Abstract
Lateral gene transfer (LGT) is an important mechanism of evolution for protists adapting to oxygen-poor environments. Specifically, modifications of energy metabolism in anaerobic forms of mitochondria (e.g., hydrogenosomes) are likely to have been associated with gene transfer from prokaryotes. An interesting question is whether the products of transferred genes were directly targeted into the ancestral organelle or initially operated in the cytosol and subsequently acquired organelle-targeting sequences. Here, we identified key enzymes of hydrogenosomal metabolism in the free-living anaerobic amoebozoan Mastigamoeba balamuthi and analyzed their cellular localizations, enzymatic activities, and evolutionary histories. Additionally, we characterized 1) several canonical mitochondrial components including respiratory complex II and the glycine cleavage system, 2) enzymes associated with anaerobic energy metabolism, including an unusual D-lactate dehydrogenase and acetyl CoA synthase, and 3) a sulfate activation pathway. Intriguingly, components of anaerobic energy metabolism are present in at least two gene copies. For each component, one copy possesses an mitochondrial targeting sequence (MTS), whereas the other lacks an MTS, yielding parallel cytosolic and hydrogenosomal extended glycolysis pathways. Experimentally, we confirmed that the organelle targeting of several proteins is fully dependent on the MTS. Phylogenetic analysis of all extended glycolysis components suggested that these components were acquired by LGT. We propose that the transformation from an ancestral organelle to a hydrogenosome in the M. balamuthi lineage involved the lateral acquisition of genes encoding extended glycolysis enzymes that initially operated in the cytosol and that established a parallel hydrogenosomal pathway after gene duplication and MTS acquisition.
Keywords: acetylCoA synthetase, sulfate activation pathway, hydrogenase, PFO, glycine cleavage system, succinate dehydrogenase
Introduction
In heterotrophic eukaryotes, the cytosol and mitochondrion are the main cellular compartments where energy metabolism takes place. Glycolysis is typically a cytosolic pathway that converts 1 mole of glucose to 2 moles of pyruvate with a net yield of 2–5 moles of ATP. In most aerobic eukaryotes, pyruvate is imported into mitochondria and then metabolized to CO2 and H2O. This pathway synthesizes 25 additional moles of ATP per mol of glucose, 23 of which are generated by oxidative phosphorylation. In contrast, the mitochondria of organisms that inhabit anaerobic niches have dramatically reduced or completely lost ATP synthesis (Müller et al. 2012). Anaerobic mitochondria have been characterized in some invertebrates (Müller et al. 2012); however, the most “reduced” mitochondria, such as hydrogenosomes and mitosomes, have been identified exclusively in unicellular eukaryotes (Embley and Martin 2006). Hydrogenosomal energy metabolism involves at its core the conversion of pyruvate to CO2, acetate, and H2 with the concomitant synthesis of a single mole of ATP per mole of pyruvate by substrate level phosphorylation. In mitosomes, ATP is not synthesized (Müller et al. 2012). Both hydrogenosomes and mitosomes lack a pyruvate dehydrogenase complex (PDH), the tricarboxylic acid cycle (TCA), a cytochrome c-dependent respiratory chain, and, consequently, ATP synthesis by chemiosmotic coupling (Müller et al. 2012). However, most of these organelles studied to date have retained the main components of the mitochondrial protein import machinery and the iron–sulfur cluster (ISC) assembly (Dolezal et al. 2006; Tachezy and Dolezal 2007), with the notable exceptions of the nitrogen fixation (NIF)- and putative sulfur mobilization (SUF)-type Fe-S cluster assembly systems in the Archamoebae and the breviate Pygsuia biforma, respectively (Nyvltova et al. 2013; Stairs et al. 2014). Anaerobic protists compensate for the lack of “typical” aerobic mitochondrial pyruvate metabolism (i.e., metabolism that is coupled to the TCA cycle) using the anaerobic pathway of extended glycolysis. Rather than undergoing oxidative decarboxylation via the PDH complex, pyruvate is converted to acetyl-CoA and CO2 by pyruvate:ferredoxin oxidoreductase (PFO) or by pyruvate:NADP+ oxidoreductase (PNO). Electrons that are generated during these reactions are transferred to an [FeFe] hydrogenase via ferredoxin to form molecular hydrogen or directly to NADP+ to form NADPH. Acetyl-CoA is either directly converted to acetate, CoA, and ATP by acetyl-CoA synthetase (ACS; ADP-forming) or the CoA moiety of acetyl-CoA is transferred to succinate by acetate:succinate CoA-transferase (ASCT), whereupon succinyl-CoA serves as a substrate for ATP synthesis by succinyl-CoA synthetase (SCS). Alternatively, acetyl-CoA is produced by pyruvate formate lyase (PFL) that catalyzes the nonoxidative conversion of pyruvate. In organisms that contain hydrogenosomes, extended glycolysis is compartmentalized within these organelles, and ATP is generated via ASCT and SCS; however, in organisms that contain mitosomes, pyruvate-metabolizing enzymes and hydrogenase operate in the cytosol, and the cytosolic activity of ACS generates ATP (Müller et al. 2012).
Hydrogenosomes have been found in parasitic protists such as the human pathogen Trichomonas vaginalis (Lindmark et al. 1975) and the fish parasite Spironucleus salmonicida (Jerlstrom-Hultqvist et al. 2013), commensals such as rumen ciliates and chytrid fungi (de Graaf et al. 2011), and the free-living heteroloboseids (de Graaf et al. 2009; Barbera et al. 2010). Mitosomes have been described only in parasitic protists, including Entamoeba histolytica (Tovar et al. 1999), Giardia intestinalis (Tovar et al. 2003), Cryptosporidium parvum (Riordan et al. 1999), Mikrocytos mackini (Burki et al. 2013), and microsporidia (Williams et al. 2002). The punctuate distribution of hydrogenosomes and mitosomes across diverse eukaryotic lineages suggests that these organelles evolved repeatedly from mitochondria (Yarlett and Hackstein 2005). Phylogenetic analysis of components required for extended glycolysis (PFO and hydrogenase) coupled with their patchy distribution in the eukaryotic tree suggests that the corresponding genes were acquired most likely by lateral gene transfer (LGT) initially from bacteria and then subsequently transferred amongst eukaryotes (Hug et al. 2010). Alternatively, it is also possible that anaerobic energy-producing pathways in anaerobic forms of mitochondria might also reflect the anaerobic history of eukaryotes (Martin 2011).
Archamoebae are an interesting group of protists for understanding the evolutionary transitions between different types of anaerobic mitochondria as it includes closely related members with mitosomes, such as the parasitic E. histolytica, as well as the free-living amoeba Mastigamoeba balamuthi that contains putative hydrogenosomes (Nyvltova et al. 2013). Unlike most other eukaryotes, these archamoebae have entirely lost the mitochondrial ISC type of Fe-S cluster assembly machinery. Instead, these organisms possess a simpler NIF system that was most likely acquired by LGT from ε-proteobacteria (van der Giezen et al. 2004; Gill et al. 2007; Nyvltova et al. 2013). In M. balamuthi, duplicate NIF systems function within the hydrogenosomes and the cytosol, whereas in E. histolytica, the NIF system is present only in the cytosol, although mitosomal localization has also been suggested (Maralikova et al. 2009; Nyvltova et al. 2013).Although the mitosomes of E. histolytica are unable to synthesize ATP, they possess a unique sulfate activation pathway that is dependent on ATP supply (Mi-ichi et al. 2009). This pathway is not present in any known mitochondria, and phylogenetic analysis of its two key components, ATP sulfurylase (AS) and adenosine-5′-phosphosulfate kinase (APSK), suggests that the corresponding genes were acquired laterally from a bacterial source. Interestingly, in Entamoeba’s relative M. balamuthi, PFO and hydrogenase are localized both in the cytosol and in the putative hydrogenosomes (Nyvltova et al. 2013). A previous survey of M. balamuthi expressed sequence tags suggested that these organelles might also possess some components of the TCA cycle, such as the succinate dehydrogenase (SDH) complex, and components of the glycine cleavage system (GCS; Gill et al. 2007). However, to date, the presence of the sulfate activation pathway, ATP synthesis and the fate of acetyl-CoA in the M. balamuthi organelles have not been investigated.
Here, we analyze the M. balamuthi genome sequence to identify the components of extended glycolysis and energy metabolism in this organism and to investigate the role of LGT in shaping the metabolism of its putative hydrogenosomes. We found that M. balamuthi possesses two complete sets of enzymes for extended glycolysis, including ACS, PFO, and a hydrogenase that allows the acetyl CoA-dependent synthesis of ATP in both the cytosol and the hydrogenosomes. We also found that hydrogenosomes possess D-lactate dehydrogenase (D-LDH), components of the sulfate activation pathway, SDH, and the GCS. Phylogenetic analyses suggest that all components of extended glycolysis and two components of the sulfate activation pathway were acquired by LGT. Importantly, each component of extended glycolysis is present in at least two copies that differ in the presence/absence of mitochondrial targeting sequences (MTS), consistent with their dual localization. Based on these phylogenetic and biochemical data, we propose that LGT followed by gene duplication was an important evolutionary mechanism during the transition from an ancestral archamoebal organelle to the hydrogenosome of M. balamuthi.
Results
Purification of M. balamuthi Hydrogenosomes and the Cellular Localization of Malate Dehydrogenase
A hydrogenosome-rich fraction was separated from M. balamuthi homogenate using discontinuous OptiPrep gradient centrifugation. Homogenate centrifugation resulted in the formation of five distinct protein fractions (fig. 1A), which were separated and analyzed. Two proteins, mitochondrial type Hsp70 and Cpn60, were selected as hydrogenosomal markers. In addition, we used malate dehydrogenase (MDH) as a putative enzymatic marker based on the prediction of a mitochondrial targeting sequence in its N-terminal domain (Gill et al. 2007). Using immunoblot analysis, homologous antibodies raised against these three Mastigamoeba proteins recognized proteins of the predicted molecular weight in fractions 4 and 5 (fig. 1B and supplementary fig. S1B, Supplementary Material online). The observed specific activities for MDH across fractions correlated with these immunoblot analyses; fractions 4 (0.96 ± 0.024 µmol/min/mg; n = 12) and 5 (1.12 ± 0.084 µmol/min/mg; n = 12) contained 33 and 67% of the total MDH activity, respectively (fig. 1C). The majority of phosphatase activity, a lysosomal marker, was found in fraction 3 (51%); only 33% and 10% of total phosphatase activity was detected in fractions 4 and 5, respectively (fig. 1C). Thus, fraction 5 was used for further analysis. The presence of hydrogenosomes in fraction 5 was confirmed using electron microscopy, which showed numerous electron-dense organelles surrounded by a double membrane; however, this fraction also contained contaminating vesicles of similar sizes (fig. 1A). The integrity of the hydrogenosomes in fraction 5 was confirmed by a determination of MDH latency. Over 76% of MDH activity was liberated when the organellar membranes were disintegrated by addition of 0.1% Triton X-100, which indicates that most organelles remain intact through all steps of isolation.
