Summary
Pseudoalteromonas antarctica NF3 is an Antarctic psychrotolerant Gram-negative bacterium that accumulates large amounts of an extracellular polymeric substance (EPS) with high protein content. Transmission electron microscopy analysis after high-pressure freezing and freeze substitution (HPF-FS) shows that the EPS is composed of a capsular polymer and large numbers of outer membrane vesicles (OMVs). These vesicles are bilayered structures and predominantly spherical in shape, with an average diameter of 25–70 nm, which is similar to what has been observed in OMVs from other Gram-negative bacteria. Analyses of lipopolysaccharide (LPS), phospholipids and protein profiles of OMVs are consistent with the bacterial outer membrane origin of these vesicles. In an initial attempt to elucidate the functions of OMVs proteins, we conducted a proteomic analysis on 1D SDS-PAGE bands. Those proteins putatively identified match with outer membrane proteins and proteins related to nutrient processing and transport in Gram-negative bacteria. This approach suggests that OMVs present in the EPS from P. antarctica NF3, might function to deliver proteins to the external media, and therefore play an important role in the survival of the bacterium in the extreme Antarctic environment.
Introduction
Cold ecosystems typically constitute a reservoir of novel and interesting microorganisms (Cavicchioli, 2006). In recent years, cold-adapted microorganisms have received greater attention for their potential biotechnological applications (Gerday et al., 2000; Cavicchioli et al., 2002; 2006). Several studies seeking to characterize cold-adapted microorganisms in the relatively unexplored Arctic and Antarctic environments have been conducted (Sullivan and Palmisano, 1984; Bowman et al., 1997; Krembs and Engel, 2001; Junge et al., 2002). Our group has isolated several microorganisms from Antarctic samples, which have been characterized as new species belonging to different taxonomic groups (Llarch et al., 1997; Montes et al., 1999; 2004; Bozal et al., 2002; 2003). One of these bacteria is Pseudoalteromonas antarctica NF3, a psychrotolerant Gram-negative bacterium isolated from mucous material near the inlet to Admiralty Bay (King George Island, South Shetland Islands, Antarctica) (Bozal et al., 1997). The bacterium accumulated large amounts of an extracellular polymeric substance (EPS), with chemical analysis revealing that the primary feature of this new polymer was the presence of high protein content (Bozal et al., 1994). This, however, was not the only feature that caught our attention; it also exhibited a number of other interesting properties, such as the ability to coat phosphatidylcholine liposomes and to protect these vesicles against different surfactants (Bozal et al., 1996; de la Maza et al., 1998a,b).
Extracellular polymers are produced by many marine bacteria and it has been demonstrated that EPS production enhances the growth and survival of microbes in natural systems (Costerton, 1999; Wolfaardt et al., 1999). In recent years, there has been a growing interest in isolating new exopolysaccharide-producing bacteria from extreme marine environments. Chemical characterization of these polymers has demonstrated that marine bacterial isolates can produce a variety of exopolysaccharides (Rougeaux et al., 1996; Raguénès et al., 1997; Mancuso Nichols et al., 2005a). A recent study by Mancuso Nichols and colleagues (2005b) has shown that closely related strains of Antarctic Pseudoalteromonas are able to produce abundant EPS. Moreover, chemical characterization of these polymers has shown that they were diverse, despite the close taxonomic proximity of these strains. Although polysaccharides are the most abundant component of the EPSs (Flemming and Wingender, 2001) other macromolecules such as proteins were found in EPSs from different Pseudoalteromonas strains (Bozal et al., 1994; Mancuso Nichols et al., 2005b). Further studies, however, are necessary to elucidate the structure of Antarctic marine bacterial exopolymers. In particular, little is known about the ultrastructure of EPS from Pseudoalteromonas strains, nor are the patterns and functions of the proteins found in such exopolymers well understood.
Previous electron microscopy studies explored the possibility that part of the protein content detected in the EPS from P. antarctica NF3 was released to the media through membrane vesicles (Nevot et al., 2006). Outer membrane vesicles (OMVs) are common to a wide variety of Gram-negative bacteria and are produced during the course of normal metabolism and cell growth (Mayrand and Grenier, 1989; Allan et al., 2003). They are spherical bilayered lipid vesicles ranging in size from 50 to 200 nm. These vesicles are extruded from regions of the bacterial outer membrane and they contain lipopolysaccharide (LPS), periplasmic proteins, outer membrane proteins and phospholipids (Zhou et al., 1998). The function of OMVs is unclear but different studies confirm they can be viewed as a new form of secretion (Beveridge, 1999; Wai et al., 2003). To date, studies of OMVs from Gram-negative bacteria have focused on the delivery of virulence factors (Kadurugamuwa and Beveridge, 1995; 1999; Beveridge and Kadurugamuwa, 1996; Kolling and Matthews, 1999; Horstman and Kuehn, 2000; Yaron et al., 2000; Kato et al., 2002; Khandelwal and Banerjee-Bhatnagar, 2003), however, little information is available about the secretion of specialized proteins through OMVs from non-pathogenic bacteria (Beveridge et al., 1997).
The aim of this study was to analyse the presence and function of OMVs produced from the non-pathogenic psychrotolerant Antarctic bacterium P. antarctica NF3. Outer membrane vesicles were visualized by electron microscopy following high-pressure freezing and freeze substitution (HPF-FS) methods. Analysis of the protein profile via PAGE electrophoresis, LPS profile and phospholipid content was carried out to confirm the outer membrane origin of OMVs. In an initial attempt to elucidate their potential functions, a proteomic analysis of the main proteins of vesicles was performed.
Results
Transmission electron microscopy (TEM)
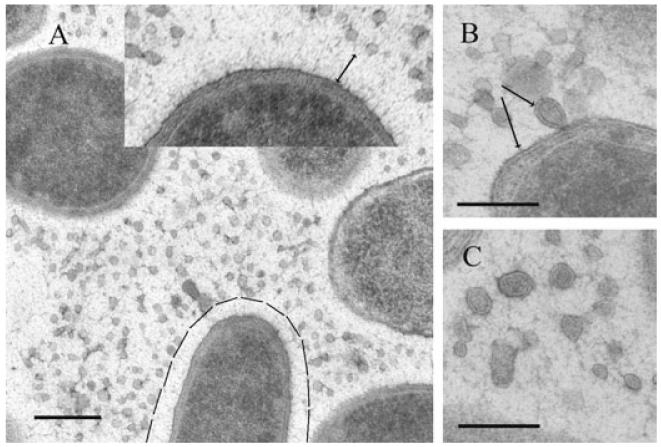
An examination of P. antarctica NF3 cells by TEM following HPF-FS revealed interesting ultrastructural features about the EPS secreted by this bacterium. Cell surfaces appeared to be covered by a halo approximately 60 nm thick, consisting of fine fibres standing perpendicularly to the cell wall structure (marked area on a cell and inset in Fig. 1A). In vicinity of this halo a net-like meshwork of more randomly distributed fibres was observed extending far beyond the individual cells. These two polymeric arrangements of fibres could correspond to the capsular material of P. antarctica. In addition, on thin sections a huge number of spherical structures were observed resembling OMVs of Gram-negative bacteria. These spherical structures were mainly interspersed among cells within the randomly distributed fibres, but they were also observed adhered to bacterial cell surfaces (Fig. 1B). The surface of these vesicles, with diameters ranging from 25 nm to 70 nm, consisted of a lipid bilayer. This was clearly visible at higher magnifications (Fig. 1B and C). Of note was (i) the presence of a unique membrane structure in these OMVs possessing a uniform width (10 nm ± 2) similar to that observed in bacterial outer membranes (Fig. 1B arrows) and (ii) the presence of an electrondense matter in the vesicles similar to that observed in the bacterial periplasmic space (Fig. 1B).
