Abstract
Derivatives of 3-amino-3,6-dideoxyhexoses are widespread in Nature. They are part of the repeating units of lipopolysaccharide O-antigens, of the glycan moiety of S-layer (bacterial cell surface layer) glycoproteins and also of many antibiotics. In the present study, we focused on the elucidation of the biosynthesis pathway of dTDP-α-d-Quip3NAc (dTDP-3-acetamido-3,6-dideoxy-α-d-glucose) from the Gram-positive, anaerobic, thermophilic organism Thermoanaerobacterium thermosaccharolyticum E207-71, which carries Quip3NAc in its S-layer glycan. The biosynthesis of dTDP-α-d-Quip3NAc involves five enzymes, namely a transferase, a dehydratase, an isomerase, a transaminase and a transacetylase, and follows a pathway similar to that of dTDP-α-d-Fucp3NAc (dTDP-3-acetamido-3,6-dideoxy-α-d-galactose) biosynthesis in Aneurinibacillus thermoaerophilus L420-91T. The ORFs (open reading frames) of interest were cloned, overexpressed in Escherichia coli and purified. To elucidate the enzymatic cascade, the different products were purified by HPLC and characterized by NMR spectroscopy. The initiating reactions catalysed by the glucose-1-phosphate thymidylyltransferase RmlA and the dTDP-d-glucose-4,6-dehydratase RmlB are well established. The subsequent isomerase was shown to be capable of forming a dTDP-3-oxo-6-deoxy-d-glucose intermediate from the RmlB product dTDP-4-oxo-6-deoxy-d-glucose, whereas the isomerase involved in the dTDP-α-d-Fucp3NAc pathway synthesizes dTDP-3-oxo-6-deoxy-d-galactose. The subsequent reaction steps of either pathway involve a transaminase and a transacetylase, leading to the specific production of nucleotide-activated 3-acetamido-3,6-dideoxy-α-d-glucose and 3-acetamido-3,6-dideoxy-α-d-galactose respectively. Sequence comparison of the ORFs responsible for the biosynthesis of dTDP-α-d-Quip3NAc revealed homologues in Gram-negative as well as in antibiotic-producing Gram-positive bacteria. There is strong evidence that the elucidated biosynthesis pathway may also be valid for LPS (lipopolysaccharide) O-antigen structures and antibiotic precursors.
Keywords: 3-amino-3,6-dideoxyhexose; bacterial cell surface layer (S-layer) glycoprotein; dTDP-3-acetamido-3,6-dideoxy-α-d-glucose (dTDP-α-d-Quip3NAc); 3,4-oxoisomerase; nucleotide-activated sugar precursor; Thermoanaerobacterium thermosaccharolyticum
INTRODUCTION
S-layer (bacterial cell surface layer) proteins frequently constitute the outermost cell envelope of archaea and bacteria, and many of them are glycosylated. Typically, bacterial S-layer glycoproteins consist of a long heteropolymeric glycan chain composed of individual repeating units and of a short oligosaccharide core, making them structurally similar to the O-antigens of LPS (lipopolysaccharide) (for a review, see [1]). The assembly of the sugar chains of LPS is quite well characterized [2], whereas the biosynthesis of S-layer glycoproteins remains still to be explored in detail. Several reports have shown that the biosynthesis pathways of nucleotide-activated sugar precursors follow identical routes, both in Gram-positive and Gram-negative organisms [3–5]. The genetic information of S-layer glycan biosynthesis is organized in slg (S-layer glycosylation) gene clusters [6]. Thermoanaerobacterium thermosaccharolyticum E207-71 is a Gram-positive, anaerobic, thermophilic organism. It possesses a glycosylated S-layer protein with hexasaccharide repeating units consisting of d-glucose, d-galactopyranose, d-galactofuranose, d-mannose, l-rhamnose and α-d-Quip3NAc (3-acetamido-3,6-dideoxy-α-d-glucose) [7]. Among the biosynthesis pathways of nucleotide sugar precursors required for the biosynthesis of the T. thermosaccharolyticum E207-71 S-layer glycan, the reaction sequences leading to TDP-l-rhamnose [8], UDP-d-Galp [9] and UDP-d-Galf [10] are well described in the literature. The biosynthetic route for dTDP-α-d-Quip3NAc, in contrast, is still unknown. d-Quip3NAc and other 3-amino-substituted sugars, such as d-Fucp3NAc (3-acetamido-3,6-dideoxy-α-d-galactose), have already been identified as part of LPS O-antigens of several Gram-negative organisms [11–14]. Only for d-Fucp3NAc occurring in the Aneurinibacillus thermoaerophilus L420-91T S-layer glycan, the biosynthetic route has been elucidated so far [15].
Perelle et al. [16] identified the gene cluster responsible for the biosynthesis of the O-antigen in Escherichia coli O91, which contains a modified d-Quip3N among other sugar residues. These authors identified a gene that may possibly code for an isomerase catalysing the conversion of dTDP-4-oxo-6-deoxy-d-glucose into dTDP-3-oxo-6-deoxy-d-glucose. However, neither biochemical experiments nor complementation experiments with knockout mutants have been performed to demonstrate the proposed protein function. Derivatives of d-Quip3N have been described not only as components of S-layer glycoproteins and LPS O-antigens, but also as important biosynthetic precursors of d-desosamine and d-mycaminose [17], both being sugar residues that are frequently found in antibiotics. Among these 3-amino-6-deoxy-sugar-containing antibiotics are erythromycin [18], oleandomycin [19,20] and tylosin [21]. There are several reports on the biosynthesis of the respective sugar precursors. For instance, in Streptomyces fradiae, which produces the antibiotic tylosin, a 3,4-oxoisomerase was found [22]. The biosynthesis of nucleotide-activated d-mycaminose follows a similar pathway to that described for d-Fucp3NAc and as postulated for d-Quip3NAc, starting from glucose-1-phosphate. This compound is converted by RmlA- and RmlB-homologous enzymes into dTDP-4-oxo-6-deoxy-d-glucose, which is the substrate for the 3,4-isomerase. The produced 3-oxo-6-deoxy sugar is subsequently aminated and methylated. Recently, the 3,4-isomerase of S. fradiae was characterized as a dTDP-4-oxo-6-deoxy-d-glucose-3,4-oxoisomerase (designated Tyl1a) [23]. A comparable pathway is also proposed for the biosynthesis of dTDP-d-desosamine. The putative isomerase involved is designated OleP1 and belongs to the cytochrome P450 mono-oxygenase family [19]. The occurrence of d-Quip3N derivatives as part of macrolide antibiotics and the presence of similar isomerases in organisms that produce these antibiotics illustrate the importance of the elucidation of the biosynthesis pathway of dTDP-α-d-Quip3N. The implication of these studies for the development of novel antibiotics was demonstrated recently. Replacement of the dTDP-4-oxo-6-deoxy-d-glucose-3,4-oxoisomerase (Tyl1a) activity with FdtA from A. thermoaerophilus L420-91T, synthesizing non-naturally occurring dTDP-4-epi-d-mycaminose, resulted in the formation of a derivative of the macrolide antibiotic tylosin [24].
