Abstract
Oncocytic cell tumors are characterized by the accumulation of morphologically abnormal mitochondria in their cells, suggesting a role for abnormal mitochondrial biogenesis in oncocytic cell transformation. Little is known about the reason for the dysmorphology of accumulated mitochondria. The proteins regulating the morphology of mitochondria, the "mitochondria-shaping" proteins, can modulate their size and number; however, nothing is known hitherto about a possible involvement of mitochondrial dynamics in oncocytic cell transformation in tumors. Our aim was to assess the status of the mitochondria morphology and its role in oncocytic cell transformation. We therefore evaluated the expression pattern of the main mitochondrial fusion and fission proteins in a series of thyroid cell tumor samples, as well as in thyroid tumor cell lines, with and without oncocytic cell features. The expression of mitochondrial fusion (Opa1, Mfn1 and Mfn2) and fission (Drp1 and Fis1) proteins were evaluated by immunohistochemistry (IHC) in a series of 88 human thyroid tumors. In vitro studies, for comparative purposes and to deepen the study, were performed using TPC1 - a papillary thyroid carcinoma derived cell line—and XTC.UC1, an oncocytic follicular thyroid carcinoma-derived cell line. Both IHC and in vitro protein analyses showed an overall increase in the levels of "mitochondrial-shaping" proteins in oncocytic thyroid tumors. Furthermore, overexpression of the pro-fission protein Drp1 was found to be associated with malignant oncocytic thyroid tumors. Interestingly, genetic and pharmacological blockage of Drp1 activity was able to influence thyroid cancer cells’ migration/invasion ability, a feature of tumor malignancy. In this study we show that unbalanced mitochondrial dynamics characterize the malignant features of thyroid oncocytic cell tumors, and participate in the acquisition of the migrating phenotype.
Introduction
Cancer cells are known to undergo a shift in their basal metabolic pathways, a process often described as the “Warburg effect”, whereby even under high oxygen tension they produce most of their ATP by glycolysis[1]. This shift in metabolism has been reported to be accompanied by a change in mitochondrial morphology and size, although the molecular details accompanying the morphological changes associated with the Warburg effect remain unexplained, also since little is known on how mitochondrial morphology is regulated[2,3]. Recent years have seen the identification of the core components of the mitochondrial dynamics machinery, a growing group of proteins which, among other cellular functions, regulate mitochondrial fusion and fission. Mitochondrial dynamics dysfunction has been progressively unveiled in a wide number of pathologies, especially neurodegenerative diseases and cancer[4].
A particular group of uncommon tumors has been reported in virtually every organ, with a higher frequency in thyroid and kidney[5,6]. Such tumors have cells with a distinct highly eosinophilic, grainy and swollen cytoplasm, with large nuclei and prominent nucleoli [7,8,9,10]. Electron microscopy has shown that this swollen appearance is due to an anomalous accumulation of mitochondria displaying abnormal morphology[11,12,13]. These tumors are described as oncocytic-, oxyphilic-, eosinophilic, “Hürthle-cell (when referring to the thyroid) tumors” or as “oncocytomas”. For the sake of simplicity, we will designate them as “oncocytomas” or “oncocytic cell tumors”[10]. In current clinical practice, the criteria for diagnosis of the oncocytic cell variants of papillary thyroid carcinoma (PTC) and follicular thyroid carcinoma (FTC) include nuclear characteristics, signs of capsular and/or vascular invasion which are shared with the non-oncocytic cell tumors of the same kind, as well as the mitochondrial proliferation typical for oncocytic cell tumors[5,6,7]. Indeed, this idea that the oncocytic transformation occurs on top of “conventional” tumors is backed up by the similar genetic alterations of oncocytic carcinomas and their non-oncocytic counterparts, which display a comparable incidence of the various genetic abnormalities, such as RET/PTC rearrangements and BRAFV600E mutations[6,14,15,16,17,18]. Although the first recommendations considered all oncocytic thyroid tumors malignant, later studies have demonstrated that as long as the cases were correctly stratified according to their clinico-pathological features-such as patients’ age, staging of the tumors and surgical procedure- the prognosis of patients with oncocytic cell PTC and FTC variants is similar to that of patients with respective conventional, non-oncocytic carcinomas[3,5,17,19].
The main differences between oncocytic and non-oncocytic tumors reside in the frequency of variations found in mitochondrial DNA (mtDNA) and nuclear DNA genes encoding mitochondrial proteins[2,5,20,21]. Of these, a large 5kb deletion, often referred to as the "common deletion" that eliminates a number of mtDNA genes and disrupts mtDNA replication and transcription, is frequently found in oncocytic thyroid tumors and is associated with an increase in mitochondrial mass[20,22,23,24].
The data on record suggest that mutations in oxidative phosphorylation (OXPHOS) genes, namely in Complex I genes, may result in OXPHOS system impairment and may lead to the accumulation of abnormal mitochondria[20,25]. Using bioinformatic tools, we have recently demonstrated that high pathogenicity mtDNA mutations, namely in genes for Complex I, lead to the oncocytic phenotype[21].
Variants of the Translocase of Inner Mitochondrial Membrane 44 homolog (TIMM44) that is part of the mitochondrial protein import system have also been described in familial forms of oncocytic thyroid tumors; this suggests that deregulation of the mitochondrial import machinery and consequently of the mitochondrial function are associated with abnormal mitochondrial biogenesis[26].
A striking feature of the accumulated mitochondria in oncocytic cell tumors is their abnormal ultrastructure and morphology. This indicates a deregulation also in the proteins that control mitochondrial dynamics, the "mitochondria-shaping" proteins, a process dependent on mitochondrial fusion and fission and inextricably linked to mitochondrial biogenesis, distribution, signaling and apoptosis[27,28,29]. Interestingly, imbalance of the mitochondria phenotype toward fragmentation orchestrates the migratory capability in cells where migration represents a crucial physiological function, such as T-cell lymphocytes and tumoral metastatic cells[30,31]. Indeed, in order to make movement possible, mitochondria need to be fragmented to favor their relocalization and recruitment to specific subcellular regions. In the case of tumoral cells, a direct correlation has been demonstrated between the metastatic degree and the Drp1-dependent fragmentation of mitochondria in breast cancer[31]. Mitochondrial fission requires the cytosolic dynamin related protein 1 (Drp1) and its mitochondrial receptors fission 1 (Fis1), mitochondrial fission factor (Mff) and Mitochondrial Division (Mid) 49 and 51. Conversely, the outer membrane mitofusin (Mfn) 1 and 2 and the inner membrane dynamin-like protein optic atrophy 1 (Opa1), mediate mitochondrial fusion[32,33]. Despite the known mitochondrial morphological alterations observed in oncocytic cell tumors, knowledge about the levels and the role of these mitochondria-shaping proteins is scanty. We therefore set out to investigate if mitochondrial dynamics is altered in oncocytic thyroid cell tumors. Our data indicate that mitochondrial fission proteins Drp1 and Fis1 are overexpressed in oncocytic cell tumours, and that Drp1 expression levels correlates with oncocytic cell tumor malignancy ex vivo and with the migration ability of an oncocytic thyroid tumor cell line in vitro. Interestingly, genetic and pharmacological blockage of Drp1 reduces the migration of the oncocytic tumor cell line, offering a novel therapeutic approach to tackle metastatic oncocytic cell tumors.