Fig. 1.
TEM micrographs of ultrathin sections from Pseudoalteromonas antarctica NF3 prepared by HPF-FS.
A. Shows a general view of cells as well as large amounts of outer membrane vesicles (OMVs). Cells are surrounded by an inner layer of oriented fibres (see marked area and magnified inset) and an extended meshwork of randomly distributed fine fibres.
B. Provides a magnified view of an OMV attached to the cell surface. The same bilayered structure is observed around the vesicle and for the bacterial outer membrane (see arrows).
C. Provides a magnified view in which OMVs released from the cells can be observed. Bar in A is 200 nm, and bars in B, C are 100 nm.
Isolation of OMVs
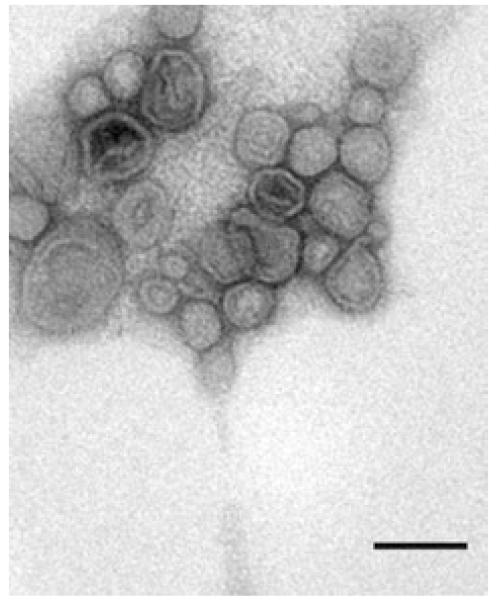
Outer membrane vesicles were isolated from clarified culture supernatant, as described in Experimental procedures. The pellet obtained after ultracentrifugation was resuspended and a small aliquot was negatively stained and examined by TEM. In these preparations bacterial flagella were observed together with OMVs. To remove flagella and other contaminating material, we purified OMVs on a sucrose density gradient. After ultracentrifugation we obtained one diffuse band near the bottom of the gradient and a pellet at the bottom of the tube. Each fraction was negatively stained and analysed by electron microscopy. We confirmed that not only the diffuse band (ρ = 1.19 g ml−1) contained exclusively OMVs without associated flagella (Fig. 2), but also that the pellet was mainly composed of flagella (data not shown).
Fig. 2.
Negative staining micrograph from purified P. antarctica NF3-OMVs. Bar is 50 nm.
Analysis of OMV protein and lipid content
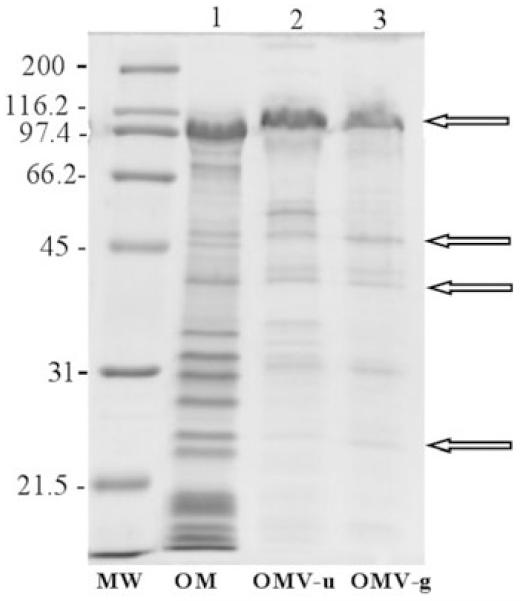
Outer membrane vesicles appear to be generated by budding from the outer membrane of the bacterium. To assess their outer membrane origin, the protein profiles of sedimented (OMV-u) and purified (OMV-g) vesicles, as well as the protein profile of outer membrane samples, were compared by SDS-PAGE (Fig. 3). Vesicles preparations exhibited at least four major polypeptides of 109, 48, 42 and 24 kDa which comigrated with polypeptides that were present in outer membrane preparations (Fig. 3 arrows). We observed that the outer membrane samples contained additional proteins that were not present or were undetectable in vesicles (compare Fig. 3, lane 1 with lanes 2 and 3). However, two bands of 44 and 31 kDa were present in sedimented and purified vesicles but absent in outer membrane preparations. Proteins with an apparent molecular mass of 52.5, 34.5 and 33 kDa were only detectable in sedimented vesicles and disappeared after vesicle purification. Some of these bands could correspond to flagella proteins which were eliminated during the purification process.
Fig. 3.
Coomassie-stained SDS-PAGE (12%) protein profiles of P. antarctica NF3 outer membrane fraction (OM); outer membrane vesicles isolated from supernatant by ultracentrifugation (OMV-u) and OMVs purified by sucrose gradient (OMV-g). Molecular mass marker (MW) is expressed in kilodaltons. Arrows indicate the four major polypeptides present in vesicles preparations and in the outer membrane fraction.
In order to confirm that P. antarctica NF3 OMVs were derived from the bacterial outer membrane, we analysed the presence of LPS in vesicles by SDS-PAGE and compared it with whole cell LPS (Fig. 4A). Vesicles phospholipid content (PL-OMV) was also examined for the same purpose (Fig. 4B). The electrophoretic pattern of LPS from vesicles was similar to that extracted from whole cells (W). In both samples, the extensive morphological heterogeneity typical of smooth LPS was not detected, although a faintly stained band near the top of the gel was present (Fig. 4A arrow). Phospholipid analysis by thin layer chromatography showed that vesicles were enriched in phosphatidylethanolamine (PE) relative to phosphatidylglycerol (PG). The same was observed in the outer membrane preparations of P. antarctica (Fig. 4B).
Fig. 4.
A. Silver-stained SDS-PAGE gel of LPS extracted from purified P. antarctica NF3-OMVs (OMV-LPS), and LPS obtained from P. antarctica NF3 whole cells (W-LPS). Arrow indicates the faintly stained band in both preparations.
B. Thin layer chromatography of phospholipids extracted from the outer membrane fraction (PL-OM) and from P. antarctica OMVs (PL-OMV).
