A325P MreB no longer samples that region of the cell, and because MreB is not present to direct rod-like elongation, further compensatory expansion continues to occur at that same location. This positive feedback loop restricts additional expansion to the already-formed bulge. Additionally, wide regions in FtsZ-depleted A325P cells often grow to > 1 μm in width (Fig. 5C), while the widest parts of non-depleted A325P cells are usually < 1 μm (Fig. 3). This supports the view that once an individual cell contains one wide region, further expansion occurs in the same place: all the thin growth occurring in FtsZ-depleted cells is compensated for by one giant bulge, whereas in a group of dividing cells many smaller bulges – but still usually one per cell – develop as individual cells move toward a wild-type-like surface area to volume ratio. Importantly, the observation that the local absence of MreB leads to unconstrained growth outward agrees with previous studies showing that genetic or chemical depletion of MreB leads to an increase in cell radius and cell bulging (Wachi et al., 1987; Doi et al., 1988; Figge et al., 2004; Gitai et al., 2004).
Wild-type MreB, in contrast to the A325P mutant, localizes throughout cells or at midcell prior to division and promotes rod-like growth at a uniform width. Significantly, however, wild-type cells are able to adopt different uniform widths in different growth conditions (Schaechter et al., 1958; Zaritsky and Pritchard, 1973; Woldringh et al., 1977). This implies that wild-type MreB is able to flexibly respond to the surface area to volume ratio requirements of the cell, and promote growth at whatever uniform width is necessary (see Fig. 7D). In other words, wild-type MreB alone does not dictate the width of cells, and instead is crucial for elongating uniformly at whatever width is required to achieve a certain surface area to volume ratio. How MreB is able to promote uniform elongation at such a wide variety of different widths remains an important open question. Interestingly, hydrolysis-deficient MreB mutants in Bacillus subtilis have been shown to adopt non-uniform, banded localization patterns and produce bulging, variable-width cells (Defeu Soufo and Graumann, 2006; Garner et al., 2011). This class of mutants, similar to A325P, is unable to redistribute and sample the entire length of cells, resulting in a variable-width phenotype. We take this as further support for the model that MreB must be able to perform its rod-maintaining function wherever new lateral wall material is being incorporated and at whatever width is dictated by the surface area to volume ratio; otherwise cells will not be able to grow at a uniform width.
Surface area to volume ratio maintenance as a morphological determinant
Perhaps the most surprising finding to emerge from this study was the observation that individual cells move toward and maintain a characteristic surface area to volume ratio, even altering their shape when necessary. Many individual proteins, including MreB, MreC (Lee et al., 2003; Kruse et al., 2005; Leaver and Errington, 2005), and RodZ (Shiomi et al., 2008; Alyahya et al., 2009; Bendezú et al., 2009), have been implicated in determining the shape of cells, but our findings suggest that the cell-scale, physiological property of surface area to volume ratio also plays a major role in determining cell morphology. The growth pattern associated with this variable-width mutant provided a unique opportunity to observe cells with incorrect ratios returning to wild-type levels through the development and expansion of wide regions. It is currently unclear how wide regions develop in this mutant, but the process is both MreB- and FtsZ-independent, and exactly complements growth in thin regions to achieve a wild-type-like surface area to volume ratio. It is also unclear how cells are able to achieve such tight control over their surface area to volume ratios, but two possibilities seem likely: either cells have a system for sensing the current surface area to volume ratio and responding accordingly, or growth is somehow intrinsically compensatory. Future work will focus on distinguishing between these possibilities and understanding the interplay between surface area to volume ratio maintenance and bacterial cell shape.
