Abstract
Immunoglobulin (IG) gene rearrangement and expression are central to disease resistance and health maintenance in animals. The IG kappa (IGK) locus in swine (Sus scrofa domestica) contributes to approximately half of all antibody molecules, in contrast to many other Cetartiodactyla, whose members provide the majority of human dietary protein and in which kappa locus utilization is limited. The porcine IGK variable locus is 27.9 kb upstream of five IG kappa J genes (IGKJ) which are separated from a single constant gene (IGKC) by 2.8 kb. Fourteen variable genes (IGKV) were identified, of which nine are functional and two are open reading frame (ORF). Of the three pseudogenes, IGKV3-1 contains a frameshift and multiple stop codons, IGKV7-2 contains multiple stop codons, and IGKV2-5 is missing exon 2. The nine functional IGKV genes are phylogenetically related to either the human IGKV1 or IGKV2 subgroups. IGKV2 subgroup genes were found to be dominantly expressed. Polymorphisms were identified on overlapping BACs derived from the same individual such that 11 genes contain amino acid differences. The most striking allelic differences are present in IGKV2 genes, which contain as many as 16 amino acid changes between alleles, the majority of which are in complementarity determining region (CDR) 1. In addition, many IGKV2 CDR1 are shared between genes but not between alleles, suggesting extensive diversification of this locus through gene conversion.
Keywords: Sus scrofa, Porcine, Light chain, kappa, Polymorphism, Gene conversion
Introduction
The adaptive humoral immune response must be able to generate a nearly infinite arsenal of antibodies to provide protection against an incredibly diverse array of pathogens and toxins. In pigs, the three immunoglobulin (IG) loci comprise the IG heavy (IGH) locus on chromosome 7q25–q26, the IG lambda (IGL) locus on 14q16–q21, and the IG kappa (IGK) locus on 3q12–q14 (Yerle et al. 1997). The variable domain of each IG chain is encoded by a variable (V) gene, a diversity (D) gene (heavy chain-only) and a joining (J) gene which rearrange through recognition of recombination signal (RS) sequences by the RAG1 and RAG2 complex and subsequent double strand break repair (McBlane et al. 1995; Kim and Oettinger 2000). The highly variable complementarity determining regions (CDR) 1 and 2 are encoded by the V region and differ in sequence between genes. CDR3 is the result of junctional diversity between genes which rearrange and is largely generated from exonuclease trimming of the gene ends and random nucleotide addition via terminal deoxynucleotidyl transfer-ase (TdT) during gene rearrangement early in B cell development. Thus, the combined three CDR of the V heavy domains (VH) and three CDR of the V light domains (VL) provide antibodies with an immensely diverse antigen-binding repertoire (Wu and Kabat 1970; Lefranc and Lefranc 2001).
The use of either kappa or lambda light chain is first determined by the somatic rearrangement of the kappa locus. However, if both homologous chromosomes fail to produce functional antibody, they are ablated through recombination between a 3′ kappa deleting element (KDE) and a recombining element in the J–C intron. The lambda locus is then able to undergo rearrangement until the B cell either produces a functional light chain or is deleted (Siminovitch et al. 1985). While most members of Cetartiodactyla rely heavily on the lambda locus, pigs are unusual in that their expressed repertoires are nearly 1:1 kappa–lambda (Butler et al. 2005). This variation between species might be due to 1) the ability of either locus to produce functional antibody (i.e. locus complexity) or 2) regulation of kappa locus ablation. However, while the kappa joining (IGKJ) and constant (IGKC) genes are previously described in pigs, the variable (IGKV) gene organization and complexity are unknown (Butler et al. 2006a). Here, we interrogated available porcine genetic and genomic sequence data (Schook et al. 2005; Humphray et al. 2007; Archibald et al. 2010) to more fully characterize the organization, complexity, and expression of the IGK locus. Our findings suggest that the locus is organized similarly to, but is more complex than, other mammalian species that rely primarily on the lambda locus for light chain expression. Interestingly, the most highly expressed IGKV subgroup (IGKV2) is highly polymorphic between alleles from the same animal, with diversity due largely to variation within CDR1. However, while many CDR1 differed between alleles, they were shared between genes, a phenomenon that is best explained by evolution through homologous recombination between genes (i.e. gene conversion). This phenomenon was not, however, observed among members of the similarly sized population of IGKV1 genes which, by comparison, are poorly expressed.
Materials and methods
Identification and sequencing of the bacterial artificial chromosomes
Sus scrofa genome build 9 was queried to identify the bacterial artificial chromosomes (BACs) containing IGKV sequences using the Basic Local Alignment Search Tool (BLAST) within the Ensembl database (Altschul et al. 1990; Hubbard et al. 2002). Nucleotide sequences for each BAC containing the region of interest were downloaded for further analysis from GenBank. The CHORI (Children's Hospital Oakland Research Institute)-242 BAC library used was derived from a single Duroc sow (http://bacpac.chori.org/porcine242.htm). Additionally, two BAC clones (CH242-221I5 and CH242-227G10) were acquired and expanded overnight, and BAC DNA was purified using the Qiagen Plasmid Midi Prep with Qiagen-tip 500 columns. Purified DNA was submitted to the University of Minnesota Biomedical Genomics Center for library preparation and paired-end sequencing using the Illumina GAIIx platform.
