Abstract
Cyclins Bl and B2 are frequently elevated in human cancers and are associated with tumour aggressiveness and poor clinical outcome; however, whether and how B-type cyclins drive tumorigenesis is unknown. Here we show that cyclin Bl and B2 transgenic mice are highly prone to tumours, including tumour types where B-type cyclins serve as prognosticators. Cyclins Bl and B2 both induce aneuploidy when overexpressed but through distinct mechanisms, with cyclin Bl inhibiting separase activation, leading to anaphase bridges, and cyclin B2 triggering aurora-A-mediated Plkl hyperactivation, resulting in accelerated centrosome separation and lagging chromosomes. Complementary experiments revealed that cyclin B2 and p53 act antagonistically to control aurora-A-mediated centrosome splitting and accurate chromosome segregation in normal cells. These data demonstrate a causative link between B-type cyclin overexpression and tumour pathophysiology, and uncover previously unknown functions of cyclin B2 and p53 in centrosome separation that may be perturbed in many human cancers.
Genomic instability provides the genetic diversity through which preneoplastic cells acquire the capabilities to become tumorigenic and eventually malignant. Loss of genomic integrity in human tumours involves both nucleotide changes and chromosomal instability (CIN; refs 1-3). CIN increases the rate of numerical or structural karyotypic change and promotes formation of cells with an abnormal chromosome content, a state defined as aneuploidy. As many as 500 human genes may cause CIN when defective, but which of these so-called CIN genes are deregulated in human tumours and responsible for the chromosomal heterogeneity associated with cancer lethality remains unknown4-6. To identify such CIN gene alterations we focused on two related genes, Ccnbl and Ccnb2, whose overexpression is a characteristic of multiple molecular signatures that predict tumour aggressiveness and poor prognosis in a broad spectrum of cancers7-10.
Human cyclins B1 and B2 have unique amino-terminal segments but are highly homologous over the last 300 residues11-13. Both proteins activate cyclin-dependent kinase 1 (Cdk1) and are expressed in most cell lines, tissues and organs with a high mitotic index14,15. B-type cyclin levels are low in G1, gradually increase during S and G2, and peak during mitosis16,17. Cdk1–cyclin B1 activation at the G2/M transition initiates the phosphorylation of various proteins that control key early mitotic events, including chromosome condensation, nuclear envelope breakdown and spindle assembly18,19. Furthermore, Cdk1–cyclin B1 coordinates anaphase onset by controlling the activity of separase, a protease that cleaves cohesion complexes that hold sister chromatids together 20. By phosphorylating separase, Cdkl–cyclin Bl prevents cohesin cleavage until proper attachment of the last kinetochore to spindle microtubules in metaphase silences the mitotic checkpoint and activates APC/CCdG20, an E3 ubiquitin ligase that targets cyclin Bl for proteasomal degradation. Little is known about cyclin B2 other than that it is localized to centriolar satellites in somatic cells and implicated in spindle formation in Xenopus oocytes21–24. Here, we show that both B-type cyclins cause chromosome missegregation and tumour formation when overexpressed in mice. Furthermore, we show that endogenously expressed cyclin B2 and the p53 tumour-suppressor protein act antagonistically on aurora A to control proper timing of centrosome separation and correct chromosome segregation.
RESULTS
Cyclin Bl and B2 transgenic mice become aneuploid through distinct segregation errors
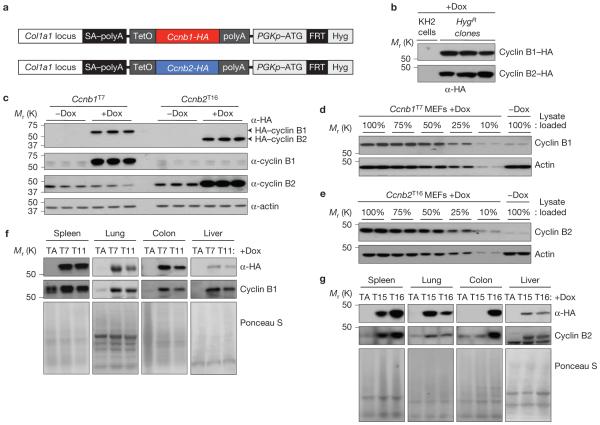
To explore whether and how overexpressed B-type cyclins promote tumorigenesis, we generated doxycycline (dox) inducible cyclin Bl and B2 transgenic mice using site-directed integration in KH2 embryonic stem (ES) cells25,26 (Fig. 1a,b). Two cyclin B1 (Ccnb1T7 and Ccnb1T11) and two cyclin B2 (Ccnb2T15 and Ccnb2T16) transgenic lines obtained were further characterized. Western blot analysis of mouse embryonic fibroblast (MEF) lysates confirmed that dox tightly controlled transgene expression (Fig. 1c), with dox-treated Ccnb1T7 and Ccnb2T16 MEFs showing ~10-fold cyclin B1 and ~20-fold cyclin B2 overexpression, respectively (Fig. 1d,e). Dox-treated animals expressed considerably more transgenic than endogenous protein in a broad spectrum of tissues, although the fold overexpression varied slightly among tissues and transgenic lines (Fig. 1f,g and Supplementary Fig. 1a–c).
Figure 1.
Ccnbl and Ccnb2 transgene production and expression analyses. (a) Generation of wild-type Ccnbl and Ccnb2 transgenic mice: schematic representation of the transgenic strategy. Using recombination at Flp recognition target (FRT) sites, DNA sequences encoding mouse wild-type cyclin Bl or cycl in B2 were inserted downstream of the 3′ untranslated region of the Co/lal locus in KH2 ES cells. SA-polyA, splice acceptor-polyA; Teto, tetracycline operator; PGKp, phosphoglycerate kinase promoter; ATG, start codon. (b) Western blots of lysates from hygromycin B-resistant KH2 ES clones with targeted insertion of HA-Ccnbl or HA-Ccnb2 probed for haemagglutinin (HA). (c) Western blots of extracts from HA-Ccnb1 and HA-Ccnb2 MEFs grown in the presence or absence of dox for 72 h. (d,e) Quantitation of western blots for cyclin B1 or cyclin B2 overexpression (fold change) in MEFs derived from HA-Ccnb1T7 or HA-Ccnb2T16 transgenic strains. (f,g) Western blot analysis of tissue extracts from six-week-old dox-treated HA-Ccnb1 (strains T7 and T11), HA-Ccnb2 (strains T15 and T16) and control (TA) transgenic mice. Ponceau S staining of blotted proteins served as a loading control. Blots in c are representative for three independent experiments, and blots in b and d-g for two independent experiments. Uncropped images of blots are shown in Supplementary Fig. 9.
Late in G2, cyclin Bl raises Cdkl kinase activity above a threshold level required for mitotic entry20 . At mid-mitosis, this burst of Cdkl activity is rapidly eliminated through proteosomal degradation of cyclin B 1 to initiate sister chromatid separation and anaphase onset. Consistent with this, endogenous cyclin Bl levels of Ccnb1T7 MEFs cultured without dox were low in Gl, sharply increased from G2 until prometaphase, and then progressively declined in meta phase and anaphase (Supplementary Fig. 2a,b). This pattern of expression was maintained on transgene induction, although absolute cyclin Bl levels were now substantially increased at each mitotic stage. Cdk activity, as measured using an antibody against phosphorylated Cdk substrates, changed in accordance with cyclin Bl levels and was thus elevated in dox-treated Ccnb1T 7 MEFs during mitosis (Supplementary Fig. 2c,d). The temporal expression pattern of endogenous cyclin B2 was similar to that of cyclin Bl (Supplementary Fig. 3a,b). However, cyclin B2 levels and Cdk activity in dox-treated Ccnb2T16 cells were elevated not only in mitosis, but also in Gland G2 (Supplementary Fig. 3a-d).
To determine whether cyclin Bl and cyclin B2 overexpression might affect karyotype stability, we analysed metaphase spreads of MEFs cultured with or without dox for 48 h. Without dox, aneuploidy rates of Ccnb1T17, Ccnb1T11, Ccnb2T15 and Ccnb2T166 MEFs were 12, 15, 15 and 16%, respectively (Table 1a). In contrast, on transgene induction, these rates markedly increased to 33, 31, 25 and 36%, respectively. Both near-diploid and near-tetraploid aneuploidies were elevated in cyclin-B 1-overexpressing MEFs. To measure in vivo-acquired aneuploidy, we counted chromosomes of splenocytes from 5-month-old Ccnb1T7, Ccnblm, Ccnb2T15 and Ccnb2T16 mice maintained on dox after weaning. Splenocytes from dox-treated transactivator (TA) mice served as controls and contained 6% aneuploidy (Table lb). Aneuploidy substantially increased on transgene induction, with Ccnb1T7, Ccnb1T11, Ccnb2T15 and Ccnb2T16 splenocytes with 20, 12, 18and 18% aneuploidy, respectively (Table 1b).
