Colloidal particles have become valuable tools in biotechnology and medicine. Even a small sample provides a huge surface which can be engineered in many ways. One of the most versatile ways to engineer the surface of colloidal particles is to coat them with a polyelectrolyte multilayer by means of the layer-by-layer (lbl) technology.[1] Multifunctional composite colloidal devices can be fabricated in this way with nanometer precision in a radial direction. Since most of the important biomolecules—nucleic acids, proteins, and many carbohydrates—are essentially polyelectrolytes, the lbl technology has the potential to generate complex colloidal multilayer composites consisting of biological and artificial building blocks. This technology can thus be seen as a promising platform for the fabrication of bio-nanocomposite systems with inbuilt biological, chemical, and physical functions.[2]
Recently, lbl technology has been extended by fusing a virus membrane with a lipid layer on top of a polyelectrolyte multilayer cushion with the purpose of fabricating particles carrying virions on their surface.[3] Specifically, rubella virus like particles (RLPs) lacking the viral genome were crafted onto the colloidal surface of lipid-coated lbl colloids by employing membrane fusion at decreased pH values. To demonstrate the general aspect of this approach and, especially, to outline the versatility of possible surface modifications by the use of genetically modified virions, baculoviruses displaying foreign peptides on their surface were used as building blocks.
Baculoviruses, here Autographa californica nuclear polyhedrosis virus (AcNPV), have been developed to a surface display system for the screening of genetic libraries.[4,16] By displaying foreign peptides fused with the major-envelope glycoprotein gp64 at the surface of the virus,[5] it was possible to select surface-modified virions with a desired binding property. Colocalization of the displayed polypeptide and the corresponding gene makes it possible to propagate the “binders” and to further enrich these viruses, thus leading to a pool or a single clone decorated with a desired functional polypeptide. The fact that no a priori knowledge of the genetic sequence is necessary and that entire pools can be screened explains the impact of the several surface display systems developed in biotechnology[6] and, recently, also in materials science.[7]
Herein, we show how combining the baculovirus surface display system with colloidal particles has a number of advantages. For example, further functions can be added to the particles without interfering with those provided by the virus, by either engineering the multilayers underneath the lipid layer or the particle core in a desired way. This situation would not be possible with the virus display system alone. In particular, it would be hard to add functions to the virus other than those of the displayed peptide, simply because of the lack of available space and engineering possibilities. Furthermore, the handling of colloidal particles by modern sorting and analytical machines is well established.
A lipid bilayer adsorbed to the polyelectrolyte multilayer support[8] provides the starting point for further functionalization. The advantage of this approach is that the versatility of the lbl technology in regards to the composition and properties of the polyelectrolyte multilayer is combined[9] with an interface similar to biological systems.
Given the proper composition of the lipid layer on top one can attempt fusion with enveloped viruses at low pH values, that is, under conditions comparable to those inside endosomes.[3] This approach mimics the second step of the infection mechanism of many lipid-enveloped viruses.[10] After fusion, the virions present their membrane proteins on the colloidal surface. If virions which display engineered polypeptides on their surface are employed, these epitopes are also presented on the colloidal surface.
Figure 1 shows electron microscopy images of virus-decorated silica particles at different stages of the coating protocol. While the presence of the polyelectrolyte supporting layer in Figure 1a is not directly visible, a closer inspection of the radial intensity distribution (see inset) revealed its presence. A polyelectrolyte layer thickness of about 15 ± 2 nm was inferred from intensity line scans. The layer is rather smooth, probably as a result of drying it under the vacuum in the electron microscope. The heavy staining by the uranyl ions is explained by their strong interaction with the cations of the layer. The presence of the lipid layer in Figure 1b is immediately evidenced by a less-stained layer with a thickness of about 15–20 nm at the surface of the silica particle. This layer is composed of both the polyelectrolyte support and the adjacent lipid layer. The less-efficient staining in Figure 1b compared to Figure 1a is presumably caused by the limited permeability of the uranyl ions through the lipid layer.
Figure 1.