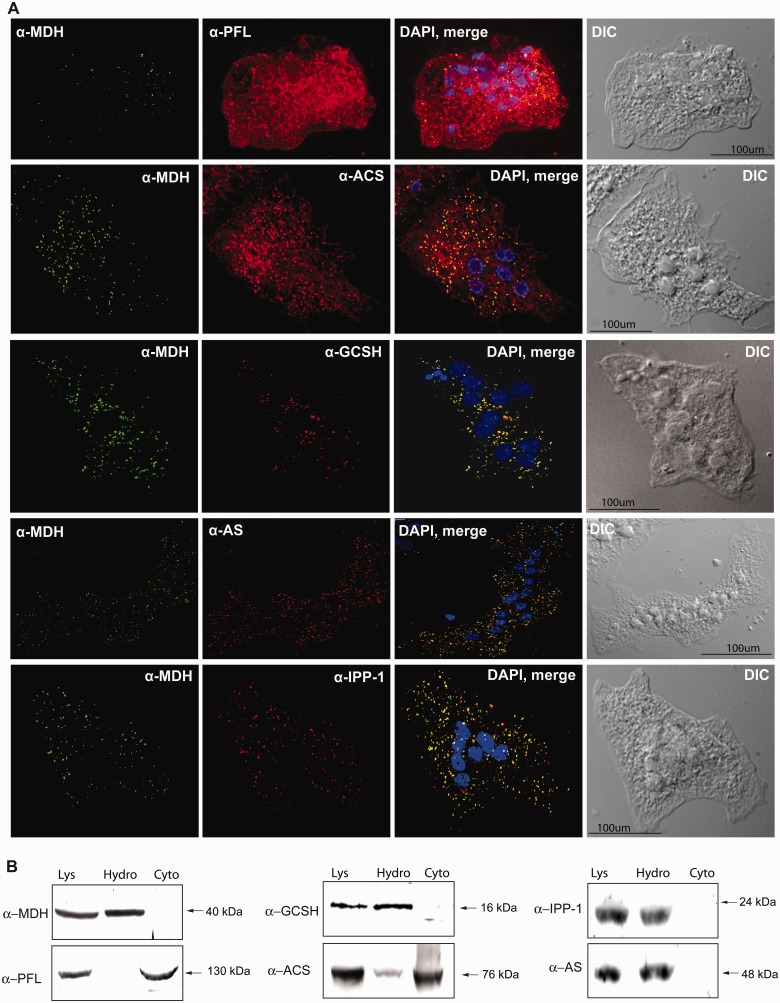
Fig. 1.
Isolation of the hydrogenosome-enriched fraction from Mastigamoeba balamuthi cell lysate. (A) Separation of the cell lysate (Lys) on a discontinuous OptiPrep gradient (15–40%) resulted in five distinct fractions (B1-5). The presence of double membrane-bound organelles in fraction B5 was confirmed using electron microscopy. (B) Immunoblot analysis of Lys and fractions B1-5 using polyclonal antibodies against the marker proteins mitHsp70, Cpn60, and MDH. (C) Distribution of marker enzymes (acid phosphatase for lysosomes and MDH for M. balamuthi hydrogenosomes) in fractions B1-5 after OptiPrep gradient centrifugation. (D) Immunofluorescence microscopy. Signal for mitochondrial marker protein Cpn60 (red) colocalized with hydrogenosomal MDH (green; PCC = 0.878 ± 0.036, n = 45).
The cellular distribution of MDH was investigated by immunofluorescence microscopy (fig. 1D). Specific anti-MDH antibodies identified MDH in approximately 100–200 organelles in cells that were colabeled with antibodies against Cpn60. To determine whether MDH activity was present exclusively in the organelles or was also present in the cytosol, we isolated the soluble cellular fraction of M. balamuthi using high-speed centrifugation (the supernatant obtained after centrifugation at 100,000 × g). We detected high NADH oxidase (a cytosolic marker enzyme) activity (table 1) but no MDH activity in this fraction. Collectively, these results suggest that MDH is exclusively localized in organelles. Therefore, we employed MDH as an enzymatic marker for M. balamuthi hydrogenosomes in the following experiments.
Table 1.
Specific Activities of Selected Enzymes in Cellular Fractions of Mastigamoeba balamuthi.
| Enzyme | Activity [µmol/min mg] | Localization |
|---|---|---|
| D-LDH (EC 1.1.1.28) | 0.721 ± 0.019 (15) | Hydrogenosomes |
| MDH (EC 1.1.1.37) | 1.009 ± 0.021 (18) | Hydrogenosomes |
| NAD malic enzyme (EC 1.1.1.37) | 0.039 ± 0.008 (15) | Cytosol |
| NADH oxidase (EC 1.6.99.3) | 0.213 ± 0.007 (18) | Cytosol |
| NADP malic enzyme (EC 1.1.1.40) | 0.041 ± 0.008 (15) | Cytosol |
| PNO (EC 1.2.1.51) | 0.447 ± 0.019 (13) | Cytosol |
| TDH (EC 1.1.1.103) | 0.004 ± 0.0003 (4) | Cytosol |
Note.—Mean ± standard deviation (n, number of experiments).
Remnants of Mitochondrial Pathways
Much like other anaerobic protists, Mastigamoeba has retained only a limited set of genes encoding enzymes that are involved in energy metabolism of aerobic mitochondria. In addition to MDH, we found all four subunits of Complex II (CII, the SDH complex [SDHA-D]). No other electron-transporting complexes were detected except the electron-transferring flavoprotein dehydrogenase (ETFDH). In aerobes, CII catalyzes the oxidation of succinate to fumarate with the concomitant reduction of ubiquinone (UQ) to ubiquinol (UQH2), which is then reoxidized by Complex III. However, in some facultative anaerobes (e.g., Ascaris suum), CII catalyzes the reverse reaction, reducing fumarate to succinate and oxidizing rhodoquinol (RQH2) to rhodoquinone (RQ). We therefore attempted to assay SDH or fumarate reductase (FRD) activity extensively in whole-cell extracts and hydrogenosome-enriched fractions using UQ and RQ substrates, without success (data not shown). We suspect that our inability to detect CII activity might be because it was 1) irreversibly inactivated during the preparation of cellular fractions and/or 2) it catalyzes a different reaction. Although we were not able to demonstrate this activity, all critical residues for substrate and cofactor binding are conserved in all four Mastigamoeba CIIs. Furthermore, homology-modeling analyses suggest that the Mastigamoeba sequence folds similarly to other, previously published structures (supplementary fig. S2, Supplementary Material online). The two largest SDH subunits (SDHA and SDHB) were subjected to phylogenetic analysis. The Mastigamoeba SDHA sequence emerged as a long branching basal taxon within eukaryotes and its SHDB sequence branched in an unresolved position in a eukaryote and α-proteobacteria (SDHB) clade (supplementary fig. S3A and B, Supplementary Material online). Both analyses are consistent with these proteins being ancestral “mitochondrial” homologs that were vertically inherited within the eukaryote lineage. Failure of the Mastigamoeba SDH subunits to branch within the closest amoebozoan relatives of Mastigamoeba (i.e., Dictyostelium) is likely due to their extreme sequence divergence, leading to long-branch attraction to the long internal branches in the tree. The marked sequence divergence of these proteins is most likely related to changing constraints due to altered structure or function relative to their aerobic counterparts. We were unable to detect a homolog of the assembly factor SDH5/SDHAF that acts in flavin attachment to SDHA (Hao et al. 2009), suggesting that the CII of Mastigamoeba is unusual.
The only other genes identified encoding quinone-utilizing enzymes were ETFa/b (table 3 and supplementary table S1B and fig. S4B and C, Supplementary Material online) and ETFDH (table 3 and supplementary table S1B and fig. S3C, Supplementary Material online). These proteins are typically involved in fatty acid oxidation reactions in which electrons are shuttled from ETFa/b to UQ via ETFDH. In some anaerobic bacteria, ETF and ETFDH are involved in butyryl-CoA metabolism (Herrmann et al. 2008). We identified partial sequences for ETFa/b and a full-length sequence of ETFDH encoding a putative MTS; however, no obvious homologs of butyryl-CoA metabolism enzymes were found. Furthermore, in phylogenetic analysis, the Mastigamoeba ETFDH emerges from within a typical mitochondrial ETFDH clade (supplementary fig. S3C, Supplementary Material online).
Table 3.
Accession Numbers of Selected Genes with Predicted Mitochondrial N-Terminal Targeting Presequences by PSORT II.
| Accesion Numbers | Annotation | N-Terminal Sequence |
|---|---|---|
| KF927023 | ACS-M1 | Yes |
| KF927022 | ACS-C2 | No |
| KF941318 | AK-M | Yes |
| KP030851 | AK-C | No |
| 0KF927026 | APS kinase | Yes |
| KF927024 | AS | Yes |
| KF941316 | D-LDH-M | Yes |
| AAP76314 | D-LDH-C | No |
| KP006414 | ETF α | Yes |
| KP006415 | ETF β | Yes |
| KJ993879 | ETFDH | Yes |
| ABV54211 | GCS-H | Yes |
| KJ993878 | GCS-L1 | Yes |
| KJ993877 | GCS-L2 | No |
| KJ993875 | GCS-P | Yes |
| KJ993876 | GCS-T | Yes |
| KJ558425 | Hydrogenase 1 | No |
| AGA37393 | Hydrogenase 2 | No |
| AGA37394 | Hydrogenase 3 | No |
| KJ558426 | Hydrogenase 4 | No |
| KJ558427 | Hydrogenase 5 | No |
| KJ920286 | Hydrogenase 8 | Yes |
| KJ920287 | Hydrogenase 9 | No |
| AGA37391 | Hydrogenase 10 | Yes |
| KF927025 | IPP-1 | Yes |
| KJ558423 | IPP-2 | No |
| KJ558424 | IPP-3 | No |
| KP030853 | LPLA | Yes |
| KJ616392 | PFO 1 | No |
| AAM53401 | PFO 2 | No |
| AGA37395 | PFO 3 | Yes |
| KJ616393 | PFO 4 | No |
| KJ616394 | PFO 5 | No |
| AGA37396 | PFO 6 | Yes |
| AGA37390 | PNO | No |
| KJ993872 | SdhA | Yes |
| AGA37361 | SdhB | Yes |
| KJ993873 | SdhC | Yes |
| KJ993874 | SdhD | Yes |
| KP057244 | SHMT | Yes |
| KM979287 | TDH | No |
Note.—ETF α/β, electron transfer flavoprotein; Sdh, succinate dehydrogenase.
The GCS pathway is conserved in diverse aerobic mitochondria (Kikuchi 1973). This multienzyme system is critical for the methylenation of the one carbon-transferring cofactor, N5, N10-methylenetetrahydrofolate (CH2-THF), using glycine as a substrate. Glycine could be formed in mitochondria from threonine in two steps by the activities of mitochondrial threonine dehydrogenase (TDH) and α-amino-β-ketobutyrate CoA ligase (AKL; Bird et al. 1984). The CH2-THF that is generated by mitochondrial GCS is used in the methylenation and subsequent hydroxylation of glycine to form serine (this reaction is catalyzed by serine hydroxymethyltransferase [SHMT]). The GCS comprises four enzymes: glycine decarboxylase (GCSP, P-protein), aminomethyltransferase (GCST, T-protein), lipoamide carrier protein (GCSH, H-protein), and dihydrolipoamide dehydrogenase (GCSL, L-protein). In Mastigamoeba, we identified homologs of all GCS components with MTS, except one of two L-proteins (GSCL2) in which MTS was not predicted (table 3). Using immunofluorescent microscopy, antibodies raised against human GCSH colocalized with antibodies directed against the hydrogenosomal marker MDH (fig. 4A) suggesting the GCS likely functions in these organelles. By immunoblotting analysis, the antihuman GCSH antibodies also recognized a protein of the predicted molecular weight in the hydrogenosome-enriched fraction (fig. 4B and supplementary fig. S1B, Supplementary Material online).