In the present study, we show that the biosynthesis of dTDP-α-d-Quip3NAc follows a similar pathway to that described for dTDP-α-d-Fucp3NAc [15]. This pathway starts from glucose 1-phosphate, representing a compound available from the general sugar metabolism. Glucose 1-phosphate is activated by the glucose 1-phosphate thymidylyltransferase RmlA to form dTDP-d-glucose and dehydrated by the dTDP-d-glucose-4,6-dehydratase RmlB to form dTDP-4-oxo-6-deoxy-d-glucose. Both proteins are key enzymes in the biosynthetic pathway of dTDP-β-l-rhamnose [8]. We could demonstrate that the isomerases of A. thermoaerophilus L420-91T and T. thermosaccharolyticum E207-71 use the same precursor – dTDP-4-oxo-6-deoxy-d-glucose – but the synthesized reaction products are dTDP-3-oxo-6-deoxy-d-hexose epimers that are further processed by subsequent amination and acetylation reactions.
EXPERIMENTAL
Materials
l-Glutamate and acetyl-CoA were obtained from Sigma (Vienna, Austria) and dTDP-d-glucose was from Fluka (Buchs, Switzerland). GSTrap, HiTrap Chelating, MonoQ HR5/5 and Sephadex G-10 were purchased from Amersham Biosciences (Uppsala, Sweden). For RP-HPLC (reversed-phase HPLC) analysis, a Nucleosil 120-3C18 column (Macherey-Nagel, Düren, Germany) was obtained from ARC (Seibersdorf, Austria). All primers used in the present study were synthesized by Invitrogen (Lofer, Austria). GSH and imidazole were obtained from ICN Chemicals (Eschwege, Germany).
Bacterial strains and culture conditions
T. thermosaccharolyticum E207-71 was grown under anaerobic conditions in S-medium at 60°C [7]. E. coli DH5α (Invitrogen) was used for cloning purposes. Enzyme overexpression was performed in E. coli BL21(DE3). For selective growth, ampicillin (Sigma) and kanamycin (Invitrogen) were used at a concentration of 100 and 50 μg/ml respectively.
DNA manipulations, DNA sequencing, PCR and gene identification
Standard DNA manipulation procedures were performed using the methods described by Sambrook and Russell [25]. To identify the genes involved in dTDP-d-Quip3NAc biosynthesis on the chromosome of T. thermosaccharolyticum E207-71, PCR with degenerate primers was performed (Table 1). Primer design was based on sequence comparison of known transaminases of organisms containing isomerase-like enzymes. For further sequencing, chromosome walking [15] and inverse PCR [25] were used. The obtained PCR products were purified by agarose gel elution (Qiagen, Hilden, Germany) and sequenced (AGOWA, Berlin, Germany). PCR was performed using a PCR Sprint thermocycler from Hybaid (Ashford, Kent, U.K.). The nucleotide sequences were analysed with the sequence analysis program OMIGA™ (Accelrys, Unterhaching-München, Germany). Homology searches and sequence alignments were performed with the BLAST tool at NCBI (National Center for Biotechnology Information) [26] and Multalign [27] respectively.
Table 1. Strains, primers and vectors used in the present study.
| Description | Source | |
|---|---|---|
| Strains | ||
| T. thermosaccharolyticum E207-71 | Wild-type strain | |
| A. thermoaerophilus L420-91T | Wild-type strain | |
| E. coli DH5α | ϕ80 lac ZΔM15 Δ(lac ZYA-argF) U169 recA1 endA1 hsdR17 (rk−, mk+) phoA sup E44 thi-1 gyr A96 relA1λ− | Invitrogen |
| E. coli BL21(DE3) | [F− ompT hsdSB (rB− mB−) gal dcm (DE3)] | Invitrogen |
| Primers | ||
| wQdtB_for1 | GARGGNGAYGARGTNAT | |
| wQdtB_rev1 | GCNGCYTGNARYTCRTC | |
| gQdtA_for1 | GCATTTTGTATAACGTTGCGTTAATA | |
| gQdtA_rev1 | TTCATTTTTGCACTCTCTCC | |
| gQdtB_for1 | GCATGAAAATATCATTTGCAAGCT | |
| gQdtB_rev1 | GCTAATAACACTTCCTAATCCG | |
| gQdtC_for1 | GCATGCCAAATAATATTTCTAAAAGTG | |
| gQdtC_rev1 | CGTTATACAAAATTCTTCCCCC | |
| Vectors | ||
| pBCKS II | Chromosome walking | Stratagene |
| pDonr201 | Cloning vector | Invitrogen |
| pDest15 | Expression vector (GST tag) | Invitrogen |
| pDest17 | Expression vector (His tag) | Invitrogen |
| gFdtA15 | FdtA in pDest15 | [15] |
| gFdtB17 | FdtB in pDest17 | [15] |
| gFdtC15 | FdtC in pDest15 | [15] |
| gQdtA | QdtA in pDonr201 | The present study |
| gQdtB | QdtB in pDonr201 | The present study |
| gQdtC | QdtC in pDonr201 | The present study |
| gQdtA17 | QdtA in pDest17 | The present study |
| gQdtB17 | QdtB in pDest17 | The present study |
| gQdtC15 | QdtC in pDest15 | The present study |
Cloning, overexpression and purification of the ORFs (open reading frames) of interest
To characterize the newly identified enzymes QdtA, QdtB and QdtC, they were cloned and overexpressed in E. coli. To clone the ORFs, the GatewayTM system from Invitrogen was used. Briefly, the genes of interest were amplified by PCR with primers containing defined nucleotide sequences at their ends (Table 1) in order to allow insertion into the vectors. For qdtA and qdtB, pDest17 vectors containing a His6 tag and, for qdtC, pDest15 vector containing a GST (glutathione transferase) tag were used. E. coli BL21(DE3) was used as expression host. Cells carrying the plasmids were grown in 0.7 litre culture volumes of LB (Luria–Bertani) medium containing 50 μg/ml ampicillin at 37°C. At an attenuance at 600 nm (D600) of 0.6, the cells were induced for expression with 1 mM IPTG (isopropyl β-d-thiogalactoside) for 3 h. The cells were harvested, washed and resuspended in 10 ml of 20 mM phosphate buffer (pH 7.4). To lyse the cells, ultrasonication on ice was performed and cell debris was removed by ultracentrifugation at 331000 g for 45 min. Protein purification was performed as recommended by the manufacturer, by using HiTrap chelating columns with bound Ni2+ ions for His6-tagged proteins or GSTrap columns for GST-tagged proteins. Protein-containing fractions were pooled and concentrated with Ultrafree-MC centrifugal devices (cut-off 10 kDa; Millipore, Vienna, Austria). The protein concentrations were adjusted to 1 mg/ml and DTT (dithiothreitol) and glycerol were added as stabilizers to a final concentration of 1 mM and 20% respectively. The purity of the enzymes was checked by SDS/PAGE [28] and the protein content was determined by the method of Bradford [29], using the Bio-Rad protein assay (Bio-Rad, Vienna, Austria).