Materials and Methods
Ethics Statement
The present study was performed under the national ethical and regulative law for the use of biological specimens. The material used in this study comes from the “Banco de Tecidos e Tumores “(Tumors and Tissues Bank) of the Hospital de São João, Porto, which collects all samples upon written informed consent of the patients, or their guardians in case of the minors. To have access to these samples, researchers must submit a project to the Ethics Committee of the Hospital de São João for approval. Our project has been approved.
Patient selection
A series of 88 thyroid tumors were retrieved from the files of Department of Pathology of Hospital S. João (HSJ), Porto. Thyroid samples were obtained from patients with ages ranging from 15 to 83 years of age. The histology of all tumor samples was revised independently by two pathologists (ER and MSS) and the final classification was made according to the WHO criteria[34]. The diagnosis of the thyroid tumors were the following: follicular thyroid adenoma (FA, n = 19; 12 oncocytic and 7 non-oncocytic), follicular thyroid carcinoma (FTC, n = 7; 1 oncocytic and 6 non-oncocytic), follicular variant of papillary thyroid carcinoma (FVPTC, n = 30; 8 oncocytic and 22 non-oncocytic), conventional papillary thyroid carcinoma (cPTC, n = 32; 9 oncocytic and 23 non-oncocytic).
Due to the short follow-up of the cases included in this series and to the consequent limited number of death events, overall survival was not analyzed. The present study was performed under the national ethical and regulative law for the use of biological specimens.
Tissue Microarray (TMA)
Representative areas of different lesions were carefully selected on haematoxylin and eosin-stained sections and were marked on individual paraffin blocks. Two tissue cores (3 mm in diameter each) were obtained from all the selected specimens and were precisely deposited into duplicate TMA recipient paraffin blocks using a tissue-array instrument (Beecher Instruments, Silver Spring, MD) as described elsewhere[35]. In each tissue microarray block, areas of non-neoplastic thyroid tissue were also included as controls. Multiple 3 μm sections were cut from the TMA blocks and transferred to ultrafrost slides.
Immunohistochemical analysis
Immunohistochemical analysis (IHC) was performed in 3μm formalin-fixed, paraffin-embedded sections. Sections were deparaffinized and rehydrated in a series of decreasing concentration of ethanol solutions. Deparaffinized single specimens as well as TMA sections were subject to heat-induced antigen retrieval either in 10mM pH6 citrate buffer (sodium citrate) or in 1mM pH8 ethylenediamine tetraacetic acid buffer (EDTA) (LabVision Corporation, Fremont, CA, USA), in a microwave set at 300watts for 10 minutes.
After retrieval solutions had cooled, tissues were subject for 10 minutes to blocking of endogenous peroxidase activity and of non-specific binding with 3% of H2O2 and with the Large Volume Ultra V Block reagent (Thermo Scientific/Lab Vision, Fremont, USA) depending on the detection system used.
The sections were then incubated in a humidified chamber with the following primary antibodies and accordingly with manufacturer’s specifications: rabbit polyclonal antibody specific for Fis1 (1:100) ref. ALX-210-907 from Alexis Biochemicals, now Enzo Life Sciences (Farmingdale, New York, USA), mouse monoclonal antibody specific for Drp1 (1:100) ref. 611112 and mouse monoclonal antibody specific for Opa1 (1:100) ref. 612607, both from BD Biosciences (Franklin Lakes, New Jersey, USA), rabbit polyclonal specific for Mfn1 (1:250) ref. sc-50330 (Santa Cruz Biotechnology Santa Cruz, USA), mouse monoclonal antibody specific for Mfn2 (1:400) ref. 9927-M03 from Abnova, (Jhongli City, Taiwan). As a control for mitochondrial content mouse polyclonal antibody specific for SDHA (1:2000) ref. MS203, from Mitosciences, now abcam (Cambridge, UK) was used. Previously tested positive controls from thyroid, breast and muscle were included for all the antibodies in the series. Negative controls were carried out by replacing the primary antibody with nonimmune mouse serum.
The immunohistochemistry technique was performed using a labeled streptavidin-biotin immunoperoxidase detection system (Thermo Scientific/Lab Vision, Fremont, USA) or the Envision G/2 System/AP (K5355, Dako, Denmark) according to the manufacturer's instructions.
For MFN1 and SDHA the immunohistochemical staining was developed with DAB (3,3'-diaminobenzidine) substrate. For the remaining antibodies the immunostaining was performed with or the Envision G/2 System/AP (K5355, Dako, Denmark), and the samples were developed with a permanent red chromogen.
Immunostaining was blindly semi-quantitatively evaluated by two observers (VM and AFS) without knowledge of any clinical information of the cases; an IHC score was obtained through the sum of intensity of staining (0- absent; 1- faint; 2- moderate; 3- strong) by the extension of stained tumor cells (1–0 to 25%; 2–25 to 50%; 3–50 to 75%; 4- >75%). Evaluation of the percentage of immunoreactive cells for all the antibodies was made by counting 300 tumor cells in random fields. In all discrepant cases a consensus was reached. As mitochondrial marker, and as a mean to assess the amount of mitochondria, SDHA expression levels were accessed and used to normalize our results. In every sample, we compared the expression of the proteins of interest, both in normal and tumor tissue, against SDHA staining. Thus, for each case, we were able to evaluate in an individual and precise manner the changes in expression of the protein in the study independently of the amount of mitochondria, indicated by the SDHA expression.
Cell lines
Thyroid cancer cell lines: TPC1 (kindly provided by Dumont JE and Mareel M—National Cancer Center Research Institute, Tokyo, Japan), a PTC-derived cell line, and XTC.UC1 (from Wong MG and Savagner F—Surgery Service, Veterans Affairs Medical Center, San Francisco, California), a cell line obtained from an oncocytic variant of follicular carcinoma, were used in this study. Both cell lines had been previously characterized at the molecular and genotypic level[36,37]. TPC1 cells were cultured with RPMI medium with Glutamax supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (v/v) Pen Strep and 0.5% (v/v) fungizone (all from GIBCO, Invitrogen, Carlsbad, USA); XTC.UC1 was maintained with DMEM/F12 (GIBCO, Invitrogen, Carlsbad, USA) supplemented with 10% (v/v) FBS, insulin at 10μg/mL, TSH at 10mU/mL (Sigma-Aldrich, St. Louis, USA), 1% Pen Strep (v/v) and 0.5% (v/v) fungizone. Both cell lines were authenticated using DNA profile analysis, obtained with the PowerPlex 16 system (Promega, Madison, USA), according to the DNA profiles available in American Type Culture Collection and Health Science Research Resource Bank.
Constructs and transfections
Cytoplasmatic GFP (ctGFP), mitochondrially targeted dsRED (mtRFP), mitochondrially targeted YFP (mtYFP) and pcDNA3.1-HA-K38A-DRP1 plasmids were previously described and were a gift from T. Pozzan (Venetian Institute of Molecular Medicine, Padua, Italy)[33]. The empty pcDNA3.1 was obtained from BD-Clontech and pcDNA3.1-HA-K38A-DRP1 plasmid generation was previously described[33]. XTC.UC1 cell lines were transiently transfected by electroporation using the Neon Transfection System (Life Technologies, Carlsbad, CA, USA). In co-transfections experiments, 1.5μg of marker carrier (ctGFP in transwell experiments) plus 3μg of pcDNA 3.1 or pcDNA3.1-HA-K38A-DRP1 were used per 2.0x106 cells. The specific combination of plasmids transfected in each experiment is indicated in the figure legends. After 24h, transfected cells were sorted by FACS and used for experiments 24h later.