Fig. 4.
Cellular localization of PFL, ACS, AS, and IPP in Mastigamoeba balamuthi. (A) Immunofluorescence microscopy. Signals for GCSH, AS, and IPP clearly colocalized with hydrogenosomal MDH (PCC = 0.907 ± 0.023, n = 42; 0.902 ± 0.025, n = 41; and 0.876 ± 0.023, n = 38, respectively). ACS colocalized with MDH. However, some signal was observed in the cytosol (PCC = 0.659 ± 0.047, n = 43). Signal for PFL was consistent with its cytosolic localization (PCC = 0.411 ± 0.063, n = 39). Proteins were visualized using antibodies against human GCSH and M. balamuthi AS, ACS, IPP, PFL, and MDH. Alexa Fluor 488 donkey α-rabbit and 594 donkey α-mouse and α-rat were used as secondary antibodies. (B) Immunoblot analysis of cellular fractions. Cell lysate (Lys) was separated into cytosolic (Cyto) and hydrogenosome-enriched (Hydro) fractions using differential centrifugation and probed with the appropriate antibodies.
To assess the evolutionary history of the Mastigamoaeba GCS, phylogenetic analysis was performed on the larger proteins GCSP, GCST, and GCSL. In both the GCSP and GCST analyses, the Mastigamoeba GCS homologs branched with other mitochondrial sequences suggesting that these enzymes were likely inherited vertically from the common ancestor of mitochondria (supplementary fig. S3D and E, Supplementary Material online). GCSL1 branches with other eukaryotic homologs with weak support, whereas GCSL2 is a very long branch that clusters with bacteria and Trichomonas (supplementary fig. S3F, Supplementary Material online). This obvious sequence divergence might be explained by the different evolutionary pressures on GCSL in hydrogenosome-bearing taxa relative to mitochondriate model organisms, such as yeast and Arabidopsis. Moreover, GCSL is known to function in a variety of other protein complexes (Spalding and Prigge 2010). It is possible therefore, that GCSL2 is involved in a pathways unrelated to glycine cleavage. Finally phylogenetic analysis of SHMT revealed eukaryotes branching as a monophyletic clade to the exclusion of bacteria (of mixed taxonomic groupings). The Mastigamoeba SHMT sequence branches toward the base of the eukaryotic clade (supplementary fig. S3G, Supplementary Material online). The lack of resolution in the phylogeny makes it difficult to determine if the Mastigamoeba sequence is more similar to cytosolic or mitochondrial homologs. However, the presence of a MTS combined with the hydrogenosomal localization of proteins known to function in cooperation with SHMT (i.e., GCS) suggests that this protein functions in the hydrogenosome. In addition to the GCS and SHMT proteins, we identified a putatively organellar lipoamide protein ligase (LPLA) protein (responsible for attaching lipoamide to GCSH and related proteins) with an MTS (table 3 and supplementary table S1B, Supplementary Material online).
However, we did not confirm presence of predicted threonine degradation pathway (Gill et al. 2007). Although, we identified genes coding for TDH and AKL, none of them possessed putative MTS (table 3). In addition, we tested TDH activity in subcellular fractions, which revealed presence of the active enzyme in the cytosol (0.004 ± 0.0003 µmol/min/mg; n = 4) while no activity was associated with the hydrogenosomal fraction (table 1). Interestingly, using phylogenetic analysis, the Mastigamoeba TDH sequence branches sister to mitochondrial TDH sequences (supplementary fig. S3H, Supplementary Material online) suggesting this protein could have been ancestrally localized to the mitochondria but has since been relocalized exclusively to the cytoplasm.
Collectively, remnants of electron transport chain (CII, ETFa/b and ETFDH) and amino acid metabolism (GCS, SHMT, LPLA) likely represent common ancestral traits that are shared between hydrogenosomes of Mastigamoeba and mitochondria of aerobic amoebozoans.
Pyruvate Metabolism
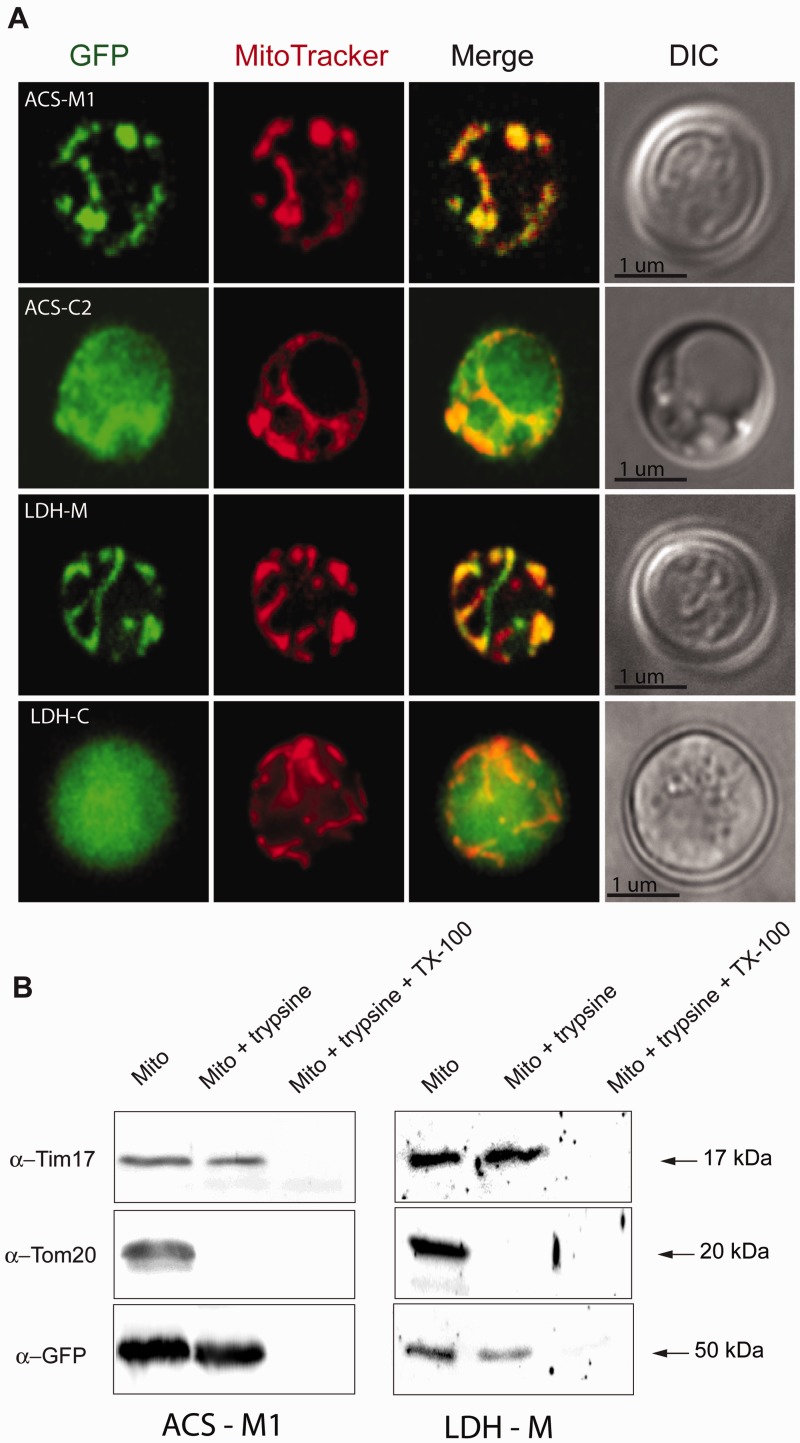
Pyruvate is a glycolytic product that is imported to mitochondria where it is converted to acetyl-CoA by PDH. However, in the hydrogenosomes of M. balamuthi, pyruvate is converted to acetyl-CoA and CO2 by PFO (Nyvltova et al. 2013), and we did not find any evidence for genes encoding PDH components. Pyruvate can also be oxidatively decarboxylated by PNO, a homolog of which we identified in the M. balamuthi genome (Nyvltova et al. 2013). Thus, we measured the PNO activity of cytosolic and hydrogenosomal fractions using a spectrophotometric assay as well as histochemical in-gel staining. Both assays revealed presence of PNO exclusively in the cytosolic fraction (table 1 and fig. 2). Malate is an alternative hydrogenosomal substrate that is oxidatively decarboxylated to pyruvate by an NAD(P)+-dependent malic enzyme in T. vaginalis hydrogenosomes (Müller et al. 2012). Surprisingly, we did not detect either NAD+- or NADP+-dependent activities of malic enzymes in M. balamuthi hydrogenosomes; however, both activities were found in the cytosolic fraction. D- or L-lactate is converted to pyruvate by cytochrome-dependent inner membrane-bound D-LDH or soluble NAD+-dependent L-LDH; thus, lactate is another possible source of pyruvate. Surprisingly, in M. balamuthi hydrogenosomes, we exclusively found a soluble NAD+-dependent D-LDH activity that has not previously been described in mitochondrion-related organelles (table 1). The activity displayed a latency (75%) similar to that of MDH, indicating that D-LDH is localized inside the organelles. Searches for the corresponding gene in the M. balamuthi genome revealed the presence of two D-LDH paralogs: D-LDH-M, which possesses a 75-amino acid N-terminal MTS with a predicted cleavage site between I75 and A76, and D-LDH-C, which lacks a presequence (supplementary fig. S4A, Supplementary Material online). Both paralogs possess conserved residues that are required for D-LDH function, including a coenzyme-binding domain GXGXXG17D sequence and Phe394, which is important for substrate specificity. To test whether D-LDH can be targeted to mitochondria, we expressed both D-LDH paralogs fused with green fluorescent protein (GFP)-tags at the carboxy-terminus in Saccharomyces cerevisiae, a model system that we have previously used successfully for testing the cellular localization of M. balamuthi proteins (Nyvltova et al. 2013). As expected, D-LDH-M was delivered into yeast mitochondria, a finding that is consistent with our measurements of organellar D-LDH activity in M. balamuthi; in contrast, D-LDH-C was localized in the cytosol (fig. 3).
Fig. 2.
