Abstract
Clefts of the lip and palate are thought to be caused by genetic and environmental insults but the role of epigenetic mechanisms underlying this common birth defect are unknown. We analyzed the expression of over 600 microRNAs in the murine medial nasal and maxillary processes isolated on GD10.0-GD11.5 to identify those expressed during development of the upper lip and analyzed spatial expression of a subset. A total of 169 microRNAs were differentially expressed across gestation days 10.0 to 11.5 in the medial nasal processes, and 77 in the maxillary processes of the first branchial arch with 49 common to both. Of the microRNAs exhibiting the largest percent increase in both facial processes were 5 members of the Let-7 family. Among those with the greatest decrease in expression from GD10.0 to GD11.5 were members of the microRNA-302/367 family that have been implicated in cellular reprogramming. The distribution of expression of microRNA-199a-3p and Let-7i was determined by in situ hybridization and revealed widespread expression in both medial nasal and maxillary facial processes while that for microRNA-203 was much more limited. MicroRNAs are dynamically expressed in the tissues that form the upper lip and several were identified that target mRNAs known to be important for its development, including those that regulate the two main isoforms of p63 (microRNA-203 and microRNA-302/367 family). Integration of these data with corresponding proteomic data sets will lead to a greater appreciation of epigenetic regulation of lip development and provide a better understanding of potential causes of cleft lip.
Keywords: microRNA, cleft lip, p63, craniofacial development, mouse
Introduction
Orofacial clefts (cleft lip with or without cleft palate, CL/P, or cleft palate only, CPO) occur with a frequency as high as 1 in 700 live births and are the most prevalent birth defects affecting humans (Stuppia et al., 2011, Rahimov et al., 2012). CL/P and CPO are distinct developmental defects with different underlying causes. The secondary palate is formed from bilateral projections originating from the oral aspect of each maxillary processes and is characterized by initial vertical growth followed by reorientation and fusion to form the continuous palate separating the oral and nasal cavities. In humans, this occurs between weeks 7–10 of gestation. Failure of the palatal processes to reorient or fuse will result in a cleft palate. The upper lip, however, is formed by the bilateral fusion of the maxillary process (MxP) of the first branchial arch (BA1) with the medial nasal process (MNP) (Fig. 1) and occurs in humans during week 4 of gestation (reviewed in (Jiang et al., 2006)). Failure of fusion between these facial processes will result in either unilateral or bilateral cleft lip. In the human, the lateral nasal process develops into the ala of the nose and failure of fusion between this process and the maxilla may result in the rare oblique facial cleft (Eppley et al., 2005). Gene knockout mouse models for CL/P have demonstrated the importance of Wnt9b and FGF (Jin et al., 2012), p63 (Ferretti et al.), Pax9 and Msx1 (Nakatomi et al., 2010), and IRF6 (Ingraham et al., 2006) in development of the upper lip. Few studies, however, have addressed the potential involvement of epigenetic factors, such as expression and function of microRNAs (miRNAs) in facial development, and most of these have focused on the developing secondary palate (Powder et al., 2012, Mukhopadhyay et al., 2010, Gatto et al., 2010, Gessert et al., 2010, Eberhart et al., 2008).
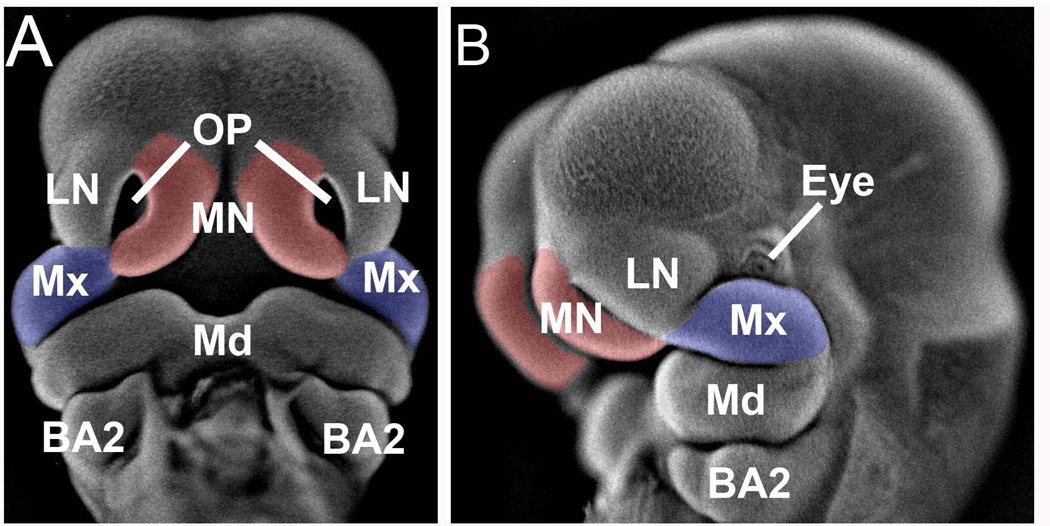
Figure 1. Facial prominences dissected for miRNA expression analysis.
Shown is a GD11.5 mouse fetus that has been incubated with DAPI and photographed under epi-fluorescence according to the method detailed in (Sandell et al., 2012). Panel A, frontal view and panel B, lateral view. The medial nasal processes are indicated in red and the maxillary processes in blue. MN, medial nasal process; LN, lateral nasal process; Mx, maxillary process of the first branchial arch; OP, olfactory pit; Md, mandibular process of first branchial arch; BA2, second branchial arch.
While multiple molecular mechanisms underlie epigenetic regulatory networks, one of the most actively investigated to date is the action of miRNAs. MiRNAs regulate expression of genes post-transcriptionally by binding to and then inhibiting the translation of, and/or destabilizing, their target mRNAs (Bartel, 2009, Bazzini et al., 2012, Djuranovic et al., 2012).
Currently there are over 2,000 known human miRNAs and 1281 identified in mice, according to release 19 of miRBase (mirbase.org). In the current study, we describe miRNA temporal expression profiles during the development of the murine medial nasal and maxillary processes of the first branchial arch. These data form the foundation necessary to understand the complex epigenetic regulatory networks governing normal and abnormal development of the upper lip.
Materials and Methods
Animals
Hsd:ICR (CD-1®) breeder mice were purchased from Harlan Laboratories, Inc. (Indianapolis, IN) and housed at an ambient temperature of 22 °C with a 12 h on/12h off light cycle and access to food and water ad libitum. Timed matings were achieved by caging one mature male and two nulliparous female mice. For gestation day (GD) 10.5 and 11.5, overnight matings (6 p.m to 9 a.m.) were set up. Females were examined for a vaginal plug and this was designated as GD 0.5. For GD 10.0 and 11.0, a short, two-hour mating window was set up between 11 a.m and 1 p.m. For these, detection of a vaginal plug was designated as GD 0. Dams were euthanized by carbon dioxide asphyxiation/cervical dislocation and embryos isolated by cesarean section and immediately placed in ice-cold PBS. All procedures for the humane use and handling of mice were approved by the University of Louisville Institutional Animal Care and Use Committee and encompass guidelines as set out in the EC Directive 86/609/EEC for animal experimentation.
Fetal staging
Because there can be significant variation in the extent of development among littermates, it was necessary to operationally define the stages of lip development. The criterion by which we defined each gestation day is detailed in Table 1. Tail somites were determined from the position of the hind limb, which forms between somites 23–28, therefore, somite 29 was taken as tail somite no. 1 and used as a measure of total number of somite pairs (Slack, 2012).
TABLE 1.
Staging of mouse fetuses
| Gestation Day | Theiler Stage | Tail Somites | Total Somites |
|---|---|---|---|
| 10.0 | 16 | 2–6 | 30–34 |
| 10.5 | 17 | 7–11 | 35–39 |
| 11.0 | 18 | 12–16 | 40–44 |
| 11.5 | 19 | 17–19 | 45–47 |
Mouse fetuses were staged according to the number of tail somites and are divided into four developmental time points (GD10.0-GD11.5). For comparison, the total number of somites and the corresponding Theiler stage is also provided.
Isolation of facial processes
Following rinses in ice-cold PBS, embryos were immediately placed in RNAlater (Qiagen, Valencia, CA) and stored at −20°C. Dissections were then performed directly in RNAlater solution. The bilateral maxillary processes of the first branchial arch and the medial nasal processes were dissected with a tungsten needle and scalpel and maintained in RNAlater solution until processed for RNA purification.
RNA isolation/cDNA synthesis
Total RNA was purified using the mirVana miRNA isolation kit (Life Technologies, Carlsbad, CA) and cDNAs prepared using Megaplex™ reverse transcription primers (rodent pool A, v2 and pool B, v3) and the Taqman microRNA reverse transcription kit (Life Technologies). Real-time PCR reactions consisted of 1X Taqman PCR master mix (Life Technologies) and 6 µl cDNA in a final volume of 900 µl. One-hundred µl was applied to each of 8 ports of a TaqMan array rodent microRNA microfluidic card (Life Technologies). For each sample, an “A” and “B” card was used. The “A” card contained 335 miRNAs unique to the mouse and the “B” card, 306 mouse-specific miRNAs for a combined 641 miRNAs. The remainder was unique to rat. There were also three endogenous controls (snoRNA135, snoRNA202 and U6) and one negative control (ath-miR159a). The cards were analyzed using the ViiA7 real-time PCR system (Life Technologies) and data were reduced as detailed below.