Enzyme assays and substrate synthesis
dTDP-4-oxo-6-deoxy-d-glucose III, which is the substrate of the isomerase QdtA, and dTDP-d-Fucp3N VIII, for the incubation with the transacetylase QdtC, were obtained in milligram amounts as described previously [15]. Standard assays contained 50 nmol of the nucleotide-activated sugar and 5 nmol of enzyme and were performed in 50 mM K2HPO4 buffer (pH 7.4) with 5 mM MgCl2. Mixtures for the transamination and transacetylation reactions additionally contained 50 nmol of l-glutamate, 5 nmol of pyridoxal phosphate and 50 nmol of acetyl-CoA respectively. The assays were performed at 37°C for 30 min. The reaction products were analysed by RP-HPLC with 0.5 M NaH2PO4 (pH 6.0) as the mobile phase at a flow rate of 0.6 ml/min and monitored by UV detection at 254 nm [30]. Large-scale purification as required for NMR analysis was performed by RP-HPLC using 0.2 M triethylammonium acetate, pH 6.0, as the eluent.
To investigate the substrate specificity of the isomerase QdtA and the transaminase QdtB, and due to the impossibility to isolate the 3-oxo substrate IV, cross-incubations of these enzymes with the corresponding enzymes from the dTDP-d-Fucp3NAc pathway were performed. In detail, the isomerase FdtA from A. thermoaerophilus L420-91T was incubated with QdtB, the transaminase from the dTDP-d-Quip3NAc pathway of T. thermosaccharolyticum E207-71. The reaction mixtures contained the same cofactors and co-substrates as described above for the single incubations.
NMR analysis
Spectra were recorded at 300 K at 300.13 MHz for 1H and at 75.47 MHz for 13C with a Bruker AVANCE 300 spectrometer equipped with a 5 mm QNP (quadrupole nuclear probehead) with z-gradients. Data acquisition and processing were performed with the standard XWINNMR software (Bruker). 1H spectra were referenced to internal 2,2-dimethyl-2-silapentane-5-sulfonic acid (δ = 0) and 13C spectra were referenced externally to 1,4-dioxane (δ = 67.40). 1H/13C HMQC (heteronuclear single quantum correlation) spectra were recorded in the phase-sensitive mode using TPPI (time-proportional phase increments) and pulsed field gradients for coherence selection. Spectra resulted from a 256 × 4096 data matrix, with 1800 or 800 scans per t1 value respectively. For in situ experiments of the isomerase reaction, samples containing dTDP-4-oxo-6-deoxy-d-glucose III (dTDP-6-deoxy-d-xylo-hex-4-ulose; 9 mg) in 20 mM K2HPO4 buffer (in 0.6 ml of 2H2O, pD 7.4) were prepared and kept at 318 K. After adding QdtA (0.113 mg) or FdtA (0.1 mg) respectively and mixing in the NMR vial, NMR measurements were immediately started and recorded at distinct time intervals.
RESULTS AND DISCUSSION
Identification of the dTDP-α-d-Quip3NAc gene locus
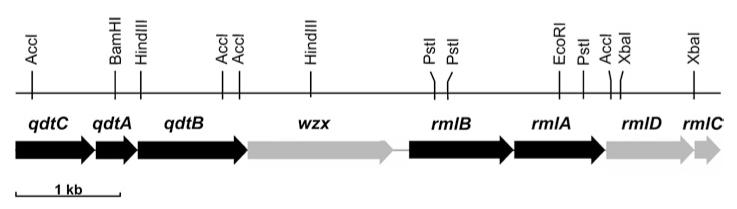
Based on protein homology of the isomerase FdtA involved in the biosynthesis of dTDP-α-d-Fucp3NAc in A. thermoaerophilus L420-91T [15] and the proposed isomerase involved in dTDP-d-Quip3NAc biosynthesis in E. coli O91 [16], it is conceivable that both nucleotide-sugar pathways follow similar routes. The crucial step of either pathway is the transition of the 4-oxo educt, dTDP-4-oxo-6-deoxy-d-glucose III, into a 3-oxo product, dTDP-3-oxo-6-deoxy-d-hexose IV, which is catalysed by a 3,4-oxoisomerase. Thus an isomerase similar to that involved in the biosynthesis of dTDP-α-d-Fucp3NAc was very likely to be present in the dTDP-α-d-Quip3NAc biosynthesis pathway of T. thermosaccharolyticum E207-71. To obtain entry into the chromosomal locus encoding the isomerization event, several aminotransferases of different organisms, which are located in close proximity to the respective 3,4-oxoisomerase on the chromosome, were aligned. Especially the amino acid composition of the hypothetical pyridoxal-phosphate-binding site seemed to be a typical feature of the compared transaminases. Based on the amino acid sequence next to the N-terminus (EGDVI) and the C-terminus (DELQAA) of the compared enzymes, two degenerate primers (wQdtB_for1 and wQdtB_rev1) were designed. A PCR fragment of 450 bp was amplified using DNA from T. thermosaccharolyticum E207-71 as a template, whose sequence revealed an incomplete ORF, similar to that encoding FdtB of the dTDP-α-d-Fucp3NAc pathway. Further sequencing starting from this fragment by chromosome walking and inverse PCR revealed the complete ORF for QdtB and, in addition, the presence of the genes encoding an isomerase and a transacetylase upstream of qdtB (Figure 1). The rml genes, which are responsible for the biosynthesis of dTDP-β-l-rhamnose as well as the 4-oxo precursor III of dTDP-α-d-Quip3NAc, were identified downstream of qdtB. Between the clustered genes encoding the enzymes of either nucleotide-sugar biosynthesis pathway, an ORF coding for a Wzx homologous protein, possibly involved in polysaccharide export, was found. In summary, sequencing experiments revealed a genetic locus that contains eight ORFs (Figure 1). It can be assumed that this 7000-bp sequence represents a partial slg gene cluster of T. thermosaccharolyticum E207-71 (GenBank® accession number AY422724). Following the nomenclature for dTDP-α-d-Fucp3NAc biosynthesis [15], the identified genes, whose gene products are responsible for the biosynthesis of dTDP-α-d-Quip3NAc, were designated QdtA (dTDP-4-oxo-6-deoxy-d-glucose-3,4-oxoisomerase), QdtB (transaminase) and QdtC (transacetylase). In this nomenclature, ‘Q’ stands for quinovosamine, ‘d’ for the d-configuration and ‘t’ for the C-3 atom of the sugar ring.