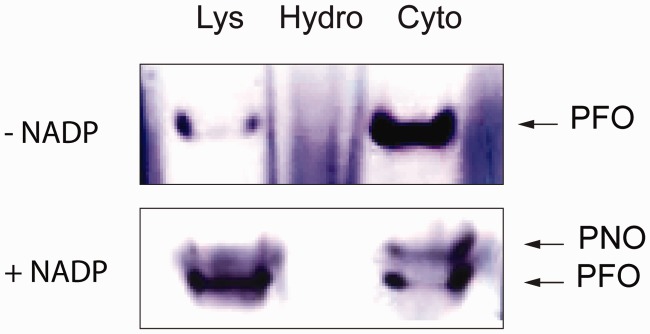
Histochemical detection of PFO and PNO in Mastigamoeba balamuthi cell fractions. Cell lysate (Lys) was separated into cytosolic (Cyto) and hydrogenosome-enriched (Hydro) fractions using differential centrifugation. Samples were separated under native conditions in a 6% polyacrylamide gel and stained for the oxidative decarboxylation of pyruvate in the presence and absence of NADP+ as an electron acceptor to distinguish PFO and PNO activity, respectively.
Fig. 3.
Expression and cell localization of Mastigamoeba balamuthi ACS and LDH in Saccharomyces cerevisiae. (A) Immunofluorescence microscopy. Mitochondrial (-M) and cytosolic (-C) versions of ACS and LDH were fused at the C-terminus with GFP and expressed in yeast. Mitochondria were visualized using MitoTracker. DIC, differential interference contrast. (B) Protease protection assay. Mitochondria were isolated from yeast expressing M. balamuthi ACS-M and LDH-M and treated with trypsin to remove proteins that are associated with the outer membrane; the mitochondria were treated with a combination of trypsin and the detergent Triton-X100 to disintegrate both membranes. Transblotted samples were probed using antibodies against Tom20 (an outer membrane marker), Tim17 (an inner membrane marker), and GFP, which is present at the C-terminus of ACS and LDH.
The D-LDH protein belongs to a larger protein family of dehydrogenases, including hydroxyacid dehydrogenases (HDHs). Phylogenetic analyses of D-LDHs and HDHs indicate that the M. balamuthi homologs D-LDH-M1 and D-LDH-C2 branch together in a paraphyletic clade of bacteria and eukaryotes that is distinct from the HDHs. Although support across the phylogeny is poor, the putative M. balamuthi D-LDH homolog does not branch with the Entamoeba and Trichomonas HDH proteins (supplementary fig. S3I, Supplementary Material online). Subsequent analyses involving removal of the HDH sequences did not increase support within the D-LDH clade (data not shown). Bacterial sequences from a variety of phyla were interspersed within the D-LDH clade, suggesting the existence of multiple LGTs between bacteria and eukaryotes (even though the interrelationships between these sequences were largely unresolved).
Collectively, these data indicate that pyruvate but not malate serves as a substrate in M. balamuthi hydrogenosomes. In addition, these organelles possess NAD+-dependent D-LDH of bacterial origin that converts lactate to pyruvate.
ATP Synthesis
Next, we investigated ATP synthesis in the organelles. Incubation of hydrogenosomal extract with ADP resulted in the formation of ATP, which was slightly increased when acetyl-CoA was added (table 2). This observation suggested the presence of adenylate kinase (AK), which catalyzes the interconversion of adenine nucleotides, and ADP-dependent ACS, which generates ATP and acetate. To distinguish these two ADP-dependent activities, we added the specific AK inhibitor, Ap5A [P1,P5-di(adenosine-5′)pentaphosphate] to the reaction. This inhibitor completely abolished AK activity, which allowed us to determine formation of ADP-dependent activity of ACS (table 2). We also tested for ASCT and SCS; however, these enzymatic activities were not detected, consistent with no evidence to date for corresponding genes in any genomic surveys.
Table 2.
Acetyl-CoA-Dependent ATP Production in Cellular Fractions of Mastigamoeba balamuthi.
| Substrates |
ATP Production [pmol/min mg] |
|
|---|---|---|
| Hydrogenosomes | Cytosol | |
| ADP | 2.11 ± 0.34 (11) | 16.67 ± 0.73 (6) |
| ADP + pyr + CoA | 2.21 ± 0.18 (9) | 19.58 ± 0.48 (6) |
| ADP + acetylCoA | 2.29 ± 0.28 (9) | 31.25 ± 0.51 (6) |
| ADP + inhibitor | 0.05 ± 0.01 (9) | 0.85 ± 0.09 (6) |
| ADP + inhibitor + acetylCoA | 1.27 ± 0.24 (9) | 20.86 ± 0.36 (4) |
| ADP + inhibitor + pyr + CoA | 0.79 ± 0.26 (9) | 18.91 ± 0.47 (4) |
Note.—pyr, pyruvate; inhibitor, P1,P5-di(adenosine-5′) pentaphosphate. Mean ± standard deviation (n = number of experiments).
Two ACS Paralogs with Dual Cellular Localization
Searches of the M. balamuthi genome for AK and ACS revealed the presence of the corresponding genes, corroborating our biochemical data. Both AK and ACS possessed an MTS, as predicted by the PSORT II software (table 3 and supplementary table S1B, Supplementary Material online). Of the four homologs of ACS we identified in the genome of Mastigamoeba, only one (ACS-M1) has a predicted MTS. Expression of ACS fused with a GFP-tag at its carboxy-terminus in yeast revealed that ACS-M1 was targeted to mitochondria, whereas the product of ACS-C2 was found in yeast cytosol (fig. 3). Next, we compared ACS activity in the cytosolic and hydrogenosomal fractions. We found that the specific activity of ACS in the cytosol was approximately eight times higher than that in hydrogenosomes (table 2). This dual localization was confirmed using immunoblot analyses: the signal for ACS was considerably stronger in the cytosolic fraction (fig. 4B and supplementary fig. S1B, Supplementary Material online). Using immunofluorescence microscopy, antibodies raised against ACS colocalized with antibodies raised against the hydrogenosomal marker MDH. However, ACS staining was also observed in the cytosol (fig. 4A).
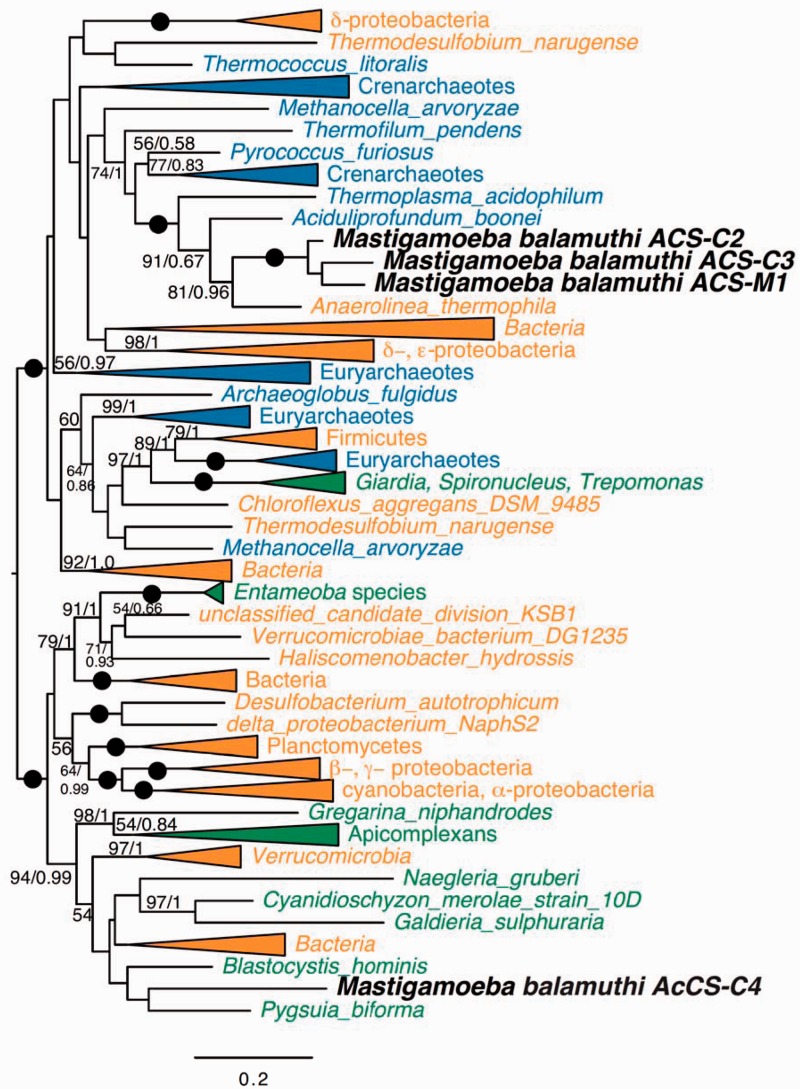
Phylogenetic analysis of these ACS homologs revealed that Mastigamoeba encodes two distinct types of ACS-like proteins (fig. 5 and supplementary fig. S3J, Supplementary Material online). Mastigamoeba balamuthi ACS proteins, including M. balamuthi ACS-M1, ACS-C2, and ACS-C3, cluster together in a group encoded by organisms consisting mostly of Archaea (crenarcheotes and euarchaeotes; bootstrap value [BV] = 100, posterior probability [PP] = 1.0). The close relationship between the three M. balamuthi ACSs suggests that these homologs result from two sequential gene duplications, after which one copy (ACS-M1) likely acquired an MTS. The remaining eukaryotic taxa branched in similar positions, as reported previously (Jerlstrom-Hultqvist et al. 2013). The second type of ACS-like proteins includes the putatively cytosolic Acyl-CoA synthetase (AcCS-C4), which groups within proteins encoded by a mixed clade of eukaryotes and bacteria (BV = 94, PP = 0.99); this group does not include Entamoeba orthologs.
Fig. 5.
Phylogeny of ACS. ML tree of ACS (187 taxa and 497 sites). Bootstrap support (BP) and PP values were calculated for each branch using RAxML and PhyloBayes, respectively. Only BV and PP values greater than 50% and 0.5, respectively, are shown. Branches with maximum support (BV = 100%; PP = 1.0) are depicted using black circles. Bacterial and archaeal clades are shown in blue and orange, respectively.
In general, the topology of ACS indicates that eukaryotic ACS proteins were separately acquired by eukaryotes though at least four independent LGT events including: 1) from Archaea to Mastigamoeba giving rise to ACS-M1, ACS-C2, and ACS-C3, 2) from firmicutes to the diplomonads, Giardia and Spironucleus (BP = 97, PP = 1.0), 3) from an unclear donor to the Entamoeba lineage (BV = 91, PP = 1.0) and 4) from an unclear donor to other eukaryotes, including AcCS-C3 (BV = 95, PP = 1.0).
Sulfate Activation Pathway
Searches of the M. balamuthi genome revealed the presence of genes coding for all main components of the sulfate activation pathway, including AS, APSK, and inorganic pyrophosphatase (IPP; table 3). Interestingly, M. balamuthi possesses three paralogs of IPP, only one of which contains a predicted MTS (IPP-1) (supplementary table S1B, Supplementary Material online). To investigate the cellular localization of the sulfate activation pathway, we generated polyclonal antibodies against recombinant AS and IPP-1. Western blot analysis of M. balamuthi fractions revealed corresponding signals exclusively in the hydrogenosomal fractions (fig. 4B and supplementary fig. S1B, Supplementary Material online). Accordingly, we observed colocalization of AS and IPP1 with the hydrogenosomal marker MDH in numerous organelles using immunofluorescence microscopy (fig. 4A).
