Abstract
Dimorphic mating-type chromosomes in fungi are excellent models for understanding the genomic consequences of recombination suppression. Their suppressed recombination and reduced effective population size are expected to limit the efficacy of natural selection, leading to genomic degeneration. Our aim was to identify the sequences of the mating-type chromosomes (a1 and a2) of the anther-smut fungi and to investigate degeneration in their nonrecombining regions. We used the haploid a1 Microbotryum lychnidis-dioicae reference genome sequence. The a1 and a2 mating-type chromosomes were both isolated electrophoretically and sequenced. Integration with restriction-digest optical maps identified regions of recombination and nonrecombination in the mating-type chromosomes. Genome sequence data were also obtained for 12 other Microbotryum species. We found strong evidence of degeneration across the genus in the nonrecombining regions of the mating-type chromosomes, with significantly higher rates of nonsynonymous substitution (dN/dS) than in nonmating-type chromosomes or in recombining regions of the mating-type chromosomes. The nonrecombining regions of the mating-type chromosomes also showed high transposable element content, weak gene expression, and gene losses. The levels of degeneration did not differ between the a1 and a2 mating-type chromosomes, consistent with the lack of homogametic/heterogametic asymmetry between them, and contrasting with X/Y or Z/W sex chromosomes.
Keywords: Y chromosome, Silene latifolia, Microbotryum violaceum, PAR, evolutionary strata, autosomes, allosomes, genetic map
Introduction
Loci determining which genotypes can mate sometimes reside within genomic regions that display exceptional characteristics, with extensive suppression of homologous recombination and cytological differentiation between members of the diploid chromosome pair (e.g., in sex chromosomes; Charlesworth 1991; Marais et al. 2008; Bergero and Charlesworth 2009; Bachtrog et al. 2011). Despite being fundamental to sexual reproduction, the suppression of recombination renders these genomic regions particularly prone to degeneration (Bachtrog 2005). Degenerative changes in the coding sequences (CDS) of sex chromosomes include higher nonsynonymous substitution rates and/or lower expression levels than in recombining regions (e.g., Filatov and Charlesworth 2002; Bartolome and Charlesworth 2006). Degeneration may even extend to the point of gene loss, illustrated for example by the small size and limited gene content of the human Y chromosome (Repping 2006). Nonrecombining regions also tend to accumulate transposable elements (TEs) whose insertions can cause deleterious mutations (e.g., Steinemann M and Steinemann S 1992; Erlandsson et al. 2000; Bachtrog 2003; Marais et al. 2008).
In plants and animals in which one sex is heterogametic (e.g., XY males in systems with XX females), degeneration of the sex chromosomes is asymmetric (Bachtrog 2013). Deleterious recessive mutations on the Y are sheltered, whereas mutations on the X can be exposed to selection when in the homogametic sex. The efficacy of selection on sex chromosomes is further limited because of their reduced effective population size, exacerbated by the process of hitchhiking by deleterious mutations, for example, linked to positively selected Y-linked mutations (Bergero and Charlesworth 2009; Bachtrog 2013). As with the sheltering of mutations, these population-level effects influence the Y chromosome more severely than the X.
Some fungal mating-type chromosomes can also display recombination suppression and size dimorphism analogous to sex chromosomes (Fraser et al. 2004; Menkis et al. 2008; Hood et al. 2013; Grognet et al. 2014). Mating-type chromosomes carry the genes regulating mating compatibility (e.g., through premating pheromones and receptors and postmating homeodomain proteins) but not those determining male/female functions (Billiard et al. 2011). Mating occurs in the haploid stage in fungi, and consequently their mating-type chromosomes are always in a heterogametic condition in diploids. Mating-type chromosomes are therefore expected, like sex chromosomes, to display phenomena of sheltering of deleterious mutations and smaller effective population size than nonmating-type (non-MAT) chromosomes, which may lead to degeneration. However, in contrast to sex chromosomes, the sheltering and reduced effective population size effects are expected to be symmetrical in the mating-type chromosome pair (Bull 1978).
The evolutionary consequences of suppressed recombination on fungal mating-type chromosomes have not been extensively studied. Nevertheless, footprints of degeneration have been reported in the form of high nonsynonymous substitution rates and degeneration of codon usage in the fungus Neurospora tetrasperma (Ellison et al. 2011; Whittle and Johannesson 2011; Whittle et al. 2011). Also, the accumulation of TEs has been shown in the mating-type chromosomes of N. tetrasperma and Microbotryum lychnidis-dioicae (Hood et al. 2004; Ellison et al. 2011). Further work is needed to assess whether reduced expression or loss of genes also occurs, and whether degeneration is similar in the two mating-type chromosomes. In the case of the M. lychnidis-dioicae mating-type chromosomes, opposite conclusions have been reached about the extent of suppressed recombination and whether evolutionary strata exist. Evolutionary strata are distinct regions of different levels of divergence between sex chromosomes, probably reflecting successive stages in the expansion of the region with suppressed recombination (Lahn and Page 1999; Bergero et al. 2007). Some results suggest a small region with distinct strata in M. lychnidis-dioicae (Votintseva and Filatov 2009), whereas others suggest that much of the chromosome pair is nonrecombining (Hood et al. 2013).
Microbotryum lychnidis-dioicae is a fungus causing anther-smut disease on Silene latifolia. It was the first fungus in which dimorphic fungal mating-type chromosomes (a1, ∼3.3 Mb, and a2, ∼4.0 Mb) were described (Hood 2002; Hood et al. 2013). The suppression of recombination between mating-type chromosomes is thought to be favored under a highly selfing mating system as occurs in this fungus (supplementary fig. S1, Supplementary Material online) (Antonovics and Abrams 2004; Hood and Antonovics 2004; Johnson et al. 2005; Giraud et al. 2008). Suppression of recombination linking the centromere, premating pheromones/receptors, and postmating homeodomain proteins renders a higher rate of gamete compatibility under intratetrad selfing (Hood and Antonovics 2000; Nieuwenhuis et al. 2013). Segregation analyses of progeny (Hood and Antonovics 2004) and the lack of colinearity found by analysis of optical maps, that is, ordered, chromosome-wide restriction maps (Hood et al. 2013), indicate that nonrecombining regions (NRRs) make up most of the M. lychnidis-dioicae mating-type chromosomes (up to 90%). In contrast, Votintseva and Filatov (2009) estimated that only 25% of the markers from the mating-type chromosomes segregated with the mating type. The NRRs of the mating-type chromosomes in M. lychnidis-dioicae are flanked at either end by recombining regions (MATRRs), as shown by segregation analyses of progeny (Votintseva and Filatov 2009) and by colinearity in the optical maps (Hood et al. 2013). Because these distal regions retain the ability to recombine, the transmission and population genetics of these chromosomes are expected to be similar to non-MAT chromosomes (Ellis et al. 1990).
Very ancient trans-specific polymorphism has been observed at the mating-pheromone receptor gene (370 My old; Devier et al. 2009) and, to a lesser extent, at other genes, not involved in mating but linked to the mating-type genes (Abbate and Hood 2010a; Petit et al. 2012). Such ancient trans-specific polymorphism indicates that cessation of recombination began before divergence of the Microbotryum anther-smut fungi. Numerous Microbotryum species have dimorphic mating-type chromosomes (Perlin 1996; Hood and Antonovics 2004), thus providing a unique opportunity to study the dynamics, roles, and patterns of degeneration in the NRRs in fungal mating-type chromosomes. However, the sequence of the a1 genome of the reference strain of M. lychnidis-dioicae, available at the Broad Institute, was insufficiently assembled to identify the a1 mating-type chromosome. Furthermore, no reference a2 genome was available. The aims of this work were therefore: 1) To identify the sequences of the a1 and a2 mating-type chromosomes and of their NRRs in M. lychnidis-dioicae, and 2) to investigate TE accumulation and degeneration in the NRRs of the mating-type chromosomes in the Microbotryum genus, including increased nonsynonymous substitution rates (dN/dS), reduced expression, and gene loss.
Results
Identification of Mating-Type Chromosomes, MATRRs, and NRRs in M. lychnidis-dioicae
We used the reference specimen whose a1 haploid genome was sequenced by the Broad Institute. Both a1 and a2 mating-type chromosomes from this reference specimen were isolated on pulsed-field electrophoretic gels and sequenced at very high coverage (1,175×). Mapping the a1 chromosome-specific DNA against the a1 reference assembly available at the Broad Institute allowed the identification of scaffolds corresponding to the a1 mating-type chromosome. We conducted de novo assembly of the sequences of the karyotype-isolated a2 mating-type chromosome.
The Broad Institute Supercontigs 37 and 43 could be anchored on previously established optical maps of the M. lychnidis-dioicae mating-type chromosomes (Hood et al. 2013). Each of these two scaffolds anchored on one of the two distal regions of structural colinearity between the a1 and a2 mating-type chromosomes (Hood et al. 2013). This allowed the sequences of the mating-type chromosome recombining regions (MATRRs) to be identified. As expected for recombining DNA in a selfing species, the sequences assigned to MATRRs showed less than 0.1% nucleotide divergence between the a1 and a2 chromosomes sequenced.