Proteomic analysis of OMVs
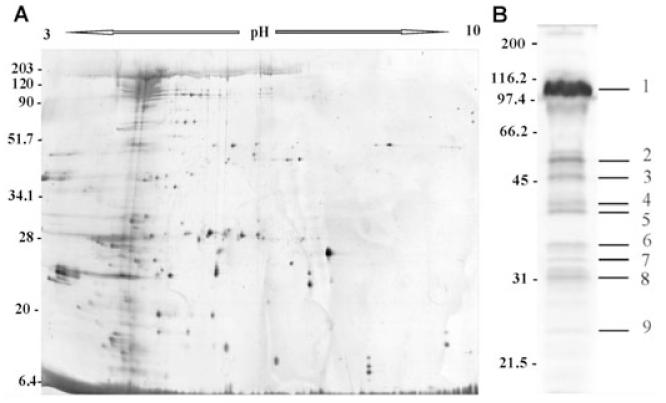
To characterize the main proteins present in OMVs of P. antarctica NF3, we performed a proteomic analysis. Due to the lack of P. antarctica sequenced genes, we used cross-species peptide mass finger printing (Molloy et al., 2001) to identify OMVs proteins. Outer membrane vesicles proteins were separated by 1D SDS-PAGE, and most of their identities were determined either by MALDI-TOF MS, or by N-terminal sequence analysis. At least nine prominent bands were visible by Coomassie blue staining (Fig. 5B). This protein profile was representative of OMVs isolated by ultracentrifugation without further purification. Each band was excised from the gel and digested with trypsin. The resulting peptide mass data were used for database searching. Protein Prospector, Profound (Proteomics) and Mascot search engines were used. These searches indicated that most of the bands consisted of a mixture of proteins, identifying 1–11 proteins for each band (Table 1). These results were confirmed after separating sample proteins by 2D electrophoresis, resolving one band of 1D SDS-PAGE in several protein spots with different isoelectric points (Fig. 5A). MALDI-TOF analysis of the bands with apparent molecular masses of 109, 48, 42 and 24 kDa identified proteins located mainly in the bacterial outer membrane. These data confirmed results obtained when comparing the electrophoretic profiles of the bacterial outer membrane fraction with those of OMVs. The band of 52.5 kDa showed a high percentage of similarity to a flagellin, as did the bands of 34.5 and 33 kDa. These data are in agreement with the absence of such bands in the protein profile of the sucrose gradient purified OMVs in which contaminating flagella had been eliminated. The other proteins were probably derived from the periplasm and/or sites of close apposition to outer membrane/inner membrane. Putative functions of the proteins identified in OMVs are summarized in Table 1.
Fig. 5.
A. Two-dimensional electrophoresis gel in the range of pH 3–10 silver-stained from P. antarctica-OMVs.
B. Protein profile of P. antarctica-OMVs using 12% 1D SDS-PAGE. Molecular mass markers (MW) in kDa are indicated at the left of each gel.
Table 1.
Proteins identified by 1D SDS-PAGE from P. antarctica OMV-u.
| Band No.a | Description | Accession number | MW (Da)b | pIc | Peptides Matches | Coverage (%) | Species |
|---|---|---|---|---|---|---|---|
| 1 | Hypothetical TonB-dependent receptor | gi|24372887 | 110.093 | 4.4 | 4 | 10 | Shewanella oneidensis |
| Hypothetical TonB-dependent receptor | gi|24374013 | 97.761 | 4.5 | 9 | 23 | Shewanella oneidensis | |
| Hypothetical TonB-dependent receptor | gi|24373387 | 101.775 | 4.6 | 7 | 17 | Shewanella oneidensis | |
| Probable RND efflux transporter | gi|15599571 | 110.533 | 5.2 | 9 | 23 | Pseudomonas aeruginosa | |
| Peptidase, M16 family | gi|24375509 | 104.287 | 5.3 | 9 | 15 | Shewanella oneidensis | |
| Organic solvent tolerance protein OstA | gi|15595792 | 104.273 | 5.4 | 6 | 15 | Pseudomonas aeruginosa | |
| AcrB/AcrD/AcrF family protein | gi|24373487 | 113.613 | 5.6 | 9 | 23 | Shewanella oneidensis | |
| Polysaccharide biosynthesis protein | gi|24374706 | 100.376 | 5.7 | 7 | 17 | Shewanella oneidensis | |
| Probable glycosyl hydrolase | gi|15597358 | 103.694 | 5.9 | 9 | 23 | Pseudomonas aeruginosa | |
| Probable ferredoxin | gi|15599966 | 102.625 | 6.2 | 6 | 15 | Pseudomonas aeruginosa | |
| IgA1 protease | gi|12597970 | 110.153 | 7.5 | 8 | 20 | Neisseria meningitidis | |
| Probable adhesin | gi|15599277 | 100.434 | 7.7 | 10 | 25 | Pseudomonas aeruginosa | |
| 2 | Flagellin | gi|16901494 | 50.853 | 4.7 | 23 | 67 | Escherichia coli |
| FliC | gi|30059880 | 48.107 | 4.8 | 20 | 58 | Escherichia coli | |
| Flagellin | gi|38049856 | 52.274 | 5.0 | 15 | 71 | Salmonella enterica | |
| 3d | Membrane-associated Zn-dependent protease | gi|71738080 | 48.561 | 5.0 | – | – | Pseudomonas syringae |
| Possible peptidoglycan-binding LysM | gi|68181658 | 51.641 | 6.8 | – | – | Jannaschia sp. | |
| 4 | Survival protein surA | gi|24375136 | 48.914 | 5.7 | 6 | 16 | Shewanella oneidensis |
| TolB protein | gi|24372890 | 46.280 | 5.8 | 4 | 11 | Shewanella oneidensis | |
| PobA protein | gi|6682326 | 44.121 | 5.9 | 9 | 25 | Pseudomonas putida | |
| Serine protease, HtrA/DegQ/DegS family | gi|24375430 | 46.551 | 6.0 | 4 | 11 | Shewanella oneidensis | |
| Flagellar hook-associated protein FliD | gi|24374747 | 47.914 | 7.7 | 7 | 19 | Shewanella oneidensis | |
| Periplasmic sulfate-binding protein | gi|24375099 | 37.109 | 8.5 | 4 | 11 | Shewanella oneidensis | |
| RND multidrug efflux membrane protein MexE | gi|15597689 | 45.031 | 8.5 | 8 | 22 | Pseudomonas aeruginosa | |
| 5 | Putative outer membrane porin | gi|24371910 | 42.311 | 4.5 | 5 | 12 | Shewanella oneidensis |
| Maltoporin | gi|16131862 | 49.913 | 4.8 | 9 | 25 | Escherichia coli | |
| Putative adhesin/invasin | gi|21427120 | 41.307 | 4.9 | 12 | 30 | Neisseria meningitidis | |
| TolB domain protein | gi|24372890 | 46.280 | 5.8 | 5 | 12 | Shewanella oneidensis | |
| Cation efflux system protein cusB precursor | gi|2495562 | 44.305 | 5.9 | 13 | 32 | Escherichia coli | |
| HlyF | gi|32470076 | 41.022 | 6.8 | 8 | 20 | Escherichia coli | |
| Probable porin | gi|15595437 | 46.125 | 7.8 | 7 | 17 | Pseudomonas aeruginosa | |
| RND multidrug efflux membrane fusion protein | gi|15597689 | 45.031 | 8.5 | 8 | 20 | Pseudomonas aeruginosa | |
| Membrane-bound lytic transglycosylase | gi|24372749 | 39.715 | 8.5 | 5 | 12 | Shewanella oneidensis | |
| 6d | Flagellin | gi|3386642 | 39.507 | 4.7 | – | – | Pseudomonas aeruginosa |
| Flagellin | gi|94771 | 40.041 | 5.4 | – | – | Pseudomonas aeruginosa | |
| 7d | Flagellin | gi|3098299 | 40.377 | 4.5 | – | – | Pseudomonas fluorescens |
| FliC | gi|30692080 | 39.432 | 4.8 | – | – | Pseudomonas aeruginosa | |
| 8 | Structural outer membrane porin OprF | gi|15596974 | 37.640 | 5.0 | 4 | 12 | Pseudomonas aeruginosa |
| Probable hydrolase | gi|15600370 | 25.629 | 5.6 | 6 | 18 | Pseudomonas aeruginosa | |
| Probable FKBP-peptidyl-prolyl cis-trans isomerase | gi|12231005 | 25.278 | 8.0 | 4 | 12 | Pseudomonas aeruginosa | |
| Periplasmic ABC-type phosphate transport | gi|46320586 | 33.141 | 9.0 | 6 | 22 | Burkholderia cepacia | |
| Putative glycosyltransferase | gi|5545325 | 35.778 | 9.3 | 4 | 12 | A. actinomycetemcomitans | |
| Chaperone protein fimC | gi|26247791 | 26.158 | 9.5 | 4 | 12 | Escherichia coli | |
| Peptidase, M23/M37 family | gi|24371648 | 38.450 | 9.6 | 4 | 12 | Shewanella oneidensis | |
| 9 | MSHA biogenesis protein MshJ | gi|24350496 | 24.838 | 5.1 | 6 | 42 | Shewanella oneidensis |
| Probable outer membrane protein | gi|11351562 | 21.748 | 5.3 | 5 | 35 | Pseudomonas aeruginosa | |
| Sodium-type flagellar protein MotX | gi|24350265 | 23.210 | 8.4 | 6 | 42 | Shewanella oneidensis | |
| Outer membrane lipoprotein (lipocalin) | gi|15804743 | 19.852 | 8.9 | 4 | 28 | Escherichia coli | |
| Intimin | gi|1703488 | 29.751 | 9.3 | 4 | 28 | Escherichia coli | |
| Pertactin | gi|49243344 | 28.759 | 9.7 | 5 | 35 | Bordetella bronchiseptica |
Band numbers are as indicated in Fig. 5B.