Phylogenetic analysis of IPP sequences from diverse eukaryotes and prokaryotes reveals two distinct clades. One clade is exclusively eukaryotic, whereas the other is predominantly bacterial (supplementary fig. S3K, Supplementary Material online). Mastigamoeba IPP-1 is grouped with the previously reported Entamoeba mitosomal IPP (Mi-ichi et al. 2009; BP = 90 PP = 1.0) and homologs from Amoebozoa (BP = 35 PP = 0.79) and eukaryotes (BP = 100 PP = 1.0), suggesting that this is the ancestral IPP common to all eukaryotes. The other two Mastigamoeba proteins (IPP2 and IPP3) cluster together and emerge from within a strongly supported clade comprising sequences from the Bacteroidetes and miscellaneous other bacterial groups (BP = 99 PP = 1.0). The absence of other eukaryotic sequences in this clade, combined with the strong support values found, suggests that Mastigamoeba acquired an ancestor of the genes encoding the IPP2 and IPP3 proteins via LGT from an unknown prokaryotic donor. After the transfer, a gene duplication event clearly occurred that gave rise to both versions. Analysis of the predominantly bacterial IPP clade alone did not resolve the exact position of the Mastigamoeba IPP2/3 group (data not shown).
Phylogenetic analyses of AS and APSK (supplementary fig. S3L and M, Supplementary Material online) were broadly congruent with previous studies (Mi-ichi et al 2009). As expected, the Mastigamoeba AS sequence clustered with the Entamoeba homolog, branching as a sister to a α-proteobacterial clade with high support (BV = 100, PP = 1.0). However, the overall resolution of the APSK phylogeny was poor. A collection of sequences from bacteria, fungi, and amoebozoans (Entamoeba, Mastigamoeba, Acanthamoeba, and dictyostelids) form a well-supported clade (BP = 83, PP = 1.0) excluding α-proteobacteria. Interestingly, all four lineages of Amoebozoa did not group together in maximum likelihood (ML) or Bayesian analyses, although resolution is extremely poor throughout the phylogeny (i.e., very low bootstrap values occur). The presence of APSK in these four amoebozoans suggests that this gene could have been present in the common ancestor of the lineage. However, due to poor resolution, the exact evolutionary history of this gene remains unclear.
Phylogenetic Analysis of PFO and Hydrogenase
Phylogenetic analyses of ACS and D-LDH (fig. 5 and supplementary fig. S3J and I, Supplementary Material online), as well as NIF components (Nyvltova et al. 2013), revealed that for each of these components, recent gene duplications preceded the acquisition of MTS by one of the duplicate genes. Thus, we investigated whether similar evolutionary patterns are evident in hydrogenase or PFO phylogenies. Our searches in the M. balamuthi genome revealed at least ten predicted splice variants of [FeFe]-hydrogenase that are encoded by eight different genes. Of these hydrogenases, only one (hydrogenase-10 and isoform hydrogenase-7) has an MTS (table 3 and supplementary table S1B, Supplementary Material online). The predicted organellar localization of hydrogenase-10 is further supported by heterologous GFP-fusion protein expression in yeast, where hydrogenase-10 was shown to colocalize with the mitoreactive stain, MitoTracker (supplementary fig. S1A, Supplementary Material online).
Phylogenetic analysis of [FeFe] hydrogenase reveals two distinct clades of paralogs. Mastigamoeba hydrogenase-1 and -2 form a clade within a larger, well-supported clade comprising mainly bacterial sequences but also sequences of Entamoeba and the anaerobic protist Trimastix pyriformis (BV = 100, PP = 1.0) (supplementary fig. S3N, Supplementary Material online). The remaining Mastigamoeba sequences, including the mitochondrial-targeting homolog, branch together with strong Bayesian support (PP = 0.76). These sequences are nested within a larger clade comprising Thermotogae and eukaryotic taxa. Previous reports have noted the presence of additional C-terminal domains on some eukaryotic hydrogenase sequences (Stechmann et al. 2008; Stairs et al. 2014). Similar to the hydrogenases of Trichomonas, Blastocystis species, and P. biforma, at least four of the Mastigamoeba sequences encode C-terminal FAD-binding and/or flavodoxin domains (see the mapped domain architecture, supplementary fig. S3N, Supplementary Material online).
We identified six PFO homologs in Mastigamoeba, but only one including an MTS (PFO-M3). In phylogenetic analysis, the PFO sequences are dispersed in a monophyletic clade of eukaryotes (BP = 60, PP = 0.94) (supplementary fig. S3O, Supplementary Material online). Although most internal branches are poorly supported, it appears that PFO-M3, PFO-C4, and PFO-C5 are the result of a sequence of two gene duplication events. Curiously, a PFO paralog from the diplomonad protist branch within this Mastigamoeba cluster is most closely related to PFO-M3. It is unclear if this finding is due to the “long branches” of the diplomonad sequences branching with the most divergent of the Mastigamoeba sequences or if it instead reflects a eukaryote-to-eukaryote LGT event. Together, these results suggest that, similarly to the ACS, D-LDH, and NIF proteins, hydrogenosomal forms of hydrogenase and PFO evolved by gene duplications of cytosolic enzymes and the subsequent acquisition of MTSs.
Discussion
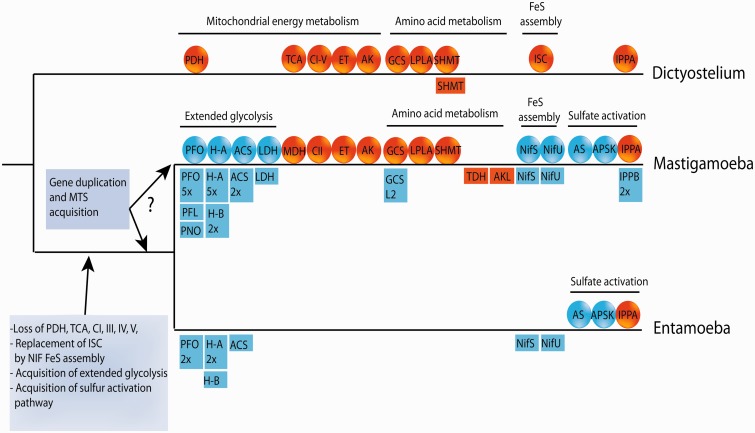
Our investigations unequivocally confirmed that mitochondrion-related organelles in M. balamuthi match the current definition of “hydrogenosomes,” which are class 4 mitochondrial family organelles (Müller et al. 2012); these organelles generate ATP by substrate-level phosphorylation and use protons as electron acceptors to form molecular hydrogen. Moreover, the hydrogenosomes of M. balamuthi display four unique features that are not found in the hydrogenosomes of other eukaryotes: 1) ATP is generated by the activity of an ACS (ADP-forming), 2) the organelles possess a sulfate activation pathway, 3) the organelles harbor homologs of CII (SDH) subunits (members of the electron transport chain), and 4) Fe-S clusters are formed by a bacterial-like NIF system (Nyvltova et al. 2013; fig. 6).
Fig. 6.
Evolution of selected biochemical pathways in amoebae. A common ancestor of mastigamoeba and entamoeba possibly lost majority of canonical mitochondrial enzymes/pathways such as PDH complex, majority of enzymes of TCA cycle, and respiration chain complexes with exception of CII. Mitochondrial ISC assembly was replaced by bacterial NIF machinery. Oxidative phosporylation was replaced by anaerobic substrate level phosporylation catalyzed by enzymes of extended glycolysis that were acquired by LGT. A common ancestor also acquired two components of sulfur activation pathway (AS and APSK). Gene duplication and acquisition of MTS resulted in dual localization of NIF system and enzymes of extended glycolysis in mastigamoeba. However, it is not clear whether these events happened in a common ancestor or only in mastigamoeba lineage. CI-V, respiratory complexes; ET, electron-transferring flavoprotein dehydrogenase (ETFDH), ETFa, and ETFb; GCS, glycin cleavage system; H-A/B, hydrogenase clade A/B; IPPA/B, inorganic pyrophosphatase clade A/B PFL, pyruvate formate lyase. Genes acquired by LGT are given in blue, genes of mitochondrial origin that were vertically inherited are highlighted by orange; squares and circles indicate cytosolic and mitochondrial localization, respectively; the numbers represent the number of cytosolic genes.
In hydrogenosomes, pyruvate and malate are the typical substrates for energy metabolism and are decarboxylated by PFO/PNO/PFL and malic enzyme, respectively. However, the absence of a malic enzyme suggests that pyruvate, not malate, is the main substrate metabolized by M. balamuthi hydrogenosomes. In mitochondria, pyruvate can be generated from D- or L-lactate via a cytochrome dependent, inner membrane-bound D-LDH or a soluble NAD+-dependent L- D-LDH (de Bari et al. 2002). Interestingly, we did not detect activities of these enzymes in M. balamuthi hydrogenosomes; instead, we found soluble NAD+-dependent D-LDH activity. In mammalian cells, D-lactate that forms in the cytosol via the methylglyoxal pathway is transported into mitochondria by three translocators, including a D-lactate/oxoacid antiporter, which mediates both the D-lactate/pyruvate and D-lactate/oxaloacetate exchanges (de Bari et al. 2002). Mastigamoeba balamuthi hydrogenosomes exhibit MDH activity that converts malate to oxaloacetate. However, neither a gene nor an activity corresponding to an oxaloacetate-metabolizing citrate synthase was identified. Thus, oxaloacetate might be available for D-lactate import. Future studies will endeavor to define which translocator is responsible for D-lactate import into hydrogenosomes and how D-lactate is formed in M. balamuthi. It has been proposed that M. balamuthi possesses a threonine degradation pathway that provides an alternative, intraorganellar means of forming pyruvate (Gill et al. 2007). The central reaction in this pathway is catalyzed by SHMT, which converts glycine and methylene-THF to serine and THF. The regeneration of THF to methyl-THF is mediated by the GCS. The presence of putative MTSs in SHMT and GCS components (and in the cofactor component, LPLA) strongly suggests that GCS operates in M. balamuthi hydrogenosomes. Furthermore, the phylogenetic analysis of GCS proteins supports a mitochondrial, and therefore ancestral, origin of these pathways. However, we found no evidence for the organellar localization of other components of threonine degradation pathways, including TDH, α-amino-β-ketobutyrate CoA ligase, and serine dehydrogenase. Nevertheless, the interconversion of glycine and serine, as catalyzed by the canonical mitochondrial complex of SHMT/GCS in hydrogenosomes, represents a putative source of active one-carbon units, which are required for various biosynthetic pathways (e.g., glutathione synthesis).
