Abstract
Major challenges for illuminating the genetic basis of phenotypic evolution are to identify causative mutations, to quantify their functional effects, to trace their origins as new or preexisting variants, and to assess the manner in which segregating variation is transduced into species differences. Here, we report an experimental analysis of genetic variation in hemoglobin (Hb) function within and among species of Peromyscus mice that are native to different elevations. A multilocus survey of sequence variation in the duplicated HBA and HBB genes in Peromyscus maniculatus revealed that function-altering amino acid variants are widely shared among geographically disparate populations from different elevations, and numerous amino acid polymorphisms are also shared with closely related species. Variation in Hb-O2 affinity within and among populations of P. maniculatus is attributable to numerous amino acid mutations that have individually small effects. One especially surprising feature of the Hb polymorphism in P. maniculatus is that an appreciable fraction of functional standing variation in the two transcriptionally active HBA paralogs is attributable to recurrent gene conversion from a tandemly linked HBA pseudogene. Moreover, transpecific polymorphism in the duplicated HBA genes is not solely attributable to incomplete lineage sorting or introgressive hybridization; instead, it is mainly attributable to recurrent interparalog gene conversion that has occurred independently in different species. Partly as a result of concerted evolution between tandemly duplicated globin genes, the same amino acid changes that contribute to variation in Hb function within P. maniculatus also contribute to divergence in Hb function among different species of Peromyscus. In the case of function-altering Hb mutations in Peromyscus, there is no qualitative or quantitative distinction between segregating variants within species and fixed differences between species.
Keywords: adaptation, gene conversion, gene duplication, high altitude, hemoglobin, standing variation
Introduction
A central question in evolutionary genetics concerns the sources of adaptive genetic variation in natural populations: Are adaptive phenotypic changes primarily fueled by the fixation of new mutations or preexisting allelic variants (standing genetic variation)? A corollary question concerns the extent to which segregating variation can be extrapolated to explain the genetic basis of phenotypic differences between species: Are allelic differences that contribute to adaptive phenotypic variation within species qualitatively or quantitatively different from fixed differences that contribute to interspecific divergence? These questions can be addressed most effectively by examining the functional effects of specific mutations that contribute to adaptive phenotypic variation within and among species. The joint analysis of intraspecific polymorphism and interspecific divergence can provide insights into the mutational origins of causative alleles (Mitchell-Olds et al. 2007; Barrett and Schluter 2008; Wray 2013) and the manner in which segregating variation is transduced into species differences (Stern 2000; Wittkopp et al. 2009).
To investigate the relationship between intraspecific polymorphism and interspecific divergence in an adaptively important biochemical phenotype, we examined the molecular basis of variation in hemoglobin (Hb)-O2 affinity within and among species of Peromyscus mice that are native to different elevations. The most geographically widespread and abundant species in the genus, the deer mouse (Peromyscus maniculatus), harbors a highly complex, multilocus Hb polymorphism that plays a well-documented role in physiological adaptation to high-altitude hypoxia (Snyder 1981, 1985; Snyder et al. 1982, 1988; Chappell and Snyder 1984; Chappell et al. 1988; Storz 2007; Storz, Sabatino, et al. 2007; Storz and Kelly 2008; Storz et al. 2009; Storz, Runck, et al. 2010; Storz, Natarajan, et al. 2012; Natarajan et al. 2013). Peromyscus maniculatus has the broadest altitudinal distribution of any North American mammal (Hock 1964) and occurs in high alpine environments at elevations greater than 4,300 m (the highest elevations in the contiguous United States) where the partial pressure of O2 (PO2) is less than 60% of the sea level value. Functional studies revealed that high-altitude deer mice from the Rocky Mountains have evolved an increased Hb-O2 affinity (which enhances pulmonary O2-loading under hypoxia) relative to lowland mice from the Great Plains (Storz et al. 2009; Storz, Runck, et al. 2010). Causative mutations have been identified, site-directed mutagenesis experiments have quantified their additive and epistatic effects (Natarajan et al. 2013), and crystallographic analyses of deer mouse Hbs have yielded insights into the structural mechanisms responsible for the allelic variation in oxygenation properties (Inoguchi et al. 2013). Complementing these experimental results, population genetic analyses of electrophoretic variation (Snyder et al. 1988) and nucleotide variation at the underlying α- and β-globin genes (Storz, Sabatino, et al. 2007; Storz and Kelly 2008; Storz et al. 2009; Storz, Runck, et al. 2010; Storz, Natarajan, et al. 2012) have provided indirect, corroborative evidence for a history of divergent selection on Hb polymorphism between deer mouse populations that are native to different elevations.
The purpose of the present study was to address questions about the origins and effects of causative mutations: How many mutations contribute to evolutionary changes in Hb function? What are the relative contributions of standing variation and new mutations? Are the mutational changes that contribute to divergence between conspecific populations distinct from those that contribute to divergence between species? The analysis of intraspecific polymorphism and interspecific divergence also enables us to address questions about phenotypic parallelism at different levels: Given that deer mice inhabit numerous different mountain ranges across western Northern America, are altitude-related changes in Hb function replicated across the species’ range? Are patterns of altitudinal differentiation among conspecific populations paralleled by patterns of divergence among species that have different altitudinal distributions?
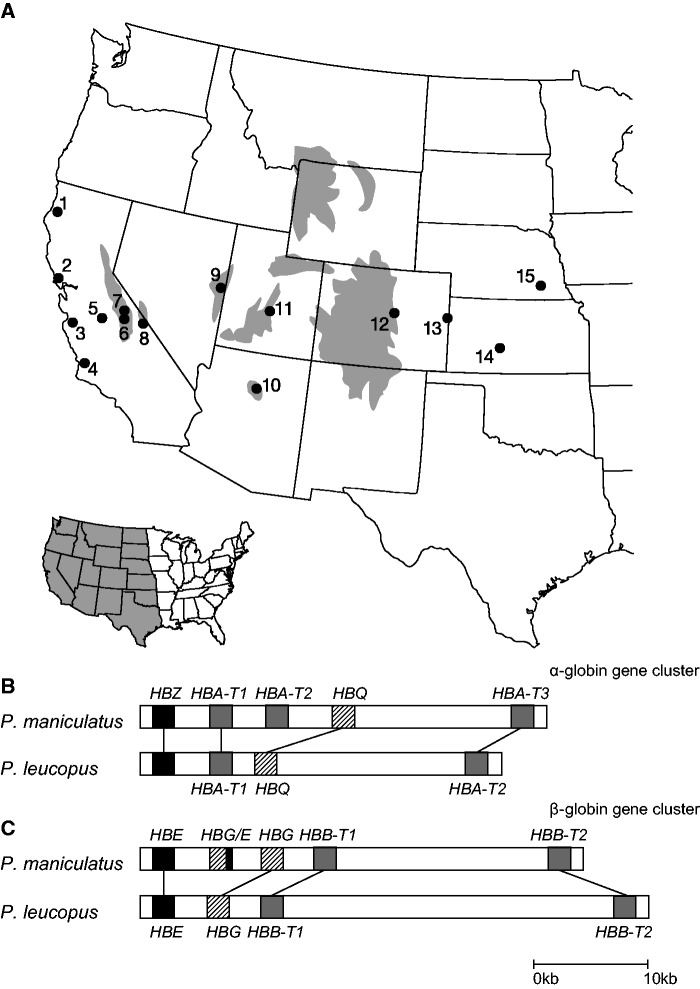
We conducted an analysis of Hb polymorphism and functional variation in deer mice that we collected from 15 geographic localities across the western United States. These localities span the elevational range of P. maniculatus, ranging from sea-level to alpine summits at elevations greater than 4,300 m. The high-altitude localities were distributed across numerous mountain ranges, including alpine and subalpine collection sites in the Southern Rocky Mountains (Colorado), the Aquarius Plateau (Utah), the San Francisco Peaks (Arizona), the Snake Range (Nevada), the White Mountains (California), and the Sierra Nevada (California; fig. 1A and supplementary table S1, Supplementary Material online). The distribution of low-altitude collecting localities was equally disparate, ranging from coastal California to prairie grassland in eastern Colorado, Kansas, and Nebraska (fig. 1A and supplementary table S1, Supplementary Material online). We surveyed sequence variation in the duplicated globin genes of all collected specimens, a subset of which were used as subjects for functional studies of purified Hb variants. To establish an historical framework for interpreting patterns of Hb polymorphism and functional variation, specimens from all 15 localities were included in a range-wide survey of mtDNA variation to characterize the phylogeographic population structure of the P. maniculatus species complex. To complement the phylogeographic analysis of mtDNA variation, we generated genome-wide nucleotide polymorphism data for a subset of high- and low-altitude population samples to gain more refined insights into relationships among populations. In addition to the survey of Hb polymorphism and functional variation within P. maniculatus, we also surveyed globin sequence variation in numerous other species of Peromyscus, and we characterized functional properties of purified Hbs from five other Peromyscus species that have different elevational range limits.
Fig. 1.
Geographic distribution of collection localities and genomic structure of the globin gene clusters in Peromyscus. (A) Collection localities for the survey of Hb polymorphism in Peromyscus maniculatus across the central and western United States. Gray shading shows subalpine and alpine terrain above the 3,000 m elevational isocline. All high-altitude collection localities (6–12) were at elevations greater than 3,000 m (details about collecting localities are provided in supplementary table S1, Supplementary Material online). (B) Genomic structure of the α-globin gene clusters in P. maniculatus and P. leucopus. The α-chain subunits of adult Hb are encoded by the HBA-T1 and HBA-T2 genes in P. maniculatus, and by the single-copy HBA-T2 gene in P. leucopus. HBA-T3 and HBA-T2 are orthologous pseudogenes in P. maniculatus and P. leucopus, respectively (see text for details). (C) Genomic structure of the β-globin gene clusters in P. maniculatus and P. leucopus. The β-chain subunits of adult Hb are encoded by the HBB-T1 and HBB-T2 genes in both species.
Our functional analysis of naturally occurring Hb variants in P. maniculatus revealed that genetic variation in Hb-O2 affinity is attributable to multiple α- and β-chain mutations that have individually small effects. These affinity-altering mutations are widely shared among geographically disparate populations of P. maniculatus that are native to a broad range of elevations, and numerous amino acid polymorphisms are also shared with closely related species. Remarkably, a sizable fraction of standing genetic variation in the duplicated α-globin genes of P. maniculatus is attributable to interparalog gene conversion from a tandemly linked pseudogene. Partly as a result of concerted evolution between duplicated globin genes, the same amino acid changes that contribute to variation in Hb function within species also contribute to divergence in Hb function between species.
Results and Discussion
Genomic Organization of the Globin Gene Clusters of Peromyscus
Tetrameric Hbs (α2β2) of adult P. maniculatus incorporate α-chain subunits that are encoded by two tandem gene duplicates, HBA-T1 and HBA-T2 (separated by 5.0 kb on Chromosome 8), and β-chain subunits that are encoded by two other tandem duplicates, HBB-T1 and HBB-T2 (separated by 16.2 kb on Chromosome 1; Storz et al. 2008; Hoffmann et al. 2008b). For the purpose of comparison with P. maniculatus, we sequenced bacterial artificial chromosome (BAC) clones containing the complete α- and β-globin gene clusters of the closely related species, P. leucopus (fig. 1B and C). Comparisons between homologous gene clusters of the two Peromyscus species revealed that P. leucopus possesses a single adult-expressed α-globin gene (HBA-T1) that is orthologous to the HBA-T1 gene of P. maniculatus (fig. 1B and supplementary fig. S1A, Supplementary Material online). Both species also share orthologous HBA pseudogenes (“HBA-T3” in P. maniculatus, ”HBA-T2” in P. leucopus) that are located downstream of their transcriptionally active paralogs (fig. 1B and supplementary fig. S1A, Supplementary Material online). No product of HBA-T3 has been detected in functional α2β2 Hb tetramers of P. maniculatus (Storz, Runck, et al. 2010), so it can be considered a pseudogene with regard to Hb synthesis. However, the gene has an intact reading frame and we cannot rule out the possibility that it is expressed in nonerythroid tissues. The pattern of pairwise sequence matches provides clear evidence for 1:1 orthology of the HBB gene pair in P. maniculatus and P. leucopus (supplementary fig. S1B, Supplementary Material online).
After characterizing the genomic structure of the globin gene clusters, we then surveyed nucleotide polymorphism at all four of the adult-expressed α- and β-globin genes in geographically disparate populations of P. maniculatus across central and western North America (fig. 1A and supplementary table S1, Supplementary Material online). For comparative purposes, we also surveyed polymorphism at orthologous genes in P. leucopus and another closely related species, P. keeni, which possesses the same complement of adult-expressed globin genes as P. maniculatus.
Historical Backdrop: Phylogeographic Structure of the P. maniculatus Species Complex
In order to interpret altitudinal patterns of Hb polymorphism and functional variation in an historical framework, we conducted an analysis of mitochondrial cytochrome b (cytb) sequence variation to characterize the range-wide phylogeographic structure of the P. maniculatus species complex. Building on results of previous studies (Dragoo et al. 2006; Gering et al. 2009; Kalkvik et al. 2012), we analyzed a total of 454 cytb sequences from P. maniculatus and closely related species, including 74 new sequences. All specimens used in the experimental analysis of Hb function and in the survey of sequence variation in the globin genes were also represented in the cytb data set.