Transmission electron microscopy images of coated silica colloids: a) coated with 5 polyelectrolyte layers (PAH/PSS). A radial intensity scan is shown in the inset. The layer is identified as a slightly less dense outer region. b) Sample (a) with an additional lipid bilayer. c) RLPs fused with sample (b). d) and e): Sample (b) decorated with baculovirions. A virion is fused at its pole with the composite surface in (d), while a virion lies flat on the surface of the colloid in (e).
If these lipid-coated lbl colloids are incubated with virions in a buffer at low pH values (pH 4.5), fusion between the viral membrane and the lipid layer takes place to give virus-decorated particle surfaces. Baculoviruses can be seen attached to the surface of the colloidal particles in Figure 1d and e. Interestingly, the baculoviruses are quite often found in an upright position (Figure 1d), thus indicating the inherent asymmetry of the fusion protein distribution over their surface.[11] The membrane fusion proteins are thought to be preferentially found at one end of the virus, but viruses lying flat on the surface can also be found.
This approach is by no means restricted to baculoviruses. There are many degrees of freedom to subclone engineered polypeptides into other virions or viruslike particles which can subsequently be fused with the lipid layer attached to the colloid.[12] For example, Figure 1c shows lipid-coated lbl colloids fused with rubella virus like particles (RLPs).[13]
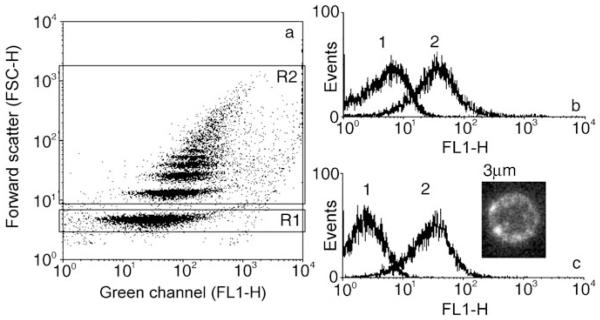
The set of proteins displayed on the colloidal surface can be monitored by immunofluorescence, which is a quantitative measure if analyzed by flow cytometry (fluorescence-assisted cell sorting, FACS). The fluorescence intensities of particles with a diameter of 1 μm displaying wild-type AcNPV are shown in Figure 2a and b. The presence of the baculovirus membrane fusion protein gp64 was detected with B12D5, a monoclonal, mouse-derived anti-gp64 antibody,[14] followed by incubation with a secondary anti-mouse conjugate labeled with fluorescein isothiocyanate (FITC), which actually provided the fluorescence signal in the FL1-H channel. Figure 2a shows a dot plot of the fluorescence intensity of colloids coated with wild-type AcNPV against the respective forward-scattering intensity. The scatter plot R1 of the FACS signal represents the population of single coated colloids, whereas region R2 summarizes the aggregates of order two and higher. The resolution is sufficient to resolve aggregates up to the sixth order. The quality of the preparations in terms of the presence of aggregates depends largely on the lipid-coating step and decreases slightly as the size of the colloids decrease. Whereas the preparation of 1-μm lbl colloids in Figure 2a shows 40% single colloids, the percentage of single particles in the case of 3-μm colloids is about 75%. Single particles corresponding to R1 in Figure 2a have been gated and are shown in terms of fluorescence intensity versus particle counts in Figure 2b. Curve 2 refers to 1-μm lipid lbl colloids coated with wild-type AcNPV. Curve 1 represents the control measured before the virus was added. In Figure 2c baculoviruses were employed which had a 17 amino acid long epitope of HIV-1 gp120 glycoprotein engineered to their surface. The inset in Figure 2c shows a confocal laser scanning microscopy (CLSM) image obtained with immunofluorescence. Although the resolution of the CLSM device is not sufficient to determine the size and shape of the coated baculoviruses exactly, the fluorescent spots on the colloidal surface indicate the presence of the viral elements. Figure 2c compares the immunofluorescence of particles carrying a virus displaying a HIV-1 gp120 glycoprotein fragment[15] (curve 2) with the fluorescence of particles covered with the wild-type virus (curve 1). The engineered HIV-1 gp120 glycoprotein fragment is clearly detectable.