Because there were numerous repetitive sequences, the remaining scaffolds assigned to the mating-type chromosome NRRs could not be satisfactorily anchored on the optical maps. Nevertheless, their assignment to the mating-type chromosomes appeared reliable because, after filtering out repetitive elements: 1) These scaffolds were highly represented in the reads from the karyotype-isolated mating-type chromosomes, and 2) significant BLASTn hits were found between the unique genes from the a1 and a2 NRR scaffolds, with substantial divergence between most alleles, consistent with expectations for NRRs. Some unique genes within scaffolds assigned to NRRs appeared highly similar between a1 and a2. However, these genes were embedded within scaffolds that carried other genes with more divergence between the two mating types (supplementary fig. S2, Supplementary Material online). Their assignment to NRRs therefore appears to be sound. Existing genetic maps were used for further validation of our scaffold assignments to NRRs and MATRRs. All 11 loci previously shown to be completely linked to the mating-type in segregation analyses of a single progeny (Abbate and Hood 2010b; Petit et al. 2012) were found on the scaffolds we assigned to NRRs. The sequences we assigned to NRRs included 11 additional loci shown to be linked to the mating type in another segregation analysis (Votintseva and Filatov 2009). One of the NRR loci (accession BZ782397 in Votintseva and Filatov 2009) corresponded to one of our NRR fragments with zero a1–a2 sequence divergence. Thus, even nondiverged regions may belong to the NRR. Five mating-type chromosome loci previously shown to be unlinked to mating type and with zero sequence divergence between a1 and a2 (Votintseva and Filatov 2009) were found in scaffolds we assigned to MATRR contigs (accessions BZ782138, BZ782443, BZ782547, BZ782204, and BZ782044).
In the a1 M. lychnidis-dioicae reference genome, 6,891 genes were predicted in non-MAT chromosomes, 349 genes in the NRRs, and 99 genes in the MATRRs. The NRRs thus carry 78% of the genes of the mating-type chromosome. Genes typically determining mating types in basidiomycetes were found in the NRRs: 1) The genes encoding the mating pheromones and receptors, which determine conjugation compatibility, and 2) the genes encoding mating homeodomains regulating growth of the dikaryon after conjugation. Other than genes previously identified as belonging to the NRRs and involved in mating (Petit et al. 2012), no other genes in the NRRs had any putative mating function.
Substantial Divergence between NRRs and Investigation of the Possibility of Evolutionary Strata in M. lychnidis-dioicae
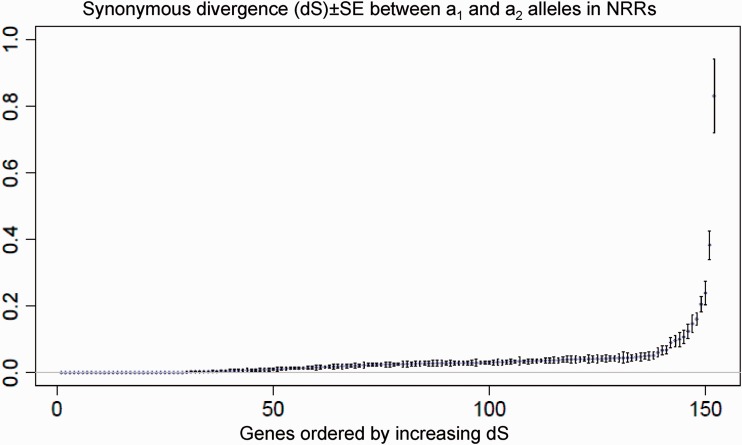
Using our de novo assembly of the a2 mating-type chromosome, we identified the reciprocal best hits between predicted NRR genes on the a1 and a2 sequences. We estimated the divergence between these alleles at synonymous coding positions, using alignments of concatenated exons per gene longer than 1,000 bp to avoid stochastic effects due to short sequences (fig. 1). Synonymous divergence was substantial, indicating that the suppression of recombination between the mating-type chromosomes of M. lychnidis-dioicae was very ancient, with some extreme values and divergence varying from 0 to 3.4, and a mean of 0.06. There were no marked gaps in the distribution of divergence values (fig. 1), as would be expected if different evolutionary strata had ceased recombining at different times. Because the assembly was incomplete, however, we could not plot the divergence levels against physical distance, which would be the most powerful approach to test for the existence of evolutionary strata. Nevertheless, the large scaffolds included genes displaying high variation in levels of a1–a2 divergence (supplementary fig. S2, Supplementary Material online), indicating that heterogeneity in divergence occurs at small scales of physical distance.
Fig. 1.
Synonymous divergence between a1 and a2 alleles in predicted genes in the NRRs of the mating-type chromosomes of Microbotryum lychnidis-dioicae, ranked by increasing divergence. Only trimmed alignments of concatenated exons longer than 1,000 bp were retained. Synonymous divergence was estimated as the numbers of synonymous substitutions (dS), divided by the number of synonymous sites in the sequence; standard errors are shown. A single dS value at 3.4 is not shown for optimal visualization of the other values. The divergence between the pheromone receptor alleles is not shown either as they could not be aligned in nucleotides.
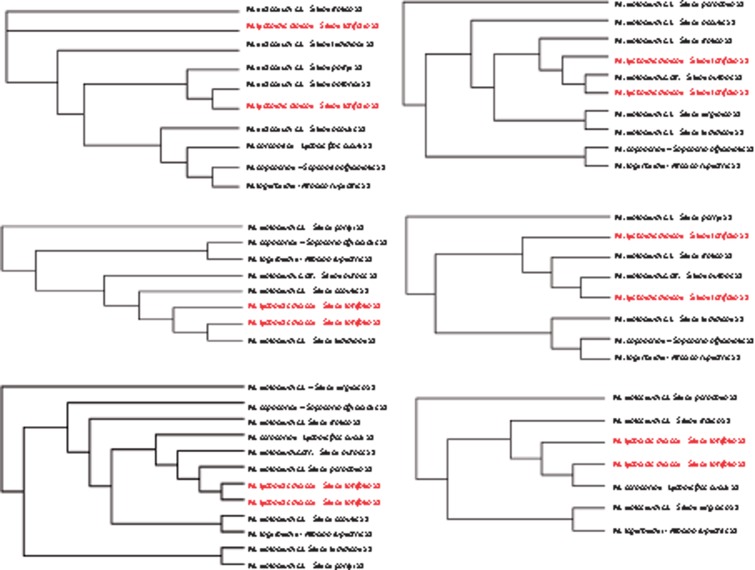
We then assessed when recombination suppression between the mating-type chromosomes of M. lychnidis-dioicae emerged relative to speciation events. We built trees of the predicted genes assigned to NRRs. Trees were built from alignments of putative orthologous groups (POGs) from the whole haploid genome sequences of either mating type (obtained by 454 technology) from 11 other Microbotryum species, as well as the a1 and a2 alleles of M. lychnidis-dioicae. In principle, recent recombination events would place the a1 and a2 alleles of M. lychnidis-dioicae close together relative to alleles of other species, whereas suppression of recombination established before speciation events will place M. lychnidis-dioicae a2 alleles closer to a2 alleles of other species than to M. lychnidis-dioicae a1 alleles (Devier et al. 2009; Petit et al. 2012). Among the 349 predicted NRR genes, only 45 reliable alignments could be obtained that included the alleles of both a1 and a2 of M. lychnidis-dioicae and of at least five other species. In 19 of the 45 trees built with confidently aligned NRR genes, the a2 alleles of M. lychnidis-dioicae clustered with a2 alleles of other species: Microbotryum from S. notarsii, from L. flos-cuculi, S. nutans and/or S. lemmoni (fig. 2). Such trans-specific polymorphism indicates that recombination was suppressed before the corresponding speciation events. The other 26 trees showed clustering of M. lychnidis-dioicae a1 and a2 alleles in a specific clade, with high similarity, suggesting that there had been recent gene conversion (fig. 2).
Fig. 2.
Examples of gene trees in NRRs of the mating-type chromosomes. Examples are shown of different placements of the a1 and a2 alleles of Microbotryum lychnidis-dioicae genes (in gray), relative to orthologs in other Microbotryum species (in black). The name of the host plant is indicated next to the Microbotryum species name.
Gene conversion footprints were investigated for predicted genes in NRRs. We used the 196 POG alignments from various Microbotryum species and assigned to NRR (see below). A total of 11 gene conversion events were detected within these NRR genes, corresponding to 1.9 × 10−4 events per base pair, affecting 3.6% of the genes.
Identification of POGs and Species Tree Reconstruction
We mapped the whole genome sequences of the 11 other Microbotryum species and the outgroup (haploid genomes of either mating type; fig. 3) onto the M. lychnidis-dioicae reference genome. We thereby identified 5,726 POGs of loci in which at least 6 of the 12 species were represented and that were longer than 150 bp. These sequences were filtered to remove intron positions and surrounding regions that did not align reliably. Based on their positions in the M. lychnidis-dioicae reference genome, 5,453 POGs were assigned to non-MAT chromosomes, 196 to NRRs, and 69 to the MATRRs.
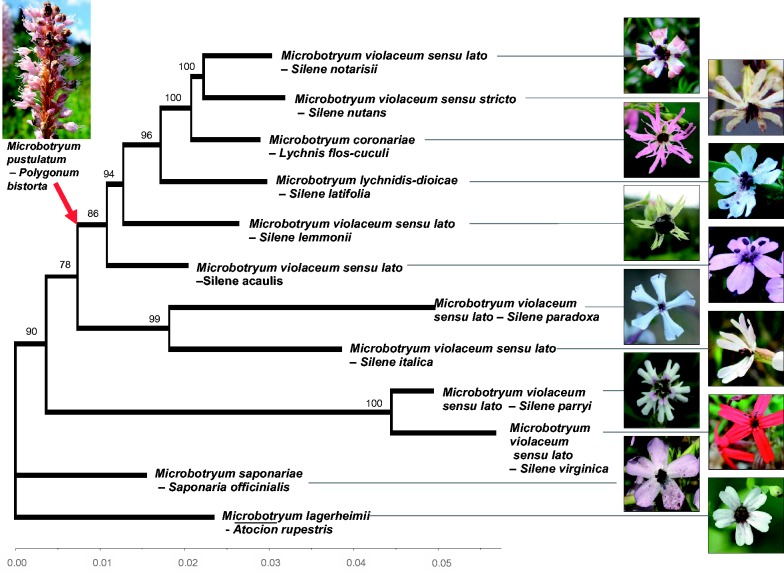
Fig. 3.