Figure 1. Chromosomal organization of the genes encoding dTDP-α-d-Quip3NAc and dTDP-β-l-rhamnose biosynthesis in T. thermosaccharolyticum E207-71 (partial slg gene cluster; GenBank® accession number AY422724).
Sequence comparison of QdtA with proteins from the database at NCBI revealed high homology with more than 80 ORFs exhibiting pfam05523 domain (pfam05523: FdtA, WxcM-like, C-terminal). This family includes FdtA from A. thermoaerophilus, which has been characterized as a dTDP-6-deoxy-3,4-oxohexulose isomerase, and WxcM from Xanthomonas campestris pv. campestris [31]. Some of these homologous proteins originate from organisms that are known to synthesize derivatives of d-Quip3N and d-Fucp3N, such as E. coli, X. campestris and A. thermoaerophilus. High similarity was observed with FdtA from A. thermoaerophilus L420-91T [identity/similarity (%) 55/69, GenBank® accession number AAO06351] and WbsB from E. coli O91 (48/64, GenBank® accession number AAK60451). Biochemical data are available for FdtA that demonstrate that this enzyme catalyses the formation of dTDP-3-oxo-6-deoxy-d-galactose VII from the substrate dTDP-4-oxo-6-deoxy-d-glucose III. WbsB from E. coli O91, harbouring d-Quip3NAc residues in its O-antigen, was identified by sequence comparison to be an isomerase [14]. Interestingly, S. fradiae is known to be a producer of tylosin, which contains the 3-amino-6-deoxy sugar residue d-mycaminose in its structure. While the QdtA-like enzyme Tyl1a was not annotated in the original GenBank® submission [22], the in vitro activity of this enzyme was demonstrated recently [23].
QdtB homologues are present in organisms that contain isomerase-like proteins. QdtB shares highest homology with the transaminase FdtB from A. thermoaerophilus L420-91T (60/74, AAO06353). Among other enzymes presumably involved in the biosynthesis of nucleotide-activated d-Quip3N and d-Fucp3N, the transaminases from the d-desosamine (OleN2 47/67, AAD55458 and DesV 45/63, AAC68680) and d-mycaminose (TylB 46/64) biosynthesis pathways share high similarity with QdtB. All these enzymes are members of the DegT/DnrJ/EryC1/StrS family of aminotransferases.
Alignment of QdtC with enzymes from the database revealed that this enzyme shows to some extent homology to other known acetyltransferases, but almost no homology to acetyltransferases catalysing the acetylation of 3-amino-6-deoxy sugar residues. Nevertheless, like all other aligned acetyltransferases, QdtC possesses a hexapeptide repeat motif [32], which unambiguously identifies it as a transferase.
The rml genes of T. thermosaccharolyticum E207-71 responsible for the biosynthesis of dTDP-β-l-rhamnose are highly homologous with the corresponding enzymes identified in Clostridium acetobutylicum [33] and A. thermoaerophilus DSM 10155 [3], and occur in the order rmlB, rmlA, rmlD and rmlC.
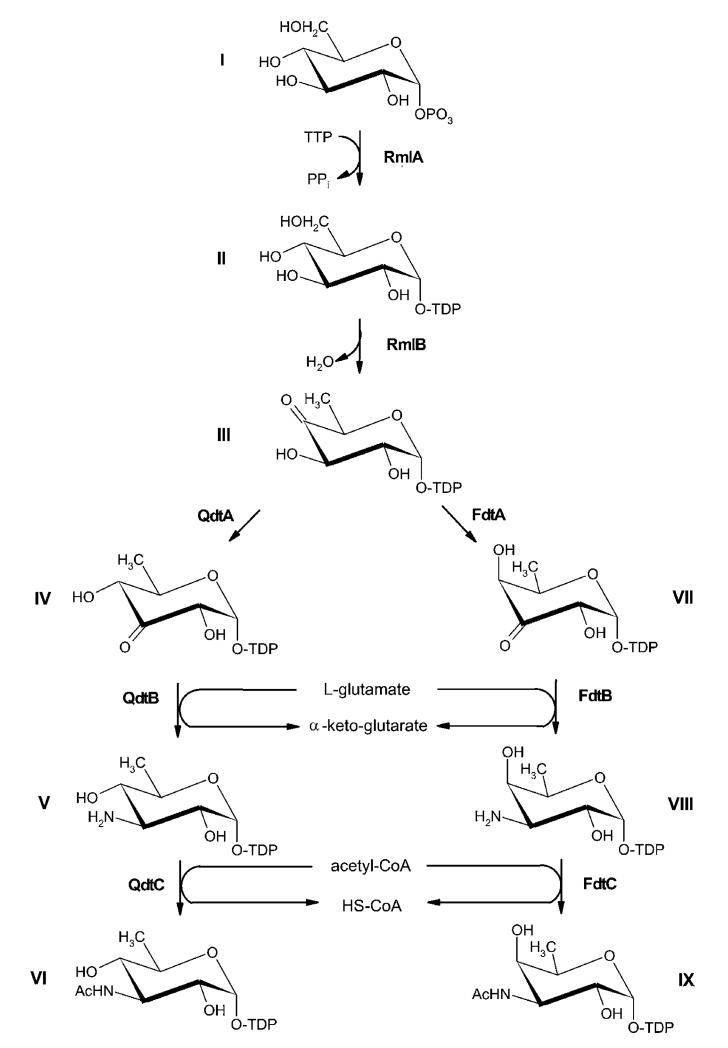
Based on the genetic data obtained in the present study, we propose the biosynthetic pathway for dTDP-α-d-Quip3NAc VI as depicted in Figure 2.