Figure 2.
a) and b): Immunofluorescence against 1-μm lipid-coated colloids fused with wild-type baculovirions (wt-AcNPV). a) Dot plot, where each dot provides a measured event, for which the forward scattering (FSC-H) and the fluorescence in the FITC channel (FL1-H) were recorded and used for subsequent analysis. Region R1 is assigned to single colloids, and R2 to particle aggregates. b) Single colloids have been gated and their fluorescence versus particle counts are shown. Curve 1: Lipid-coated lbl colloids, curve 2: lipid-coated lbl colloids fused with wt-AcNPV. c) Inset: CLSM scan of a 3-μm colloid coated with AcNPV displaying an antibody-binding fragment of HIV-1 gp120 (curve 2). Curve 1: colloids with wt-AcNPV; curve 2: colloids displaying the HIV-1 gp120 epitope on their surface.
This presence of the engineered HIV-1 gp120 antibody-binding fragment on the colloidal surface can be regarded as a “proof of principle”, which consists of linking a combinatorial surface display strategy, that is, the so-called epitope mapping approach,[16,4] with the engineering of a functional colloidal surface.[1]
The direct use of the viral building elements, which had been previously screened for functional polypeptide, has the major advantage that a loss of the functionality of the screened epitope is rather unlikely to occur. In phage display systems, the functional polypeptide is, for example, selected on a phage. When it is subsequently used attached to another surface there is always a potential risk that functionality is lost, since the latter was provided in part by the phage surface itself or by the peptide–phage interaction and not by the peptide alone.
Fusion of viruses with the lipid-coated lbl composites, which is the key step of coating, is closely related to the infectivity of the virions. This ensures that the virus surface properties can be transferred to the lbl composites as long as the virions can still be produced and propagated in the cell culture despite the introduced surface-protein modifications. Therefore it is possible to equip the lipid-coated colloids with a number of functions if virus populations displaying different epitopes or different virus species are mixed before coating.
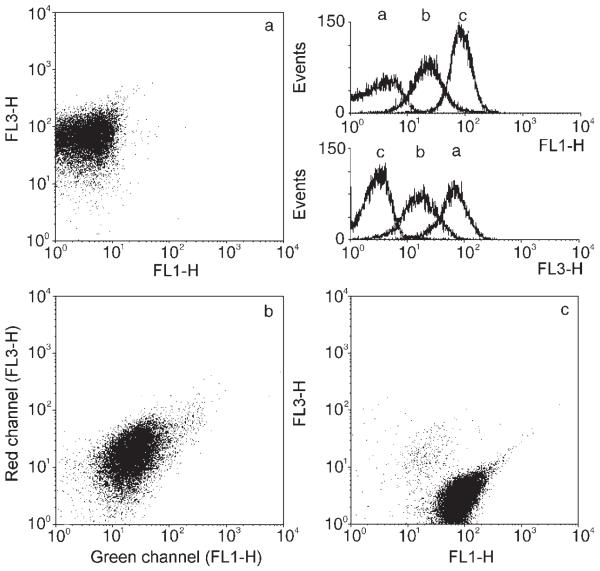
To prove this idea, a 1:1 mixture of two different baculovirus populations—one displaying the streptag peptide, a molecular mimic of biotin, the other displaying an epitope derived from HIV-1 gp41[17]—was prepared. Figure 3a shows the results obtained with the colloids coated with only the streptag-displaying baculoviruses, Figure 3c corresponds to colloids coated with viruses displaying only the HIV-1 gp41 epitope, and Figure 3b corresponds to colloids coated with a mixture of both baculo clones. All three samples were incubated with streptavidin labeled with Quantum Red for the detection of the streptag epitope, with the anti HIV-1 gp41 antibody 3D6, and with an anti-human, FITC-labeled antibody conjugate, respectively. The dot plots shown in Figure 3a, b, and c demonstrate the specific binding of the fluorescent conjugates on the three different colloidal surfaces. The streptag epitope carrying colloids show up as the right-most fluorescence distribution in the red channel (FL3-H) and as the left-most distribution in the green FITC channel (FL1-H). The appearance of the HIV-1 gp41 epitope carrying colloids is the opposite way round. Colloids carrying both epitopes, which were prepared from the mixture, show an intermediate distribution in both channels, thus indicating that sample b indeed displays both engineered epitopes on its surface, however, as expected, at a diminished frequency.
Figure 3.