Unrooted phylogenetic tree of the Microbotryum species studied, with a gene partition model under the maximum-likelihood framework. The topologies obtained with the different data sets were all identical: The full set of 5,726 putative orthologous genes (POGs), the 46 POGs including the outgroup, the 5,453 non-MAT chromosome POGs, and the set of 288 non-MAT chromosome POGs with high individual bootstraps. Bootstraps obtained using the full set of 5,726 POGs are indicated at the nodes. The root obtained with the 46 concatenated POGs including the outgroup species, M. pustulatum parasitizing Polygonum bistorta, is indicated with a red arrow. The scale indicates the total number of substitutions per base accumulated in each lineage. Fungal species names, and host species names, are indicated. Microbotryum violaceum sensu lato applies to all host-specific lineages for which no Latin name has been given, whereas Microbotryum violaceum sensu stricto indicates the host-specific lineage to which this Latin name was originally given. Pictures of diseased host plants are shown.
We built a phylogeny of the Microbotryum species based upon the set of 5,453 POGs from non-MAT chromosomes (fig. 3); genes on the mating-type chromosomes were excluded because they may display trans-specific polymorphism that could blur species relationships (Devier et al. 2009; Abbate and Hood 2010b; Petit et al. 2012). The relationships between species were consistent with those reported previously for this fungal genus (Kemler et al. 2006). The phylogeny was rooted using a subset of 46 POGs from the outgroup M. pustulatum parasitizing Polygonum bistorta (Kemler et al. 2006); the topology of the resulting rooted phylogeny was identical to the unrooted version based on the data set from non-MAT chromosomes, although with smaller bootstrap values. The topology obtained using the full set of 5,726 POGs (including from the mating-type chromosome loci) was also identical and with smaller bootstrap values, due to trans-specific polymorphisms of genes in the NRRs.
Bootstrap support of 100% for nodes in phylogenomic trees may nevertheless mask disagreement between trees for individual genes. The phylogeny was therefore reconstructed using only the 288 POGs from non-MAT chromosomes displaying strong bootstrap values in their individual gene trees as previously recommended (Salichos and Rokas 2013). This yielded the same topology as the rooted tree, with 100% bootstrap support for all nodes (fig. 3).
Increased Nonsynonymous Substitution Rates in NRRs
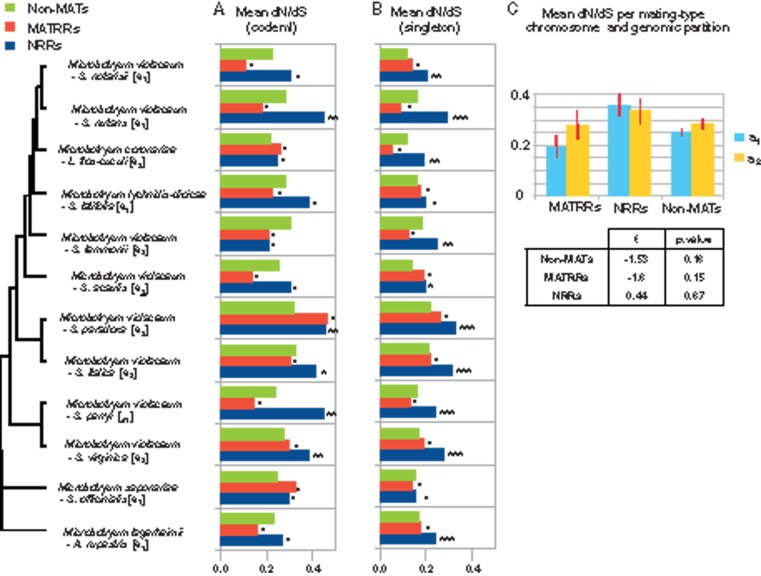
Footprints of less effective selection were identified as the accumulation of nonsynonymous substitutions (dN, assumed to be deleterious) at a faster rate than of synonymous substitutions (dS) in the 12-species POG data set. The comparison of median ω (=dN/dS) values for terminal branches (fig. 4A) indicated that rates of nonsynonymous substitution in NRRs were higher than those in non-MAT chromosomes in all species except in the branch of the Microbotryum species parasitizing S. lemmonii. The differences in ω values between NRRs and non-MAT chromosomes were significant in four Microbotryum species, those parasitizing S. nutans, S. paradoxa, S. parryi, and S. virginica. In contrast, ω values did not differ between the MATRRs on the mating-type chromosomes and those on the non-MAT chromosomes; ω values were, however, lower in the MATRR than in non-MAT chromosomes in 8 of the 12 species (fig. 4A). Sequencing errors are unlikely to be responsible for the differences in the estimated substitution rates between compartments (NRR, MATRR, or non-MAT chromosomes), as the mean read qualities for the different compartments were similar.
Fig. 4.
Degeneration estimated by comparing mean nonsynonymous with synonymous substitutions (ω = dN/dS) across CDS in each of non-MAT chromosomes, nonrecombining regions of the mating-type chromosomes (NRRs), and recombining regions of the mating-type chromosomes (MATRRs) in 12 haploid Microbotryum genomes (either of a1 or a2 mating type): (A) With the full frequency spectrum of substitutions used to compute dN/dS through a free-ratio branch model (Codeml, PAML), or (B) with only singleton substitutions taken into account to focus on recent events. The significance of the differences between NRR and non-MAT chromosomes and between MATRRs and non-MAT chromosomes was assessed by bootstrapping. Significance levels of 1% (***), 5% (**) and 10% (*) and nonsignificance (-) are reported on barplots. (C) Mean ω (dN/dS) per mating-type chromosome (a1 and a2), for NRRs MATRRs and non-MAT chromosomes. Red bars indicate standard deviations. Associated two-sample t-tests are presented in the table: No significant difference in mean dN/dS was detected between a1 and a2.
The NRRs of both the a1 and a2 mating-type chromosomes had higher rates of nonsynonymous substitutions than in non-MAT chromosomes across the genus as a whole (fig. 4C). We then compared degeneration between the a1 and a2 NRR within a single species, M. lychnidis-dioicae. We computed the ω values for the 45 NRR POGs with reliable alignments between the a1 and a2 alleles of M. lychnidis-dioicae. The difference between a1 and a2 was significant for only one of these 45 NRR genes, and thus there was no evidence for asymmetry of degeneration between a1 and a2 mating-type chromosomes.
To study recent, phylogenetically independent substitutions, we also analyzed ω values for mutations found in only one of the species (singletons): ω ratios were again significantly higher in NRRs than in non-MAT chromosomes (fig. 4B) in 10 of the 12 species (for all except M. lychnidis-dioicae parasitizing S. latifolia, and M. saponariae parasitizing Saponaria officinalis). MATRR ω values again were not significantly different from those in non-MAT chromosomes (fig. 4B).
Distinguishing Decreased Efficacy of Purifying Selection from Positive Selection
The higher nonsynonymous substitution rates in NRRs than in non-MAT chromosomes may reflect decreased efficacy of purifying selection, as would be expected under sheltering and reduced effective population size. Indeed, the results of codon-model tests with and without selection were not consistent with the alternative explanation of positive selection (supplementary table S1, Supplementary Material online). The proportions of loci inferred to evolve under positive selection in the 12-species POG data set appeared to be similar in NRRs (6%, 12 predicted genes), MATRRs and non-MAT chromosomes (4% each). Furthermore, discarding the POGs falling in the “nearly neutral selection” (0.8 < ω < 1.2) and “positive selection” (ω > 1.2) classes led to similarly higher mean ω in NRR than in non-MAT chromosomes and MATRR compartments (supplementary fig. S3A and B, Supplementary Material online). This suggests that the generally higher ω in NRRs than other compartments is a consequence of a lower efficacy of purifying selection rather than the effects of positive selection on genes.
Higher TE Accumulation in NRRs than Recombining Regions
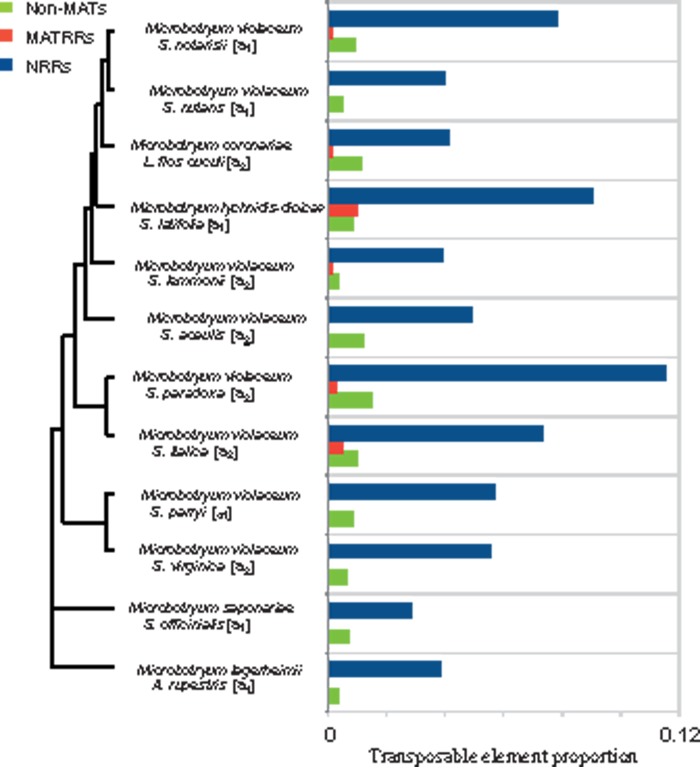
TE content was significantly higher in the NRRs than in non-MAT chromosomes of all Microbotryum species and this was true for both a1 and a2 mating-type chromosomes (fig. 5): The TE content was at least four times greater in NRR than in non-MAT chromosomes in all cases, and 26 times more in the case of M. violaceum sensu stricto parasitizing S. nutans. We examined separately the three most frequent TE families in M. lychnidis-dioicae: Copia-like, Gypsy-like, and rolling circle Helitron-like elements (Hood 2005; Hood et al. 2005; Yockteng et al. 2007). The greater TE content in NRR than non-MAT chromosomes applied to all the three families (supplementary fig. S3D, Supplementary Material online). MATRRs tended to harbor fewer TEs than the non-MAT chromosomes (fig. 5), but the difference was not significant.