Figure 2. Comparison of the biosynthesis pathways for dTDP-α-d-Quip3NAc and dTDP-α-d-Fucp3NAc.
(I) d-Glucose-1-phosphate; (II) dTDP-d-glucose; (III) dTDP-4-oxo-6-deoxy-d-glucose; (IV) dTDP-3-oxo-6-deoxy-d-glucose; (V) dTDP-d-Quip3N; (VI) dTDP-d-Quip3NAc; (VII) dTDP-3-oxo-6-deoxy-d-galactose; (VIII) dTDP-d-Fucp3N, (IX) dTDP-d-Fucp3NAc. RmlA, glucose-1-phosphate thymidylyltransferase; RmlB, dTDP-d-glucose 4,6-dehydratase; QdtA, dTDP-4-oxo-6-deoxy-d-glucose-3,4-oxoisomerase; QdtB, dTDP-3-oxo-6-deoxy-d-glucose aminase; QdtC, dTDP-d-Quip3N acetylase; FdtA, dTDP-4-oxo-6-deoxy-d-glucose-3,4-oxoisomerase; FdtB, dTDP-3-oxo-6-deoxy-d-galactose aminase; FdtC, dTDP-d-Fucp3N acetylase.
Cloning of qdtA, qdtB and qdtC and overexpression of the encoded proteins of T. thermosaccharolyticum E207-71 in E. coli
All enzymes involved in the biosynthesis of dTDP-α-d-Quip3NAc were cloned with the GatewayTM system from Invitrogen. Prior to heterologous expression, the plasmids were verified by nucleotide sequencing. In order to allow rapid and efficient purification, QdtA and QdtB were expressed as His6-fusions and QdtC was expressed as a GST-tagged protein. Since affinity chromatography resulted in enzymes of high purity, no further purification steps were applied. According to the SDS/PAGE evidence, the molecular mass of the enzymes QdtA, QdtB and QdtC of 18.6, 42.1 and 56.9 kDa respectively corresponded to the masses predicted from the amino acid sequences (results not shown).
The isomerase QdtA forms dTDP-3-oxo-6-deoxy-d-glucose
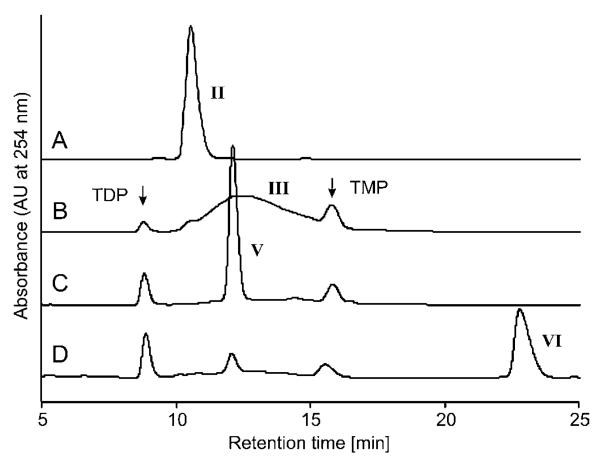
BLAST search revealed the high similarity of QdtA to FdtA, which is the isomerase involved in the biosynthesis of dTDP-α-d-Fucp3NAc IX. Thus it was concluded that QdtA may also act as an isomerase. The dTDP-4-oxo-6-deoxy-d-glucose III educt of this enzyme is obtained by the sequential action of RmlA and RmlB. Initial experiments performed with QdtA, QdtB and QdtC, acting on the required substrates and co-substrates, revealed a rather fast conversion of dTDP-4-oxo-6-deoxy-d-glucose III into dTDP-α-d-Quip3NAc VI. In contrast, experiments without addition of the isomerase to the reaction mixture showed only poor yields of the final product dTDP-α-d-Quip3NAc VI. This observation is in agreement with previously published data [15] and clearly shows the necessity of QdtA in the pathway. Conversion of the 4-oxo substrate III into the 3-oxo product IV was initially monitored by RP-HPLC. The elution profile showed a similar chromatographic pattern to that described for the FdtA-catalysed reaction, implicating that this method was not suitable for the separation of educt and product due to chemical similarities of the substances (Figure 3).
Figure 3. HPLC analysis of the products of the enzymatic reactions catalysed by QdtA, QdtB and QdtC.
(A) dTDP-glucose II; (B) dTDP-4-oxo-6-deoxy-d-glucose III, synthesized from II by RmlB; (C) dTDP-d-Quip3N V, synthesized from III by sequential action of QdtA and QdtB; (D) dTDP-d-Quip3NAc VI, final reaction product of the concerted action of RmlB, QdtA, QdtB and QdtC using II as substrate. The numbers used for designation of enzymatic products are identical with those used in Figure 2.
To verify the function of the enzyme, online-NMR monitoring of the enzymatic reaction was applied. Although highest enzyme activities were obtained between 60 and 65°C (results not shown), the direct conversion of dTDP-4-oxo-6-deoxy-d-glucose III into the 3-oxo product IV by the action of QdtA was studied by NMR measurements recorded at 45°C in order to obtain detectable quantities of the isomerization product and to prevent its degradation. Formation and increase of a new anomeric signal at 5.87 p.p.m. (3JH–1,H–2 = 4.3, 3JH–1,P, = 7.1 Hz) was observed within 10 min and reached a maximum signal intensity of ~25% after ~3 h. The signal of H-1 was correlated with a solvent-hidden signal of H-2 at 4.65 p.p.m. by a COSY experiment lending support to the formation of a oxo group at C-3, thus causing the observed downfield shift of H-2. Due to concomitant degradation of the material, a full spectral assignment of the 3-oxo intermediate IV was not possible. The spectral characteristics, however, are in agreement with those of the reaction product reported for a Tyl1a-catalysed conversion of III [23] and with the NMR data published for dTDP-3-oxo-6-deoxy-α-d-galactose VII (dTDP-6-deoxy-d-ribo-hex-3-ulose), obtained from a reversed transamination reaction [34] and prepared via a non-enzymatic conversion of the substrate [35]. For comparison, a control experiment without the addition of enzyme was performed, which revealed the formation and increase of the new anomeric 1H signal within 30 min to a maximum of ~5% of the mixture, followed by a subsequent decrease in its signal intensity and appearance of signals arising from degradation of the sample (Figure 4). Due to inherent instability of the 3-oxo intermediate, direct conversion was not attempted.