Dot plots of 3-μm colloids fused with two different genetically engineered baculoviruses: a) with baculoviruses displaying streptag, c) with virions displaying an HIV-1 gp41 epitope, b) with a mixture of both. Whereas in (a) and (c) only one epitope is evident, both epitopes show up in (b). The FITC-channel (FL1-H) was used for antibody detection, while the Quantum Red fluorescence sensitive channel (FL3-H) was used for streptag detection.
Baculoviruses displaying the streptag peptide were chosen since they substantially extend the display possibilities of the system. The streptag peptide binds with great specificity to streptavidin which, as a result of its four biotin binding sites, can be used as a molecular linker between the viruslike surface of the lbl composite and biotinylated species. If a desired sequence, for example, cannot be displayed, because it interferes with fusion, it can nevertheless be added afterwards by utilizing the streptag linker.
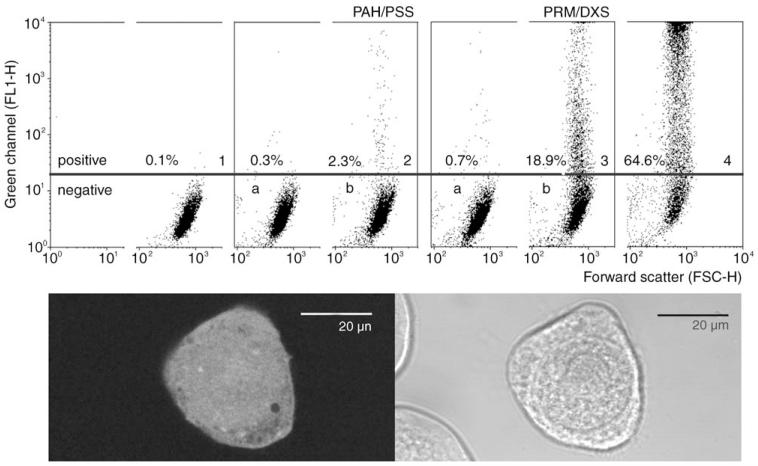
When using viruses as modules for the colloidal display system, it is of great interest as to whether they can still be propagated after they have become part of the colloidal composite. On one hand virus viability is crucial in regards to the issue of biological safety,[18] while on the other hand it has a large impact on the technology itself. For example, if viruses attached to colloids can be propagated, this would allow screening of the engineered functionalities at the level of the composites themselves. Thus, experiments were performed with baculoviruses which contain an expression cassette on their genome encoding for the green fluorescent protein (GFP).[19] They were coated onto 3-μm particles and added to Spodoptera frugiperda (Sf9) cells. The coated colloids were readily endocytosed and could be detected inside the cells. Particles inside endosomes and also free particles in the cytoplasma have been observed. The possibility of propagation is closely connected to the stability of the composite within a biological environment. Two different samples were prepared, one carrying rather stable poly(allylamine hydrochloride) and poly(sodium 4-styrenesulfonate) (PAH/PSS) layers and the other containing the biologically degradable protamine/dextrane sulfate couple. Protamine release has been shown previously for the latter sample.[20] The cells were tested after 72 h for GFP expression by means of FACS analysis. The CLSM image in Figure 4 indeed shows a virus-infected, GFP-expressing Sf9 cell with one visible particle appearing as a dark sphere in the bottom right region of the cell. The image recorded in the transmission mode shows that not all cells express GFP. GFP expression is quantified in Figure 4 (panels 1–4). The control images 1, 2a, and 3a show no transfection, thus proving the absence of viruses in the supernatant. Only a small rate of transfection (2.3%) was observed when lbl colloids with a PAH/PSS support were applied. Baculoviruses on colloids with protamine/dextrane sulfate layers underneath the lipid layer, however, led to considerable transfection of 18.9%, which is about ten times more than with PAH/PSS. The transfection rate achieved by adding free virus to the cell culture reached a value of 64.6%. Although more insight into the mechanism of decomposition of such composites is needed, the stability of the polyelectrolytes used for the assembly seems to play an important role for the decomposition of the viruslike surface. It cannot be excluded either that the lipid polyelectrolyte interaction may contribute to the difference in transfection efficiencies.
Figure 4.