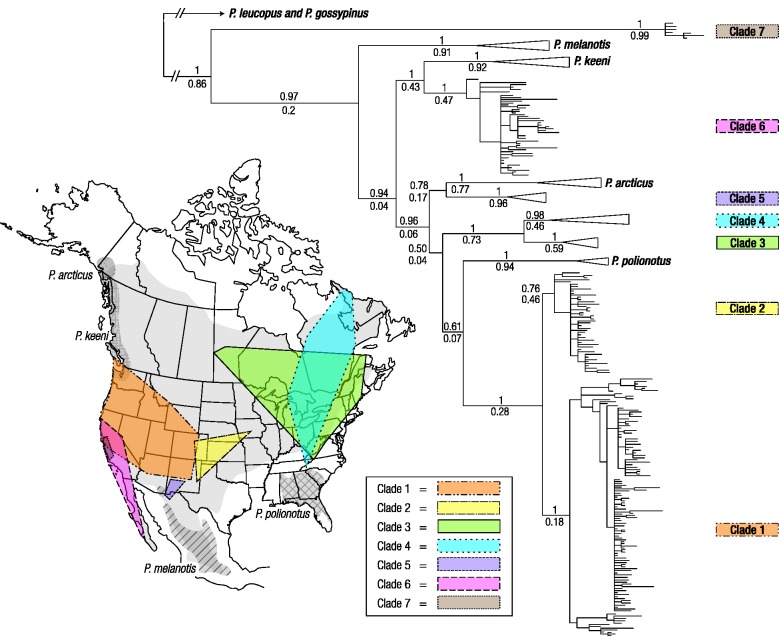
The range-wide phylogeographic analysis recovered seven main clades of cytb sequences from P. maniculatus (fig. 2). The analysis also revealed that four nominal species (P. melanotis, P. keeni, P. arcticus, and P. polionotus) are nested within P. maniculatus. This pattern of paraphyly—combined with a consideration of geographic distributions (fig. 2)—suggests that the above-mentioned species originated as peripheral isolates of the P. maniculatus parent species, possibly during Pleistocene glacial-interglacial cycles (Blair 1950). Our results corroborate the placement of P. polionotus as sister to P. maniculatus clades 1 and 2 and the placement of P. keeni as sister to P. maniculatus clade 6 (Gering et al. 2009; Kalkvik et al. 2012; fig. 2). In broad outline, six of the P. maniculatus clades have been characterized previously (Dragoo et al. 2006; Gering et al. 2009; Kalkvik et al. 2012), but our additional sampling uncovered a highly distinct group (“clade 7”) comprising specimens from the central coast of California (San Luis Obispo and Monterey counties). Bayesian and maximum-likelihood trees supported the same main clades and many similar relationships among clades, including sister relationships between clades 1 + 2, clades 3 + 4, and clade 6 + P. keeni, respectively.
Fig. 2.
Bayesian phylogeny of cytb sequences from 454 specimens of Peromyscus maniculatus and closely related species based on a GTR+I+G model of nucleotide substitution. Bayesian posterior probabilities (above) and maximum-likelihood bootstrap support values (below) are shown for all major nodes in the tree. The inset map of North America shows the geographic range of P. maniculatus and the superimposed distributions of seven main cytb clades. There is a broad zone of admixture between two highly distinct phylogroups (clades 1 and 6) that extends from coastal California to the western periphery of the Great Basin. Consequently, cytb sequences from northern Californian mice are distributed between clade 1 (which includes specimens from geographically disparate high-altitude localities across the western United States) and clade 6 (see supplementary fig. S2, Supplementary Material online). Also shown are geographic distributions of four other Peromyscus species (P. arcticus, P. keeni, P. melanotis, and P. polionotus) that are paraphyletic relative to P. maniculatus in the cytb phylogeny.
The P. maniculatus specimens used in the survey of Hb polymorphism all fell in clades 1, 2, 6, and 7. The majority of specimens from the Southern Rocky Mountains and the Great Basin (including montane regions of Colorado, Utah, Arizona, and Nevada) fell in clade 1, the majority of specimens from the Great Plains (eastern Colorado, Kansas, and Nebraska) fell in clade 2, and—as mentioned above—specimens from Central California fell in clade 7 (including all specimens from San Luis Obispo Co.; supplementary fig. S2, Supplementary Material online). Specimens from northern California were the most widely dispersed phylogenetically, as they all fell into one of two highly distinct clades (1 and 6) that are distinguished from one another by a net sequence divergence of 0.1788 at silent sites. Among specimens collected from high-altitude localities in the Sierra Nevada and the White Mountains of eastern California, one-third fell in clade 1 (along with mice from far-flung localities across the Rocky Mountains and the Great Basin) and the remaining two-third fell in clade 6 (along with mice from the coastal lowlands and Central Valley of California; supplementary fig. S2, Supplementary Material online). Specimens collected from geographically disparate low-altitude localities in northern California were almost evenly split between clades 1 and 6 (supplementary fig. S2, Supplementary Material online).
Phyletic Continuity among Alpine Populations
The fact that high-altitude mice from geographically disparate mountain ranges were intermingled in the same clade of the cytb phylogeny suggests possible historical connections among alpine populations. To test this using patterns of genome-wide nucleotide variation, we surveyed 4,344 restriction-site associated DNA (RAD) polymorphisms in a total of 60 deer mouse specimens sampled from two geographically proximal pairs of high- and low-altitude localities in Colorado (Mt. Evans [n = 15] and Yuma Co. [n = 15]) and in California (White Mountain Peak [n = 15] and Humboldt Co., [n = 15]). We then performed a four-population test of “treeness” (Reich et al. 2009; Patterson et al. 2012) to assess whether spatial proximity counts for more than native elevation in explaining phylogenetic affinities. Consistent with the phylogenetic patterns of mtDNA variation, the constraint tree ([Mt. Evans, Yuma Co.], [White Mountain Peak, Humboldt Co.]) failed the four-population test (f4 [ ± SE] = 0.0025 ± 0.007, Z = 3.88, P < 0.05). This result appears to be attributable to an historical connection between the two geographically disparate high-altitude populations, as genomic differentiation based on all 4,344 nt polymorphisms was roughly 2-fold lower in the comparison between Mt. Evans and White Mountain Peak than in the comparison between Yuma Co. and Humboldt Co. (FST = 0.0509 vs. 0.1235). Moreover, the level of genomic differentiation between the two distant alpine populations was also lower than that between either population and the geographically closer lowland population: FST (Mt. Evans vs. Yuma Co.) = 0.0638 and FST(White Mountain Peak vs. Humboldt Co.) = 0.0982.
Intraspecific Polymorphism and Interspecific Divergence: HBA Genes
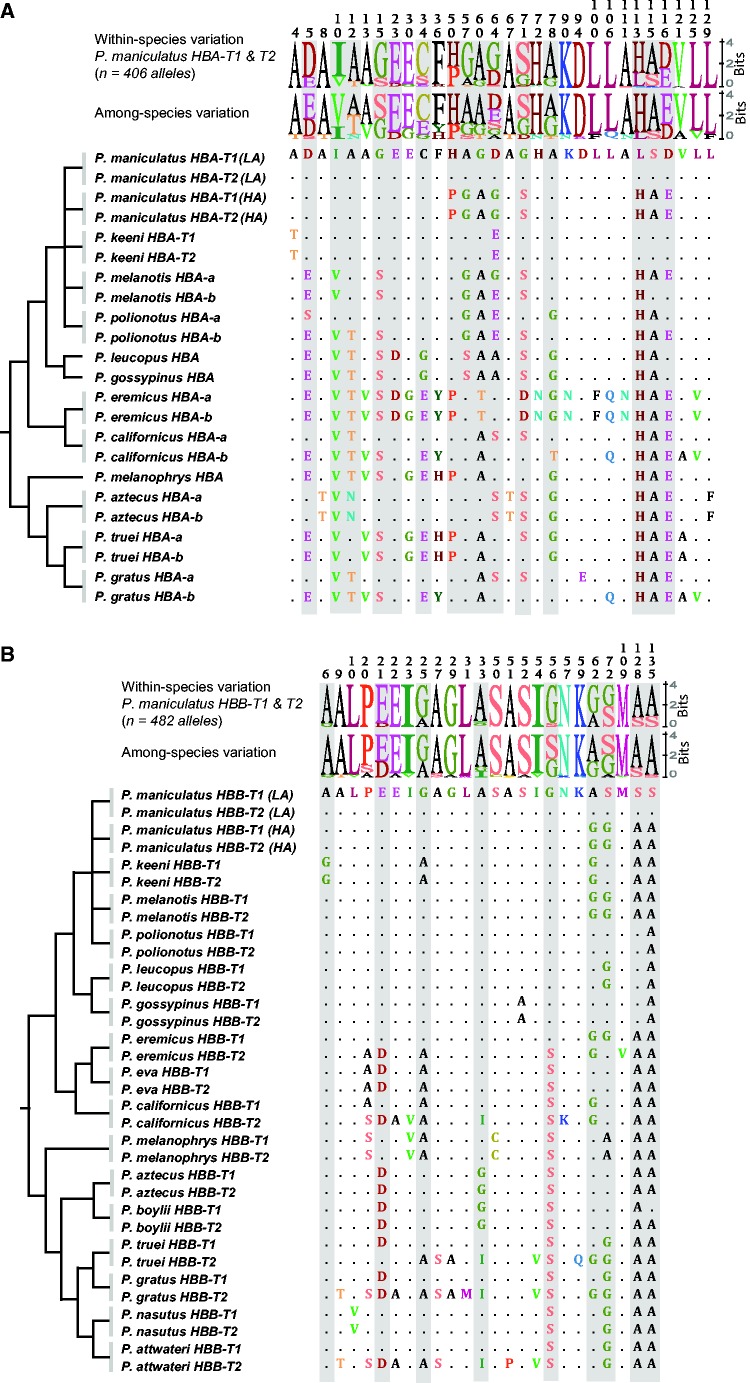
The survey of nucleotide polymorphism in the HBA genes of P. maniculatus from the central and western United States (n = 406 alleles) revealed 15 intermediate-frequency amino acid polymorphisms with minor allele frequencies (MAFs) greater than 0.05 (fig. 3A). We separately cloned and sequenced the tandemly linked HBA-T1 and HBA-T2 genes in a subset of specimens to obtain complete tetraploid genotypes ( = 228 experimentally phased alleles). Likewise, we generated tetraploid HBA-T1/HBA-T2 genotypes for P. keeni and diploid genotypes for the single-copy HBA-T1 gene of P. leucopus ( = 24 and 22 experimentally phased alleles, respectively). For each of these same three species we also generated polymorphism data for the downstream pseudogene (HBA-T3 in P. maniculatus and P. keeni, HBA-T2 in P. leucopus). Data for the HBA-T1 and HBA-T2 genes of P. maniculatus confirmed previous reports that the same alleles are segregating at both genes due to a history of interparalog gene conversion (Storz and Kelly 2008; Storz, Runck, et al. 2010). However, P. maniculatus specimens from one particular locality, Humboldt Co., CA, possess structurally distinct HBA paralogs that are distinguished by fixed or nearly fixed differences at six amino acid sites (supplementary fig. S3, Supplementary Material online). Amino acid replacements at most of the sites that distinguish HBA-T1 and HBA-T2 in the Humboldt mice are identical to sequence differences between alternative alleles that are segregating at both paralogs in mice from other regions (supplementary fig. S3, Supplementary Material online). HBA-T1 and HBA-T2 paralogs in the Humboldt mice are also characterized by far fewer shared polymorphisms in comparison with other population samples of P. maniculatus (supplementary table S2, Supplementary Material online), suggesting a lower rate of HBA-T1↔HBA-T2 gene conversion (and, hence, a lower rate of concerted evolution). However, overall sequence divergence between the HBA-T1 and HBA-T2 paralogs in the Humboldt mice was not unusually high compared with levels of interparalog divergence in other populations of P. maniculatus (supplementary table S2, Supplementary Material online).
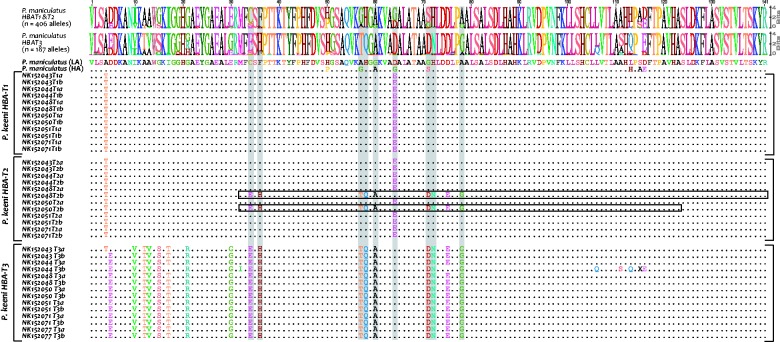
Fig. 3.
Intraspecific polymorphism and interspecific divergence in the HBA and HBB genes of Peromyscus. (A) Alignment of 30 variable amino acid sites in 12 Peromyscus species. Shaded columns denote sites that harbor intermediate-frequency polymorphisms (MAF >0.05) in Peromyscus maniculatus. At each site, sizes of the letter codes for alternative amino acids are proportional to their representation in the sample, and therefore provide a graphical depiction of site-specific allele frequency variation within P. maniculatus (top) and conservation among species (bottom). The HBA-T1 and HBA-T2 paralogs are individually identified in P. maniculatus and P. keeni. For P. maniculatus, the “LA” and “HA” alleles refer to the predominant amino acid haplotypes sampled from lowland (Great Plains) and highland (Rocky Mountain) localities, respectively. Some species (P. leucopus, P. gossypinus, and P. melanophrys) possess a single-copy HBA gene. In the remaining species, “HBA-a” and “HBA-b” refer to paralogous sequences with unknown linkage order in the α-globin gene cluster. (B) Alignment of 30 variable amino acid sites in the HBB-T1 and HBB-T2 genes of 16 Peromyscus species. As in panel A, shaded columns denote sites that harbor intermediate-frequency polymorphisms in P. maniculatus. Sequence logos depict site-specific allele frequency variation within P. maniculatus (top) and conservation among species (bottom). In both panels, the tree topologies are based on a consensus of recent phylogenetic studies (Bradley et al. 2007; Gering et al. 2009).