Fig. 5.
TE contents in contigs assigned to non-MAT chromosomes, nonrecombining regions of the mating-type chromosomes (NRRs), and recombining regions of the mating-type chromosomes (MATRRs), in 12 haploid Microbotryum genomes (either of a1 or a2 mating type). The results reported are proportions of sequences corresponding to TEs in 1,000 random 200-bp fragments in each compartment (non-MAT chromosomes, NRRs, and MATRRs).
Differential Expression or Gene Loss between the Mating-Type Chromosomes in M. lychnidis-dioicae
We then examined gene expression using RNA sequencing (RNA-Seq) in separated a1 and a2 haploid cultures of M. lychnidis-dioicae, under conducive (low nutrient water agar) and nonconducive (nutrient-rich medium) mating environments. The gene expression profiles (supplementary fig. S4A, Supplementary Material online) revealed 177 genes differentially expressed (cutoff 1E−10) in M. lychnidis-dioicae between a1 and a2 haploid cultures in either low or high nutrient environments. Among these 177 genes, most of them (93) were differentially expressed under both conditions. Eighty-four of the 177 differentially expressed genes were located in the NRRs and two in the MATRRs. This represents a 49% enrichment of differentially expressed genes in the NRRs, which is highly significant (Pearson χ2 = 52.61, P < 0.001).
Differential expression of NRR genes between a1 and a2 cultures can be caused by 1) deleterious mutations impairing proper expression but sheltered in the dikaryotic/diploid stage by permanent heterozygosity in NRRs, or 2) selection favoring linkage to one mating type because of antagonistic effects in cells of the other mating type; this would be similar to genes on sex chromosomes with antagonistic effects in males and females (Charlesworth et al. 2005; Bergero and Charlesworth 2009). Were this second possibility the case, the differentially expressed genes in NRR would be expected to be associated with mating-type functions and thus potentially upregulated during mating. However, the differentially expressed genes in NRRs were not associated with any putative functions involved in mating, except for the mating-type pheromone receptor and homeodomain loci. Furthermore, we did not detect a higher percentage of genes upregulated during mating in NRRs than in non-MAT chromosomes or MATRRs: 2.63% of NRR genes were upregulated during mating, versus 2.02% in MATRRs and 1.71% in non-MAT chromosomes. Moreover, only 10 of the 130 genes upregulated during mating were also differentially expressed between separated a1 and a2 cells. Thus, few, if any, of the genes identified as differentially expressed between mating types are involved in mating-type determination, arguing against the idea that numerous genes have antagonistic effects between the two mating types. These observations suggest that most of the genes with weak or no expression in one mating type correspond to genes with deleterious mutations rather than mating-type specific roles.
In our RNA-Seq analysis, fewer genes appear to be expressed specifically in a2 than a1 haploid cells. Sixty-eight of the 86 differentially expressed genes mapping to the a1 mating-type chromosome were significantly more strongly expressed in a1 cultures and 18 in a2 cultures (supplementary fig. S3, Supplementary Material online). This was probably because reads were mapped against the gene set from the a1 reference genome, so genes lost from the a1 mating-type chromosome (and thus only expressed in a2 cells) would not have been identified. To recover those genes, RNA-Seq reads from the a1 and a2 cell libraries were separately assembled de novo. The resulting transcripts were mapped against both the a1 reference genome and a de novo assembly of the a2 mating-type chromosome from the reference specimen. For some genes lacking transcripts in cultures of one mating type, there was evidence of gene loss from the corresponding mating-type chromosome. Sixteen of the 30 genes expressed only in cultures of the a1 mating type were located in the NRR and not found among a2 genome sequences. Similarly, 9 of the 21 genes expressed only in cultures of the a2 mating type were located in the NRR and not found among a1 genome sequences. The number of gene losses from a1 and a2 NRRs were not significantly different (χ2 = 0.2; P = 0.35). We confirmed gene loss from either one of the mating-type chromosomes by polymerase chain reaction (PCR) detection using gene-specific primers in the M. lychnidis-dioicae Lamole reference strain (supplementary table S2A, Supplementary Material online). We also screened 18 other M. lychnidis-dioicae or M. silenes-dioicae strains by PCR using the same primer pairs for the presence of these genes. Overall, the pattern of gene loss observed was same as that in the Lamole reference strain. However, a couple of genes were found in both mating types in several strains, indicating that the gene losses are not in all cases fixed within the species (supplementary table S2B, Supplementary Material online).
Discussion
Identification of DNA Sequences of the Mating-Type Chromosomes in M. lychnidis-dioicae, and Its Large NRRs
Sequencing the chromosomes extracted from pulse-field gels allowed the identification of the a1 and a2 mating-type chromosomes within the reference genome sequence, as well as their MATRRs and NRRs. Several lines of evidence validated our assignments of scaffolds to NRRs, MATRRs, and non-MAT chromosomes, noting that any failure to properly assign a few sequences would obscure the differences between the mating-type chromosomes and non-MAT chromosomes. The tests for differences between NRRs and recombining regions should therefore be conservative.
Orthologs between the two mating-type chromosomes (a1 and a2) were found in the NRRs. This finding supports the view that the dimorphic mating-type chromosomes in fungi were initially derived from a homologous pair of non-MAT chromosomes, as observed for sex chromosomes and other fungal mating-type chromosomes (Charlesworth 1991; Charlesworth et al. 2005; Menkis et al. 2008). Several hundred genes were identified in the NRRs of the mating-type chromosomes, representing most of the predicted genes of the mating-type chromosomes. This is consistent with suppressed recombination extending over most of the mating-type chromosome pair in M. lychnidis-dioicae, as indicated by analysis of optical maps (Hood et al. 2013). Votintseva and Filatov (2009), in contrast, reported that the NRRs covered a much shorter section of the mating-type chromosomes in M. lychnidis-dioicae. That result, however, was partly due to the inclusion of loci that were repetitive sequences or were missassigned to MATRRs (Hood et al. 2013).
Divergence between a1 and a2 Alleles in the NRRs of the Mating-Type Chromosomes in M. lychnidis-dioicae
The high divergence of synonymous substitutions between the a1 versus a2 alleles in many of the genes in the NRR indicates that the cessation of recombination is ancient. We found no evidence for allele divergence in discrete stages, which could have been expected were there evolutionary strata in mating-type chromosomes of M. lychnidis-dioicae. A high heterogeneity in divergence levels was found, which could result from the evolutionary strata, although the heterogeneity was observed at small physical scale. Physical mapping of loci may be required before a definitive conclusion about the existence of evolutionary strata or other processes is possible.
Dating the suppression of recombination in M. lychnidis-dioicae is limited by the inability to use absolute calibration. Indeed, calibration points are rare; exploitable calibrations points require the alignment of numerous genes in fungal species much more distant than the genus level examined here. Using fossil records for calibration, we estimated that the suppression of recombination at the highly conserved mating pheromone receptor gene in M. lychnidis-dioicae occurred approximately 370 Ma (Devier et al. 2009). Here, we could only estimate a relative date for the suppression of recombination by investigating the placement of the a1 and a2 alleles in the broader gene trees that contained several Microbotryum species. Among the subset of NRR genes that could reliably be aligned with other species, their placements often indicated that recombination was suppressed before the speciation of the sister species M. lychnidis-dioicae and M. silenes-dioicae. This speciation event has been estimated to have been 420,000 years ago (Gladieux et al. 2011). For some predicted genes, however, a1 and a2 alleles were very similar, indicating recent cessation of recombination or gene conversion. The heterogeneity of divergence times may be the result of an expansion of the region with suppressed recombination (i.e., evolutionary strata), transposition of genes, and/or the influence of gene conversion. Further studies that provide more complete assembly will aid in disentangling these various possibilities.
Evidence of Degeneration in the Regions of Suppressed Recombination in the Mating-Type Chromosomes of Multiple Microbotryum Species
Consistent with the reduced efficacy of selection expected in the nonrecombining mating-type chromosomes, we detected signs of genetic degeneration in the NRRs of Microbotryum species, in the form of higher rates of nonsynonymous than synonymous substitutions in NRRs than in MATRRs or non-MAT chromosomes. Note that the nonsynonymous substitutions detected here may represent rare variants within the species, as we did not have polymorphism data to check whether the substitutions had been fixed at the species level. Even in this case, however, the finding of higher numbers of nonsynonymous substitutions in NRRs than in non-MAT chromosomes would still be a strong indication of a lower efficacy of purifying selection, leading to deleterious substitutions being maintained at higher frequencies in the NRRs than in non-MAT chromosomes.