In situ hybridization
Fetuses were isolated on GD11.5 and fixed overnight in 4% paraformaldehyde (PFA) (dissolved in PBS) and dehydrated through a graded methanol series. Fetuses were digested with 10 µg/ml proteinase K (Roche, Indianapolis, IN) for 45 min at 37°C and post fixed in PBT (PBS + 0.1% Tween-20) that contained 4% PFA and 0.2% glutaraldehyde (Fisher Scientific, Waltham, MA). Samples were acetylated with 0.1M triethanolamine, pH 8.0 and 0.25% (v/v) acetic anhydride (each from Sigma Chemical Co., St. Louis, MO) for 10 min at room temperature. Fetuses were then hybridized with digoxigenin-labelled locked nucleic acid (LNA) probes purchased from Exiqon, Inc. (Woburn, MA) and used at 10nM, 30°C below the calculated RNA melting temperature (Tm) for 24h. From this point, fetuses were processed as described by Kloosterman et al. (Kloosterman et al., 2006). A control, scrambled LNA probe (Exiqon, Inc.) was included for each experiment.
Data analysis
Three independent, biological replicates for each tissue were used for final determination of the data where each was processed and loaded onto the paired “A” and “B” cards. The experimental design consisted of four time-points (GD 10.0, 10.5, 11.0, and 11.5), two tissue types (MNP and MxP), and three replicates for a total of 24 arrays for each of the “A” and “B” cards. All Ct values declared as “undetermined (Ct values >40) were treated as missing and raw Ct values for each card were first processed in the following manner: All miRNAs with missing values in greater than 50% of the arrays were removed, after ensuring that missing values were not occurring solely within certain experimental groups. Any remaining missing values were subsequently imputed using k-nearest-neighbors imputation (Troyanskaya et al., 2001). Finally, Ct values on each array were normalized by subtracting the global median of the array. This normalization method was selected after trying alternative normalization methods based on subtracting control RNAs (e.g., snoRNA135) and determined to provide the most stable measurement of miRNA expression across all samples. Normalized Ct values were then analyzed to determine differential expression between time points within each tissue using a linear regression model as implemented in the Bioconductor package limma (Gentleman et al., 2004, Smyth, 2005). To maximize power, one model for each miRNA was fit to the entire set of 24 arrays and linear trends were tested separately within each tissue type. To account for the paired nature of the data (i.e. the same tissue was used to produce one MNP and one MxP sample), we included tissue source as a blocking factor in the analysis and fit a generalized least squares model after estimating the correlation between paired samples using the duplicateCorrelation function in limma. P-values were adjusted for multiple comparisons using the Benjamini-Hochberg method to control the overall false discovery rate at the 5% level (Benjamini & Hochberg, 1995).
Single-tube real time PCR assays
To validate the results of the array cards, the expression of a selected set of miRNAs was independently determined by real-time PCR (Taqman, Life Technologies). cDNAs were synthesized as detailed above and the expression of individual miRNAs was determined using pre-designed probe:primer sets from Life Technologies. The expression of each miRNA tested was normalized to that for SnoRNA135 by the ΔΔCt method (Livak & Schmittgen, 2001).
Results
To determine the expression of miRNAs involved in development of the upper lip in mice, the bilateral medial nasal processes (MNPs) and maxillary processes (MxPs) (Fig. 1), contributors to the formation of the upper lip, were microdissected from three independent litters and total RNA extracted and used to synthesize cDNAs from expressed miRNAs. Fetuses were staged according to somite number and grouped into four gestational days (GD10, 10.5, 11 and 11.5) (see Table 1) for analysis. The expression of 641 murine miRNAs was assayed by PCR microarray (see Methods) from the MXP and MNP from three independent litters of 4–5 fetuses from each of GD10.0, 10.5, 11.0, and 11.5. While constitutively expressed miRNAs were noted during the developmental time frame examined (defined as adjusted p-values for linear trend >0.05 from GD10.0–11.5, see additional files 1 [Table S1] and 2 [Table S2]), focus was directed toward the identification of miRNAs that exhibited differential expression (defined as an adjusted p-value for linear trend <0.05 across GDs 10.0 to 11.5). A linear model (rather than an ANOVA) was utilized to detect miRNAs that exhibited a linear trend over the time course examined. Analysis of the data using ANOVA resulted in a virtually identical list of genes (80–90% overlap). The linear model actually detected more genes exhibiting significant changes in expression due, in part, to greater power to detect a linear trend. Moreover, the vast majority of genes exhibited a linear trend over the time course examined.
Many miRNAs were classified as “not detected”, defined as those with an average Ct value >35 in each of the GDs examined (see additional files 3 [Table S3] and 4 [Table S4). Careful examination of the expression of the 641 miRNAs on our array cards revealed not a single instance of a miRNA that was not expressed on GD10.0, but then expressed on any/all of the subsequent days of gestation. This provided justification for using GD10 as our reference point (temporal changes relative to GD10).
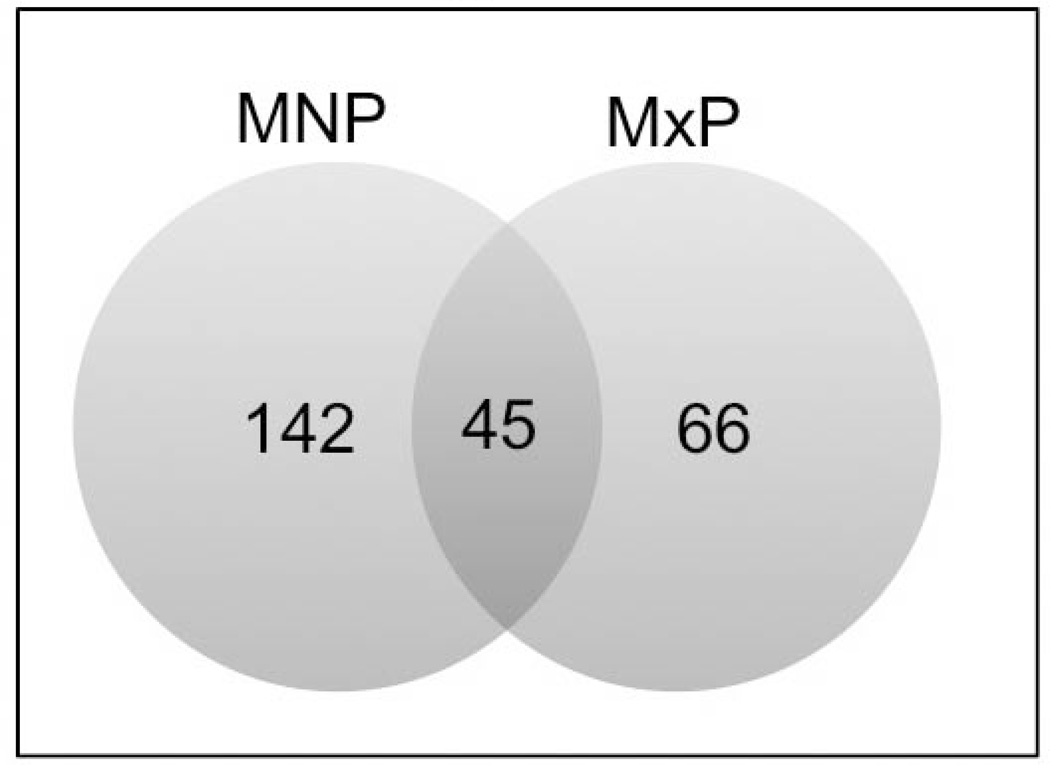
The miRNAs in the MNP meeting the criteria of differential expression (defined as an adjusted p-value for linear trend <0.05 across GDs 10.0 to 11.5) are reported in Table 2 and those in the MxP are reported in Table 3. In each table, the miRNAs are ordered according to p-value and separated into those that are up-regulated from GD10.0 to 11.5 and then those that are down-regulated. As shown in Table 2, 142 miRNAs, whose expression ranged from a 13-fold decrease to an over 12-fold increase on GD11.5 compared to GD10.0 were identified in the MNPs. Interestingly, of the top 10 miRNAs exhibiting the largest increase in expression, 5 were members of the Let-7 family (Let-7i, -7a, -7c, -7e, and -7g). Two others (Let-7d and Let-7f) were expressed >3.4-fold. Thus, 7 of the 12 member Let-7 family exhibited a significant increase in expression during development of the MNP. Of the 10 miRNAs exhibiting the largest fold-decrease in expression in the MNPs were 4 members of the 9 member miR-302/367 cluster. The number of expressed miRNAs that were identified in the MxP (66) was significantly less than the total number differentially expressed in the MNP. Nevertheless, as with the MNP, members of the Let-7 family (Let-7i, -7d, -7a, -7g, and -7c) were highly represented among miRNAs in the MxP exhibiting the greatest increase in expression. Let-7e and Leg-7f were also significantly increased (>2.2-fold). Similar to expression data from the MNPs, miRNAs exhibiting the greatest fold-decrease in expression from GD10.0-GD11.5 were members of the miR-302/367 cluster (302-a, -b, -c, -d, and 367). The Venn diagram in Fig. 2 illustrates that 45 differentially-expressed miRNAs were common to both facial processes, 97 were unique to the MNP and 21 were unique to the MxP. The list of miRNAs in each of these categories is also reported in additional files 5–7 (Table S5–Table S7, respectively).