Phylogenetic analysis of adult HBA genes from P. maniculatus, P. keeni, and P. leucopus revealed that a history of interparalog gene conversion has produced a typical pattern of concerted evolution where paralogs from the same species cluster together to the exclusion of orthologs from different species (supplementary fig. S4, Supplementary Material online). However, orthologous relationships are revealed by sequence matches in noncoding flanking regions that are largely unaffected by interparalog gene conversion (supplementary fig. S1A, Supplementary Material online). We successfully cloned orthologous HBA genes from multiple individuals of 12 different Peromyscus species. A multiple alignment of HBA sequences revealed that the 15 amino acid sites that are polymorphic within P. maniculatus are also variable among species (fig. 3A).
In P. maniculatus, the HBA-T1 and HBA-T2 genes share low- to intermediate-frequency amino acid polymorphisms at six sites (α5, α10, α12, α15, α23, and α34) where the shared, minor alleles are fixed or are present at high frequency at paralogous sites in the HBA-T3 pseudogene. This pattern suggests that these variants were introduced via HBA-T3→HBA-T1/T2 gene conversion. In the pooled sample of P. maniculatus HBA alleles, overall levels of nucleotide divergence between HBA-T3 and the other two HBA paralogs are sufficiently high that T3-derived conversion tracts are readily detectable against the T1/T2 background, and vice versa. Maximum-likelihood analysis of polymorphism data for all three HBA paralogs of P. maniculatus (n = 593 alleles, see Materials and Methods) indicated that 7% of HBA-T1/HBA-T2 alleles had coding sequences that were partially converted by HBA-T3, and these T3-derived conversion tracts accounted for low-frequency amino acid variants segregating at each of four triallelic sites. The average conversion tract length was 181 bp excluding gaps (range = 2–725 bp) and the average per-site probability of detecting interparalog gene conversion (ψ) was 0.054 (n = 97 informative sites).
Overall levels of nucleotide variation were exceptionally high for both HBA-T1 and HBA-T2 paralogs in P. maniculatus, with silent-site diversities greater than 0.025 in all population samples other than Humboldt Co. (table 1). The HBA-T1 gene of P. keeni was monomorphic but HBA-T2 of P. keeni and the single-copy HBA-T1 gene of P. leucopus both exhibited high levels of nucleotide diversity commensurate with those observed for the HBA-T1 and HBA-T2 genes of P. maniculatus (table 1).
Table 1.
DNA Polymorphism and Intragenic LD in the HBA Genes of Peromyscus maniculatus, P. keeni, and P. leucopus.
| Population | Paralog | Length (bp)a | N | S | θπ(silent) | θW(silent) | 4Nc | ZnS | ZZ |
|---|---|---|---|---|---|---|---|---|---|
| P. maniculatus | |||||||||
| Humboldt Co., CA | HBA-T1 | 1511.9 | 16 | 85 | 0.0183 | 0.0158 | 0.0020 | 0.4191 | 0.0765 |
| Mono Co., CA | HBA-T2 | 1520.6 | 16 | 66 | 0.0111 | 0.0125 | 0.0029 | 0.2810 | 0.1623 |
| HBA-T1 | 1499.5 | 16 | 252 | 0.0330 | 0.0472 | 0.0018 | 0.2568 | 0.1891 | |
| HBA-T2 | 1489.7 | 16 | 242 | 0.0367 | 0.0461 | 0.0000 | 0.4154 | 0.2192 | |
| Clear Creek Co., CO | HBA-T1 | 1505.4 | 34 | 241 | 0.0268 | 0.0343 | 0.0036 | 0.1538 | 0.1648 |
| HBA-T2 | 1488.7 | 34 | 214 | 0.0252 | 0.0297 | 0.0016 | 0.1648 | 0.2038 | |
| Yuma Co., CO | HBA-T1 | 1493.2 | 34 | 531 | 0.0589 | 0.0806 | 0.0004 | 0.1705 | 0.2834 |
| HBA-T2 | 1493.8 | 34 | 346 | 0.0392 | 0.0517 | 0.0065 | 0.1682 | 0.1860 | |
| P. keeni | |||||||||
| Wrangell Island, AK | HBA-T1 | 1548.8 | 12 | 0 | 0.0000 | 0.0000 | — | — | — |
| HBA-T2 | 1530.7 | 12 | 60 | 0.0074 | 0.0100 | 0.0000 | 0.6824 | 0.2746 | |
| P. leucopus | |||||||||
| Saunders Co., NE | HBA-T1 | 712.8 | 22 | 114 | 0.0297 | 0.0439 | 0.0027 | 0.1675 | 0.2160 |
Note.—For P. maniculatus and P. keeni, polymorphism data are from population samples in which both alleles of the tandemly duplicated HBA-T1 and HBA-T2 genes were separately cloned and sequenced. Peromyscus leucopus possesses a single adult-expressed α-globin gene, HBA-T1.
aExcluding alignment gaps.
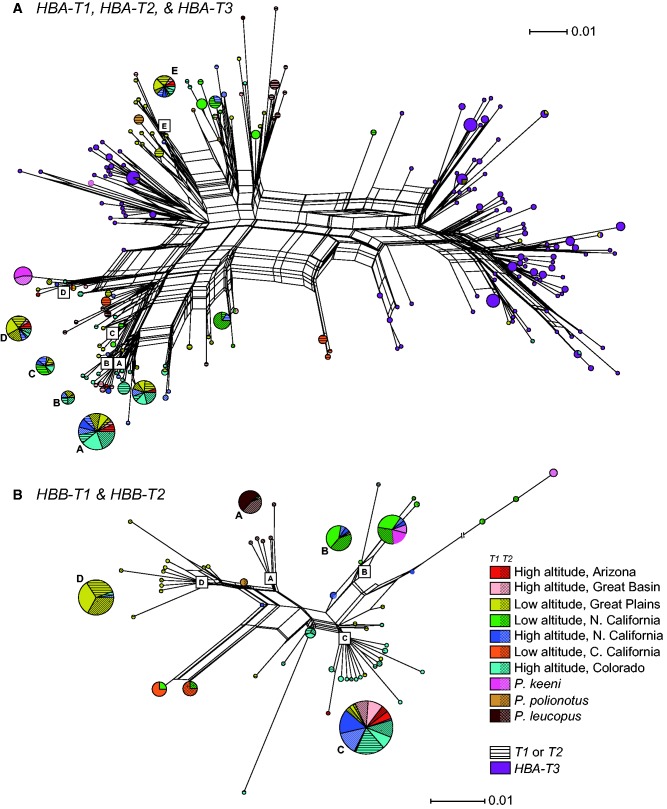
The history of interparalog gene conversion among the three HBA genes is reflected in the highly reticulate network of amino acid haplotypes (fig. 4A). HBA-T1 and HBA-T2 sequences sampled from the same geographic locality are typically far more similar to one another than to alleles of the same gene that were sampled from more distant localities, a pattern that reflects different histories of concerted evolution in different geographic populations (fig. 4A).
Fig. 4.
Reconstructed networks of HBA and HBB amino acid haplotypes. (A) Network of HBA amino acid sequences from Peromyscus maniculatus (n = 406), P. keeni (n = 24), P. leucopus (n = 22), and P. polionotus (n = 8). Also included are 187 HBA-T3 sequences from a geographically diverse sample of P. maniculatus. The HBA-T3 pseudogene sequences were included in the reconstruction because HBA-T3↔HBA-T1/T2 gene conversion accounts for many reticulate connections in the network. The sizes of pie diagrams are proportional to the numerical representation of each amino acid haplotype in the total sample. Pie diagrams labeled A–E are set apart from their corresponding nodes in the network to avoid obscuring the topology. (B) Network of HBB amino acid sequences from Peromyscus maniculatus (n = 482), P. keeni (n = 24), P. leucopus (n = 40), and P. polionotus (n = 4). As in panel A, pie diagrams labeled A–D are set apart from their corresponding nodes in the network. One of the salient features of both networks is that identical amino acid haplotypes are shared between geographically disparate high-altitude populations of P. maniculatus (e.g., node A in the HBA network, and node C in the HBB network, both of which show extensive haplotype-sharing between high-altitude populations in Colorado and California). Another clearly discernible pattern relates to semi-independent histories of concerted evolution across the species range: in both networks, T1 sequences typically show a closer affinity to T2 sequences from the same population than to T1 sequences from different populations and vice versa.
The history of interparalog gene conversion also contributes to shared polymorphism between species. For example, there are nine amino acid polymorphisms shared between the HBA-T1 and HBA-T2 genes of P. maniculatus and the HBA-T2 gene of P. keeni and shared minor alleles at each site are either fixed or are present at high frequency at paralogous sites of the HBA-T3 pseudogenes in each species. In two distinct HBA-T2 alleles of P. keeni, novel amino acid variants are contained within T3-derived conversion tracts (515–1,022 bp in length, excluding gaps) that span all of exon 2 and 57–100% of exon 3 (fig. 5). For the ten HBA-T2 sites segregating T3-derived amino acid variants (fig. 5), the average persite probability of detecting interparalog gene conversion was ψ = 0.996. These results indicate that shared polymorphisms between closely related species are not necessarily attributable to incomplete lineage sorting or introgressive hybridization; in the HBA genes of P. maniculatus and P. keeni, transpecific polymorphism is clearly attributable to recurrent HBA-T3→HBA-T1/T2 gene conversion that has occurred independently in each species.
Fig. 5.
Allele-sharing between Peromyscus maniculatus and P. keeni is attributable to interparalog HBA-T3→HBA-T1/T2 gene conversion that has occurred independently in each species. Site-specific patterns of allele frequency variation in the transcriptionally active HBA-T1 and HBA-T2 genes and the HBA-T3 pseudogene of P. maniculatus (above) and sequence variation in the corresponding orthologs of P. keeni (below). Shaded columns depict nine sites (34, 36, 57, 58, 60, 64, 71, 72, and 78) that harbor shared amino acid polymorphisms between the HBA-T1 and HBA-T2 genes of P. maniculatus and the HBA-T2 gene of P. keeni. At each of the nine sites, the shared minor allele is fixed or present at high frequency in the HBA-T3 pseudogenes of both species. At site 34, for example, Cys (C) is fixed or nearly fixed in the HBA-T1 and HBA-T2 paralogs of both P. maniculatus and P. keeni, whereas Glu (E) is the major allele at this site in the HBA-T3 paralog of both species. The same pattern is evident at the remaining eight sites. In the HBA-T2 gene of P. keeni, conversion tracts from the HBA-T3 donor sequence (shown in boxes) span all of exon 2 and 57–100% of exon 3 in the adjacent HBA-T2 gene. In P. maniculatus, the same variants were independently derived via HBA-T3→HBA-T1/T2 gene conversion and are present at low frequency across the species’ range. Although the conversion tracts are depicted in the alignment of amino acid sequences, the gene conversion tracts were identified on the basis of the underlying nucleotide variation; see Materials and Methods).
Intraspecific Polymorphism and Interspecific Divergence: HBB Genes
The survey of nucleotide polymorphism in the tandemly linked HBB-T1 and HBB-T2 genes of P. maniculatus revealed nine intermediate frequency polymorphisms (n = 482 alleles). We separately cloned and sequenced both alleles from each of the two genes in a subset of specimens to obtain complete tetraploid HBB genotypes ( = 396 experimentally phased alleles). Likewise, we generated tetraploid HBB-T1/HBB-T2 genotypes for both P. keeni and P. leucopus ( = 24 and 40 experimentally phased alleles, respectively). Data for HBB-T1 and HBB-T2 in P. maniculatus confirmed previous reports that the same alleles are segregating at both genes due to interparalog gene conversion (Storz et al. 2009; Storz, Runck, et al. 2010; Storz, Natarajan, et al. 2012) similar to the case with the HBA genes. The P. maniculatus specimens from San Luis Obispo Co., CA, constituted the only exception to this pattern, as the HBB-T1 and HBB-T2 paralogs in mice from that particular locality were distinguished by a single amino acid substitution at β135. The alternative residues at this site are segregating as allelic variants at both paralogs in other populations (supplementary fig. S5, Supplementary Material online), similar to the case of the HBA paralogs in mice from Humboldt Co., CA. These findings indicate that rates of gene conversion are variable among populations of P. maniculatus, resulting in nonuniform patterns of concerted evolution across the species’ range. This is reflected in the variable levels of interparalog divergence in different populations of P. maniculatus (supplementary table S3, Supplementary Material online).
Levels of nucleotide variation were generally high for both HBB paralogs in P. maniculatus and P. leucopus. In P. keeni, in contrast, silent-site diversity at HBB-T1 was an order of magnitude lower than that at HBB-T2 (table 2). Similar to the case with the HBA genes, the highly reticulate network of HBB amino acid haplotypes reflects a history of HBB-T1↔HBB-T2 gene conversion and intragenic recombination (fig. 4B).
Table 2.