Quantitative PCR
For cDNA preparation, 1ng of total RNA was reverse transcribed using the RevertAid first strand cDNA synthesis kit (Fermentas, Burlington, ON, Canada). Reverse transcription products were amplified for all the aforementioned genes by qPCR using TaqMan PCR Master Mix (Applied Biosystems, Foster City, CA, USA). For data analysis Actin and hCyclophilin were used as endogenous controls (data shown only for actin). Data are expressed as relative expression for each individual gene normalized to the corresponding controls.
The ABI PRISM 7500 Fast Sequence Detection System (Applied Biosystems) was used to detect the amplification level and was programmed to an initial step of 2 min at 50°C, 10 min at 95°C, followed by 45 cycles of 95°C for 15 sec and 60°C for 1 min. 50ng of cDNA for each sample was used and all the amplifications were done in triplicate. The relative quantification of target genes was determined using the ∆∆CT method, which was previously validated by Livak’s Linear Regression Method (slope¼0.0696) (Sequence Detector User Bulletin 2; Applied Biosystems).
Subcellular fractionation and immunoblotting
Subcellular fractionation was performed as described in Frezza et al.[38]. Mitochondrial and cytosolic fractions were obtained and were blotted with a mouse monoclonal antibody anti-TOM20 (1:5000) ref. FL-145 (Santa Cruz Biotechnology Santa Cruz, USA) and a goat polyclonal antibody anti-Actin (1:2000) ref sc1616 (Santa Cruz Biotechnology Santa Cruz, USA). Cells were lysed in RIPA buffer, in the presence of phosphatase and protease inhibitors. Similar amounts of total protein were resolved by SDS-PAGE and transferred onto a nitrocellulose membrane (GE Healthcare, Little Chalfont, England). We have used as primary antibodies, the ones already referred in the immunohistochemistry section (all at a dilution of 1:1000). For protein detection, a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, USA) and a luminescence system (Perkin-Elmer, Foster City, USA) were used. Membranes were re-probed with a mouse monoclonal antibody anti-TOM20 (1:5000) ref. FL-145 (Santa Cruz Biotechnology Santa Cruz, USA) for control of the protein loading. Protein expression was quantified using the Bio-Rad Quantity One 1-D Analysis software.
Electron microscopy
Cells were prepared using a conventional electron microscopy fixation procedure and images were taken using a Technai 20 (FEI, Eindhoven, Netherlands). Differences in mitochondrion size between cell lines were calculated using the whole mitochondria surface area. A minimum of 15 mitochondria were measured per cell and at least 5 different cells per specimen were randomly used for the size measuring. All experiments were made in triplicate.
Mitochondrial network imaging
To quantify structural mitochondrial network fragmentation, cells were grown in glass-bottom dishes, transfected with mitochondrial targeted yellow and red fluorescent protein, mtYFP and mtRFP respectively, and after 24h were fixed with ice-cold 3.7% formaldehyde for 30 minutes. Cells expressing mtRFP were imaged with the Zeiss 510 META confocal laser scanning microscope using a Plan-Apochromat 100/1.46NA objective with a 2x digital zoom (excitation at 561 nm, emission recorded above 575nm. For every cell line or condition at least 25 randomly selected cells were imaged and quantified according to its mitochondrial network. A 100x Plan-Apochromat (1.46NA) objective and 2x zoom were used to image single cells. Digital images were processed using ImageJ 1.47 (U.S. National Institutes of Health—NIH, Bethesda, MD, USA).
Scratch-wound assay
Cells were grown to near 90% confluence in 6 well plates. Under aseptically conditions a thin "wound" was introduced by scratching with a 10μl pipette tip and detached cells were rinsed with PBS. Using a phase contrast set up and a 40x total magnification pictures were taken every 5 minutes for a total of 15 hours. Five different measurements per field were made using the pictures taken at 0, 3, 6, 9, 12 and 15 hours using ImageJ software. More detailed information on this methodology has been previously published[39]. Images were processed with ImageJ software.
Migration/invasion assays
Transiently transfected TPC-1 and XTC.UC1 cells were resuspended in serum-free RPMI and with DMEM/F12, respectively, with 0.1% FBS and seeded in the upper chamber of a transwell 12-well plate (Corning Costar). The transwell plates were pre-coated with 5μg/ml fibronectin (F2006-1MG, Sigma Aldrich) and incubated at 37°C 5% CO2 for 4h. Plates were then rinsed with PBS1x at pH7.4 before adding the 100.000 cells per well. The lower chambers were filled with the respective medium supplemented with 10% FBS. After 12 hours of assay, the transwell membranes were excised, inverted and stained with DAPI on a glass slide. The nuclei visible at the facing-down side of the membrane were counted. A minimum of 5 different fields were counted for each slide and a triplicate was made for each experimental setting. Mdivi1 inhibitor was used at 50μM based on previous published data and in the absence of observable toxic or stress cell changes[40]. No differences were found between cells treated with Mdivi1 from time zero or pre-treated 1hour before the beginning of the assay.
Statistical analysis
The statistical analysis of the IHC results was performed with STAT VIEW-J 5.0 (SAS Institute, Inc., Cary, NC). The relationship between the average expression level (score) of the immunohistochemical markers and the histopathology of the different cases were evaluated by ANOVA; results were considered statistically significant whenever p value < 0.05. When appropriate, multiple comparison corrections were performed using the post hoc Bonferroni/Dunn test and, upon program indication, comparisons were only considered significant whenever p value < 0.0018.
Every in vitro analysis was performed at least in triplicate and the mean thus obtained was used for independent Student’s t-test. All the data are expressed as mean ± SE, as indicated in the legends of the figures.
Results
Mfn2, Opa1, Drp1 and Fis1, are overexpressed in oncocytic cell tumors and Drp1 overexpression is associated with malignant oncocytic cell tumors
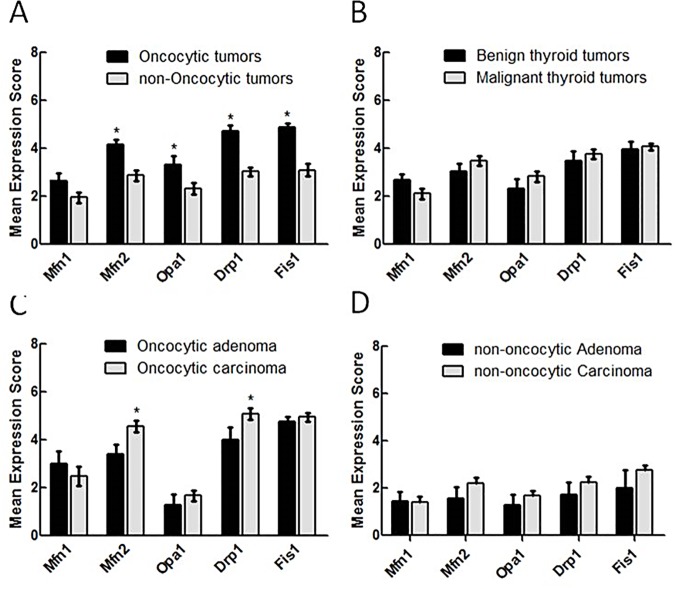
The so-called "mitochondria-shaping" proteins are key regulators of the mitochondria shape and of their dynamics. The mitochondrial network, indeed, is defined by the highly regulated balance between fusion and fission processes, depending on the cell’s physiological needs. Among several cellular processes and patho-physiological conditions, the mitochondria-shaping proteins also act on the homeostasis of the organelles size and number[4,28,29,32]. Once the oncocytic cell tumors differ from the non-ococytic ones by an abnormal accumulation of mitochondria with an altered morphology in the cell cytoplasm, we hypothesized that mitochondrial dynamics might have a role in the phenotypic and physiological oncocytic definition[11,12,13]. Thus, we have performed a histological analysis of the main "mitochondria-shaping" protein levels on several histological human thyroid tumor samples. Fig 1 shows representative immunostaining micrographs of the "mitochondria-shaping" proteins observed in different histological types of thyroid tumors, including 1 oncocytic follicular thyroid carcinoma (FTC), 6 non-oncocytic FTC, 8 oncocytic follicular variant of papillary thyroid carcinoma (FVPTC), 22 non-oncocytic FVPTC, 9 oncocytic conventional papillary thyroid carcinoma (cPTC), 23 non-oncocytic cPTC, 12 oncocytic follicular thyroid adenoma (FA) and 7 non-oncocytic FA. IHC micrographs representative of a benign oncocytic lesion (oFA), a malignant oncocytic lesion (oPTC) and a non oncocytic lesion (the noFVPTC), the histotypes of main interest in this work, are presented in Fig 1A, 1B and 1C. Since the expression of "mitochondria-shaping" proteins in the normal adjacent parenchyma was zero, or only detectable below the score 2, we omitted in the histograms the expression levels of such proteins in normal tissue. With the exception of Mfn1, we identified an increase in the expression levels of the other proteins studied, in the oncocytic cell tumors vs. the non-oncocytic ones and in all four histo-types analyzed (FVPTC, FTC, cPTC and FA) (Figs 1 and 2A).