Theoretical molecular mass of the proteins.
Theoretical charge of the proteins.
Peptide sequences obtained using N-terminal as described in Experimental procedures.
Discussion
Extreme environments like the Antarctica offer novel microbial biodiversity. The bacterium P. antarctica NF3 has been isolated from mucous material located a few centimetres beneath the water surface at the base of a glacier in King George Island in Antarctica. This highly mucoid psychrotolerant bacterium produced abundant extracellular matter. Most microbial cells in the marine environment are surrounded by extracellular polymers and have usually been characterized as high molecular weight carbohydrate polymers. The high protein content of P. antarctica’s EPS (78–86%) proved remarkable (Bozal et al., 1994; 1997). Mancuso and coworkers have recently demonstrated that other Antarctic strains of the genus Pseudoalteromonas also produce large amounts of EPSs. Moreover, the crude chemical composition of EPS produced by these Antarctic marine bacteria, showed the presence of proteins in different percentages (Mancuso Nichols et al., 2004; 2005b). However, it is not clear how these proteins are integrated in the EPS or by which methods they are released into the environment. To address these questions, we first conducted electron microscopy studies. Ultrastructural characterization of EPSs secreted by bacteria remains quite limited, in part due to the difficulty of preserving these highly hydrated structures (Graham et al., 1991). High-pressure freezing and freeze substitution techniques have greatly improved the ultrastructural preservation of cell morphology and bacterial EPSs (Kellenberger, 1987; Graham et al., 1991). Transmission electron microscopy analysis following HPF-FS showed that the EPS of P. antarctica includes large quantities of OMVs. These vesicles are bilayered structures and predominantly spherical in shape, with an average diameter of 25–70 nm, which is in agreement with other OMVs from different Gram-negative bacteria (Beveridge, 1999). The presence of OMVs with high protein content is well documented in Gram-negative bacteria, although it has never been reported for psychrotolerant organisms of the genus Pseudoalteromonas. When viewed at higher magnifications, vesicles are mainly immersed in a net-like meshwork of fibres. For P. antarctica cells, the presence of an inner layer of oriented packed fibres extending perpendicularly away from the cell envelope, as well as an outer layer of randomly distributed fine fibres could be regarded as a capsule. These two polymeric arrangements have also been reported for capsules from Escherichia coli K1 and Klebsiella pneumoniae K (Amako et al., 1988). Other encapsulated bacteria have been observed by TEM following HPF-FS (Graham et al., 1991; Korenevsky et al., 2002). These authors showed with this technique that capsules are visualized as packaged fibres standing perpendicular to the cell envelope, although polymer length, diameter and the arrangement of fibres can vary between strains. For P. antarctica, a similar arrangement has been observed.
Lipopolysaccharide, phospholipid and protein profile analyses confirmed that OMVs are derived predominantly from the bacterial outer membrane. Whole cells and OMVs display the same LPS profile, with low molecular weight LPS or lipooligosaccharide and a weakly staining high molecular mass band, all of which suggest either the presence of an LPS O-polymer of relatively constant length, or the existence of a polysaccharide capsule. This LPS pattern has also been reported by Korenevsky and colleagues (2002) in several species of Shewanella, where it was impossible to distinguish S-LPS from capsular material on the basis of the LPS profile. Although more studies will be necessary to elucidate the P. antarctica NF3 LPS structure, the observed fibrous fringe surrounding P. antarctica cells would indicate the presence of such a capsule in this strain.
The phospholipid content of OMVs enriched in PE relative to PG also proved characteristic of the bacterial outer membrane (Osborn et al., 1972) and indicated that the OMVs from P. antarctica NF3 are derived from it.
SDS-PAGE profiles revealed that protein patterns of OMVs from P. antarctica NF3, and of the bacterial outer membrane, are highly similar though not identical. For OMVs, the absence of certain proteins of the outer membrane, and the enrichment of other proteins in secreted vesicles, supports the model of vesicle formation described by Kadurugamuwa and Beveridge (1995). They suggest that specific components of the outer membrane and periplasm may be selectively included or excluded during vesicle formation. In the same way, Horstman and Kuehn (2000) suggested that differences in protein profiles are due to a vesiculation process that takes place at outer membrane sites with specific protein compositions.
Outer membrane vesicles have been observed in organisms growing under very different conditions, such as in in vitro biofilms, on solid or liquid media, or in natural environments (Beveridge, 1999; Khandelwal and Banerjee-Bhatnagar, 2003). The functions of OMVs are not well defined, but they have been mainly related to adherence and delivery of virulence factors to bacterial or eukaryotic cells (Meyer and Fives-Taylor, 1993; Fiocca et al., 1999; Kadurugamuwa and Beveridge, 1999; Horstman and Kuehn, 2000; Wai et al., 2003). For the psychrotolerant bacterium P. antarctica NF3, the role and functions of OMVs released in this manner remain unknown. In an attempt to address these questions, we conducted a proteomic analysis of OMVs from P. antarctica. The full analysis of a bacterial proteome is a promising approach for characterizing individual proteins and their functions (Pandey and Mann, 2000). For P. antarctica this proved a challenging task as no peptide mass data have yet been generated for this genus. For this reason, the approach chosen for this study was to perform cross-species identification using peptide sequencing by ESI-MS/MS or MALDI-TOF/MS, supplemented with Edman-sequencing in some cases.
Lipopolysaccharide, phospholipids and protein composition analysis have confirmed that vesicles from P. antarctica are derived from the bacterial outer membrane. Proteome analysis of membrane and cell surface proteins is complex due to their intrinsic hydrophobic nature, alkaline pI and transmembrane spanning regions. Although 2D electrophoresis provides high-resolution separation of complex mixtures, the efficiency of membrane protein identification is not always satisfactory because of their hydrophobicity and low abundance (Santoni et al., 2000; Herbert et al., 2001). The proteomic analysis approach for P. antarctica vesicles using 1D SDS-PAGE proved to be effective. 1D SDS gels have shown that vesicles only contain a fraction of the total bacterial proteins; thus, we initially chose to use this method for better recovery of membrane proteins. As expected, 2D electrophoresis showed that a single 1D SDS band contained several proteins that yield to a mixture of peptides upon in-gel digestion. However, the liquid chromatography set-up prior to nanospray-MS/MS analysis (Cap-LC-nano-ESI-Q-TOF) has provided the high resolution peptide separation and detection sensitivity required for analysing complex peptide mixtures. The same approach had been very effective for characterizing membrane vesicles from Neisseria meningitidis (Post et al., 2005), and for proteomic profiling of the membrane constituents of Mycobacterium tuberculosis (Gu et al., 2003).