MreB as a radius-maintaining factor
Together, our observations suggest that MreB might be functioning as a radius-maintaining factor rather than a radius-determining factor. A325P MreB localizes to thin regions and promotes elongation at that radius; wild-type MreB localizes throughout cells and promotes elongation at that radius. An integral question, then, is how do wild-type cells flexibly change their width in response to changing environmental conditions? We believe that the answer to this question might lie in our finding that individual cells can alter their width in order to achieve a specific surface area to volume ratio. We have demonstrated that a cellular mechanism exists that allows individual cells to change their width in an MreB- and FtsZ-independent way in order to attain a particular surface area to volume ratio. This mechanism is likely also at play in wild-type cells: when a wild-type cell is switched from one growth condition to another and alters its width accordingly, this mechanism could drive that change in width, with MreB acting downstream to ensure that new elongation occurs uniformly at the modified width. In this way, MreB need only function as a radius-maintaining factor provided there is some mechanism upstream of it that can alter the width of cells. In the A325P mutant, this process breaks down when A325P cells begin to grow wider, because A325P MreB samples wide regions less frequently, allowing them to grow outward in a geometrically unconstrained manner, leading to the development of wide bulges. Because wild-type MreB continues to sample the length of cells, small increases in width are not magnified into bulges but are rather smoothed throughout entire cells, resulting in a uniform increase in width. This view of MreB is both consistent with our data and sufficient to explain the rod shape-promoting properties of MreB as previously characterized in Caulobacter and other rod-shaped bacterial cells.
Experimental Procedures
Strain construction
Strains were constructed by transducing fluorescent fusions and FtsZ depletion constructs into recipient strains using phage transduction (ϕCR30), and selected for using appropriate antibiotics as described (Ely, 1991) (Table 1).
Table 1.
Strains used in this study.
| Name | Background or construction |
Description | Resistance | Source |
|---|---|---|---|---|
| LS101 | CB15N | Synchronizable derivative of Caulobacter
crescentus CB15 |
None | Evinger and Agabian, 1977 |
| JAT702 | CB15N | mreB::A325PmreB | None | Dye et al., 2011 |
| JAT783 | CB15N | xylX::Venus-mreB | Kan | Dye et al., 2011 |
| JAT798 | CB15N |
mreB::A325PmreB; xylX::Venus- A325PmreB |
Kan | Dye et al., 2011 |
| CJW2951 | CB15N | ftsZ::pVMCS-6ftsZ5' | Chlor | Alyahya et al., 2009 |
| JAT 1202 | CJW2951 transduced into JAT783 |
ftsZ::pVMCS-6ftsZ5', xylX::Venus-mreB | Chlor, Kan | This work |
| JAT 1203 | CJW2951 transduced into JAT798 |
ftsZ::pVMCS-6ftsZ5', mreB::A325PmreB, xylX::Venus-A325PmreB |
Chlor, Kan | This work |
| LS3205 | CB15N |
divJ::DivJ-CFP pleC::PleC-YFP |
Kan, Spec/Strep | Wheeler and Shapiro, 1999 |
| JAT1017 | LS3205 transduced into CB15N | divJ::DivJ-CFP | Kan | This work |
| JAT1018 | LS3205 transduced into JAT702 | divJ::DivJ-CFP, mreB::A325PmreB | Kan | This work |
| LS4527 | CB15N | tipN::TipN-GFP | Kan | Huitema et al., 2006 |
| JAT1016 | LS4527 transduced into JAT702 | tipN::TipN-GFP, mreB::A325PmreB | Kan | This work |
| JAT878 | CB15N | vanA::mCherry-Pbp2 | Spec/Strep | Hocking et al., 2012 |
| JAT880 | JAT878 transduced into JAT783 | vanA::mCherry-Pbp2, xylX::Venus-MreB | Kan, Spec/Strep | This work |
| JAT870 | CB15N | xylX::Venus-A325PmreB | Kan | Dye et al., 2011 |