TABLE 2.
Differential expression of miRNAs in the medial nasal processes from GD10.0-GD11.5 mouse fetuses
| miRNA | GD 10.5 | GD 11.0 | GD 11.5 | Adjusted P-value |
|---|---|---|---|---|
| Increased Expression vs. GD10.0 | ||||
| miR-24 | 1.78 | 3.16 | 5.58 | <0.001 |
| let-7i | 2.68 | 10.06 | 12.22 | <0.001 |
| miR-203 | 2.47 | 6.58 | 7.64 | <0.001 |
| let-7c | 2.65 | 4.78 | 8.16 | <0.001 |
| miR-199a-3p | 2.80 | 5.19 | 5.93 | <0.001 |
| let-7g | 2.48 | 6.06 | 6.46 | <0.001 |
| miR-429 | 2.35 | 3.17 | 4.62 | <0.001 |
| miR-200a | 1.74 | 2.17 | 2.52 | <0.001 |
| miR-27b | 2.42 | 4.77 | 4.83 | <0.001 |
| let-7e | 3.62 | 5.62 | 6.96 | <0.001 |
| miR-145 | 1.95 | 3.13 | 3.75 | <0.001 |
| miR-200b | 1.96 | 2.52 | 3.12 | <0.001 |
| miR-27a | 2.45 | 3.34 | 4.42 | <0.001 |
| let-7d | 1.39 | 3.49 | 3.91 | <0.001 |
| let-7a | 4.41 | 8.99 | 10.19 | <0.001 |
| miR-199a-5p | 2.11 | 4.52 | 4.20 | <0.001 |
| miR-140 | 1.43 | 1.78 | 2.94 | <0.001 |
| miR-26a | 1.40 | 1.97 | 1.91 | <0.001 |
| miR-182 | 1.80 | 2.35 | 3.19 | <0.001 |
| miR-141 | 1.91 | 2.89 | 3.38 | <0.001 |
| miR-205 | 2.08 | 4.01 | 3.91 | <0.001 |
| miR-708 | 1.65 | 1.95 | 2.68 | <0.001 |
| miR-322 | 1.76 | 2.60 | 2.56 | <0.001 |
| miR-30e | 1.17 | 1.40 | 1.74 | <0.001 |
| miR-143 | 1.98 | 3.40 | 2.84 | <0.001 |
| miR-183 | 2.27 | 3.06 | 3.59 | <0.001 |
| miR-23b | 0.94 | 2.25 | 3.48 | <0.001 |
| miR-200c | 1.55 | 1.77 | 2.49 | <0.001 |
| miR-152 | 1.46 | 1.88 | 2.03 | <0.001 |
| miR-199b | 3.78 | 5.72 | 6.17 | <0.001 |
| miR-24-2* | 1.57 | 3.47 | 4.51 | <0.001 |
| miR-125b-5p | 1.32 | 2.49 | 2.96 | <0.001 |
| miR-322* | 2.76 | 3.27 | 5.26 | <0.001 |
| let-7f | 1.28 | 2.29 | 3.42 | 0.001 |
| miR-301a | 1.55 | 1.99 | 1.89 | 0.001 |
| miR-301b | 1.37 | 1.72 | 1.75 | 0.002 |
| miR-148a | 2.06 | 2.44 | 2.40 | 0.002 |
| miR-224 | 1.54 | 2.38 | 3.55 | 0.002 |
| miR-532-5p | 1.44 | 1.56 | 1.62 | 0.003 |
| miR-1839-5p | 1.59 | 2.20 | 2.43 | 0.003 |
| miR-214 | 1.29 | 2.03 | 2.91 | 0.003 |
| miR-676 | 1.14 | 1.90 | 2.30 | 0.004 |
| miR-21 | 2.75 | 4.58 | 3.26 | 0.004 |
| miR-195 | 1.19 | 1.71 | 1.75 | 0.008 |
| miR-138 | 1.52 | 2.00 | 2.57 | 0.008 |
| miR-31* | 1.48 | 2.25 | 2.38 | 0.009 |
| miR-1839-3p | 1.22 | 1.73 | 2.15 | 0.014 |
| miR-30c | 1.28 | 1.30 | 1.72 | 0.014 |
| miR-7b | 2.71 | 3.20 | 2.72 | 0.015 |
| miR-16 | 1.21 | 1.30 | 1.54 | 0.015 |
| miR-30b | 1.23 | 1.52 | 1.48 | 0.015 |
| miR-872* | 1.21 | 1.65 | 1.93 | 0.016 |
| miR-96 | 1.42 | 3.43 | 3.29 | 0.017 |
| miR-28 | 1.70 | 1.73 | 1.73 | 0.017 |
| miR-133a | 1.88 | 1.97 | 2.75 | 0.022 |
| miR-28* | 1.45 | 1.53 | 2.49 | 0.023 |
| miR-503* | 1.84 | 1.97 | 2.36 | 0.026 |
| miR-181c | 1.10 | 2.00 | 2.05 | 0.029 |
| miR-872 | 1.64 | 1.76 | 1.70 | 0.031 |
| miR-340-3p | 1.45 | 1.48 | 1.70 | 0.035 |
| miR-25 | 1.73 | 1.75 | 1.81 | 0.041 |
| miR-669l | 2.68 | 3.10 | 3.22 | 0.046 |
| Decreased expression vs. GD 10.0 | ||||
|
miR-323-3p |
1.31 | 2.54 | 2.92 | <0.001 |
| miR-367 | 2.61 | 6.09 | 13.02 | <0.001 |
| miR-292-3p | 1.57 | 3.11 | 6.96 | <0.001 |
| miR-409-3p | 1.76 | 2.93 | 2.83 | <0.001 |
| miR-302d | 1.41 | 3.36 | 8.55 | <0.001 |
| miR-335-3p | 1.28 | 2.17 | 3.22 | <0.001 |
| miR-376b | 1.46 | 2.89 | 3.68 | <0.001 |
| miR-494 | 1.60 | 2.49 | 4.03 | <0.001 |
| miR-376a | 1.48 | 2.20 | 2.90 | <0.001 |
| miR-495 | 1.21 | 2.06 | 2.82 | <0.001 |
| miR-370 | 1.26 | 2.23 | 2.52 | <0.001 |
| miR-410 | 1.54 | 2.30 | 2.83 | <0.001 |
| miR-431 | 1.38 | 2.09 | 2.26 | <0.001 |
| miR-9 | 1.75 | 4.28 | 6.48 | <0.001 |
| miR-543 | 1.20 | 2.02 | 3.18 | <0.001 |
| miR-302b | 1.91 | 3.57 | 10.99 | <0.001 |
| miR-302a | 1.24 | 3.88 | 4.01 | <0.001 |
| miR-127 | 1.38 | 1.74 | 1.94 | <0.001 |
| miR-434-3p | 1.37 | 2.27 | 2.19 | <0.001 |
| miR-129-3p | 2.84 | 4.54 | 8.20 | <0.001 |
| miR-667 | 1.86 | 2.66 | 3.37 | <0.001 |
| miR-135b | 1.41 | 2.22 | 3.43 | <0.001 |
| miR-433 | 2.07 | 3.59 | 2.92 | <0.001 |
| miR-302c | 1.51 | 4.32 | 7.34 | <0.001 |
| miR-221 | 1.43 | 2.16 | 3.36 | <0.001 |
| miR-20b | 1.25 | 1.93 | 2.24 | <0.001 |
| miR-485-3p | 1.33 | 2.42 | 2.99 | <0.001 |
| miR-135a | 1.32 | 3.23 | 5.55 | 0.001 |
| miR-539 | 1.41 | 2.06 | 2.03 | 0.001 |
| miR-411 | 1.22 | 1.61 | 1.60 | 0.001 |
| miR-487b | 1.24 | 1.69 | 2.12 | 0.002 |
| miR-291a-3p | 1.30 | 1.62 | 2.78 | 0.002 |
| miR-293 | 1.47 | 2.29 | 3.39 | 0.002 |
| miR-132 | 2.06 | 1.96 | 2.42 | 0.002 |
| miR-509-3p | 2.28 | 2.96 | 4.05 | 0.002 |
| miR-376c | 1.08 | 1.73 | 1.85 | 0.002 |
| miR-337-5p | 1.38 | 1.92 | 2.17 | 0.003 |
| miR-540-3p | 1.71 | 2.13 | 2.44 | 0.004 |
| miR-380-5p | 1.04 | 1.75 | 1.73 | 0.004 |
| miR-134 | 1.36 | 1.87 | 1.89 | 0.005 |
| miR-382 | 1.12 | 1.30 | 1.75 | 0.005 |
| miR-369-5p | 2.19 | 2.64 | 3.29 | 0.005 |
| miR-222 | 1.29 | 2.12 | 2.04 | 0.005 |
| miR-92a | 1.26 | 1.71 | 1.63 | 0.006 |
| miR-363 | 1.47 | 2.64 | 3.87 | 0.007 |
| miR-1905 | 1.71 | 2.03 | 3.85 | 0.008 |
| miR-712 | 0.86 | 2.35 | 2.78 | 0.008 |
| miR-434-5p | 1.28 | 1.57 | 2.14 | 0.008 |
| miR-2135 | 1.39 | 1.98 | 2.74 | 0.009 |
| miR-696 | 1.78 | 2.93 | 2.24 | 0.01 |
| miR-665 | 2.56 | 3.98 | 3.00 | 0.011 |
| miR-331-5p | 1.55 | 2.86 | 2.11 | 0.013 |