Table 1.
B-type cyclin overexpression induces aneuploidy.
| (a) Overexpression of B-type cyclins in MEFs leads to increased aneuploidy | ||||||||||||||
| MEF genotype (n) |
Mitotic
cells inspected |
%
aneuploid figures (s.d.) |
%
(near) tetraploid figures (s.d.) |
Karyotypes with the indicated chromosome number
|
||||||||||
| 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 71–79 | 80 | >80 | ||||
|
| ||||||||||||||
| Ccnb1T7 −Dox (4) | 200 | 12(3) | 4(4) | 1 | 4 | 9 | 174 | 3 | 1 | 6 | 2 | |||
| Ccnb1T7 +Dox (4) | 200 | 33(7)** | 11(3)* | 4 | 10 | 21 | 131 | 8 | 3 | 1 | 13 | 3 | ||
| Ccnb1T11 −Dox (3) | 150 | 15(3) | 6(5) | 4 | 3 | 7 | 125 | 1 | 1 | 4 | 2 | |||
| Ccnb1T11 +Dox(3) | 150 | 31(4)** | 17(5) | 3 | 6 | 10 | 96 | 5 | 4 | 15 | 8 | 3 | ||
| Ccnb2T15 −Dox (3) | 150 | 15(3) | 5(3) | 5 | 6 | 123 | 4 | 3 | 2 | 2 | 4 | 1 | ||
| Ccnb2T15 +Dox (3) | 150 | 25(1)** | 8(2) | 1 | 4 | 10 | 108 | 12 | 2 | 1 | 4 | 5 | 3 | |
| Ccnb2T16 −Dox (3) | 150 | 16(3) | 7(1) | 4 | 1 | 2 | 3 | 122 | 3 | 3 | 1 | 7 | 4 | |
| Ccnb2T16 −Dox (3) | 150 | 36(8)* | 10(2) | 3 | 2 | 8 | 19 | 91 | 11 | 11 | 8 | 5 | 2 | |
|
| ||||||||||||||
| (b) Splenocytes of transgenic mice with high levels of cyclin B1 or B2 are prone to aneuploidization | ||||||||||||||
| Mouse genotype (n) |
Mitotic
cells inspected |
%
aneuploid figures (s.d.) |
% (near)
tetraploid figures (s.d.) |
Karyotypes with the indicated chromosome number
|
||||||||||
| 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 79 | 80 | ||||
|
| ||||||||||||||
| TA control (3) | 150 | 6(2) | 0 | 3 | 5 | 141 | 1 | |||||||
| Ccnb1T7 (3) | 150 | 20(3)** | 1(1) | 4 | 2 | 4 | 9 | 119 | 8 | 2 | 1 | 1 | ||
| Ccnb1T11 (3) | 150 | 12(0)** | 0 | 5 | 3 | 4 | 2 | 132 | 1 | 2 | 1 | |||
| Ccnb2T15 (3) | 150 | 18(4)** | 0 | 3 | 5 | 4 | 7 | 123 | 5 | 3 | ||||
| Ccnb2T16 (3) | 150 | 18(3)** | 0 | 2 | 7 | 9 | 123 | 5 | 3 | 1 | ||||
Asterisks in a mark values statistically different from −dox counterparts. Asterisks in b mark values statistically different from dox-treated transactivator (TA) control values.
P<0.05;
P<0.01.
In live-cell imaging experiments, dox-treated Ccnb1T7 and Ccnb2T16 MEFs both showed markedly increased chromosome missegregation rates when compared with untreated counterparts (Fig. 2a,b). However, the main mitotic errors resulting from each transgene showed no substantial overlap, with cyclin Bl overexpression inducing chromatin bridges or anaphase failure, and cyclin B2 overexpression causing lagging chromosomes (Fig. 2a,b). Thus, although the extents to which cyclin Bl and B2 perturb the fidelity of chromosome segregation are very similar, the fundamental basis for the inaccuracies is unique in each case.
Figure 2.
B-type cyclins induce distinct chromosome missegregation when overexpressed. (a) Representative images of cells with indicated chromosome segregation errors. Scale bar, 10μm. (b) Live-eel I imaging analysis of chromosome segregation errors in Ccnb1r7 and Ccnb2n6 transgenic primary MEFs with and without transgene induction for 72 h. Anaphase failure refers to complete failure of sister chromatid separation producing one daughter with 4N DNA and one with out DNA. The analysis was carried out on three independent MEF I ines per genotype (>20 cells per line were analysed). Statistical significance was determined by a two-tailed, unpaired t-test. Statistics source data are provided in Supplementary Table 2. *P < 0.05; **P <0.01. (c) Schematic representation of the separase activity biosensor (top) and time-lapse images of biosensor-positive Ccnb1r7 MEFs progressing through mitosis in the absence or presence of transgenic eye I in Bl. hSCCl, human sister chromatid cohesion protein l. Sea le bars, 5μm. (d) Quantification of biosensor fluorescence at chromosomes as cells progress through mitosis. Data represent mean ± s.e.m. from five independent experiments. We note that the level of overexpression varied among dox-treated Ccnbzr7 MEFs, with those showing anaphase bridges expressing significantly higher levels of HA-cyclin B1 than those without bridges.
Cyclin Bl overexpression perturbs separase activation
The anaphase failure and chromatin bridging phenotypes of cyclin B1 transgenic cells were reminiscent of the mitotic defects of cells lacking theseparasen 27,28 . This led to the idea that cyclin B1-Cdk1, which inhibits separase through phosphorylation at Ser 1121 (ref. 29) and is hyperactive when cyclin B1 is overexpressed (Supplementary Fig. 2c,d), might interfere with the timely activation of separase on chromosome bi-orientation. To test this hypothesis, we monitored separase activity in live cells using a biosensor consisting of two separase cleavage sites flanked by mCherry-histone H2B and enhanced green fluorescent protein (eGFP) at the amino- and carboxy-terminal ends, respectively (Fig. 2c). In non-induced Ccnb1T7 MEFs, this biosensor resided at chromatin during interphase and the first part of mitosis, producing a yellow colour in merged images of eGFP and mCherry fluorescence (Fig. 2c). Cleavage of the biosensor on separase activation in late metaphase released eGFP from chromosomes, thus converting yellow to red. Quantitation of eGFP fluorescence at chromosomes revealed that activation of separase starts 6-8 min before onset of anaphase (Fig. 2d). A similar pattern of Seel cleavage was observed in dox-induced Ccnb1T7 MEFs undergoing accurate chromosome separation (Fig. 2c,d). In contrast, chromosomes of cells with anaphase bridges, which we find have higher levels of cyclin B1 overexpression than their counterparts without bridges (Supplementary Fig. 4a), released only relatively small amounts of eGFP, remaining yellow throughout anaphase and telophase. Cells experiencing anaphase failure showed even less separase activity than those with bridges (Fig. 2d), indicating that the severity of mitotic phenotypes inversely correlates with separase activity. Biosensor cleavage was normal in dox-treated Ccnb2T16 MEFs (Supplementary Fig. 4b,c).
Incomplete decatenation of centromeric DNA and replication fork stalling are alternative causes of chromatin bridge formation30,31. Top2a, the decatenase that disentangles centromeric DNA, was normally expressed and properly targeted to inner centromeres of mitotic MEFs with elevated cyclin B1 (Supplementary Fig. 4d,e). The number of stalled replication forks, as determined by immunostaining for the replication protein RPA2 (ref 30), was not increased in cyclin B1 overexpressing MEFs (Supplementary Fig. 4f). These MEFs also showed no evidence for increased double strand DNA breaks (Supplementary Fig. 4g). Thus, DNA replication and decatenation problems seemed to play a limited or no role in the chromatin bridging phenotype. Timing of S/G2, prophase, prometaphase and metaphase also seemed normal in cyclin B1 overexpressing MEFs (Supplementary Fig. 4h,i). Classical growth rate analysis revealed a slight, yet significant, reduction in cell number at high cyclin B!levels (Supplementary Fig. 4j), which is consistent with the modest rate of anaphase failure observed in these cells (Fig. 2b).
Cyclin B2 overexpression hyperactivates Plkl through aurora A to deregulate mitosis
Cyclin B2 overexpression primarily causes lagging chromosomes. Although this type of segregation error has been associated with mitotic checkpoint failure32, no such defect was observed (Supplementary Fig. 5a). Lagging chromosomes are due to merotelic attachments, a type of kinetochore-microtubule malattachment that can be caused by defects in attachment error correction, microtubule dynamics or spindle geometry33-36. However, monastrol washout assays indicated that correction of merotelic attachments was unperturbed in cyclin B2 overexpressing cells (Supplementary Fig. 5b). Microtubule dynamics also seemed normal in these cells, as measured using photo-activatable GFP-cHubulin (Supplementary Fig. 5c). To evaluatespindle geometry, Ccnb2T16 MEFs were stained for γ- and cHubulin. At high cyclin B2 levels, there were noticeably more metaphases in which the spindle poles were not lying perpendicular to the metaphase plate (Fig. 3a). We quantitated this defect by determining the percentage of cells that had an acute angle between the spindle pole axis and the metaphase plate ofless than 85° (Fig. 3a). According to these criteria, 44% of dox-induced Ccnb2Tl6 metaphases were abnormal, compared with 23% of control metaphases (Fig. 3b). Cyclin B1 overexpression was not associated with spindle geometry defects (Fig. 3b).