| JAT973 | JAT878 transduced into JAT702 | vanA::mCherry-Pbp2, mreB::A325PmreB | Spec/Strep | This work |
| JAT979 | JAT870 transduced into JAT973 |
vanA::mCherry-Pbp2, mreB::A325PmreB, xylX::Venus-A325PmreB |
Kan, Spec/Strep | This work |
| CJW3288 | CB15N | xylX::GFP-cc1516 | Tet | Hocking et al., 2012 |
| JAT1003 | CJW3288 transduced into JAT702 |
xylX::GFP-cc1516, mreB::A325PmreB | Tet | This work |
| EG258 | CB15N | xylX::MurG-mCherry | Kan/Gent | Goley et al., 2010 |
| JAT1002 | EG258 transduced into JAT702 | xylX::MurG-mCherry, mreB::A325PmreB | Kan | This work |
| YB1585 | CB15N | ftsZ::pBJM1 | Kan | Wang et al., 2001 |
| JAT1005 | YB1585 transduced into JAT702 | ftsZ::pBJM1, mreB::A325PmreB | Kan | This work |
Bacterial growth
Caulobacter strains were grown at 30°C in PYE rich medium (0.2% Bacto peptone, 0.1% yeast extract, 1 mM MgSO4, 0.5 mM CaCl2) unless growth in M2G minimal medium was specified (6.1 mM Na2HPO4, 3.9 mM KH2PO4, 9.3 mM NH4Cl, 0.5 mM MgSO4, 0.01 mM FeSO4, 0.5 mM CaCl2, 0.2% [wt/vol] glucose). FtsZ depletion was achieved using the ftsZ::pBJM1 construct where FtsZ is induced in the presence of xylose and repressed in the presence of glucose (Wang et al., 2001). When Venus-MreB or Venus-MreBA325P were expressed in the same strain, however, we used the ftsZ::pVMCS-6ftsZ5' construct to induce FtsZ with vanillic acid (Alyahya et al., 2009).
All strains were grown overnight in media containing the appropriate selective antibiotics. FtsZ depletion strains requiring xylose or vanillic acid to produce FtsZ were supplemented with 0.3% xylose or 0.5 mM vanillic acid. Strains expressing fluorescent proteins under the xylose promoter were supplemented with 0.2% glucose except JAT 1202 and JAT 1203 which did not grow well in the presence of glucose. In the morning, strains were diluted from the overnight culture into fresh media of the same composition but lacking antibiotics and grown for several hours to achieve log phase growth. To deplete FtsZ, FtsZ depletion strains were washed in media lacking inducer and in the case of xylose-inducible FtsZ, 0.2% glucose was added to repress FtsZ expression. FtsZ was depleted for 3 hours prior to imaging. Fluorescent proteins expressed under the xylose promoter were induced by washing out glucose and adding 0.03% xylose for 1-2 hours. For TIRF imaging (Fig. 4), however, MreB was induced with 0.0002% xylose. Additionally, MreB was induced during imaging in Fig. 5D-E. Fluorescent proteins expressed under the vanillic acid promoter were induced with 0.5 mM vanillic acid for 1-2 hours.
Imaging conditions
Strains grown in PYE were imaged on 1% agarose/PYE pads except strains harboring polar markers (Fig. S3), which were imaged on 1% agarose/M2G pads to reduce background fluorescence. Strains grown in M2G were imaged on 1% agarose/M2G pads. Xylose (0.03%) was added to the agarose for continued induction of fluorescent MreB during time-lapse imaging (Fig. 2) or to induce expression (Fig. 5D-E); glucose (0.2%) was added for continued repression of FtsZ during time-lapse imaging (Fig. 5B-C). In MP265 wash in/out experiments (Fig. 2C), cells were imaged with an ONIX Microfluidic Perfusion Platform (CellASIC, Hayward, CA, USA). Cells were induced with xylose for 1 hour prior to but not during imaging, and fresh PYE either with or without 150 μg ml−1 MP265 was constantly flowed into the chamber. In 3D deconvolution imaging, cell membranes were stained with the lipophilic dye FM 4-64 (Life Technologies, Grand Island, NY, USA). Cells in PYE were incubated with 1 μg/ml FM 4-64 for 10 minutes prior to imaging and imaged on a 1% agarose/PYE pad containing 2 μg/ml FM 4-64.