| miR-376a* | 1.03 | 1.96 | 2.26 | 0.014 |
| miR-1193 | 0.95 | 1.40 | 2.43 | 0.016 |
| miR-295 | 0.82 | 2.85 | 2.70 | 0.019 |
| miR-493 | 1.07 | 2.82 | 2.69 | 0.019 |
| miR-337-3p | 1.31 | 1.93 | 1.99 | 0.019 |
| miR-544 | 1.54 | 2.28 | 3.75 | 0.02 |
| miR-486 | 1.82 | 2.85 | 2.63 | 0.02 |
| miR-380-3p | 1.60 | 1.74 | 2.05 | 0.025 |
| miR-383 | 2.27 | 5.40 | 2.97 | 0.025 |
| miR-217 | 1.25 | 2.04 | 2.19 | 0.025 |
| miR-1897-5p | 1.72 | 2.20 | 2.43 | 0.025 |
| miR-465c-5p | 2.50 | 3.66 | 4.45 | 0.026 |
| miR-2182 | 0.97 | 1.45 | 2.35 | 0.03 |
| miR-1971 | 1.80 | 2.09 | 2.01 | 0.033 |
| miR-412 | 1.04 | 1.43 | 2.37 | 0.033 |
| miR-1896 | 2.00 | 1.66 | 2.33 | 0.033 |
| miR-18a* | 1.53 | 2.07 | 1.97 | 0.033 |
| miR-669n | 1.62 | 2.38 | 7.85 | 0.036 |
| miR-124 | 1.36 | 2.90 | 2.59 | 0.037 |
| miR-379 | 1.19 | 1.37 | 1.42 | 0.038 |
| miR-1960 | 1.54 | 1.93 | 2.01 | 0.04 |
| miR-216b | 1.22 | 1.74 | 2.18 | 0.041 |
| miR-694 | 1.81 | 1.83 | 2.37 | 0.043 |
| miR-18b | 1.39 | 2.75 | 2.06 | 0.044 |
| miR-298 | 1.21 | 1.32 | 1.48 | 0.044 |
| miR-1191 | 0.85 | 1.84 | 1.70 | 0.045 |
| miR-335-5p | 1.07 | 1.19 | 1.65 | 0.048 |
| miR-692 | 1.00 | 1.35 | 3.12 | 0.048 |
The bilateral medial nasal processes from three independent litters of GD10.0, 10.5, 11.0, and 11.5 mouse fetuses were dissected, RNA purified, cDNAs synthesized, and analyzed by real-time PCR for the expression of 641 miRs unique to mouse. Values are fold-change relative to the level of expression on GD10.0. P-values indicate the p-value for overall linear trend as determined using general linear regression modelling in the limma package.
TABLE 3.
Differential expression of miRNAs in the maxillary processes from GD10.0-GD11.5 mouse fetuses
| miRNA | GD 10.5 | GD 11.0 | GD 11.5 | Adjusted P-value |
|---|---|---|---|---|
| Increased expression vs. GD 10.0 | ||||
| let-7i | 1.67 | 6.69 | 11.72 | <0.001 |
| let-7d | 1.87 | 6.44 | 9.27 | <0.001 |
| let-7g | 1.96 | 4.13 | 5.61 | <0.001 |
| miR-203 | 2.08 | 4.29 | 4.51 | <0.001 |
| let-7c | 1.74 | 3.23 | 4.58 | <0.001 |
| miR-218 | 1.35 | 2.25 | 2.79 | <0.001 |
| miR-27b | 1.52 | 2.82 | 3.54 | <0.001 |
| miR-125b-5p | 1.39 | 3.23 | 4.13 | <0.001 |
| miR-145 | 1.13 | 2.05 | 2.59 | <0.001 |
| let-7a | 1.79 | 6.11 | 6.73 | <0.001 |
| let-7e | 1.56 | 2.77 | 3.60 | <0.001 |
| miR-181a | 1.51 | 2.19 | 3.15 | <0.001 |
| miR-143 | 1.12 | 2.03 | 2.59 | 0.002 |
| miR-199a-3p | 1.20 | 2.04 | 2.33 | 0.003 |
| miR-181c | 1.70 | 2.13 | 3.90 | 0.003 |
| miR-24 | 1.29 | 1.80 | 1.80 | 0.003 |
| miR-1839-5p | 1.67 | 2.26 | 2.75 | 0.003 |
| miR-184 | 1.19 | 3.43 | 7.13 | 0.004 |
| miR-322* | 1.80 | 3.72 | 3.74 | 0.004 |
| miR-199b | 1.71 | 3.31 | 4.03 | 0.005 |
| miR-214 | 1.52 | 2.24 | 3.06 | 0.006 |
| miR-148a | 1.16 | 1.94 | 2.01 | 0.007 |
| miR-672 | 1.22 | 1.70 | 2.65 | 0.007 |
| miR-26a | 1.08 | 1.44 | 1.50 | 0.008 |
| miR-205 | 1.60 | 2.88 | 2.50 | 0.008 |
| miR-23b | 1.63 | 2.67 | 2.70 | 0.01 |
| miR-24-2* | 2.37 | 2.61 | 3.70 | 0.013 |
| miR-27a | 1.20 | 1.90 | 1.98 | 0.015 |
| miR-322 | 1.12 | 2.50 | 3.12 | 0.017 |
| let-7f | 0.66 | 1.55 | 2.20 | 0.02 |
| miR-301a | 1.14 | 1.55 | 1.52 | 0.03 |
| miR-362-5p | 1.77 | 2.35 | 2.68 | 0.033 |
| miR-872* | 1.26 | 1.80 | 1.88 | 0.037 |
| miR-450B-3P | 0.61 | 2.32 | 2.45 | 0.038 |
| miR-497 | 1.08 | 1.57 | 1.84 | 0.038 |
| miR-301b | 1.15 | 1.46 | 1.45 | 0.039 |
| miR-224 | 0.66 | 1.67 | 2.03 | 0.041 |
| miR-676* | 1.74 | 2.62 | 3.27 | 0.046 |
| Decreased expression vs. GD10.0 | ||||
| miR-20b | 1.64 | 4.29 | 6.89 | <0.001 |
| miR-302a | 1.74 | 4.26 | 11.06 | <0.001 |
| miR-367 | 1.99 | 11.57 | 10.71 | <0.001 |
| miR-292-3p | 2.21 | 6.09 | 6.79 | <0.001 |
| miR-302d | 1.81 | 4.01 | 8.96 | <0.001 |
| miR-302b | 2.21 | 6.87 | 9.43 | <0.001 |
| miR-106a | 1.25 | 1.74 | 2.21 | <0.001 |
| miR-302c | 2.28 | 7.46 | 10.12 | <0.001 |
| miR-295 | 2.17 | 5.94 | 8.79 | <0.001 |
| miR-335-3p | 1.06 | 1.53 | 2.46 | <0.001 |
| miR-293 | 2.05 | 3.21 | 4.78 | <0.001 |
| miR-291a-3p | 1.16 | 2.31 | 2.96 | <0.001 |
| miR-296-3p | 1.46 | 2.57 | 2.75 | <0.001 |
| miR-375 | 1.27 | 3.44 | 3.57 | <0.001 |
| miR-17 | 1.19 | 1.37 | 1.80 | 0.002 |
| miR-19b | 1.07 | 1.48 | 1.65 | 0.003 |
| miR-687 | 2.42 | 6.40 | 5.36 | 0.005 |
| miR-139-5p | 0.89 | 1.72 | 2.12 | 0.005 |
| miR-19a | 1.06 | 1.43 | 1.73 | 0.007 |
| miR-493 | 1.03 | 3.50 | 3.50 | 0.007 |
| miR-685 | 1.78 | 2.79 | 2.80 | 0.007 |
| miR-294 | 2.25 | 2.10 | 4.29 | 0.009 |
| miR-92a | 1.23 | 1.60 | 1.68 | 0.009 |
| miR-696 | 1.44 | 2.18 | 2.31 | 0.022 |
| miR-216b | 0.67 | 1.74 | 2.20 | 0.024 |
| miR-546 | 1.32 | 1.70 | 3.91 | 0.038 |
| miR-449b | 1.01 | 1.40 | 1.77 | 0.05 |
| miR-449a | 0.78 | 2.02 | 3.05 | 0.05 |
The maxillary processes from three independent litters of GD10.0, 10.5, 11.0, and 11.5 mouse fetuses were dissected, RNA purified, cDNAs synthesized, and then assayed for the expression of 641 miRs unique to mouse by real-time PCR. The data are presented as fold-change (increase or decrease) relative to the level expessed on GD10.0. P-values indicate the p-value for overall linear trend as determined using general linear regression modelling in the limma package.