In M. balamuthi, we demonstrated that ACS synthesizes acetyl-CoA dependent ATP formation in the cytosol as well as in hydrogenosomes. We identified four genes in Mastigamoeba that are evolutionarily related to the ACS family. However, only one of these sequences (ACS-M1) codes for a putative MTS, suggesting that at least one ACS is present in the organelle and that multiple isoforms function in the cytoplasm. In other anaerobic eukaryotes, including chytrid fungi, parabasalids such as T. vaginalis, the ciliate Nyctotherus ovalis, and Blastocystis sp., hydrogenosomal ATP synthesis is catalyzed in two steps by the activities of ASCT and SCS (Müller et al. 2012). In contrast, ACS catalyzes the cytosolic, acetyl-CoA-dependent synthesis of ATP in organisms containing mitosomes, such as G. intestinalis and E. histolytica (Müller et al. 2012). Genes for ACS were also found in other eukaryotes with divergent lineages, such as the apicomplexan Plasmodium falciparum, the diatom Thalassiosira pseudonana (Armbrust et al. 2004), the excavate Naegleria gruberi (Fritz-Laylin et al. 2010), and the breviate P. biforma (Stairs et al. 2014). Interestingly, it has been predicted that ACS homologs of N. gruberi (Fritz-Laylin et al. 2010) and Th. pseudonana (Atteia et al. 2013) may function in mitochondria and in secondary plastids, respectively, whereas the homolog in Pygsuia appears to be targeted to its mitochondrion-related organelle (Stairs et al. 2014). However, no experimental data are available for the enzymatic activities of any of these enzymes from these organisms; thus, the hydrogenosome of M. balamuthi is the first eukaryotic organelle in which ACS activity has been experimentally demonstrated.
The dual localization of ACS activity in M. balamuthi is consistent with the dual localization of two other components used in extended glycolysis: PFO and hydrogenase (Nyvltova et al. 2013). Interestingly, in addition to PFO, M. balamuthi also possesses genes encoding two further pyruvate-metabolizing enzymes, PNO and PFL (Stairs et al. 2011; Nyvltova et al. 2013). Our enzymatic assays revealed that PNO participates in pyruvate metabolism exclusively in the cytosol. Cytosolic localization was also observed for PFL using immunodetection methods; however, our attempts to detect PFL activity were unsuccessful, possibly due to the high oxygen sensitivity of this enzyme (Lindmark et al. 1969). Three enzymes, PFO, PNO, and PFL, most likely participate in cytosolic pyruvate breakdown. These and previous results demonstrate that, uniquely among eukaryotes, M. balamuthi possesses dual extended glycolysis pathways together with dual machineries for Fe-S cluster assembly in the cytosol and in hydrogenosomes. We note that the specific enzymatic activities of PFO, hydrogenase, and ACS in the cytosol are considerably higher (by at least two orders of magnitude) than those of enzymes in the hydrogenosomal pathway. It is puzzling that such a weak hydrogenosomal pathway is retained in M. balamuthi and yet has been lost in E. histolytica.
It has been hypothesized that a common ancestor of the two amoebozoan lineages, Eumycetozoa and Archamoebae, possessed aerobic mitochondria and that these were retained in Eumycetozoa; meanwhile, Archamoebae adapted to anaerobic niches, converting their aerobic mitochondria to their anaerobic counterparts (hydrogenosomes, mitosomes; Gill et al. 2007). This organellar transition included the loss of most canonical mitochondrial pathways, such as the TCA cycle, cytochrome c-dependent respiration and oxidative phosphorylation, and the acquisition of anaerobic pathways by LGT (Hug et al. 2010). Our current and previous phylogenetic analyses of M. balamuthi support an LGT origin for the components of 1) anaerobic energetic metabolism including PFO, hydrogenase, ACS, and D-LDH, 2) ISC assembly machinery (NifS, NifU), and 3) sulfate activation (AS, APSK). Moreover, the dual cellular localization of pathways (1) and (2) raises interesting questions: What was the initial cellular localization of the components immediately after their acquisition by LGT, and what preceded their organellar localization? In general, there are two possible scenarios regarding the history of LGT-acquired components: 1) The acquired bacterial proteins were predisposed to be delivered to the mitochondrial matrix at low efficiency but later gained targeting presequences that improved the efficiency and specificity of protein delivery to the organelles. In this case, there is no need for gene duplication if the newly acquired function is not required in the cytosol. 2) Initially, the newly acquired proteins operated in the cytosol. After gene duplications and the accidental acquisition of MTSs, one copy was targeted and translocated into the organelles. The first scenario is supported by bioinformatic analysis of proteobacterial proteins showing that at least 5% of the proteins are predisposed for targeting to mitochondria (Lucattini et al. 2004). In addition, it has been shown that key proteins of extended glycolysis, including PFO, α-SCS, and malic enzyme, can be delivered into the hydrogenosomes of T. vaginalis without a targeting presequence (Zimorski et al. 2013). Moreover, no MTS has been identified in proteins targeted to hydrogenosomes in Sp. salmonicida (Jerlstrom-Hultqvist et al. 2013). The second scenario is supported by the dual localization of extended glycolysis and the ISC assembly machinery, which we observed in M. balamuthi. We found that each gene coding for ACS, D-LDH, PFO, hydrogenase, NifS, and NifU is present in at least two copies that differ in presence or absence of MTS (fig. 6). Using the S. cerevisiae model, we showed that only forms possessing an MTS could be targeted and translocated to yeast mitochondria, whereas paralogs not possessing an MTS remain in the cytosol. Accordingly, we found acetyl-CoA-dependent ATP synthesis, PFO, hydrogenase, and NifS activities both in the hydrogenosomes and in the cytosol of M. balamuthi. Dual localization of the components of extended glycolysis was also observed in Sp. salmonicida. This organism possesses genes for five PFOs and seven hydrogenases; however, only a single PFO and two hydrogenase paralogs were found to be targeted to hydrogenosomes; the other paralogs were localized to the cytosol (Jerlstrom-Hultqvist et al. 2013). In the anaerobic breviate P. biforma, multiple copies of PFO and hydrogenases also exist; however, only some appear to be localized to the mitochondrion-related organelle (Stairs et al. 2014). Therefore, dual localization of extended glycolysis is not restricted to Archamoebae, strengthening the argument for the second scenario.
Another intriguing question concerns the evolutionary transition of ATP-generating hydrogenosomes to energetically “silent” mitosomes. Considering the dual localization of extended glycolysis and NIF system enzymes in M. balamuthi and the cytosolic localization of these pathways in E. histolytica (Nyvltova et al. 2013), it is tempting to speculate that these organisms arose from an ancestor possessing a common set of LGT genes and that copies with hydrogenosomal targeting signals were lost in E. histolytica during the hydrogenosome-to-mitosome transition. Alternatively, it is possible that the gene duplication events occurred only in the M. balamuthi lineage, whereas the mitosomes of E. histolytica evolved directly through the reduction of mitochondrial pathways; that is, without their common ancestor going through a hydrogenosome-containing phase. In both scenarios, we could expect a common origin for LGT genes. However, phylogenetic analyses revealed a much more complex picture. A common origin was suggested for genes coding for AS and APSK (components of the sulfate activation pathway, which is present in M. balamuthi hydrogenosomes and in E. histolytica mitosomes) and for components of the NIF system. The phylogenies of the NIF subunits further suggests that organellar NifS and NifU in E. histolytica have lost their targeting sequences and replaced their cytosolic versions (Nyvltova et al. 2013). In contrast, our phylogenetic analyses of the duplicated components of extended glycolysis (ACS, PFO, and hydrogenase) in M. balamuthi revealed evolutionary histories different than those in E. histolytica. Three paralogs of ACS (ACS-M1, ACS-C2, and ACS-C3) in M. balamuthi were likely acquired by LGT from Archaea, whereas two further independent LGTs led to ACS acquisition in diplomonads (Giardia, Spironucleus) and in a diverse range of eukaryote lineages including E. histolytica. [FeFe]-hydrogenases that are present in multiple copies in M. balamuthi and in E. histolytica appear in distinct clusters within a large clade of eukaryotic [FeFe]-hydrogenases. Moreover, the cytosolic and hydrogenosomal hydrogenases of M. balamuthi clustered together, indicating the occurrence of recent gene duplication events. Similar patterns were observed with respect to multiple copies of PFO. Cytosolic and hydrogenosomal forms of M. balamuthi homologs clustered together in positions of the tree that are distinct from those occupied by the cytosolic PFO sequence of E. histolytica.
Collectively, these findings suggest that a common ancestor of M. balamuthi and E. histolytica possessed duplicated NIF systems and an organellar sulfate activation pathway, whereas components of extended glycolysis were duplicated only in M. balamuthi.
The complex phylogenetic histories of extended glycolysis enzymes in M. balamuthi and E. histolytica are reminiscent of the complex patterns observed for the glycolytic enzymes (Embden-Meyerhof-Parnas pathway) in these organisms (Liapounova et al. 2006). In anaerobic eukaryotes including the Archamoebae, two ATP-consuming enzymes, phosphofructokinase (PFK) and pyruvate kinase are replaced by inorganic pyrophosphate (PPi)-dependent PFK and pyruvate phosphate dikinase (PPDK), respectively, that increase the ATP yield of glycolysis (Mertens 1993; Slamovits and Keeling 2006). In addition, the class I fructose-bisphosphate aldolase (FBA) that is present in most eukaryotes is replaced by unrelated enzyme class II FBA (Sanchez et al. 2002). Previous phylogenetic analyses indicated that PPDK and FBA class II were present in the common ancestor of M. balamuthi and E. histolytica (Sanchez et al. 2002; Slamovits and Keeling 2006). PPDK was most likely acquired from gamma proteobacteria, while the phylogeny of class II FBA suggested an LGT event between T. vaginalis and the amoebae (Liapounova et al. 2006). In contrast, the phylogeny of PPi-PFK suggested that M. balamuthi and E. histolytica acquired this enzyme from different LGT events (Liapounova et al. 2006). To obtain more detailed insights into the history of LGT in Archamoebae, we will need genomic information about basal lineages such as Pelomyxa or Rhizomastix (Ptackova et al. 2013).
In conclusion, we found that M. balamuthi possesses dual pathways for extended glycolysis and the ACS-dependent synthesis of ATP; these pathways occur in parallel in the cytosol and in hydrogenosomes. Based on phylogenetic analyses of key components of extended glycolysis and the FeS cluster assembly machinery, we propose that LGT, gene duplication, and the acquisition of MTS played a key role during the evolution of the M. balamuthi hydrogenosome.
Materials and Methods
Cell Cultivation
Mastigamoeba balamuthi (ATCC 30984) was a kind gift of M. Müller (Rockefeller University, USA) and was maintained axenically in PYGC medium at 24 °C (Chavez et al. 1986). Saccharomyces cerevisiae strain YPH499 was grown in a rich medium or selective medium as previously described (Lithgow et al. 1994).
Gene Cloning and Expression
Genes coding for M. balamuthi ACS, AS, APSK, IPP, and D-LDH were obtained using TBLAST searches of the draft M. balamuthi genome sequence http://www.ebi.ac.uk/ena/data/view/CBKX00000000 (January 22, 2015) using the orthologous sequences of E. histolytica as queries, except for D-LDH, for which we used the protein sequence of Chlamydomonas reinhardtii D-LDH as the query. Coding sequences without introns were obtained by polymerase chain reaction (PCR) using specific primers (see supplementary table S1A, Supplementary Material online, for sequences) and cDNA as the template. Total RNA was isolated from an exponential culture of M. balamuthi using Tri-Reagent (Sigma) and used as template for cDNA synthesis using SuperScript III (Invitrogen). Gene sequences were deposited in the NCBI database under the following accession numbers: ACS-M (KF927023), ACS-C (KF927022), AS (KF927024), APSK (KF927026), IPP-1 (KF927025), and D-LDH (KF941316).