The different patterns of gene expression between a1 and a2 haploids also provided strong evidence for degeneration, with some genes showing reduced expression or even having been lost in one mating type. The differential expression was not likely to be due to selection associated with mating-type determination because, other than mating pheromone receptors and homeodomain loci, differentially expressed genes in NRRs did not have any obvious mating functions. Consistent with this, cells of different mating types are not very differentiated and are of similar size, and the haploid stage is very transient and restricted to the tetrad formation (Zakharov 1986; Hood and Antonovics 2000, 2004) (supplementary fig. S1, Supplementary Material online). Only the mating-type genes at the pheromone/receptor and the homeodomain loci are therefore clearly expected to be associated with mating-type determinism, as is typical of basidiomycetes (Feldbrugge et al. 2004). Under “mating” conditions, the genes in the NRRs did not appear more frequently upregulated than genes on non-MAT chromosomes. The high frequency of genes with mating-type-specific expression in the NRRs is thus likely to be associated with the accumulation of deleterious mutations impairing proper expression. Indeed, genes not required for growth during the brief haploid stage may carry loss-of-function mutations at high frequencies, as these would be sheltered by the permanent heterozygosity of these regions.
The substantial degeneration in the NRRs agrees with observations of frequent haplolethal mutations in natural populations of several Microbotryum species. Deleterious alleles have been detected that were linked to one mating type (a1 or a2 depending on the strain), which prevent sustained growth of cells of one mating type when the haploid stage is artificially extended through in vitro culturing (Oudemans et al. 1998; Hood and Antonovics 2000; Thomas et al. 2003). In the case of the Lamole reference strain, from which the reference genome was obtained, both a1 and a2 cells can grow in vitro, so genes lost from the mating-type chromosomes in this genome are not essential for haploid growth.
The TE content of the Microbotryum mating-type chromosome NRRs was found in all species to be much higher than that of their MATRRs and non-MAT chromosomes. This is an expected consequence of suppressed recombination, as has been observed in sex chromosomes (Bachtrog 2013); TE insertions may contribute to degeneration by disrupting genes or altering their expression. TE accumulation was more similar between the MATRRs and non-MAT chromosomes, consistent with recombination occurring in MATRR and allowing for more effective selection such that deleterious TE insertions are more likely to be purged. Various families of TEs accumulated differently in NRRs of different Microbotryum species, providing further evidence of ongoing and independent mating-type chromosome degeneration during the divergence of these fungal lineages. Analyses of TE transcripts and of mutations also indicate that the activity of TEs in several Microbotryum species is continuing (Garber and Ruddat 1995, 1998; Yockteng et al. 2007).
A possible bias in our analyses is that all genomes were mapped against the M. lychnidis-dioicae reference assembly. Therefore, genes identified as present in the mating-type chromosomes based on the orthology with M. lychnidis-dioicae may be present on non-MAT chromosomes in other Microbotryum species. However, our analyses are conservative, as any movements between genomic compartments would obscure differences; our results indicate that sequences assigned to the mating-type chromosome evolved in a distinctive manner across species and that it is unlikely that many of them were misassigned between NRR, non-MAT chromosomes, and MATRR categories.
Overall, there are various lines of evidence for high levels of divergence, degeneration, and TE accumulation on the mating-type chromosomes in numerous species of Microbotryum. It is striking that a nonsex chromosome system exhibits such similar phenomena to well-characterized in sex chromosomes, with extensive suppressed recombination, loss of gene content, and accumulation of a high content of repetitive elements. Importantly, similar degeneration was observed on a1 and a2 mating-type chromosomes, in contrast to XY sex determining systems, where the Y chromosome degenerates much more than the X (Bachtrog 2013). This is consistent with Bull’s prediction that degeneration is not expected to be asymmetrical in species where sex or mating compatibility is determined in the haploid stage and the chromosomes responsible are always heterogametic in the diploid stage (Bull 1978).
Materials and Methods
Reference Genome (M. lychnidis-dioicae)
Information on the a1 haploid genome of the Lamole reference strain of M. lychnidis-dioicae parasitizing S. latifolia was obtained from the Broad Institute web-server. This included the scaffolds (i.e., supercontigs) corresponding to the nonmitochondrial regions, the associated CDS annotations (gff files), and the annotations obtained with Interproscan, a powerful integrated database and diagnostic tool (Jones et al. 2014).
Resequencing of the a1 Mating-Type Chromosomes (M. lychnidis-dioicae)
The a1 mating-type chromosome from the same haploid Lamole genotype whose whole genome was sequenced by the Broad Institute was isolated using electrophoretic karyotype analysis as previously described (Hood et al. 2004). The DNA was subjected to multiple displacement amplifications with the REPLI-g Kit (QIAGEN), and then sequenced at a coverage of 1,175 × using 2- and 5-kb insert size, mate-paired libraries for Titanium version 454 technology (www.roche.com). A first assembly using Newbler (the Roche assembler) was performed with approximately 30-fold and approximately 20-fold coverage.
Identifying the Mating-Type Chromosome Scaffolds on the a1 Reference Genome
To determine which scaffolds of the M. lychnidis-dioicae genome belonged to the a1 mating-type chromosomes and which to non-MAT chromosomes, the assembled contigs from the gel-isolated mating-type chromosomes were mapped against the M. lychnidis-dioicae reference genome using NUCmer (http://mummer.sourceforge.net/). Matches were assigned to the mating-type chromosome, and scaffolds without any match were assigned to non-MAT chromosomes. To validate these results, our 454 data (Newbler) were mapped onto the Broad Institute assembly scaffolds to identify contamination from non-MAT chromosomes by their scattered weak read coverage. Finally, all nonmitochondrial Broad Institute assembly scaffolds with coverage greater than 20-fold when mapped with contigs from the gel-isolated mating-type chromosomes were assigned to mating-type chromosomes.
Sequencing and Assembly of the a2 Mating-Type Chromosome (M. lychnidis-dioicae)
The a2 mating-type chromosome from the Lamole reference strain was isolated and amplified as described above for the a1 mating-type chromosome. Four libraries were sequenced on Illumina HiSeq 2000, one paired-end library (insert size of 250 bp) and three mate-pair libraries (expected insert sizes 3, 8, 20 kb; observed insert sizes of 2.2, 9, 12.5 kb, respectively). The sequence data for the paired-end library (100 bp sequenced at both ends of fragments) represent a coverage of 1,175 × (given the expected genome size of 25 Mb). The 2.2-, 9- and 12.5-kb libraries include 34, 6.4 and 2.2 millions of base pairs, respectively.
Contigs were generated from the paired-end data, with SOAPdenovo 2.04 (Li et al. 2010) using a kmer of 91. Then, a three-step process was applied to remove artifacts from the assembly: 1) Contigs fully included in longer contigs were removed; 2) contigs containing 15-mers that were not present in raw reads were split, as they are likely to be chimeras; and 3) contigs were trimmed (46 bp ∼ 1/2 kmer length) to remove the artificial overlaps created by the large kmer used.
Paired-end and mate-pair reads were mapped on contigs using the glint software (Faraut T and Courcelle E, unpublished data; http://lipm-bioinfo.toulouse.inra.fr/download/glint; parameters - -best-score - -step 2 - -no-lc-filtering - -mmis 5 - -lrmin 80). Due to the large numbers of highly conserved repeats, additional stringent criteria were used for further hit selection (length of the hit ≥ 90 bp; identity = 100%). LYNX scaffolder (Gouzy J, unpublished data) was used to generate scaffolds. A minimum number of five links was required for linking information from the 250-nt and 2.2-kb libraries to be considered, and at least two links for the 9- and 12.5-kb libraries. Finally, SOAPGapCloser was used to fill in gaps in the assembly. The final assembly was 24,832,594 nucleotides long in 388 scaffolds (N50 of scaffolds longer than 1,000 nt = 299 kb; 4.75% of N).
Identification of the MATRRs (M. lychnidis-dioicae)
To identify sequence assemblies corresponding to the two MATRRs of M. lychnidis-dioicae mating-type chromosomes reported in optical maps (Hood et al. 2013), contigs were aligned with restriction-digest optical maps using the MapSolver software (OpGen) (Latreille et al. 2007). This software predicts restriction digest patterns from DNA sequence data for comparison with the observed optical maps (restriction-digest fragments sizes); alignments are based on a cumulative scoring function that rewards matching cut-site distributions and penalizes mismatch or missing sites. The software’s default settings were used to calculate alignments.
Genome Sequencing of Additional Microbotryum Species and Mapping against the Reference
The fungal material used in this study was collected as diploid teliospores from natural populations in North America and Europe. Some Microbotryum species have not received a specific Latin name yet and are referred to as M. violaceum sensu lato with the additional indication of their host plant species (fig. 3). Microbotryum species have been distinguished on the basis of a combination of multiple gene genealogies, interfertility assays, and hybrid fitness and fertility assessments (Le Gac, Hood, Fournier, et al. 2007; Le Gac, Hood, and Giraud 2007; de Vienne, Hood, et al. 2009; de Vienne, Refrégier, et al. 2009). Haploid cultures of the 11 Microbotryum species were obtained by micromanipulation of the postmeiotic yeast-like sporidial cells, which were grown on potato dextrose agar (Difco). Mating types of the haploid cultures were identified by pairing with cultures of known mating types and examining the conjugation response elicited by the alternate mating pheromone (Day 1979). Also, PCR primers that discriminate between a1 and a2 pheromone receptors (Devier et al. 2009) were used to test extracted DNA for mating type with the DNeasy Plant Mini Kit (QIAGEN). The extracted DNA from the Microbotryum species was also used to obtain shotgun sequence libraries using the Titanium version 454 technology (performed at the University of Virginia, Genomics Core Facility) with a coverage of 2–2.5 × . Only reliable sequenced positions, with sufficient qualities, were taken into account.