Characterization of the porcine IGK locus
Approximately 20 million high quality reads were sorted by molecular tag to differentiate samples and assembled using a combination of ABySS and Velvet (Simpson et al. 2009; Zerbino and Birney 2008). Generated contigs were assembled against the existing BAC sequences using Sequencher 4.10.1 (Gene Codes Corporation). The resulting complete BAC sequences were manually annotated and interrogated for immunoglobulin features such as RS (i.e. heptamers and nonamers), promoters (i.e. octamers), and gene structure using the annotation software Artemis (Rutherford et al. 2000).
The sequences of CH242-221I5, CH242-227G10, and CH242-148A13 were acquired from GenBank (accession numbers: CU694848, FP312898, and CU928807, respectively) and assessed for IGKV, IGKJ, and IGKC genes using BLAST. Phylogenetic analyses were performed in CLC Sequence Viewer (CLC Bio) and Dendroscope (Huson et al. 2007) using Unweighted Pair Group Method with Arithmetic Mean (UPGMA) with 1000 bootstrap iterations. Genes were annotated according to IMGT®, the international ImMunoGeneTics information system® (Lefranc et al. 2009). Translated amino acid sequences of the IGKV genes were compared, and CDR and framework (FR) boundaries were annotated according to the IMGT unique numbering for V domain (Lefranc et al. 2003). IGKC gene translation was annotated according to the IMGT unique numbering for C domain (Lefranc et al. 2005). Expression of germline IGKV genes was compared using 41 BLAST hits obtained from 398,837 porcine expressed sequence tags (ESTs) in GenBank and deposited at http://pigest.ku.dk/index.html (Gorodkin et al. 2007) using an E-value threshold of 10−12.
Nomenclature
The porcine IGKV genes are named according to IMGT nomenclature (Lefranc and Lefranc 2001; Lefranc 2007, 2008), using human V gene subgroup nomenclature to maintain consistency with porcine heavy chain, light chain, and cattle lambda light chain nomenclature (Eguchi-Ogawa et al. 2010; Butler et al. 2005, 2006a; Pasman et al. 2010). The porcine IGKV, IGKJ, and IGKC genes were submitted to IMGT/GENE-DB (Giudicelli et al. 2005). IgBLAST was used to organize IGKV genes into subgroups using a 75% identity threshold. IGKV genes are described based on IMGT nomenclature rules. Briefly, genes were deemed pseudogenes if they contained a truncation, stop codon, frameshift, or a defective initiation codon. Additionally, IGKV genes were described as ORF (open reading frame) if they were missing one (or more) key amino acids (1st-CYS 23, CONSERVED-TRP 41, hydrophobic 89, 2nd-CYS 104). RS were deemed non-canonical if the heptamer was anything other than “CACAGTG” for V-HEPTAMER (or “CACTGTG” for J-HEPTAMER) or if the nonamer contained at least two nucleotides which were each present in less than 10% of all RS described by Ramsden et al. (1994).
Results
Organization of the porcine immunoglobulin kappa locus
The porcine IGK locus contains a single constant gene (IGKC) 2.8 kb downstream from five IGKJ genes. The IGKV locus begins 27.9 kb upstream from IGKJ1. We identified 14 distinct IGKV genes spanning approximately 89 kb (Fig. 1). Two additional IGKV genes were identified on the BAC CH242-148A13 and are most likely orphons as the BAC maps to a different region of chromosome 3. BLAST failed to identify additional IGKV genes in the porcine genome or on other shotgun sequenced CH242 BACs (not shown). Three of the 14 identified IGKV genes within the locus are pseudogenes; IGKV3-1 contains a frameshift and multiple stop codons, IGKV7-2 contains multiple stop codons, and IGKV2-5 is missing the VEXON (Table 1).
Fig. 1.
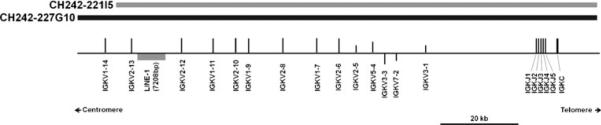
Organization of the porcine IGK locus. Overlapping BACs spanning the region are displayed as grey and black bars to indicate sequence heterogeneity. Genes are displayed along a scaffold line as vertical bars representing functional genes (long bars), pseudogenes (short bars), or ORF (intermediate bars). A nearly intact LINE-1 insertion present on CH242-221I5 but not on CH242-227G10 is depicted as a grey box. Genes above the scaffold line are transcribed left to right. Genes below the line are expressed in the opposite orientation
Table 1.