FACS dot plots (FL1-H versus FSC) of Sf9 cells incubated for 72 h with baculovirus-coated 3-μm colloids. The baculoviruses contained a GFP-expression cassette to monitor infection. GFP is recorded in FL1-H. Panel 1: control cells, panel 2b: cells incubated with virus-coated colloids carrying a PAH/PSS multilayer support; panel 3b: with protamine/dextrane sulfate support; panel 4: cells inoculated with free baculoviruses; panels 2a and 3a: controls, cells incubated with supernatants of the colloidal samples. The CLSM image shows a GFP-expressing Sf9 cell with an incorporated 3-μm colloid. The transmission image demonstrates the presence of two other cells which have not been transfected.
Experiments are currently under way to explore the technological potential of these engineered systems in diagnostics and other fields. For example, a bead array with four particle populations of different fluorescence intensity carrying AcNPV with the HIV-1 gp120 epitope, wild-type AcNPV, UV-inactivated influenza A/PR8, and influenza A type Singapore were prepared, mixed, and added to sera containing antibodies to either one of these antigenes. Figure 5 shows that the presence of the antibodies can be verified. The advantage of the bead array approach is that very small sample volumes are required. The virus-decorated lbl particles can be easily prepared and provide a great versatility in regard to the use of antigenes given the possibilities of the display approach. It is worth mentioning that the lipid layer serves not only as a means for virus fusion during the process of fabrication, but, most importantly, inhibits in a universal manner nonspecific interactions with the antibodies, a major problem associated with surface-bound antigenes.
Figure 5.
FACS dot plots of virus-decorated lbl colloids used as a bead array for antibody detection. Particles carrying different amounts of fluorescent polyelectrolyte layers in the red channel (FL2-H) and four different virus species, respectively. 1: influenza A/Singapore, 2: wild-type AcNPV, 3: influenza A/PR8, 4: AcNPV with HIV-1 gp120 epitope. In a) ARP 360 (directed against the engineered HIV-1 gp120 epitope), b) B12D5 (directed against gp64, the major envelope protein of AcNPV), c) polyclonal sera against influenza A/PR8, and d) influenza A/Singapore was added to the samples. The different colloidal populations display corresponding fluorescence signals with respect to the bound antigens on the colloidal surface.
Although there are many challenging questions left, the suggested colloid/lbl/lipid layer/virus composite may have considerable technological potential. It provides flexibility, is easy to prepare, and fits well to the currently available analysis techniques. We hope it will find its way toward applications in the interdisciplinary field of nanobiotechnology, thus offering highly versatile possibilities in the engineering of proteinogenic surfaces.
Experimental Section
Materials
Silica particles with diameters of 3 μm, 1 μm, and 500 nm were purchased from microparticles GmbH (Berlin, Germany). Poly(allylamine hydrochloride) (PAH; M.W. ca. 70000) and poly(sodium 4-styrenesulfonate) (PSS; M.W. ca. 70000) were obtained from Aldrich. Protamine sulfate (PRM) from herring and dextrane sulfate sodium salt (DXS; M.W. ca. 500000) were obtained from Sigma. l-α-Phosphatidylserine (PS; porcine brain, sodium salt; 20 mgmL−1 in chloroform) and l-α-phosphatidylcholine (PC; egg, chicken; 20 mgmL−1 in chloroform) were purchased from Avanti Polar Lipids, Inc. Quantum Red labeled streptavidin, anti-mouse IgG-FITC, and anti-human IgG-FITC (γ-chain specific) were purchased from Sigma. ARP 360[21] was obtained from The Aids Reagent Project. The hmAb 3D6[17] was kindly provided by H. Katinger (IAM, Vienna, Austria). B12D5[14] was provided by L. Volkman (University of California Berkeley, USA).