Fig 1. The expression levels’ analysis of mitochondria-shaping proteins shows an overall increase in oncocytic thyroid tumors when compared to their non-oncocytic counterparts.
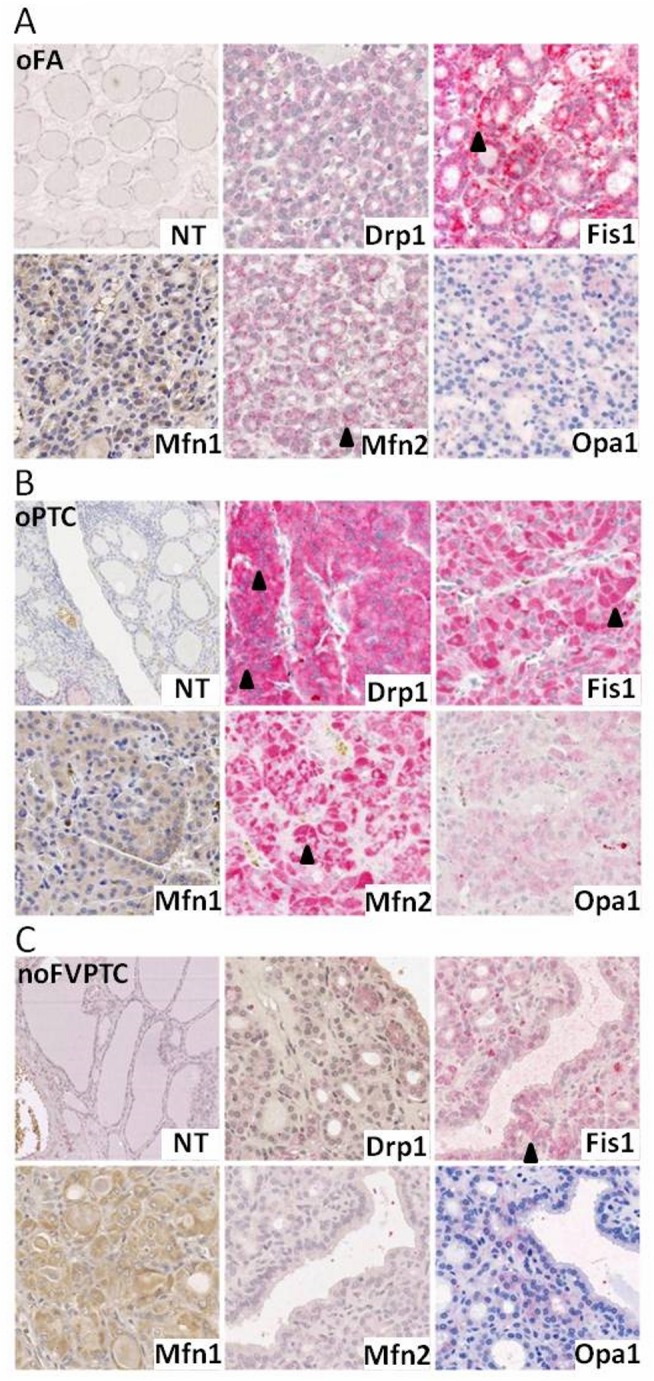
Representative micrographs showing immunostaining of Mfn1, Mfn2, Opa1, Drp1 and Fis1 antibodies in normal thyroid tissue (NT) and tumoral tissue of an oncocytic follicular adenoma—oFA) (A), an oncocytic papillary thyroid carcinoma—oPTC (B) and a non-oncocytic follicular variant of papillary thyroid carcinoma—noFVPTV (C) at a total magnification of 300x. Illustrative black arrowheads denote stained measured tumoral areas with an increased gradation from noFVPTV, to oFA and finally oFTC.” # Mfn1—Mitofusin 1, Mfn2—Mitofusin 2, Opa1—Optic atrophy 1, Drp1—Dynamin related protein 1, Fis1—Mitochondrial fission 1 protein.
Fig 2. A significant increase on Drp1 expression is associated with malignant oncocytic thyroid tumors.
The immunostaining of Mfn1, Mfn2, Opa1, Drp1 and Fis1 was evaluated, taking into account all thyroid tumors (A and B), the oncocytic thyroid tumors alone (C) or the non-oncocytic thyroid tumors alone (D). The mean antigen expression was calculated as described in Material and Methods. Results are shown as mean score ±SEM. * p<0.05 (unpaired t-test). # Mfn1—Mitofusin 1, Mfn2—Mitofusin 2, Opa1—Optic atrophy 1, Drp1—Dynamin related protein 1, Fis1—Mitochondrial fission 1 protein.
Mfn2 and the pro-fission Drp1 are significantly overexpressed in malignant oncocytomas
In a deeper analysis of the histological samples from human thyroid tumors, Mfn1 was the only mitochondria-shaping protein to show no significant difference in the expression levels, independently of the phenotype or tumoral types compared (Fig 2A–2D). An overall increase of the levels of the other proteins analyzed, independently of their pro-fusion or pro-fission role, were all significantly higher in the oncocytic tumors compared to in the non-oncocytic ones (4.2±0.2 vs. 2.9±0.2, p<0.0001 for Mfn2; 3.3±0.3 vs. 2.3±0.2, p = 0.01 for Opa1; 4.7±0.2 vs. 3.0±0.2, p<0.0001 for Drp1; 4.9±0.1 vs. 3.5±0.2, p<0.0001 for Fis1), as shown in the IHC quantification of Fig 2A (Fig 2A). In contrast, there was no appreciable difference for any of these proteins, when we compared malignant vs. benign tumors, without distinguishing and grouping the samples into oncocytic or non-oncocytic (3.5±0.2 vs. 3.0±0.3, p = 0.31 for Mfn2; 2.8±0.2 vs. 2.3±0.4, p = 0.28 for Opa1; 3.8±0.2 vs. 3.5±0.4, p = 0.48 for Drp1; 4.1±0.1 vs. 3.9±0.34, p = 0.71 for Fis1) (Fig 2B). Therefore, it seems that the alteration of the machinery responsible for mitochondrial morphology is strictly related to the oncocytic phenotype, rather than to the overall tumor aggressiveness. Furthermore, within the non-oncocytic tumors no differences in the expression levels of mitochondrial dynamics proteins were found between benign and malignant tumors (Fig 2D). However, if we consider the degree of severity of the tumor, only within the oncocytic phenotype, interestingly we observe that Mfn2 and Drp1 are significantly more abundant in carcinomas compared to the benign counterparts’ adenomas (4.5±0.2 vs. 3.4±0.4, p = 0.012 for Mfn2 and 5.1±0.2 vs. 4.0±0.5, p = 0.038 for Drp1 (Fig 2C). Of note, these differences are not related to differences in mitochondrial number (S1 Fig). This suggests that within oncocytic cell tumors, the mitochondrial dynamics assume a role in the definition of tumor malignancy and aggressiveness.