IGKV alleles
| IMGT subgroup | IMGT gene name | IMGT allele name | Fct | Clone names | Clone accession numbers |
|---|---|---|---|---|---|
| IGKV1 | IGKV1-7 | IGKV1-7*01 | F | CH242-227G10 | FP312898 |
| IGKV1-7*02 | F | CH242-221I5 | CU694848 | ||
| IGKV1-9 | IGKV1-9*01 | F | CH242-227G10 | FP312898 | |
| IGKV1-9*02 | F | CH242-221I5 | CU694848 | ||
| IGKV1-11 | IGKV1-11*01 | F | CH242-227G10 | FP312898 | |
| IGKV1-11*02 | F | CH242-221I5 | CU694848 | ||
| IGKV1-14 | IGKV1-14*01 | F | CH242-227G10 | FP312898 | |
| IGKV2 | IGKV2-5 | IGKV2-5*01 | Pa | CH242-227G10 | FP312898 |
| CH242-221I5 | CU694848 | ||||
| IGKV2-6 | IGKV2-6*01 | F | CH242-227G10 | FP312898 | |
| IGKV2-6*02 | F | CH242-221I5 | CU694848 | ||
| IGKV2-8 | IGKV2-8*01 | F | CH242-227G10 | FP312898 | |
| IGKV2-8*02 | F | CH242-221I5 | CU694848 | ||
| IGKV2-10 | IGKV2-10*01 | F | CH242-227G10 | FP312898 | |
| IGKV2-10*02 | F | CH242-221I5 | CU694848 | ||
| IGKV2-12 | IGKV2-12*01 | F | CH242-227G10 | FP312898 | |
| IGKV2-12*02 | F | CH242-221I5 | CU694848 | ||
| IGKV2-13 | IGKV2-13*01 | F | CH242-227G10 | FP312898 | |
| IGKV2-13*02 | F | CH242-221I5 | CU694848 | ||
| IGKV2/OR3-1 | IGKV2/OR3-1*01 | Pb | CH242-148A13 | CU928807 | |
| IGKV3 | IGKV3-1 | IGKV3-1*01 | Pc | CH242-227G10 | FP312898 |
| CH242-221I5 | CU694848 | ||||
| IGKV3-3 | IGKV3-3*01 | ORFd | CH242-227G10 | FP312898 | |
| IGKV3-3*02 | ORFe | CH242-221I5 | CU694848 | ||
| IGKV3/OR3-2 | IGKV3/OR3-2*01 | Pf | CH242-148A13 | CU928807 | |
| IGKV5 | IGKV5-4 | IGKV5-4*01 | ORFg | CH242-227G10 | FP312898 |
| IGKV5-4*02 | ORFg | CH242-221I5 | CU694848 | ||
| IGKV7 | IGKV7-2 | IGKV7-2*01 | Ph | CH242-227G10 | FP312898 |
| IGKV7-2*02 | Ph | CH242-221I5 | CU694848 |
Fct functionality, F functional, P pseudogene, ORF open reading frame
V-EXON is missing
Orphon, stop codon at position 109
Frameshift in CDR1-IMGT
L89>R
L89>C
Orphon, frameshifts in FR1-IMGT and FR2-IMGT
1st-CYS C23>R, CONSERVED-TRP W41>C
Stop codons at positions 44, 55, and 67
The kappa deleting element (KDE) was identified approximately 23.2 kb downstream from IGKC and contains a canonical RS that is identical to the cattle KDE RS (Das et al. 2009). Likewise, the recombining element in the J–C intron is comprised of a canonical heptamer (CACAGTG). The conservation of this system between pigs and other members of Cetartiodactyla suggests that ease of kappa locus ablation does not explain preferential lambda locus usage (Das et al. 2009).
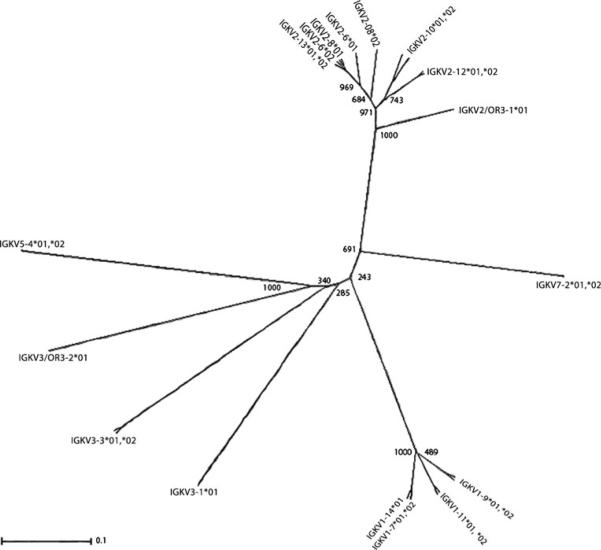
Phylogenetic analysis of IGKV genes
The first four C-proximal IGKV genes are related to the human IGKV subgroups IGKV3, IGKV7, and IGKV5. The fifth gene, IGKV2-5, is missing the V-EXON, and its membership in the IGKV2 subgroup is inferred here based solely on its leader sequence. The remaining nine genes are split among the IGKV1 and IGKV2 subgroups (Fig. 2). In general, this pattern of gene organization is mirrored in humans (Kawasaki et al. 2001; Lefranc and Lefranc 2001). Compared to humans, the swine IGKV3-1, IGKV7-2, and IGKV5-4 genes are closest to the human C-proximal IGKV3-7, IGKV7-3, and IGKV5-2 genes, respectively. However, the remaining porcine genes forming the IGKV1 and IGKV2 subgroups are all phylogenetically similar to only one or two human IGKV genes (IGKV1-9, IGKV1-27, IGKV2-28, and IGKV2-40). This suggests that substantial IGKV repertoire expansion and contraction of specific IGKV subgroups has occurred during the evolutionary divergence of swine and humans.