Sequence libraries for each of the 11 Microbotryum species were assembled individually by mapping against the M. lychnidis-dioicae reference genome using Newbler. Pairwise mapped sequences were realigned within species libraries using Prank (Loytynoja and Goldman 2005, 2008) with the “-codon” option in accordance with recent recommendations based on simulations for the use of codon-based models to detect selection (Fletcher and Yang 2010; Markova-Raina and Petrov 2011). In the pairwise alignments obtained, the positions creating gaps on the reference sequence were discarded to maintain the initial coordinates in the reference.
Sequences matching the same fragments of the reference M. lychnidis-dioicae genome were clustered together as putative orthologs using Newbler-generated coordinates of the mapped contigs on the reference genome. Noncoding regions were filtered out from POGs using the CDS annotations of the reference genome provided by the Broad Institute. Loci shorter than 300 bp and those missing from more than six Microbotryum species were discarded.
An outgroup species, M. pustulatum, parasitizing P. bistorta (Kemler et al. 2006), was also sequenced and assembled against the reference genome as described above. Given its distance from the Microbotryum species (mean number of substitutions per site between the outgroup and the ingroups is 0.085 ± 0.067), few POGs (46) were available for the phylogenetic studies. Consequently a second, larger data set was generated without the outgroup species, including 5,726 POGs, which was used for all the subsequent analyses.
Species Phylogeny Inference
To avoid potential biases in the phylogenetic signal caused by trans-specific polymorphism, as observed at some mating-type genes (Devier et al. 2009; Abbate and Hood 2010b; Petit et al. 2012), only POGs not belonging to mating-type chromosomes were used for the species tree reconstruction. The 5,453 POGs not belonging to mating-type chromosomes were concatenated and JMODELTEST (Posada 2008) was used to infer the most suitable nucleotide substitution model. Phylogenetic trees were constructed in a maximum-likelihood framework using RAxML 7.8.1 (Stamatakis et al. 2005) with a GTR + gamma substitution model and on gene-partitioned data sets (substitution model parameters were optimized for each partition of genes with JMODELTEST). Other phylogenies based on codon position partitioning (computing estimations separately for the first two codon positions, GC12, vs. the third codon position, GC3) or on other amino acid substitution models, had identical topologies. A rooted phylogeny was also produced based on the small subset of 46 POGs for which orthologs were found in the Microbotryum species and the P. bistorta outgroup species (Lutz et al. 2005; Kemler et al. 2006).
The robustness of nodes in our tree was tested using only the POGs not belonging to the mating-type chromosomes yielding individual trees with a high mean bootstrap support (MBS) or a high Tree Certainty (TC) (Salichos and Rokas 2013). For each POG, ModelTest v 2.3.1 (Darriba et al. 2012) was used to find the best model, and PhyML v3 (Guindon and Gascuel 2003) to reconstruct each tree with 100 bootstrap replicates. For each tree, the MBS and the TC (using RAxML 7.8.1) were then computed. MBS and TC values being highly correlated (R2 = 0.8276, P < 2.2e-16), only MBS was used thereafter. POGs with MBS ≥ 80 were selected: The sequences of the 288 POGs selected were concatenated. RAxML v 7.2.8 was used to reconstruct the final trees using the gtr+gamma substitution model and 100 bootstrap replicates. Note that the tree obtained when selecting the genes based on their TC was the same as that based on MBS.
Placement of NRR Alleles in a1 and a2 of M. lychnidis-dioicae in the Species Tree and Computation of Their Divergence
BLASTn and tBLASTn searches (NCBI blast 2.2.29 universal macosx) of the M. lychnidis-dioicae a1 alleles for predicted genes in NRRs were performed to search for hits in the a2 mating-type chromosome of M. lychnidis-dioicae. Both methods converged on the same hits. The a2 protein hits obtained from the tBLASTn search were added to the respective POG protein alignments obtained above and realigned using t-coffee 10.00.r1613 (Notredame et al. 2000) with default settings. The resulting alignments were trimmed with Trimal version 1.2rev59 (Capella-Gutierrez et al. 2009), removing 20% of the gaps introduced when adding the a2 sequences. We kept only the 45 reliable alignments, and used them to build the trees with RAxML version 7.4.4 (Stamatakis 2006) and the GTRGAMMA model. The placement of the a1 and a2 alleles relative to alleles of other species was visually inspected in the predicted trees. The synonymous divergence of NRR alleles between a1 and a2 of M. lychnidis-dioicae was computed as the estimated number of synonymous substitutions, including inferred multiple substitutions, divided by the number of synonymous sites in the sequence. The synonymous divergence and their associated standard error values were computed using the yn00 program in the PAML package (v. 4.7a) (Yang 2007) (fig. 1).
Detection of Selection Signatures
The efficacy of purifying selection to maintain gene function was assessed by two approaches. First classic models of codon substitution (Yang 2007) were used: Parameters were inferred from the species phylogeny to reconstruct changes in the ratios of nonsynonymous to synonymous substitutions (ω = dN/dS). The rationale is that nonsynonymous, deleterious substitutions are expected to accumulate if purifying selection is weak or relaxed. Ratios of dN/dS were estimated for each POG using codon models with CODEML in the PAML package (Yang 2007). The tree used was the species tree (fig. 3). Species not represented in these POGs were pruned from the species tree using the Ape package in R. We also ran analyses using individual gene trees for NRR POGs instead of species tree, and similar results were obtained (supplementary fig. S3C, Supplementary Material online).
The efficacy of purifying selection was also investigated using an estimation of the global ω per branch (the “free-ratio branch model” under CODEML): Mean and median ω values per branch for each terminal lineage were calculated, and genes with dS values close to 0 that would produce artifactually very large ω values were excluded. Positive selection was tested by comparing neutral and selective codon models. Genes displaying unrealistically high dN or dS values (i.e., “999” in the codeml output) were discarded from the analysis as they probably corresponded to saturated levels of substitutions and would bias ω estimations. We also excluded from the analysis those genes with null dS and dN values. More stringent filtering (discarding sequences with SdS lower than 2, 3, or 4, or filtering using branch length parameters) led to results for ω similar to those presented.
To control for the possibility of positive selection increasing dN/dS, which would not constitute degeneration, positive selection was also tested using a branch-site test based on codon models which accounts for variation of the ω ratio between both sites and lineages (Yang and Nielsen 2002; Zhang et al. 2005). A likelihood ratio test was used to compare a null model without positive selection and an alternative model allowing positive selection (Nielsen and Yang 1998). In cases where the null hypothesis was rejected, and the “selective” model retained, positions under positive selection among foreground branches were determined using a Bayesian empirical Bayes approach (Yang et al. 2005). Similar automated procedures (python + R scripts) were used to run codeml automatically and both extract and filter parameter values as in the free-ratio branch model described above.
The second approach used to assess the efficacy of purifying selection focused on recently derived substitutions; these have the advantage of being free of assumptions on phylogenic relationships and are more likely to have occurred while being in the current genomic compartment; indeed, substitutions may have occurred before the putative transfer of genes between recombining regions and NRRs (Abbate and Hood 2010b; Petit et al. 2012). Recent substitutions were identified as singletons in aligned POGs. From the singleton data set for all alignments of orthologous loci, median dN/dS ratios between different genomic compartments (NRRs, MATRRs, and non-MAT chromosomes) were computed and compared. The nonsynonymous rate was computed as dN = n/N, where n is the nonsynonymous singleton count and N is the number of nonsynonymous sites in the alignment. Similarly, the synonymous rate was computed as dS = s/S where s is the count of synonymous singleton substitutions and S the number of synonymous sites in the alignment. This approach, however, does not take into account multiple substitutions at a given site so it may underestimate substitution counts.
Gene Conversion Analyses
Geneconv v1.81 (Sawyer 1989) was used to look for footprints of gene conversion in NRRs directly. To be conservative, only results from estimations of “inner” gene converted fragments are reported. As defined in the Geneconv manual, such fragments represent evidence of possible gene conversion events between ancestors of two sequences in the alignment. Geneconv was executed through a batch script independently for the alignments of putative CDS regions (i.e., POGs) in NRRs.
TE Analyses
TE contents of contigs were estimated and assigned to non-MAT chromosomes, NRRs, and MATRRs in each Microbotryum species. Because many contigs were small, especially in the NRRs, the analysis was run on fragments of similar sizes using databases of 200-bp fragments generated in each species and genomic compartment (non-MAT chromosomes, NRR, and MATRR). To compare databases of equivalent sizes, 1,000 reads were randomly resampled without replacement from each database. A set of full-length TEs of the most common types in Microbotryum (Hood 2005; Hood et al. 2005; Yockteng et al. 2007) (Copia-like, e.g., Supercontig 147: 1,901–7,063 bp; Gypsy-like, e.g., Supercontig 151: 14,408–18,631 bp; and rolling circle Helitron-like, e.g., Supercontig 124:8,251–12,551 bp) were used as queries to search against the 1,000 read databased using tBLASTx (Basic Local Alignment Search Tool; blast.ncbi.nlm.nih.gov/Blast.cgi) with the significance threshold set at E value < 10−6. To assess the significance of the differences in TE counts between genomic compartments, Z tests were used. P values were assigned by comparing the computed Z value to the critical Z value for a two-tailed test in a standard normal distribution table. The null hypothesis was that the difference between the proportions of the two samples considered is zero.