Proteomic analysis of OMVs from P. antarctica has revealed the presence of outer membrane and periplasmic proteins qualitatively similar to other OMVs characterized in Gram-negative species (Fredrickson et al., 2002; Post et al., 2005; Rolhion et al., 2005). Proteins of the vesicles match in their sequence with proteins possessing several functions such as proteolytic enzymes, transport receptor and binding proteins, secretion proteins, polysaccharide biosynthesis proteins, enzymes involved in bacterial cell wall degradation, putative porins, proteins that participate in electron transport, adhesins and proteins involved in protein folding.
The identification of putative TonB-dependent receptors as one of the main proteins in OMVs from P. antarctica proved to be a remarkable finding. If we take in account that vesicles are derived from bacterial outer membrane, the presence of TonB-dependent receptors suggests that they could play a role in sensing nutrients and importing them into P. antarctica cells. The presence of a large number of genes coding for Ton B-dependent receptors has also been reported for Caulobacter crescentus (Nierman et al., 2001). This strain has no OmpF-type outer membrane porins, which allow passive diffusion of hydrophilic substrates. The presence of proteins from the family of TonB-dependent outer membrane channels has been postulated to represent an alternative mechanism for survival in nutrient-limiting conditions. Ireland and colleagues (2002) have also suggested that a large number of TonB-dependent receptors in Caulobacter are likely to be involved in the transport of nutrients and macromolecules into the cell. Recently, the complete genome of Pseudoalteromonas haloplanktis TAC 125 has been sequenced (Médigue et al., 2005). For this Antarctica bacterium, the authors predicted a great number of genes coding for TonB-dependent receptors, a number not found in the genome of other cold-adapted marine bacteria. The potential implications of the above-mentioned studies for the strain P. antarctica NF3 warrant investigation.
Proteomic analysis also revealed the presence of putative proteolytic degradative enzymes in OMVs. Their presence could contribute to the degradation of high molecular weight compounds present in the organic matter common to marine environments, which are largely unavailable for direct uptake by marine bacteria for catabolic and biosynthetic purposes (Chrost, 1991). In this context, the hydrated EPS produced by P. antarctica NF3 could provide a stable environment around those cells facilitating the activities of such exocellular proteins. Huston and colleagues (2004) have purified a cold-active aminopeptidase produced by a marine psychrophilic bacterium whose activity and stability significantly increased in the presence of EPS. Once liberated to the medium, OMVs from P. antarctica could provide hydrolytic enzymes or other still as yet unidentified factors in a way that may play a crucial role in the survival of the bacterium in the harsh Antarctic environment.
In summary, we have revealed that P. antarctica NF3 EPS is composed of a capsular polymer and large quantities of OMVs. Analysis of the LPS, phospholipids and proteins from vesicles has demonstrated that these structures are derived from bacterial outer membrane. Proteomic analysis of OMVs revealed that proteins in vesicles match with outer membrane and periplasmic proteins detected in vesicles from other Gram-negative species. Some of these proteins seem to be involved in degradation of organic matter and transfer of nutrients into the cell. Further proteomic analysis work will be necessary to assign functions to the released proteins and help to explain the physiological role of P. antarctica NF3 OMVs in the Antarctic environment.
Experimental procedures
Bacteria and growth conditions
Unless specified, all chemicals and biochemicals were purchased from Fluka (Switzerland). Pseudoalteromonas antarctica NF3 is a psychrotolerant Gram-negative bacterium isolated from mucous material located only a few cm beneath the water surface at the base of a glacier near the inlet of Admiralty Bay (King George Island, South Shetland Islands, Antarctica) and was deposited in the Spanish Type Culture Collection with the reference number CECT 4664. Pseudoalteromonas antarctica NF3 was grown on MM5, a minimal medium described by Bozal and colleagues (1994). This medium contains per litre 5 g Na2HPO4, 2 g KH2PO4, 1 g NaCl, 7 g NH4Cl, 0.5 g MgSO4·7H2O, 0.001 g FeSO4·7H2O, 0.05 g CaCl2 and 20 g glucose, adjusted to a pH of 7.0. For solid media, 15 g of bacteriological agar per litre was added. MM5 broth cultures were inoculated from solid cultures of P. antarctica NF3 and incubated at 15°C on an orbital shaker at 120 r.p.m.
Transmission electron microscopy
Negative staining
Purified OMVs were visualized by negative staining electron microscopy. The sample was adsorbed onto carbon-coated copper/palladium grids for 2 min, and then washed 2 times by floating it on drops of sterilized deionized water for 10 s. Finally, the sample was negatively stained by floating the grids on a drop of 1% (w/v) uranyl acetate for 1 min.
High-pressure freezing fixation and freeze substitution
Bacterial colonies were selected under a stereomicroscope and transferred to planchettes (1.5 mm in diameter and 200 μm deep) and immediately cryoimmobilized using a Leica E MPact high-pressure freezer (Leica, Vienna, Austria) in the complete absence of cryoprotectants or freezing solutions. Planchettes were then stored in liquid nitrogen until further use. Frozen samples were freeze-substituted in a Leica EMAFS (automatic freeze substitution system, Leica, Vienna, Austria), where the substitution was performed in pure acetone containing 2% (w/v) osmium tetroxide and 0.1% (w/v) uranyl acetate at −90°C for 72 h. The temperature was gradually increased (Δt = 5°C/h) to 4°C and held constant for 2 h, and then finally increased to room temperature and maintained for 1 h. Samples were washed for 1 h in acetone at room temperature and infiltrated in a graded series of Epon-acetone mixtures: 1:3 for 2 h, 2:2 for 2 h, 3:1 for 16 h and pure Epon for 30 h. Samples were embedded in fresh Epon and polymerized at 60°C for 48 h.
Sectioning and electron microscopy analysis
Ultrathin sections were cut with a Leica UCT ultramicrotome and mounted on Formvar carbon-coated copper grids. Sections were post-stained with 2% (w/v) aqueous uranyl acetate and lead citrate and examined in a JEOL 1010 TEM at an acceleration voltage of 80 keV.
Isolation of OMVs from the culture medium
Outer membrane vesicles naturally secreted into the medium were collected from the supernatant of a late log phase culture, as described by Kadurugamuwa and Beveridge (1995). The cells were pelleted by centrifugation at 17 000 g for 20 min at 4°C, and the supernatant was filtered through 0.45 and 0.22 μm pore-size filters to remove remaining bacterial cells. Outer membrane vesicles were obtained by high-speed centrifugation at 150 000 g for 3 h at 5°C in a Ti 70.1 rotor (Optima™ L-90K Ultracentrifuge, Beckman Coulter), washed and then resuspended in 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) buffer, pH 6.8 (Sigma).
Outer membrane vesicle purification
Isolated OMVs were purified on a 30–60% (w/w) sucrose gradient in 30 mM Tris-HCl (pH 7.8) and centrifuged at 183 000 g for 18 h in a SW41 rotor (Beckman Coulter). Bands were visualized by indirect light, removed using a syringe needle, and washed by centrifugation at 150 000 g, 1 h, 4°C, three times with 30 mM Tris-HCl (pH 7.8). Buoyant density (ρ’s) was calculated by measuring the refractive indices of the sucrose gradients.