Fig. 2.
Phylogenetic analysis of porcine IGKV nucleotide sequences using UPGMA with 1000 bootstrap iterations. Nucleotide sequences are derived from the V-EXON. Thus, IGKV2-5 is not represented, as it is missing this region. Most porcine IGKV genes cluster with either of two subgroups, IGKV1 or IGKV2. Nodes are labeled with bootstrap values
Allelic variation and gene conversion
The sequenced BACs CH242-221I5 and CH242-227G10 overlap across much of the characterized kappa locus. Thirteen of the 14 IGKV genes are represented on both BACs as are all IGKJ genes and IGKC (Fig. 1). Nucleotide identity across this overlap varies from approximately 99% in the IGKJ–IGKC locus to 97% in the IGKV locus. Differences of one or more nucleotides between BACs were identified in 11 IGKV genes (Table 1), and amino acid changes were found between each of these alleles. A total of 62 amino acid changes were identified and ranged from two (IGKV7-2 and IGKV5-4) to 16 (IGKV2-8) per gene (Table 2). Interestingly, all intact IGKV2 subgroup genes except IGKV2-12 were highly polymorphic in CDR1 (Table 2). The CDR1 of IGKV2-6, IGKV2-8, IGKV2-10, and IGKV2-13 contained four, seven, six, and three amino acid changes, respectively, between alleles. Interestingly, many of these CDR1 are shared between genes, while the upstream and downstream regions remain largely identical between alleles (Table 2). For example, IGKV2-13 and IGKV2-6 on CH242-221I5 possess the same CDR1 sequence which is different from their alleles on CH242-227G10 (Table 3). This suggests that the IGKV2 repertoire has diversified CDR1 through gene duplication and gene conversion. Thus, the allelic diversity of the IGKV2 subgroup may be quite large.
Table 2.
Protein display of in-frame IGKV genes using IMGT unique numbering
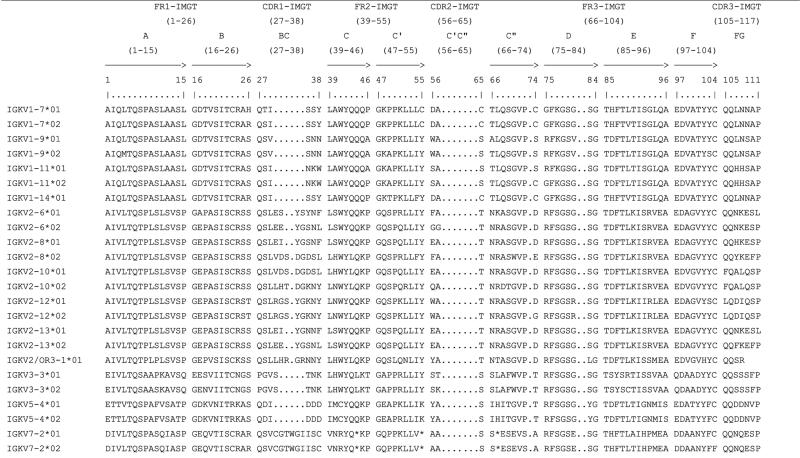
|
Table 3.
Alignment of porcine IGKV CDR1-IMGT

|
In contrast to the IGKV genes, the IGKJ genes were all identical at the amino acid level between BACs (not shown). IGKC contains a single nucleotide polymorphism resulting in a Ser-122 (IGKC*01) to Asn-122 (IGKC*02) amino acid change (Table 4). Comparison with Minnesota Miniature swine complimentary DNA sequence (GenBank: M59321.1) reveals the presence of ten amino acid differences from IGKC*01 described here, indicating substantial variation between pig breeds even within the relatively conserved constant region (Lammers et al. 1991). The IGKC Cys-126 is the terminal amino acid in cattle (GenBank: BC151500.1) and other described species in the IMGT database, including humans, mice, rats, and rabbits (Emorine and Max 1983; Giudicelli et al. 2005; Hieter et al. 1980; Sheppard and Gutman 1981). Interestingly, in both the Minnesota Miniature and Duroc pig breeds IGKC contains two additional amino acids downstream from Cys-126 (Table 4; Lammers et al. 1991).