Drp1 actively accumulates in the mitochondria and unbalance mitochondrial network towards fission
In order to clarify and investigate in depth the role of mitochondrial dynamics alteration on oncocytic cell tumors, we switched to in vitro experiments on two thyroid cancer cell lines: XTC.UC1, which is the only oncocytic cell line available, with carcinoma derivation, and TPC1, a non-oncocytic thyroid cell line, used for comparative purposes. First, we evaluated in the two cell lines the expression level of the same fusion/fission proteins studied in the human tumor samples.
By western blot analysis, we detected, in the oncocytic cell line, the same trend of expression pattern of "mitochondria-shaping" proteins, as we observed in oncocytic thyroid tumors, i.e. an increase in the mitochondrial fission proteins Drp1 (p<0.05) and Fis1 (although not statistically significant) in XTC.UC1 cell line in comparison with TPC1 cell line (Fig 3A and 3B). Regarding the fusion proteins, we observed a statistically significant downregulation of Mfn1 (p<0.05) in XTC.UC1, a non-statistically significant increased expression level of Mfn2 and no differences in Opa1 (Fig 3A and 3B). Interestingly, at variance with the significant increase of Drp1 protein expression in XTC.UC1 when comparing with TPC1 cell line, Drp1 mRNA levels were similar in both cell lines (Fig 3C).
Fig 3. In vitro characterization of mitochondria in oncocytic XTC.UC1 and non-oncocytic TPC1 cell lines.
Mitochondrial fission proteins are overexpressed in oncocytic cancer cells and Drp1 actively accumulates on mitochondria. (A and B) Representative western blot analysis of Mfn1, Mfn2, Opa1, Drp1 and Fis1 expression levels in protein extracts of XTC.UC1 and TPC1 show an overexpression of mitochondrial fission proteins and a downregulation of pro-fusion Mfn1. The levels of Mfn1, Mfn2, Opa1, Drp1 and Fis1 were quantified and normalized by the levels of control protein (TOM-20). Results are shown as mean expression value ± SEM of at least three independent experiments.* p<0.05 (unpaired t-test). (C) Drp1 mRNA levels were similar in XTC.UC1 and TPC1 cell lines. mRNA levels of Drp1, Fis1, Mfn2, Mfn1 and Opa1 were accessed by qPCR in XTC.UC1 and TPC1 cell lines. Results are expressed as relative expression for each individual gene normalized to the respective control protein (Actin) between cell lines. Results are shown as mean expression value of at least three independent experiments. Error bars represent SEM. * p<0.05 (unpaired t-test). (D) Drp1 actively accumulates at mitochondria in the XTC.UC1 cell line. Representative western blot analysis of equal protein extracts from the heavy membrane fraction and cytosolic fraction is shown in both XTC.UC1 and TPC1 cell lines. (E and F)- Mitochondrial network is fragmented in almost half of the cellular population of XTC.UC1. Representative confocal images of XTC.UC1 and TPC1 cell lines transfected with mitochondrial targeted RFP (mtRFP) showing mitochondrial fragmentation in XTC.UC1 and TPC1 at basal conditions. XTC.UC1 cell line shows a 4-fold increase in the number of cells with fragmented mitochondrial network in comparison to TPC-1. The number of cells with fragmented mitochondria was quantified by counting at least 30 cells per field. Results are shown as mean expression value ± SEM of at least three independent experiments.* p<0.05. # Mfn1—Mitofusin 1, Mfn2—Mitofusin 2, Opa1—Optic atrophy 1, Drp1—Dynamin related protein 1, Fis1—Mitochondrial fission 1 protein and TOM20—translocase of outer mitochondrial membranes 20 kDa. XTC.UC1—oncocytic cell follicular carcinoma, TPC1—Papillary thyroid carcinoma-derived cell line. (G and H) Electron microscopy showed significantly smaller mitochondria in XTC.UC1. Representative electron microscopy images of XTC.UC1 and TPC1 cells are shown, using a total magnification of 5800x. Black arrowheads denote differences in size. The mitochondrial size is less than 2.5 times lower in XTC.UC1 than in TPC1. Mitochondrial area was estimated by measuring the outer contour of a minimum of 15 mitochondria per cell. At least 5 different cells per specimen were randomly used for the size estimation. Results are shown as mean expression value ± SEM of at least three independent experiments.* p<0.05. # XTC.UC1—oncocytic cell follicular carcinoma, TPC1—Papillary thyroid carcinoma-derived cell line.
We decided to further investigate Drp1 in thyroid tumors’ oncocytic phenotype because we observed a statistically significant increase of Drp1 protein, both in oncocytic tumors and in the oncocytic cell line, and because of the known involvement of this protein in breast cancer malignancy[31].
Given that Drp1 is a cytosolic protein that translocates to mitochondria to be functionally active and take part in mitochondrial division, we wondered if Drp1 was actively recruited to the mitochondrial membrane or if it accumulated passively at the cytosol. To clarify this issue, we performed a sub-cellular fractioning in both cell lines and, we assessed the levels of Drp1 bound to the organelles on the heavy membrane fraction enriched on mitochondria. The levels of this protein were around 8 times higher in XTC.UC1 than in TPC1 (p<0.05) in the mitochondria-rich fraction, much higher than that detected in the total cytosolic protein lysates (Fig 3D) and in cytoplasm; this indicated a substantial accumulation, and thus activation, of the pro-fission protein at mitochondria. Indeed, as expected at this point, confocal microscopy analysis on single cells, with a fluorescent protein specifically targeted to mitochondria (mtRFP) overexpressed in order to visualize the organelles, showed that mitochondria of the oncocytic cell line XTC.UC1 are much more fragmented than the non-oncocytic ones (46,5% vs. 10,2%) (Fig 3E and 3F).
By electron microscopy, we were able to confirm this morphological imbalance toward fission observed by WB and qPCR in the oncocytic XTC.UC1 cells as denoted by the black arrowheads (about 2.5 times smaller mitochondria in XTC.UC1 than in TPC1, p<0.05) in Fig 3G and 3H.
Oncocytic cancer cell line faster migration is sensitive to genetic and pharmacological blockage of Drp1
Since it has been previously shown that mitochondrial dynamics, namely Drp1, play a crucial role in orchestrating lymphocyte chemotaxis and migration of metastatic breast cancer, we set out to establish if the Drp1 pro-fission protein overexpression correlated with a higher capability of migration in the oncocytic XTC.UC1 cell line[30,31]. Accordingly we decided to perform a scratch-wound assay. This is a commonly used method to measure basic cell migration parameters, by following, in timelapse, how fast confluent cells fill in a wound manually scratched in a culture plate. We observed through this assay that XTC.UC1 cells have, indeed, higher motility and need less time to fill in the wound space than TPC1 cells (S2 Fig). We then confirmed this result by transwell assay, a test frequently used to study cell migratory response and invasion ability toward a specific chemical gradient created by a fibronectin-coated artificial filter. In this case, either at a 12 or 18 hours interval, we obtained more than double the number of cells able to transpose the fibronectin coated transwell membranes in XTC.UC1 than in TPC1 (Fig 4A).