Preparation of outer membranes from P. antarctica NF3 cells
Outer membranes were prepared as described by Puig and colleagues (1993). A bacterial pellet was washed twice with 10 mM HEPES buffer, pH 7.4 and sonicated on ice using 3 × 1 min pulses at 140 W. The cell lysate was centrifuged at 17 000 g for 20 min, and the clear supernatant was subjected to ultracentrifugation at 100 000 g for 90 min in a TLA 55 rotor (Optima™ TLX Ultracentrifuge, Beckman Coulter). The crude membrane pellet was resuspended in 2% (w/v) Sarcosyl (N-lauroylsarcosine, Sigma) in 10 mM HEPES buffer, pH 7.4, for 30 min at room temperature to solubilize the inner membrane. The suspension was centrifuged at 60 000 g for 60 min, at 20°C. The pellet was then resuspended in the same buffer.
Lipopolysaccharide profiles
To determine the LPS profile of P. antarctica NF3 cells and P. antarctica NF3-MVs, a known amount of protein was denatured by boiling with SDS-containing sample buffer for 10 min. The protein was digested by adding proteinase K to a final concentration of 50 μg ml−1 at 37°C overnight. Samples were then boiled for a further 10 min, and a second treatment with proteinase K was performed. Samples were then incubated at 56°C for 3 h. Protease-treated samples were separated by SDS-PAGE and visualized by silver staining (Tsai and Frasch, 1982).
Thin layer chromatography (TLC)
Phospholipids extraction was performed according to the method described by Bligh and Dyer (1959) with some modifications. Outer membrane enriched fractions and OMVs were treated with 750 μl of methanol/chloroform (2:1, v/v) and mixed three times for 30 min. Suspensions were centrifuged at 10 000 r.p.m. for 10 min and the supernatant transferred to a fresh 1.5-ml eppendorf tube to remove insoluble material. Two hundred and fifty microlitres of water and 250 μl of chloroform were added to the supernatant and mixed three times for a period of 30 min. Phase separation was accomplished by centrifugation (5000 r.p.m. for 1 min at room temperature). The lower chloroform phase was transferred to a fresh 1.5-ml tube and dried under a stream of nitrogen. Lipids were redissolved in 100 μl of methanol/chloroform (1:1, v/v). Outer membrane vesicles phospholipids (PL-OMV) and outer membrane phospholipids (PL-OM) were resolved by TLC on silica gel plates (Merck, Germany) using a solvent system of chloroform/methanol/water/acetic acid (65:25:4:1, v:v:v:v) (Weinrauch et al., 1999). The spots were visualized by spraying with 10% (w/v) phosphomolybdic acid in ethanol solution and heating at 120°C.
Electrophoresis
One-dimensional electrophoresis
One-dimensional electrophoresis was performed under denaturing conditions on 12% SDS-PAGE according to Laemmli’s method (Laemmli, 1970). Gels were stained for protein with Coomassie blue. The protein concentration of samples was determined by the Bradford method (Bradford, 1976) using standard Bio-Rad Reagent (Bio-Rad laboratories GmbH). For the analysis, 5 μg of total protein was loaded per well.
Two-dimensional electrophoresis
Proteins were resuspended in 450 μl of rehydration buffer (7 M urea, 2 M thiourea, 80 mM DTT, 2% CHAPS, 0.5% IPG buffer, bromophenol blue) and loaded onto 24 cm, pH 3-10 NL IPG strip. The IEF separation was performed in an IPGphor system (Amersham Bioscience) according to the manufacturer’s instructions. Focused strips were equilibrated with equilibration buffer (65 mM DTT, 50 mM Tris-HCl, 6 M urea, 30% glycerol, 2% SDS, bromophenol blue) for 15 min followed by buffer (135 mM IAA, 50 mM Tris-HCl, 6 M urea, 30% glicerol, 2% SDS, bromophenol blue) for 15 min. The equilibrated strips were directly applied to 12.5% acrylamide gel and separated at 3 V gel−1 for 30 min and at 18 V gel−1 for 6 h in an Ettan DALT II system (Amersham Bioscience). Gels were fixed overnight followed by silver mass spectrometry compatible staining and scanned.
N-terminal sequencing
Analysis of protein amino acid sequence via Edman degradation was performed using a Procise cLC 492 Sequencer (Applied Biosystems, ABI). Materials, reagents and standards were obtained from ABI. The sequencing programs (operating steps) are based on the manufacturer’s recommendations. In-solution proteins were applied to a membrane pretreated with Biobrene while proteins in the PVDF membrane were destained with methanol, and cut into small pieces in the reactor as specified by the manufacturer. Data analysis was carried out using the ABI 610 A Data Analysis Program on a Mac OS 9.1 computer with a standard calibration cycle followed by analysis of the sample cycle data.
In-gel digestion
Proteins were in-gel digested with trypsin (modified sequencing grade, Promega) using the automatic Investigator ProGest robot from Genomic Solutions. Briefly, excised gels spots were washed sequentially with ammonium bicarbonate buffer and acetonitrile. Proteins were reduced and alkylated, respectively, by treatment with 10 mM DTT solution over 30 min, followed by treatment with a 100-mM solution of iodine acetamide. After sequential washings with buffer and acetonitrile, proteins were digested overnight at 37°C with 0.27 nmol of trypsin. Tryptic peptides were extracted from the gel matrix with 10% (v/v) formic acid and acetonitrile, with the extracts then pooled and dried in a vacuum centrifuge.
Acquisition of MS and MS/MS spectra
Proteins excised from the gel were either analysed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF/TOF) mass spectrometry (MS) (4700 Proteomics Analyzer, Applied Biosystems) or electrospray ionization mass spectrometry (ESI-MS-MS) (Q-TOF Global, Micromass-Waters). In the former, the digests were redissolved in 5 μl of 0.1% (v/v) trifluoroacetic acid in 50% (v/v) acetonitrile. Typically a 0.5-μl aliquot was mixed with the same volume of a matrix solution, 5 mg ml−1 α-cyano-4-hydroxycinnamic acid (CHCA) (Aldrich) in 0.1% (v/v) trifluoroacetic acid in 50% (v/v) acetonitrile. Major peaks were selected for characterization by MS/MS analysis. Spectra were submitted for database searching in a generic MASCOT format. Some of the tryptically digested peptides samples were analysed by on-line liquid chromatography tandem mass spectrometry (Cap-LC-nano-ESI-Q-TOF) (CapLC, Micromass-Waters). In those cases, samples were resuspended in 10 μl of 10% (v/v) formic acid solution and 4 μl were injected for chromatographic separation in a reverse-phase capillary C18 column (75 μm of internal diameter and 15 cm length, PepMap column, LC Packings). The eluted peptides were ionized via coated nano-ES needles (PicoTip™, New Objective). A capillary voltage of 1800–2200 V was applied together with a cone voltage of 80 V. The collision energy in the CID (collision-induced dissociation) was 20–35 eV and argon was employed as a collision gas. Data were generated in PKL file format, which were submitted for database searching in the MASCOT server.
Acknowledgements
We wish thank Dr Carmen López Iglesias (Servicios Científico Técnicos, SCT, Universidad de Barcelona) for her assistance with electron microscopy, and the Proteomic unit from SCT for assistance in proteomic analysis. We would also like to thank Robin Rycroft for editorial assistance. This work was supported by Grant 2005SGR00066 from the Generalitat de Catalunya; M. Nevot is the recipient of a fellowship from the Fundació Universitària Agustí Pedro Pons.