Figure 3.
Cyclin B2 overexpression causes aurora-A-mediated hyperactivation of Plk1. (a) Maximum intensity projection images of Ccnb2n6 metaphases immunostained for a.-tubu lin (red) and γ-tubulin (green). (b) Incidence of spindle geometry abnormalities. (c) Images of MEFs in G2 immunosta ined for γ-tubulin (visualizing centrosomes) and pH3510 (phosphorylated histone H3, used as a marker for G2). (d) Quantitation of accelerated centrosome separation. Cells with centrosome distances ≥ 3 μm were classified as accelerated. (e) Images of cells immunosta ined for p-Plkl1210 , γ-tubulin and pH3510 . Primary antibodies for γ-tubulin and pH3510 were both raised in rabbits and were visualized with the same fluorophore-conjugated secondary antibodies. This did not create a detection conflict because γ-b.Jbul in and pH3510 are confined to the cytoplasm and nucleus, respectively. (f) Quantitation of p-Plkl1210 signa I at centrosomes in Ccnb2n6 MEFs. (g) Western blots of mitotic extracts from Ccnb2n6 MEFs grown in the presence or absence of dox for 72 h. Taxol and okada ic acid were added to the culture medium 5 h before cell harvest. Blots are representative for two independent experiments. Uncropped images of blots are shown in Supplementary Fig. 9. (h) Qua ntitation of metaphases with spindle geometry abnormalities in the presence or absence of 10nM 812536. (i) Images of prophases immunostained for p-aurora and γ-tubu lin. P-aurora is a pan-aurora antibody: signal at spindle poles represents p-aurora A and signa I at inner centromeres is p-aurora B. (j) Quantification of p-aurora signa I at centrosomes in Ccnb2Tl6 MEFs at the indicated stages of mitosis. (k) The same as h except with 10nM MLN8054. (I) The same as d except with 10 nM MLN8054. DNA in a,c,e,i was visualized with Hoechst. Arrows mark centrosomes. Scale bars, 5 μm in a and 10μm in c,e,i. Quantifications in b,d ,f ,h and j-1 were done on three independent MEF cell lines (per Iine, 20 cells in b,h ,k and I, 10 eel Is in f and j, and >20 cells in d, were analysed). Data represent mean± s.e.m. Statistical significance was determined by a two-tailed, unpaired I-test. Statistics source data are provided in Supplementary Table 2. *P < 0.05, **P <0.01, ***P <0.001.
Abnormal spindle geometry has been linked to defects in centrosome separation37. As a first step to probe for such defects, we assessed whether cyclin B2 is centrosome associated. Indeed, cyclin B2 co-localized with centrin 2, a well-established centrosomal marker protein, both before and after mitosis onset (Supplementary Fig. 5d,e). Next, we stained dox-treated and untreated Ccnb2Tl6 MEFs for γ-tubulin to determine the timing of centrosome separation in G2 phase, and found that overexpression of cyclin B2 significantly accel-erated centrosome separation (Fig. 3c,d and Supplementary Fig. 5f). To decipher the underlying mechanism, we focused on Plkl, a mitotic kinase that plays a central role in disengagement and full separation of centrosomes38 . Plkl activity typically increases in late G2, resulting in phosphorylation and activation ofMst2, which in turn phosphory-lates and activates Nek2A6768 . Activated Nek2A then phosphorylates C-Napl and rootletin, two proteins that hold duplicated centrosomes, resulting in centrosome separation69,70. In cooperation with Cdkl, Plkl also regulates recruitment of the kinesin EgS to centrosomes, driving the full separation of centrosomes in prophase40 . To evaluate whether cyclin B2 overexpression alters Plkl activity at centrosomes, we stained Ccnb2T16 MEFs with an antibody specific for Plkl containing the activating phosphorylation on Thr 210 (ref. 41) (Fig. 3e). Indeed, centrosomal p-Plk1Tzto staining was nearly twice as high in dox-treated MEFs as in their untreated counterparts (Fig. 3f,g).
To determine whether the observed Plkl hyperactivity might drive the mitotic defects in cyclin B2 overexpressing cells, we used low amounts of a small-molecule inhibitor of Plkl, BI2S36, to normalize Plkl activity in dox-treated Ccnb2T16 MEFs. As shown in Supplementary Fig. 5g, 10nM BI2S36 corrected the accelerated centrosome separation phenotype of cells overexpressing cyclin B2 and had no impact in cells in which the transgene was not induced. Furthermore, spindle geometry defects in dox-treated Ccnb2T16 MEFs declined from 48 to 1S% (Fig. 3h), and chromosome missegregation rates decreased from 36 to 14% (Table 2a). Consistent with improved chromosome segregation, aneuploidy rates dropped from 39 to 21% (Table 2b). Importantly, 10 nM BI2S36 had no impact on spindle geometry, chromosome segregation or aneuploidy rates of Ccnb2T16 MEFs not expressing the transgene (Fig. 3h and Table 2a,b). These findings indicated that the mitotic phenotypes of cyclin B2 overexpressing cells were due to Plkl hyperactivity.
Table 2.
Chromosome missegregation in cyclin-B2-overexpressing MEFs is due to aurora A and PIk1 hyperactivity.
| (a) Normalization of PIk1 activity in cyclin-B2-overexpressing MEFs restores accurate chromosome segregation | |||||||
|---|---|---|---|---|---|---|---|
| MEF genotype (n) | Mitotic cells inspected |
% cells with segregation defects (s.e.m.) |
% metaphases with misaligned chromosomes (s.e.m.) |
% anaphases with chrom. bridges (s.e.m.) |
% anaphases with lagging chromosomes (s.e.m.) |
% anaphase failure (s.e.m.) |
% cytokinesis defects (s.e.m.) |
| Ccnb2T16 −Dox (3) | 65 | 13(1) | 1(1) | 5(3) | 4(2) | 2(2) | 0(0) |
| Ccnb2T16 +Dox (3) | 65 | 36(10)* | 7(4) | 9(3) | 15(4) | 3(3) | 2(2) |
| Ccnb2T16 −Dox (3) + BI | 69 | 19(3) | 0(0) | 10(3) | 7(1) | 2(2) | 0(0) |
| Ccnb2T16 +Dox (3) + BI | 77 | 14(1) | 3(1) | 6(2) | 3(3) | 3(1) | 0(0) |
| (b) Cyclin-B2-overexpressing MEFs in which PIk1 activity is normalized are not prone to aneuploidy | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MEF genotype (n) | % aneuploid figures (s.e.m.) |
% (near) tetraploid figures (s.e.m.) |
Karyotypes with the indicated chromosome number |
||||||||||||
| 37 | 38 | 39 | 40 | 41 | 42 | 43 | 77 | 78 | 79 | 80 | 81 | 82 | |||
| Ccnb2T16−Dox (3) | 18(1) | 7(2) | 2 | 5 | 5 | 118 | 6 | 3 | 1 | 2 | 2 | 5 | 1 | ||
| Ccnb2T16 + Dox (3) | 39(1)*** | 7(1) | 3 | 2 | 22 | 89 | 13 | 6 | 4 | 1 | 2 | 5 | 3 | ||
| Ccnb2T16 −Dox(3)+BI | 21(1) | 8(0) | 3 | 5 | 9 | 114 | 7 | 4 | 3 | 1 | 4 | ||||
| Ccnb2T16 +Dox(3)+BI | 21(2) | 7(1) | 2 | 6 | 11 | 112 | 7 | 1 | 1 | 1 | 1 | 6 | 2 | ||
| (c) Normalization of aurora A activity in cyclin-B2-overexpressing MEFs prevents aneuploidy | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MEF genotype (n) | % aneuploid figures (s.e.m.) |
% (near) tetraploid figures (s.e.m.) |
Karyotypes with the indicated chromosome number |
|||||||||||||||
| 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | |||
| Ccnb2T16 −Dox (3) | 17(1) | 8(1) | 1 | 2 | 8 | 119 | 5 | 1 | 1 | 1 | 1 | 2 | 2 | 6 | 1 | |||
| Ccnb2T16 +Dox (3) | 34(3)** | 11(1) | 5 | 5 | 14 | 96 | 12 | 2 | 3 | 2 | 3 | 3 | 4 | 1 | ||||
| Ccnb2T16 −Dox(3)+MLN | 23(1) | 8(1) | 5 | 4 | 13 | 109 | 6 | 1 | 1 | 2 | 6 | 2 | 1 | |||||
| Ccnb2T16 +Dox(3)+MLN | 22(2) | 8(3) | 4 | 5 | 11 | 112 | 5 | 1 | 1 | 1 | 1 | 2 | 5 | 2 | ||||
Asterisks mark values statistically different from −dox counterparts.