To obtain recombinant MDH, ACS, AS, and IPP-1, the corresponding genes were amplified by PCR (primer sequences are given in supplementary table S1A, Supplementary Material online) using cDNA as template, subcloned into the pET42b+ vector (Novagen), and expressed with a 6xHis tag in Escherichia coli BL21 (DE3). The proteins were purified by affinity chromatography under denaturing conditions according to the manufacturer’s protocol (Qiagen) and used to immunize rats (ACS, AS, IPP) or rabbits (MDH).
In the cellular localization studies, 600-bp gene fragments corresponding to the N-terminal segments of ACS-M1, ACS-C2, D-LDH-M, D-LDH-C, and hydrogenase 10-M were expressed with C-terminal GFP tags in S. cerevisiae. The gene fragments were cloned into the pUG35 plasmid using the restriction enzymes XbaI and HindIII (Niedenthal et al. 1996).
Cellular Fractionation
Subcellular fractions of Mastigamoeba were obtained by differential and OptiPrep (Axis-Shield PoC AS) gradient centrifugation of the cell homogenate. All steps were carried out at 4 °C in the presence of protease inhibitors (Complete Mini EDTA-free cocktail tablets, Roche). The cells were grown for 6 days in a tissue culture flask (Nunc). To harvest the cells, the growth medium was exchanged for fresh ice-cold medium, and the cells were carefully detached from the flask walls using a cell scraper. The cells were collected by centrifugation at 1,200 × g for 15 min, washed, and resuspended in SDM buffer (250 mM sucrose, 10 mM MOPS-KOH, 10 mM DTT, pH 7.4). The washed cells were disrupted using sonication on ice. The homogenate was centrifuged at 1,000 × g for 15 min to remove unbroken cells, membrane fragments, and nuclei, and the supernatant was carefully decanted. The supernatant was loaded onto a discontinuous OptiPrep gradient (15–40%) and five cell fractions were separated after centrifugation at 100,000 × g overnight.
Alternatively, the supernatant was centrifuged at 9,000 × g for 30 min to remove cell debris and lysosomes. The mitochondrion-enriched fraction was obtained by centrifugation of the supernatant at 100,000 × g for 25 min. The organellar fraction (pellet) was washed, resuspended, and centrifuged again at 100,000 × g for 25 min. The supernatant represented the cytosolic fraction.
Immunofluorescence Microscopy and Immunoblot Analysis
For immunofluorescence microscopy, M. balamuthi cells were fixed in 1% formaldehyde for 30 min, washed, and treated in 1% Triton TX-100 for 10 min. Fixed cells were stained on slides using polyclonal rat α-SDHB, α-AS, α-IPP, α-PFL, or mouse α-GCSH, or rabbit α-MDH and α-Cpn60 Abs (Nyvltova et al. 2013) and secondary Alexa Fluor 488 donkey α-rabbit or Alexa Fluor 594 donkey α-rat or α-mouse Abs (Life Technologies), respectively. For the colocalization of rabbits α-MDH and α-Cpn60, 250 µl of the MDH Ab was conjugated with 10 µg Alexa Fluor 488 and purified on Sephadex G-25.
Saccharomyces cerevisiae expressing GFP-tagged proteins were incubated for 15 min with the mitochondrial marker MitoTracker Red CMXRos (Molecular Probes), washed twice in PBS, and stabilized in 1% agarose. All slides were observed using an Olympus IX-81Cell R system. Colocalization of probes was evaluated using Fiji image analysis software and Coloc 2 plug-in (http://fiji.sc/Fiji, January 22, 2015) and quantified for individual cells using Pearson correlation coefficient (PCC) that is expressed as a mean ± standard deviation, where n is the number of individually measured cells. To analyze cellular fractions by immunoblotting, the proteins were resolved using 13.5% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE), transferred to nitrocellulose membranes, and probed with the corresponding antibody; the specific signals were visualized using a LAS-4000 chemiluminescent reader. Polyclonal rabbit Ab raised against mitochondrial Hsp70 of Neocallimastix was kindly provided by M. Embley. We raised polyclonal rabbit Ab against a peptide of Cpn60 M. balamuthi (CKQISSQDEVRQVATV, EZBiolab USA), and rabbit polyclonal antibodies against S. cerevisiae Tim17 and Tom20 were a kind gift from T. Lithgow and K. Gabriel, respectively. Monoclonal mouse Ab raised against human GCS-H was obtained from Abnova (H00002653).
Enzyme Assays
PFO (EC 1.2.7.1), hydrogenase (EC 1.18.3.1), NAD(P)+-dependent malic enzyme (EC 1.1.1.37 and EC 1.1.1.40), MDH (EC 1.1.1.37), and NADH oxidase were assayed spectrophotometrically as described (Drmota et al. 1997; Rasoloson et al. 2001). Acid phosphatase (EC 3.1.3.2) was measured according to (Barrett 1972). PNO (EC 1.2.1.51) was assayed using the method described in Inui et al. (1987). D-LDH (EC 1.1.1.28) was measured spectrophotometrically at 340 nm in 50 mM Tris-KCl buffer, pH 7.4, using 1 mM NAD+ as an electron acceptor and 2 mM D-lactate as a substrate. TDH (EC 1.1.1.103) was assayed spectrophotometrically at 340 nm in 50 mM Tris-KCl buffer, pH 7.4, 1 mM MgCl2, using 1 mM NAD+ as an electron acceptor and 5 mM threonine as a substrate (Boylan and Dekker 1981). Protein concentration was assayed according to Lowry’s method.
Native Gel Electrophoresis
Nondenaturing PAGE was carried out in 6% polyacrylamide gels in the absence of SDS in a BioRad Mini-Protean apparatus. Cellular fractions were diluted with a minimal volume of loading dye containing 20% glycerol and bromophenol blue reagent. Electrophoresis was performed at 110 V for 2.5 h at 4 °C. Enzymatic activity was detected using a procedure modified from that of (Meinecke et al. 1989). Gel slides were washed in assay buffer (50 mM HEPES, 2 mM TPP, 2.5 mM MgCl2) for 10 min. For the PFO reaction, the gel was incubated in assay buffer containing 2.5 mM CoA, 2.5 mM pyruvate, and 2 mg/ml NBT. For the PNO activity assays, 1 mM NADP was added to the incubation mixture. The reaction was stopped by washing with water.
ATP Production
ATP production was determined according to Drew and Leeuwenburgh (2003) using a luminometer and an ATP Bioluminescence Assay Kit CLS II (Roche Applied Science) containing luciferase. Several substrates were added to the reaction mixture (2 mM pyruvate, 2 mM ADP, 2.5 mM CoA, and 1.5 mM acetyl-CoA). ATP standard curves were generated using 1 mM, 100 μM, 10 μM, 1 μM, 100 nM, 10 nM, and 1 nM solutions. The organellar fraction obtained after differential centrifugation was used to analyze the ATP content and ATP production rate. To test the functionality of the ATP determination assay, 2.5 mM of the inhibitor P1,P5 –di (adenosine-5′) pentaphosphate was added to freshly isolated organelles, and the mixture was incubated for 10 min. The rate of inhibition of ATP production was determined by comparing the inhibitor-treated organelles to an equal portion of freshly isolated organelles.
Electron Microscopy
To examine the organellar fractions using electron microscopy, OptiPrep fractions were centrifuged and fixed in 0.1 M cacodylate buffer containing 8% glutaraldehyde at 4 °C and in 1% osmium tetroxide for 1 h, stained in block with uranyl acetate, and dehydrated in acetone. The sections were viewed using a JEOL 1011 transmission electron microscope.
Phylogenetic Analysis and Homology Modeling
Mastigamoeba PFO homologs were added to the alignment of Stairs et al. (2014). For all other proteins analyzed, Mastigamoeba amino acid sequences were used as queries to retrieve eukaryotic and at least 1,000 prokaryotic homologs using BLASTP against the nonredundant (nr) database in GenBank. These data sets were filtered by finding all sequences with greater than 98% sequence identity to another sequence and then randomly deleting one of them. Data sets were aligned using MAFFT (Katoh and Toh 2008), and regions of ambiguous alignment were automatically trimmed using BMGE (-blosum30; Criscuolo and Gribaldo 2010). Initial phylogenetic analyses were performed using FastTree (Price et al. 2010). The sequences of closely related‚ “redundant” species were removed by visual inspection of the FastTree output to generate the final data sets. To select the optimal substitution model for the final phylogenetic analyses, we performed model testing, as implemented in RAxML version 8.0.19 (Stamatakis 2014). In addition, we compared the likelihood scores of trees generated using the LG4X and LG4M models (Le et al. 2012) and the optimal model selected by RAxML. In all but one case, the LG4X model was favored over all other models according to the Akaike Information Criterion (the GCSP data set favored the LG4M model). For each gene, the percent bootstrap support from 500 bootstrap replicates was mapped onto the best-scoring ML tree (of 100 heuristic searches). Bayesian inference was conducted using PhyloBayes 3.2 with four Monte Carlo Markov Chains under the site-heterogeneous catfix C20 model of evolution (C20+poisson and gamma 4; Le et al. 2008; Lartillot et al. 2009). Chains were run until convergence (maxdiff < 0.3) and/or 300,000 generations were run for each chain. Every 100 trees were sampled after a manually determined burn-in of trees was removed. PPs were subsequently mapped onto the ML tree using Dendropy (Sukumaran and Holder 2010).
Homology models for SDH were generated using the Swiss-MODEL automated workspace server (Ye and Godzik 2003; Arnold et al. 2006).
Supplementary Material
Supplementary figures S1–S4 and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Czech Grant Foundation (P305/11/1061), the Biomedicine Center of the Academy of Sciences and Charles University (CZ.1.05/1.1.00/02.0109) from the European Regional Development Fund and GAUK101710 from Charles University in Prague. C.W.S. is supported by a Natural Science and Engineering Research Council (NSERC) Alexander Graham Bell Canadian Graduate Scholarship and a Killam Graduate Scholarship. A.J.R. acknowledges the Canadian Institute for Advanced Research Program in Microbial Biodiversity, in which he is a Senior Fellow, and the Canada Research Chairs Program (CIHR; MOP-62809). Access to computing and storage facilities was provided by ELIXIR CZ and the National Grid Infrastructure MetaCentrum.