Table 4.
Protein display of IGKC genes using IMGT unique numbering

|
Expression of IGKV genes
In addition to the presence of stop codons and frame-shifts, functionality is further restricted in some IGKV genes by non-canonical RS necessary for recombination and octamers necessary for transcription. Non-canonical RS were found in three of the four IGKV1 subgroup genes (IGKV1-7, IGKV1-9, and IGKV1-14) (Table 5). The remaining member, IGKV1-11, contained a non-canonical RS on CH242-221I5 but a canonical sequence on CH242-227G10. Additionally, non-canonical RS were also located in the first two pseudogenes (IGKV3-1 and IGKV7-2). Non-canonical octamers (i.e. other than ATTTGCAT) were present upstream of IGKV3-3 and two of the four IGKV1 subgroup genes (IGKV1-7 and IGKV1-14). Non-canonical 5′ splice sites (i.e. other than GT) were also observed for several genes, most notably among IGKV1 subgroup genes (Table 5). Combined, these data suggest that the functional repertoire of the kappa variable region is largely restricted to members of the IGKV2 subgroup.
Table 5.
Genomic features of the porcine IGKV genes
| IGKV gene | Fct | Octamer (promoter) | nt | ini. | L-PART1 (exon 1) (nt) | 5' Splice site | Intron (nt) | 3' Splice site | V-EXON (exon 2) (nt) | RS |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V-HEPTAMER | V-SPACER (nt) | V-NONAMER | ||||||||||
| IGKV3-1*01 | P | ATTTGCAT | 110 | ATG | 49 | GT | 180 | AG | 311 | CACAGTG | 12 | ATATTAACT |
| IGKV7-2*01 | P | ATTTGCAT | 94 | ATG | 49 | GC | 234 | AG | 316 | CACAATG | 10 | CAAAAAACC |
| IGKV3-3*01 | ORF | ATTTGCAC | 98 | ATG | 49 | GT | 208 | AG | 301 | CACAGTG | 12 | ACAAAAACC |
| IGKV5-4*01 | ORF | ATTTGCAT | 94 | ATG | 49 | GT | 420 | AG | 301 | CACAGTG | 12 | ACAAAAACT |
| IGKV2-5*01 | P | ATTTGCAT | 98 | ATG | 49 | GT | - | - | - | - | - | - |
| IGKV2-6*01 | F | ATTTGCAT | 98 | ATG | 49 | GT | 368 | AG | 310 | CACAGTG | 12 | ACACAAACC |
| IGKV1-7*01 | F | ATCTGCTT | 93 | ATG | 49 | GG | 196 | AG | 298 | CAGAGTG | 12 | ACCTAACCC |
| IGKV2-8*01 | F | ATTTGCAT | 98 | ATG | 49 | GT | 383 | AG | 313 | CACAGTG | 12 | ACACAAACC |
| IGKV1-9*01 | F | ATTTGCAT | 93 | ATG | 49 | GT | 193 | AG | 298 | CACAGTG | 12 | ACAAACCCC |
| IGKV2-10*01 | F | ATTTGCAT | 98 | ATG | 49 | GT | 383 | AG | 313 | CACAGTG | 12 | ACACAAACC |
| IGKV1-11*01 | F | ATTTGCAT | 93 | ATG | 49 | GT | 194 | AG | 298 | CACAGTG | 12 | CCAAAAACC |
| IGKV1-11*02 | F | ATCTGCTT | 93 | ATG | 49 | GG | 194 | AG | 298 | CACAGTG | 12 | CCAAAAACC |
| IGKV2-12*01 | F | ATTTGCAT | 98 | ATG | 49 | GG | 375 | AG | 313 | CACAGTG | 12 | ACACAAACC |
| IGKV2-13*01 | F | ATTTGCAT | 98 | ATG | 49 | GT | 367 | AG | 310 | CACAGTG | 12 | ACACAAACC |
| IGKV1-14*01 | F | ATCTGCTT | 93 | ATG | 49 | GG | 196 | AG | 298 | CAGAGTG | 12 | ACCTAACCC |
| IGKV2/OR3-1 | P | ATTTGCAT | 98 | ATG | 49 | GT | 377 | AG | 313 | CACAGTG | 12 | ACACAAACC |
| IGKV3/OR3-2 | P | GTTTGCAT | 91 | ATG | 49 | GT | 194 | AG | 273 | CACAGTG | 12 | ACAGAAACC |
IMGT standardized labels are written in capital letters
Fct functionality, F functional, P pseudogene, ORF open reading frame
Indeed, BLAST analysis of a porcine EST database revealed that all five functional IGKV2 subgroup genes (IGKV2-6, IGKV2-8, IGKV2-10, IGKV2-12, and IGKV2-13) were expressed, with the most highly expressed genes being IGKV2-10 and IGKV2-13) (Fig. 3). Relatively low level expression (a combined 19% of EST hits) of IGKV1-9 and IGKV1-11 was also observed. They are the only two IGKV1 subgroup genes possessing both a canonical RS and promoter octamer. These findings agree with an earlier report that IGKV2 represents approximately 90% of the expressed porcine pre-immune repertoire (Butler et al. 2004). The gene IGKV5-4 was not found in the database despite having an intact open reading frame and both canonical RS and octamer (Table 5). This confirms that the amino acid changes (C23>R and W41>C) that assign the functionality of that gene to “ORF” instead of “functional”, have a detrimental effect on the variable domain structure.