All epifluorescence and phase contrast images except MP265 wash in/out and 3D deconvolution experiments were collected on an upright fluorescence microscope (Zeiss, Thornwood, NY, USA) with a Plan-Apo 1.4 100x phase objective, conventional epifluorescence filter set, and 1024 × 1024 back illuminated EM-CCD camera (Andor, South Windsor, CT, USA), giving an effective pixel size of 65 nm. During time-lapse imaging, the slide was heated with an objective warmer (Warner, Hamden, CT, USA) set to 33.1°C. MP265 addition and removal experiments were performed on a Nikon Eclipse TiE equipped with a microscope incubator (Haison Technology, Taipei, Taiwan) set to 30°C, and acquired with a Plan-Apo 1.45 100x phase objective, conventional epifluorescence filter set, and 1024 × 1024 back illuminated EM-CCD camera (Andor, South Windsor, CT, USA), giving an effective pixel size of 86 nm. TIRF microscopy was performed on an Axio Observer Z1 with TIRF 3 attachment (Zeiss, Thornwood, NY, USA), Plan-Apo 1.46 100x phase objective, 514 nm laser, single-band laser filter set optimized for 514 nm laser (Semrock, Rochester, NY, USA), and 1024 × 1024 back illuminated EM-CCD camera (Hamamatsu, Bridgewater, NJ, USA), giving an effective pixel size of 82 nm. 3D deconvolution microscopy was performed on a DeltaVision OMX BLAZE system (Applied Precision-GE, Issaquah, WA, USA) with a UPlanApo 1.4 100x objective (Olympus, Center Valley, PA, USA), InsightSSI™ illuminator (568 nm) and standard emission filter set equipped with an EM-CCD camera (Evolve, Photometrics, Tucson, AZ, USA), giving an effective pixel size of 81 nm. A z-step of 150 nm was used and iterative deconvolution was performed using softWoRx software (Applied Precision-GE, Issaquah, WA, USA).
Image analysis
All segmentation of phase contrast images was performed using the MATLAB-based MicrobeTracker software (Sliusarenko et al., 2011) and verified by hand. MicrobeTracker divides cells into approximately 1-pixel-thick segments along the cell backbones, and the average width of each segment was determined by dividing the area of that segment by its thickness. Volume was calculated using MicrobeTracker and surface area was calculated by assuming that individual segments were cylindrically symmetric. Integrated fluorescence signal in each segment was also measured using MicrobeTracker, and fluorescence intensity was calculated by dividing integrated fluorescence in each segment by that segment’s area. Background fluorescence intensity was calculated by using MicrobeTracker to create a mock cell outline in a region of the image that did not contain any cells and taking the average fluorescence intensity of the mock cell’s segments. The reported value of fold above background was found by dividing each segment’s fluorescence intensity by the background intensity.
Automatic detection of Venus-MreB spots in TIRF images for MreB tracking was done using the MicrobeTracker function SpotFinderZ. Parameters for detection were set using several training images and held constant for all analyses. Coordinates and amplitudes of spots were then passed to U-Track (Jaqaman et al., 2008), which performed frame-to-frame linking with no track merging, splitting, or gap closing, a minimum track length of 3 frames, and a maximum search radius of 2 pixels. The line perpendicular to the local axis of the cell used for calculating perpendicularity (Fig. 4B) was found by determining the cell segment as defined by MicrobeTracker closest to each length of track and taking a line through the center of that segment. In Fig. 4F, distance is defined as the displacement between the final and initial track coordinates, and speed in Fig. 4C and Fig. S4 is this distance divided by track duration.
Analysis of deconvolved z-stacks was performed using ImageJ and MATLAB software. Z-stacks were first converted to binary in ImageJ and then holes were filled and pixels not connected to the main cell body deleted. Images were then read into MATLAB and an isosurface around the cell was created and used as the initial starting point for a 3D snake algorithm (Kroon, 2010) that was used to refine the 3D mesh. Deconvolved image stacks exhibited stretching in the z-direction that was corrected for by performing the same analysis on spherical 1 μm amino beads (Polysciences, Warrington, PA, USA) coated with Alexa Fluor 568 NHS esters (Life Technologies, Grand Island, NY, USA). After finding 3D meshes for 8 beads, the aspect ratios of these beads in the xz and yz directions were averaged to find that the z axis was stretched by a factor of 1.78. We therefore scaled the z-coordinates of all cell meshes by a factor of 0.56 to compress the cells in the z direction. The surface areas and volumes of the resulting meshes were calculated using geom3d MATLAB software (Legland, 2009).