P<0.05;
P<0.01;
P<0.001.
Aurora A kinase activates centromeric Plkl through phosphorylation of Thr 210 (refs 42-44), which prompted us to test whether aurora A hyperactivity is responsible for hyperactivation of Plkl in cyclin B2 overexpressing cells. When stained with an antibody specific for phosphorylated aurora proteins, which visualizes activated aurora A at centrosomes, cyclin B2 overexpressing MEFs showed markedly increased fluorescence in prophase (Fig. 3i,j). No such increase was observed in cyclin B1 transgenic MEFs (Supplementary Fig. 5h). Low amounts (lOnM) of the aurora A inhibitor MLN80S4 normalized p-PlklT210 levels and corrected the spindle geometry, centrosome separation and chromosome segregation defects in cells overexpressing cyclin B2 (Fig. 3k,l, Table 2c and Supplementary Fig. 5i,j). Collectively, these data indicate that cyclin B2 overexpression promotes Plk1 hyperactivity through hyperactivation of aurora A. Notably, Plk1 has been implicated in several other mitotic processes, including mitotic spindle positioning45 and attachment error correction46, but these functions seemed unperturbed by cyclin B2 overexpression (Supplementary Fig. 5k–m).
Endogenous cyclin B2 and p53 antagonistically regulate centrosome separation
To explore whether endogenous cyclin B2 regulates centrosome separation in normal cells, we depleted cyclin B2 from wild-type MEFs using two independent short hairpin RNAs (shRNAs). Low cyclin B2 markedly increased the percentage of G2 cells with unseparated centrosomes (Fig. 4a–c). Importantly, cyclin B2 depletion significantly reduced centrosomal levels of phosphorylated Plk1 and aurora A, and increased rates of asymmetrical mitotic spindles and chromosome lagging (Fig. 4d–h). Centrosome nondisjunction on cyclin B2 depletion was also observed in primary epithelial cell lines established from murine lung and mammary gland tissue (Fig. 4i). The same holds true for human primary fetal lung fibroblasts (IMR-90) and adult human primary skin fibroblasts (Fig. 4j,k), indicating that cyclin B2 functions in centrosome separation across mammalian species. Cyclin B2 loss, however, did not impair centrosome separation in HeLa cells (Fig. 4k).
Figure 4.
Endogenous eye Iin B2 regulates aurora-A-mediated centrosome separation. (a-h) Wild-type (WT) MEFs transduced with cycl in B2 shR NA or non-targeting sh RNA negative control lentiviruses and analysed 72 h later. (a) Western blot analysis for eye Iins B1 and B2. (b) lmmunostain ing for cycl in B2 and γ-tubulin. (c) Quantification of cells with unseparated centrosomes (<l μml in G2. (d) G2 cells immunostained for p-Plkl1210 , γ-b.Jbulin and pH3510 . We note that p-Plkl1210 levels at centrosomes of cyclin B2 overexpressing cells were increased irrespective of inter-centrosome distance. (e) Quantification of p-Plkl1210 signa I at centrosomes. (f) Quantification of centrosomal p-aurora A signa Is in prophase. p-aurora A levels at centrosomes of eye Iin B2 over expressing eel Is were increased irrespective of intercentrosome distance. (g) Analysis of spindle geometry. (h) Analysis of chromosome segregation errors by live-cell imaging. The analysis was carried out on three independent MEF lines (15-22 cells were analysed per Iine). Statistical significance was determined by a one-tailed, unpaired t-test. (i) The same as c for mouse epithelial cells. (j,k) Different human cell lines transduced with cyclin B2 shR NA or non-targeting shR NA negative control lentiviruses and analysed 72 h later (HF, human skin fibroblasts). (j) Western blot analysis of human skin fibroblast lysates for cyclins B1 and B2. (k) Quantification of cells with unseparated centrosomes in G2 phase. Colour codes in e-g and i are as in c. DNA in b and d was visualized with Hoechst. Sea le bars in b and d, 10 μm. Quantifications in c,e-g and k were done on three independent cell lines (per line, 20 cells in c,g and k and 10 cells in e and f were analysed). Quantifications in i were done on the same line in triplicate (20 cells for each analysis). Data represent mean± s.e.m. Statistical significance was determined by a two-tailed, unpaired I-test inc,g,i and k and by a one sam pie I-test against a theoretica Imean of unity in eand f. *P < 0.05; **P < 0.01. Statistics source data are provided in Supplementary Table 2. Blots in a and j are representative for three independent experiments. Uncropped images of blots are shown in Supplementary Fig. 9.
To investigate whether this might be due to cell transformation, we immortalized wild-type MEFs by expressing SV40 large T antigen, which perturbs p53 and RB function47. Indeed, cyclin B2 depletion failed to inhibit centrosome separation in immortalized MEFs (Fig. 5a). Subsequent studies revealed that cyclin B2 depletion does not impair centrosome disjunction in p53−/− MEFs (Fig. 5a). Furthermore, we noticed that the percentage of unseparated centrosomes was significantly lower in p53−/− MEFs than in wild-type MEFs (Fig. 5a). A recent report documented that p53 negatively regulates aurora A transcript and protein abundance48, which we confirmed by quantitative PCR with reverse transcription (qRT–PCR) and western blot analysis of wild-type and p53−/− MEFs (Supplementary Fig. 6a,b). This led us to hypothesize that elevated aurora A expression due to p53 loss increases levels of phosphorylated aurora A. This would lead to increased amounts of p-Plk1T210, accelerated centrosome splitting, abnormal spindle geometry and chromosome missegregation. Consistent with this notion, p53−/− MEFs had significantly higher amounts of p-aurora A and p-Plk1T210 at centrosomes, as well as significantly increased rates of spindle geometry defects and lagging chromosomes (Fig. 5b–f,i). The observation that errors in centrosome separation, spindle geometry and chromosome segregation were largely corrected by culturing p53−/− MEFs in low amounts of aurora A inhibitor further supported our hypothesis (Fig. 5g–i). Centrosomes of cyclin B2-depleted p53−/− MEFs had elevated p-aurora A and p-Plk1T210 levels, indicating that p53 loss leads to cyclin B2- independent aurora A activation (Supplementary Fig. 6c,d). Collectively, these data uncover previously unknown functions for cyclin B2 and p53 in proper timing of centrosome separation to ensure correct spindle pole positioning and chromosome segregation (Fig. 5j).
Figure 5.
Cyclin B2 and p53 antagonistically control aurora-A-mediated centrosome separation. (a) WT, WT SV40 large T antigen-transformed and p53−/− MEFs transduced with cyclin B2 sh RNA or non-targeting shR NA negative control lentiviruses and analysed for G2 cells with unseparated centrosomes (< 1μm). (b) Images of prophases immunosta ined for p-aurora and γ-tubu lin. (c) Quantification of p-aurora signa I at centrosomes in prophase. (d) Incidence of spindle geometry abnormalities. (e) Images of G2 cells immunostained for p-Plk1T210, γ-tubulin and pH3S10. (f) Quantification of p-Plk1T210 signal at centrosomes in G2 cells. (g) Quantitation of cells with unseparated centrosomes in G2 in the presence or absence of 10nM MLN8054. (h) Incidence of spindle geometry abnormalities in the presence or absence of 10nM MLN8054. (i) Analysis of chromosome segregation errors by live-cell imaging. The analysis was carried out on three independent MEF lines (14-26 cells were analysed per line). (j) Model for antagonistic control of the centrosome splitting signalling cascade by p53 and cyclin B2-Cdk1. In this model, p53 functions to limit the amount of activated (phosphorylated) aurora A by inhibiting AURKA gene transcription and promoting proteosomal degradation of aurora A protein. Cyclin B2-Cdk1, on the other hand, triggers the phosphorylation of the residual aurora A protein pool in a timely manner to induce phosphorylation of centrosome-associated Plk1 and permit centrosome disjunction38,67-70. Whether cyclin B2-Cdk1 triggers aurora A activation directly or indirectly remains to be determined. Colour codes in d and f are as in c. Colour code in h is as in g. DNA in b and e was visualized with Hoechst. Scale bars represent 10 μm. Arrows mark centrosomes. p53−/− cells with supernumerary centrosomes were excluded from the analyses in b-h. Quantifications in a,c,d and f-h were done on three independent MEF lines (per line, 20 cells in a,d,g and h and 10 cells in c and f were analysed). Data represent mean ± s.e.m. Statistical significance was determined by a two-tailed, unpaired t-test. Statistics source data are provided in Supplementary Table 2. *P < 0.05; **P <0.01; ***P <0.001.