Fig. 3.
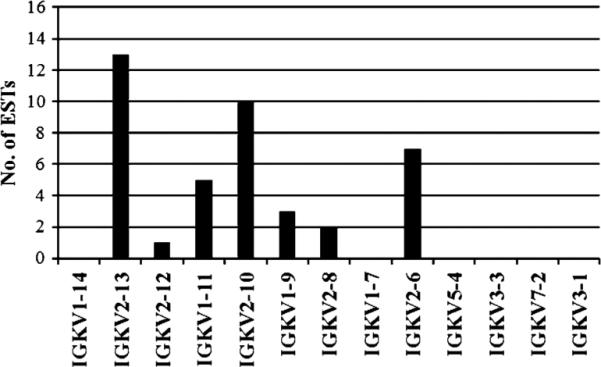
Expression of IGKV genes. BLAST analysis revealed 41 hits from 398,837 expressed sequence tags in GenBank and deposited at http://pigest.ku.dk/index.html (Gorodkin et al. 2007). The x-axis is organized by gene order in the locus with the most 5′ (most C-distal) IGKV genes on the left. The IGKV2 subgroup dominates (~ 80%) the expression profile
Discussion
The general organization of the porcine kappa locus is typical of mammals. Functional diversity of the locus is more complex than in cattle, in which only eight of 24 IGKV genes are functional (Ekman et al. 2009). It is similar to the human kappa locus, which also has high functional diversity and equivalent expression of IGK and IGL (Kawasaki et al. 2001; Lefranc and Lefranc 2001; Butler et al. 2005). The high level of functional diversity in swine and humans may facilitate efficient IGK expression, thus avoiding KDE-dependent ablation of IGKC. Conversely, low functional diversity in cattle may result in a high rate of KDE-dependent IGKC ablation and low relative IGK usage in B cells.
IGKV2 subgroup genes are dominantly expressed as previously described (Butler et al. 2004). However, we identified only five complete IGKV2 genes in the first 89 kb of the IGKV locus, which is substantially less than an estimated 61 IGKV2 genes in 250 kb of sequence identified by Southern hybridization and sequencing of IGKV2-specific PCR clones (Butler et al. 2004). The previous estimate may be an overestimation of kappa locus complexity since the density of genes across the entire locus would need to be at least twice that observed by directly sequencing BACs. The IGKV gene density observed here is similar to that of the human C-proximal IGKV cluster (75 IGKV genes per 542 kb) (Kawasaki et al. 2001; Lefranc and Lefranc 2001). The presence of additional upstream IGKV genes in the porcine locus has not been ruled out, although a BLAST search of shotgun sequenced genomic BACs did not reveal any additional upstream sequence specific to the kappa locus.
The presence of extensive IGKV allelic differences, up to 10% of sequence variation in IGKV2-8, occurs primarily within the CDR1 of IGKV2 subgroup genes. Further, CDR1 appears to be shared by some IGKV2 subgroup genes, but not necessarily between alleles. Interestingly, a similar phenomenon was observed in the porcine IGH locus, in which many IGHV genes share individual CDR (Butler et al. 2006b; Eguchi-Ogawa et al. 2010). The pattern of variation may be due to evolution through non-crossover homologous recombination events, namely gene conversion. Evidence for germline gene conversion was also observed in the human IGKV (Bentley and Rabbitts 1983) and mouse IGHV (Cohen and Givol 1983) genes and in the human IGHC genes (Flanagan et al. 1984; Lefranc et al. 1986; Huck et al. 1989). IG gene conversion is generally presented as a mechanism of somatic antibody diversification, especially in chickens and rabbits which utilize a diverse array of pseudogenes as templates for functional IG gene assembly during B cell development to diversify limited functional germline repertoires (recently reviewed by Kurosawa and Ohta (2011)). In swine, however, there is little evidence supporting somatic diversification through templated gene conversion (Butler et al. 2006a). Thus, the porcine kappa locus appears to have achieved repertoire diversity through gene duplication and germline gene conversion to increase allelic variation in CDR1.
Acknowledgements
We thank Erin Babineau for excellent technical assistance, and Dr. Jane Loveland for her generous review of the manuscript. This work was supported by a grant from the National Pork Board (10–139).
Contributor Information
John C. Schwartz, Department of Veterinary and Biomedical Sciences, University of Minnesota, 1971 Commonwealth Avenue, St. Paul, MN 55108, USA
Marie-Paule Lefranc, IMGT®, the international ImMunoGeneTics information system®, Laboratoire d'ImmunoGénétique Moléculaire and Institut de Génétique Humaine, Université Montpellier, Montpellier 34396, France.