References
- Allan ND, Kooi C, Sokol PA, Beveridge TJ. Putative virulence factors are released in association with membrane vesicles from Burkholderia cepacia. Can J Microbiol. 2003;49:613–624. doi: 10.1139/w03-078. [DOI] [PubMed] [Google Scholar]
- Amako K, Meno Y, Takade A. Fine structures of the capsules of Klebsiella pneumoniae and Escherichia coli K1. J Bacteriol. 1988;170:4960–4962. doi: 10.1128/jb.170.10.4960-4962.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge TJ, Kadurugamuwa JL. Periplasm, periplasmic spaces and their relation to bacterial wall structure: novel secretion of selected periplasmic proteins from Pseudomonas aeruginosa. Microb Drug Resist. 1996;2:1–8. doi: 10.1089/mdr.1996.2.1. [DOI] [PubMed] [Google Scholar]
- Beveridge TJ, Makin SA, Kadurugamuwa JL, Li Z. Interactions between biofilms and the environment. FEMS Microbiol Lett. 1997;20:291–303. doi: 10.1111/j.1574-6976.1997.tb00315.x. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bowman JP, McCammon SA, Brown MV, Nichols DS, McMeeking TA. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl Environ Microbiol. 1997;63:3068–3078. doi: 10.1128/aem.63.8.3068-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozal N, Manresa A, Castellví J, Guinea J. A new bacterial strain of Antarctica, Alteromonas sp. that produces a heteropolymer slime. Polar Biol. 1994;14:561–567. [Google Scholar]
- Bozal N, Guinea J, Tudela E, Congregado F, Parra JL, de la Maza A, et al. Use of a glycoprotein for liposome protection. ES 2 126 516 A1 Spanish patent. 1996
- Bozal N, Tudela E, Rossello-Mora R, Lalucat J, Guinea J. Pseudoalteromonas antarctica sp. nov., isolated from an Antarctic coastal environment. Int J Syst Bacteriol. 1997;47:345–351. doi: 10.1099/00207713-47-2-345. [DOI] [PubMed] [Google Scholar]
- Bozal N, Montes MJ, Tudela E, Jiménez F, Guinea J. Shewanella frigidimarina and Shewanella livingstonensis sp. nov. isolated from Antarctic coastal areas. Int J Syst Evol Microbiol. 2002;52:195–205. doi: 10.1099/00207713-52-1-195. [DOI] [PubMed] [Google Scholar]
- Bozal N, Montes MJ, Tudela E, Guinea J. Characterization of several Psychrobacter strains isolated from Antarctic environments and description of Psychrobacter luti sp. nov. & Psychrobacter fozii sp. nov. Int J Syst Evol Microbiol. 2003;53:1093–1100. doi: 10.1099/ijs.0.02457-0. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R. Cold adapted Archaea. Nat Rev Microbiol. 2006;4:331–343. doi: 10.1038/nrmicro1390. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R, Siddiqui KS, Andrews D, Sowers KR. Low-temperature extremophiles and their applications. Curr Opin Biotech. 2002;13:253–261. doi: 10.1016/s0958-1669(02)00317-8. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R, Curmi PMG, Siddiqui KS, Thomas T. Proteins from psychrophiles. In: Rainey FA, Oren A, editors. Methods in Microbiology – Extremophiles. Vol. 35. Elsevier; London, UK: 2006. in press. [Google Scholar]
- Chrost RJ. Environmental control of the synthesis and activity of aquatic microbial ectoenzymes. In: Chrost RJ, editor. Microbial Enzymes in Aquatic Environments. Springer-Verlag; New York, USA: 1991. pp. 84–95. [Google Scholar]
- Costerton JW. The role of bacterial exopolysaccharides in nature and disease (Reprinted from Developments in Industrial Microbiology, vol 26, pp. 249-261, 1985) J Ind Microbiol Biotechnol. 1999;22:551–563. [Google Scholar]
- Fiocca R, Necchi V, Sommi P, Ricci V, Telford J, Cover TL, Solcia E. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles: uptake of released toxin and vesicles by gastric epithelium. J Pathol. 1999;188:220–226. doi: 10.1002/(SICI)1096-9896(199906)188:2<220::AID-PATH307>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. Relevance of microbial extracellular polymeric substances (EPSs) – Part I: structural and ecological aspects. Water Sci Technol. 2001;43:1–8. [PubMed] [Google Scholar]
- Fredrickson J, Gorby Y, McLean J, Lipton MS, PasaTolic L, Tsapin A, et al. Global characterization of proteins associated with S. oneidensis MR-1 outer membrane vesicles; Ninth Doe Genome Contractor and Grantee Workshop; Oakland, CA, USA. 2002.Jan 27-31, [Google Scholar]
- Gerday C, Aittaleb M, Bentahir M, Chessa J-P, Claverie P, Collins T, et al. Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol. 2000;18:103–107. doi: 10.1016/s0167-7799(99)01413-4. [DOI] [PubMed] [Google Scholar]
- Graham LL, Harris R, Villiger W, Beveridge TJ. Freeze-substitution of gram-negative eubacteria: general cell morphology and envelope profiles. J Bacteriol. 1991;173:1623–1633. doi: 10.1128/jb.173.5.1623-1633.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Chen J, Dobos KM, Bradbury EM, Belisle JT, Chen X. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol Cell Proteomics. 2003;2:1284–1296. doi: 10.1074/mcp.M300060-MCP200. [DOI] [PubMed] [Google Scholar]
- Herbert BR, Harry JL, Packer NH, Gooley AA, Pedersen SK, Williams KL. What place for polyacrylamide in proteomics? Trends Biotechnol. 2001;19:S3–S9. doi: 10.1016/S0167-7799(01)01796-6. [DOI] [PubMed] [Google Scholar]
- Horstman AL, Kuehn MJ. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J Biol Chem. 2000;275:12489–12496. doi: 10.1074/jbc.275.17.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston AL, Methe B, Deming JW. Purification, characterization, and sequencing of an extracellular cold-active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea strain 34H. Appl Environ Microbiol. 2004;70:3321–3328. doi: 10.1128/AEM.70.6.3321-3328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland MME, Karty JA, Quardokus EM, Reilly JP, Brun YV. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol Microbiol. 2002;45:1029–1041. doi: 10.1046/j.1365-2958.2002.03071.x. [DOI] [PubMed] [Google Scholar]
- Junge K, Imhoff JF, Staley JT, Deming JW. Phylogenetic diversity of numerically important bacteria in Arctic sea. Microbial Ecol. 2002;43:315–328. doi: 10.1007/s00248-001-1026-4. [DOI] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other gram-negative bacteria. Microbiology. 1999;145:2051–2060. doi: 10.1099/13500872-145-8-2051. [DOI] [PubMed] [Google Scholar]
- Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog. 2002;32:1–13. doi: 10.1006/mpat.2001.0474. [DOI] [PubMed] [Google Scholar]
- Kellenberger E. The response of biological macromolecules and supramolecular structures to the physics of specimen cryopreparation. In: Steinbrecht RA, Zierold K, editors. Cryotechniques in Biological Electron Microscopy. Springer-Verlag; Berlin, Germany: 1987. pp. 35–63. [Google Scholar]
- Khandelwal P, Banerjee-Bhatnagar N. Insecticidal activity associated with the outer membrane vesicles of Xenorhabdus nematophilus. Appl Environ Microbiol. 2003;69:2032–2037. doi: 10.1128/AEM.69.4.2032-2037.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolling GL, Matthews KR. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157: H7. Appl Environ Microbiol. 1999;65:1843–1848. doi: 10.1128/aem.65.5.1843-1848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenevsky AA, Vinogradov E, Gorby Y, Beveridge TJ. Characterization of the lipopolysaccharides and capsules of Shewanella spp. Appl Environ Microbiol. 2002;68:4653–4657. doi: 10.1128/AEM.68.9.4653-4657.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krembs C, Engel A. Abundance and variability of microorganisms and transparent exopolymer particles across ice–water interface of melting first-year sea ice in the Laptev Sea (Artic) Mar Biol. 2001;138:173–185. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. (London) [DOI] [PubMed] [Google Scholar]