Figure 2. Venn diagram illustrating the number of miRNAs differentially regulated.
There were 143 and 67 miRNAs differentially regulated in the MNP (blue) and MxP (yellow), respectively. Of those, 46 miRNAs were common to both facial processes.
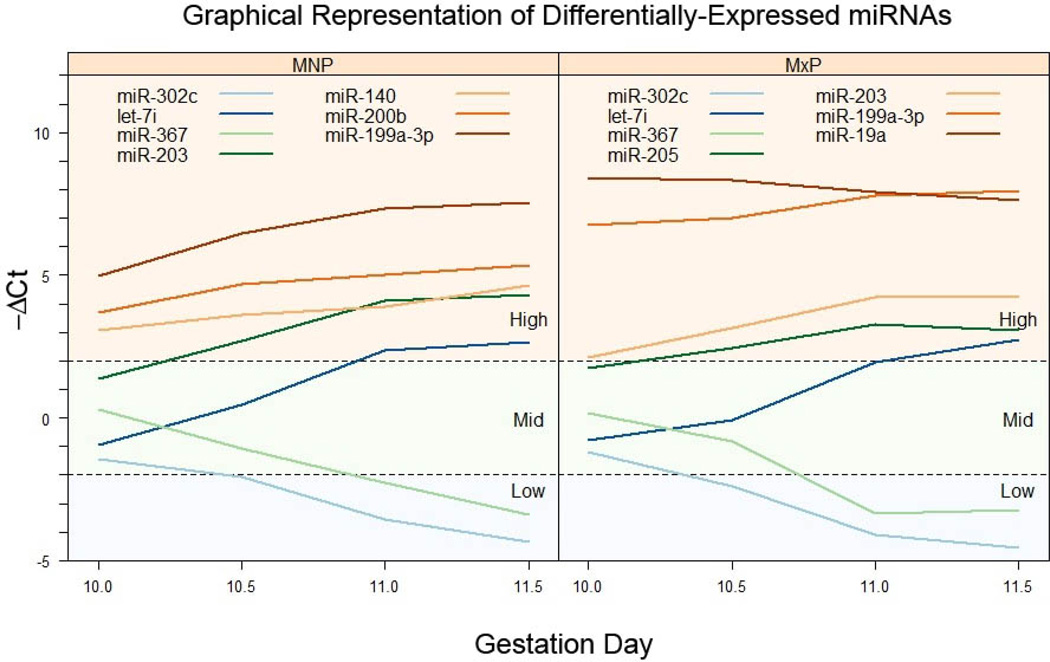
Because the expression level of individual miRNAs varied on GD10.0 from relatively low to relatively high, analyzing fold-change values alone (relative to GD10.0) may mask overall levels of expression on each gestation day. Therefore, we also report the expression of differentially-expressed miRNAs as shown in Figure 3. This provides a graphical representation of the data reported in Table 2 and Table 3 as changes in −ΔCt values from GD10.0 to GD11.5. (Note: all ΔCt values were multiplied by −1 so that an increase in expression was reflected as a line with a positive slope and decreased expression, a negative slope). The differentially-expressed miRNAs in the MNP and MxP were divided into three groups based on their expression level on GD10.0 as: low (−ΔCt < −2), mid (−ΔCt between −2 and 2), and high (−ΔCt >2) level expressers. When the data is presented in this manner, it can be seen that the expression of some miRNAs, despite having large fold-change values, are still expressed at a lower level than other miRNAs with small fold-changes, but higher initial concentrations. For example, in both tissues the expression of Let-7i increased ~12-fold from GD10.0 to GD11.5 compared to a 2–6-fold increase for miR-199a-3p (Tables 2 and 3). However, on GD10.0 the expression of miR-199a-3p was initially relatively high whereas the expression of Let-7i was classified as low (Fig. 3), with the end result that there was a higher concentration of miR-199a-3p than Let-7i in each tissue. Frequently, miRNAs with large fold-changes in expression are assumed to be more biologically relevant than those with more modest changes that actually yield a much higher concentration of miRNAs and possibly leading to a greater biological effect.
Figure 3. Longitudinal profiles of differentially-expressed miRNAs.
Differentially-expressed miRNAs in MNP (left panel) or MxP (right panel) tissue were categorized by their initial ΔCt values (those on GD10) as low, mid, or high expressers, as indicated in each panel. Each line represents the ΔCt value for a specific miRNA on GD10.0-GD11.5. For clarity, only a selected few miRNAs are shown on each category.
To independently validate the results of the microarray analysis, the expression of 10 miRNAs from each list was assayed by real-time PCR (Taqman). PCR results, reported in Table 4 (MNP) and Table 5 (MxP), were directionally consistent with those from the microarray cards. While some variability in the magnitude of changes in miRNA expression was seen, this is likely due to the differences in the amount of input cDNA between the array cards and the single tube assays.
TABLE 4.
Validation of PCR array results by independent Taqman assay (medial nasal processes)
| miRNA | GD10.5 | GD11.0 | GD11.5 |
|---|---|---|---|
| 203 | 3.9 ± 0.3 | 16.3 ± 0.8 | 15.1 ± 0.5 |
| Let-7i | 5.7 ± 1.1 | 10.2 ± 0.7 | 20.1 ± 1.0 |
| 367 | −1.8 ± 0.6 | −6.0 ± 0.8 | −8.3 ± 2.5 |
| 302a | −3.2 ± 0.2 | −4.3 ± 0.1 | −7.2 ± 0.6 |
| 20b | 1.0 ± 1.0 | −61.4 ± 0.1 | −89.9 ± 4.7 |
| 199a-3p | 5.1 ± 0.1 | 3.9 ± 0.2 | 6.5 ± 0.1 |
| 200b | 1.9 ± 0.3 | 2.3 ± 0.3 | 3.0 ± 0.3 |
| 292-3p | −2.6 ± 0.4 | −3.6 ± 0.4 | −4.9 ± 0.5 |
| 140 | 1.6 ± 0.2 | 1.7 ± 0.2 | 2.8 ± 0.2 |
| 127 | −2.4 ± 0.2 | −1.8 ± 0.3 | −3.1 ± 0.4 |
Total RNA was purified from the paired medial nasal processes of GD10.0, GD10.5, GD11.0, and GD11.5 mouse fetuses and cDNAs prepared as detailed in the Materials and Methods section. The expression of individual miRNAs was determined by real-time PCR and normalized to the expression of SnoRNA135. The data are presented as fold-change ± S.D. (n=3) relative to the level on GD10.0. Positive values are fold-increase and negative values are fold-decrease.
TABLE 5.
Validation of PCR array results by independent Taqman assay (maxillary processes)
| miRNA | GD10.5 | GD11.0 | GD11.5 |
|---|---|---|---|
| 203 | 2.6 ± 0.3 | 7.7 ± 0.4 | 8.3 ± 0.4 |
| Let-7i | 5.5 ± 1.4 | 13.6 ± 1.8 | 23.1 ± 0.1 |
| 367 | −3.1 ± 0.5 | −10.9 ± 1.4 | −17.5 ± 1.1 |
| 302a | −2.2 ± 0.4 | −3.6 ± 0.5 | −7.3 ± 0.7 |
| 20b | 1.0 ± 1.0 | −104.6 ± 3.9 | −254.2 ± 6.4 |
| 199a-3p | 1.6 ± 1.3 | 1.9 ± 0.3 | 2.1 ± 0.5 |
| 200b | 1.3 ± 0.6 | 1.8 ± 0.5 | 1.8 ± 0.4 |
| 292-3p | −1.6 ± 0.2 | −2.7 ± 0.2 | −3.3 ± 0.2 |
| 143 | 2.6 ± 0.4 | 3.6 ± 0.4 | 5.4 ± 0.4 |
Total RNA was purified from the maxillary processes of GD10.0, GD10.5, GD11.0, and GD11.5 mouse fetuses and analyzed as detailed in the Materials and Methods section and in the legend to Table 4. Positive values fold-increase and negative, fold-decrease.