Cyclins B1 and B2 both drive tumorigenesis when overexpressed
To determine whether cyclin B1 or cyclin B2 overexpression drives tumorigenesis, we established cohorts of Ccnb1, Ccnb2 and TA control mice that were on dox-containing water from weaning on. These mice were killed and screened for tumours at 14 months of age. Tumours were collected and analysed by histopathology. Both B- type cyclin transgenic strains showed marked increases in tumour incidence (Fig. 6a–c). Fluorescence in situ hybridization (FISH) analysis confirmed that the tumours had increased numbers of aneuploid cells (Supplementary Table 1). Lung tumours were the most prevalent tumour type in both models (Fig. 6b,c), which is interesting because several studies have documented elevated cyclin B1 expression in human lung cancer10,49–51. To corroborate these studies and to determine the frequency of cyclin B2 overexpression in human lung cancers, we measured CCNB1 and CCNB2 transcript levels of primary tumour samples of lung cancer patients and compared these with normal lung tissue (Supplementary Fig. 7a,b). CCNB1 transcript levels were more than 10-fold elevated in 43% of adenocarcinomas and 37% of squamous cell carcinomas, whereas CCNB2 transcript levels were more than 10-fold increased in 20% of adenocarcinomas and 23% of squamous cell carcinomas, indicating that subsets of human lung cancers overexpress CCNB1 and CCNB2. Measurements of BUB1 and Ki67 transcript levels confirmed that the observed increases in CCNB1 and CCNB2 expression were not simply a consequence of increased numbers of cycling cells (Supplementary Fig. 7c,d).
Figure 6.
Cyclins B1 and B2 drive tumorigenesis when overexpressed. (a) Spontaneou s tumour incidence of 14-month-old mice. (b) Spectrum of spontaneous tumour types in 14-month-old mice. (c) Representative images and histological analysis of representative spontaneous tumours from transgenic mice Sea le bar, 100μm. (d-g) Overexpressed B-type eye Iins synergize with APC+/min to develop aggressive colonic tumours. Colour codes and cohort sizes in f and g are as indicated in e. (d) Haematoxylin-eosinsta ined sections of colonic polyps in 90-daγ-old mice continuously treated with dox. Enlargements (right) of the Ccnb1; APC+/Min and Ccnb2; APC+/Min tumours show areas where intramucosal invasion has occurred to illustrate that these tumours have a more aggressive tumour phenotype. The TA; APC+/Min tumour is less advanced as it lacks invasive properties. Black and red scale bars are 1mm and 100 μm respectively. (e) Colon tumour incidence of mice that were continuously on dox (3 months (mo) +dox) or taken off dox for the last 30 days of the experiment (2 months +dox and 1 month −dox). (f) Colon tumour burden. (g) Average colon tumour size. (h) Tumour incidence of DMBA-treated mice. (i) Lung and skin tumour burden of DM BA-treated mice. n in a,e and h represents number of mice per genotype. Data represent mean ± s.e.m. in f ,gand i. Statistica I signiiica nee was determined by Fisher's exact test in a and h, a one-tailed chi-squared test in b and e, a two-tailed unpaired t-test in f and g and a two-tailed Mann-Whitney test in i. *P < 0.05; **P<0.01; ***P<0.001.
Next we focused on colon cancer, a human tumour type characterized by elevated B-type cyclin levels52–54. Ccnb1T7, Ccnb2T16 and TA control mice were bred onto a APC+/Min genetic background, which predisposes mice to intestinal tract polyps55, placed on dox water at weaning age and screened for intestinal tumours at day 90. Colon tumour incidence and multiplicity were significantly increased in Ccnb1T7;APC+/Min and Ccnb2T16;APC+/Min mice when compared with TA;APC+/Min control mice (Fig. 6d–f). Furthermore, colon tumours from Ccnb1T7;APC+/Min and Ccnb2T16;APC+/Min mice were significantly larger than those of TA;APC+/Min mice, and showed marked intramucosal invasion (Fig. 6d–g), indicating that B-type cyclin overexpression promotes a more aggressive tumour phenotype. There were no significant differences in small intestinal tumour multiplicity between the various cohorts (Supplementary Fig. 8a). When we repeated the above experiments but stopped dox treatment 30 days before tumour analysis, colon tumour incidence, multiplicity and size did not significantly decline when compared with counterparts that were continuously treated with dox (Fig. 6e– g). Tumour histopathology was similar regardless of dox treatment regimen (Supplementary Fig. 8b). These data indicate that cyclin B1 and B2 overexpression is important for initial tumour formation but dispensable for continued cancer cell viability and tumour growth in this model.
The carcinogen 7,12-dimethylbenz(a)anthracene (DMBA) predis- poses wild-type mice to lung and skin tumours when it is applied to the dorsal skin at low dose31. We found that the occurrence of DMBA- induced skin tumours was significantly higher in cyclin B1 and B2 transgenic mice than in TA control mice (Fig. 6h). However, DMBA- induced lung tumours were only elevated in cyclin B2 transgenic mice. The observation that both lung and skin tumour multiplicity were elevated in cyclin B2 transgenic mice underscored that cyclin B2 is the more potent proto-oncogene in this tumour model (Fig. 6i).
DISCUSSION
Cyclins B1 and B2 are frequently elevated in human cancers and are associated with tumour aggressiveness and poor clinical outcome. Here, we demonstrate that both these cyclins robustly promote tumour development when overexpressed in mice. Analysis of MEFs from these mice revealed that each cyclin perturbs chromosome segregation through a distinct mechanism, with cyclin B1 inhibiting separase activation and cyclin B2 accelerating aurora-A-mediated Plk1 activation and centrosome separation. Our data strongly indicate that cyclin B1 overexpression induces chromatin bridging through continued inhibition of separase after chromosome biorientation and mitotic checkpoint silencing have occurred. However, contributions of other mechanisms cannot be fully excluded, given the complex nature of the sister chromatid separation process and the multitude of mitotic substrates that cyclin B1–Cdk1 is thought to phosphorylate.
The role of endogenous cyclin B2 has remained elusive for decades, but the collective results from the cyclin B2 knockdown and overexpression studies presented here now indicate that cyclin B2– Cdk1 acts to initiate centrosome separation in late G2 by triggering the phosphorylation of aurora A. Overexpression of cyclin B2 accelerates this process, resulting in early centrosome disjunction and formation of aberrant spindles that tend to separate chromosomes inaccurately. Although activated aurora A is readily detectable at centromeres in prophase, we were unable to demonstrate this during G2. One interpretation of this result is that aurora-A-mediated Plk1 activation in G2 does not require the activator to be stably centrosome bound. The same could hold true for cyclin B2-mediated activation of aurora A, although our finding that cyclin B2 localizes to centrosomes in G2 leaves open the possibility that the centrosome-associated pool of cyclin B2 establishes a local increase in soluble aurora A activity.
Three observations reported here indicate that p53 mediates proper timing of centrosome disjunction by negatively regulating aurora A kinase activity. First, cells lacking p53 are prone to rapid centrosome separation in G2 phase. Similarly to cyclin B2 overexpression, p53 loss results in formation of asymmetrical spindles that promote chromosome lagging. Second, p53 null cells have increased amounts of phosphorylated aurora A and Plk1 at centrosomes. Third, normalization of aurora A activity in p53 null cells with a low amount of aurora A inhibitor restores timely centrosome separation, spindle geometry and accurate chromosome segregation. The implication of these findings is that tumours with p53 defects, which represent a relatively large fraction of human tumours, may acquire chromosomal plasticity that promotes tumour formation through a mechanism that involves aberrant centrosome segregation. Collectively, our data best fit a model whereby p53 and cyclin B2–Cdk1 act antagonistically to control aurora-A-mediated centrosome disjunction, with p53 keeping aurora A protein levels in check through transcriptional and post- translational inhibitory mechanisms and cyclin B2–Cdk1 activating the residual aurora A pool in a timely manner (Fig. 5j). Our data further indicate that p53 inactivation bypasses cyclin B2-dependent initiation of centrosome disjunction, whereas cyclin B2 overexpression overrides p53-mediated inhibition of aurora A kinase activity.
METHODS
Methods and any associated references are available in the online version of the paper.