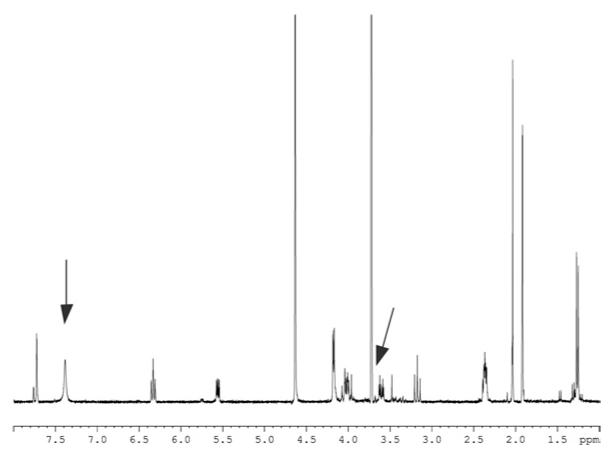
Figure 4. 300 MHz 1H NMR spectrum of dTDP-Quip(3)NAc VI recorded at 300 K in 2H2O.
The signal at 3.7 p.p.m. (indicated by an arrow) corresponds to residual Tris buffer; the signal at ~7.4 p.p.m. (indicated by an arrow) is due to residual imidazole.
QdtB aminates the 3-oxo-6-deoxy-gluco-epimer forming dTDP-α-d-Quip3N
The formation of dTDP-α-d-Quip3N V was observed in a combined reaction of QdtB with the isomerase QdtA and, in another reaction, without the isomerase. In an assay containing the isomerase, quantitative turnover of the dTDP-4-oxo-6-deoxy-d-glucose III substrate was obtained over a time period of 30 min at 37°C. However, also without addition of QdtA a turnover of approx. 25% was observed after 12 h of incubation. These results are in agreement with previous data from the biosynthesis of dTDP-d-Fucp3NAc IX, where the transaminase FdtB was able to convert dTDP-4-oxo-6-deoxy-d-glucose III directly into dTDP-d-Fucp3N VIII (over 36 h) [15]. Obviously, over a certain time span, both transaminases, QdtB and FdtB, are capable of synthesizing the respective 3-aminated 6-deoxy sugar product V and VIII without the involvement of an isomerase.
To investigate the specificity of QdtB, enzyme assays were also performed with the substrate VII for the transaminase FdtB, formed by the isomerase FdtA from the dTDP-α-d-Fucp3NAc biosynthesis pathway. Incubations were done at 37°C for 3 h and the reaction products were monitored by RP-HPLC. Inspection of the HPLC chromatograms showed that QdtB has a clear preference for the substrate provided by the isomerase QdtA from T. thermosaccharolyticum E207-71. The amination of the 3-oxo-gluco-epimer IV is approx. 3 times more efficient than that of the 3-oxo-galacto-epimer VII.
The introduction of the 3-amino-group into the product of the enzymatic reaction was clearly visible in the 300 MHz 1H NMR spectrum of V. The spectrum revealed two upfield-shifted triplets for H-3 and H-4 respectively, with large values of the vicinal coupling constants and with H-3 being correlated to a nitrogen-bearing carbon occurring at 56.04 p.p.m. (Table 2).
Table 2. NMR spectroscopic data* of dTDP-α-d-Quip3N V and of dTDP-α-d-Quip3NAc VI.
|
V
|
VI
|
|||||
|---|---|---|---|---|---|---|
| Atom | 1H (p.p.m.) | J, [Jhet] (Hz) | 13C (p.p.m.) | 1H (p.p.m.) | J, [Jhet] (Hz) | 13C (p.p.m.) |
| 1″ | 5.58 | 3.5/[7.4] | 95.00 | 5.56 | 3.4/[7.0] | 95.32 |
| 2″ | 3.78 | [3.4] | 69.57 | 3.60 | [3.0] | 70.85 |
| 3″ | 3.42 | 10.0 | 56.04 | 4.04 | 10.5 | 54.62 |
| 4″ | 3.35 | 9.2 | 72.01 | 3.17 | 9.8 | 74.28 |
| 5″ | 4.01 | 6.4 | 69.73 | 4.02 | 6.2 | 70.39 |
| 6″ | 1.28 | – | 17.16 | 1.26 | – | 17.12 |
| NH(Ac) | 2.05 | 22.45 | ||||
| 1′ | 6.34 | 7.0 | 85.83 | 6.33 | 7.2 | 85.65 |
| 2a′/2′b | 2.39/2.35 | 7.0 | 39.38 | 2.39/2.34 | 7.2 | 38.92 |
| 3′ | 4.62 | n.d.† | 71.79 | 4.63 | n.d. | 71.94 |
| 4′ | 4.17 | n.d. | 86.03 | 4.18 | n.d. | 86.06 |
| 5a′/5′b | 4.17/4.15 | n.d. | 66.27 | 4.17 | n.d. | 66.09 |
| 2 | 152.55 | n.d. | ||||
| 4 | 167.38 | n.d. | ||||
| 5 | 112.53 | n.d. | ||||
| 6 | 7.72 | n.d. | 138.15 | 7.72 | 1.2 | 138.37 |
| Me | 1.92 | – | 12.44 | 1.92 | – | 12.01 |
Spectra were recorded at 300 K for solutions in 2H2O at pD 7.4.
n.d., not determined.
Acetylation is catalysed by QdtC
QdtC turned out to be responsible for the transfer of the acetyl group from acetyl-CoA to dTDP-α-d-Quip3N V. dTDP-α-d-Quip3NAc VI obtained in this reaction was purified by RP-HPLC (Figure 3D) and further analysed by NMR spectroscopy. The product VI of the N-acetylation reaction (Figure 4) displayed signals of an N-acetyl group at 2.05 p.p.m./22.45 p.p.m. for the 1H- and 13C NMR signals of the methyl group and a substantial downfield shift of the proton signal of H-3 (Table 2). In addition, small downfield shifts were observed for the neighbouring carbons. Signals of the pyranose unit were in good agreement with the data for a terminal α-d-Quip3NAc unit observed in an O-antigenic bacterial polysaccharide [36].
The substrate specificity of the transacetylase QdtC was determined by incubating the enzyme also with dTDP-α-d-Fucp3N VIII. According to the chromatographic profile, the transacetylation reaction catalysed by QdtC turned out to be bifunctional, acetylating both epimers, dTDP-α-d-Quip3N V and dTDP-α-d-Fucp3N VIII. Interestingly, alignment of QdtC from T. thermosaccharolyticum E207-71 and FdtC from A. thermoaerophilus L420-91T revealed that their amino acid sequences are not highly homologous.