Supplementary Material
Acknowledgements
We would like to thank members of the Theriot lab as well as Lucy Shapiro, KC Huang, and members of their labs for technical assistance and intellectual feedback. We are particularly grateful to members of the Shapiro lab as well as Christine Jacobs-Wagner’s lab for providing strains. Thanks also to Xiaoxue Zhou for help with 3D imaging and analysis. Sarah Carden, Alexandre Colavin, and KC Huang provided helpful comments on the manuscript. This work was supported by NIH Training Grant T32-GM008294 (Molecular Biophysics), a National Science Foundation (NSF) Graduate Research Fellowship to L.K.H., NIH ROI AI067712, and the Howard Hughes Medical Institute. 3D deconvolution imaging was supported, in part, by Award Number 1S10OD01227601 from the National Center for Research Resources (NCRR).
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C. RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci. 2009;106:1239–1244. doi: 10.1073/pnas.0810794106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú FO, Hale CA, Bernhardt TG, Boer PAJ. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat Rev Micro. 2005;3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- Carballido-López R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Charbon G, Cabeen MT, Jacobs-Wagner C. Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin. Genes Dev. 2009;23:1131–1144. doi: 10.1101/gad.1795509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavin A, Hsin J, Huang KC. Effects of polymerization and nucleotide identity on the conformational dynamics of the bacterial actin homolog MreB. Proc Natl Acad Sci U S A. 2014;111:3585–3590. doi: 10.1073/pnas.1317061111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Graumann PL. Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB. Mol Microbiol. 2006;62:1340–1356. doi: 10.1111/j.1365-2958.2006.05457.x. [DOI] [PubMed] [Google Scholar]
- Doi M, Wachi M, Ishino F, Tomioka S, Ito M, Sakagami Y, et al. Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli cells. J Bacteriol. 1988;170:4619–4624. doi: 10.1128/jb.170.10.4619-4624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domónguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Söldner R, Carballido-López R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Fisher IC, Shapiro L, Theriot JA. Mutations in the nucleotide binding pocket of MreB can alter cell curvature and polar morphology in Caulobacter. Mol Microbiol. 2011;81:368–394. doi: 10.1111/j.1365-2958.2011.07698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. van den. [DOI] [PubMed] [Google Scholar]
- Ent F, Izoré T, Bharat TA, Johnson CM, Löwe J. Bacterial actin MreB forms antiparallel double filaments. eLife. 2014;3:e02634. doi: 10.7554/eLife.02634. van den. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favini-Stabile S, Contreras-Martel C, Thielens N, Dessen A. MreB and MurG as scaffolds for the cytoplasmic steps of peptidoglycan biosynthesis. Environ Microbiol. 2013;15:3218–3228. doi: 10.1111/1462-2920.12171. [DOI] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci U S A. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Goley ED, Dye NA, Werner JN, Gitai Z, Shapiro L. Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol Cell. 2010;39:975–987. doi: 10.1016/j.molcel.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking J, Priyadarshini R, Takacs CN, Costa T, Dye NA, Shapiro L, et al. Osmolality-dependent relocation of penicillin-binding protein PBP2 to the division site in Caulobacter crescentus. J Bacteriol. 2012;194:3116–3127. doi: 10.1128/JB.00260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev MMBR. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S-TD, Behrens C, Cabrita LD, Dobson CM. 1H, 15N and 13C assignments of yellow fluorescent protein (YFP) Venus. Biomol NMR Assign. 2009;3:67–72. doi: 10.1007/s12104-009-9143-y. [DOI] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Iwai N, Nagai K, Wachi M. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci Biotechnol Biochem. 2002;66:2658–2662. doi: 10.1271/bbb.66.2658. [DOI] [PubMed] [Google Scholar]
- Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJ, Carballido-López R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol. 2009;71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc Natl Acad Sci. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL. What size should a bacterium be? A question of scale. Annu Rev Microbiol. 1996;50:317–348. doi: 10.1146/annurev.micro.50.1.317. [DOI] [PubMed] [Google Scholar]
- Kroon D-J. Snake?: Active Contour - File Exchange - MATLAB Central. 2010 http://www.mathworks.com/matlabcentral/fileexchange/file_infos/28149-snake---active-contour. Accessed August 19, 2014.