DNA Polymorphism and Intragenic LD in the Tandemly Linked HBB-T1 and HBB-T2 Genes of Peromyscus maniculatus, P. keeni, and P. leucopus.
| Population | Paralog | Length (bp)a | N | S | θπ(silent) | θW(silent) | 4Nc | ZnS | ZZ |
|---|---|---|---|---|---|---|---|---|---|
| P. maniculatus | |||||||||
| Humboldt Co., CA | HBB-T1 | 1303.7 | 16 | 30 | 0.0066 | 0.0067 | 0.0001 | 0.448 | 0.1674 |
| HBB-T2 | 1284.9 | 16 | 56 | 0.0156 | 0.0129 | 0.0023 | 0.4262 | 0.1077 | |
| Marin Co., CA | HBB-T1 | 1298.6 | 10 | 63 | 0.0152 | 0.0169 | 0.0045 | 0.4389 | 0.042 |
| HBB-T2 | 1290.4 | 10 | 94 | 0.0283 | 0.0252 | 0.0114 | 0.3869 | 0.1455 | |
| Monterey Co., CA | HBB-T1 | 1295.6 | 10 | 115 | 0.0316 | 0.0292 | 0.0037 | 0.6844 | 0.0043 |
| HBB-T2 | 1287.5 | 10 | 161 | 0.0405 | 0.0395 | 0.0051 | 0.4893 | 0.0616 | |
| San Luis Obispo Co., CA | HBB-T1 | 1302 | 12 | 1 | 0.0001 | 0.0003 | — | — | — |
| HBB-T2 | 1291.8 | 12 | 0 | 0 | 0 | — | — | — | |
| Merced Co., CA | HBB-T1 | 1278.6 | 12 | 101 | 0.0157 | 0.0228 | 0 | 0.4302 | 0.1634 |
| HBB-T2 | 1299.6 | 12 | 104 | 0.0255 | 0.0232 | 0.0016 | 0.4309 | 0.2139 | |
| Tuolemne Co, CA | HBB-T1 | 1284.6 | 12 | 86 | 0.0195 | 0.0214 | 0.0169 | 0.2751 | 0.1678 |
| HBB-T2 | 1297.1 | 12 | 127 | 0.029 | 0.0309 | 0.0175 | 0.2658 | 0.1707 | |
| Mariposa Co., CA | HBB-T1 | 1287.4 | 12 | 99 | 0.0186 | 0.0247 | 0.004 | 0.3343 | 0.157 |
| HBB-T2 | 1293.2 | 12 | 75 | 0.0205 | 0.0184 | 0.0024 | 0.5504 | 0.0878 | |
| Mono Co., CA | HBB-T1 | 1246.1 | 16 | 139 | 0.0223 | 0.0327 | 0.0041 | 0.1859 | 0.1511 |
| HBB-T2 | 1262.3 | 16 | 93 | 0.0196 | 0.0213 | 0.0001 | 0.4984 | 0.1456 | |
| White Pine Co., NV | HBB-T1 | 1285.6 | 10 | 62 | 0.0146 | 0.0165 | 0.0045 | 0.3519 | 0.2395 |
| HBB-T2 | 1293.5 | 10 | 86 | 0.0246 | 0.023 | 0.0059 | 0.4612 | 0.0628 | |
| Coconino Co., AZ | HBB-T1 | 1283.2 | 10 | 71 | 0.0198 | 0.0196 | 0.014 | 0.3735 | 0.2284 |
| HBB-T2 | 1296.5 | 10 | 76 | 0.0215 | 0.0205 | 0.0063 | 0.4487 | 0.1274 | |
| Wayne Co., UT | HBB-T1 | 1282.8 | 10 | 57 | 0.0142 | 0.0157 | 0.0083 | 0.3606 | 0.1594 |
| HBB-T2 | 1296.6 | 8 | 62 | 0.0167 | 0.0184 | 0.0009 | 0.4556 | −0.0104 | |
| Clear Creek Co., CO | HBB-T1 | 1200.9 | 28 | 94 | 0.0141 | 0.0178 | 0.0065 | 0.2095 | 0.0723 |
| HBB-T2 | 1232.4 | 28 | 97 | 0.0211 | 0.0194 | 0.0026 | 0.3185 | 0.0674 | |
| Yuma Co., CO | HBB-T1 | 1212.7 | 30 | 88 | 0.013 | 0.0169 | 0 | 0.4184 | −0.0067 |
| HBB-T2 | 1204.4 | 30 | 78 | 0.0139 | 0.0149 | 0 | 0.4585 | 0.0107 | |
| Pawnee Co., KS | HBB-T1 | 1296.9 | 10 | 14 | 0.0022 | 0.0033 | 0.0364 | 0.375 | 0 |
| HBB-T2 | 1291.1 | 10 | 26 | 0.0059 | 0.0063 | 0.0027 | 0.3113 | 0.3902 | |
| P. keeni | |||||||||
| Wrangell Island, AK | HBB-T1 | 1302.3 | 12 | 6 | 0.0011 | 0.0015 | 0 | 1 | 0 |
| HBB-T2 | 1224.6 | 12 | 91 | 0.0237 | 0.0195 | 0.0015 | 0.5554 | 0.151 | |
| P. leucopus | |||||||||
| Saunders Co., NE | HBB-T1 | 1271.6 | 20 | 40 | 0.0115 | 0.0089 | 0.0078 | 0.364 | 0.2219 |
| HBB-T2 | 1268.3 | 20 | 68 | 0.0075 | 0.014 | 0 | 0.4252 | 0.0265 | |
Note.—For each species, polymorphism data are from population samples in which both alleles of the HBB-T1 and HBB-T2 genes were separately cloned and sequenced.
aExcluding alignment gaps.
We successfully cloned both HBB-T1 and HBB-T2 from multiple individuals of 16 different Peromyscus species. Similar to the case with the HBA-T1 and HBA-T2 paralogs, phylogeny reconstructions of HBB sequences in Peromyscus revealed the classic pattern of concerted evolution where paralogs from the same species grouped together to the exclusion of orthologs from different species (fig. 6A–C). In contrast, phylogeny reconstructions based on 3′-flanking sequences (which are largely unaffected by gene conversion) resolved reciprocally monophyletic groups of HBB-T1 and HBB-T2 sequences (fig. 6D). Similar to the case of the HBA genes, the nine HBB sites that are polymorphic within P. maniculatus are also variable among species (fig. 3B).
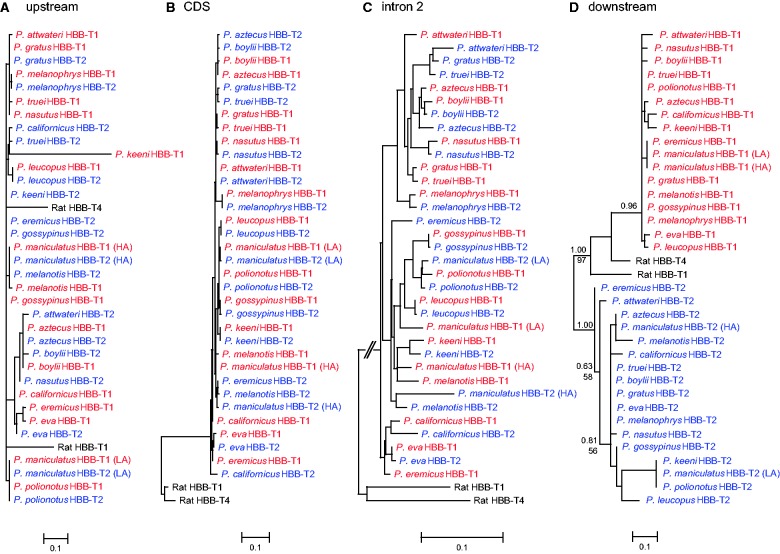
Fig. 6.
Maximum-likelihood phylogenies depicting relationships among the HBB-T1 and HBB-T2 genes of 16 Peromyscus species. Phylogeny reconstructions were based on four discrete data partitions: (A) 5′-flanking region (upstream), (B) the complete coding sequence (cds), (C) intron 2, and (D) 3′-flanking region (downstream). Phylogenies in (A–C) show the characteristic pattern of concerted evolution where paralogs from the same species group together to the exclusion of positional orthologs in other species. In contrast, downstream flanking sequence is largely unaffected by interparalog gene conversion, so the phylogeny in D depicts relationships that reflect the true history of species divergence. For the tree in panel D (which is the only tree that is informative about orthologous relationships), measures of support for relevant nodes are presented as Bayesian posterior probabilities (above) and bootstrap values (below).
Altitudinal Patterning of Hb Polymorphism within P. maniculatus
In the case of P. maniculatus, altitudinal patterns of allele frequency variation at 12 of the 24 polymorphic sites in the HBA and HBB genes have been described previously for population samples from the Rocky Mountains and Great Plains (α-globin sites 50, 57, 60, 64, 71, 113, 115, and 116, and β-globin sites 62, 72, 128, and 135; (Storz, Sabatino, et al. 2007; Storz and Kelly 2008; Storz et al. 2009; Storz, Runck, et al. 2010; Storz, Natarajan, et al. 2012) and site-directed mutagenesis experiments have quantified their additive and epistatic effects on Hb function (Natarajan et al. 2013). Results of the present survey reveal that the same allelic variants that contribute to variation in Hb-O2 affinity in the Rocky Mountains and the Great Plains are also segregating in highland and lowland populations across central and western North America.
Twelve of the 24 intermediate-frequency polymorphisms exhibited significant correlations with altitude across all 15 population samples of P. maniculatus (α-chain sites 5, 10, 12, 15, 34, 50, 64, 78, 113, and 115, and β-chain sites 25 and 72); correlations for two unlinked sites (α64 and β72) remained significant at a Bonferroni-adjusted critical value of α′ = 0.0021. In broad outline, the networks of HBA and HBB amino acid haplotypes reflect the same underlying pattern of geographic population structure that was revealed by the analysis of mtDNA variation (figs. 2 and 4). HBA and HBB haplotypes harboring multisite combinations of characteristically high-altitude amino acid variants are shared among mice from geographically disparate high-altitude localities across the western United States (e.g., node “A” in fig. 4A and node “C” in fig. 4B). This pattern of allele-sharing among highland mice from different mountain ranges is consistent with their shared phylogenetic heritage, as reflected in the mtDNA phylogeny (supplementary fig. S2, Supplementary Material online) and patterns of genome-wide nucleotide variation, as discussed above.
Variation in Hb Function within P. maniculatus
Evolutionary changes in Hb-O2 affinity may be caused by changes in intrinsic O2 affinity and/or changes in the sensitivity of Hb to the inhibitory effects of endogenously produced allosteric cofactors (Mairbäurl and Weber 2012). To gain insight into the mechanistic basis of variation in Hb-O2 affinity, we measured oxygenation properties of purified Hbs in the presence and absence of two main allosteric effectors that regulate Hb-O2 affinity in mammalian red blood cells: Cl− ions (added as KCl; 0.1 mol l−1) and 2,3-diphosphoglycerate (DPG, at 2-fold molar excess over tetrameric Hb). Measurements of Hb-O2 affinity in the simultaneous presence of Cl− and DPG are most relevant to in vivo conditions in the red blood cells of rodents (Mairbäurl and Weber 2012; Tufts et al. 2013).
Consistent with previous studies of Hb function in P. maniculatus from the Rocky Mountains and Great Plains (Storz et al. 2009; Storz, Runck, et al. 2010), mice from highland and lowland localities in California exhibited a high level of interindividual variation in Hb-O2 affinity (as indexed by P50, the PO2 at which Hb is half-saturated) and there was considerable overlap in the range of values for high- and low-altitude specimens (table 3). In contrast to previous studies which revealed that highland mice from the Rocky Mountains have evolved an increased Hb-O2 affinity relative to their lowland counterparts from the Great Plains (Storz et al. 2009; Storz, Runck, et al. 2010; Natarajan et al. 2013), the set of California specimens that we examined showed the opposite pattern of differentiation: Mice from the sea-level localities (Humboldt Co. and San Luis Obispo Co.) exhibited slightly higher Hb-O2 affinities and lower cooperativity coefficients (indicating less sigmoidal O2-equilbrium curves) relative to conspecifics collected from an elevation of 3,800 m in the White Mountains (table 3). These experimental results for purified Hbs are consistent with results of a previous survey of blood-O2 affinity in deer mice from different elevations in northern California (Sawin 1970).
Table 3.