Expression Analysis in the Reference Species (M. lychnidis-dioicae)
Expression data were generated under five different conditions using the Lamole reference strain, with two biological replicates per condition. Haploid fungal cells of Lamole a1 sporidial cells (p1A1Rich) and a2 sporidial cells (p1A2Rich) were grown separately for 5 days on yeast peptone dextrose media (1% yeast extract, 10% dextrose, 2% peptone, 2% agar; rich growth conditions) at room temperature, and then harvested for RNA extraction. Similarly, haploid cells grown separately on 2% water agar for 2 days were harvested for RNA extraction and named p1A1Water and p1A2Water; these samples allowed comparison between the gene expression when haploid cells of different mating types were subjected to a nutrient-free environment without a mating partner. We also studied a “mated” condition: An equal mixture of a1 and a2 cells was kept at 14 °C on 2% water agar for 2 days, which induces conjugation, the first stage of mating (Day 1979).
RNA was extracted from samples of the cells cultured in these various conditions using the RNeasy Plant Mini Kit (QIAGEN, cat no: 74904) according to the manufacturer’s instructions. Ambion’s TURBO DNA-free (Applied Biosystems, cat no: AM1907) DNase was used according to the manufacturer’s instruction. For quality assessment before Illumina sequencing, 5 μg of DNase-treated RNA was reverse transcribed with SuperScript III First Strand Synthesis System for reverse transcription PCR (Life Technologies, cat no: 18080-051). PCR was performed using TaKaRa Ex Taq Hot-Start DNA Polymerase (Takara, cat. no: RR001B) in a reaction volume of 25 μl. To check for DNA contamination and possible inhibitory substances in the RNA, two sets of housekeeping primers (Eurofins/MWG/Operon) were used. Targeting the Microbotryum mepA gene, the forward primer, 5′-CTTTTGCGTAGGAAGAATGC-3′ and the reverse primer, 5′-AGCACTGAACACCCCAACTT-3′ were used; this combination yielded a 532-bp fragment from cDNA, and a 1,039-bp fragment from genomic DNA. The other primer combination targeted the Microbotryum beta-tubulin gene, with forward primer, 5′-CGGACACCGTTGTCGAGCCT-3′, and reverse primer, 5′-TGAGGTCGCCGTGAGTCGGT-3’, yielding a 150-bp fragment from cDNA and a 215-bp fragment from genomic DNA. The PCR program was 30 s at 94 °C, 30 s at 60 °C, and 1 min at 72 °C for 35 cycles. An Agilent BioAnalyzer was also used to determine the quality of the RNA prior to Illumina RNA-Seq.
De novo assemblies of both the a1 and a2 RNA-Seq data were generated with Trinity (Grabherr et al. 2011) to identify genes specific to each mating chromosome. Assembled transcripts were compared between a1 and a2 using blast to identify transcripts specific for each cell type: a1 transcripts were mapped to the gene set based on the whole genome assembly; the a2 assembly is available at the Broad website, at http://www.broadinstitute.org/annotation/genome/Microbotryum_violaceum/assets/Mvio_A2_trinity.fa.gz.
Differential expression scripts in the Trinity pipeline version r2013-02-25 (Grabherr et al. 2011) were run to process RNA-Seq reads from the ten libraries (two biological replicates of each of p1A1Water, p1A2Water, p1A1Rich, p1A2Rich, and Mated). The RNA-Seq reads were aligned by bowtie (Langmead et al. 2009) against protein CDS extracted from the reference genome, adding 100 bases of flanking sequence to approximate UTRs. The read alignment files were then processed with RSEM (Li and Dewey 2011) to quantify transcript abundances. To identify genes differentially expressed between each pair of conditions, edgeR with the trimmed mean of M-value normalization method (TMM) (Robinson et al. 2010; Kadota et al. 2012) was run using an adjusted P value cutoff of 1E-10; TMM normalization is a method for estimating relative RNA production levels from RNA-Seq data by estimating scale factors between samples. The agglomerative hierarchical clustering method implemented in the R package cluster (Maechler et al. 2013) was then used to identify coexpressed gene clusters of those genes with significant differential expression between conditions. Expression data are available at the Broad Institute website, www.broadinstitute.org/annotation/genome/Microbotryum_violaceum.
Some genes displayed mating-type-specific expression profiles, which could be the result of a lack of expression of these genes on one of the mating-type chromosomes or of hemizygosity (there is only a single copy of the gene, on only one of the mating-type chromosomes). To distinguish these possibilities, the presence of the genes expressed only from one mating-type chromosome was investigated using BLASTn by aligning the assembled cDNA against the genomic sequence of the opposite mating type (the published a1 haploid genome for a2-specific transcripts, and the a2 mating type chromosome for a1-specific ones).
The absence of some genes from one of the mating-type chromosomes was then checked with primers specific for these genes (supplementary table S2, Supplementary Material online): DNAs extracted from a2 and a1 cells from the reference Lamole strain, 20 other M. lychnidis-dioicae strains, and one strain from each other species studied here, were tested for amplification with these primers.
Statistical Tests Using Monte Carlo Resampling of the Genomic Background
To test for the significance of the differences in the values of several variables described above (dN/dS, GC content, preferred/unpreferred codons) between the NRRs, MATRRs, and non-MAT chromosomes, a null genomic distribution of the variable values was constructed through randomization. To do this, 1,000 sets of non-MAT chromosome loci were randomly sampled and the mean and median values were computed for the variables of interest. The size of each of these sets was selected to be equal to the number of loci observed in the region of interest (NRR or MATRR). Significance thresholds of 0.01 or 0.05 were used to score the mating-type chromosome data set (MATRR or NRR) as significantly different from the non-MAT chromosome set.
Supplementary Material
Supplementary tables S1 and S2 and figures S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Pierre Fontanillas, Laurent Duret, Gabriel Marais, and Antoine Branca for valuable discussions. This work was supported by the Agence Nationale de la Recherche (ANR-09-BLAN-064 to T.G.), the European Research Council (starting grant GenomeFun 309403 to T.G.), and the National Science Foundation (DEB 0747222 to M.E.H., 0947963 to M.H.P. and C.A.C.). The authors thank the IT service at the Station Biologique Marine de Roscoff and the Centre National de Séquencage (Génoscope) for computing cluster facilities. They are grateful to Stéphanie Le Prieur, Alodie Snirc, and Odile Jonot for help with molecular biology experiments. They also thank Janis Antonovics, Mel Harte, and Steve Matson for some of the pictures in figure 2. The accession numbers are PRJEB6323 for sequences of the a2 mating type chromosome of M. lychnidis-dioicae and PRJEB6548 for the genomes of the other species. T.G. and M.E.H. designed the research project; J.P., P.W., E.P., M.E.H., Z.C., S.T., C.A.C., and M.H.P. obtained the genomic sequences and transcriptome data; E.F., V.B., Z.C., C.A.C., H.B., J.G., and M.P. performed assembly and mapping; E.F., H.B., E.P., J.G., V.B., C.A.C., M.P., D.M.d.V., H.B., P.G., and G.A. performed analyses; E.F., T.G., and M.E.H. wrote the paper; all authors approved the final manuscript.