Fig 4. Pharmacological and genetic blockage of Drp1 prevent oncocytic cell migration/invasion.
(A) XTC.UC1 cells show higher basal migration ability on a transwell assay. A minimum of 5 different fields were counted for each slide and triplicates were made for each experimental setting. (B) Pharmacological blockage of Drp1 significantly restrains oncocytic cell (XTC.UC1) migration in a wound-healing assay. Representative frames acquired at the indicated time points (every 1 hour) during a 15h wound-healing assay performed in XTC.UC1 untreated, XTC.UC1+DMSO and XTC.UC1+Mdivi1 (a specific Drp1 inhibitor) cells are here shown. Results are shown as mean expression value ± SEM of at least three independent experiments. * denotes p < 0.05 in a paired sample t-test. (C) Pharmacological or genetically blocking Drp1 results in diminished XTC.UC1 cell migration/invasion ability in a transwell assay. Representative images taken at the end of a 12hour time point transwell assay showing a decrease in the migration/invasion ability of cells treated both with a genetic (XTC.UC1+dnDRP) and pharmacological (XTC.UC1+Mdivi1) inhibition of Drp1. Conditions: XTC.UC1 untreated, GFP-transfected XTC.UC1 (as a transfection control), GFP+dnDRP co-transfected XTC.UC1, XTC.UC1+DMSO (as a pharmacological blockage control) and XTC.UC1+Mdivi1 (a specific Drp1 inhibitor). Results are shown as mean expression value ± SEM of at least three independent experiments. * p < 0.05 in a paired sample t-test. # XTC.UC1—oncocytic cell follicular carcinoma; GFP—green fluorescent protein; dnDRP—dominant negative form of dynamin related protein 1 (Drp1) Drp1-K38A; DMSO—Dimethyl sulfoxide; Mdivi1—selective cell-permeable inhibitor of mitochondrial division Drp1.
To evaluate if the aforementioned higher ability to move and migrate could be attributed to the overexpression of the pro-fission Drp1 found in XTC.UC1, we interfered, both pharmacologically and genetically, with this cell line’s fission machinery by blocking the Drp1 activity. First, we performed a scratch-wound assay in the presence of Mdivi1, a specific Drp1 inhibitor blocking its assembly and translocation to mitochondria[40]. The pharmacological block of Drp1 did indeed reduce the cell motility by almost 50% in comparison to untreated cells. This was accompanied by a reduction in the number of cells with fragmented mitochondria in favor of an increase of cells with an interconnected mitochondrial network (Fig 4B and S3 Fig). A transwell assay performed under the same conditions confirmed our observations on the migration reduction (Fig 4C). To reinforce this interesting result, we also assessed the migration/invasion competence of XTC.UC1 cells by genetically interfering with the fission machinery, specifically targeting Drp1 activity. We performed a co-transfection with a mitochondrial targeted fluorescent protein mtGFP and a dominant negative form of Drp1 (dnDrp1) Drp1-K38A, a form of Drp1 with a mutation in its GTPase catalytic domain that blocks its activity. We then analyzed cell migration by transwell assay. Again, we observed the same trend of cell chemotaxis inhibition (Fig 4C), thus indicating the dependence on Drp1 of the migratory process. This set of experiments unraveled a role for Drp1 and mitochondrial fragmentation on modulating the migratory capability in oncocytic thyroid cancer cells.
Discussion
Our study on a subset of human thyroid oncocytic tumors sheds light on a pivotal role of mitochondrial dynamics in determining this specific cancer phenotype and, at least partially, its malignant features. Our data highlight an alteration in the mitochondrial network regulation, particularly relevant in oncocytic tumors, and culminating in a weakened fusion and enhanced fission of the organelles.
The fact that both the pro-fusion proteins Mfn2 and Opa1 were more expressed in oncocytic thyroid tumors than in their non-oncocytic counterparts may be related to other known cellular of these proteins: Mfn2-dependent ER-mitochondria tethering and Opa1 anti-apoptotic role in maintaining a proper cristae structure[27,28]. We can speculate that Mfn2 could regulate calcium signaling and ER stress-induced cell death, thus delaying apoptosis. Alternatively, it may facilitate ATP generation in the thyroid model as in the case of cardiac and skeletal muscle setting[41,42]. Although Opa1 was overexpressed in oncocytic thyroid tumor samples, no significant difference was found in cell lines. This apparent discrepancy may reflect tumor cells’ adaptive response to specific tumor microenvironmental conditions that might not be related to Opa1’s fusion activity[43].
Moreover, the decrease in Mfn1 levels in the oncocytic cell line, could have the functional meaning of delaying the oxidative stress-induced loss of mitochondrial membrane in such a highly-rich ROS environment as in the case of mitochondria-rich tumors (as observed in cardiomyocytes[44]), although this might not justify per se the observed increase in the mitochondrial network fragmentation. Such a finding is most likely related to alterations in the fission machinery. Indeed, both ex vivo and in vitro data also showed an overexpression of the fission proteins Drp1 and Fis1 in the oncocytic thyroid tumors histotype, favoring fission over fusion. Thus, the high amount of small mitochondria in these cells most probably results from a combination of both increased fission and decreased fusion processes. Fis1 is permanently anchored to mitochondria and, besides its role in fission, “behaves” like a mitochondrial structural protein. It therefore makes sense that mitochondrion-rich cells present an increase in Fis1 expression.
Regarding Drp1, we found that the mean expression levels in oncocytic thyroid tumors are higher than in non-oncocytic ones. No statistically significant differences between benign and malignant thyroid tumors were observed in the expression levels of Drp1; however, when we considered in our analysis only the oncocytic tumors, Drp1 expression levels were higher in carcinomas than in adenomas. This finding suggests that Drp1 may play a role in malignant transformation within the oncocytic thyroid tumor setting. Additionally, in regard to the non-oncocytic tumors, no differences in the expression levels of any protein were found between benign and malignant thyroid tumors. We believe that when we consider all thyroid tumors histotypes, these differences might become diluted; a possible correlation between mitochondrial dynamics proteins expression levels might thereby become lost, thus reinforcing the importance of these proteins in the oncocytic group.
In our in vitro results it is interesting to note that, although no differences were found in Drp1 mRNA levels when comparing the two thyroid cell lines, by western blot analysis the oncocytic XTC.UC1 presented significantly higher expression Drp1 levels than did the non-oncocytic TPC1. Drp1 exists as a cytosolic protein which actively needs to translocate to the mitochondrial outer membrane to promote organelle division. Several forms of post-translational regulation of Drp1 have been reported; it is possible that in oncocytic tumors, Drp1 could be stabilized, namely by Ser-616 phosphorylation or Ser-637 dephosphorylation, and translocated to mitochondria, thus promoting an unbalanced organelle division as previously described[45,46,47,48]. We saw that the oncocytic cell line displays mitochondria which are particularly small and often with an aberrant shape. We also observed a significant co-localization of Drp1 with the mitochondrial membrane upon heavy membrane fraction analysis in the same model. We then logically went on to investigate on these cells’ migratory capability.