O2 Affinities (P50, torr) and Cooperativity Coefficients (n50) of Purified Hbs from Different Populations of Peromyscus maniculatus and Different Species of Peromyscus (mean ± 1SD).
| Stripped |
+DPG |
+KCl |
DPG + KCl |
|||||
|---|---|---|---|---|---|---|---|---|
| P50 | n50 | P50 | n50 | P50 | n50 | P50 | n50 | |
| P. maniculatus | ||||||||
| Humboldt Co., CA (n = 4) | 6.04 ± 0.71 | 1.82 ± 0.19 | 7.81 ± 1.20 | 1.87 ± 0.22 | 10.60 ± 1.69 | 1.90 ± 0.24 | 10.42 ± 1.33 | 1.84 ± 0.28 |
| San Luis Obispo Co., CA (n = 6) | 7.55 ± 0.85 | 1.57 ± 0.17 | 9.46 ± 1.39 | 1.79 ± 0.31 | 10.61 ± 1.24 | 1.68 ± 0.15 | 11.34 ± 0.85 | 1.75 ± 0.22 |
| Mono Co., CA (n = 4) | 8.88 ± 0.92 | 2.35 ± 0.06 | 9.66 ± 0.52 | 2.49 ± 0.08 | 14.10 ± 0.58 | 2.50 ± 0.12 | 14.08 ± 0.35 | 2.44 ± 0.09 |
| Clear Creek Co., CO (n = 3/20)a | 6.96 ± 0.24 | 2.01 ± 0.28 | 8.10 ± 0.76 | 2.27 ± 0.23 | 11.62 ± 0.73 | 2.21 ± 0.15 | 12.34 ± 0.55 | 2.30 ± 0.21 |
| 7.38 ± 0.52 | 2.12 ± 0.25 | 9.00 ± 1.12 | 2.21 ± 0.19 | 12.64 ± 1.13 | 2.25 ± 0.13 | 13.65 ± 1.48 | 2.36 ± 0.19 | |
| Yuma Co., CO (n = 3/15)a | 7.39 ± 0.22 | 2.31 ± 0.03 | 9.53 ± 0.30 | 2.48 ± 0.10 | 13.70 ± 0.40 | 2.49 ± 0.14 | 14.32 ± 0.54 | 2.45 ± 0.20 |
| 7.91 ± 0.54 | 2.27 ± 0.07 | 9.60 ± 0.99 | 2.44 ± 0.12 | 14.47 ± 1.05 | 2.50 ± 0.10 | 13.90 ± 0.52 | 2.46 ± 0.14 | |
| P. keeni (n = 4) | 10.03 ± 1.10 | 2.24 ± 0.18 | 12.96 ± 0.75 | 2.50 ± 0.08 | 15.04 ± 1.09 | 2.41 ± 0.19 | 15.40 ± 1.27 | 2.29 ± 0.24 |
| P. polionotus (n = 2) | 8.52 ± 0.74 | 2.09 ± 0.35 | 11.68 ± 0.37 | 2.37 ± 0.20 | 13.62 ± 0.35 | 2.60 ± 0.04 | 14.13 ± 0.49 | 2.49 ± 0.03 |
| P. leucopus (n = 2) | 7.63 ± 0.55 | 2.09 ± 0.32 | 12.30 ± 0.79 | 2.19 ± 0.19 | 14.42 ± 0.56 | 2.26 ± 0.03 | 14.94 ± 0.10 | 2.38 ± 0.04 |
| P. aztecus (n = 2) | 7.96 ± 0.54 | 2.17 ± 0.27 | 10.94 ± 0.63 | 2.36 ± 0.11 | 13.16 ± 1.32 | 2.47 ± 0.18 | 14.00 ± 0.59 | 2.48 ± 0.17 |
| P. melanophrys (n = 2) | 7.71 ± 0.54 | 1.75 ± 0.12 | 11.51 ± 1.07 | 2.18 ± 0.03 | 15.65 ± 0.81 | 2.18 ± 0.18 | 13.71 ± 0.10 | 2.10 ± 0.12 |
Note.—P50 is the PO2 at which Hb is 50% saturated, and is therefore an inverse measure of O2 affinity. O2 equilibria were measured in 0.1 M HEPES buffer at pH 7.40 ( ± 0.02), 37 °C, in the absence of allosteric effectors (stripped), in the presence of DPG alone (DPG/Hb tetramer ratio, 2.0), in the presence of Cl− alone (0.10 M KCl), and in the simultaneous presence of both effectors ([heme], 0.2–0.3 mM). P50 and n50 values were obtained by fitting the Hill equation to the experimental O2 saturation data using a nonlinear regression model.
aSample sizes refer to values in upper/lower rows. For each population, the upper row reports P50 and n50 values for individual specimens with HBA and HBB genotypes that represent the most common genotypes in the total sample. The lower row reports values for individual specimens and pooled samples of 2–8 specimens with identical genotypes that represent all sampled Hb variants; mean P50 and n50 values are not weighted according to the frequency of each variant in the population sample.
With respect to intrinsic O2 affinity and mode of allosteric regulation, Hbs of mice from the White Mountains (Mono Co., CA) were qualitatively similar to those of mice from the Rocky Mountains (Clear Creek Co., CO) and Great Plains (Yuma Co., CO). In contrast, Hbs of lowland California mice from Humboldt and San Luis Obispo counties had relatively high O2-affinities in the presence of allosteric effectors (i.e., low values of P50(DPG+KCl)) and relatively low cooperativities (i.e., Hill coefficients, n50, were < 2.00 across all treatments; table 3). In the case of Hbs from the Humboldt mice, the low values of P50(DPG+KCl) were mainly attributable to high intrinsic O2-affinities. This is indicated by the especially low P50 for “stripped” Hb (purified Hb that is stripped of allosteric effectors) relative to values for other P. maniculatus populations or other Peromyscus species (table 3). In spite of the unusually high intrinsic O2-affinity (lower P50(stripped)), Hbs of the Humboldt mice exhibit DPG and Cl− sensitivities in the normal range for the species (supplementary fig. S6, Supplementary Material online), indicating that the mode of allosteric regulation was not qualitatively or quantitatively distinct. In the case of Hbs from the San Luis Obispo mice, by contrast, the low P50(DPG+KCl) is mainly attributable to a suppressed sensitivity to Cl− ions, as indicated by unusually small differences in log-transformed P50 values in the presence and absence of 0.1 M KCl: Hbs of these mice have lower values of Δlog-P50(KCl-str) and Δlog-P50([KCl+DPG]-str) than any P. maniculatus population or any other Peromyscus species (supplementary fig. S6, Supplementary Material online). Thus, the unexpectedly high Hb-O2 affinities of the two lowland populations of California mice are attributable to different functional mechanisms, suggesting that each must have a distinct structural basis.
Genotype–Phenotype Associations
The causative effects of individual amino acid replacements can be most readily discerned in comparisons of Hb function among specimens from the same locality or, in some cases, among those from geographically proximal localities, as the Hbs of such mice often differ by a relatively small number of amino acid changes. Previous analyses of native Hb variants and recombinant Hb mutants yielded clear insights into the structural basis of allelic differences in Hb function that distinguish mice from the Rocky Mountains and Great Plains (Storz et al. 2009; Storz, Runck, et al.2010; Inoguchi et al. 2013; Natarajan et al. 2011). New comparisons involving select specimens from highland and lowland California localities confirmed the functional effects of previously identified sites and revealed several additional amino acid polymorphisms that contribute to variation in Hb-O2 affinity. For example, comparisons among three specimens from Humboldt and Mono counties isolated the net effect of three β-chain mutations against otherwise identical genotypic backgrounds at the HBA and HBB genes: P50(KCl+DPG) was 32.7% lower for the triple-mutant β6 G-β25 A-β72 S genotype relative to the alternative β6 A-β25 G-β72 G genotype (9.35 vs. 13.89 torr, respectively).
To complement the comparisons involving small numbers of genotypically similar specimens, we conducted an association analysis in a Bayesian framework to estimate the quantitative effects of all segregating amino acid sites and the cumulative proportion of phenotypic variation that they explained in a larger set of P. maniculatus specimens from multiple populations. In spite of the fact that we have a full accounting of all segregating amino acid variants that could potentially contribute to genetically based variation in the functional properties of purified Hbs, and in spite of the fact that the analysis controlled for the effects of population structure, estimates of heritability (h2) for most measures of Hb function were substantially less than unity (h2 = 0.22–0.72; supplementary table S4, Supplementary Material online). This is likely due to a combination of small sample size, the low magnitude of overall phenotypic variation, heterogeneity of variance among populations, and pervasive epistasis among affinity-altering mutations (Natarajan et al. 2013). It may also be partly attributable to recurrent gene conversion, as the resultant allelic heterogeneity would diminish measured associations with particular causative mutations.
Sites with the largest effects on intrinsic Hb-O2 affinity (as indexed by P50(stripped)) were restricted to the α-chain, including previously implicated sites (α64, α71, α113, α115, and α116; Inoguchi et al. 2013; Natarajan et al. 2013;) (supplementary table S5, Supplementary Material online). In contrast, sites with the largest effects on Cl− sensitivity (as indexed by Δlog-P50(KCl-str)) were mainly restricted to the β-chain, including β6 in addition to several previously implicated sites (β62, β72, β128, and α50; supplementary table S5, Supplementary Material online). DPG sensitivity (as indexed by Δlog-P50(DPG-str)) exhibited the largest genetic variance (supplementary table S4, Supplementary Material online). Four closely linked α-globin sites (α12, α15, α23, and α34) made the largest site-specific contributions to variation in DPG sensitivity, with individual effects that were greater than 10-fold higher than the median value across all sites (supplementary table S5, Supplementary Material online). These results suggest a modular architecture of Hb-O2 affinity, with distinct sets of mutations affecting intrinsic O2-affinity and different modes of anion-mediated allosteric regulation.
Which sites are responsible for the unexpectedly high Hb-O2 affinities in mice from Humboldt and San Luis Obispo counties? In the presence of allosteric effectors, the elevated Hb-O2 affinity of the Humboldt mice is associated with derived variants at six α-globin sites that span the A- and B-helices of the encoded α-chain polypeptide (α5, α10, α12, α15, α23, and α34; supplementary table S5, Supplementary Material online). As described above, these derived variants appear to have been introduced via gene conversion from the closely linked HBA-T3 pseudogene. These findings demonstrate that HBA-T3→HBA-T1/T2 gene conversion provides an important source of standing genetic variation in Hb function.
Variation in Hb Function among Species
To complement the analysis of variation in Hb function within P. maniculatus, we also measured oxygenation properties of purified Hbs from five other Peromyscus species, including three predominantly lowland species, P. keeni, P. polionotus, and P. leucopus, and two species that occur at intermediate to high elevations in montane regions of Mexico, P. aztecus (500–3,200 m) and P. melanophrys (100–2,600 m). The range of variation in Hb-O2 affinity among these five species is commensurate with the observed range of variation among populations of P. maniculatus (table 3). The three lowland species have slightly lower mean Hb-O2 affinities than those of P. aztecus and P. melanophrys (P50(DPG+KCl) = 14.13–15.40 torr vs. 13.71–14.00 torr, respectively), but the overall magnitude of variation is too small to permit conclusions about a possible altitudinal trend. Functional properties of Hbs from these five species are qualitatively similar to those of the P. maniculatus Hb variants. Under identical experimental conditions, Hb-O2 affinities of Peromyscus species were higher than those of house mice in the genus Mus (P50(DPG+KCl) = 16.9–17.5 torr; Runck et al. 2010; Storz, Weber, et al. 2012).
Lowland P. keeni from Alaska have Hbs with especially low intrinsic O2-affinities relative to all other Peromyscus species and populations of P. maniculatus (table 3). Comparisons of interspecific sequence divergence reveal that this reduced affinity is attributable to the independent or joint effects of substitutions at eight α-chain sites: α5, α10, α15, α71, α78, α113, α115, and α116, each of which also contributes to variation in intrinsic Hb-O2 affinity within P. maniculatus (supplementary table S5, Supplementary Material online). For example, in the case of three closely linked sites, α113, α115, and α116, P. keeni is fixed for the three-site haplotype “LSD,” whereas the predominant haplotype in all other species is “HAD” or “HAE” (figs. 3 and 5). The same three-site LSD α-globin haplotype occurs at high frequency in lowland deer mice from the Great Plains, and is associated with a reduced intrinsic Hb-O2 affinity relative to high-altitude specimens from the Rocky Mountains that possess the HAE haplotype (Storz et al. 2009; Storz, Runck, et al. 2010; Natarajan et al. 2013). Thus, divergence in Hb function among species of Peromyscus is caused by the same amino acid changes that contribute to variation within P. maniculatus.
Origins of Genetic Variation in Hb Function
Having identified amino acid mutations that contribute to variation in Hb function within and among populations of the P. maniculatus species complex, and having determined that the causal variants are widely shared among geographically disparate populations of P. maniculatus and closely related species, we can now turn to questions about the origins and effects of these variants.
How Many Mutations Contribute to Evolutionary Changes in Hb Function?
Consistent with results of site-directed mutagenesis experiments (Natarajan et al. 2013) and targeted comparisons of native Hb variants that differ at just one or a few amino acid sites, results of the association analysis of naturally occurring amino acid polymorphisms suggest that variation in Hb-O2 affinity is attributable to multiple mutations that have individually small effects. Our analysis of native Hbs from mice with known genotypes suggests that there are no single amino acid mutations that account for most of the naturally occurring variation in Hb-O2 affinity. The concentration of many small-effect allelic variants distributed across tandemly linked genes results in large aggregate effects for each of the linked HBA and HBB gene pairs. This genetic architecture of Hb-O2 affinity is consistent with theoretical predictions for divergently selected traits in populations at migration-selection equilibrium (Yeaman and Whitlock 2011). More generally, models of adaptation from standing genetic variation predict a disproportionate contribution of allelic variants with small effects (Hermisson and Pennings 2005). Our results suggest that changes in biochemical phenotype can be produced by the composite effects of small-effect variants at numerous sites in the same gene or in closely linked genes, consistent with results of several other candidate gene association studies (Stam and Laurie 1996; Bickel et al. 2011; Linnen et al. 2013).
What Are the Relative Contributions of Standing Variation and New Mutations to Local Adaptation?
In cases where it is possible to identify causative alleles that have contributed to the adaptation of particular populations to geographically localized conditions, past selection on standing variation is implicated if the same alleles are segregating in other geographic populations and/or closely related species that have experienced a different selection regime (Colosimo et al. 2005; Barrett and Schluter 2008). Our survey of HBA and HBB variation in P. maniculatus demonstrated that affinity-altering variants are widely shared among geographically disparate populations, and numerous amino acid polymorphisms are also shared with P. keeni and P. leucopus. These findings suggest that divergence in Hb function between locally adapted populations of P. maniculatus is largely attributable to the sifting of standing genetic variation by spatially varying selection.