RLP preparation
RLPs were isolated according to Fischlechner et al.[3]
Baculoviruses and insect cells
All baculoviral constructs were derived from wild-type AcNPV (Autographa californica nuclear polyhedrosis virus, ATCC). Standard protocols for virus propagation as were applied described by O’Reilly et al.[22] The baculoviral clone AcCOPS-PCR-LIB-Frg10 (Frg10), which displays a fragment of HIV-1 (IIIB) gp120 on its surface, was constructed as described in the literature.[16]
The baculoviral clone AcCOIN-3D6 was constructed in a similar fashion.[5] The construction of AcOmega-GFP and AcCOIN-streptag is described by Ernst et al.[5]
The total protein content of ultracentrifuged baculoviruses was determined by using the Bio-Rad Protein Assay. Sf9 cells (Spodoptera frugiperda, ATCC) were grown in modified IPL-41 (Sigma–Aldrich, Munich, Germany) as described by Toellner.[16]
Particle preparation
Silica particles were coated with 5 poly-electrolyte layers. PAH and PSS were dissolved in 0.5m NaCl in a concentration of 1 mgmL−1 for the coating of 3-μm particles (3 mgmL−1 and 6 mgmL−1 for 1 μm and 500 nm particles, respectively). After each adsorption step (10 min at room temperature) the colloidal suspension was washed by centrifuging 3 times in 0.1m NaCl. Coating with the PRM/DXS couple (first layer PAH) was performed in an analogous manner, except that PRM was dissolved in 0.1m NaCl and 7 multilayers were adsorbed.
Addition of the lipid layer
Small unilamellar liposomes consisting of PS and PC (75% PS, 25% PC (mol mol−1), in 0.1m NaCl) were prepared by extrusion through a polycarbonate membrane with a pore diameter of 50 nm. The lipid bilayer was formed by adsorption of unilamellar vesicles onto multilayer-coated colloids with PAH or PRM as the top layer. The vesicles and the substrates were stirred for 30 min in 0.1m NaCl at room temperature. After incubation, the samples were washed 3 times with 0.1m NaCl or phosphate-buffered saline (PBS). Liposomes made from 1 mg phospholipids were used for the coating with (100 μL, 5% (v/v) stock solution) 3-μm lbl colloids (3 mg and 6 mg for 1-μm and 500-nm particles, respectively).
Fusion procedure
The fusion of baculovirions onto the lipid-coated colloids was conducted in 0.2m phosphate/0.1m citrate buffer at pH 4.5 by incubation of lipid-coated colloids with the virions for 10 mins at room temperature followed by 3 washings with PBS. Typically, viral protein (8 μg) was used for coating the lipid-coated 3-μm LbL colloids (10 μL, 5% (v/v) stock solution; 25 μg and 50 μg viral protein were used for 1-μm and 500-nm particles, respectively).
Electron microscopy
Colloids of 500 nm size were prepared as described before and suspended in PBS. Colloids were adsorbed onto the microscope grid by floating it on a drop of sample suspension for 1 min. The adsorbed colloids were then negatively stained by floating the grid on a drop of 1% aqueous uranyl acetate solution for another minute. Images were taken on a Philips CM12 electron microscope, operated at 80 keV acceleration voltage.
Immunofluorescence testing
Typically, colloidal sample (10 μL; 5% (v/v)) was incubated with the primary antibody, diluted 1:50 in PBS/5% FCS for 1 h followed by two washings with PBS. The secondary antibody was added in similar fashion. After washing the samples twice with PBS, they were analyzed by flow cytometry (FACSCalibur (Becton Dickinson)), with 10000 single colloids measured. The data were analyzed using the WinMDI software written by J. Trotter.
Confocal laser scanning microscopy (CLSM)
CLSM images were obtained using a Leica TCS SP2 microscope. Images were analyzed using ImageJ software (Rasband, W.S., ImageJ, U.S. National Institutes of Health, Bethesda, Maryland, USA).
Viability assay
3-μm lipid-coated colloids, one sample made up of 5 layers of PAH/PSS, the other made up of PAH following 6 layers PRM/DXS, were fused with AcOmega-GFP as explained above. The colloids were incubated with Sf9 cells for 72 h in a ratio of 5 colloids per cell, then analyzed by means of FACS. As negative controls, the supernatants of the last washing steps of the colloidal samples were given to Sf9 cells. As a positive control, native AcOmega-GFP (0.02 μg total protein) was applied.
Footnotes
This research was supported by a grant from the Volkswagenstiftung within the framework of the program “Complex Materials” (VW foundation I/78 168). We thank Wolfgang Ernst for advice and discussions, Alexandra Spenger for the construction of AcCOIN-3D6, Nicole Borth for help with FACS analysis, and Harald Berger for support with CLSM imaging. Many thanks to Julia Romanova and Boris Ferko for influenza A viruses and Bernhard Benke for designing Figure 1.