References
- Abbate JL, Hood ME. Dynamic linkage relationships to the mating-type locus in automictic fungi of the genus Microbotryum. J Evol Biol. 2010;23:1800–1805. doi: 10.1111/j.1420-9101.2010.02036.x. [DOI] [PubMed] [Google Scholar]
- Antonovics J, Abrams J. Intratetrad mating and the evolution of linkage relationships. Evolution. 2004;58:702–709. doi: 10.1111/j.0014-3820.2004.tb00403.x. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Accumulation of Spock and Worf, two novel non-LTR retrotransposons, on the neo-Y chromosome of Drosophila miranda. Mol Biol Evol. 2003;20:173–181. doi: 10.1093/molbev/msg035. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Sex chromosome evolution: molecular aspects of Y chromosome degeneration in Drosophila. Genome Res. 2005;15:1393–1401. doi: 10.1101/gr.3543605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, Rice WR, Valenzuela N. Are all sex chromosomes created equal? Trends Genet. 2011;27:350–357. doi: 10.1016/j.tig.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Bartolome C, Charlesworth B. Evolution of amino-acid sequences and codon usage on the Drosophila miranda neo-sex chromosomes. Genetics. 2006;174:2033–2044. doi: 10.1534/genetics.106.064113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Bergero R, Forrest A, Kamau E, Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics. 2007;175:1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiard S, Lopez-Villavicencio M, Devier B, Hood M, Fairhead C, Giraud T. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol Rev. 2011;86:421–442. doi: 10.1111/j.1469-185X.2010.00153.x. [DOI] [PubMed] [Google Scholar]
- Bull JJ. Sex chromosomes in haploid dioecy—unique contrast to Mullers theory for diploid dioecy. Am Nat. 1978;112:245–250. [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada G, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DW. Mating type and morphognesis in Ustilago violacea. Bot Gaz. 1979;140:94–101. [Google Scholar]
- de Vienne D, Hood M, Giraud T. Phylogenetic determinants of potential host shifts in fungal pathogens. J Evol Biol. 2009;22:2532–2541. doi: 10.1111/j.1420-9101.2009.01878.x. [DOI] [PubMed] [Google Scholar]
- de Vienne DM, Refrégier G, Hood M, Guigue A, Devier B, Vercken E, Smadja C, Deseille A, Giraud T. Hybrid sterility and inviability in the parasitic fungal species complex Microbotryum. J Evol Biol. 2009;22:683–698. doi: 10.1111/j.1420-9101.2009.01702.x. [DOI] [PubMed] [Google Scholar]
- Devier B, Aguileta G, Hood M, Giraud T. Ancient trans-specific polymorphism at pheromone receptor genes in basidiomycetes. Genetics. 2009;181:209–223. doi: 10.1534/genetics.108.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis N, Taylor A, Bengtsson BO, Kidd J, Rogers J, Goodfellow P. Population-structure of the human pseudoautosomal boundary. Nature. 1990;344:663–665. doi: 10.1038/344663a0. [DOI] [PubMed] [Google Scholar]
- Ellison CE, Stajich JE, Jacobson DJ, Natvig DO, Lapidus A, Foster B, Aerts A, Riley R, Lindquist EA, Grigoriev IV, et al. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics. 2011;189:55–69. doi: 10.1534/genetics.111.130690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandsson R, Wilson JF, Paabo S. Sex chromosomal transposable element accumulation and male-driven substitutional evolution in humans. Mol Biol Evol. 2000;17:804–812. doi: 10.1093/oxfordjournals.molbev.a026359. [DOI] [PubMed] [Google Scholar]
- Feldbrugge M, Kamper J, Steinberg G, Kahmann R. Regulation of mating and pathogenic development in Ustilago maydis. Curr Opin Microbiol. 2004;7:666–672. doi: 10.1016/j.mib.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Filatov DA, Charlesworth D. Substitution rates in the X- and Y-linked genes of the plants, Silene latifolia and S. dioica. Mol Biol Evol. 2002;19:898–907. doi: 10.1093/oxfordjournals.molbev.a004147. [DOI] [PubMed] [Google Scholar]
- Fletcher W, Yang Z. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol. 2010;27:2257–2267. doi: 10.1093/molbev/msq115. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2004;2:2243–2255. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ED, Ruddat M. Genetics of Ustilago violacea. XXXII. Genetic evidence for transposable elements. Theor Appl Genet. 1995;89:838–846. doi: 10.1007/BF00224506. [DOI] [PubMed] [Google Scholar]
- Garber ED, Ruddat M. Genetics of Ustilago violacea. XXXIV. Genetic evidence for a transposable element functioning during mitosis and txo transposable elements functioning during meiosis. Int J Plant Sci. 1998;159:1018–1022. [Google Scholar]
- Giraud T, Yockteng R, Lopez-Villavicencio M, Refregier G, Hood ME. The mating system of the anther smut fungus, Microbotryum violaceum: selfing under heterothallism. Eukaryot Cell. 2008;7:765–775. doi: 10.1128/EC.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladieux P, Vercken E, Fontaine M, Hood M, Jonot O, Couloux A, Giraud T. Maintenance of fungal pathogen species that are specialized to different hosts: allopatric divergence and introgression through secondary contact. Mol Biol Evol. 2011;28:459–471. doi: 10.1093/molbev/msq235. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng QD, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grognet P, Bidard F, Kuchl C, Chan Ho Tong L, Coppin E, Benkhali JA, Couloux A, Wincker P, Debuchy R, Silar P. Maintaining two mating types: structure of the mating type locus and its role in heterokaryosis in Podospora anserina. Genetics. 2014;197:421–432. doi: 10.1534/genetics.113.159988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hood ME. Dimorphic mating-type chromosomes in the anther-smut fungus. Genetics. 2002;160:457–461. doi: 10.1093/genetics/160.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood ME. Repetitive DNA in the automictic fungus Microbotryum violaceum. Genetica. 2005;124:1–10. doi: 10.1007/s10709-004-6615-y. [DOI] [PubMed] [Google Scholar]
- Hood ME, Antonovics J. Intratetrad mating, heterozygosity, and the maintenance of deleterious alleles in Microbotryum violaceum (=Ustilago violacea) Heredity. 2000;85:231–241. doi: 10.1046/j.1365-2540.2000.00748.x. [DOI] [PubMed] [Google Scholar]
- Hood ME, Antonovics J, Koskella B. Shared forces of sex chromosome evolution in haploids-mating and diploids-mating organisms: Microbotryum violaceum and other model organisms. Genetics. 2004;168:141–146. doi: 10.1534/genetics.104.029900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood ME, Antonovics JA. Mating within the meiotic tetrad and the maintenance of genomic heterozygosity. Genetics. 2004;166:1751–1759. doi: 10.1534/genetics.166.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood ME, Katawezik M, Giraud T. Repeat-induced point mutation and the population structure of transposable elements in Microbotryum violaceum. Genetics. 2005;170:1081–1089. doi: 10.1534/genetics.105.042564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood ME, Petit E, Giraud T. Extensive divergence between mating-type chromosomes of the anther-smut fungus. Genetics. 2013;193:309–315. doi: 10.1534/genetics.112.146266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LJ, Antonovics J, Hood ME. The evolution of intratetrad mating rates. Evolution. 2005;59:2525–2532. [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota K, Nishiyama T, Shimizu K. A normalization strategy for comparing tag count data. Algorithms Mol Biol. 2012;7:5. doi: 10.1186/1748-7188-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler M, Goker M, Oberwinkler F, Begerow D. Implications of molecular characters for the phylogeny of the Microbotryaceae (Basidiomycota : Urediniomycetes) BMC Evol Biol. 2006;6:35. doi: 10.1186/1471-2148-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latreille P, Norton S, Goldman BS, Henkhaus J, Miller N, Barbazuk B, Bode HB, Darby C, Du ZJ, Forst S, et al. Optical mapping as a routine tool for bacterial genome sequence finishing. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gac M, Hood ME, Fournier E, Giraud T. Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evolution. 2007;61:15–26. doi: 10.1111/j.1558-5646.2007.00002.x. [DOI] [PubMed] [Google Scholar]
- Le Gac M, Hood ME, Giraud T. Evolution of reproductive isolation within a parasitic fungal complex. Evolution. 2007;61:1781–1787. doi: 10.1111/j.1558-5646.2007.00144.x. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320:1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- Lutz M, Goker M, Piatek M, Kemler M, Begerow D, Oberwinkler F. Anther smuts of Caryophyllaceae: molecular characters indicate host-dependent species delimitation. Mycol Prog. 2005;4:225–238. [Google Scholar]
- Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. 2013 cluster: cluster analysis basics and extensions. R package version 1.14.4. [Google Scholar]
- Marais GAB, Nicolas M, Bergero R, Chambrier P, Kejnovsky E, Moneger F, Hobza R, Widmer A, Charlesworth D. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr Biol. 2008;18:545–549. doi: 10.1016/j.cub.2008.03.023. [DOI] [PubMed] [Google Scholar]
- Markova-Raina P, Petrov D. High sensitivity to aligner and high rate of false positives in the estimates of positive selection in the 12 Drosophila genomes. Genome Res. 2011;21:863–874. doi: 10.1101/gr.115949.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkis A, Jacobson DJ, Gustafsson T, Johannesson H. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 2008;4:e1000030. doi: 10.1371/journal.pgen.1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis BPS, Billiard S, Vuilleumier S, Petit E, Hood ME, Giraud T. 2013 doi: 10.1038/hdy.2013.67. Evolution of uni- and bifactorial sexual compatibility systems in fungi. Heredity 111:445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins D, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Oudemans PV, Alexander HM, Antonovics J, Altizer S, Thrall PH, Rose L. The distribution of mating-type bias in natural populations of the anther-smut Ustilago violacea on Silene alba in Virginia. Mycologia. 1998;90:372–381. [Google Scholar]
- Perlin MH. Pathovars or formae speciales of Microbotryum violaceum differ in electrophoretic karyotype. Int J Plant Sci. 1996;157:447–452. [Google Scholar]
- Petit E, Giraud T, de Vienne DM, Coelho M, Aguileta G, Amselem J, Kreplak J, Poulain J, Gavory F, Wincker P, et al. Linkage to the mating-type locus across the genus Microbotryum: insights into non-recombining chromosomes. Evolution. 2012;66:3519–3533. doi: 10.1111/j.1558-5646.2012.01703.x. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Repping S. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichos L, Rokas A. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature. 2013;497:327–331. doi: 10.1038/nature12130. [DOI] [PubMed] [Google Scholar]
- Sawyer SA. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Steinemann M, Steinemann S. Degenerating Y chromosome of Drosophila miranda: a trap for retrotransposons. Proc Natl Acad Sci U S A. 1992;89:7591–7595. doi: 10.1073/pnas.89.16.7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Shykoff J, Jonot O, Giraud T. Mating-type ratio bias in populations of the phytopathogenic fungus Microbotryum violaceum from several host species. Int J Plant Sci. 2003;164:641–647. [Google Scholar]
- Votintseva AA, Filatov DA. 2009 ‘Evolutionary Strata’ in a small mating type-specific region of the smut fungus Microbotryum violaceum. Genetics 182:1391–1396. [Google Scholar]
- Whittle CA, Johannesson H. Evidence of the accumulation of allele-specific non-synonymous substitutions in the young region of recombination suppression within the mating-type chromosomes of Neurospora tetrasperma. Heredity. 2011;107:305–314. doi: 10.1038/hdy.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CA, Sun Y, Johannesson H. Degeneration in codon usagewithin the region of suppressed recombination in the mating-type chromosomes of Neurospora tetrasperma. Eukaryot Cell. 2011;10:594–603. doi: 10.1128/EC.00284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. Bayes Empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yockteng R, Marthey S, Chiapello H, Hood M, Rodolphe F, Devier B, Wincker P, Dossat C, Giraud T. Expressed sequence tags of the anther smut fungus, Microbotryum violaceum, identify mating and pathogenicity genes. BMC Genomics. 2007;8:272. doi: 10.1186/1471-2164-8-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharov IA. Some principles of the gene localization in eukaryotic chromosomes. Formation of the problem and analysis of nonrandom localization of the mating-type loci in some fungi. Genetika. 1986;22:2620–2624. [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.