- Llarch A, Logan NA, Castellví J, Prieto MJ, Guinea J. Isolation and characterization of thermophilic Bacillus spp. from geothermal environments on Deception Island, South Shetland Archipelago. Microb Ecol. 1997;34:58–65. doi: 10.1007/s002489900034. [DOI] [PubMed] [Google Scholar]
- Mancuso Nichols CA, Garon S, Bowman JP, Raguénès G, Guézennec J. Production of exopolysaccharides by Antarctic marine bacterial isolates. J Appl Microbiol. 2004;96:1057–1066. doi: 10.1111/j.1365-2672.2004.02216.x. [DOI] [PubMed] [Google Scholar]
- Mancuso Nichols CA, Guézennec J, Bowman JP. Bacterial exopolysaccharides from extreme marine environments with special consideration of the Southern Ocean, sea ice, and deep-sea hydrothermal vents: a review. Mar Biotechnol. 2005a;7:253–271. doi: 10.1007/s10126-004-5118-2. [DOI] [PubMed] [Google Scholar]
- Mancuso Nichols CA, Garon Lardière S, Bowman JP, Nichols PD, Gibson JAE, Guézennec J. Chemical characterization of exopolysaccharides from Antarctic marine bacteria. Microb Ecol. 2005b;49:578–589. doi: 10.1007/s00248-004-0093-8. [DOI] [PubMed] [Google Scholar]
- Mayrand D, Grenier D. Biological activities of outer membrane vesicles. Can J Microbiol. 1989;35:607–613. doi: 10.1139/m89-097. [DOI] [PubMed] [Google Scholar]
- de la Maza A, Lopez O, Parra JL, Sabés M, Guinea J. Aggregation state of the glycoprotein excreted by Pseudoalteromonas antarctica NF3, on a support of phosphatidylcholine liposomes. Langmuir. 1998a;14:5680–5684. [Google Scholar]
- de la Maza A, Parra JL, Congregado F, Bozal N, Guinea J. Interaction of the glycoprotein excreted by Pseudoalteromonas antarctica NF3, with phosphatidylcholine liposomes. Colloids Surfaces A. 1998b;137:181–188. [Google Scholar]
- Médigue C, Krin E, Pascal G, Barbe V, Bernsel A, Bertin PN, et al. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005;15:1325–1335. doi: 10.1101/gr.4126905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DH, Fives-Taylor PM. Evidence that extracellular components function in adherence of Actinobacillus actinomycetemcomitans to epithelial cells. Infect Immun. 1993;61:4933–4936. doi: 10.1128/iai.61.11.4933-4936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy MP, Phadke ND, Maddock JR, Andrews PC. Two-dimensional electrophoresis and peptide mass fingerprinting of bacterial outer membrane proteins. Electrophoresis. 2001;22:1686–1696. doi: 10.1002/1522-2683(200105)22:9<1686::AID-ELPS1686>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Montes MJ, Belloch C, Galiana M, García MD, Andrés C, Ferrer S, et al. Polyphasic taxonomy of a novel yeast isolated from Antarctic environment; description of Cryptococcus victoriae sp. nov. Syst Appl Microbiol. 1999;22:97–105. doi: 10.1016/S0723-2020(99)80032-0. [DOI] [PubMed] [Google Scholar]
- Montes MJ, Mercadé E, Bozal N, Guinea J. Paenibacillus antarcticus sp. nov. a novel psychrotolerant organism from the Antarctic environment. Int J Syst Evol Microbiol. 2004;54:1521–1526. doi: 10.1099/ijs.0.63078-0. [DOI] [PubMed] [Google Scholar]
- Nevot M, Deroncele V, López-Iglesias C, Bozal N, Ghinea J, Mercade E. Ultrastructural analysis of the extracellular matter secreted from the psychrotolerant bacterium Pseudoalteromonas antarctica NF3. Microb Ecol. 2006 doi: 10.1007/s00248-006-9065-5. in press. [DOI] [PubMed] [Google Scholar]
- Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen J, et al. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci USA. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Gander JE, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- Post D, Zhang D, Eastvold J, Teghanemt A, Gibson BW, Weiss PW. Biochemical and functional characterization of membrane blebs purified from Neisseria meningitidis serogroup B. J Biol Chem. 2005;280:38383–38394. doi: 10.1074/jbc.M508063200. [DOI] [PubMed] [Google Scholar]
- Puig M, Fusté C, Viñas M. Outer membrane proteins from Serratia marcescens. Can J Microbiol. 1993;39:108–111. doi: 10.1139/m93-015. [DOI] [PubMed] [Google Scholar]
- Raguénès GHC, Peres A, Ruimy R, Pignet P, Christen R, Loaec M, et al. Alteromonas infernus sp. nov., a new polysaccharide-producing bacterium isolated from a deep-sea hydrothermal vent. J Appl Microbiol. 1997;82:422–430. doi: 10.1046/j.1365-2672.1997.00125.x. [DOI] [PubMed] [Google Scholar]
- Rolhion N, Barnich N, Claret L, Darfeuille-Michaud A. Strong decrease in invasive ability and outer membrane vesicle release in Crohn’s disease-associated adherent-invasive Escherichia coli strain LF82 with the yfgL gene deleted. J Bacteriol. 2005;187:2286–2296. doi: 10.1128/JB.187.7.2286-2296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeaux H, Pichon R, Kervarec N, Raguénès GHC, Guézennec JG. Novel bacterial exopolysaccharides from deep-sea hydrothermal vents. Carbohydr Polym. 1996;31:237–242. [Google Scholar]
- Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis. 2000;21:1054–1070. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Sullivan CW, Palmisano AC. Sea ice microbial communities: distribution, abundance and diversity of ice bacteria in McMurdo Sound, Antarctica, in 1980. Appl Environ Microbiol. 1984;47:788–795. doi: 10.1128/aem.47.4.788-795.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CM, Frasch CE. A sensitive silver stain for detecting lipopolysaccharide in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- Wai SN, Lindmark B, Söderblom T, Takade A, Westermark M, Oscarsson J, et al. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115:25–35. doi: 10.1016/s0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- Weinrauch Y, Katz SS, Munford RS, Elsbach P, Weiss J. Deacylation of purified lipopolysaccharides by cellular and extracellular components of a sterile rabbit peritoneal inflammatory exudates. Infect Immun. 1999;67:3376–3382. doi: 10.1128/iai.67.7.3376-3382.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfaardt GM, Lawrence JR, Korber DR. Function of EPS. In: Wingender J, Neu TR, Flemming HC, editors. Microbial Extracellularpolymeric Substances: Characterization, Structure and Function. Springer-Verlag; New York, NY, USA: 1999. pp. 171–200. [Google Scholar]
- Yaron S, Kolling GL, Simon L, Matthews KR. Vesicle-mediated transfer of virulence genes from Escherichia coli O157: H7 to other enteric bacteria. Appl Environ Microbiol. 2000;66:4414–4420. doi: 10.1128/aem.66.10.4414-4420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Srisatjaluk R, Justus DE, Doyle RJ. On the origin of membrane vesicles in gram-negative bacteria. FEMS Microbiol Lett. 1998;163:223–228. doi: 10.1111/j.1574-6968.1998.tb13049.x. [DOI] [PubMed] [Google Scholar]