METHODS
Mouse strains
All mice were housed in a pathogen-free barrier environment. Mouse protocols were reviewed and approved by the Mayo Clinic Institutional Animal Care and Use Committee. All animals were mixed in a 129/Sv x C57BL/6 genetic background. Ccnb1–HA and Ccnb2–HA transgenic mice were generated using KH2 ES cells (Origene Technologies) according to a previously described method26. Briefly, wild-type Ccnb1 and Ccnb2 complementary DNA sequences with an HA tag at the 3′ end were PCR-amplified from established murine cDNA clones11 and cloned into the EcoRI site of Flp-in vector pBS31. The resulting plasmids were electroporated into KH2 ES cells with pCAGGS-FLPe-puro to target integration of Ccnb1–HA and Ccnb2–HA to an Flp recognition target site downstream of Col1a1. KH2 ES clones properly expressing Ccnb1–HA or Ccnb2–HA were injected into blastocysts, and chimaeras from two independent Ccnb1–HA (Ccnb1T7 and Ccnb1T11 ) and Ccnb2–HA (Ccnb2T15 and Ccnb2T16 ) clones achieved germline transmission. These transgenes were maintained on an M2-rtTA (TA) hemizygous background (ROSA26 ). Both sexes were used for experimentation. After weaning, transgenic mice were continuously administered 2 mg ml−1 dox in drinking water containing 5% sucrose. Mice in spontaneous tumour susceptibility studies were monitored daily. At 14 months, mice were killed and major organs were screened for overt tumours. Tumours were processed for histopathology by standard procedures. Mice on an APC+/Min genetic background56 received dox either from day 21 to 90 or from day 21 to 60. All mice were killed at 90 days to screen for tumours. All tumour histology is representative for at least three independent animals. For carcinogen-induced tumorigenesis, mice were given a single application of 50 μl 0.5% DMBA to the dorsal skin on postnatal day 3–5 and killed 4 months later.
Generation and culture of MEFs
MEFs were generated at embryonic day 13.5 and cultured as previously described57. Primary MEFs were frozen at passage 2 (P2) or P3 and used for experimentation between P4 and P6. At least three independently generated MEF lines per genotype were used. In experiments in which the impact of cyclin B1 or cyclin B2 overexpression was examined, 1 μg ml−1 dox was added to the culture medium 72 h before analysis, unless otherwise stated.
Statistical analyses
GraphPad Prism software was used for all statistical analyses. A two-tailed unpaired t -test was used for comparisons in the following figures: Figs 2b,d, 3b,d,f,h,j–l, 4c,g,i,k, 5a,c,d,f–i, 6f,g; Supplementary Figs 2b,d, 3b,d, 4a,c,f–j, 5a–c, g–j,m, 6b–d, 8a; Tables 1 and 2b,c. A one-sample t -test against a theoretical mean of unity was used for comparisons in Fig. 4e,f. A one-tailed unpaired t -test was used for Fig. 4h and Table 2a. A two-tailed Mann–Whitney test was used for pairwise significance analysis in Fig. 6i and Fisher’s exact tests were used in Fig. 6a,h. A one-tailed chi-squared test was used in Fig. 6b,e. Graphs are indicated with the significance score of *P < 0.05, **P < 0.01 and ***P < 0.001. We note that no power calculations were used. Sample sizes were based on previously published experiments where differences were observed. No samples were excluded. The experiments were not randomized and the investigator not blinded to allocation during experiments and outcome assessment.
Western blot analysis
Western blot analysis was carried out as previously described58. Tissue lysates were prepared by snap-freezing tissues and grinding into powder. Ten milligrams of the powder was suspended in 100 μl PBS and boiled for 10 min after the addition of 100 μl 2× Laemmli lysis buffer. Primary antibodies used for western blotting were mouse anti-cyclin B1 (1:1,000, SC-245/clone CNS-1, Santa Cruz), rabbit anti-cyclin B2 (1:2,000, SC-22776, Santa Cruz), mouse anti-tubulin (1:10,000, T9026/clone DM1A, Sigma), mouse anti-actin (1:10,000, A5441/clone AC-15, Sigma-Aldrich) and mouse anti-HA (1:1,000, 12013819001/clone 3F10, Roche). Ponceau S staining served as a loading control for tissue blots. All western data are representative for two or three independent experiments.
Immunostaining and confocal microscopy
Indirect immunofluorescence was carried out as described previously59. Primary MEFs were cultured in the presence or absence of dox for 72 h. For cyclin B1, cyclin B2, p-CDK substrate and p-Plk1T210 staining, cells were fixed in PBS/3% paraformaldehyde for 12 min at room temperature (RT), permeabilized in PBS/0.2% Triton X-100 for 10 min and blocked in PBS/1% BSA for 30 min at RT. For γ-tubulin, cyclin B2 and p-PlkT210 staining at centrosomes, cells were first permeabilized in PHEM buffer (25 mM HEPES, 10 mM EGTA, 60 mM PIPES and 2 mM MgCl2 at pH 6.9) containing 0.5% Triton X-100 for 5 min at RT and then fixed in ice cold methanol for 10 min at RT. For p-aurora and centrosome staining, cells were fixed with PBS/1% paraformaldehyde for 5 min and then incubated in ice-cold methanol for 10 min at RT. A laser-scanning microscope (LSM 510 or LSM 780; Carl Zeiss) with an inverted microscope (Axiovert 100M; Zeiss) was used to analyse immunostained cells and capture images. Quantification of p-Cdk substrates, cyclin B1, cyclin B2, p-Plk1T210 and p-aurora signals was carried out using ImageJ software (NIH). Confocal microscopy images were converted to eight-bit grey scale, cell edges were traced and the mean pixel intensity density (arbitrary units) within the marked area was calculated. G2 cells were identified using rabbit anti-pH3S10 , which selectively stains heterochromatin foci in G2 phase. In triple stainings for p-Plk1T210 , γ-tubulin and pH3S10 , primary antibodies for γ-tubulin and pH3S10 used were both raised in rabbits. We visualized both these rabbit antibodies with the same fluorophore-conjugated secondary antibody. This did not create a detection conflict because γ-tubulin and pH3S10 are confined to the cytoplasm and nucleus, respectively. G2 cells in which the centrosomes were localized in the cytoplasmic area flanking the nucleus were selected for p-Plk1T210 quantitation. All confocal microscopy images are representative of at least three independent experiments. Primary antibodies for immunostaining were as follows: mouse anti-cyclin B1 (1:100, 554178/clone GNS-11, BD); rabbit anti-cyclin B2 (1:200, SC-22776, Santa Cruz); rabb t anti-p-Cdk substrates (1:1,000, 9447, Cell Signaling Technology); mouse anti-centrin 2 (1:1,000)60; mouse or rabbit anti-γ-tubulin (1:300, T6557/clone GTU-88 or T5192, Sigma); mouse anti-α-tubulin (1:1,000, T9026/clone DM1A, Sigma); rabbit anti-HA (1:100, 3724, Cell Signaling Technology); mouse anti-p- Plk1T210 (1:300, ab39068/clone 2A3, Abcam); rabbit anti-p-aurora (1:100, 2914, Cell Signaling Technology); rabbit anti-Top2a (1:300, TG2011-1, Topogen); human anti- centromeric antibody (1:100, 15-234-0001, Antibodies, Inc.); and rabbit anti-pH3S10 (1:10,000, 06-570, Millipore).
Aneuploidy analyses
Chromosome counts were carried out on metaphase spreads from colcemid-treated transgenic MEFs grown in the presence or absence of dox for 48 h or on splenocytes of 5-month-old transgenic mice treated with dox since weaning57. Interphase FISH analysis with probes for chromosomes 4 and 7 was carried out on single cell suspensions of various tissues and tumours as previously described61. To evaluate aneuploidy rates after correction of aurora A or Plk1 hyperactivity, MEFs were cultured in the presence or absence of dox for 24 h. 10 nM BI2536 or 10 nM MLN8054 was then added to the culture medium and metaphase spreads prepared 48 h later.