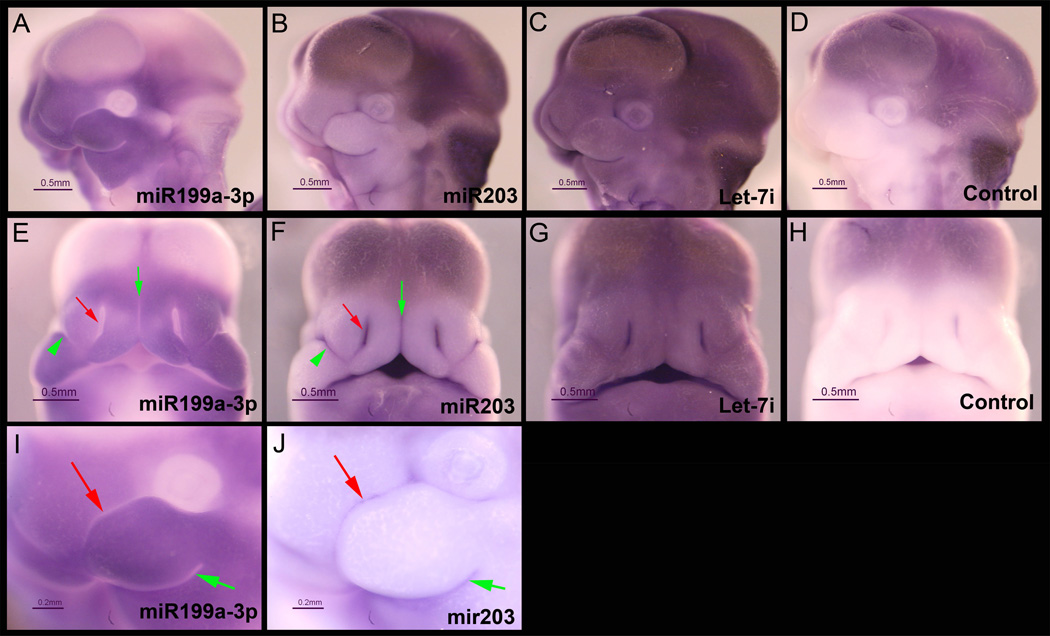
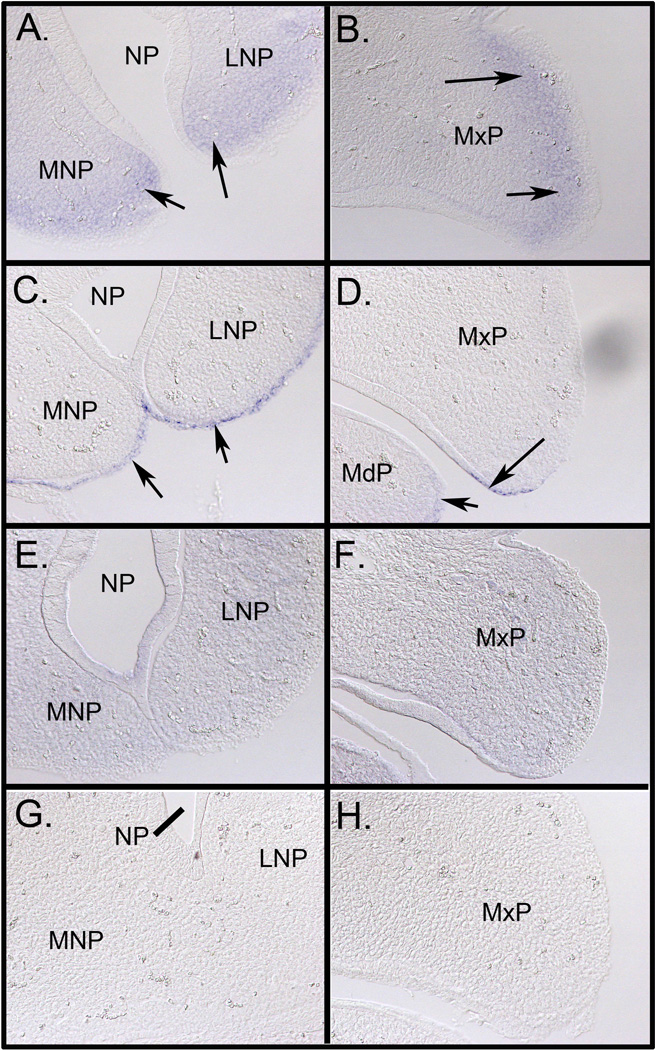
Distribution of miR-199a-3p, miR-203, and Let-7i in the craniofacial region of GD11.5 mouse fetuses was determined by in situ hybridization utilizing locked nucleic acid-modified DNA probes (LNA-probes) (Fig. 4). As expected from the RT-PCR results, each of these miRNAs was observed to be expressed in both the MNP and MxP. Expression patterns for each miRNA were, however, unique. miR-199a-3p was expressed in each facial process but was excluded from the rostral (red arrow) and caudal (green arrow) surfaces of the MxP (Fig. 4, panel I), either side of the nasolacrimal groove (green arrowhead, Fig. 4, panel E), lining of the nasal pit (red arrow, Fig. 4, panel E) and in the junction between the bilateral MNPs (green arrow, Fig. 4, panel E). The eye was also completely devoid of staining for miR199a-3p as was the entire neural tube (not shown). MiR-203 was expressed in the most restrictive pattern of the three miRNAs tested by in situ hybridization (Fig 4., panels B, F, and J). In contrast to the expression of miR-199a-3p, the expression of miR-203 was expressed only on the rostral (red arrow, panel J) and caudal (green arrow, panel J) surfaces of the MxP. MiR-203 was additionally expressed in the tissue adjacent to the nasolacrimal groove (green arrowhead, panel F), lining the nasal pit (red arrow, panel J) and the medial surfaces of the paired MNPs (green arrow, panel J). Staining in the developing brain in panels B and C may be trapping of the LNA-probe and/or anti-digoxigenin antibodies because a similar pattern was observed with a scrambled LNA-probe (panels D and H). It is not clear why the pattern with the miR199-a-3p probe differs in the area of the developing brain. Importantly, there was no non-specific binding detected in the facial prominences (panel H). The expression of Let-7i was ubiquitous throughout the entire fetus including both the MNP and MxP (Fig 4., panels C and G). These embryos were also subjected to cryosectioning which confirmed mesenchyme-specific expression of miR-199a-3p, epithelium-specific expression of miR-203, and uniform expression of Let-7i (Fig. 5).
Figure 4. Expression of miR-199a-3p, miR-203, and Let-7i in GD11.5 fetuses.
Mouse fetuses were dissected on GD11.5 and fixed overnight in 4% paraformaldehyde and then processed for in situ hybridization as described in the Methods section. Fetuses were incubated with locked nucleic acid (LNA)-modified DNA probes (Exiqon) targeting miR-199a-3p (panels A, E, and I), miR-203 (panels B, F, and J), Let-7i (panels C and G), or a scrambled sequence control probe (Control, panels D and H). Images shown in panels A-H were taken under identical lighting conditions and exposure time. Panels A-D and I-J are lateral views and Panels E-F are the corresponding frontal views. Panels I and J are higher magnification images of the same fetuses shown in A and B, respectively. There was variable trapping of the LNA probes in the ventricles of the brain, however, there was no non-specific binding detected in the medial and lateral nasal processes or the maxillary and mandibular processes of the first branchial arch (Panels D and H). miR-199a-3p was expressed in both the medial and lateral nasal processes and in the maxillary process (Panels A and E). In contrast, the expression of miR-203 was more restricted (arrows in Panels B and F) and appeared to be mutually exclusive with miR-199a-3p. The expression of Let-7i was ubiquitous (Panels C and G). Scales bars are 0.5 mm.
Figure 5. Expression of miR-199a-3p, miR-203, and Let-7i in GD11.5 mouse facial processes.
To determine the specific cellular expression pattern of each miR analyzed, the fetuses shown in Figure 3 were equilibrated in 30% sucrose/PBS, embedded and frozen in OCT medium, and sectioned at a thickness of 16 µm. Panel A shows the expression of miR-199a-3p in the mesenchyme of the MNP and LNP (arrows) and in the MxP (Panel B, arrows). Panels C and D reveal epithelium-specific expression of miR-203 in the MNP and LNP and MxP, respectively (arrows). Expression was also noted in the MdP (panel D, arrow). The expression of Let-7i was more uniform and widespread than that for miR-199a-3p and miR-203 (panels E and F). The negative control probe is shown in panels G and H. MNP, medial nasal process; LNP, lateral nasal process; MxP, maxillary and MdB, mandibular aspect of the first branchial arch; NP, nasal pit. All images were taken at 200X magnification.
Discussion
Every year in the United States 150,000 to 200,000 babies are born with a structural birth defect. Despite unprecedented intellectual and technological strides in the biomedical sciences, including sequencing of the human genome and advances in prenatal care/diagnostics, the underlying causes of nearly 70 percent of all birth defects and developmental disabilities remain unknown. Isolated oral-facial clefts are among the most common birth defects in this country. Failure of proper fusion between the maxillary and nasal processes results in cleft lip. While there have been significant advancements in indentifying genes important for secondary palate development, similar studies, specifically focused on cleft lip, are few in number.
Exploration of the epigenome—the epigenetic variability in cells (epimutations)—holds the hope for precise diagnosis of disease and congenital anomalies such as cleft lip. While multiple molecular mechanisms underlie epigenetic regulatory networks, one of the most actively investigated to date is the action of miRNAs. MiRNAs regulate expression of genes post-transcriptionally by binding to, and then inhibiting the translation of (Djuranovic et al., 2012, Bazzini et al., 2012), and/or destabilizing, their target mRNAs (Bartel, 2009). While their role in embryonic development is well documented and now widely accepted (Fjose & Zhao, 2010, Ivey & Srivastava, 2010), their role in development of the midface, particularly the upper lip, is virtually unknown and results of scientific inquiry in this area have only recently begun to emerge (Radhakrishna, 2012, Izzo et al., 2013, Wang et al., 2013). Indeed, the importance of miRNAs for craniofacial development in general, is supported by the severe craniofacial malformations seen in mouse embryos with a conditional deletion of Dicer in Wnt-1 expressing neural crest cells (Zehir et al., 2010). Loss of Dicer expression in Pax2-expressing cells also leads to craniofacial defects, including cleft palate and midfacial hypoplasia (Barritt et al., 2012). We have thus conducted the current study to identify miRNAs that are expressed in the facial processes that contribute to the formation of the upper lip. This represents the first systematic analysis of the temporal expression of miRNAs in this tissue.