Michael P. Murtaugh, Department of Veterinary and Biomedical Sciences, University of Minnesota, 1971 Commonwealth Avenue, St. Paul, MN 55108, USA
References
- Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Archibald AL, Bolund L, Churcher C, Fredholm M, Groenen MAM, Harlizius B, Lee KT, Milan D, Rogers J, Rothschild MF, Uenishi H, Wang J, Schook LB, the Swine Genome Sequencing Consortium Pig genome sequence—analysis and publication strategy. BMC Genomics. 2010;11:438. doi: 10.1186/1471-2164-11-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL, Rabbitts TH. Evolution of V genes: evidence indicating that recently duplicated human VK sequences have diverged by gene conversion. Cell. 1983;32:181–189. doi: 10.1016/0092-8674(83)90508-1. [DOI] [PubMed] [Google Scholar]
- Butler JE, Sun J, Wertz N, Sinkora M. Antibody repertoire development in swine. Dev Comp Immunol. 2006a;30:199–221. doi: 10.1016/j.dci.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Butler JE, Weber P, Wertz N. Antibody repertoire development in fetal and neonatal piglets. XIII. Hybrid VH genes and the preimmune repertoire revisited. J Immunol. 2006b;177:5459–5470. doi: 10.4049/jimmunol.177.8.5459. [DOI] [PubMed] [Google Scholar]
- Butler JE, Wertz N, Sun J, Wang H, Lemke C, Chardon P, Piumi F, Wells K. The pre-immune variable kappa repertoire of swine is selectively generated from certain subfamilies of Vκ2 and one Jκ gene. Vet Immunol Immunop. 2005;108:127–137. doi: 10.1016/j.vetimm.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Butler JE, Wertz N, Sun J, Wang H, Chardon P, Piumi F, Wells K. Antibody repertoire development in fetal and neonatal pigs.VII. Characterization of the preimmune kappa light chain repertoire. J Immunol. 2004;173:6794–6805. doi: 10.4049/jimmunol.173.11.6794. [DOI] [PubMed] [Google Scholar]
- Cohen JB, Givol D. Allelic immunoglobulin VH genes in two mouse strains: possible germline gene recombination. EMBO J. 1983;2:2013–2018. doi: 10.1002/j.1460-2075.1983.tb01693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Nikolaidis N, Nei M. Genomic organization and evolution of immunoglobulin kappa gene enhancers and kappa deleting element in mammals. Mol Immunol. 2009;46:3171–3177. doi: 10.1016/j.molimm.2009.05.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi-Ogawa T, Wertz N, Sun XZ, Puimi F, Uenishi H, Wells K, Chardon P, Tobin GJ, Butler JE. Antibody repertoire development in fetal and neonatal piglets. XI. The relationship of variable heavy chain gene usage and the genomic organization of the variable heavy chain locus. J Immunol. 2010;184:3734–3742. doi: 10.4049/jimmunol.0903616. [DOI] [PubMed] [Google Scholar]
- Ekman A, Niku M, Liljavirta J, Iivanainen A. Bos taurus genome sequence reveals the assortment of immunoglobulin and surrogate light chain genes in domestic cattle. BMC Immunol. 2009;10:22. doi: 10.1186/1471-2172-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emorine L, Max EE. Structural analysis of a rabbit immunoglobulin kappa 2 J–C locus reveals multiple deletions. Nucl Acids Res. 1983;11:8877–8890. doi: 10.1093/nar/11.24.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V, Chaume D, Lefranc M-P. IMGT/GENE-DB: a comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucl Acids Res. 2005;33:D256–261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JG, Lefranc M-P, Rabbitts TH. Mechanisms of divergence and convergence of the human immunoglobulin alpha1 and alpha2 constant region gene sequences. Cell. 1984;36:681–688. doi: 10.1016/0092-8674(84)90348-9. [DOI] [PubMed] [Google Scholar]
- Gorodkin J, Cirera S, Hedegaard J, Gilchrist MJ, Panitz F, Jørgensen C, Scheibye-Knudsen K, Arvin T, Lumholdt S, Sawera M, Green T, Nielsen BJ, Havgaard JH, Rosenkilde C, Wang J, Li H, Li R, Liu B, Hu S, Dong W, Li W, Yu J, Wang J, Stærfeldt H, Wernersson R, Madsen LB, Thomsen B, Hornshøj H, Bujie Z, Wang X, Wang X, Bolund L, Brunak S, Yang H, Bendixen C, Fredholm M. Porcine transcriptome analysis based on 97 non-normalized cDNA libraries and assembly of 1,021,891 expressed sequence tags. Genome Biol. 2007;8:R45. doi: 10.1186/gb-2007-8-4-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter PA, Max EE, Seidman JG, Maizel JV, Leder P. Cloned human and mouse kappa immunoglobulin constant and J region genes conserve homology in functional segments. Cell. 1980;22:197–207. doi: 10.1016/0092-8674(80)90168-3. [DOI] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Huminiecki L, Kasprzyk A, Lehvaslaiho H, Lijnzaad P, Melsopp C, Mongin E, Pettett R, Pocock M, Potter S, Rust A, Schmidt E, Searle S, Slater G, Smith J, Spooner W, Stabenau A, Stalker J, Stupka E, Ureta-Vidal A, Vastrik I, Clamp M. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck S, Lefranc G, Lefranc M-P. A human immunoglobulin IGHG3 allele (Gmb0, b1, c3, c5, u) with an IGHG4 converted region and three hinge exons. Immunogenetics. 1989;30:250–257. doi: 10.1007/BF02421328. [DOI] [PubMed] [Google Scholar]