Several other candidate gene studies have documented evidence for adaptation from standing variation in natural populations of animals and plants (Colosimo et al. 2005; Steiner et al. 2007; Tishkoff et al. 2007; Feldman et al. 2009; Rebeiz et al. 2009; Wittkopp et al. 2009; Domingues et al. 2012; Jones et al. 2012; Prasad et al. 2012), although in many cases the causative mutations have not been conclusively identified. One of the unique features of the deer mouse Hb polymorphism is that an appreciable fraction of functional standing variation in the two transcriptionally active HBA paralogs is attributable to recurrent gene conversion from a linked pseudogene (HBA-T3). This finding contributes to a growing body of data documenting a role for ectopic gene conversion as a source of functional genetic variation (Casola et al. 2012). Whether the functional variation contributed by HBA-T3→HBA-T1/T2 gene conversion is beneficial, deleterious, or neutral remains an open question, although there is no evidence to suggest that T3-derived amino acid variants contribute to high-altitude adaptation. In high-altitude deer mice from the Rocky Mountains, none of the HBA-T1/T2 mutations that have contributed to an increased Hb-O2 affinity are contained within T3-derived conversion tracts (Storz, Runck, et al.2010).
Are the Mutational Changes that Contribute to Divergence between Conspecific Populations Qualitatively or Quantitatively Distinct from Those That Contribute to Divergence between Species?
Reciprocal-transplant experiments involving wild-derived strains of P. maniculatus revealed trade-offs in physiological performance associated with different Hb-O2 affinities at different elevations: When tested under hypoxic conditions at high altitude, mice with high-affinity HBA genotypes exhibited superior aerobic performance relative to mice with the alternative low-affinity genotypes (due to enhanced pulmonary O2-loading at low alveolar PO2 that safeguards arterial O2 saturation), whereas mice with low-affinity HBA genotypes exhibited superior performance at sea-level (due to enhanced O2 unloading to respiring tissues under normoxic conditions when arterial O2 saturation is maximal; Chappell and Snyder 1984; Chappell et al. 1988). Similar types of trade-offs have been documented for allelic variants that contribute to local adaptation in natural populations of animals and plants (Mitchell-Olds et al. 2007; Anderson et al. 2011; Colautti et al. 2012). According to one school of thought (Jeong et al. 2008; Stern and Orgogozo 2008, 2009) such variants may contribute to adaptive phenotypic differences between conspecific populations but are not likely to make important contributions to phenotypic divergence between species. The reasoning is that locally adaptive alleles with antagonistically pleiotropic effects in different environments are unlikely to become fixed at the species level. Contrary to this expectation, however, our results demonstrate that the same amino acid changes that contribute to variation in Hb function within P. maniculatus also contribute to divergence in Hb function among different species of Peromyscus. In the case of affinity-altering HBA and HBB alleles in Peromyscus, there is no qualitative or quantitative distinction between segregating variants within species and fixed differences between species.
No Evidence for Phenotypic Parallelism among Highland Populations or Species
Although affinity-enhancing variants are present at high-frequency in many high-altitude deer mouse populations across western North America, these variants clearly do not have independent mutational origins, as they often occur in identical haplotype backgrounds in geographically disparate localities (fig. 4). On the basis of physiological experiments, Snyder and colleagues argued that the altitudinal patterning of Hb polymorphism in P. maniculatus reflects a history of spatially varying selection that favors different Hb-O2 affinities in different elevational zones (Snyder 1981, 1985; Snyder et al. 1982, 1988; Chappell and Snyder 1984; Chappell et al. 1988). The evidence for local adaptation to different elevational zones is strongest in comparisons of population samples from the Southern Rocky Mountains and the Great Plains, as indicated by differentiation in Hb function and steep altitudinal clines in allele frequency at causative sites (Snyder et al. 1988; Storz 2007; Storz, Sabatino, et al. 2007; Storz and Kelly 2008; Storz et al. 2009; Storz, Runck, et al. 2010; Storz, Natarajan, et al. 2012). At face value, the altitudinal patterns of allele frequency variation across the western United States suggest that repeated adaptation to high-altitude could have involved the parallel reuse of standing variation. However, our insights into patterns of population structure across the species’ range suggest that the altitudinal patterning of Hb polymorphism may also be largely explained by population history, as alpine mice from geographically disparate mountain ranges show closer genetic affinities with one another than with geographically closer lowland populations.
With regard to functional properties of Hb, our data provide no strong evidence for a replicated pattern of altitudinal differentiation across the species range in western North America. The altitudinal differentiation in Hb function that has been documented between deer mice from the Rocky Mountains and the Great Plains (Storz et al. 2009; Storz, Runck, et al. 2010) is not apparent in comparisons between highland and lowland population samples from California (table 3). The original electrophoretic surveys of Hb polymorphism in deer mice also documented that levels of altitudinal differentiation were highest in comparisons between populations from the Rocky Mountains and the Great Plains (Snyder et al. 1988). Lower levels of altitudinal differentiation in the mountain ranges of California and elsewhere was attributed to the swamping effects of gene flow from surrounding lowland populations. In comparison, the Southern Rocky Mountains of Colorado are characterized by a much vaster expanse of terrain in the alpine and subalpine zones, a geographic circumstance more conducive to local adaptation, as selection that favors an increased Hb-O2 affinity in alpine mice would be less constrained by countervailing gene flow from lowland populations.
Our experimental results did not reveal any dramatic variation in Hb function among Peromyscus species with different elevational distributions. Comparative studies of other vertebrate taxa have documented positive associations between Hb-O2 affinity and native elevation in some cases but not in others (Projecto-Garcia et al. 2013; Revsbech et al. 2013; Cheviron et al. 2014; Tufts et al. 2015). In cases where genetically based modifications of Hb-O2 affinity are not implicated in physiological adaptation to high-altitude hypoxia, it is of interest to determine whether adjustments at other steps in the O2 cascade have obviated the need for such changes (Storz, Scott, et al. 2010).
In the absence of experimental measures of Hb function, the observed patterns of amino acid sequence variation within and among species could be uncritically interpreted in purely adaptive terms, especially given a priori expectations for a positive association between native elevation and Hb-O2 affinity (Storz and Moriyama 2008; Storz, Scott, et al. 2010). Interpretation of the sequence data in light of experimental results forces a more nuanced view. Given that variation in the respiratory properties of deer mouse Hb stems from a combination of common polymorphisms and population-specific substitutions, and given the pervasiveness of epistasis among affinity-altering mutations (Natarajan et al. 2013), patterns of functional variation within and among species cannot be accurately predicted on the basis of sequence data alone. Our results highlight the importance of obtaining experimental data on protein function in order to interpret the evolutionary significance of amino acid variation within and among species (Jensen et al. 2007; Storz and Wheat 2010; Storz and Zera 2011), an approach that is central to the functional synthesis of molecular biology, biochemistry, and evolution (Golding and Dean 1998; Dean and Thornton 2007; Harms and Thornton 2013).
Materials and Methods
Samples
The analyses of intraspecific polymorphism and Hb variation was based on a total of 148 specimens of P. maniculatus from 15 localities in North America, as well as 7 P. keeni from Wrangell Island, AK, and 11 P. leucopus from Saunders Co, NE (supplementary table S1, Supplementary Material online). For the comparisons among species, we collected specimens of P. melanotis from the Chiricahua Mountains of southern Arizona (Cochise Co.), and we obtained blood samples and frozen tissues of P. polionotus, P. aztecus, and P. melanophrys (two individuals per species) from the Peromyscus Genetic Stock Center at the University of South Carolina. For the remaining Peromyscus species that were used in the analysis of sequence variation, we obtained frozen tissues (at least two individuals per species) from the Museum of Vertebrate Zoology (Berkeley, CA). All field-collected mice used in this study were deposited as vouchered specimens in the zoological collections of the University of Nebraska State Museum (Lincoln, NE), the Denver Museum of Nature and Science (Denver, CO), the Natural History Museum of Utah (Salt Lake City, UT), the University of Arizona Museum of Natural History (Tucson, AZ), the Museum of Southwestern Biology (Albuquerque, NM), and the Museum of Vertebrate Zoology (Berkeley, CA).
All field-collected mice were live-trapped and handled in accordance with guidelines approved by the University of Nebraska Institutional Animal Care and Use Committee (IACUC no. 07-07-030D). After killing each mouse, heart and liver were snap-frozen in liquid nitrogen and were stored at −80 °C prior to DNA or RNA extraction. For mice used in the experimental analysis of Hb function, blood was collected via cardiac puncture and red cell fractions were snap-frozen in liquid nitrogen.
BAC Clone Sequencing
We isolated, subcloned, sequenced, and assembled separate BAC clones that contained the entire α- and β-globin gene clusters of P. leucopus using the same approach that we used previously to characterize the orthologous globin gene clusters of P. maniculatus (Storz et al. 2008). The P. leucopus contigs containing the α- and β-globin gene clusters were 187.4 and 179.8 kb in length, respectively.
Polymerase Chain Reaction, Cloning, and Sequencing
Protocols for amplifying and sequencing cytb are provided in Gering et al. (2009). Polymerase chain reaction (PCR) protocols, cloning protocols, and paralog-specific primer sequences for the HBA-T1 and HBA-T2 genes were described in Storz and Kelly (2008), those for the HBB-T1 and HBB-T2 genes were described in Storz et al. (2009) and Storz, Natarajan, et al. (2012), and RT-PCR protocols and primer sequences for both sets of gene duplicates were described in Storz, Runck, et al. (2010). For several Peromyscus species, we used a different set of paralog-specific primers for the HBA-T1 and HBA-T2 genes: HBA-T1, forward: GATGGGCACTGCTCAGGCTGGTC; HBA-T2, reverse: ACCCAGCCAAGTACTGCTGGAC; HBA-T2, forward: GATGGACACTGCTCAGGCTGGTC; HBA-T2, reverse: AGGCCAGGACAAGAGGCTCTCCAC. The genes were PCR-amplified using the following thermal cycling protocol: 94 °C (120 s) initial denaturing, [94 °C (10 s), 50–60 °C (70 s), 68 °C (120 s)] 10 cycles, [94 °C (15 s), 50–60 °C (30 s), 68 °C (120 s)] 20 cycles, and a final extension step at 72 °C (7 min). Automated DNA sequencing of cloned PCR products was performed on an ABI 3730 capillary sequencer using Big Dye chemistry (Applied Biosystems, Foster City, CA). For all P. maniculatus specimens from Lancaster Co., NE (n = 12), and a subset of specimens from Clear Creek Co., CO (n = 17), HBA and HBB cDNAs were derived from P. maniculatus transcriptome assemblies generated via RNA-seq (Cheviron et al. 2012, 2014). Sequence data from 58 P. maniculatus specimens from Clear Creek Co., CO, Yuma Co., CO, and Pawnee Co., KS, have been published previously (Storz, Sabatino, et al. 2007; Storz and Kelly 2008; Storz et al. 2009; Storz, Natarajan, et al. 2012).
Phylogeographic Analysis
We used cytb sequences from a total of 454 museum-vouchered specimens of P. maniculatus and other Peromyscus species to conduct an analysis of phylogeographic structure using Bayesian and maximum-likelihood phylogenetic analyses. All analyses were run using the best-fit model of nucleotide substitution, GTR+I+G, with sequences from P. leucopus and P. gossypinus as outgroups. We used MrBayes 3.2 (Ronquist and Huelsenbeck 2003) to estimate tree topologies and Bayesian posterior probabilities for each node. Analyses were run for 10 million generations, sampling every 1,000 generations, with the first 1,000 trees discarded as burn-in. A plot of –ln L against generations was used as a guide to assess convergence. Maximum-likelihood tree searches and 100 bootstrap replicates were conducted in Garli 2.0 (Zwickl 2006), which employs a genetic algorithm to effectively search large tree space. Analyses were run until –ln L values converged (changing <0.02 in 5,000 generations), and consensus values for bootstraps were summarized in PAUP* (Swofford 2002). Redundant haplotypes were identified using a distance matrix and were removed at random to leave one individual per haplotype in the resulting tree.
RAD-Seq Analysis
To survey genome-wide patterns of nucleotide variation among high- and low-altitude populations of deer mice, we produced multiplexed, reduced-representation Illumina libraries following Parchman et al. (2012). The analysis was based on a total of 60 individuals (15 individuals each from four population samples: Humboldt Co., CA, White Mountain Peak, CA, Yuma Co., CO, and Mt. Evans, CO [collected from localities 1, 8, 12, and 13 in fig. 1A; see Supplementary table S1, Supplementary Material online]). We digested genomic DNA samples with two restriction endonucleases, EcoRI and Mse1. Following digestion, we ligated double-stranded adaptor oligonucleotides to the resulting DNA fragments that contained Illumina sequencing binding sites and a unique 10 bp barcode for individual identification, and we used PCR to amplify the adaptor-ligated fragments. Details on the adaptor sequences as well as the digestion and PCR conditions can be found in Parchman et al. (2012). Following amplification, barcoded amplicons from each individual were pooled in equimolar concentrations, and electrophoresed on 2% agarose gels for size selection. We excised fragments that were between 350 and 500 bp in length and purified these gel-extracted fragments using a Qiaquick gel extraction kit (Qiagen Inc.). The pooled library was sequenced in a single lane on the Illumina HiSeq 1000 platform as 100 nt single-end reads. The resulting RAD-seq data were processed using the program STACKS (Catchen et al. 2013).