Among the 10 miRNAs exhibiting the largest increase in expression in both the MNP and MxP from GD10.0 to GD11.5 were five members of the Let-7i family. The Let-7 miRNA precursor was initially identified in C. elegans (Rougvie, 2001) and later shown to be part of a much larger class of non-coding RNAs (Ambros, 2001). As such, the Let family of miRNAs is among the oldest known miRNAs and thus has been amongst the most intensely studied. Let-7i family members impact a wide variety of developmental processes including stem cell differentiation (Melton et al.), developmental timing (Reinhart et al., 2000), cell motility (Hu et al., 2013) and proliferation (Johnson et al., 2007), all of which have been implicated in orofacial development.
The expression of miR-203 was also among the miRNAs with significant increases in expression in both the MNP and MxP. As with Let-7i, miR-203 has been shown to regulate proliferation (Chen et al., 2013) and stem cell differentiation (Nissan et al., 2011). In addition, miR-203 has been shown to be involved in epithelial-mesenchymal transdifferentiation (Ding et al., 2013), a process thought to be critical for fusion of the facial processes (Jiang et al., 2006). Interestingly, of the 10 miRNAs exhibiting the largest fold-decrease in expression during upper lip development were 4 (MNP) and all 5 (MxP) members of the miR-302/367 family. Of particular interest, and perhaps relevance to known causes of cleft lip, at least in mouse models, is the fact that miR-203 and members of the miR-302/367 family target different isoforms of the p63 transcription factor. p63, a homologue of the p53 tumor suppressor, is transcribed from two different promoters, one leading to expression of the transactivating form (TA-p63, a target of miR-302/267) and the other missing the transactivating domain (ΔN-p63, a target of miR-203), each with significantly different activities (Yang et al., 1998). In both the MNP and MxP, the expression of miR-203 increased and that for miR-302/367 decreased. These data allow the suggestion that the regulated expression of p63 isoforms is critical to normal development of the upper lip. This notion draws support from the observation that complete loss of p63 expression leads to cleft lip and palate (Thomason et al., 2008). The importance of precise control of p63 expression is further emphasized by the demonstration that Wnts and BMP-4, both critical for proper midfacial morphogenesis and/or lip fusion (Jiang et al., 2006), are capable of regulating p63 expression (Ferretti et al., 2011; Medawar et al., 2008). Moreover, the expression patterns for p63 and miR-203 overlap, demonstrating one criterion for functional interaction (Thomason et al., 2008). Irf6, another gene linked to lip development, has been shown to be regulated by ΔNp63, but not by TA-p63 (Moretti et al., 2010). The expression of p63 is thus critical for craniofacial, and in particular lip and palate, development and we hypothesize that miR203 and miR302/367 coordinate to tightly control the expression of both ΔNp63 and TA-p63.
Several previous studies, most using an avian model system, have provided characterization of miRNA expression patterns in midfacial development. Darnell et al. performed a large-scale in situ hybridization screen for the expression of 135 miRNAs expressed during chicken development (Darnell et al., 2006). Of these, 13 were found to be expressed in the craniofacial region. We also identified several of these in our screen, including miRs-125b, -135b, and -217 in the medial nasal process and miRs-125b, -184, -205, and -218 in the maxillary process. These results suggest conserved function in these two species. miR-140 was also detected in the facial processes (Darnell et al., 2006). In zebrafish miR-140 has been implicated in development of the structure equivalent of the secondary palate through regulation of Pdgf expression (Eberhart et al., 2008). Powder et al. analyzed the expression of miRNAs expressed in the frontonasal process in three avian species by deep sequencing (Powder et al., 2012). In all three studies, the expression of several members of the miR-302 family were significantly down-regulated and members of the Let-7 family upregulated, consistent with the results in the current study, suggesting critical functions for these two groups of miRNAs (as described above). Sheehy et al. reported that miR-452 was enriched in neural crest cells and important for the regulation of a SHH-FGF8-DLX2 signaling axis during development of the first branchial arch (Sheehy et al., 2010). We did not, however, detect the expression of miR-452 in our screen, suggesting that its expression was down-regulated before the earliest developmental time point we examined.
In order to develop hypotheses on the mechanisms of epigenetic regulation of gene expression in the developing upper lip, based upon the expression profiles of the miRNAs detected in the current study, it is necessary to have a complete understanding of the proteome. However, there are no current datasets of the proteome in the developing facial processes. There have been a few studies systematically analyzing the expression of mRNAs in the developing mouse face (Feng et al., 2009). However, one must be careful in merging miRNA expression and mRNA expression profiles because the mechanism of miRNA silencing of mRNA expression involves not only mRNA destabilization, but also inhibition of translation (Djuranovic et al., 2012). Therefore, there may not be a direct correlation between miRNA expression and target mRNA expression. Furthermore, it must be demonstrated that the miRNA and mRNA are present in the same cell at the same time. By focusing on a limited number of miRNAs with experimentally validated targets using the Tarbase 6.0 database (Vergoulis et al., 2012), we can propose that mRNAs previously identified in the facial processes, and predicted to be important for mid-facial development, are regulated by miRNAs identified in the current study. For example, the expression of Zeb2 (a regulator of epithelial to mesenchymal transdifferentiation), Dkk1 (a Wnt inhibitor), and Mylip (myosin light chain interacting protein) were all increased in the maxillary process on GD11.5 when compared to the expression level of GD10.5 (Feng et al., 2009). These mRNAs are known to be targeted by miR-200c (Zeb2), miRs-292-3p and 291a-3p (Dkk1), and miRs-92a and -20b (Mylip). While the expression of miR-200c is constant across the GD10.0-GD11.5 window, the expression of miRs 292-3p, 291a-3p, 92a, and 20b are all decreased, consistent with the regulation of the above mRNAs during upper lip development.
Several members of the miR-17–92 cluster were found to be expressed in the MNP and MxP, some differentially-expressed (miR-92a in the MNP and miRs-19a, -19b, and -92a in the MxP) and other constitutively expressed (miRs-17 and -20a in the MNP and miRs-18a and -20a in the MxP). The miR-17–92 cluster is located in an area of human chromosome 13, that, when deleted, leads to craniofacial abnormalities, a phenotype that can be modeled in the mouse (de Pontual et al., 2011). This miRNA cluster has been been previously shown to be regulated by BMP2 and BMP4 (Wang et al., 2010). Indeed, mice lacking miR-17–92 expression have cleft palate and bilateral cleft lip, possibly through dysregulation of direct targets, Tbx3, Osr1, Fgf10, and Shox2 (Wang et al., 2013).
Forty-five miRNAs were differentially regulated and common to both the MNP and MxP (Additional file 1, Table S1). Using Tarbase 6.0 to identify validated mRNA targets of these miRNAs, it was revealed that three target Runx1 (miR-27b, miR-27a, and miR199b). Runx1 has been previously shown to be important for the fusion of the primary and secondary palates (Charoenchaikorn et al., 2009). The BMP-regulated Smads were also targeted by more than one miRNA expressed in both tissues (miR-27b targets Smad-3 and -5; miR-199b targets Smad1). Five miRNAs that were differentially expressed in the MNP but not the MxP targeted both Zeb1 and Zeb2 mRNA (miRs-429-3p, -200a, -200b, -200c, and -141-3p). Zeb1 and Zeb2 have been shown regulate epithelial-mesenchymal transdifferentiation in a number of systems, including fusion of the secondary palate, however, there have been no studies to examine its potential role in fusion of the facial prominences (Liu et al., 2008). Two miRNAs (miR195c-3p and miR16-5p) target the pro-apoptotic gene, Bcl-2. MiR-16-5p also targets Wnt-3a, a Wnt isoform that has been linked to human cases of non-syndromic cleft lip (Chiquet et al., 2008). MiR-363-3p regulates the expression of Hand2 and Tbx3, both previously shown to be important for development of the secondary palate, but their role in upper lip development is not known (Zirzow et al., 2009, Xiong et al., 2009). Of the miRNAs differentially expressed in the MxP, none targets any previously identified gene important for craniofacial development. Some of the targets, however, include CD69, the Wnt receptor Fzd4, MAP kinase 14, the transcription factor Stat3, and the Notch signaling inhibitor, Atxn1. Further pathway analyses using Ingenuity pathway analysis (Ingenuity Systems, Redwood City, CA) demonstrated that a broad array of cellular processes are predicted to be regulated by the miRNAs identified in the current study (and in the MNP and MxP), including the cell cycle, cell death, cell movement, BMP, FGF, sonic hedgehog, and Wnt signaling.