Contributor Information
Martin Fischlechner, Institute of Medical Physics and Biophysics, Leipzig University, Härtelstrasse 16–18, 04107 Leipzig (Germany).
Dr. Lars Toellner, Institute of Medical Physics and Biophysics, Leipzig University, Härtelstrasse 16–18, 04107 Leipzig (Germany)
Prof. Dr. Paul Messner, Center for NanoBiotechnology, University of Natural Resources and Applied Life Sciences, 1180 Vienna (Austria)
Prof. Dr. Reingard Grabherr, Institute of Applied Microbiology, University of Natural Resources and Applied Life Sciences, 1190 Vienna (Austria)
Prof. Dr. Edwin Donath, Institute of Medical Physics and Biophysics, Leipzig University, Härtelstrasse 16–18, 04107 Leipzig (Germany).
References
- [1].a) Donath E, Sukhorukov GB, Caruso F, Davis SA, Möhwald H. Angew. Chem. 1998;110:2324. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2201::AID-ANIE2201>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 1998;37:2202. [Google Scholar]; b) Caruso F, Caruso RA, Möhwald H. Science. 1998;282:1111. doi: 10.1126/science.282.5391.1111. [DOI] [PubMed] [Google Scholar]; c) Decher G. Science. 1997;277:1232. [Google Scholar]
- [2].a) Peyratout CS, Dähne L. Angew. Chem. 2004;116:3850. doi: 10.1002/anie.200300568. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2004;43:3762. [Google Scholar]; b) Sukhorukov GB, Rogach AL, Zebli B, Liedl T, Skirtach AG, Köhler K, Antipov AA, Gaponik N, Susha AS, Winterhalter M, Parak WJ. Small. 2005;1:194. doi: 10.1002/smll.200400075. [DOI] [PubMed] [Google Scholar]
- [3].Fischlechner M, Zschörnig O, Hofmann J, Donath E. Angew. Chem. 2005;117:2952. doi: 10.1002/anie.200460763. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2005;44:2892. [Google Scholar]
- [4].a) Grabherr R, Ernst W, Oker-Blom C, Jones I. Trends Biotechnol. 2001;19:231. doi: 10.1016/s0167-7799(01)01610-9. [DOI] [PubMed] [Google Scholar]; b) Grabherr R, Ernst W. Comb. Chem. High Throughput Screening. 2001;4:185. doi: 10.2174/1386207013331165. [DOI] [PubMed] [Google Scholar]
- [5].a) Ernst W, Grabherr R, Wegner D, Borth N, Grassauer A, Katinger H. Nucleic Acids Res. 1998;26:1718. doi: 10.1093/nar/26.7.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ernst WJ, Spenger A, Toellner L, Katinger H, Grabherr RM. Eur. J. Biochem. 2000;267:4033. doi: 10.1046/j.1432-1327.2000.01439.x. [DOI] [PubMed] [Google Scholar]; c) Spenger A, Grabherr R, TMllner L, Katinger H, Ernst W. Eur. J. Biochem. 2002;269:4458. doi: 10.1046/j.1432-1033.2002.03135.x. [DOI] [PubMed] [Google Scholar]
- [6].a) Smothers JF, Henikoff S, Carter P. Science. 2002;298:621. doi: 10.1126/science.298.5593.621. [DOI] [PubMed] [Google Scholar]; b) Benhar I. Biotechnol Adv. 2001;19:1. doi: 10.1016/s0734-9750(00)00054-9. [DOI] [PubMed] [Google Scholar]
- [7].a) Sarikaya M, Tamerler C, Jen AKY, Schulten K, Baneyx F. Nat. Mater. 2003;2:577. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]; b) Lee SW, Mao C, Flynn CE, Belcher AM. Science. 2002;296:892. doi: 10.1126/science.1068054. [DOI] [PubMed] [Google Scholar]; c) Mao C, Flynn CE, Hayhurst A, Qi R, Sweeney J, Georgiou G, Iverson B, Belcher AM. Proc. Natl. Acad. Sci. USA. 2003;100:6946. doi: 10.1073/pnas.0832310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Moya S, Donath E, Sukhorukov GB, Auch M, Bäumler H, Lichtenfeld H, Möhwald H. Macromolecules. 2000;33:4538. [Google Scholar]; b) Moya S, Richter W, Leporatti S, Bäumler H, Donath E. Biomacromolecules. 2003;4:808. doi: 10.1021/bm034013r. [DOI] [PubMed] [Google Scholar]