Sequences were parsed by individual barcodes and trimmed of adaptor sequences using the program process_radtags in STACKS resulting in a final mean read length of 87 nt. Individual reads were assembled de novo using the following input parameters for ustacks:—m 3, -M 2, -N 4, ––max_locus_stacks 3. Twenty individuals (five randomly chosen individuals from each population) were included when compiling the single nucleotide polymorphism catalog in cstacks. We restricted downstream analysis to loci that were genotyped in at least ten individuals per population with a minimum sequencing depth of 3 reads/locus/individual and an MAF of 0.05 using the sstacks and populations programs in STACKS resulting in a final data set of 4,344 unique loci. Parsed Illumina reads have been deposited in the NCBI short read archive (SRA PRJNA271314).
Population Genomic Analysis of Population Structure
To evaluate the relative importance of geographic distance and native elevation in explaining phylogenetic relationships among deer mouse populations, we performed the four-population test of treeness (Reich et al. 2009; Patterson et al. 2012) using RAD-Seq data from the 60 mice sampled from geographically proximal high- and low-altitude localities in California (White Mountain Peak, CA, and Humboldt Co., CA) and Colorado (Mt. Evans and Yuma Co.). Using the program TreeMix v. 1.12 (Pickrell and Pritchard 2012), we calculated the f4 test statistic for the tree ([White Mountain Peak, Humboldt Co.], [Mt. Evans, Yuma Co.]). Testing whether f4 is consistent with zero constitutes a test of whether each pair of geographically proximal highland and lowland populations are clades in the tree. The standard error of f4 was computed using a weighted block jackknife and statistical significance was assessed using a Z-score (Patterson et al. 2012). We computed genome-wide FST values using the populations program in STACKS.
Assigning Orthologous Relationships
To infer gene orthology and to characterize the extent of interparalog gene conversion in the α- and β-globin gene clusters, we used the program Pipmaker (Schwartz et al. 2000) to make pairwise comparisons of sequence similarity between the P. maniculatus and P. leucopus BAC clones. To complement this approach, we used sequence data from a more inclusive set of Peromyscus species to conduct phylogenetic analyses on four discrete data partitions of the HBA and HBB genes: 5′-flanking sequence immediately upstream of the initiation codon (350 and 93 bp for the HBA and HBB genes, respectively), the complete coding sequence, the complete intron 2 sequence, and 3′-flanking sequence immediately downstream of the termination codon (125 and 52 bp for the HBA and HBB genes, respectively). Comparisons among phylogenies that were based on different data partitions allowed us to infer orthologous relationships and to detect past histories of interparalog gene conversion (Storz, Baze, et al. 2007; Hoffmann et al. 2008a, 2008b; Opazo et al. 2008a, 2008b, 2009; Runck et al. 2009; Gaudry et al. 2014). We used HBA and HBB paralogs from Rattus novegicus as outgroup sequences in all analyses.
We aligned sequences using the L-INS-i strategy from Mafft v7 (Katoh and Standley 2013) and we performed maximum-likelihood phylogenetic analyses in Treefinder, version March 2011 (Jobb et al. 2004), evaluating support for the nodes with 1,000 bootstrap pseudoreplicates. We used the “propose model” tool of Treefinder to select the best-fitting models of nucleotide substitution based on the Akaike information criterion with correction for small sample size. We estimated Bayesian phylogenies in Mr. Bayes v.3.1.2 (Ronquist and Huelsenbeck 2003), running six simultaneous chains for 2 × 107 generations, sampling every 2.5 × 103 generations, and using default priors. A given run was considered to have reached convergence once the likelihood scores reached an asymptotic value and the average standard deviation of split frequencies remained less than 0.01. We discarded all trees that were sampled prior to convergence, and we evaluated support for the nodes and parameter estimates from a majority rule consensus of the last 2,500 trees.
Population Genetic Analysis of the Globin Genes
Summary statistics of nucleotide polymorphism and linkage disequilibrium (LD) were computed with the programs SITES (Hey and Wakeley 1997) and DnaSP v. 5.0 (Librado and Rozas 2009). To characterize levels of nucleotide variation, we computed two different estimators of the scaled mutation rate at silent sites ( = 4Nμ, where N is the effective population size and μ is the mutation rate per nucleotide): θπ, based on nucleotide diversity and Watterson’s θW, based on the number of segregating sites. To characterize levels of intralocus LD, we computed Hudson’s (1987) estimator of the scaled recombination rate, 4Nc, where c is the rate of crossing-over between adjacent nucleotides. We also measured Kelly’s (1997) ZnS, a measure of intralocus LD based on the average squared correlation in allele frequencies (r2) between pairs of sites. To summarize the effect of recombination on intralocus LD, we computed the ZZ test statistic of Rozas et al. (2001), which measures the difference in r2 between adjacent nucleotide polymorphisms and the average of pairwise r2 values across the entire gene. To test for significance of ZnS and ZZ values we conducted 10,000 coalescent simulations (no recombination) that were conditioned on the number of segregating sites. Finally, to measure nucleotide divergence between tandemly duplicated globin genes, we computed the average number of nucleotide substitutions per site, Dxy (Nei 1987, eq. 10.20), the number of net nucleotide substitutions per site, Da (Nei 1987, eq. 10.21), and silent-site divergence with Jukes–Cantor correction (Nei 1987, eq. 5.3). We used a maximum-likelihood method (Betrán et al. 1997) to detect evidence of interparalog gene conversion in both directions between HBA-T3 and the HBA-T1/HBA-T2 gene pair. We estimated observed tract lengths and the persite probability of detecting an ectopic conversion event (ψ) using equations A1 and A4, respectively, in Betrán et al. (1997). We reconstructed networks of amino acid haplotypes using a force-directed algorithm, as implemented in the program HapStar v.0.5 (Teacher and Griffiths 2011).
Protein Purification and Experimental Analysis of Hb Function
Hbs were stripped of organic phosphates and other ionic cofactors by means of a mixed bed resin MB-1 AG501-X8 column (BioRad, Hercules, CA). The purified samples were concentrated by ultrafiltration (cutoff 10,000), dialyzed in CO-equilibrated 10 mM HEPES buffer, pH 7.6, and stored at −80 °C as CO-derivatives. The Hb composition of hemolysates from each individual mouse was confirmed by using thin-layer isoelectric focusing (PhastSystem, GE Healthcare Biosciences, Piscataway, NJ).
After eliminating heme-bound CO by cycles of N2- and O2-equilibrium of the purified Hb solutions, O2-equilibrium curves were measured on 3 μl thin-film samples (0.2–0.3 mM heme concentration, 0.1 M HEPES buffer) at pH 7.4 and 37 °C. Using standard experimental conditions (Imai 1982; Mairbäurl and Weber 2012), we measured oxygenation properties of purified Hb solutions under four treatments: 1) in the absence of allosteric effectors (stripped), 2) in the presence of Cl− ions (0.1 M KCl), 3) in the presence of DPG (DPG/Hb tetramer ratio = 2.0), and 4) in the simultaneous presence of both effectors, as described previously (Storz, Runck, et al. 2010; Storz, Weber, et al. 2012). O2-equilibrium curves were measured using a modified O2 diffusion chamber and absorption at 436 nm was monitored while subjecting thin-film samples to varying O2 tensions of gas mixtures (prepared using Wösthoff gas-mixing pumps) that perfuse the chamber (Weber 1981, 1992). P50 (O2 tension at half-saturation) and n50 (Hill’s cooperativity coefficient at half-saturation) values were obtained by fitting the Hill equation Y = PO2n/(P50n+ PO2n) to the experimental O2 saturation data using a nonlinear regression model (Y = fractional saturation and n = n50, cooperativity coefficient). Measurements of Hb function in P. maniculatus from Clear Creek Co. and Yuma Co., CO, were reported previously (Storz, Runck, et al. 2010). Free Cl−concentrations were controlled with a model 926 S Mark II chloride analyzer (Sherwood Scientific Ltd, Cambridge, UK).
Association Analysis
To estimate the individual effects of segregating amino acid variants on respiratory properties of Hb, we performed an association analysis based on a BayesB algorithm (Kizilkaya et al. 2010) using GenSel software (Fernando and Garrick 2009). The phenotypic trait values represent in vitro measurements of ligand affinities on purified Hbs under standardized experimental conditions. Thus, there is essentially no scope for environmental variance or background genetic variance in trait values that is not accounted for by sequence differences in the HBA-T1, HBA-T2, HBB-T1, and HBB-T2 genes. To minimize the impact of among-group variance, and to ensure that we were assessing variation among potentially interbreeding (conspecific) individuals, we restricted the association analysis to a set of 43 P. maniculatus specimens from Humboldt, CA, White Mountains, CA, Mt. Evans, CO, and Yuma Co., CO. The association analysis was restricted to a total of 21 amino acid polymorphisms (15 α-globin sites and 6 β-globin sites) in specimens for which we had complete α- and β-globin genotypes. Specimens from San Luis Obispo Co., CA, were not included in the association analyses due to the fact that they formed a highly divergent clade in the mtDNA phylogeny (fig. 2). Consequently, the association analysis excluded three segregating sites that were monomorphic in all population samples outside of San Luis Obispo Co. To account for the effects of population structure, population-of-origin was fitted in the model as a fixed effect. The Markov Chain Monte Carlo chain length was 150,000 iterations with the first 50,000 discarded as burn-in. The proportion of sites assumed to have a null effect was set equal to 0.5, such that at each iteration only half of the sites were fitted in the model to avoid overparameterizing, given the small sample size. The results represent the mean over all 100,000 post burn-in iterations.
Supplementary Material
Supplementary figures S1–S6 and tables S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank M.-B. Hemmingsen (Aarhus) and E. J. Gering, S. J. Sabatino, and A. M. Runck (Lincoln) for assistance in the lab, A. M. Runck and D. M. Tufts for assistance with field collections, J. Demboski (Denver Museum of Nature and Science), S. Gardner (University of Nebraska State Museum), and F. Powell (White Mountain Research Station) for logistical support with field work, J. Crossland (Peromyscus Genetic Stock Center) and C. Conroy (Museum of Vertebrate Zoology) for sending tissue and/or blood samples of select specimens, and C. Ahlberg for computational assistance with the association analysis. The authors also thank two anonymous reviewers for helpful comments. This work was funded by grants from the National Institutes of Health (HL087216 to J.F.S.) and the National Science Foundation (DEB-0614342 and IOS-1354390 to J.F.S, and IOS-1354934 to Z.A.C.). All new sequences reported in this paper have been deposited in GenBank under accession numbers KJ826529, EU053203, EU559333, EU204642 (BAC clones), KJ725381–KJ726381, KJ826530–KJ826570 (globin sequences), and KP308008–KP308085 (cytb sequences). Illumina RAD-Seq reads were deposited in the NCBI Short Read Archive PRJNA271314.