- Humphray SJ, Scott CE, Clark R, Marron B, Bender C, Camm N, Davis J, Jenks A, Noon A, Patel M, Sehra H, Yang F, Rogatcheva MB, Milan D, Chardon P, Rohrer G, Nonneman D, de Jong P, Meyers SN, Archibald A, Beever JE, Schook LB, Rogers J. A high utility map of the pig genome. Genome Biol. 2007;8:R139. doi: 10.1186/gb-2007-8-7-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinform. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Asakawa S, Sasaki T, Klobeck HG, Combriato G, Zachau HG, Shimizu N. Evolutionary dynamics of the human immunoglobulin κ locus and the germline repertoire of the Vκ genes. Eur J Immunol. 2001;31:1017–1028. [PubMed] [Google Scholar]
- Kim DR, Oettinger MA. V(D)J recombination: site-specific cleavage and repair. Mol Cells. 2000;10:367–374. [PubMed] [Google Scholar]
- Kurosawa K, Ohta K. Genetic diversification by somatic gene conversion. Genes. 2011;2:48–58. doi: 10.3390/genes2010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers BM, Beaman KD, Kim YB. Sequence analysis of porcine immunoglobulin light chain cDNAs. Mol Immunol. 1991;28:877–880. doi: 10.1016/0161-5890(91)90051-k. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P. WHO-IUIS nomenclature subcommittee for immunoglobulins and T cell receptors report. Immunogenetics. 2007;59:899–902. doi: 10.1007/s00251-007-0260-4. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P. WHO-IUIS Nomenclature Subcommittee for Immunoglobulins and T cell receptors report. Immunoglobulins and T cell receptors report August 2007, 13th International Congress of Immunology, Rio de Janeiro, Brazil. Dev Comp Immunol. 2008;32:461–463. doi: 10.1016/j.dci.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P, Lefranc G. The immunoglobulin FactsBook. Academic; London: 2001. pp. 1–458. [Google Scholar]
- Lefranc M-P, Helal AN, De Lange G, Chaabani H, Van Loghem E, Lefranc G. Gene conversion in human immunoglobulin gamma locus shown by unusual location of IgG allotypes. FEBS Letters. 1986;196:96–102. doi: 10.1016/0014-5793(86)80221-6. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P, Pommié C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P, Pommié C, Kaas Q, Duprat E, Bosc N, Guiraudou D, Jean C, Ruiz M, Da Piédade I, Rouard M, Foulquier E, Thouvenin V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev Comp Immunol. 2005;29:185–203. doi: 10.1016/j.dci.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Lefranc M-P, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT®, the international ImMunoGeneTics information system®. Nucl Acids Res. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane JF, Van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- Pasman Y, Saini SS, Smith E, Kaushik AK. Organization and genomic complexity of bovine λ-light chain gene locus. Vet Immunol Immunop. 2010;135:306–313. doi: 10.1016/j.vetimm.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Baetz KA, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 1994;22:1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Schook LB, Beever JE, Rogers J, Humphray S, Archibald A, Chardon P, Milan D, Rohrer G, Eversole K. Swine Genome Sequencing Consortium (SGSC): a strategic roadmap for sequencing the pig genome. Comp Funct Genom. 2005;6:251–255. doi: 10.1002/cfg.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard HW, Gutman GA. Allelic forms of rat kappa chain genes: evidence for strong selection at the level of nucleotide sequence. Proc Natl Acad Sci USA. 1981;78:7064–7068. doi: 10.1073/pnas.78.11.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminovitch KA, Bakhshi A, Goldman P, Korsmeyer SJ. A uniform deleting element mediates the loss of kappa genes in human B cells. Nature. 1985;316:260–262. doi: 10.1038/316260a0. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Kabat EA. An analysis of the sequences of the variable regions of Bence Jones proteins and their myeloma light chains and their implications for antibody complementarity. J Exp Med. 1970;132:211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerle M, Lahbib-Mansais Y, Pinton P, Robic A, Goureau A, Milan D, Gellin J. The cytogenetic map of the domestic pig (Sus scrofa domestica). Mamm Genome. 1997;8:592–607. doi: 10.1007/s003359900512. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]