The results presented here are the first systematic analysis of the expression of miRNAs in the developing upper lip in the mouse and provide a framework from which to further investigate the role of individual miRNAs and miRNA families during lip development. In addition to identifying many miRNAs that were differentially regulated, we have also identified two miRNAs with opposing expression patterns and that regulate the expression of different isoforms of p63 (miR-203 and the miR-302/367 family), previously shown to be critical for both palate and lip development. Furthermore, miRNAs were identified that potentially regulate many of the signaling pathways known to be essential for normal lip development. A complete understanding of the regulation of expression of all genes necessary for normal upper lip development will require the integration of miRNA expression profiles with other datasets. These include the transcriptome, proteome, methylome, and histone modifications. How these varied regulatory mechanisms lead to the coordinated morphogenetic processes necessary for normal upper lip development is a prerequisite for developing strategies for the treatment or prevention of orofacial clefts.
Supplementary Material
Acknowledgements
This study was supported by NIH grants DE018215, HD053509 and P20 RR017702 (to RMG) from the COBRE program of the NIGMS.
Footnotes
Authors’ contributions
DRW participated in the study design, tissue procurement, data collection and analysis, and helped to draft the manuscript. PM participated in data analysis and drafting of the manuscript. GB performed all statistical analyses. CLW participated in the tissue collection. MMP and RMG provided the idea for the study and participated in the design, data analysis and manuscript preparation.
References
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Barritt LC, Miller JM, Scheetz LR, et al. Conditional deletion of the human ortholog gene Dicer1 in Pax2-Cre expression domain impairs orofacial development. Indian J Hum Genet. 2012;18:310–319. doi: 10.4103/0971-6866.107984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Charoenchaikorn K, Yokomizo T, Rice DP, et al. Runx1 is involved in the fusion of the primary and the secondary palatal shelves. Dev Biol. 2009;326:392–402. doi: 10.1016/j.ydbio.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Chen J, Tran UM, Rajarajacholan U, Thalappilly S, Riabowol K. ING1b–inducible microRNA203 inhibits cell proliferation. Br J Cancer. 2013 doi: 10.1038/bjc.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquet BT, Blanton SH, Burt A, et al. Variation in WNT genes is associated with non-syndromic cleft lip with or without cleft palate. Hum Mol Genet. 2008;17:2212–2218. doi: 10.1093/hmg/ddn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell DK, Kaur S, Stanislaw S, Konieczka JH, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- De Pontual L, Yao E, Callier P, et al. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Park SI, Mccauley LK, Wang CY. Signaling between TGF-beta and Transcription factor SNAI2 Represses Expression of microRNA miR-203 to Promote Epithelial-Mesenchymal Transition and Tumor Metastasis. J Biol Chem. 2013 doi: 10.1074/jbc.M112.443655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart JK, He X, Swartz ME, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40:290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppley BL, Van Aalst JA, Robey A, Havlik RJ, Sadove AM. The spectrum of orofacial clefting. Plast Reconstr Surg. 2005;115:101e–114e. doi: 10.1097/01.prs.0000164494.45986.91. [DOI] [PubMed] [Google Scholar]
- Feng W, Leach SM, Tipney H, et al. Spatial and temporal analysis of gene expression during growth and fusion of the mouse facial prominences. PLoS One. 2009;4:e8066. doi: 10.1371/journal.pone.0008066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Li B, Zewdu R, et al. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev Cell. 21:627–641. doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Li B, Zewdu R, et al. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev Cell. 2011;21:627–641. doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjose A, Zhao XF. Inhibition of the microRNA pathway in zebrafish by siRNA. Methods Mol Biol. 2010;629:239–255. doi: 10.1007/978-1-60761-657-3_15. [DOI] [PubMed] [Google Scholar]
- Gatto S, Della Ragione F, Cimmino A, et al. Epigenetic alteration of microRNAs in DNMT3B–mutated patients of ICF syndrome. Epigenetics. 2010;5:427–443. doi: 10.4161/epi.5.5.11999. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessert S, Bugner V, Tecza A, Pinker M, Kuhl M. FMR1/FXR1 and the miRNA pathway are required for eye and neural crest development. Dev Biol. 2010;341:222–235. doi: 10.1016/j.ydbio.2010.02.031. [DOI] [PubMed] [Google Scholar]
- Hu X, Guo J, Zheng L, et al. The Heterochronic microRNA let-7 Inhibits Cell Motility by Regulating the Genes in the Actin Cytoskeleton Pathway in Breast Cancer. Mol Cancer Res. 2013 doi: 10.1158/1541-7786.MCR-12-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Izzo G, Freitas EL, Krepischi AC, et al. A microduplication of 5p15.33 reveals CLPTM1L as a candidate gene for cleft lip and palate. Eur J Med Genet. 2013;56:222–225. doi: 10.1016/j.ejmg.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Dev Dyn. 2006;235:1152–1166. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YR, Han XH, Taketo MM, Yoon JK. Wnt9b–dependent FGF signaling is crucial for outgrowth of the nasal and maxillary processes during upper jaw and lip development. Development. 2012;139:1821–1830. doi: 10.1242/dev.075796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, De Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development. 2008;135:579–588. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Medawar A, Virolle T, Rostagno P, et al. DeltaNp63 is essential for epidermal commitment of embryonic stem cells. PLoS One. 2008;3:e3441. doi: 10.1371/journal.pone.0003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, Marinari B, Lo Iacono N, et al. A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J Clin Invest. 2010;120:1570–1577. doi: 10.1172/JCI40267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay P, Brock G, Pihur V, Webb C, Pisano MM, Greene RM. Developmental microRNA expression profiling of murine embryonic orofacial tissue. Birth Defects Res A Clin Mol Teratol. 2010;88:511–534. doi: 10.1002/bdra.20684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatomi M, Wang XP, Key D, et al. Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev Biol. 2010;340:438–449. doi: 10.1016/j.ydbio.2010.01.031. [DOI] [PubMed] [Google Scholar]
- Nissan X, Denis JA, Saidani M, Lemaitre G, Peschanski M, Baldeschi C. miR-203 modulates epithelial differentiation of human embryonic stem cells towards epidermal stratification. Dev Biol. 2011;356:506–515. doi: 10.1016/j.ydbio.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Powder KE, Ku YC, Brugmann SA, et al. A cross-species analysis of microRNAs in the developing avian face. PLoS One. 2012;7:e35111. doi: 10.1371/journal.pone.0035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna U. Small players with a big role: MicroRNAs in pathophysiology of cleft lip and palate. Indian J Hum Genet. 2012;18:272–273. doi: 10.4103/0971-6866.107973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F, Jugessur A, Murray JC. Genetics of nonsyndromic orofacial clefts. Cleft Palate Craniofac J. 2012;49:73–91. doi: 10.1597/10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rougvie AE. Control of developmental timing in animals. Nat Rev Genet. 2001;2:690–701. doi: 10.1038/35088566. [DOI] [PubMed] [Google Scholar]
- Sandell LL, Kurosaka H, Trainor PA. Whole mount nuclear fluorescent imaging: convenient documentation of embryo morphology. Genesis. 2012;50:844–850. doi: 10.1002/dvg.22344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy NT, Cordes KR, White MP, Ivey KN, Srivastava D. The neural crest-enriched microRNA miR-452 regulates epithelial-mesenchymal signaling in the first pharyngeal arch. Development. 2010;137:4307–4316. doi: 10.1242/dev.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JMS. Essential Developmental Biology. Wiley-Blackwell: 2012. [Google Scholar]
- Smyth G. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Stuppia L, Capogreco M, Marzo G, et al. Genetics of syndromic and nonsyndromic cleft lip and palate. J Craniofac Surg. 2011;22:1722–1726. doi: 10.1097/SCS.0b013e31822e5e4d. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Dixon MJ, Dixon J. Facial clefting in Tp63 deficient mice results from altered Bmp4, Fgf8 and Shh signaling. Dev Biol. 2008;321:273–282. doi: 10.1016/j.ydbio.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Vergoulis T, Vlachos IS, Alexiou P, et al. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40:D222–229. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Bai Y, Li H, et al. MicroRNA-17-92, a direct Ap-2alpha transcriptional target, modulates T-box factor activity in orofacial clefting. PLoS Genet. 2013;9:e1003785. doi: 10.1371/journal.pgen.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Greene SB, Bonilla-Claudio M, et al. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell. 2010;19:903–912. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, He F, Morikawa Y, et al. Hand2 is required in the epithelium for palatogenesis in mice. Dev Biol. 2009;330:131–141. doi: 10.1016/j.ydbio.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Zehir A, Hua LL, Maska EL, Morikawa Y, Cserjesi P. Dicer is required for survival of differentiating neural crest cells. Dev Biol. 2010;340:459–467. doi: 10.1016/j.ydbio.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirzow S, Ludtke TH, Brons JF, Petry M, Christoffels VM, Kispert A. Expression and requirement of T-box transcription factors Tbx2 and Tbx3 during secondary palate development in the mouse. Dev Biol. 2009;336:145–155. doi: 10.1016/j.ydbio.2009.09.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.