References
- Anderson JT, Willis JH, Mitchell-Olds T. Evolutionary genetics of plant adaptation. Trends Genet. 2011;27:258–266. doi: 10.1016/j.tig.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Betrán E, Rozas J, Navarro A, Barbadilla A. The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics. 1997;146:89–99. doi: 10.1093/genetics/146.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel RD, Kopp A, Nuzhdin SV. Composite effects of polymorphisms near multiple regulatory elements create a major-effect QTL. PLoS Genet. 2011;7:e1001275. doi: 10.1371/journal.pgen.1001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair WF. Ecological factors in speciation of Peromyscus. Evolution. 1950;24:253–275. [Google Scholar]
- Bradley RD, Durish ND, Rogers DS, Miller JR, Engstrom MD, Kilpatrick CW. Toward a molecular phylogeny for Peromyscus: evidence from mitochondrial cytochrome-b sequences. J Mammal. 2007;88:1146–1159. doi: 10.1644/06-MAMM-A-342R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola C, Zekonyte U, Phillips AD, Cooper DN, Hahn MW. Interlocus gene conversion events introduce deleterious mutations in at least 1% of human genes associated with inherited disease. Genome Res. 2012;22:429–435. doi: 10.1101/gr.127738.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell MA, Hayes JP, Snyder LRG. Hemoglobin polymorphisms in deer mice (Peromyscus maniculatus), physiology of β-globin variants and α-globin recombinants. Evolution. 1988;42:681–688. doi: 10.1111/j.1558-5646.1988.tb02486.x. [DOI] [PubMed] [Google Scholar]
- Chappell MA, Snyder LRG. Biochemical and physiological correlates of deer mouse α-chain hemoglobin polymorphisms. Proc Natl Acad Sci U S A. 1984;81:5484–5488. doi: 10.1073/pnas.81.17.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, Bachman GC, Connaty AD, McClelland GB, Storz JF. Regulatory changes contribute to the adaptive enhancement of thermogenic capacity in high-altitude deer mice. Proc Natl Acad Sci U S A. 2012;109:8635–8640. doi: 10.1073/pnas.1120523109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, Connaty AD, McClelland GB, Storz JF. Functional genomics of adaptation to hypoxic cold-stress in high-altitude deer mice: transcriptomic plasticity and thermogenic performance. Evolution. 2014;68:48–62. doi: 10.1111/evo.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, Natarajan C, Projecto-Garcia J, Eddy DK, Jones J, Carling MD, Witt CC, Moriyama H, Weber RE, Fago A, et al. Integrating evolutionary and functional tests of adaptative hypotheses: a case study of altitudinal differentiation in hemoglobin function in an Andean sparrow, Zonotrichia capensis. Mol Biol Evol. 2014;31:2948–2962. doi: 10.1093/molbev/msu234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colautti RI, Lee CR, Mitchell-Olds T. Origin, fate, and architecture of ecologically relevant genetic variation. Curr Opin Plant Biol. 2012;15:199–204. doi: 10.1016/j.pbi.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues VS, Poh YP, Peterson BK, Pennings PS, Jensen JD, Hoesktra HE. Evidence of adaptation from ancestral variation in young populations of beach mice. Evolution. 2012;66:3209–3223. doi: 10.1111/j.1558-5646.2012.01669.x. [DOI] [PubMed] [Google Scholar]
- Dragoo JW, Lackey JA, Moore KE, Lessa EP, Cook JA, Yates TL. Phylogeography of the deer mouse (Peromyscus maniculatus) provides a predictive framework for research on hantaviruses. J Gen Virol. 2006;87:1997–2003. doi: 10.1099/vir.0.81576-0. [DOI] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED, Brodie ED, Pfrender ME. The evolutionary origins of beneficial alleles during the repeated adaptation of garter snakes to deadly prey. Proc Natl Acad Sci U S A. 2009;106:13415–13420. doi: 10.1073/pnas.0901224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando RL, Garrick DJ. 2009 GenSel—user manual for a portfolio of genomic selection related analyses. Animal breeding and genetics. Ames (IA): Iowa State University. [Google Scholar]
- Gaudry MJ, Storz JF, Butts GT, Campbell KL, Hoffman FG. Repeated evolution of chimeric fusion genes in the β-globin gene family of laurasiatherian mammals. Genome Biol Evol. 2014;6:1219–1233. doi: 10.1093/gbe/evu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gering EJ, Opazo JC, Storz JF. Molecular evolution of cytochrome b in high- and low-altitude deer mice (genus Peromyscus) Heredity. 2009;102:226–235. doi: 10.1038/hdy.2008.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding GB, Dean AM. The structural basis of molecular adaptation. Mol Biol Evol. 1998;15:355–369. doi: 10.1093/oxfordjournals.molbev.a025932. [DOI] [PubMed] [Google Scholar]
- Harms MJ, Thornton JW. Evolutionary biochemistry: revealing the historical and physical causes of protein function. Nat Rev Genet. 2013;14:559–571. doi: 10.1038/nrg3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Wakeley J. A coalescent estimator of the population recombination rate. Genetics. 1997;145:833–846. doi: 10.1093/genetics/145.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock RJ. Physiological responses of deer mice to various native altitudes. In: Wiehe WH, editor. The physiological effects of high altitude. New York: Macmillan; 1964. [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. Rapid rates of lineage-specific gene duplication and deletion in the β-globin gene family. Mol Biol Evol. 2008a;25:591–602. doi: 10.1093/molbev/msn004. [DOI] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. New genes originated via multiple recombinational pathways in the β-globin gene family of rodents. Mol Biol Evol. 2008b;25:2589–2600. doi: 10.1093/molbev/msn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Estimating the recombination parameter of a finite population model without selection. Genetics Res. 1987;50:245–250. doi: 10.1017/s0016672300023776. [DOI] [PubMed] [Google Scholar]
- Imai K. Allosteric Effects in Haemoglobin. Cambridge (UK): Cambridge University Press; 1982. [Google Scholar]
- Inoguchi N, Oshlo JR, Natarajan C, Weber RE, Fago A, et al. Deer mouse hemoglobin exhibits a lowered oxygen affinity owing to mobility of the E helix. Acta Crystallogr F. 2013;69:393–398. doi: 10.1107/S1744309113005708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JD, Wong A, Aquadro CF. Approaches for identifying targets of positive selection. Trends Genet. 2007;23:568–577. doi: 10.1016/j.tig.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4 doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MV, White S, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkvik HM, Stout IJ, Doonan TJ, Parkinson CL. Investigating niche and lineage diversification in widely distributed taxa: phylogeography and ecological niche modeling of the Peromyscus maniculatus species group. Ecography. 2012;35:54–64. [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK. A test of neutrality based on interlocus associations. Genetics. 1997;146:1197–1206. doi: 10.1093/genetics/146.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizilkaya K, Fernando RL, Garrick DJ. Genomic prediction of simulated multibreed and purebred performance using observed fifty thousand single nucleotide polymorphism genotypes. J Anim Sci. 2010;88:544–551. doi: 10.2527/jas.2009-2064. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Poh YP, Peterson BK, Barrett RDH, Larson JG, Jensen JD, Hoekstra HE. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science. 2013;339:1312–1316. doi: 10.1126/science.1233213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairbäurl H, Weber RE. Oxygen transport by hemoglobin. Compr Physiol. 2012;2:1463–1489. doi: 10.1002/cphy.c080113. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- Natarajan C, Inoguchi N, Weber RE, Fago A, Moriyama H, Storz JF. Epistasis among adaptive mutations in deer mouse hemoglobin. Science. 2013;340:1324–1327. doi: 10.1126/science.1236862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan C, Jiang XB, Fago A, Weber RE, Moriyama H, Storz JF. Expression and purification of recombinant hemoglobin in Escherichia coli. PLoS One. 2011;6:e20176. doi: 10.1371/journal.pone.0020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF. Genomic evidence for independent origins of β-like globin genes in monotremes and therian mammals. Proc Natl Acad Sci U S A. 2008a;105:1590–1595. doi: 10.1073/pnas.0710531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc Natl Acad Sci U S A. 2008b;105:12950–12955. doi: 10.1073/pnas.0804392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Sloan AM, Campbell KL, Storz JF. Origin and ascendancy of a chimeric fusion gene: the β/δ-globin gene of paenungulate mammals. Mol Biol Evol. 2009;26:1469–1478. doi: 10.1093/molbev/msp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchman TL, Gompert Z, Mudge J, Schilkey FD, Benkman CW, Buerkle CA. Genome-wide association genetics of an adaptive trait in lodgepole pine. Mol Ecol. 2012;21:2991–3005. doi: 10.1111/j.1365-294X.2012.05513.x. [DOI] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Song BH, Olson-Manning C, Anderson JT, Lee CR, Schranz ME, Windsor AJ, Clauss MJ, Manzaneda AJ, Naqvi I, et al. A gain-of-function polymorphism controlling complex traits and fitness in nature. Science. 2012;337:1081–1084. doi: 10.1126/science.1221636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projecto-Garcia J, Natarajan C, Moriyama H, Weber RE, Fago A, Cheviron ZA, Dudley R, McGuire JA, Witt CC, Storz JF. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc Natl Acad Sci U S A. 2013;110:20669–20674. doi: 10.1073/pnas.1315456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science. 2009;326:1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revsbech IG, Tufts DM, Projecto-Garcia J, Moriyama H, Weber RE, Storz JF, Fago A. Hemoglobin function and allosteric regulation in semi-fossorial rodents (family Sciuridae) with different altitudinal ranges. J Exp Biol. 2013;216:4264–4271. doi: 10.1242/jeb.091397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rozas J, Gullaud M, Blandin G, Aguade M. DNA variation at the rp49 gene region of Drosophila simulans: evolutionary inferences from an unusual haplotype structure. Genetics. 2001;158:1147–1155. doi: 10.1093/genetics/158.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runck AM, Moriyama H, Storz JF. Evolution of duplicated β-globin genes and the structural basis of hemoglobin isoform differentiation in Mus. Mol Biol Evol. 2009;26:2521–2532. doi: 10.1093/molbev/msp165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runck AM, Weber RE, Fago A, Storz JF. Evolutionary and functional properties of a two-locus β-globin polymorphism in Indian house mice. Genetics. 2010;184:1121–1131. doi: 10.1534/genetics.109.113506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin CF. Hematology of sea-level and high-altitude native Sonoran deer mice. Am J Physiol. 1970;218:1701–1703. doi: 10.1152/ajplegacy.1970.218.6.1701. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, et al. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder LRG. Deer mouse hemoglobins—is there genetic adaptation to high-altitude? Bioscience. 1981;31:299–304. [Google Scholar]
- Snyder LRG. Low P50 in deer mice native to high-altitude. J Appl Physiol. 1985;58:193–199. doi: 10.1152/jappl.1985.58.1.193. [DOI] [PubMed] [Google Scholar]
- Snyder LRG, Born S, Lechner AJ. Blood-oxygen affinity in high-altitude and low-altitude populations of the deer mouse. Respir Physiol. 1982;48:89–105. doi: 10.1016/0034-5687(82)90052-4. [DOI] [PubMed] [Google Scholar]
- Snyder LRG, Hayes JP, Chappell MA. α-chain hemoglobin polymorphisms are correlated with altitude in the deer mouse, Peromyscus maniculatus. Evolution. 1988;42:689–697. doi: 10.1111/j.1558-5646.1988.tb02487.x. [DOI] [PubMed] [Google Scholar]
- Stam LF, Laurie CC. Molecular dissection of a major gene effect on a quantitative trait: the level of alcohol dehydrogenase expression in Drosophila melanogaster. Genetics. 1996;144:1559–1564. doi: 10.1093/genetics/144.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 2007;5:1880–1889. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. Evolutionary developmental biology and the problem of variation. Evolution. 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. Is genetic evolution predictable. Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF. Hemoglobin function and physiological adaptation to hypoxia in high-altitude mammals. J Mammal. 2007;88:24–31. [Google Scholar]
- Storz JF, Baze M, Waite JL, Hoffmann FG, Opazo JC, et al. Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics. 2007;177:481–500. doi: 10.1534/genetics.107.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Hoffmann FG, Opazo JC, Moriyama H. Adaptive functional divergence among triplicated α-globin genes in rodents. Genetics. 2008;178:1623–1638. doi: 10.1534/genetics.107.080903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Kelly JK. Effects of spatially varying selection on nucleotide diversity and linkage disequilibrium: insights from deer mouse globin genes. Genetics. 2008;180:367–379. doi: 10.1534/genetics.108.088732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Moriyama H. Mechanisms of hemoglobin adaptation to high altitude hypoxia. High Alt Med Biol. 2008;9:148–157. doi: 10.1089/ham.2007.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Natarajan C, Cheviron ZA, Hoffmann FG, Kelly JK. Altitudinal variation at duplicated β-globin genes in deer mice: effects of selection, recombination, and gene conversion. Genetics. 2012;190:203–216. doi: 10.1534/genetics.111.134494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Runck AM, Moriyama H, Weber RE, Fago A. Genetic differences in hemoglobin function between highland and lowland deer mice. J Exp Biol. 2010;213:2565–2574. doi: 10.1242/jeb.042598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Runck AM, Sabatino SJ, Kelly JK, Ferrand N, Moriyama H, Weber RE, Fago A. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci U S A. 2009;106:14450–14455. doi: 10.1073/pnas.0905224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Sabatino SJ, Hoffmann FG, Gering EJ, Moriyama H, Ferrand N, Monteiro B, Nachman MW. The molecular basis of high-altitude adaptation in deer mice. PLoS Genet. 2007;3:448–459. doi: 10.1371/journal.pgen.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Scott GR, Cheviron ZA. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J Exp Biol. 2010;213:4125–4136. doi: 10.1242/jeb.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Weber RE, Fago A. Oxygenation properties and oxidation rates of mouse hemoglobins that differ in reactive cysteine content. Comp Biochem Physiol A Mol Integr Physiol. 2012;161:265–270. doi: 10.1016/j.cbpa.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Wheat CW. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution. 2010;64:2489–2509. doi: 10.1111/j.1558-5646.2010.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Zera AJ. Experimental approaches to evaluate the contributions of candidate protein-coding mutations to phenotypic evolution. In: Orgogozo V, Rockman MV, editors. Molecular methods for evolutionary genetics (Methods in molecular biology. Vol. 772) New York: Springer Science; 2011. pp. 377–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (and other methods) Sunderland (MA): Sinauer Associates; 2002. [Google Scholar]
- Teacher A, Griffiths D. HapStar: automated haplotype network layout and visualisation. Mol Ecol Resour. 2011;11:151–153. doi: 10.1111/j.1755-0998.2010.02890.x. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufts DM, Natarajan C, Revsbech IG, Projecto-Garcia J, Hoffmann FG, Weber RE, Fago A, Moriyama H, Storz JF. Epistasis constrains mutational pathways of hemoglobin adaptation in high-altitude pikas. Mol Biol Evol. 2015 doi: 10.1093/molbev/msu311. doi: 10.1093/molbev/msu311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufts DM, Revsbech IG, Cheviron ZA, Weber RE, Fago A, et al. Phenotypic plasticity in blood-oxygen transport in highland and lowland deer mice. J Exp Biol. 2013;216:1167–1173. doi: 10.1242/jeb.079848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber RE. Cationic control of O2 affinity in lugwork erythrocruorin. Nature. 1981;292:386–387. [Google Scholar]
- Weber RE. Use of ionic and zwitterionic (Tris bistris and Hepes) buffers in studies on hemoglobin function. J Appl Physiol. 1992;72:1611–1615. doi: 10.1152/jappl.1992.72.4.1611. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Stewart EE, Arnold LL, Neidert AH, Haerum BK, et al. Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila. Science. 2009;326:540–544. doi: 10.1126/science.1176980. [DOI] [PubMed] [Google Scholar]
- Wray GA. Genomics and the evolution of phenotypic traits. Annu Rev Ecol Evol Syst. 2013;44:51–72. [Google Scholar]
- Yeaman S, Whitlock MC. The genetic architecture of adaptation under migration-selection balance. Evolution. 2011;65:1897–1911. doi: 10.1111/j.1558-5646.2011.01269.x. [DOI] [PubMed] [Google Scholar]
- Zwickl D. 2006 Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin (TX): University of Texas. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.