Conclusions
Biosynthesis of dTDP-α-d-Quip3NAc and dTDP-α-d-Fucp3NAc follows similar pathways. The crucial reaction of either pathway is the conversion of dTDP-4-oxo-6-deoxy-d-glucose into the respective dTDP-3-oxo-6-deoxy-d-hexose epimer by the 3,4-oxoisomerases QdtA and FdtA respectively. In a previous report, it was shown that FdtA is able to provide a 3-oxo substrate in the galacto-configuration at C-4 [15]. In the present study, we could show that QdtA forms a similar sugar residue, but retains the gluco-configuration at C-4. The transition of the 4-oxo group into the 3-oxo group is the crucial event for a component to be recognized as a substrate for the transaminases (QdtB and FdtB), followed by the last step of the cascade, the acetylation by the transacetylases (QdtC and FdtC).
Only recently, Davis et al. [37] have elucidated the X-ray structure of FdtA. These authors have determined the active site of the enzyme to be built by conserved His49 and His51 residues. The third His95 of the active site is supposed to be specific for 3,4-oxoisomerases, which convert the configuration at C-4. Furthermore, they speculate that His95 is replaced by an arginine residue in 3,4-oxoisomerases with function being retained. All of these relevant amino acids can be found in the QdtA sequence (Figure 5).
Figure 5. Amino acid sequence alignment of T. thermosaccharolyticum E207-71 QdtA, A. thermoaerophilus L420-91T FdtA and S. fradiae Tyl1a.
The amino acids of the active site are in boldface letters and the characteristic signature sequence is highlighted in grey. The positions of the conserved histidine residues (His49 and His51 in FdtA) are indicated by black dots. Note that His95 (indicated by a grey dot) in FdtA is an arginine residue in QdtA and in Tyl1a.
Acknowledgments
This work was supported by the Austrian Science Fund [project numbers P18013-B10 (to P.M.) and P19047-B12 (to C.S.)].
Abbreviations used
- α-d-Fucp3NAc
3-acetamido-3,6-dideoxy-α-d-galactose
- α-d-Quip3NAc
3-acetamido-3,6-dideoxy-α-d-glucose
- dTDP-α-d-Fucp3NAc
dTDP-3-acetamido-3,6-dideoxy-α-d-galactose
- dTDP-α-d-Quip3NAc
dTDP-3-acetamido-3,6-dideoxy-α-d-glucose
- GST
glutathione transferase
- LPS
lipopolysaccharide
- ORF
open reading frame
- RP-HPLC
reversed-phase HPLC
- S-layer
bacterial cell surface layer
- slg
S-layer glycosylation
REFERENCES
- 1.Messner P, Schäffer C. Prokaryotic glycoproteins. In: Herz W, Falk H, Kirby GW, editors. Progress in the Chemistry of Organic Natural Products. Volume 85. Springer, Wien, Austria, and New York: 2003. pp. 51–124. [DOI] [PubMed] [Google Scholar]
- 2.Keenleyside WJ, Whitfield C. Genetics and biosynthesis of lipopolysaccharides. In: Brade H, Opal SN, Vogel SN, Morrison DC, editors. Endotoxins in Health and Disease. Marcel Dekker; New York: 1999. pp. 331–358. [Google Scholar]
- 3.Graninger M, Kneidinger B, Bruno K, Scheberl A, Messner P. Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-β-l-rhamnose biosynthesis in the gram-positive thermophile Aneurinibacillus thermoaerophilus DSM 10155. Appl. Environ. Microbiol. 2002;68:3708–3715. doi: 10.1128/AEM.68.8.3708-3715.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kneidinger B, Marolda C, Graninger M, Zamyatina A, McArthur F, Kosma P, Valvano MA, Messner P. Biosynthesis pathway of ADP-l-glycero-β-d-manno-heptose in Escherichia coli. J. Bacteriol. 2002;184:363–369. doi: 10.1128/JB.184.2.363-369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zayni S, Steiner K, Pföstl A, Hofinger A, Kosma P, Schäffer C, Messner P. The dTDP-4-dehydro-6-deoxyglucose reductase encoding fcd gene is part of the surface layer glycoprotein glycosylation gene cluster of Geobacillus tepidamans GS5-97T. Glycobiology. 2007;17:433–443. doi: 10.1093/glycob/cwl084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novotny R, Pföstl A, Messner P, Schaffer C. Genetic organization of chromosomal S-layer glycan biosynthesis loci of Bacillaceae. Glycoconj. J. 2004;20:435–447. doi: 10.1023/B:GLYC.0000038290.74944.65. [DOI] [PubMed] [Google Scholar]
- 7.Altman E, Schäffer C, Brisson J-R, Messner P. Characterization of the glycan structure of a major glycopeptide from the surface layer glycoprotein of Clostridium thermosaccharolyticum E207-71. Eur. J. Biochem. 1995;229:308–315. doi: 10.1111/j.1432-1033.1995.tb20470.x. [DOI] [PubMed] [Google Scholar]
- 8.Giraud M-F, Naismith JH. The rhamnose pathway. Curr. Opin. Struct. Biol. 2000;10:687–696. doi: 10.1016/s0959-440x(00)00145-7. [DOI] [PubMed] [Google Scholar]
- 9.Frey PA. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10:461–470. [PubMed] [Google Scholar]
- 10.Köplin R, Brisson J-R, Whitfield C. UDP-galactofuranose precursor required for formation of the lipopolysaccharide O antigen of Klebsiella pneumoniae serotype O1 is synthesized by the product of the rfbDKPO1 gene. J. Biol. Chem. 1997;272:4121–4128. doi: 10.1074/jbc.272.7.4121. [DOI] [PubMed] [Google Scholar]
- 11.Aspinall GO, McDonald AG, Pang H, Kurjanczyk LA, Penner JL. Lipopolysaccharide of Campylobacter coli serotype O:30. Fractionation and structure of liberated core oligosaccharide. J. Biol. Chem. 1993;268:6263–6268. [PubMed] [Google Scholar]
- 12.Perry MB, MacLean LL. Structural characterization of the antigenic O-chain of the lipopolysaccharide of Escherichia coli serotype O65. Carbohydr. Res. 1999;322:57–66. doi: 10.1016/s0008-6215(99)00212-8. [DOI] [PubMed] [Google Scholar]