- Kruse T, Bork-Jensen J, Gerdes K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol. 2005;55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- Kruse T, Møller-Jensen J, Løbner-Olesen A, Gerdes K. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver M, Errington J. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol. 2005;57:1196–1209. doi: 10.1111/j.1365-2958.2005.04736.x. [DOI] [PubMed] [Google Scholar]
- Lee J-C, Cha J-H, Zerbv DB, Stewart GC. Heterospecific expression of the Bacillus subtilis cell shape determination genes mreBCD in Escherichia coli. Curr Microbiol. 2003;47:146–152. doi: 10.1007/s00284-002-3937-9. [DOI] [PubMed] [Google Scholar]
- Legland D. geom3d - File Exchange - MATLAB Central. 2009 http://www.mathworks.com/matlabcentral/fileexchange/file_infos/24484-geom3d. Accessed August 18, 2014.
- Meisenzahl AC, Shapiro L, Jenal U. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J Bacteriol. 1997;179:592–600. doi: 10.1128/jb.179.3.592-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, Karczmarek A, Crouvoisier M, Bouhss A, Mengin-Lecreulx D, Blaauwen T. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol Microbiol. 2007;65:1106–1121. doi: 10.1111/j.1365-2958.2007.05851.x. den. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp D, Narita A, Maeda K, Fujisawa T, Ghoshdastider U, Iwasa M, et al. Filament structure, organization, and dynamics in MreB sheets. J Biol Chem. 2010;285:15858–15865. doi: 10.1074/jbc.M109.095901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner LD, Eswaramoorthy P, Ramamurthi KS, Weibel DB. Studying biomolecule localization by engineering bacterial cell wall curvature. PloS One. 2013;8:e84143. doi: 10.1371/journal.pone.0084143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje J, den Ent F, Boer P, Löwe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43:478–487. doi: 10.1016/j.molcel.2011.07.008. van. de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaechter M, Maaløe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Shiomi D, Sakai M, Niki H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 2008;27:3081–3091. doi: 10.1038/emboj.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swulius MT, Jensen GJ. The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-Terminal yellow fluorescent protein tag. J Bacteriol. 2012;194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C. MreB Drives De Novo Rod Morphogenesis in Caulobacter crescentus via Remodeling of the Cell Wall. J Bacteriol. 2010;192:1671–1684. doi: 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. van. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2011;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, Ouzounov N, et al. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc Natl Acad Sci U S A. 2014;111:E1025–1034. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Young KD. In Escherichia coli, MreB and FtsZ direct the synthesis of lateral cell wall via independent pathways that require PBP 2. J Bacteriol. 2009;191:3526–3533. doi: 10.1128/JB.01812-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats P, Rothfield L. Duplication and segregation of the actin (MreB) cytoskeleton during the prokaryotic cell cycle. Proc Natl Acad Sci U S A. 2007;104:17795–17800. doi: 10.1073/pnas.0708739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmer B, Heinemann M. Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PloS One. 2011;6:e23126. doi: 10.1371/journal.pone.0023126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol. 1987;169:4935–4940. doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jones BD, Brun YV. A set of ftsZ mutants blocked at different stages of cell division in Caulobacter. Mol Microbiol. 2001;40:347–360. doi: 10.1046/j.1365-2958.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- White CL, Kitich A, Gober JW. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol. 2010;76:616–633. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- Woldringh CL, Jong MA, Berg W, Koppes L. Morphological analysis of the division cycle of two Escherichia coli substrains during slow growth. J Bacteriol. 1977;131:270–279. doi: 10.1128/jb.131.1.270-279.1977. de. van den. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky A, Pritchard RH. Changes in cell size and shape associated with changes in the replication time of the chromosome of Escherichia coli. J Bacteriol. 1973;114:824–837. doi: 10.1128/jb.114.2.824-837.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.