By scratch-wound and transwell assay we observed that oncocytic XTC.UC1 cells have higher migration/invasion capacity. XTC.UC1 being a cell line derived from a metastatic tumor, it makes sense that it has a higher motility and migration ability than TPC1. These observations are not biased by differences in cell proliferation rates since the proliferation rate is higher in TPC1 than in XTC.UC1 cell line[49].
Given that we observed Drp1 overexpression in the oncocytic cell line as well in the oncocytic tumors, where it associates with malignancy, we hypothesize that the Drp1 protein may have a role in the observed higher migration/invasion abilities of the XTC.UC1 cells. Our hypothesis was supported by the observation that the inhibition of Drp1 function, both pharmacologically and genetically, leads to a decrease in the migration/invasion capacity of the oncocytic XTC.UC1 cells. Our results match previously published data showing that mitochondrial dynamics regulates lymphocyte and breast cancer cell migration[30,31]. In both these experimental models, either inhibiting the fission process by Drp1 downregulation or promoting a hiperfused mitochondrial network phenotype, by overexpressing Mfn1, led to limited cell migration or invasion. In our case also, a block of Drp1 was sufficient to reduce the oncocytic cell migration.
Our hypothesis would be better verified if we could use other cell lines, namely a cell line derived from an oncocytic adenoma and one from a non-metastatic primary oncocytic carcinoma. However these cell lines do not exist. Alternatively, we cannot rely on primary cultures of oncocytic and non-oncocytic, benign or malignant thyroid tumors, since unfortunately the culture of primary thyroid cells has been very difficult to obtain and do not last in the laboratory long enough for us to perform the studies. Those cell lines, if available, would be valuable tools for understanding not only thyroid tumors’ oncocytic transformation, but also their malignant transformation.
To sum up, our data show that the mitochondrial fission proteins, Drp1 and Fis1, are overexpressed in thyroid oncocytic cell tumors. This suggests that the mitochondrial fission process upregulation might contribute to mitochondrial accumulation in tumor cells and thus be responsible, at least partially, for the oncocytic cell phenotype. Furthermore, we found that mitochondrial fission protein expression, in particular Drp1 overexpression, correlates with oncocytic thyroid tumor malignancy ex vivo and also with a higher migration/invasion ability of the XTC.UC1 cell line, an oncocytic cell line derived from a metastatic malignant thyroid tumor.
Further studies on the mitochondrial dynamics are needed to better dissect some of the cellular dysfunctions observed in tumor cells in general and in oncocytic tumor cells in particular.
Supporting Information
(TIF)
UC1 cells have higher motility and need less time to fill in the wound.
(TIF)
(TIF)
Acknowledgments
This study was partially supported by the project PIC/IC/83037/2007 (to VM), the project MFAG-12120 from AIRC and GR-2011-02351643 from the Italian Ministry of Health (to SC). Further funding was obtained from the project ‘Microenvironment, metabolism and cancer’ based at IPATIMUP and partially supported by Programa Operacional Regional do Norte (ON.2—O Novo Norte), under the Quadro de Referência Estratégico Nacional (QREN), and through the Fundo Europeu de Desenvolvimento Regional (FEDER). IPATIMUP is an Associate Laboratory of the Portuguese Ministry of Science, Technology and Higher Education and is partially supported by the Portuguese Foundation for Science and Technology (FCT). The study was also FCT through a PhD grant to AFS (Ref.: SFRH/BD/39104/2007).
We thank FR Mariotti for technical help.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was partially supported by the project PIC/IC/83037/2007 (to VM), the project MFAG-12120 from AIRC and GR-2011-02351643 from the Italian Ministry of Health (to SC). Further funding was obtained from the project ‘Microenvironment, metabolism and cancer’ based at IPATIMUP and partially supported by Programa Operacional Regional do Norte (ON.2—O Novo Norte), under the Quadro de Referência Estratégico Nacional (QREN), and through the Fundo Europeu de Desenvolvimento Regional (FEDER). IPATIMUP is an Associate Laboratory of the Portuguese Ministry of Science, Technology and Higher Education and is partially supported by the Portuguese Foundation for Science and Technology (FCT). The study was also FCT through a PhD grant to AFS (Ref.: SFRH/BD/39104/2007).
References
- 1. Warburg O (1956) On the Origin of Cancer Cells. Science 123: 309–314. [DOI] [PubMed] [Google Scholar]
- 2. Maximo V, Botelho T, Capela J, Soares P, Lima J, Taveira A, et al. (2005) Somatic and germline mutation in GRIM-19, a dual function gene involved in mitochondrial metabolism and cell death, is linked to mitochondrion-rich (Hurthle cell) tumours of the thyroid. Br J Cancer 92: 1892–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maximo V, Lima J, Soares P, Sobrinho-Simoes M (2009) Mitochondria and cancer. Virchows Arch 454: 481–495. 10.1007/s00428-009-0766-2 [DOI] [PubMed] [Google Scholar]
- 4. Corrado M, Scorrano L, Campello S (2012) Mitochondrial dynamics in cancer and neurodegenerative and neuroinflammatory diseases. Int J Cell Biol 2012: 729290 10.1155/2012/729290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maximo V, Lima J, Prazeres H, Soares P, Sobrinho-Simoes M (2012) The biology and the genetics of Hurthle cell tumors of the thyroid. Endocr Relat Cancer 19: R131–147. 10.1530/ERC-11-0354 [DOI] [PubMed] [Google Scholar]
- 6. Maximo V, Sobrinho-Simoes M (2000) Hurthle cell tumours of the thyroid. A review with emphasis on mitochondrial abnormalities with clinical relevance. Virchows Arch 437: 107–115. [DOI] [PubMed] [Google Scholar]
- 7. Tallini G (1998) Oncocytic tumours. Virchows Arch 433: 5–12. [DOI] [PubMed] [Google Scholar]
- 8. Ito K, Tsukuda M, Kawabe R, Nakagawa C, Matsushita K, Kubota A, et al. (2000) Benign and malignant oncocytoma of the salivary glands with an immunohistochemical evaluation of Ki-67. ORL J Otorhinolaryngol Relat Spec 62: 338–341. [DOI] [PubMed] [Google Scholar]
- 9. Gudbjartsson T, Hardarson S, Petursdottir V, Thoroddsen A, Magnusson J, Einarsson GV. (2005) Renal oncocytoma: a clinicopathological analysis of 45 consecutive cases. BJU Int 96: 1275–1279. [DOI] [PubMed] [Google Scholar]
- 10. Lima J, Máximo V, Soares P, Portugal R, Guimarães S, Sobrinho-Simões M. (2009) Mitochondria and Oncocytomas In: Singh KK, Costello LC, editors. Mitochondria and cancer. New York: Springer,. [Google Scholar]
- 11. Roth SI, Olen E, Hansen LS (1962) The eosinophilic cells of the parathyroid (oxyphil cells), salivary (oncocytes), and thyroid (Huerthle cells) glands. Light and electron microscopic observations. Lab Invest 11: 933–941. [PubMed] [Google Scholar]
- 12. Sobrinho-Simoes MA, Nesland JM, Holm R, Sambade MC, Johannessen JV (1985) Hurthle cell and mitochondrion-rich papillary carcinomas of the thyroid gland: an ultrastructural and immunocytochemical study. Ultrastruct Pathol 8: 131–142. [DOI] [PubMed] [Google Scholar]
- 13. Nesland JM, Sobrinho-Simoes MA, Holm R, Sambade MC, Johannessen JV (1985) Hurthle-cell lesions of the thyroid: a combined study using transmission electron microscopy, scanning electron microscopy, and immunocytochemistry. Ultrastruct Pathol 8: 269–290. [DOI] [PubMed] [Google Scholar]