- [9].a) Caruso F, Schüler C. Langmuir. 2000;16:9595. [Google Scholar]; b) Peyratout CS, Dähne L. Angew. Chem. 2004;116:3850. doi: 10.1002/anie.200300568. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2004;43:3762. [Google Scholar]
- [10].Jardetzky TS, Lamb RA. Nature. 2004;427:307. doi: 10.1038/427307a. [DOI] [PubMed] [Google Scholar]
- [11].Chapple SDJ, Jones IM. J. Biotechnol. 2002;95:269. doi: 10.1016/s0168-1656(02)00023-8. [DOI] [PubMed] [Google Scholar]
- [12].a) Briggs JAG, Wilk T, Fuller SD. J. Gen. Virol. 2003;84:757. doi: 10.1099/vir.0.18779-0. [DOI] [PubMed] [Google Scholar]; b) Schnell MJ, Buonocore L, Kretzschmar E, Johnson E, Rose JK. Proc. Natl. Acad. Sci. USA. 1996;93:11359. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hobman TC, Lundstrom ML, Mauracher CA, Woodward L, Gillam S, Farquhar MG. Virology. 1994;202:574. doi: 10.1006/viro.1994.1379. [DOI] [PubMed] [Google Scholar]
- [14].Keddie BA, Aponte GW, Volkman LE. Science. 1989;243:1728. doi: 10.1126/science.2648574. [DOI] [PubMed] [Google Scholar]
- [15].In brief, a library consisting of 60 overlapping PCR-derived DNA fragments of the HIV-1 gp120 sequence was cloned into the baculovirus genome in a way that the corresponding polypeptides were presented as part of an additional gp64 membrane-fusion protein. Baculovirus-infected cells containing the binding sequence for a selected antibody at their membrane were identified by their immunofluorescence and sorted by means of FACS. Subsequent virus propagation yielded a monoclonal virus population displaying the selected epitope sequence.
- [16].Toellner L. Establishment of a baculovirus surface display library using the major envelope protein gp120 from human immunodeficiency virus 1 as a model. University of Natural Resources and Applied Life Sciences; Vienna, Austria: 2002. http://edocs.tu-berlin.de/diss/2002/toellner_lars.htm PhD Thesis. [Google Scholar]
- [17].a) Grunow R, Jahn S, Porstmann T, Kiessig SS, Steinkellner H, Steindl F, Mattanovich D, Gurtler L, Deinhardt F, Katinger H. J. Immunol. Methods. 1988;106:257. doi: 10.1016/0022-1759(88)90206-2. [DOI] [PubMed] [Google Scholar]; b) Spenger A, Grabherr R, Töllner L, Katinger H, Ernst W. Eur. J. Biochem. 2002;269:4458. doi: 10.1046/j.1432-1033.2002.03135.x. [DOI] [PubMed] [Google Scholar]
- [18].a) Viruses with pathogenic features probably can also be used as building blocks, because fusion mechanisms appear to be quite general;; Dimitrov DS. Nat. Rev. Microbiol. 2004;2:109. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ernst W, Grabherr R, Wegner D, Borth N, Grassauer A, Katinger H. Nucleic Acids Res. 1998;26:1718. doi: 10.1093/nar/26.7.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hiller S, Leporatti S, Schnäckel A, Typlt E, Donath E. Biomacromolecules. 2004;5:1580–1587. doi: 10.1021/bm049875m. [DOI] [PubMed] [Google Scholar]
- [21].Moore JP, Sattenau QJ, Yoshiyama H, Thali M, Charles M, Sullivan N, Poon SW, Fung MS, Traincard F, Pinkus M. J. Virol. 1993;67:6136. doi: 10.1128/jvi.67.10.6136-6151.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].O’Reilly DR, Miller LK, Luckow VA. Baculovirus expression vectors: a laboratory manual. W. H. Freeman; New York: 1992. [Google Scholar]