Live-cell imaging
For chromosome missegregation analysis, chromosome movements of monomeric red fluorescent protein (mRFP)–H2B-positive primary MEFs progressing through an unchallenged m tosis were followed at interframe intervals of 3 min as previously described32,62. MEFs were seeded onto 35 mm glass bottom dishes (MatTek). All experiments were carried out using a microscope system (Axio Observer; Zeiss) w th CO2 Module S, TempModule S, Heating Unit XL S, a Plan Apo ×63 NA 1.4 oil differential interference contrast III objective (Zeiss), camera (AxioCam MRm; Zeiss) and AxioVision 4.6 software (Zeiss). Imaging medium was kept at 37 ? C. BI2536 or MLN8054 was then added to a final concentration of 10 nM and MEFs immediately analysed for chromosome segregation errors by live-cell imaging. All chromosome missegregation error images are representative of three independent experiments. Nocodazole and taxol challenge assays were carried out as previously described58. To generate the biosensor to measure separase activity, a 390-base pair (bp) BglII–SacI H2B cDNA fragment was ligated into peGFP-N1 (Clontech). In two consecutive rounds of cloning, a 720-bp HindIII–SalI mCherry and a 982 bp KpnI–ApaI human SCC1 fragment were cloned into the resulting plasmid. Finally, the H2B–mCherry–hSCC1–eGFP fragment from this vector was excised with BglII–NotI, blunted, and ligated into a blunted XhoI site in pTSIN. Two days after transduction with biosensor containing pTSIN lentivirus, cells were followed through an unchallenged mitosis by live-cell imaging 24 h later. Time-lapse images of mCherry and eGFP fluorescent signals were captured every 3 min and quantified using ImageJ. Specifically, the DNA area was outlined in mCherry images using the freehand tool. The integrated fluorescence density within this area was measured after background subtraction. The same area was similarly analysed for eGFP (ref. 63). The ratio (eGFP— background)/(mCherry—background) was calculated for all images aligned on the time axis according to anaphase onset, as determined by sister chromatid separation. The value corresponding to the −12 min time point before anaphase onset was considered as 100% and subsequent time-lapse values were normalized against it. The plots represent the average data for the indicated number of cells for each genotype. All separase biosensor fluorescence images are representative of five independent experiments. Error bars represent s.e.m. For analysis of cell cycle duration, Ccnb1T7 MEFs were co-transduced with mKO2–hCdt1(30/120)/pCSII–EF and mAG–hGeminin(1/110)/pCSII–EF lentiviruses64 grown in the presence or absence of dox for 48 h, and then followed by live-cell imaging for 36 h. Time-lapse images of differential interference contrast (DIC), mCherry and Venus fluorescent signals were captured every 5 min using a Plan Apochromat ×20/0.8 M27 D = 0.55 objective. Fluorescence intensity was quantified semi-automatically for each mCherry and Venus individual image using the mean region of interest (ROI) tool within the ZEN software. Specifically, a constant circular ROI was selected to encompass the cell of interest and the same ROI area size was used for background subtraction. The G1/S time was defined as the time with the lowest Venus intensity. The S/G2 time was defined as the time for which mCherry ends the high intensity plateau and begins a descending slope. Nuclear envelope breakdown (mitosis entry) was identified by DIC and by the diffusion of nuclear Venus into mitotic cytosol.
Measurement of centrosome distance, spindle geometry and spindle position
For centrosome distance measurements, serial optical sections (z-series) were collected from pH3S10 /γ-tubulin stained MEFs using a laser-scanning microscope (LSM 780 with an Axiovert 100M inverted microscope, Zeiss). Following maximum intensity projection, the distance between centrosomes (γ-tubulin signals) was measured using ZEN software (Zeiss). For spindle geometry analysis, serial optical sections were collected from γ-tubulin/α-tubulin stained MEFs using a laser-scanning microscope. Following maximum intensity projection, the angle between the spindle and the metaphase plate was measured using ZEN software (Zeiss). G2 cells with centrosome distances ≥ 3 μm were classified as accelerated. Abnormal spindle geometry experiments were conducted in the presence of 10 nM BI2536 or 10 nM MLN8054 (Selleck Chemicals), which was added 6 h before immunostaining. p53−/− MEFs with supernumerary centrosomes were excluded from spindle geometry and centrosome distance analysis. For analysis of spindle position, cells were stained for γ-tubulin antibodies to mark spindle poles and with Hoechst to mark chromosomes. Images were acquired using a laser-scanning microscope (LSM 780; Zeiss) under a ×100 1.4 NA oil-immersion objective, and spindle pole-to-cell cortex distance was measured using ZEN software. Spindle pole-to-cell cortex distance was calculated with ZEN software by marking the centrosome positions, drawing a line between the centrosomes and then marking the points of intersection with the cell edges, as determined under the DIC image. The corresponding two distances between the spindle pole and the point of intersection with the cell edge were measured for 20 cells of each group, and the ratio (smaller/larger value) was averaged and plotted in Excel.
GFP–α-tubulin photoactivation and analysis
MEFs transduced with photoactivatable GFP (PA-GFP)–α tubulin lentivirus were grown in the presence or absence of dox and photoactivated as previously described36,65. Briefly, mitotic cells were identified by DIC microscopy using a ×100 1.4 NA oil-immersion objective on a Zeiss LSM780 microscope. PA-GFP–α tubulin in a thin area near chromosomes (500 nm wide and as long as the metaphase plate) was photoactivated with ten pulses from a 405 nm laser at 100% power. 3 × 1 μm stacks of fluorescent images were acquired less than 1 s before and after photoactivation. Subsequent images were acquired every 30 s up to 4 min and saved as maximum projection time-lapse images. To calculate fluorescence dissipation after photoactivation, we used ImageJ to measure pixel intensities within the photoactivation area. Background pixel intensity was subtracted using an equal ROI from the non-activated opposite spindle. The fluorescence values were corrected for photobleaching by determining the percentage of fluorescence loss during 4 min in activated cells treated with 10 μM taxol. For each cell, fluorescence values were normalized to the first time after photoactivation. Normalized fluorescence was then averaged for multiple cells at each time and plotted against time using Excel 2011.
Ccnb2 knockdown using lentiviral shRNA
Non-targeting shRNA TRC2 negative control vector (SCH202), Ccnb2 shRNA TRC2 clones NM_007630.2- 1098s21c1 (referred to as Ccnb2 shRNA 1), NM_007630.2-1332s21c1 (Ccnb2 shRNA 2), NM_007630.2-454s21c1 (CCNB2 shRNA 1) and NM_004701.2-1190s21c1 (CCNB2 shRNA 2) were purchased from Sigma and lentiviruses prepared according to the manufacturer’s manual. Cells were infected with cyclin B2 shRNA or non-targeting shRNA as a control for 48 h (every 24 h supernatants were refreshed), selected with puromycin (2 μg ml−1 ) for 24 h and analysed over the next 48 h.
Quantitative real-time PCR
Total RNA was extracted from cryosections of normal human lung tissue (n = 17), human adenocarcinomas (n = 35) and human lung squamous cell carcinomas (n = 35), or from cultured wild-type and p53−/− MEFs. cDNA synthesis was carried out with random hexamers and SuperScript III reverse transcription (Invitrogen). PCR reactions on human cDNA samples were carried out in duplicate with Taqman probes using the ABI PRISM Sequence Detection System 7900 Biosystems (Applied Biosystems). For b.b.Ct calculations, TBP was used for normalization. The relative fold change was determined after normalizing to normal lung samples. TaqMan primer IDs were as follows: Hs01030097 and Hs00270424 for CCNB1 and CCNB2, respectively, 4332659 for TBP, Hs01032443 for Ki-67 and Hs00177821 for BUB1. PCR reactions on MEF cDNA samples used SYBR green PCR Master Mix (Applied Biosystems) to a final volume of 12 μl, with each cDNA sample measured in triplicate in the ABI PRISM 7,900 Sequence Detection System (Applied Biosystems) according to the protocol of the manufacturer. The expression of genes was normalized to glyceraldehyde- 3-phosphate dehydrogenase (GAPDH ). Primer sequences were forward 5′ -CTTA CTGCTTGGCTCAAACG-3′ and reverse 5′ -TCAATCATCTCTGGGGGC-3′ for Aurka set 1, forward 5′ -GAGAGTTGAAGATTGCAGACTTC-3′ and reverse 5′ -AA GTCTCCTGGTACGTGTGTGCC-3′ for Aurka set 2, forward 5′ -CCAAGCACAT CAACCCAGTG-3′ and reverse 5′ -TGAGGCAGGTAATAGGGAGACG-3′for Plk1 set 1 and forward 5′ -CCATTGAGTGCCACCTTAGTGA-3′ and reverse 5′ -TGTCC GAATAGTCCACCCACT-3′ for Plk1 set 2.
Supplementary Material
ACKNOWLEDGEMENTS
We thank W. Zhou and M. Li of Mayo Clinic’s Gene Knockout Mouse Core Facility for ES cell microinjection and chimaera breeding, and L. Malureanu, K. Jeganathan and D. Baker for help with live-cell imaging, karyotyping and tumour analyses, respectively. We are grateful to P. Galardy, R. Ricke, R. Naylor, D. Baker and L. Malureanu for discussions and critical evaluation of this manuscript, J. Sali5bury for sharing antibody, J-M. Peters for sharing the Scc1 construct, D. Compton and L. Kabeche for sharing photoactivable GFP–α-tubulin construct and help with measurements of microtubule dynamics, V. Shridhar for human lung (tumour) samples and the Mayo Clinic Cytogenetics shared resource for FISH analysis. This work was supported by the National Institutes of Health (CA126828 to J.M.v.D.).