- 14. Cheung CC, Ezzat S, Ramyar L, Freeman JL, Asa SL (2000) Molecular basis off hurthle cell papillary thyroid carcinoma. J Clin Endocrinol Metab 85: 878–882. [DOI] [PubMed] [Google Scholar]
- 15. Trovisco V, Vieira de Castro I, Soares P, Maximo V, Silva P, Magalhães J, et al. (2004) BRAF mutations are associated with some histological types of papillary thyroid carcinoma. J Pathol 202: 247–251. [DOI] [PubMed] [Google Scholar]
- 16. Hoos A, Stojadinovic A, Singh B, Dudas ME, Leung DH, Shaha AR, et al. (2002) Clinical significance of molecular expression profiles of Hurthle cell tumors of the thyroid gland analyzed via tissue microarrays. Am J Pathol 160: 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sobrinho-Simoes M, Maximo V, Rocha AS, Trovisco V, Castro P, Preto A, et al. (2008) Intragenic mutations in thyroid cancer. Endocrinol Metab Clin North Am 37: 333–362, viii. 10.1016/j.ecl.2008.02.004 [DOI] [PubMed] [Google Scholar]
- 18. de Vries MM, Celestino R, Castro P, Eloy C, Maximo V, van der Wal JE, et al. RET/PTC rearrangement is prevalent in follicular Hurthle cell carcinomas. Histopathology 61: 833–843. 10.1111/j.1365-2559.2012.04276.x [DOI] [PubMed] [Google Scholar]
- 19. Evans HL, Vassilopoulou-Sellin R (1998) Follicular and Hurthle cell carcinomas of the thyroid: a comparative study. Am J Surg Pathol 22: 1512–1520. [DOI] [PubMed] [Google Scholar]
- 20. Maximo V, Soares P, Lima J, Cameselle-Teijeiro J, Sobrinho-Simoes M (2002) Mitochondrial DNA Somatic Mutations (Point Mutations and Large Deletions) and Mitochondrial DNA Variants in Human Thyroid Pathology: A Study with Emphasis on Hurthle Cell Tumors. Am J Pathol 160: 1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pereira L, Soares P, Maximo V, Samuels DC (2012) Somatic mitochondrial DNA mutations in cancer escape purifying selection and high pathogenicity mutations lead to the oncocytic phenotype: pathogenicity analysis of reported somatic mtDNA mutations in tumors. BMC Cancer 12: 53 10.1186/1471-2407-12-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Attardi G, Yoneda M, Chomyn A (1995) Complementation and segregation behavior of disease-causing mitochondrial DNA mutations in cellular model systems. Biochim Biophys Acta 1271: 241–248. [DOI] [PubMed] [Google Scholar]
- 23. Heddi A, Faure-Vigny H, Wallace DC, Stepien G (1996) Coordinate expression of nuclear and mitochondrial genes involved in energy production in carcinoma and oncocytoma. Biochim Biophys Acta 1316: 203–209. [DOI] [PubMed] [Google Scholar]
- 24. Maximo V, Sobrinho-Simoes M (2000) Mitochondrial DNA 'common' deletion in Hurthle cell lesions of the thyroid. J Pathol 192: 561–562. [DOI] [PubMed] [Google Scholar]
- 25. Gasparre G, Porcelli AM, Bonora E, Pennisi LF, Toller M, Iommarini L, et al. (2007) Disruptive mitochondrial DNA mutations in complex I subunits are markers of oncocytic phenotype in thyroid tumors. Proc Natl Acad Sci U S A 104: 9001–9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bonora E, Evangelisti C, Bonichon F, Tallini G, Romeo G (2006) Novel germline variants identified in the inner mitochondrial membrane transporter TIMM44 and their role in predisposition to oncocytic thyroid carcinomas. Br J Cancer 95: 1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, et al. (2006) OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126: 177–189. [DOI] [PubMed] [Google Scholar]
- 28. de Brito OM, Scorrano L (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456: 605–610. 10.1038/nature07534 [DOI] [PubMed] [Google Scholar]
- 29. Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J 26: 2175–2186. 10.1096/fj.11-196543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Campello S, Lacalle RA, Bettella M, Manes S, Scorrano L, Viola A. (2006) Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J Exp Med 203: 2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW, et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene 32: 4814–4824. 10.1038/onc.2012.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Westermann B Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11: 872–884. 10.1038/nrm3013 [DOI] [PubMed] [Google Scholar]
- 33. Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L (2004) OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A 101: 15927–15932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeLellis RA LR, Heitz PU, Eng C, editor (2004) World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Endocrine Glands. Lyon: IARC Press. [Google Scholar]
- 35. Packeisen J, Korsching E, Herbst H, Boecker W, Buerger H (2003) Demystified…tissue microarray technology. Mol Pathol 56: 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meireles AM, Preto A, Rocha AS, Rebocho AP, Maximo V, Pereira-Castro I, et al. (2007) Molecular and genotypic characterization of human thyroid follicular cell carcinoma-derived cell lines. Thyroid 17: 707–715. [DOI] [PubMed] [Google Scholar]
- 37. Ricarte-Filho JC, Ryder M, Chitale DA, Rivera M, Heguy A, Ladanyi M, et al. (2009) Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Res 69: 4885–4893. 10.1158/0008-5472.CAN-09-0727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frezza C, Cipolat S, Scorrano L (2007) Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2: 287–295. [DOI] [PubMed] [Google Scholar]
- 39. Cory G Scratch-wound assay. Methods Mol Biol 769: 25–30. 10.1007/978-1-61779-207-6_2 [DOI] [PubMed] [Google Scholar]
- 40. Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, et al. (2008) Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell 14: 193–204. 10.1016/j.devcel.2007.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ngoh GA, Papanicolaou KN, Walsh K. Loss of mitofusin 2 promotes endoplasmic reticulum stress. J Biol Chem 287: 20321–20332. 10.1074/jbc.M112.359174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eisner V, Csordas G, Hajnoczky G Interactions between sarco-endoplasmic reticulum and mitochondria in cardiac and skeletal muscle—pivotal roles in Ca(2)(+) and reactive oxygen species signaling. J Cell Sci 126: 2965–2978. 10.1242/jcs.093609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155: 160–171. 10.1016/j.cell.2013.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Papanicolaou KN, Ngoh GA, Dabkowski ER, O'Connell KA, Ribeiro RF Jr., Stanley WC, et al. Cardiomyocyte deletion of mitofusin-1 leads to mitochondrial fragmentation and improves tolerance to ROS-induced mitochondrial dysfunction and cell death. Am J Physiol Heart Circ Physiol 302: H167–179. 10.1152/ajpheart.00833.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Gräber S, et al. (2006) Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J 25: 3900–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K (2007) Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem 282: 11521–11529. [DOI] [PubMed] [Google Scholar]
- 47. Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, et al. (2008) Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A 105: 15803–15808. 10.1073/pnas.0808249105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Braschi E, Zunino R, McBride HM (2009) MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep 10: 748–754. 10.1038/embor.2009.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goncalves AP, Videira A, Maximo V, Soares P. Synergistic growth inhibition of cancer cells harboring the RET/PTC1 oncogene by staurosporine and rotenone involves enhanced cell death. J Biosci 36: 639–648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
UC1 cells have higher motility and need less time to fill in the wound.
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.