References
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web- based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Atteia A, van LR, Tielens AG, Martin WF. Anaerobic energy metabolism in unicellular photosynthetic eukaryotes. Biochim Biophys Acta. 2013;1827:210–223. doi: 10.1016/j.bbabio.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Barbera MJ, Ruiz-Trillo I, Tufts JY, Bery A, Silberman JD, Roger AJ. Sawyeria marylandensis (Heterolobosea) has a hydrogenosome with novel metabolic properties. Eukaryot Cell. 2010;9:1913–1924. doi: 10.1128/EC.00122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AT. Lysosomal enzymes. In: Dingle JT, editor. Lysosomes. Amsterdam: North-Holland Publishing Company; 1972. pp. 46–135. [Google Scholar]
- Bird MI, Nunn PB, Lord LA. Formation of glycine and aminoacetone from L-threonine by rat liver mitochondria. Biochim Biophys Acta. 1984;802:229–236. doi: 10.1016/0304-4165(84)90166-1. [DOI] [PubMed] [Google Scholar]
- Boylan SA, Dekker EE. L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12. J Biol Chem. 1981;256:1809–1815. [PubMed] [Google Scholar]
- Burki F, Corradi N, Sierra R, Pawlowski J, Meyer GR, Abbott CL, Keeling PJ. Phylogenomics of the intracellular parasite Mikrocytos mackini reveals evidence for a mitosome in rhizaria. Curr Biol. 2013;23:1541–1547. doi: 10.1016/j.cub.2013.06.033. [DOI] [PubMed] [Google Scholar]
- Chavez LA, Balamuth W, Gong T. A light and electron microscopical study of a new, polymorphic free-living amoeba, Phreatamoeba balamuthi n. g., n. sp. J Protozool. 1986;33:397–404. doi: 10.1111/j.1550-7408.1986.tb05630.x. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bari L, Atlante A, Guaragnella N, Principato G, Passarella S. D-lactate transport and metabolism in rat liver mitochondria. Biochem J. 2002;365:391–403. doi: 10.1042/BJ20020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf RM, Duarte I, van Alen TA, Kuiper JW, Schotanus K, Rosenberg J, Huynen MA, Hackstein JH. The hydrogenosomes of Psalteriomonas lanterna. BMC Evol Biol. 2009;9:287. doi: 10.1186/1471-2148-9-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf RM, Ricard G, van Alen TA, Duarte I, Dutilh BE, Burgtorf C, Kuiper JW, van der Staay GW, Tielens AG, Huynen MA, et al. The organellar genome and metabolic potential of the hydrogen-producing mitochondrion of Nyctotherus ovalis. Mol Biol Evol. 2011;28:2379–2391. doi: 10.1093/molbev/msr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Drew B, Leeuwenburgh C. Method for measuring ATP production in isolated mitochondria: ATP production in brain and liver mitochondria of Fischer-344 rats with age and caloric restriction. Am J Physiol Regul Integr Comp Physiol. 2003;285:R1259–R1267. doi: 10.1152/ajpregu.00264.2003. [DOI] [PubMed] [Google Scholar]
- Drmota T, Tachezy J, Kulda J. Isolation and characterization of cytosolic malate dehydrogenase from Trichomonas vaginalis. Folia Parasitol. 1997;44:103–108. [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Gill EE, Diaz-Trivino S, Barbera MJ, Silberman JD, Stechmann A, Gaston D, Tamas I, Roger AJ. Novel mitochondrion-related organelles in the anaerobic amoeba Mastigamoeba balamuthi. Mol Microbiol. 2007;66:1306–1320. doi: 10.1111/j.1365-2958.2007.05979.x. [DOI] [PubMed] [Google Scholar]
- Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann G, Jayamani E, Mai G, Buckel W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol. 2008;190:784–791. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Stechmann A, Roger AJ. Phylogenetic distributions and histories of proteins involved in anaerobic pyruvate metabolism in eukaryotes. Mol Biol Evol. 2010;27:311–324. doi: 10.1093/molbev/msp237. [DOI] [PubMed] [Google Scholar]
- Inui H, Ono K, Miyatake K, Nakano Y, Kitaoka S. Purification and characterization of pyruvate:NADP+ oxidoreductase in Euglena gracilis. J Biol Chem. 1987;262:9130–9135. [PubMed] [Google Scholar]
- Jerlstrom-Hultqvist J, Einarsson E, Xu F, Hjort K, Ek B, Steinhauf D, Hultenby K, Bergquist J, Andersson JO, Svard SG. Hydrogenosomes in the diplomonad Spironucleus salmonicida. Nat Commun. 2013;4:2493. doi: 10.1038/ncomms3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kikuchi G. The glycine cleavage system: composition, reaction mechanism, and physiological significance. Mol Cell Biochem. 1973;1:169–187. doi: 10.1007/BF01659328. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Le SQ, Dang CC, Gascuel O. Modeling protein evolution with several amino acid replacement matrices depending on site rates. Mol Biol Evol. 2012;29:2921–2936. doi: 10.1093/molbev/mss112. [DOI] [PubMed] [Google Scholar]
- Le SQ, Lartillot N, Gascuel O. Phylogenetic mixture models for proteins. Philos Trans R Soc Lond B Biol Sci. 2008;363:3965–3976. doi: 10.1098/rstb.2008.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liapounova NA, Hampl V, Gordon PM, Sensen CW, Gedamu L, Dacks JB. Reconstructing the mosaic glycolytic pathway of the anaerobic eukaryote Monocercomonoides. Eukaryot Cell. 2006;5:2138–2146. doi: 10.1128/EC.00258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindmark DG, Müller M, Shio H. Hydrogenosomes in Trichomonas vaginalis. J Parasitol. 1975;61:552–554. [Google Scholar]
- Lindmark DG, Paolella P, Wood NP. The pyruvate formate-lyase system of Streptococcus faecalis. I. Purification and properties of the formate-pyruvate exchange enzyme. J Biol Chem. 1969;244:3605–3612. [PubMed] [Google Scholar]
- Lithgow T, Junne T, Suda K, Gratzer S, Schatz G. The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc Natl Acad Sci U S A. 1994;91:11973–11977. doi: 10.1073/pnas.91.25.11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucattini R, Likic VA, Lithgow T. Bacterial proteins predisposed for targeting to mitochondria. Mol Biol Evol. 2004;21:652–658. doi: 10.1093/molbev/msh058. [DOI] [PubMed] [Google Scholar]
- Maralikova B, Ali V, Nakada-Tsukui K, Nozaki T, van der Giezen M, Henze K, Tovar J. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell Microbiol. 2009;12:331–342. doi: 10.1111/j.1462-5822.2009.01397.x. [DOI] [PubMed] [Google Scholar]
- Martin WF. Early evolution without a tree of life. Biol Direct. 2011;6:36. doi: 10.1186/1745-6150-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke B, Bertram J, Gottschalk G. Purification and characterization of the pyruvate-ferredoxin oxidoreductase from Clostridium acetobutylicum. Arch Microbiol. 1989;152:244–250. doi: 10.1007/BF00409658. [DOI] [PubMed] [Google Scholar]
- Mertens E. ATP versus pyrophosphate: glycolysis revisited in parasitic protists. Parasitol Today. 1993;9:122–126. doi: 10.1016/0169-4758(93)90169-g. [DOI] [PubMed] [Google Scholar]
- Mi-ichi F, Abu YM, Nakada-Tsukui K, Nozaki T. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci U S A. 2009;106:21731–21736. doi: 10.1073/pnas.0907106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu RY, van der Giezen M, Tielens AG, Martin WF. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 2012;76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal RK, Riles L, Johnston M, Hegemann JH. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Nyvltova E, Sutak R, Harant K, Sedinova M, Hrdy I, Paces J, Vlcek C, Tachezy J. NIF-type iron-sulfur cluster assembly system is duplicated and distributed in the mitochondria and cytosol of Mastigamoeba balamuthi. Proc Natl Acad Sci U S A. 2013;110:7371–7376. doi: 10.1073/pnas.1219590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptackova E, Kostygov AY, Chistyakova LV, Falteisek L, Frolov AO, Patterson DJ, Walker G, Cepicka I. Evolution of Archamoebae: morphological and molecular evidence for pelobionts including Rhizomastix, Entamoeba, Iodamoeba, and Endolimax. Protist. 2013;164:380–410. doi: 10.1016/j.protis.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Rasoloson D, Tomková E, Cammack R, Kulda J, Tachezy J. Metronidazole-resistant strains of Trichomonas vaginalis display increased susceptibility to oxygen. Parasitology. 2001;123:45–56. doi: 10.1017/s0031182001008022. [DOI] [PubMed] [Google Scholar]
- Riordan CE, Langreth SG, Sanchez LB, Kayser O, Keithly JS. Preliminary evidence for a mitochondrion in Cryptosporidium parvum: phylogenetic and therapeutic implications. J Eukaryot Microbiol. 1999;46:52S–55S. [PubMed] [Google Scholar]
- Sanchez L, Horner D, Moore D, Henze K, Embley T, Müller M. Fructose-1,6-bisphosphate aldolases in amitochondriate protists constitute a single protein subfamily with eubacterial relationships. Gene. 2002;295:51–59. doi: 10.1016/s0378-1119(02)00804-1. [DOI] [PubMed] [Google Scholar]
- Slamovits CH, Keeling PJ. Pyruvate-phosphate dikinase of oxymonads and parabasalia and the evolution of pyrophosphate-dependent glycolysis in anaerobic eukaryotes. Eukaryot Cell. 2006;5:148–154. doi: 10.1128/EC.5.1.148-154.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MD, Prigge ST. Lipoic acid metabolism in microbial pathogens. Microbiol. Mol Biol Rev. 2010;74:200–228. doi: 10.1128/MMBR.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stairs CW, Eme L, Brown MW, Mutsaers C, Susko E, Dellaire G, Soanes DM, van der Giezen M, Roger AJ. A SUF Fe-S cluster biogenesis system in the mitochondrion-related organelles of the anaerobic protist Pygsuia. Curr Biol. 2014;24:1176–86. doi: 10.1016/j.cub.2014.04.033. [DOI] [PubMed] [Google Scholar]
- Stairs CW, Roger AJ, Hampl V. Eukaryotic pyruvate formate lyase and its activating enzyme were acquired laterally from a Firmicute. Mol Biol Evol. 2011;28:2087–2099. doi: 10.1093/molbev/msr032. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechmann A, Hamblin K, Perez-Brocal V, Gaston D, Richmond GS, van der Giezen M, Clark CG, Roger AJ. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr Biol. 2008;18:580–585. doi: 10.1016/j.cub.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. DendroPy: a Python library for phylogenetic computing. Bioinformatics. 2010;26:1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- Tachezy J, Dolezal P. Iron-sulfur proteins and iron-sulfur cluster assembly in organisms with hydrogenosomes and mitosomes. In: Martin WF, Müller M, editors. Origin of mitochondria and hydrogenosomes. Berlin: Springer; 2007. pp. 105–133. [Google Scholar]
- Tovar J, Fischer A, Clark CG. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- Tovar J, Leon-Avila G, Sanchez LB, Sutak R, Tachezy J, van der Giezen M, Hernandez M, Müller M, Lucocq JM. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426:172–176. doi: 10.1038/nature01945. [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Cox S, Tovar J. The iron-sulfur cluster assembly genes iscS and iscU of Entamoeba histolytica were acquired by horizontal gene transfer. BMC Evol Biol. 2004 doi: 10.1186/1471-2148-4-7. 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BAP, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- Yarlett N, Hackstein JHP. Hydrogenosomes: one organelle, multiple origins. BioScience. 2005;55:657–667. [Google Scholar]
- Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl 2):ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- Zimorski V, Major P, Hoffmann K, Bras XP, Martin WF, Gould SB. The N-terminal sequences of four major hydrogenosomal proteins are not essential for import into hydrogenosomes of Trichomonas vaginalis. J Eukaryot Microbiol. 2013;60:89–97. doi: 10.1111/jeu.12012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.