Footnotes
Note: Supplementary Information is available in the online version of the paper
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Holland AJ, Cleveland DW. Losing balance: The origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13:501–514. doi: 10.1038/embor.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfau SJ, Amon A. Chromosomal instability and aneuploidy in cancer: From yeast to man. EMBO Rep. 2012;13:515–527. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: Mouse modelling of the human disease. Nat. Rev. Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stirling PC, et al. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 2011;7:e1002057. doi: 10.1371/journal.pgen.1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: Therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–538. doi: 10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ricke RM, van Deursen JM. Aneuploidy in health, disease and aging. J. Cell Biol. 2013;201:11–21. doi: 10.1083/jcb.201301061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death- from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parris TZ, et al. Clinical implications of gene dosage and gene expression patterns in diploid breast carcinoma. Clin. Cancer Res. 2010;16:3860–3874. doi: 10.1158/1078-0432.CCR-10-0889. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa T, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One. 2008;3:e2318. doi: 10.1371/journal.pone.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi ZA. signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 11.Chapman DL, Wolgemuth DJ. Isolation of the murine cyclin B2 cDNA and characterization of the lineage and temporal specificity of expression of the B1 and B2 cyclins during oogenesis, spermatogenesis and early embryogenesis. Development. 1993;118:229–240. doi: 10.1242/dev.118.1.229. [DOI] [PubMed] [Google Scholar]
- 12.Draviam VM, Orrechia S, Lowe M, Pardi R, Pines J. The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J. Cell Biol. 2001;152:945–958. doi: 10.1083/jcb.152.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pines J, Hunter T. The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B. EMBO J. 1994;13:3772–3781. doi: 10.1002/j.1460-2075.1994.tb06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackman M, Firth M, Pines J. Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandeis M, et al. Cyclin B2-null mice develop normally and are fertile whereas cyclin B1-null mice die in utero. Proc. Natl Acad. Sci. USA. 1998;95:4344–4349. doi: 10.1073/pnas.95.8.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailly E, Pines J, Hunter T, Bornens M. Cytoplasmic accumulation of cyclin B1 in human cells: association with a detergent-resistant compartment and with the centrosome. J. Cell Sci. 1992;101:529–545. doi: 10.1242/jcs.101.3.529. Pt 3. [DOI] [PubMed] [Google Scholar]
- 17.Pines J. Cyclins: wheels within wheels. Cell Growth Differ. 1991;2:305–310. [PubMed] [Google Scholar]
- 18.Nigg EA. Cellular substrates of p34(cdc2) and its companion cyclin-dependent kinases. Trends Cell Biol. 1993;3:296–301. doi: 10.1016/0962-8924(93)90011-o. [DOI] [PubMed] [Google Scholar]
- 19.Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell. 2010;18:533–543. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 21.Spalluto C, Wilson DI, Hearn T. Evidence for centriolar satellite localization of CDK1 and cyclin B2. Cell Cycle. 2013;12:1802–1803. doi: 10.4161/cc.24840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshitome S, Furuno N, Hashimoto E, Sagata N. The C-terminal seven amino acids in the cytoplasmic retention signal region of cyclin B2 are required for normal bipolar spindle formation in Xenopus oocytes and embryos. Mol. Cancer Res. 2003;1:589–597. [PubMed] [Google Scholar]
- 23.Yoshitome S, Furuno N, Sagata N. Overexpression of the cytoplasmic retention signal region of cyclin B2, but not of cyclin B1, inhibits bipolar spindle formation in Xenopus oocytes. Biol. Cell. 1998;90:509–518. [PubMed] [Google Scholar]
- 24.Kotani T, Yoshida N, Mita K, Yamashita M. Requirement of cyclin B2, but not cyclin B1, for bipolar spindle formation in frog (Rana japonica) oocytes. Mol. Reprod. Dev. 2001;59:199–208. doi: 10.1002/mrd.1023. [DOI] [PubMed] [Google Scholar]
- 25.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 26.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 27.Kumada K, et al. The selective continued linkage of centromeres from mitosis to interphase in the absence of mammalian separase. J. Cell Biol. 2006;172:835–846. doi: 10.1083/jcb.200511126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wirth KG, et al. Separase: A universal trigger for sister chromatid disjunction but not chromosome cycle progression. J. Cell Biol. 2006;172:847–860. doi: 10.1083/jcb.200506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107:715–726. doi: 10.1016/s0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- 30.Kawabata T, et al. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell. 2011;41:543–553. doi: 10.1016/j.molcel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dawlaty MM, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIα. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J. Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganem NJ, Godinho SA, Pellman DA. mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis- segregation in cancer cells. PLoS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J. Clin. Invest. 2012;122:4362–4374. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabeche L, Compton DA. Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Curr. Biol. 2012;22:638–644. doi: 10.1016/j.cub.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silkworth WT, Cimini D. Transient defects of mitotic spindle geometry and chromosome segregation errors. Cell Div. 2012;7:19–26. doi: 10.1186/1747-1028-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruinsma W, Raaijmakers JA, Medema RH. Switching Polo-like kinase-1 on and off in time and space. Trends Biochem. Sci. 2012;37:534–542. doi: 10.1016/j.tibs.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Smith E, et al. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. Embo J. 2011;30:2233–2245. doi: 10.1038/emboj.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Yeh PC, Huang JC, Yeh CC, Juang YL. The spindle checkpoint protein MAD1 regulates the expression of E-cadherin and prevents cell migration. Oncol. Rep. 2012;27:487–491. doi: 10.3892/or.2011.1519. [DOI] [PubMed] [Google Scholar]
- 42.Macurek L, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 43.Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carmena M, et al. The chromosomal passenger complex activates Polo kinase at centromeres. PLoS Biol. 2012;10:e1001250. doi: 10.1371/journal.pbio.1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suijkerbuijk SJ, Vleugel M, Teixeira A, Kops GJ. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore- microtubule attachments. Dev. Cell. 2012;23:745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 47.Hahn WC, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu CC, et al. p53 negatively regulates Aurora A via both transcriptional and posttranslational regulation. Cell Cycle. 2012;11:3433–3442. doi: 10.4161/cc.21732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soria JC, et al. Overexpression of cyclin B1 in earlγ-stage non-small cell lung cancer and its clinical implication. Cancer Res. 2000;60:4000–4004. [PubMed] [Google Scholar]
- 50.Yoshida T, Tanaka S, Mogi A, Shitara Y, Kuwano H. The clinical significance of Cyclin B1 and Wee1 expression in non-small-cell lung cancer. Ann. Oncol.: Official J. Eur. Soc. Med. Oncol./ESMO. 2004;15:252–256. doi: 10.1093/annonc/mdh073. [DOI] [PubMed] [Google Scholar]
- 51.Cooper WA, et al. Expression and prognostic significance of cyclin B1 and cyclin A in non-small cell lung cancer. Histopathology. 2009;55:28–36. doi: 10.1111/j.1365-2559.2009.03331.x. [DOI] [PubMed] [Google Scholar]
- 52.Park SH, et al. NF-γ-dependent cyclin B2 expression in colorectal adenocarcinoma. Clin. Cancer Res. 2007;13:858–867. doi: 10.1158/1078-0432.CCR-06-1461. [DOI] [PubMed] [Google Scholar]
- 53.Wang A, Yoshimi N, Ino N, Tanaka T, Mori H. Overexpression of cyclin B1 in human colorectal cancers. J. Cancer Res. Clin. Oncol. 1997;123:124–127. doi: 10.1007/BF01269891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarafan-Vasseur N, et al. Overexpression of B-type cyclins alters chromosomal segregation. Oncogene. 2002;21:2051–2057. doi: 10.1038/sj.onc.1205257. [DOI] [PubMed] [Google Scholar]
- 55.Dove WF, et al. The intestinal epithelium and its neoplasms: genetic, cellular and tissue interactions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1998;353:915–923. doi: 10.1098/rstb.1998.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moser MJ, et al. Genetic instability and hematologic disease risk in Werner syndrome patients and heterozygotes. Cancer Res. 2000;60:2492–2496. [PubMed] [Google Scholar]
- 57.Babu JR, et al. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J. Cell Biol. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kasper LH, et al. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hart PE, Glantz JN, Orth JD, Poynter GM, Sali5bury JL. Testis-specific murine centrin, Cetn1: genomic characterization and evidence for retroposition of a gene encoding a centrosome protein. Genomics. 1999;60:111–120. doi: 10.1006/geno.1999.5880. [DOI] [PubMed] [Google Scholar]
- 61.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J. Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malureanu L, et al. Cdc20 hypomorphic mice fail to counteract de novo synthesis of cyclin B1 in mitosis. J. Cell Biol. 2010;191:313–329. doi: 10.1083/jcb.201003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell- cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 65.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKenzie L, et al. p53-dependent repression of polo-like kinase-1 (PLK1) Cell Cycle. 2010;9:4200–4212. doi: 10.4161/cc.9.20.13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mardin BR, et al. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 2010;12:1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schiebel E. Plk1. controls the Nek2A-PP1γ antagonism in centrosome disjunction. Curr. Biol. 2011;21:1145–1151. doi: 10.1016/j.cub.2011.05.047. [DOI] [PubMed] [Google Scholar]
- 69.Fry AM, et al. C Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.