- 13.Kocharova NA, Blaszczyk A, Zatonsky GV, Torzewska A, Bystrova OV, Shashkov AS, Knirel YA, Rozalski A. Structure and cross-reactivity of the O-antigen of Providencia stuartii O18 containing 3-acetamido-3,6-dideoxy-d-glucose. Carbohydr. Res. 2004;22:409–413. doi: 10.1016/j.carres.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 14.Zych K, Perepelov AV, Siwinska M, Knirel YA, Sidorczyk Z. Structures of the O-polysaccharides and classification of Proteus genomospecies 4, 5 and 6 into respective Proteus serogroups. FEBS J. 2005;272:5536–5543. doi: 10.1111/j.1742-4658.2005.04958.x. [DOI] [PubMed] [Google Scholar]
- 15.Pfoestl A, Hofinger A, Kosma P, Messner P. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-α-d-galactose in Aneurinibacillus thermoaerophilus L420-91T. J. Biol. Chem. 2003;278:26410–26417. doi: 10.1074/jbc.M300858200. [DOI] [PubMed] [Google Scholar]
- 16.Perelle S, Dilasser F, Grout J, Fach P. Identification of the O-antigen biosynthesis genes of Escherichia coli O91 and development of a O91 PCR serotyping test. J. Appl. Microbiol. 2002;93:758–764. doi: 10.1046/j.1365-2672.2002.01743.x. [DOI] [PubMed] [Google Scholar]
- 17.He XM, Liu HW. Formation of unusual sugars: mechanistic studies and biosynthetic applications. Annu. Rev. Biochem. 2002;71:701–754. doi: 10.1146/annurev.biochem.71.110601.135339. [DOI] [PubMed] [Google Scholar]
- 18.Summers RG, Donadio S, Staver MJ, Wendt-Pienkowski E, Hutchinson CR, Katz L. Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in l-mycarose and d-desosamine production. Microbiology. 1997;143:3251–3262. doi: 10.1099/00221287-143-10-3251. [DOI] [PubMed] [Google Scholar]
- 19.Aguirrezabalaga I, Olano C, Allende N, Rodriguez L, Brana AF, Mendez C, Salas JA. Identification and expression of genes involved in biosynthesis of l-oleandrose and its intermediate l-olivose in the oleandomycin producer Streptomyces antibioticus. Antimicrob. Agents Chemother. 2000;44:1266–1275. doi: 10.1128/aac.44.5.1266-1275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doumith M, Legrand R, Lang C, Salas JA, Raynal MC. Interspecies complementation in Saccharopolyspora erythraea: elucidation of the function of oleP1, oleG1 and oleG2 from the oleandomycin biosynthetic gene cluster of Streptomyces antibioticus and generation of new erythromycin derivatives. Mol. Microbiol. 1999;34:1039–1048. doi: 10.1046/j.1365-2958.1999.01666.x. [DOI] [PubMed] [Google Scholar]
- 21.Cundliffe E, Bate N, Butler A, Fish S, Gandecha A, Merson-Davies L. The tylosin-biosynthetic genes of Streptomyces fradiae. Antonie van Leeuwenhoek. 2001;79:229–234. doi: 10.1023/a:1012065300116. [DOI] [PubMed] [Google Scholar]
- 22.Melançon CE, III, Yu WL, Liu HW. TDP-mycaminose biosynthetic pathway revised and conversion of desosamine pathway to mycaminose pathway with one gene. J. Am. Chem. Soc. 2005;127:12240–12241. doi: 10.1021/ja053835o. [DOI] [PubMed] [Google Scholar]
- 23.Melançon CE, III, Hong L, White JA, Liu YN, Liu HW. Characterization of TDP-4-oxo-6-deoxy-d-glucose-3,4-oxoisomerase from the d-mycaminose biosynthetic pathway of Streptomyces fradiae: in vitro activity and substrate specificity studies. Biochemistry. 2007;46:577–590. doi: 10.1021/bi061907y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melançon CE, III, Liu HW. Engineered biosynthesis of macrolide derivatives bearing the non-natural deoxysugars 4-epi-d-mycaminose and 3-N-monomethylamino-3-deoxy-d-fucose. J. Am. Chem. Soc. 2007;129:4896–4897. doi: 10.1021/ja068254t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli UK. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 30.Tonetti M, Sturla L, Bisso A, Benatti U, De Flora A. Synthesis of GDP-l-fucose by the human FX protein. J. Biol. Chem. 1996;271:27274–27279. doi: 10.1074/jbc.271.44.27274. [DOI] [PubMed] [Google Scholar]
- 31.Marchler-Bauer A, Bryant SH. CD-search: protein domain annotations on the fly. Nucleic Acid Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaara M. Eight bacterial proteins, including UDP-N-acetylglucosamine acyltransferase (LpxA) and three other transferases of Escherichia coli, consist of a six-residue periodicity theme. FEMS Microbiol. Lett. 1992;76:249–254. doi: 10.1016/0378-1097(92)90344-n. [DOI] [PubMed] [Google Scholar]
- 33.Nölling J, Breton G, Omelchenko MV, Makarova KS, Zeng Q, Gibson R, Lee HM, Dubois J, Qiu D, Hitti J, et al. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 2001;183:4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Yeung S-M, Que NLS, Müller T, Schmidt RR, Liu H-W. Expression, purification, and characterization of TylB, an aminotransferase involved in the biosynthesis of mycaminose. J. Am. Chem. Soc. 1999;121:7166–7167. [Google Scholar]
- 35.Naundorf A, Klaffke W. Substrate specificity of native dTDP-d-glucose-4,6-dehydratase: chemo-enzymatic syntheses of artificial and naturally occurring deoxy sugars. Carbohydr. Res. 1996;285:141–150. doi: 10.1016/s0008-6215(96)90180-9. [DOI] [PubMed] [Google Scholar]
- 36.Toukach FV, Bartodziejska B, Senchenkova SN, Wykrota M, Shashkov AS, Rozalski A, Knirel YA. Structure of a new acidic O-antigen of Proteus vulgaris O22 containing O-acetylated 3-acetamido-3,6-dideoxy-d-glucose. Carbohydr. Res. 1999;318:146–153. doi: 10.1016/s0008-6215(99)00088-9. [DOI] [PubMed] [Google Scholar]
- 37.Davis ML, Thoden JB, Holden HM. The X-ray structure of dTDP-4-oxo-6-deoxy-d-glucose-3,4-oxoisomerase. J. Biol. Chem. 2007;282:19227–19236. doi: 10.1074/jbc.M702529200. [DOI] [PubMed] [Google Scholar]